Abstract
We recently identified Grainyhead-like 2 (GRHL2), a mammalian homolog of Grainyhead in Drosophila, to be a novel transcription factor that regulates hTERT gene expression and enhances proliferation of normal human epidermal keratinocytes (NHEK). In the current study, we show that GRHL2 impairs keratinocyte differentiation through transcriptional inhibition of the genes clustered at the epidermal differentiation complex (EDC), located at chromosome 1q21. Gene expression profiling and subsequent in vitro assays revealed consistent downregulation of EDC genes, for example, IVL, KRT1, FLG, LCEs, and SPRRs, in NHEK expressing exogenous GRHL2. In vivo binding assay by chromatin immunoprecipitation revealed GRHL2 association at the promoter regions of its target genes, many of which belong to EDC. Exogenous GRHL2 expression also inhibited recruitment of histone demethylase Jmjd3 to the EDC gene promoters and enhanced the level of histone 3 Lys 27 trimethylation enrichment at these promoters. Survey of GRHL2 expression in human skin tissues demonstrated enhanced protein and mRNA levels in chronic skin lesions with impaired keratinocyte differentiation, for example, atopic dermatitis and psoriasis, compared with normal epidermis. These data indicate that GRHL2 impairs epidermal differentiation by inhibiting EDC gene expression through epigenetic mechanisms and support its role in the hyperproliferative skin diseases.
Keywords: epidermal differentiation complex, epigenetic, GRHL2, histone methylation, keratinocyte
Grainyhead-like 2 (GRHL2) is one of the three known mammalian homologs of Drosophila Grainyhead (GRH), along with GRHL1 and GRHL3, all of which are involved in epithelial morphogenesis.1 GRH is a developmental transcriptional factor that affects epithelial barrier function.2, 3, 4 GRH also positively regulates post-embryonic cell proliferation5 and controls expression of proliferating cell nuclear antigen (PCNA) gene, suggesting its involvement in DNA replication.6 In mammalian cells, GRHL proteins orchestrate development of epithelial impermeability through transcriptional activation of genes required for cell adhesion.7, 8 Little is known about the role of GRHL2 in keratinocyte differentiation, which is one of the key elements required of epithelial impermeability barrier.
Keratinocyte differentiation requires expression of genes clustered in chromosome 1q21, collectively referred to as epidermal differentiation complex (EDC). The EDC cluster consists of at least 43 genes that primarily make up the structural elements of cornfield layer, for example, involucrin (IVL), filaggrin (FLG), loricrin (LOR), late cornifying envelope (LCE) genes, and small proline-rich (SPRR) genes.9, 10 Coordinated expression of the EDC genes is required for intact keratinocyte terminal differentiation and epidermal function. As such, defective expression or mutation of EDC genes is linked with various skin disorders, in part, from impaired epidermal barrier. This connection is exemplified in atopic dermatitis (AD), also known as eczema, which affects nearly 20% of population, especially in children in developed countries.11 Gene linkage studies found mutations and polymorphisms of EDC genes, for example, FLG, LCE1D, and SPRR3, as strong predisposing factors for AD.10, 12 Reduced expression of LOR and deletion mutations of LCE3B and LCE3C are linked with psoriasis (PS), an inflammatory skin disorder exhibiting keratinocyte hyperproliferation and aberrant differentiation.13, 14, 15 It appears that multiple EDC genes are involved in the pathogenesis of chronic skin diseases. Therefore, mechanistic understanding of EDC gene regulation would help to identify potential therapeutic targets for such skin disorders. A recent study revealed an epigenetic mechanism elicited by the histone demethylase Jmjd3 and polycomb group (PcG) proteins.16 Jmjd3 is recruited to the promoter regions of EDC genes during Ca++-induced keratinocyte differentiation to remove the methyl groups from the histone 3 Lys 27 trimethylation (H3K27Me3). Depletion of Jmjd3 led to inhibition of EDC gene expression and impaired epidermal differentiation, suggesting an active role of histone modification in EDC gene regulation.
We previously showed that GRHL2 is a novel transcription regulator of hTERT, (human telomerase reverse transcriptase) gene and enhances replicative potential of normal human epidermal keratinocytes (NHEK).17 In the current study, we show that GRHL2 inhibits keratinocyte differentiation through epigenetic suppression of EDC gene expression. Ectopic expression of GRHL2 blocked recruitment of Jmjd3 to the EDC gene promoters and enhanced H3K27Me3 enrichment levels in gene promoter. Immunohistochemical staining of GRHL2 and gene expression analyses showed increased GRHL2 expression in epidermal tissues exhibiting AD or PS histopathology compared with normal skin. These lesions also demonstrated hyperproliferative epithelium and impaired EDC gene expression, suggesting potential involvement of GRHL2 in epithelial pathology of skin.
Results
Gene expression profiling reveals EDC genes as potential GRHL2 targets
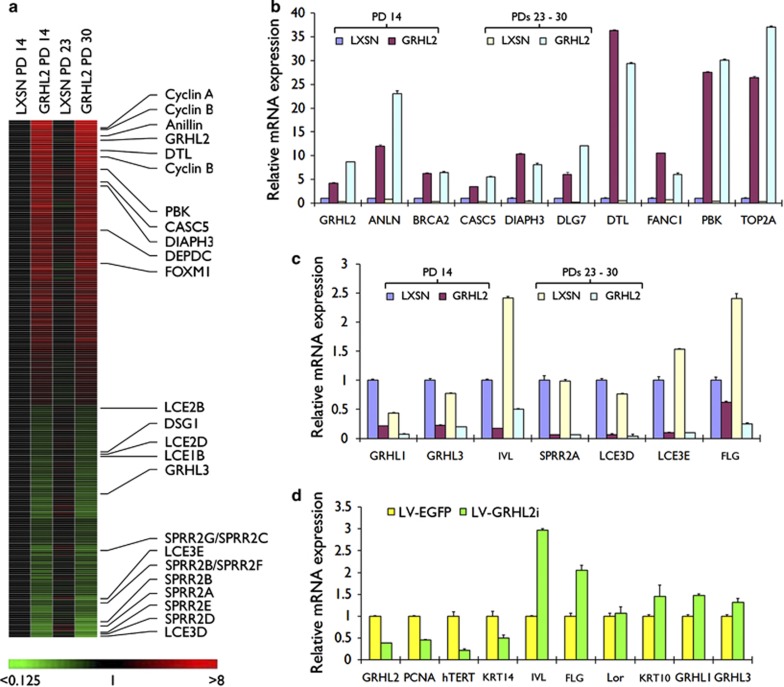
Gene expression profiling was performed between NHEK infected with retroviral vector expressing GRHL2 (NHEK/GRHL2) and the empty vector (NHEK/LXSN) at early passage (population doubling (PD) 14) or late passage (PDs 23 and 30) (Figure 1a). Among the genes differentially expressed by GRHL2 transduction, the genes that belong to EDC, for example, IVL, FLG, LCEs, and SPRRs, were consistently downregulated in NHEK/GRHL2 compared with the controls. On the contrary, the genes involved in cell proliferation, for example, Cyclins A/B, FoxM1, and DNA topoisomerases, were strongly induced by GRHL2. Microarray data were validated by quantitative reverse transcription–PCR (qRT-PCR), which confirmed suppression of the EDC genes in NHEK/GRHL2 compared with the control (Figures 1b and c). Endogenous GRHL2 is highly expressed in oral squamous cell carcinoma (SCC) cell lines.18 When GRHL2 expression was knocked down in oral SCC15 cell line, EDC gene expression was induced (Figure 1d). Interestingly, we found that GRHL1 and GRHL3, which are the two isoforms of GRHL2,19, 20 were strongly suppressed in NHEK by GRHL2 transduction and induced in SCC15 with endogenous GRHL2 knocked down (Figures 1c and d). Thus, GRHL2 may have negative regulatory roles for the expression of GRHL1 and GRHL3, both of which may facilitate epithelial differentiation.
Figure 1.
Genes differentially regulated by GRHL2 in NHEK. (a) Microarray experiment was performed between NHEK/LXSN and NHEK/GRHL2 cells at early (PD 14 for both cultures) and late (PDs 23 and 30, respectively) passages. Heat map was generated for each gene data in reference to those of NHEK/LXSN at PD 14. (b) Parallel cultures of NHEK/LXSN and NHEK/GRHL2 were harvested for qRT-PCR to validate the genes upregulated by GRHL2 transduction. Pairwise comparison is made between GRHL2 and LXSN at different PD levels (P<0.01 for all data sets). (c) Validation qRT-PCR was performed for gene downregulated by GRHL2 transduction using NHEK/LXSN and NHEK/GRHL2. All pairwise comparison between GRHL2 and LXSN were significant (P<0.01). (d) Endogenous GRHL2 was knocked down in SCC15 cells using lentiviral vector expressing shRNA against GRHL2 (LV-GRHL2i). Cells infected with control empty vector (LV-EGFP) were included for comparison. Bars represent the means of triplicates and standard deviation for each gene. Pairwise comparison between the two groups revealed significant difference (P<0.05) for all tested genes except for LOR
GRHL2 directly binds and regulates the EDC gene promoters
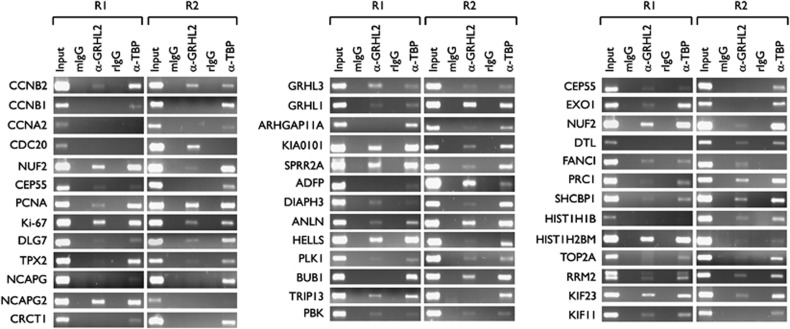
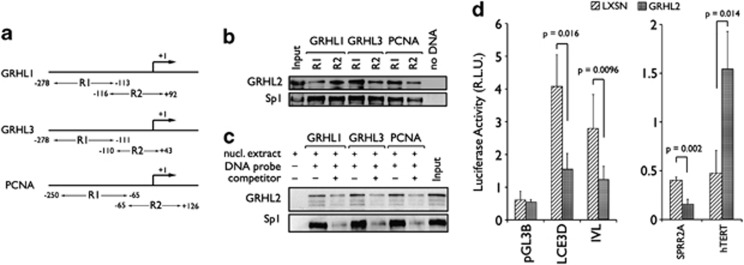
We examined whether GRHL2 directly binds the promoter regions of the differentially regulated genes in vivo. Chromatin immunoprecipitation (ChIP) assay was performed and the DNA-protein complexes from SCC4 were immunoprecipitated using anti-GRHL2 antibody or non-specific IgG and anti-TBP antibody as controls. GRHL2 co-immunoprecipitated with most of the promoters tested, including those of CCNB2, PCNA, Ki-67, GRHL1, GRHL3, and SPRR2A (Figure 2), suggesting that GRHL2 physically associates with these promoters in vivo. We confirmed binding of GRHL2 to its target promoters by promoter magnetic precipitation (PMP) assay using the promoter fragments immobilized onto Dynabead magnetic particles, followed by western blotting. GRHL2 binding to the promoter regions of GRHL1, GRHL3, and PCNA was tested at the two proximal regions of each promoter, that is, R1 and R2 (Figure 3a). For comparison, we also assessed the binding of Specificity Protein 1 (Sp1), a transcription factor known to bind PCNA promoter.21 As shown in Figure 3b, GRHL2 and Sp1 binding was noted in both R1 and R2 of the promoters tested, whereas the absence of promoter DNA abolished the binding signal. GRHL2-binding intensity differed in R1 and R2 for each of the tested promoters, and this variation mirrored the changes in GRHL2-binding activity in vivo as shown in the ChIP analysis (Figure 2). We also confirmed the specificity of GRHL2 binding to the promoter by addition of non-biotinylated DNA competitor, which reduced the binding of GRHL2 and Sp1 to the immobilized DNA fragments (Figure 3c).
Figure 2.
GRHL2 binds to the proximal promoter regions of its target genes. ChIP assays were performed using SCC4 cells, which were cross-linked with 1% formaldehyde and lysed. Immunoprecipitation (IP) was performed with primary antibodies against tata-binding protein (TBP) as positive control or GRHL2. For negative controls, we included mouse and rabbit IgG (mIgG and rIgG, respectively). We selected 39 genes that were differentially regulated by GRHL2 transduction, according to the results shown in Figure 1. Two regions of the proximal promoters were amplified: R1 spanning approximately from −300 to −150 and R2 from −100 to +50. The PCR products were separated by electrophoresis through 2.0% agarose gels. Input is the DNA from the SCC4 cells without IP, serving as positive control
Figure 3.
GRHL2 binds to the proximal regions and regulates the activity of its target gene promoters. (a) PMP assay was performed with two different regions of each promoter denoted as R1 and R2, location of which is shown in the diagram with respect to the TSS (+1). (b) PMP was performed using 50 μg nuclear extracts of NHEK with R1 and R2 fragments of each promoter immobilized onto Dynabeads. With the protein samples that co-precipitated with the promoter fragments, western blotting was performed for GRHL2 and Sp1. (c) PMP was performed with the promoter fragments of GRHL1, GRHL3, and PCNA in the presence or absence of non-immobilized competitor DNA, and western blotting was done for GRHL2 and Sp1. Data illustrates the specificity of interaction between the proteins (GRHL2 and Sp1) and the promoter fragments. (d) Promoter activity was measured in NHEK/LXSN and NHEK/GRHL2 for LCE3D, IVL, SPRR2A, and hTERT, which were differentially regulated by GRHL2. Firefly luciferase reporter gene expression was driven by the promoters of EDC genes or hTERT, and normalized against Renilla luciferase driven by SV40 promoter
Then we utilized luciferase reporter activity systems, which are driven by the LCE3D, IVL, or SPRR2A promoters, to confirm whether GRHL2 binding has inhibitory effects on EDC gene promoter activity (Figure 3d). As comparison, hTERT promoter linked to luciferase gene was included. The promoter activities of LCE3D, IVL, and SPRR2A were notably reduced in NHEK/GRHL2 compared with those of NHEK/LXSN, whereas hTERT promoter activity was induced in GRHL2-transduced cells, as we previously reported.17 The data demonstrate that GRHL2 physically binds at gene promoters of EDC and other GRHL2 target genes, and such binding leads to alter gene promoter activity.
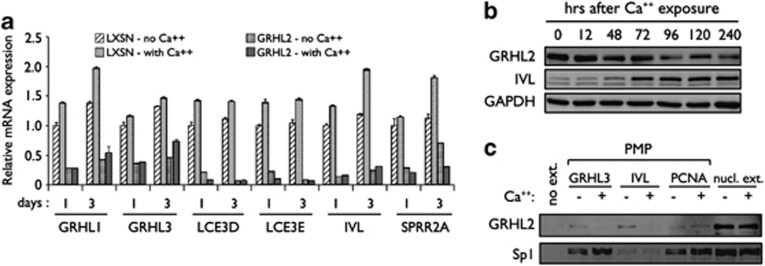
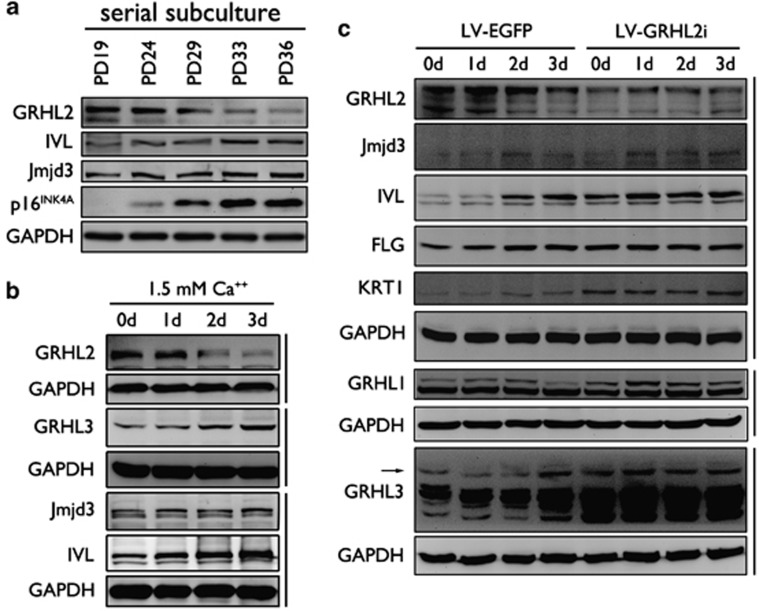
To assess the inhibitory roles of GRHL2 on EDC gene expression in differentiating conditions, NHEK/LXSN and NHEK/GRHL2 cells were exposed to Ca++, and expression of LCE3D, LCE3E, IVL, and SPRR2A, as well as GRHL1 and GRHL3 were measured by qRT-PCR (Figure 4a). Expression of the EDC genes and GRHL1 and GRHL3 were increased upon Ca++ exposure but strongly suppressed by GRHL2 transduction with or without Ca++ treatment. We then assessed the changes of GRHL2 binding in cells during differentiation. Rapidly proliferating HaCaT cells were then exposed to 2 mM Ca++ for up to 3 days, and PMP was performed using GRHL3, IVL, and PCNA promoter fragments (Figures 4b and c) to assess GRHL2 binding to the promoters in vitro. GRHL2 binding was noted on all the three promoters in the absence of Ca++ treatment, and strong Sp1 binding was noted on GRHL3 and PCNA promoters, while IVL promoter exhibited weak association with Sp1. Upon Ca++ exposure, GRHL2 expression level decreased in cells in time-dependent manner, and the binding intensity decreased from the GRHL3 and IVL promoters; on the contrary, Ca++ level did not affect GRHL2 binding at the PCNA promoter and Sp1 binding in all the three promoters tested. These data suggest that EDC gene expression is induced during keratinocyte differentiation, presumably involving the loss of GRHL2 binding at the promoter regions.
Figure 4.
GRHL2 transduction inhibits EDC gene expression in differentiating keratinocytes. (a) Rapidly proliferating NHEK/LXSN and NHEK/GRHL2 were exposed to 1.5 mM Ca++ for 3 days, and qRT-PCR was performed for EDC genes as well as GRHL1 and GRHL3. Bars represent the means of triplicates and S.D. for each gene. Gene expression data in the GRHL2 and LXSN groups (with or without Ca++ treatment) were compared in pairwise manner (P<0.01 for all tested genes). (b) HaCaT cells were exposed to 2 mM Ca++ for up to 10 days and harvested for western blotting after various time points for GRHL2 and IVL. GAPDH (glyceraldehyde 3-phosphate dehydrogenase) was probed as loading control. (c) PMP assay was performed for GRHL3, IVL, and PCNA promoter regions using the HaCaT nuclear extracts collected with or without 72 h exposure to Ca++
GRHL2 inhibits the EDC gene expression by altering histone modification at the gene promoter regions
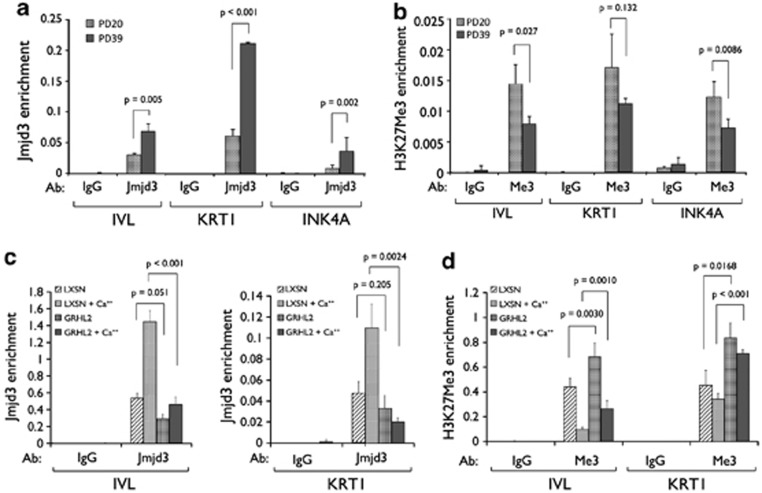
Epidermal differentiation can be regulated epigenetically through Jmjd3-mediated histone modification.16 To understand the mechanism by which GRHL2 inhibits EDC gene expression and keratinocyte differentiation, we explored the functional relationship between GRHL2 and epigenetic gene regulation by Jmjd3. In NHEK with increasing PDs, GRHL2 levels progressively decreased while the levels of IVL and p16INK4A increased (Figure 5a). Ca++-induced terminal differentiation in NHEK led to induction of IVL and decreased GRHL2 expression (Figure 5b). In both serially subcultured cells and those exposed to Ca++, changes in Jmjd3 level were remote. In another experiment, expression of endogenous GRHL2 was knocked down in HaCaT cells by lentiviral vector expressing shRNA against GRHL2 (LV-GRHL2i). HaCaT cells were used herein because these cells express high level of endogenous GRHL2 and undergo normal keratinocyte differentiation when exposed to physiological Ca++ level (Figure 4b). As a control, HaCaT cells were infected with an empty viral vector (LV-EGFP). GRHL2 knockdown in HaCaT led to induction of IVL, FLG, and KRT1 even in the absence of Ca++ treatment to the levels similar to Ca++-exposed control cells (Figure 5c). Interestingly, Ca++ exposure led to gradual increase in the level of GRHL3 and transient increase of GRHL1 level. GRHL2 knockdown in HaCaT cells led to further increase in the levels of these proteins, suggesting the potential negative gene regulation by GRHL2. Jmjd3 expression level was remotely detected in HaCaT cells, and Ca++ treatment led to small increment of the protein level. When we performed ChIP assay, Jmjd3 bindings at the promoter regions of IVL, KRT1, and INK4A were found notably increased in NHEK at PD 39 compared with the replicating culture at PD 20 (Figure 6a). Conversely, H3K27Me3 recruitment levels decreased at these promoters in NHEK with higher PD value (Figure 6b), suggesting that such repressive histone mark was removed by increased Jmjd3 binding. Next, Jmjd3 binding was compared in NHEK/LXSN and NHEK/GRHL2 cells before and after Ca++ exposure. At the IVL and KRT1 promoters, Jmjd3 enrichment was enhanced by Ca++ exposure in NHEK/LXSN, but there was strong repression of Jmjd3 binding at these promoters when GRHL2 was transduced even after Ca++ treatment (Figures 6c and d). As a result, H3K27Me3 enrichment at the promoter sites was increased in NHEK/GRHL2 compared with those of NHEK/LXSN (Figure 6e). These data suggest that GRHL2 inhibits EDC gene expression, in part, by modulating Jmjd3 enrichment to the gene promoters, resulting in increased repressive histone methylation.
Figure 5.
Reduced GRHL2 level triggers expression of EDC proteins in differentiating keratinocytes. (a) Rapidly proliferating NHEK were serially subcultured until senescence, and western blotting was performed at varying PD levels for GRHL2, IVL, Jmjd3, and p16INK4A. (b) NHEK were exposed to 1.5 mM Ca++ for up to 3 days and collected for western blotting for GRHL2, Jmjd3, IVL, and GRHL3. (c) HaCaT cells were infected with LV-EGFP or LV-GRHL2i to knock down endogenous GRHL2. The cells were then exposed to 2.0 mM Ca++ for 3 days and harvested for western blotting for GRHL2, Jmjd3, IVL, FLG, KRT1, GRHL1, GRHL3, and Jmjd3. Arrow indicates the expected size of GRHL3 (∼ 60 kDa). For all experiments, GAPDH (glyceraldehyde 3-phosphate dehydrogenase) was used as a loading control
Figure 6.
GRHL2 inhibits recruitment of Jmjd3 to the EDC gene promoters. (a) NHEK were collected at PDs 20 and 39, and ChIP assay was performed using Jmjd3 antibody. Co-precipitation of the at the IVL, KRT1, and INK4A gene promoter fragments were assessed by qPCR. (b) To assess the histone methylation status at the gene promoters, ChIP assay was performed using H3K27Me3 antibody. NHEK/LXSN and NHEK/GRHL2 were cultured in the presence or absence of 1.5 mM Ca++ for 3 days. (c) ChIP was performed for IVL and KRT1 promoters to assess recruitment of Jmjd3 and (d) to determine H3K27Me3 status. In all ChIP experiments, immunoglobulin G (IgG) was included as negative control, and the promoter PCR amplification for each sample was normalized against input DNA without immunoprecipitation
GRHL2 expression is enhanced in hyperproliferative skin lesions
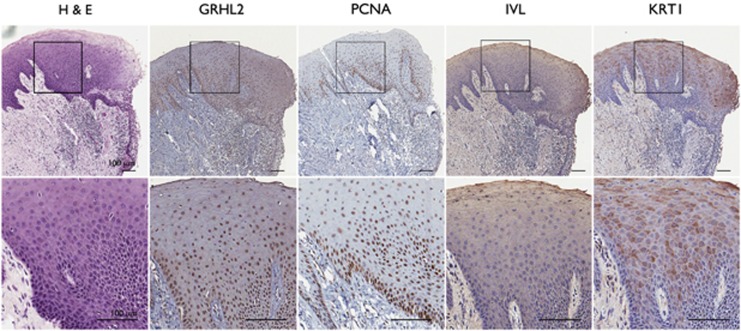
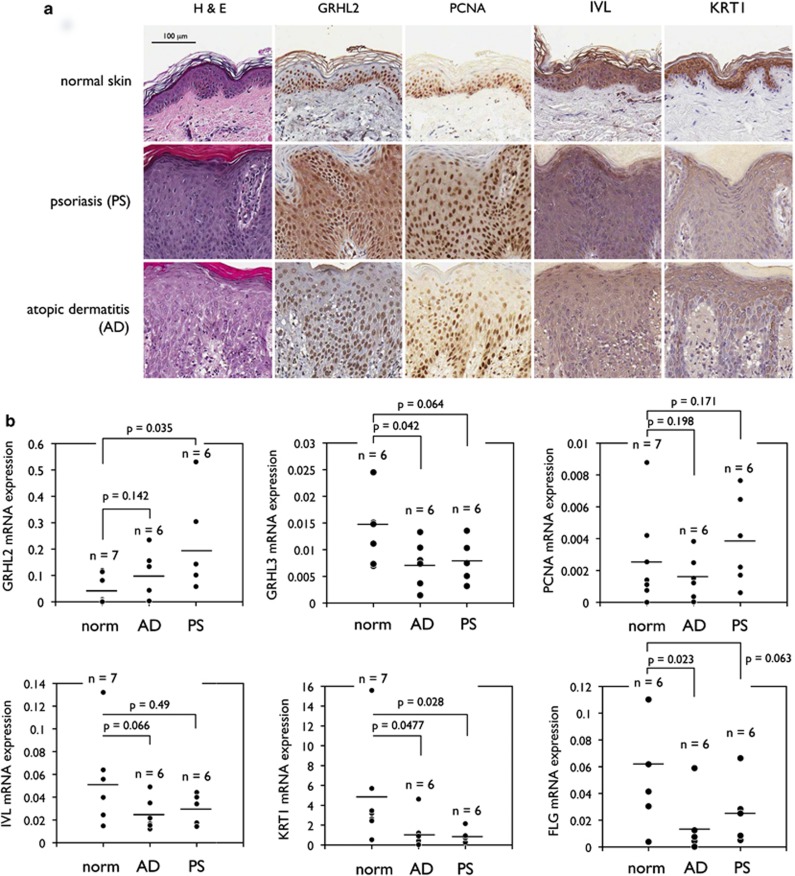
Our data above show that GRHL2 inhibits keratinocyte differentiation through transcriptional suppression of key target genes. Then the disease relevance of this finding is explored by assessing the expression of GRHL2 in human tissue samples. Immunohistochemistry was performed with oral mucosal tissues (Figure 7). In situ signal of GRHL2 staining was noted primarily at the basal cell layer of gingival epithelium and coincided well with PCNA staining. We confirmed the specificity of the GRHL2 antibody by staining the cells in which GRHL2 levels were altered by overexpression or knockdown (Supplementary Figure S1). IVL and KRT1 staining signals were found mostly at the suprabasal layer away from the basal cells with GRHL2 and PCNA staining. When we compared GRHL2 staining in normal skin and hyperproliferative skin lesions, for example, PS, it was found that GRHL2 staining was strongly induced in the thickened epithelial layer of the psoriatic tissue, whereas normal skin displayed GRHL2 staining limited to the basal layer (Figure 8). PCNA staining followed that of GRHL2 in both normal and psoriatic tissues. IVL and KRT1 staining was noted in the suprabasal layer of normal skin epithelium, but the staining intensity was notably reduced in psoriatic lesions. We also examined the GRHL2 expression in skin tissues presenting with AD, a chronic inflammatory skin disorder with epithelial hyperproliferation.22 AD tissues demonstrate thickened epithelium with inflammatory infiltrates in underlying connective tissue. Individual cells in the thickened rete pegs were positively stained for GRHL2 and PCNA, while the levels of IVL and KRT1 staining were reduced compared with those of normal skin. To quantitate the differences in the gene expression, we excised the epithelium from normal skins and those with AD or PS histopathology by laser-captured microdissection (LCM) and performed qRT-PCR. GRHL2 mRNA expression was increased in AD and PS samples, while those of GRHL3, IVL, KRT1, and FLG were notably reduced compared with normal skin (Figure 8b). These results demonstrate association of GRHL2 with epithelial hyperproliferation and impaired EDC gene expression in human skin lesions.
Figure 7.
GRHL2 expression is enriched at the basal cell layer of oral mucosal epithelium. Oral mucosal tissues isolated from gingiva were embedded in paraffin and sectioned for histological staining. Consecutively prepared thick (5 μm) sections of oral mucosa were stained for hematoxylin and eosin (H&E) or processed for immunohistochemistry (IHC) for GRHL2, PCNA, IVL, and KRT1. Upper panel shows the panoramic view of the mucosa, including the submucosal layer; islet is shown at higher magnification in the lower panel
Figure 8.
GRHL2 level is enhanced in hyperproliferative diseases of the skin. (a) Skin biopsy specimens presenting with normal, PS, and AD histopathology were stained with hematoxylin and eosin (H&E) or processed for immunohistochemistry (IHC) for GRHL2, PCNA, IVL, and KRT1. (b) Epithelial tissue, excluding acellular cornified layer, was excised from the skin tissues displaying normal, PS, and AD histopathology by LCM. mRNA levels of GRHL2, GRHL3, PCNA, IVL, KRT1, and FLG were then determined from each dissected tissue specimen by qRT-PCR
Discussion
Through high throughput proteomic profiling, we previously identified GRHL2 as a novel transcription factor of hTERT that controls the level of telomerase activity in NHEK and enhances replicative lifespan of cells.17 In the current study, we demonstrate that GRHL2 negatively regulates the expression of large subset of genes that belong to the EDC clustered at chromosome 1q21. This was found initially from microarray array gene profiling and subsequently confirmed through biochemical assays in NHEK with and without GRHL2 gene transduction. ChIP experiment revealed that GRHL2 interacts in vivo with the promoter regions of its numerous target genes identified from the microarray data. In vitro binding analyses by PMP assay demonstrated the specificity of such interaction. Studies of luciferase reporter constructs of several EDC gene promoters consistently showed that GRHL2 can reduce these promoter activities. On the contrary, GRHL2 transduction led to activation of the hTERT promoter, suggesting that GRHL2 may function as either positive or negative transcriptional regulator, presumably based on additional factors assembled on the promoter region.
During keratinocytes differentiation, whether through serial subculture until senescence or exposure to high Ca++ level, there was a progressive loss of endogenous GRHL2 protein level in time-dependent manner. When endogenous GRHL2 was knocked down in HaCaT cells, the basal levels of IVL, FLG, and KRT1 proteins were induced to the level similar to those of Ca++-treated control cells; also, Ca++ exposure did not cause further induction of these proteins in cells after GRHL2 knockdown. These data suggest that loss of GRHL2 alone is sufficient to trigger keratinocyte differentiation. Upon modulation of endogenous GRHL2 level in cells, there were concordant changes in the EDC gene expression to reflect the inhibitory effects of GRHL2. Underlying mechanism appears to involve epigenetic modification of H3K27 at the target gene promoters, causing increased histone methylation in the presence or absence of Ca++ treatment in NHEK. Consequently, this led to suppression of EDC gene expression in cells transduced with GRHL2, as H3K27Me3 is associated with epigenetic gene silencing.23 Epigenetic gene regulation by Jmjd3 and PcG proteins is a widespread phenomenon for multitude of biological pathways.24 H3K27Me3 is established by polycomb repressive complex 2 (PRC2) containing EZH2. PRC1 is then recruited to H3K27Me3 by polycomb (Pc) and inhibits RNA polymerase II, thereby causing gene silencing.25 Our current study showed that keratinocyte differentiation is inhibited by GRHL2, which suppressed many EDC genes expression through transcriptional silencing of the target promoters. Furthermore, GRHL2 enhanced the expression of EZH2 and Bmi-1 levels in NHEK (data not shown), whereas it inhibited recruitment of Jmjd3 to the EDC gene promoter regions. Consequently, the repressive H3K27Me3 histone mark is elevated and the genes remain silenced. The molecular mechanism by which GRHL2 inhibits Jmjd3 recruitment remains unknown and needs to be elucidated in further studies.
In vitro studies suggested the biological roles of GRHL2 in regulation of keratinocyte proliferation and differentiation presumably through regulating large number of potential ‘effector' genes of GRHL2. In situ staining of GRHL2 in stratified epithelia, as those found in oral mucosa and the skin, revealed intense staining of GRHL2 concentrated in the regenerative basal cell layer, in parallel with PCNA staining. This is the first reported in situ expression of GRHL2 in human tissues. The finding is in accordance with our data that GRHL2 binds to the promoter of PCNA and controls the gene expression (Figure 3). Also, GRHL2 staining pattern appears to be inversely correlated with those of differentiation markers, such as IVL and KRT1, again supporting its inhibitory role in keratinocyte differentiation. Investigation of GRHL2 levels in pathological skin lesions, for example, PS and AD, revealed enhanced protein and mRNA expression levels compared with normal tissue specimens. PS and AD present with enhanced basal cell proliferation coupled with impaired epithelial differentiation, which occurs, in part, through abnormal EDC gene expression.26, 27 For instance, KRT1 and IVL expression is suppressed in psoriatic and AD lesions.28 Loss-of-function variants of FLG gene were identified as a predisposing factor for AD;12 LCE3C and LCE3B deletions were identified as risk factors for PS through multicenter meta-analysis involving 9389 PS patients and 9477 control subjects.29 Our current study identified GRHL2 as a novel transcriptional regulator for numerous EDC genes, many of which are directly involved in the skin barrier formation. As GRHL2 also activates the expression of genes necessary for cell division, GRHL2 may contribute to increased basal cell proliferation and aberrant keratinocytes differentiation in PS and AD.
A recent study about GRHL2 knockout (KO) mice demonstrated the role of GRHL2 in regulation of E-cadherin and claudin 4 expressions, which make up the epithelial junctional complex.30 Deregulation of adhesion molecules is believed to cause gross structural defects, such as incomplete embryonic closure of neural tube, cranium, and body wall in GRHL2 KO mice.31 Besides its role in embryonic development, GRHL2 can also constitute Oct4 interactome along with other proteins in mouse embryonic stem cells, presumably regulating cellular plasticity and pluripotency.32 Using pooled shRNA screening, Dompe et al.33 identified GRHL2 as anti-apoptotic gene; knock down of endogenous GRHL2 in various breast cancer cell lines conferred sensitivity to death receptor-mediated apoptosis. Likewise, GRHL2 gene is frequently amplified in hepatocellular carcinoma and linked with early recurrence of the disease.34 We recently demonstrated the GRHL2 expression is enhanced in head and neck SCC and is required for telomerase activity and head and neck SCC cell proliferation.17, 18 These studies suggest additional roles of GRHL2 in cellular homeostasis and function other than ectodermal embryogenesis. The current study is the first demonstration that GRHL2 regulates epidermal skin keratinocyte differentiation through epigenetic transcriptional regulation of its target genes. Our current data also show that GRHL2 controls the expression of EDC genes in oral SCC cell lines, like SCC4 and SCC15, suggesting its role in cancer cell differentiation. Further study is needed to elucidate its involvement in the pathogenesis of epithelial hyperproliferative disorders, for example, PS, AD, and SCC and to test the possibility that GRHL2 may be a therapeutic target for these common diseases.
Materials and Methods
Cell culture
Primary NHEK were prepared from keratinized foreskin according to the methods described elsewhere.35 and maintained in EpiLife medium supplemented with the growth factors (Invitrogen, Carlsbad, CA, USA). The cells were serially subcultured until senescence, and the replication kinetics was determined by noting the PD levels with respect to time in culture.36 Exogenous GRHL2 was transduced into rapidly proliferating NHEK by retroviral vector expressing GRHL2 (LXSN-GRHL2). As a control, sister culture of NHEK were infected with empty viral vector (LXSN). The infected cells were selected with 200 μg/ml G418 (Sigma, St. Louis, MO, USA), and the resulting cells were designated as NHEK/GRHL2 or NHEK/LXSN. The cells were then maintained in serial subcultures as described above. Methods for retroviral construction, preparation, and infection can be found elsewhere.37
Microarray analysis
NHEK/LXSN and NHEK/GRHL2 cells were harvested at early (PDS 14 for both cell types) and late (PDs 23 and 30, respectively) stages of serial passage. Total cellular RNA was isolated using Qiagen RNeasy total RNA isolation kit (Qiagen, Chatsworth, CA, USA). cDNA was synthesized from 10 μg total RNA and the biotin-labeled antisense cRNA was produced by in vitro transcription using the BioArray Highyield kit (ENZO Life Sciences, Farmingdale, NY, USA). cRNA (15 μg) was fragmented and hybridized to GeneChip Human Gene 1.0 ST Array (Affymetrix, Santa Clara, CA, USA). The chip was stained with streptavidin–phycoerythrin and analyzed using GeneChip Operating Software (GCOS 1.2; Affymetrix) for pair wise comparison of gene expression. The statistical significance for each gene was evaluated by analysis of variance single-factor analysis and the fold difference >5.0 at P<0.05 were considered significant. Gene chip hybridization and data analyses were performed at the Microarray Core at the David Geffen School of Medicine at UCLA (Los Angeles, CA, USA).
qRT-PCR
Total RNA was isolated from the cultured cells using the RNeasy Mini kit (Qiagen). DNA-free total RNA (5 μg) was used for RT reaction in the buffer containing 300U Superscript II (Invitrogen), 10 mM dithiotrietol, 0.5 μg random hexamer (Promega), and 125 μM dNTP mixture (Promega, Madison, WI, USA). qPCR was performed in triplicates for each sample with LC480 SYBR Green I master using universal cycling conditions in LightCycler 480 (Roche, South San Francisco, CA, USA). The primer sequences were obtained from the Universal Probe Library (Roche)—the primer sequences are shown in Supplementary Table S1. The PCR cycling conditions were 45 cycles of 10 s at 95 °C, 45 s at 55 °C, and 20 s at 72 °C. Second-derivative Cp value determination method was used to compare the fold differences, and Cp is the cycle at which the threshold is crossed. PCR product quality was monitored using post-PCR melt curve analysis.
ChIP assay
ChIP assay was performed according to the methods described earlier.38 Briefly, nuclear proteins were cross-linked to DNA by adding 1% formaldehyde for 10 min, and cross-linking was stopped by adding glycine (to final 0.15 M). The cells were then collected in ice-cold phosphate-buffered saline supplemented with a protease inhibitor mixture and lysed in a buffer (1% SDS, 10 mM EDTA, protease inhibitors, 50 mM Tris-HCl, pH 8.1), and the genomic DNA was fragmented to 300–1000 bp in length by sonication. Cellular debris was removed by centrifugation and the lysates were diluted 1 : 10 in ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM NaCl, protease inhibitors, 16.7 mM Tris-HCl, pH 8.1). Chromatin solutions were incubated with 5 μl primary antibodies (or 5 μl of control IgG) overnight at 4 °C with rotation. Immunocomplexes were precipitated with 60 μl of protein G-agarose slurry (Upstate Biotechnology, Lake Placid, NY, USA) by centrifugation and eluted in 500 μl of buffer (1% SDS, 100 mM NaHCO3). Precipitated DNA was recovered by phenol extraction and used for PCR amplification for gene promoter regions. We chose two regions of the promoter for analysis: approximately from −300 to −150 (Region 1) and from −100 to+50 (Region 2). The PCR products were separated by electrophoresis through 2.0% agarose gels. The sequences for the PCR primers are shown in Supplementary Table S2. ChIP was followed with qPCR to determine enrichment of H3K27Me3 and Jmjd3 at the IVL and KRT1 gene promoters in NHEK. qPCR was performed with the purified DNA using LightCycler 480 (Roche). As a negative control, we included samples pulled down with IgG. qPCR readout was normalized relative to the amount of amplification from input.
Western blotting
Whole-cell extracts from the cultured cells were isolated using lysis buffer (1% Triton X-100, 20 mM Tris –HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 2.5 mM sodium pyrophosphate, 1 μM β-glycerophosphate, 1 mM sodium orthovanadate, 1 mg/ml PMSF). whole-cell extractswere then fractionated by SDS-PAGE, and proteins were transferred onto Immobilon protein membranes (Millipore, Billerica, MA, USA). The membranes were incubated successively with the primary and the secondary antibodies as indicated above, and the immunocomplexes were visualized using the chemiluminescence reagent (Amersham Pharmacia Biotech, Piscataway, NJ, USA). We used the following primary antibodies for this study: GAPDH from Santa Cruz Biotech (Santa Cruz, CA, USA); GRHL2 and GRHL3 from Abnova (Taipei City, Taiwan); PCNA from Calbiochem (Gibbstown, NJ, USA); IVL and GRHL1 from Sigma; FLG from Covance (Emeryville, CA, USA); H3K27Me3 from Millipore; Jmjd3 and TBP from Abcam (Cambridge, MA, USA). Horseradish peroxidase (HRP)-conjugated secondary antibodies were obtained from Santa Cruz Biotech.
PMP assay
PMP was performed using the streptavidin-coated Dynabeads (Invitrogen) according to the methods described elsewhere.18 Biotinylated gene promoter DNA fragments of GRHL1, GRHL3, PCNA, and IVL were amplified in a PCR reaction with biotin conjugation on the 5′-end of the forward primer. For GRHL1, GRHL3, and PCNA, two regions were amplified as detailed in Supplementary Table S2. For IVL promoter, one region from −121 to +23 was amplified, where +1 denotes TSS. The PCR products were immobilized onto the beads and then incubated with 50 μg nuclear extract isolated from HaCaT cells. After 1 h incubation at room temperature, the beads were collected by the magnetic apparatus and washed with the washing buffer containing 0.5% nonidet P-40. Proteins that bound the gene promoters were co-precipitated with the beads, and fractionated by SDS-PAGE. Subsequently, western blotting was performed to determine the presence of GRHL2 and Sp1 in the protein samples that co-precipitated with the magnetic beads.
Gene promoter luciferase assay
Promoter regions of LCE3D (−1000 to +104), IVL (−1000 to +107), SPRR2A (−999 to +279), and hTERT (−1665 to +5) were cloned into pGL3B-Luc reporter plasmid (Promega) expressing firefly luciferase. The promoter-luciferase constructs were transfected into NHEK (1 × 105 cells per well) using Lipofectin Reagent (Invitrogen), along with pRL-SV40 containing Renilla luciferase cDNA under the control of SV40 enhancer/promoter. Cells were collected after 48 h post-transfection, and the lysates were prepared using Dual Luciferase Reporter Assay System (Promega). Firefly and Renilla luciferase activities were measured using a luminometer (Turner Designs, Sunnyvale, CA, USA). Renilla luciferase activity was used to control for the varied transfected efficiency. The gene promoter activity, reflected by the firefly luciferase activity, was determined as the mean of at least triplicates per experiment.
Immunohistochemistry
In situ expression of GRHL2, IVL, KRT1, and PCNA were determined in tissue specimens by immunohistochemistry. Normal skin tissues and those diagnosed with AD or PS were obtained from the Translational Pathology Core Laboratory (TPCL) of the Department of Pathology and Laboratory Medicine at the David Geffen School of Medicine, UCLA (Los Angeles, CA, USA). Oral mucosal tissues were obtained from the UCLA Dental Center (Los Angeles, CA, USA). immunohistochemistry was carried out with appropriated primary antibodies on a 5-μm thick slides according to the methods described.39
LCM
Following histological examination of haematoxylin and eosin sections, samples of the normal skin (n=7), AD tissues (n=6), and PS tissues (n=6) were used for LCM. Epithelium from the paraffin-embedded tissue samples were isolated using the Leica laser microdissection (LMD) 7000 system (Leica Microsystems, Richmond, IL, USA) at the California NanoSystems Institute (CNSI, UCLA). Subsequently, total RNA was extracted using the High Pure RNA Paraffin Kit (Roche), according to the manufacturer's guidelines. RNA samples were reverse-transcribed using a Superscript II RT kit (Invitrogen) with random hexamer primers (Promega). Transcript expression was analyzed by qPCR with the LightCycler 480 system (Roche). Thermocycle conditions for all PCR reactions contained an initial denaturation stage of 95 °C for 10 min, followed by 50 cycles of 95 °C for 15 s and 60 °C for 1 min.40
Statistical analysis
Means were compared using one way analysis of variance (ANOVA) methods and pair wise t-tests under the ANOVA model (post hoc t-tests).
Acknowledgments
This study was supported, in part, by the grants (R01DE18295 and K02DE18959) from the NIDCR/NIH and through the UCLA School of Dentistry Faculty Seed Grant Program. MKK is also supported by the Jack A. Weichman Endowed Fund. Authors thank Dr Jeff Gornbein for assistant with statistical analyses of data.
Glossary
- GRHL2
Grainyhead-like 2
- GRH
Grainyhead
- hTERT
human telomerase reverse transcriptase
- PMP-MS
promoter magnetic precipitation and mass spectrometry
- PCNA
proliferating cell nuclear antigen
- NHEK
normal human epidermal keratinocytes
- EDC
epidermal differentiation complex
- SPRR
small proline-rich
- PD
population doubling
- PS
psoriasis
- SCC
squamous cell carcinoma
- Sp1
Specificity Protein 1
- H3K27Me3
histone 3 Lys 27 trimethylation
- ChIP
chromatin immunoprecipitation
- IVL
involucrin
- FLG
filaggrin
- AD
atopic dermatitis
- LCE
late cornifying envelope
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by E Candi
Supplementary Material
References
- Stramer B, Martin P. Cell biology: master regulators of sealing and healing. Curr Biol. 2005;15:R425–R427. doi: 10.1016/j.cub.2005.05.034. [DOI] [PubMed] [Google Scholar]
- Bray SJ, Kafatos FC. Developmental function of Elf-1: an essential transcription factor during embryogenesis in Drosophila. Genes Dev. 1991;5:1672–1683. doi: 10.1101/gad.5.9.1672. [DOI] [PubMed] [Google Scholar]
- Huang JD, Dubnicoff T, Liaw GJ, Bai Y, Valentine SA, Shirokawa JM, et al. Binding sites for transcription factor NTF-1/Elf-1 contribute to the ventral repression of decapentaplegic. Genes Dev. 1995;9:3177–3189. doi: 10.1101/gad.9.24.3177. [DOI] [PubMed] [Google Scholar]
- Narasimha M, Uv A, Krejci A, Brown NH, Bray SJ. Grainy head promotes expression of septate junction proteins and influences epithelial morphogenesis. J Cell Sci. 2008;121:747–752. doi: 10.1242/jcs.019422. [DOI] [PubMed] [Google Scholar]
- Cenci C, Gould AP. Drosophila grainyhead specifies late programmes of neural proliferation by regulating the mitotic activity and Hox-dependent apoptosis of neuroblasts. Development. 2005;132:3835–3845. doi: 10.1242/dev.01932. [DOI] [PubMed] [Google Scholar]
- Hayashi Y, Yamagishi M, Nishimoto Y, Taguchi O, Matsukage A, Yamaguchi M. A binding site for the transcription factor grainyhead/nuclear transcription factor-1 contributes to regulation of the Drosophila proliferating cell nuclear antigen gene promoter. J Biol Chem. 1999;274:35080–35088. doi: 10.1074/jbc.274.49.35080. [DOI] [PubMed] [Google Scholar]
- Yu Z, Lin KK, Bhandari A, Spencer JA, Xu X, Wang N, et al. The Grainhead-like epithelial transactivator Get-1/Grhl3 regulates epidermal terminal differentiation and interacts functionally with LMO4. Dev Biol. 2006;299:122–136. doi: 10.1016/j.ydbio.2006.07.015. [DOI] [PubMed] [Google Scholar]
- Wilanowski T, Caddy J, Ting SB, Hislop NR, Cerruti L, Auden A, et al. Perturbed desmosomal cadherin expression in grainy head-like 1-null mice. EMBO J. 2008;27:886–897. doi: 10.1038/emboj.2008.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marenholz I, Volz A, Ziegler A, Davies A, Ragoussis I, Korge B, et al. Genetic analysis of the epidermal differentiation complex (EDC) on human chromosome 1q21: Chromosomal orientation, new markers, and a 6-Mb YAC contig. Genomics. 1996;37:295–302. doi: 10.1006/geno.1996.0563. [DOI] [PubMed] [Google Scholar]
- Marenholz I, Gimenez Rivera VA, Esparza-Gordillo J, Bauerfeind A, Lee-Kirsch MA, Ciechanowicz A, et al. Association screening in the epidermal differentiation complex (EDC) identifies an SPRR3 repeat number variant as a risk factor for eczema. J Invest Dermatol. 2001;131:1644–1649. doi: 10.1038/jid.2011.90. [DOI] [PubMed] [Google Scholar]
- Spergel JM. Epidermiology of atopic dermatitis and atopic march in children. Immunol Allergy Clin North Am. 2010;30:269–280. doi: 10.1016/j.iac.2010.06.003. [DOI] [PubMed] [Google Scholar]
- Palmer CAN, Irvine AD, Terron-Kwiatkowski A, Zhao Y, Liao H, Lee SP, et al. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat Genet. 2006;38:441–446. doi: 10.1038/ng1767. [DOI] [PubMed] [Google Scholar]
- Giardina E, Sinibaldi C, Chini L, Moschese V, Marulli G, Provini A, et al. Co-localization of susceptibility loci for psoriasis (PSORS4) and atopic dermatitis (ATOD2) on human chromosome 1q21. Hum Hered. 2006;61:229–236. doi: 10.1159/000095059. [DOI] [PubMed] [Google Scholar]
- De Cid R, Riveira-Munoz E, Zeeuwen PL, Robarge J, Liao W, Dannhauser EN, et al. Deletion of the late cornified envelope LCE3B and LCE3C genes as a susceptibility factor for psoriasis. Nat Genet. 2009;41:211–215. doi: 10.1038/ng.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergboer JGM, Zeeuwen PLJM, Schalkwijk J. Pathogenesis of atopic dermatitis and psoriasis: Focus on the epidermal differentiation complex. Open Dermatol J. 2010;4:48–51. [Google Scholar]
- Sen GL, Webster DE, Barragan DI, Chang HY, Khavari PA. Control of differentiation in a self-renewing mammalian tissue by the histone demethylase JMJD3. Genes Dev. 2008;22:1865–1870. doi: 10.1101/gad.1673508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Dong Q, Shin KH, Kim RH, Oh JE, Park NH, et al. Grainyhead-like 2 enhances the human telomerase reverse transcriptase gene expression by inhibiting DNA methylation at the 5'-CpG island in normal human keratinocytes. J Biol Chem. 2010;285:40852–40863. doi: 10.1074/jbc.M110.103812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang X, Chen W, Kim RH, Kang MK, Park NH. Regulation of the hTERT promoter activity by MSH2, the hnRNPs K and D, and GRHL2 in human oral squamous cell carcinoma cells. Oncogene. 2009;28:565–574. doi: 10.1038/onc.2008.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilanowski T, Tuckfield A, Cerruti L, O'Connell S, Saint R, Parekh V, et al. A highly conserved novel family of mammalian developmental transcription factors related to Drosophila grainyhead. Mech Dev. 2002;114:37–50. doi: 10.1016/s0925-4773(02)00046-1. [DOI] [PubMed] [Google Scholar]
- Ting SB, Wilanowski T, Cerruti L, Zhao LL, Cunningham JM, Jane SM. The identification and characterization of human Sister-of-Mammalian Grainyhead (SOM) expands the grainyhead-like family of developmental transcription factors. Biochem J. 2003;370:953–962. doi: 10.1042/BJ20021476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paunesku T, Mittal S, Protic M, Oryhon J, Korolev V, Joachimiak A, et al. Proliferating cell nuclear antigen (PCNA): ringmaster of the genome. Int J Radiat Biol. 2001;77:1007–1021. doi: 10.1080/09553000110069335. [DOI] [PubMed] [Google Scholar]
- Proksch E, Folster-Holst R, Jensen JM. Skin barrier function, epidermal proliferation and differentiation in eczema. J Dermatol Sci. 2006;43:159–169. doi: 10.1016/j.jdermsci.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Kondo Y, Shen L, Chen AS, Ahmed S, Boumber Y, Charo C, et al. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat Genet. 2008;40:741–750. doi: 10.1038/ng.159. [DOI] [PubMed] [Google Scholar]
- Prezioso C, Orlando V. Polycomb proteins in mammalian cell differentiation and plasticity. FEBS Lett. 2011;585:2067–2077. doi: 10.1016/j.febslet.2011.04.062. [DOI] [PubMed] [Google Scholar]
- Eckert RL, Adhikary G, Rorke EA, Chew YC, Balasubramanian S. Polycomb group proteins are key regulators of keratinocyte function. J Invest Dermatol. 2011;131:295–301. doi: 10.1038/jid.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovenschen HJ, Seyger MM, van de Kerkhof PC. Plaque psoriasis vs atopic dermatitis and lichen planus: a comparison for lesional T cell subsets, epidermal proliferation and differentiation. Br J Dermatol. 2005;153:72–78. doi: 10.1111/j.1365-2133.2005.06538.x. [DOI] [PubMed] [Google Scholar]
- Lowes MA, Bowcock AM, Krueger JG. Pathogenesis and therapy of psoriasis. Nature. 2007;445:866–873. doi: 10.1038/nature05663. [DOI] [PubMed] [Google Scholar]
- Jensen JM, Folster-Holst R, Baranowsky A, Schunck M, Winoto-Morbach S, Neumann C, et al. Impaired sphingmyelinase activity and epidermal differentiation in atopic dermatitis. J Invest Dermatol. 2004;122:1423–1431. doi: 10.1111/j.0022-202X.2004.22621.x. [DOI] [PubMed] [Google Scholar]
- Riveira-Munoz E, He SM, Escaramís G, Stuart PE, Hüffmeier U, Lee C, et al. Meta-analysis confirms the LCE3C LCE3B deletion as a risk factor for psoriasis in several ethnic groups and finds interaction with HLA-Cw6. J Invest Dermatol. 2011;131:1105–1109. doi: 10.1038/jid.2010.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werth M, Walentin K, Aue A, Schonheit J, Wuebken A, Pode-Shakked N, et al. The transcription factor grainyhead-like 2 regulates the molecular composition of the epithelial apical junctional complex. Development. 2010;137:3835–3845. doi: 10.1242/dev.055483. [DOI] [PubMed] [Google Scholar]
- Pyrgaki C, Liu A, Niswander L. Grainyhead-like 2 regulates neural tube closure and adhesion molecule expression during neural fold fusion. Dev Biol. 2011;353:38–49. doi: 10.1016/j.ydbio.2011.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg DL, Snoek T, Mullin NP, Yates A, Bezstarosti K, Demmers J, et al. An Oct4-centered protein interaction network in embryonic stem cells. Cell Stem Cell. 2011;6:369–381. doi: 10.1016/j.stem.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dompe N, Rivers CS, Li L, Cordes S, Schwickart M, Punnoose EA, et al. A whole-genome RNAi screen identifies an 8q22 gene cluster that inhibits death receptor-mediated apoptosis. Proc Natl Acad Sci. 2011;108:E943–E951. doi: 10.1073/pnas.1100132108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Kanai F, Tada M, Tateishi R, Sanada M, Nannya Y, et al. Gain of GRHL2 is associated with early recurrence of hepatocellular carcinoma. J Hepatol. 2008;49:746–757. doi: 10.1016/j.jhep.2008.06.019. [DOI] [PubMed] [Google Scholar]
- Kang MK, Bibb C, Baluda MA, Rey O, Park NH. In vitro replication and differentiation of normal human oral keratinocytes. Exp Cell Res. 2000;258:288–297. doi: 10.1006/excr.2000.4943. [DOI] [PubMed] [Google Scholar]
- Kang MK, Guo W, Park NH. Replicative senescence of normal human oral keratinocytes is associated with the loss of telomerase activity without shortening of telomeres. Cell Growth Differ. 1998;9:85–95. [PubMed] [Google Scholar]
- Kang MK, Park NH. Extension of cell life span using exogenous telomerase. Methods Mol Biol. 2007;371:151–165. doi: 10.1007/978-1-59745-361-5_12. [DOI] [PubMed] [Google Scholar]
- Dong Q, Oh JE, Chen W, Kim R, Kim RH, Shin KH, et al. Radioprotective effects of Bmi-1 involve epigenetic silencing of oxidase genes and enhanced DNA repair in normal human keratinocytes. J Invest Dermatol. 2011;131:1216–1225. doi: 10.1038/jid.2011.11. [DOI] [PubMed] [Google Scholar]
- Kang MK, Kim RH, Kim SJ, Yip FK, Shin KH, Dimri GP, et al. Elevated Bmi-1 expression is associated with dysplastic cell transformation during oral carcinogenesis and is required for cancer cell replication and survival. Br J Cancer. 2007;96:126–133. doi: 10.1038/sj.bjc.6603529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budczies J, Weichert W, Noske A, Müller BM, Weller C, Wittenberger T, et al. Genome-wide gene expression profiling of formalin-fixed paraffin-embedded breast cancer core biopsies using microarrays. J Histochem Cytochem. 2011;59:146–157. doi: 10.1369/jhc.2010.956607. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.