Abstract
Epigenetic regulation of the genome is critical for the emergence of diverse cell lineages during development. To understand the role of DNA methylation during retinal network formation, we generated a mouse retinal-specific Dnmt1 deletion mutation from the onset of neurogenesis. In the hypomethylated Dnmt1-mutant retina, neural progenitor cells continue to proliferate, however, the cell cycle progression is altered, as revealed by an increased proportion of G1 phase cells. Despite production of all major retinal neuronal cell types in the Dnmt1-mutant retina, various postmitotic neurons show defective differentiation, including ectopic cell soma and aberrant dendritic morphologies. Specifically, the commitment of Dmnt1-deficient progenitors towards the photoreceptor fate is not affected by DNA hypomethylation, yet the initiation of photoreceptor differentiation is severely hindered, resulting in reduction and mislocalization of rhodopsin-expressing cells. In addition to compromised neuronal differentiation, Dnmt1 deficiency also leads to rapid cell death of photoreceptors and other types of neurons in the postnatal retina. These results indicate that Dnmt1-dependent DNA methylation is critical for expansion of the retinal progenitor pool, as well as for maturation and survival of postmitotic neurons.
Keywords: DNA methylation, Dnmt1, knockout, retina, development, mouse, neuronal differentiation and survival
Epigenetic regulation is essential for orchestrating tissue specific gene expression during mammalian development. Both histone modifications and DNA methylation contribute to the dynamic chromatin remodeling associated with distinct differentiation programs. Among the known DNA methyltransferases that catalyze the formation of 5-methylcytosine at the CpG dinucleotides, the activity of Dnmt1 is required for maintaining DNA methylation patterns during DNA replication in newly synthesized daughter DNA strands,1, 2 whereas Dnmt3a and Dnmt3b are responsible for de novo DNA methylation.3
Establishing and maintaining DNA methylation patterns are important for proper development and normal function of the nervous system. In humans, defective DNA methylation is associated with a number of neurological disorders.4 Mutations in Dnmt1 have been linked to neurodegeneration in a form of hereditary sensory neuropathy and late-onset cerebellar ataxia, deafness, and narcolepsy.5, 6 In age-related macular degeneration, DNA hypermethylation has been associated with decreased expression of genes involved in reducing oxidative stress.7 In the developing mouse brain, deletion of Dnmt1 in progenitor cells impairs neuronal maturation and survival, and causes precocious astroglial differentiation.8, 9 Genome-wide analyses have revealed that Dnmt3a-dependent methylation in gene body facilitates transcription of neurogenic genes in adult neural stem cells by antagonizing Polycomb repression.10 In postmitotic forebrain neurons, deletion of both Dnmt1 and Dnmt3a, but neither one alone, leads to abnormal gene expression and synaptic plasticity,11 suggesting that actively maintained DNA methylation is necessary for normal neural activities.
The vertebrate retina consists of six major types of neurons and the Muller glia derived from a common progenitor pool.12 The mature retinal neurons are highly specialized cells. For example, photoreceptors display unique transcriptomes,13, 14, 15 distinct cell morphologies, and nuclear structures.16 Molecular analyses have shown that distinct mature retinal neurons have established cell-specific DNA methylation patterns correlating with neuronal type-specific gene expression.17 In the developing zebrafish retina, morpholino knockdown studies have demonstrated that Dnmt3 and Dnmt1 have non-redundant functions and are both required for proper laminar layer formation and differentiation.18, 19 In addition, injury-induced retinal regeneration in zebrafish involves DNA demethylation in Muller glia and retinal ganglion cells (RGCs).20 To determine the function of Dnmt1 in mammalian retina development, we generated a retinal-specific Dnmt1 gene deletion at the onset of mouse retinogenesis. Our analyses show that embryonic retinal development can still proceed after Dnmt1 ablation, but maturation and differentiation of postmitotic retinal neurons are severely disrupted in the absence of Dnmt1 activity. Moreover, Dnmt1 function is essential for retinal neuron survival, as Dnmt1 conditional mutants show rapid postnatal retinal degeneration.
Results
Deletion of Dnmt1 gene causes rapid degeneration of the postnatal retina
In the developing mouse retina, Dnmt1 protein showed dynamic expression patterns as detected by immunolabeling. A higher level of Dnmt1 was expressed in embryonic retinal progenitors and lens epithelial cells than in postmitotic retinal neurons (Supplementary Figures 1A–C). Furthermore, distinct nuclear distributions of Dnmt1, either diffused or as intensely aggregated puncta, were observed. As the retina approached maturity, Dnmt1 was detected at high levels in retinal interneurons and RGCs, but nearly absent in the majority of photoreceptors (Supplementary Figures 1D–F). In adult retinas, a subset of photoreceptors, likely cone cells based on their location in the outer nuclear layer (ONL), retained a low yet detectable level of Dnmt1 (Supplementary Figure 1).
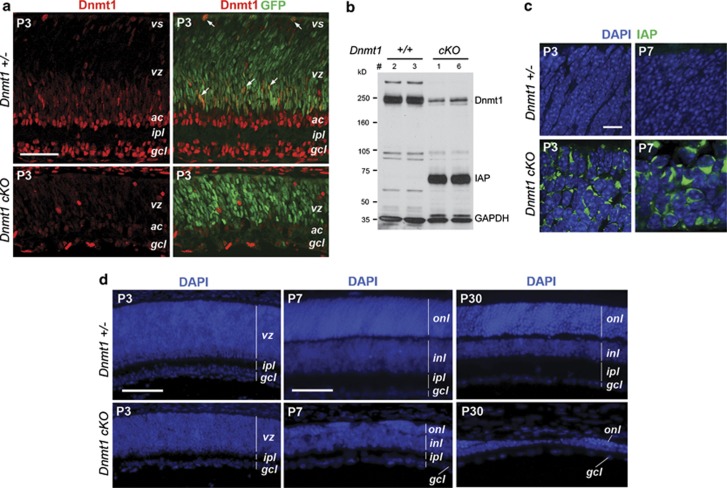
To elucidate the function of Dnmt1, we generated a retinal-specific Dnmt1 deletion using Cre-loxP recombination. Mice carrying a Dnmt1 conditional allele with loxP sites flanking exons 4 and 5 were crossed with a Chx10-Cre driver strain expressing a Cre-green fluorescent protein (GFP) fusion protein in retinal progenitors at embryonic day 10.5,21 to yield a mutant Dnmt1 protein lacking the catalytic domain.22 Immunocytochemistry showed that compared with the Dnmt1 heterozygous mutant (Dnmt1fl/+; Chx10-Cre) Dnmt1 conditional knockout retinas (Dnmt1fl/fl; Chx10-Cre, referred as cKO) contained drastically reduced Dnmt1 at postnatal day 3 (P3) (Figure 1a). A few residual Dnmt1+ cells were negative for GFP, due to the mosaic expression of Cre.23 Furthermore, western blots showed more than a 90% reduction of Dnmt1 protein in Dnmt1 cKO mutants (Figure 1b). To assess the DNA methylation status, we performed immunolabeling for intracisternal A-particle (IAP), a retroviral protein expressed only under hypomethylation conditions.8, 22 In contrast to the complete absence of IAP expression in heterozygous controls, Dnmt1 cKO mutants showed intense labeling of IAP (Figure 1c and Supplementary Figure 2).
Figure 1.
Effects of retinal-specific deletion of Dnmt1 by Chx10-Cre. (a) Confocal micrographs of retinas co-labeled for Dnmt1 (red) and GFP (green) at P3. The Dnmt1 cKO retina shows reduced Dnmt1 protein expression compared with the heterozygous control. White arrows point to cells co-expressing Dnmt1 and Cre-GFP. (b) Western blots show reduction of Dnmt1 protein in two individual P3 Dnmt1 cKO mutant retinas that also express high levels of IAP. (c) Confocal images of the outer retinas co-labeled for DAPI and IAP at P3 and P7. Dnmt1 cKO mutant retinas contain IAP+ cells with altered nuclear morphology. (d) DAPI labeling shows progressive thinning of Dnmt1-mutant retina at P3, P7 and P30. Scale bars, (a) 50 μm, (c) 10 μm, (d) P3, 100 μm; P7 and P30, 50 μm. ac, amacrine cells; gcl, ganglion cell layer; inl, inner nuclear layer; ipl, inner plexiform layer; vs, ventricular surface; vz, ventricular zone
Histological analyses showed that Dnmt1 cKO retinas were reduced in size at P0 compared with controls (Supplementary Figure 3). Although Dnmt1-mutant retinas were only 20–30% thinner than heterozygous controls at P3, between P3 and P7, the mutant retina showed rapid reduction in thickness, especially in the ONL (Figure 1d). This trend continued till P30, resulting in the loss of most retinal cells (Figure 1d and Supplementary Figure 3).
Dnmt1 gene deletion affects progenitor cell cycle progression and exit
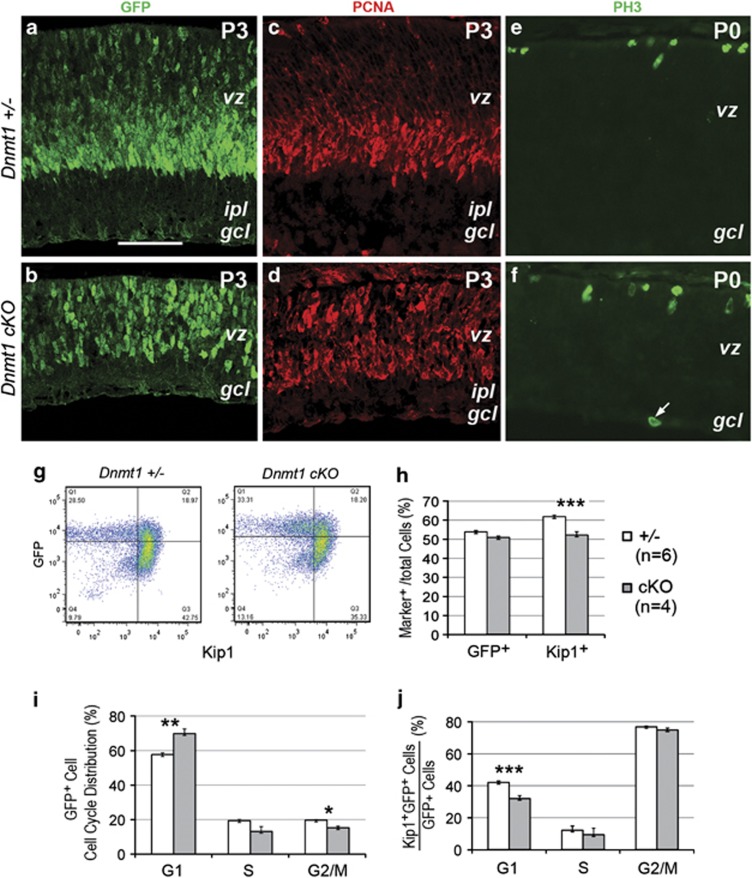
We next examined the effects of Dnmt1 deletion on retinal progenitor. Immunolabeling for GFP+ expressed by progenitors in Chx10-cre mice23 and for the proliferating cell nuclear antigen showed abnormal distributions of progenitors in the ventricular zone in Dnmt1 cKO mutants at P3 (Figures 2a–d). However, immunolabeling for the M-phase marker phospho-histone 3 (PH3) did not detect significant differences between the control and Dnmt1 mutants, except for occasional ectopic PH3+ cells (Figures 2e and f).
Figure 2.
Dnmt1 deletion alters progenitor distribution and cell cycle progression. (a–f) Immunolabeling of retinas show distribution of progenitor markers GFP (a and b) and proliferating cell nuclear antigen (PCNA, c and d) at P3, and M-phase marker PH3 (e and f) at P0. Arrow in f indicates an ectopic PH3+ cell. Scale bar, (a) for (a–f) 50 μm. gcl, ganglion cell layer; ipl, inner plexiform layer; vz, ventricular zone. (g–j) Flow cytometric analyses of cell markers and cell cycle properties at P2. (g) Representative flow cytometry profiles for GFP and cyclin-dependent protein kinase inhibitor p27Kip1. (h) Fluorescence-activated cell sorting quantification of GFP+ cells and p27Kip1+ cells among total cells. (i) Cell cycle distributions of GFP+ progenitors. (j) Quantification of p27Kip1+ GFP+ cells among GFP+ cells in different cell cycle phases. Numbers of individual retinas analyzed in (h–j) are indicated in h. *P<0.05, **P<0.01, ***P<0.001
To assess progenitor behavior during the cell cycle, we performed fluorescence-activated cell sorting analyses of GFP+ cells and the cyclin-dependent protein kinase inhibitor p27Kip1, which is upregulated in cells poised to exit or already withdrawn from the cell cycle. The percentages of GFP+ cells among total cells were not altered by the Dnmt1 mutation at P2 (Figures 2g and h). Analyses of the cell cycle showed that in Dnmt1 mutants, the distribution of GFP+ progenitors were significantly increased in G1 phase from 57.6±1.0 to 69.8±2.75% and decreased in G2/M phase from 19.6±0.38 to 15.2±0.92% (Figure 2i), revealing defects in cell cycle progression and abnormal G1 accumulation.
In Dnmt1 mutant retinas, the percentage of p27Kip1+ cells among total cells was significantly reduced from 61.9±0.72 to 52.2±1.77% (Figure 2h), suggesting a reduction of either postmitotic neurons or progenitors exiting the cell cycle. Analyses of cell cycle distribution revealed that control and mutant retinas contained similar percentages (75–77%) of p27Kip1+ cells among G2/M-phase GFP+ progenitors at P2, indicating that mutant cells were able to upregulate p27Kip1. However, Dnmt1 mutant retinas showed a significant decrease of p27Kip1+ GFP+ cells from 42.1±0.9% to 32.0±1.85% among the 2n G0 cell population (Figure 2j), suggesting the defect occurred during or soon after the M phase to G0 phase transition. Together, these results demonstrated that the Dnmt1 mutation affected progenitor cell cycle progression, and resulted in a defective cell cycle exit or faltered initial steps in becoming postmitotic neurons.
Dnmt1 deficiency affects retinal neuronal production
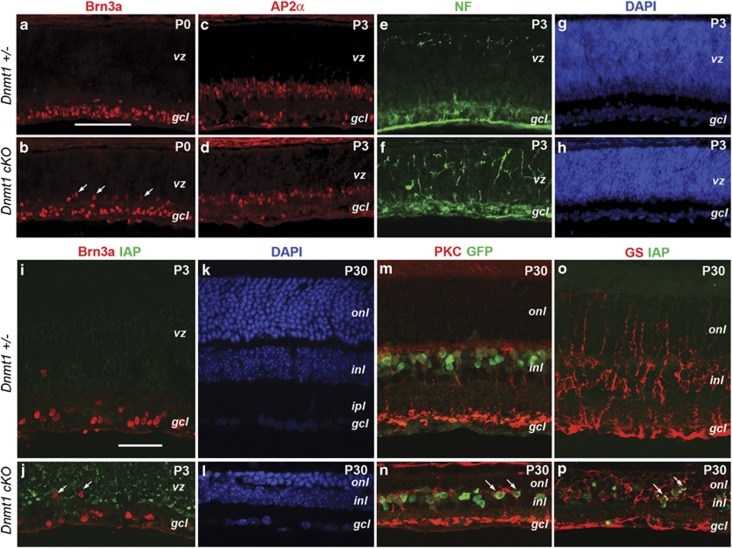
We next examined the effects of Dnmt1 deletion on the production of embryonically born retinal neurons. Immunolabeling with the RGC marker Brn3a at P0 detected fewer RGCs, and many were mislocalized in the Dnmt1 cKO mutant (Figures 3a and b). Interestingly, unlike the rest of the retina, less IAP was detected among RGCs (Figures 3i and j), indicating that either a small portion of RGCs were produced before Dnmt1 deletion or this subset of RGCs had retained a higher level of DNA methylation. Another early born retinal neuronal type, the horizontal cells, exhibited abnormally exuberant dendritic arbors in the Dnmt1 mutant as detected by neurofilament labeling (Figures 3e and f). Immunolabeling for the amacrine cell marker AP2α showed a reduced amacrine interneuron population due to Dnmt1 deficiency at P3 (Figures 3c and d).
Figure 3.
Effects of Dnmt1 deletion on retinal cell type production. (a–j) Immunolabeling of early born cell types in the neonatal retina. Cell marker labeling for RGCs (a and b), amacrine cells (c and d), horizontal cells (e and f) are presented. i and j shows co-labeling of Brn3a and IAP. White arrows (b and j) point to mislocalized RGCs. DAPI staining (g and h) shows retinal laminar structures at P3. (k–p) Confocal images of P30 retinas for late-born retinal cell types. Cell marker labeling for bipolar cells (m and n) and Muller cells (o and p) are presented. Arrows in n point to rod bipolar cells that are double labeled for Cre-GFP and protein kinase C-α (PKCα), and p indicate mutant Muller cell double labeled for IAP and glutamine synthetase (GS). Scale bars, a for a–h, 100 μm; i for i–p, 50 μm. gcl, ganglion cell layer; inl, inner nuclear layer; vz, ventricular zone
Among postnatally produced cell types, the bipolar cells were clearly present in Dnmt1-mutant retinas, as indicated by GFP signals driven by the Chx10 promoter and the rod bipolar cell marker protein kinase C-α (Figures 3m and n). We also detected the presence of cells co-labeled by the Müller glial marker glutamine synthetase and IAP, indicating that Dnmt1-deficient progenitors gave rise to Müller glia (Figures 3o and p). These results demonstrated that progenitors in Dnmt1 cKO mutant retinas were able to generate most retinal cell types.
Dnmt1 activity is essential for photoreceptor differentiation and survival
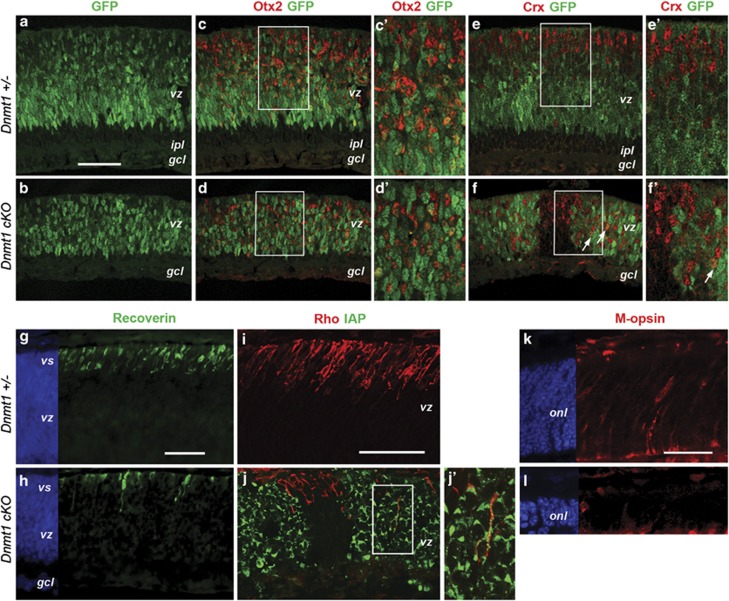
The rapid demise of the ONL in Dnmt1-mutant retinas suggested that photoreceptor development was impaired. We first investigated the commitment of progenitors toward photoreceptor fates by detecting the homeobox protein Otx2, an essential positive regulator of photoreceptor production.24 Similar expression patterns of Otx2 were detected among a subset of GFP+ progenitors in Dnmt1 heterozygous and cKO mutant retinas at P3 (Figures 4a–d). We also examined expression of Crx, a homeobox protein turned on in postmitotic photoreceptor precursors and required for both rod and cone photoreceptor differentiation.25, 26, 27 Although Crx was detected in both control and Dnmt1 cKO mutants, Crx+ cells in the mutant retina were dispersed throughout the ventricular zone rather than localized near the future ONL (Figures 4e and f). Moreover, abnormal co-expression of Crx and the progenitor cell marker GFP was also observed in Dnmt1 mutants (Figures 4f and f').
Figure 4.
Effects of Dnmt1 mutation on photoreceptor commitment and differentiation. (a–f') Confocal images of co-immunolabeling at P3 for Otx2 and GFP (a–d'), or Crx and GFP (e–f'). c', d' and e', f' show twofold magnification of framed areas in c, d and e, f, respectively. The Crx+ cells in control (e and e') and in Cre− regions of the mutant (f and f') are located near the ventricular surface; but Dnmt1-deficient Crx+ cells are distributed throughout vz (f and f'). Arrows indicate co-labeled GFP+ Crx+ cells. (g and h) Immunolabeling for recoverin and DAPI at P3. (i, j and j') Confocal images of co-labeling for Rho (red) and IAP (green) at P3. A lack of Rho+ cells is seen in Dnmt1-deficient regions expressing IAP (j). A rare Rho and IAP co-labeled cell framed in j is shown in j' at twofold magnification. (k and l) Confocal images of immunolabeling for M-opsin at P3. Scale bars, a for a–d, e and f, g for g and h, i for i and j, 50 μm; k for k and l, 25 μm. gcl, ganglion cell layer; ipl, inner plexiform layer; ventricular surface; vz, ventricular zone
We next examined the onset of photoreceptor differentiation. At P3, Dnmt1 mutant retinas showed a reduced number of cells expressing the early photoreceptor marker recoverin (Figures 4g and h). Furthermore, rod-specific rhodopsin expression was significantly reduced in Dnmt1-deficient cells that were IAP+ (Figures 4i and j). Confocal imaging detected very few rhodopsin and IAP double-positive cells (Figures 4j and j'). Similarly, M-opsin expression by cone photoreceptors was greatly diminished in Dnmt1-mutant retinas (Figures 4k and l). These data demonstrated that the commitment of progenitors toward a photoreceptor cell fate was not affected by DNA hypomethylation, but the onset and subsequent differentiation of photoreceptors were blocked.
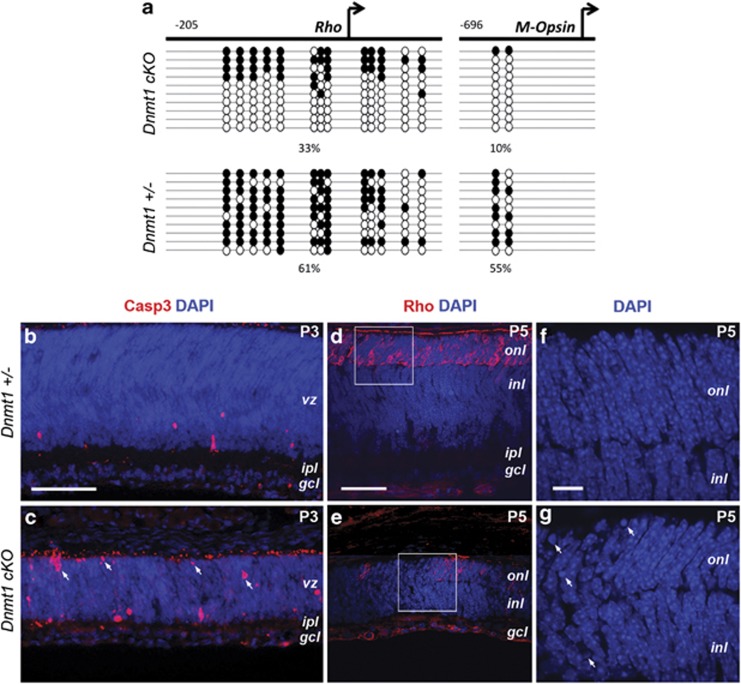
Further evaluation of DNA methylation status of photoreceptor-specific genes using bisulfite sequencing showed a significant demethylation at CpG sites in both Rhodopsin and M opsin promoter regions in Dnmt1 cKO mutants at P3 (Figure 5a). To determine whether the rapid reduction of retinal cell numbers was due to increased apoptosis, we labeled retinas for activated caspase 3. Compared with the heterozygous control retina, Dnmt1 cKO mutants showed significantly increased caspase 3 signals throughout the retina (Figures 5b and c). Furthermore, increased numbers of pyknotic nuclei were observed among Dnmt1-deficient cells, which failed to express rhodopsin (Figures 5d and g), suggesting an increased apoptosis of postmitotic photoreceptors.
Figure 5.
Dnmt1 ablation causes DNA hypomethylation and cell death in the postnatal retina. (a) DNA methylation patterns at Rhodopsin and M-opsin promoters as determined by bisulfide sequencing. Significantly reduced CpG dinucleotide methylation is detected at P3 in the mutant retina. Columns correspond to CpG sites with the relative location from the transcription-starting sites (arrows), while rows correspond to sequenced clones. Solid circles indicate methylated CpG; open circles indicate unmethylated CpG. (b and c) Immunolabeling for activated caspase 3 shows increased apoptosis in Dnmt1 cKO retinas at P3. Note that Casp3+ cells are distributed throughout the mutant retina. (d and e) Co-labeling of DAPI and Rho at P5. f and g represent magnified regions framed in d and e to show condensed nuclei (arrows) present in mutant cells. Scale bars, b for b and c, d for d and e, 50 μm; f for f and g, 10 μm. gcl, ganglion cell layer; inl, inner nuclear layer; ipl, inner plexiform layer; vz, ventricular zone
Discussion
We have addressed the role of Dnmt1 during mammalian retinogenesis using retinal-specific gene ablation. The conditional Dnmt1 mutant yields a truncated protein without most of the replication fork interaction and the entire methyltransferase catalytic domain.22 Our results demonstrate that maintaining DNA methylation status by Dnmt1 is critical for mammalian retinal development. Although progenitor proliferation persists in the embryonic retina, the proper differentiation of retinal neurons is severely impaired due to Dnmt1 deficiency. Moreover, DNA hypomethylation leads to rapid retinal degeneration.
The high levels of Dnmt1 protein expression in early retinal progenitors are consistent with its role in maintaining DNA methylation patterns as cells undergo rapid division. However, it is unexpected that mature retinal interneurons and projection neurons retain high levels of Dnmt1 expression. Our results are consistent with a recent study,28 which describes overlapping expression patterns of Dmnt3a and Dnmt3b with Dnmt1 in RGCs, amacrine cells and cone photoreceptors. It is plausible that Dnmt1 activity complements other DNA methyltransferases to maintain neuronal function. In zebrafish, both Dnmt1 and Dnmt3 are required for retinal development, but engage distinct downstream chromatin modifiers.18, 19 Knocking down Dnmt1 in zebrafish retina reduces differentiated photoreceptor marker, but without rapid ONL cell death. Accumulating evidence suggest that the activity of Dnmt1 may not be limited to maintaining methylation patterns. For example, in the adult mouse brain, Dnmt1 and Dnmt3a are both required for normal function and neural plasticity.11 Furthermore, chromatin remodeling is clearly involved in brain and retinal stem cell potentiation.10, 20 Therefore, the potential function of Dnmt1 in mature retinal neurons and during neural repair warrants further investigation.
Previous studies have shown that loss of Dnmt1 activity results in progressive hypomethylation at CpG dinucleotides as progenitor cells undergo multiple rounds of cell divisions.8, 9 Although the onset of Chx10-Cre expression coincides with the initiation of retinal neurogenesis, effects of Dnmt1 deletion may be cumulative. The low IAP expression in RGCs suggests that this early born cell type retains a certain level of DNA methylation. In contrast, another early born neuronal type, the horizontal cell, displays extensive IAP expression and dendritic abnormalities. Therefore, the lack of hypomethylation in RGCs may be due to compensation by other Dnmts expressed by RGCs.28 Our results show that despite ablation of Dnmt1 from the onset of retinogenesis, all retinal cell types are produced from Dnmt1-deficient progenitors. However, the impact of the mutation becomes increasingly severe, likely reflecting the increasing degree of DNA hypomethylation.
The effect of the Dnmt1 deletion on embryonic retinal progenitor proliferation is relatively mild. However, in the postnatal stages, Dnmt1 deficiency alters cell cycle progression resulting in an increased distribution of G1 phase progenitors. At P2, the majority of progenitors are typically ready to exit the cell cycle thus upregulating the p27Kip1 as they enter the M phase. Our results show that Dnmt1 mutant cells can upregulate p27Kip1 during M, but then quickly suffer a significant loss as they transition into the G0 phase and become postmitotic. Therefore, the overall reduction of p27Kip1+ cells in Dnmt1-mutant retinas is likely due to that many nascent neurons with hypomethylated genomes are unable to execute their specific differentiation programs and soon undergo apoptosis. This conclusion is consistent with the rapid cell loss in Dnmt1 cKO retinas within the first postnatal week.
Photoreceptor cells are highly specialized sensory neurons with distinct transcriptomes. The nuclear structure of nascent rod photoreceptors undergoes extensive reorganization as they approach maturity,16 and establish cell type-specific DNA methylation patterns.17 Establishing a rod-specific transcription program is likely to involve coordination between DNA methylation and histone modifications under the direction of photoreceptor-specific transcription factors.29 Commitment by proliferating progenitors towards a photoreceptor fate occurs in sequential steps30 and is initiated by expression of Otx2, followed by upregulation of Crx as the committed cells withdraw from the cell cycle to become photoreceptor precursors.24, 27, 31 Our results show that Dnmt1 deficiency does not affect progenitors commitment to the photoreceptor fate, as Otx2 expression appears normal. Furthermore, we demonstrate that the onset of Crx expression is also not affected. However, the co-expression of Crx and progenitor markers and mislocalization of both Crx+ and rhodopsin+ cells indicate that DNA methylation defects interfere with further differentiation of photoreceptor precursors. Interestingly, bisulfide sequencing confirms that in Dnmt1 cKO mutants, the CpG sites in both Rhodopsin and M-opsin promoters are hypomethylated at P3, and yet unable to sustain photoreceptor differentiation. This may reflect that hypomethylation alone is insufficient in sustaining and establishing photoreceptor-specific transcriptions. Moreover, failed neuronal differentiation results in rapid and massive cell death as previous observed in other neuronal cell types.8
Our study demonstrates the critical function of Dnmt1 during retinal development, especially photoreceptor differentiation and survival. Recent evidence indicates that DNMT1 mutations can lead to progressive neurodegeneration and late-onset neuropathies in humans.5, 6 Furthermore, decline in DNA methylation has also been observed in age-related macular degeneration.7 Future investigation on maintaining DNA methylation and epigenetic regulation in the mature retina may provide importance insights on disease mechanisms and treatment strategies.
Materials and Methods
Animals
Dnmt1fl/fl mice previously described22 were crossed with heterozygous males carrying the Dnmt1fl/+ allele and a Cre-GFP fusion transgene under the control of the Chx10 locus.21 The mouse lines were maintained in C57bl6 background. Genotypes were determined by PCR using primers for Dnmt1 (forward: 5′-GGG CCA GTT GTG TGA CTT GG-3′, reverse: 5′-CTT GGG CCT GGA TCT TGG GGA TC-3′) and the Cre transgene (forward: 5′-GAC GAT GCA ACG AGT GAT GA-3′, reverse: 5′-AGC ATT GCT GTC ACT TGG TC-3′), respectively. All animal procedures followed NIH guidelines and were approved by the Animal Research Committee at University of California Los Angeles.
Immunocytochemistry
Cryosections fixed in 4% paraformaldehyde were processed for immunocytochemistry as described.27 Antibodies used for immunolabeling are summarized in Supplementary Table 1. Immunofluorescent images were captured using Nikon E800 (Nikon, Tokyo, Japan) or Olympus FluoView 1000 confocal microscopes (Olympus, Tokyo, Japan). Samples from multiple animals (minimal, n≥3) of each genotype were analyzed.
Western blots
Cell lysates of P3 retinas were processed as described.27 Signals of horseradish peroxidase conjugated to secondary antibodies were detected using ECL (GE Healthcare Bio-Sciences, Pittsburgh, PA, USA). Multiple retinal extracts (minimal, n≥3) of different genotypes were analyzed.
Flow cytometry
Dissociated P2 retinal cells were immunolabeled and analyzed using an LSRII flow cytometer (BD Biosciences, San Jose, CA, USA) as described.23 Individual samples from multiple heterozygous (n=6) or cKO (n=4) retinas were analyzed. Flow cytometry data were analyzed with FlowJo (Tree Star, Ashland, OR, USA). Quantification of markers is shown as mean±S.E.M. after statistical analysis using Student's t-Test. P values<0.05 were considered significant.
Methylation analysis
Genomic DNA isolated from P3 retinas were processed with bisulfite conversion using EZ DNA Methylation Kit (Zymo) by following the manufacture instructions. The Meth-primers are described previously.17 Amplified PCR products were purified and cloned into the TOPO vector (Life Technologies, Grand Island, NY, USA). Ten individual control (n=10) and Dnmt1-mutant (n=10) clones were sequenced to detect methylation patterns.
Acknowledgments
We thank Dr. C Cepko for Chx10-cre mice and Tanya Hioe for technical assistance. This work was in part supported by funds from Research to Prevent Blindness Foundation, NIH grants EY12270 and EY019052 to XJY, and NEI core grant EY000331.
Glossary
- cKO
conditional knockout
- Dnmt1
DNA methyltransferase 1
- GFP
green fluorescent protein
- GS
glutamine synthetase
- IAP
intracisternal A-particle
- ONL
outer nuclear layer
- PCNA
proliferating cell nuclear antigen
- PH3
phospho-histone 3
- PKCα
Protein kinase C-α
- RGC
retinal ganglion cell
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by A Verkhratsky
Supplementary Material
References
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33 (Suppl:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Robertson KD, Wolffe AP. DNA methylation in health and disease. Nat Rev Genet. 2000;1:11–19. doi: 10.1038/35049533. [DOI] [PubMed] [Google Scholar]
- Klein CJ, Botuyan MV, Wu Y, Ward CJ, Nicholson GA, Hammans S, et al. Mutations in DNMT1 cause hereditary sensory neuropathy with dementia and hearing loss. Nat Genet. 2011;43:595–600. doi: 10.1038/ng.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkelmann J, Lin L, Schormair B, Kornum BR, Faraco J, Plazzi G, et al. Mutations in DNMT1 cause autosomal dominant cerebellar ataxia, deafness and narcolepsy. Hum Mol Genet. 2012;21:2205–2210. doi: 10.1093/hmg/dds035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter A, Spechler PA, Cwanger A, Song Y, Zhang Z, Ying GS, et al. DNA methylation is associated with altered gene expression in AMD. Invest Ophthalmol Vis Sci. 2012;53:2089–2105. doi: 10.1167/iovs.11-8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan G, Beard C, Chen RZ, Csankovszki G, Sun Y, Siniaia M, et al. DNA hypomethylation perturbs the function and survival of CNS neurons in postnatal animals. J Neurosci. 2001;21:788–797. doi: 10.1523/JNEUROSCI.21-03-00788.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutnick LK, Golshani P, Namihira M, Xue Z, Matynia A, Yang XW, et al. DNA hypomethylation restricted to the murine forebrain induces cortical degeneration and impairs postnatal neuronal maturation. Hum Mol Genet. 2009;18:2875–2888. doi: 10.1093/hmg/ddp222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Coskun V, Tao J, Xie W, Ge W, Yoshikawa K, et al. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science. 2010;329:444–448. doi: 10.1126/science.1190485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, et al. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci. 2010;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cepko CL, Austin CP, Yang X, Alexiades M, Ezzeddine D. Cell fate determination in the vertebrate retina. Proc Nat Acad Sci USA. 1996;93:589–595. doi: 10.1073/pnas.93.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackshaw S, Fraioli RE, Furukawa T, Cepko CL. Comprehensive analysis of photoreceptor gene expression and the identification of candidate retinal disease genes. Cell. 2001;107:579–589. doi: 10.1016/s0092-8674(01)00574-8. [DOI] [PubMed] [Google Scholar]
- Livesey FJ, Furukawa T, Steffen MA, Church GM, Cepko CL. Microarray analysis of the transcriptional network controlled by the photoreceptor homeobox gene Crx. Curr Biol. 2000;10:301–310. doi: 10.1016/s0960-9822(00)00379-1. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Mears AJ, Friedman JS, Carter T, He S, Oh E, et al. Expression profiling of the developing and mature Nrl-/- mouse retina: identification of retinal disease candidates and transcriptional regulatory targets of Nrl. Hum Mol Genet. 2004;13:1487–1503. doi: 10.1093/hmg/ddh160. [DOI] [PubMed] [Google Scholar]
- Solovei I, Kreysing M, Lanctôt C, Kösem S, Peichl L, Cremer T, et al. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell. 2009;137:356–368. doi: 10.1016/j.cell.2009.01.052. [DOI] [PubMed] [Google Scholar]
- Merbs SL, Khan MA, Hackler L, Oliver VF, Wan J, Qian J, et al. Cell-specific DNA methylation patterns of retina-specific genes. PloS One. 2012;7:e32602. doi: 10.1371/journal.pone.0032602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai K, Nadauld LD, Chidester S, Manos EJ, James SR, Karpf AR, et al. Zebra fish Dnmt1 and Suv39h1 regulate organ-specific terminal differentiation during development. Mol Cell Biol. 2006;26:7077–7085. doi: 10.1128/MCB.00312-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai K, Jafri IF, Chidester S, James SR, Karpf AR, Cairns BR, et al. Dnmt3 and G9a cooperate for tissue-specific development in zebrafish. J Biol Chem. 2010;285:4110–4121. doi: 10.1074/jbc.M109.073676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell C, Elsaeidi F, Goldman D. Injury-dependent Muller glia and ganglion cell reprogramming during tissue regeneration requires Apobec2a and Apobec2b. J Neurosci. 2012;32:1096–1109. doi: 10.1523/JNEUROSCI.5603-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan S, Cepko CL. Genetic analysis of the homeodomain transcription factor Chx10 in the retina using a novel multifunctional BAC transgenic mouse reporter. Dev Biol. 2004;271:388–402. doi: 10.1016/j.ydbio.2004.03.039. [DOI] [PubMed] [Google Scholar]
- Jackson-Grusby L, Beard C, Possemato R, Tudor M, Fambrough D, Csankovszki G, et al. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat Genet. 2001;27:31–39. doi: 10.1038/83730. [DOI] [PubMed] [Google Scholar]
- Sakagami K, Gan L, Yang XJ. Distinct effects of Hedgehog signaling on neuronal fate specification and cell cycle progression in the embryonic mouse retina. J Neurosci. 2009;29:6932–6944. doi: 10.1523/JNEUROSCI.0289-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida A, Furukawa A, Koike C, Tano Y, Aizawa S, Matsuo I, et al. Otx2 homeobox gene controls retinal photoreceptor cell fate and pineal gland development. Nat Neurosci. 2003;6:1255–1263. doi: 10.1038/nn1155. [DOI] [PubMed] [Google Scholar]
- Furukawa T, Morrow EM, Cepko CL. Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell. 1997;91:531–541. doi: 10.1016/s0092-8674(00)80439-0. [DOI] [PubMed] [Google Scholar]
- Chen S, Wang QL, Nie Z, Sun H, Lennon G, Copeland NG, et al. Crx, a novel Otx-like paired-homeodomain protein, binds to and transactivates photoreceptor cell-specific genes. Neuron. 1997;19:1017–1030. doi: 10.1016/s0896-6273(00)80394-3. [DOI] [PubMed] [Google Scholar]
- Rhee KD, Goureau O, Chen S, Yang XJ. Cytokine-induced activation of signal transducer and activator of transcription in photoreceptor precursors regulates rod differentiation in the developing mouse retina. J Neurosci. 2004;24:9779–9788. doi: 10.1523/JNEUROSCI.1785-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasonkin IO, Lazo K, Hambright D, Brooks M, Fariss R, Swaroop A. Distinct nuclear localization patterns of DNA methyltransferases in developing and mature mammalian retina. J Comp Neurol. 2011;519:1914–1930. doi: 10.1002/cne.22613. [DOI] [PubMed] [Google Scholar]
- Peng GH, Chen S. Crx activates opsin transcription by recruiting HAT-containing co-activators and promoting histone acetylation. Hum Mol Genet. 2007;16:2433–2452. doi: 10.1093/hmg/ddm200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh EC, Khan N, Novelli E, Khanna H, Strettoi E, Swaroop A. Transformation of cone precursors to functional rod photoreceptors by bZIP transcription factor NRL. Proc Natl Acad Sci USA. 2007;104:1679–1684. doi: 10.1073/pnas.0605934104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omori Y, Katoh K, Sato S, Muranishi Y, Chaya T, Onishi A, et al. Analysis of transcriptional regulatory pathways of photoreceptor genes by expression profiling of the Otx2-deficient retina. PloS One. 2011;6:e19685. doi: 10.1371/journal.pone.0019685. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.