Abstract
HOX genes have been implicated as regulators of normal and leukemic stem cell functionality, but the extent to which these activities are linked is poorly understood. Previous studies revealed that transduction of primitive mouse hematopoietic cells with a NUP98HOXA10homeodomain (NA10HD) fusion gene enables a subsequent rapid and marked expansion in vitro of hematopoietic stem cell numbers without causing their transformation or deregulated expansion in vivo. To determine whether forced expression of NA10HD in primitive human cells would have a similar effect, we compared the number of long-term culture-initiating cells (LTC-ICs) present in cultures of lenti-NA10HD versus control virus-transduced CD34+ cells originally isolated from human cord blood and chronic phase (CP) chronic myeloid leukemia (CML) patients. We found that NA10HD greatly increases outputs of both normal and Ph+/BCR-ABL+ LTC-ICs, and this effect is particularly pronounced in cultures containing growth factor-producing feeders. Interestingly, NA10HD did not affect the initial cell cycle kinetics of the transduced cells nor their subsequent differentiation. Moreover, immunodeficient mice repopulated with NA10HD-transduced CP-CML cells for more than 8 months showed no evidence of altered behavior. Thus, NA10HD provides a novel tool to enhance both normal and CP-CML stem cell expansion in vitro, without apparently altering other properties.
Keywords: hematopoietic stem cells, HOX genes, chronic myeloid leukemia, long-term culture-initiating cells
Introduction
Hematopoietic stem cells (HSCs) are responsible for the lifetime output of mature blood cells. This is achieved through the generation of progenitors that produce the different lineages of blood cells as well as daughter HSCs through self-renewal divisions. Many factors that are essential to the creation, expansion and maintenance of HSCs in vivo have now been identified.1 Prominent among these are the HOX family of transcriptional regulators.2 These include a number of HOX genes, such as HOXB4, that enhance mouse HSC expansion under specified conditions in vitro. When expressed as part of fusions with the nucleoporin 98 gene (NUP98) other HOX genes also perturb differentiation and/or contribute to the development of myeloid leukemia. As part of a larger survey of the properties of these naturally occurring and engineered genes, we identified an extremely potent HOX-derivative that consists of the 5′ sequences of a NUP98 cDNA and the homeodomain (HD) of the HOXA10 cDNA (hereafter termed NA10HD).3, 4 Intriguingly, this fusion construct stimulates a very rapid and marked expansion in vitro of transduced mouse HSCs, and yet, these cells retain normal HSC functionality in vivo.3, 4 The properties of NA10HD have thus provided a powerful new tool for manipulating and investigating the self-renewal behavior of primitive murine hematopoietic cells.
The present study was designed to investigate whether forced overexpression of NA10HD would also have an ability to promote the self-renewal/expansion of primitive human hematopoietic cells. For this purpose, we created a lenti-viral vector encoding NA10HD and used it to transduce human CD34+ cells isolated from various sources. These included samples of normal human cord blood (CB) and samples obtained from patients with chronic phase (CP) chronic myeloid leukemia (CML) in which the most primitive compartments had a variable representation of Ph+/BCR-ABL+ cells. The selection of CML patient samples with either predominantly Ph+/BCR-ABL+ or predominantly normal cells in their primitive compartments enabled effects of NA10HD on primitive normal human HSCs in both newborn and adult tissues to be evaluated and compared with the effects on primitive human cells in which a first ‘hit' (creation of the BCR-ABL oncogene) has occurred. Effects on CP-CML cells are of particular interest because of their postulated reduced self-renewal potential, as indicated by the long latent period before CP-CML becomes apparent (5–7 years),5, 6, 7 and the very slow rate at which CP-CML stem cells accumulate. A relative deficiency in the self-renewal ability of CP-CML HSCs also explains the superior, albeit short-lived, competitive repopulating activity exhibited by residual normal HSCs in CML patients immediately after they are given intensive chemotherapy,8 and the inferior retention of Ph+/BCR-ABL+ long-term culture-initiating cell (LTC-IC) activity in vitro under the same optimized conditions.9, 10 Defective self-renewal behavior is also characteristic of BCR-ABL-transformed mouse HSCs.11, 12, 13, 14
The studies presented here show that transduction of primitive normal and BCR-ABL+ (CP-CML) cells with NA10HD enables an enhanced generation of LTC-ICs of both genotypes without a detectable effect on their execution of normal differentiation programs or other evidence of further transformation upon prolonged follow-up of their progeny in transplanted immunodeficient mice. Thus, NA10HD provides a novel tool to enhance both normal and CP-CML stem cell expansion in vitro, without apparently altering other properties.
Materials and methods
Primary cells
Low-density (<1.077 g/ml) cells were isolated from anonymized samples of heparinized human CB obtained from normal newborns delivered by cesarian section and the cells then cryopreserved. Heparin-treated peripheral blood or leukapheresis cells were obtained from four CP-CML patients with elevated WBC counts (Supplementary Table S1). None had been treated with tyrosine kinase inhibitors before obtaining the cells. All human cells were obtained with consent according to procedures approved by the Research Ethics Board of the University of British Columbia. CD34+ cell-enriched populations (>80%) were isolated either by positive selection or by removal of lineage-marker-positive (Lin+) cells (using the EasySep kit or StemSep kit, respectively, from Stemcell Technologies, Vancouver, BC, Canada). Purer (>95%) subpopulations of Lin−CD34+ cells (that is, subdivided according to their expression of CD38 and rhodamine-123 (Rho)) were isolated using a fluorescence-activated cell sorter (FACSVantage or Influx, Becton Dickinson, San Jose, CA, USA).15, 16
Colony-forming cell (CFC) and LTC-IC assays
Granulopoietic (CFU-GM), erythroid (CFU-E and BFU-E) and multi-potent myeloid (CFU-GEMM) progenitors and LTC-ICs were assayed as previously described,17 the latter by determining the number of CFCs present after 6 weeks of co-culture with a mixture of irradiated M210B4 and Sl/Sl stromal cells engineered to produce human SF, interleukin-3 (IL-3) and G-CSF either in bulk assays or by limiting dilution analysis, as indicated. Morphological analyses were performed on Wright–Giemsa-stained cytospin preparations.
Colony genotyping
Individual 12–14-day-old colonies were removed with a fine pipette from CFC assays containing, when possible, <100 colonies per 35 mm petri dish. For cytogenetic analyses, cells were processed and metaphases G-banded.18 For BCR-ABL and NA10HD transcript analyses, removed colonies were washed individually in PBS, and RNA was then extracted using a Picopure kit (Life Technologies, Carlsbad, CA, USA). Reverse transcription was performed using Superscript III and random hexamers (Life Technologies). Quantitative PCR measurements of BCR-ABL, NA10HD and GAPDH transcripts were carried out using SYBR Green master mix (Life Technologies) and the following primers: forward BCR, 5′-ACTGTCCACAGCATTCCGC-3′ reverse ABL, 5′-GAGCGGCTTCACTCAGACC-3′ forward-NUP98, 5′-GGGCCCCTGGATTTAATACT-3′ reverse HOXA10HD, 5′-CCTTCTCCAGCTCCAGTGTC-3′ forward GAPDH, 5′-ACGTACTCAGCGCCAGCATC-3′ reverse GAPDH, 5′-ACCGTCAAGGCTGAGAACGG-3′. Only colonies for which the Ct value for GAPDH transcripts was <27 and the Tm obtained by a Gaussian dissociation curve was the expected Tm of the amplicon±0.2 °C were further analyzed. These were classified as BCR-ABL+ if the corresponding dCt was ⩽10, and normal if the BCR-ABL transcripts were undetectable after 45 cycles, or if the dCt was >12. Colonies for which dCt values of 10 <dCt ⩽12 were discarded.
Suspension cultures
Test cells were cultured in 96 or 24-well plates preloaded with 200 μl or 2 ml, respectively, of serum-free Iscove's medium (SFM) containing bovine serum albumin, insulin and transferrin (BIT, Stemcell Technologies), 40 μg/ml low-density lipoproteins, 10−4 ℳ 2-mercaptoethanol (Sigma, St Louis, MO, USA) and the following purified recombinant human growth factors (GFs): 20 ng/ml IL-3 (Novartis, Basel, Switzerland), 20 ng/ml IL-6 (Cangene, Mississauga, ON, Canada), 20 ng/ml granulocyte colony-stimulating factor (G-CSF, Stemcell Technologies), 100 ng/ml flt3-ligand (Immunex Corp., Seattle, WA, USA) and 100 ng/ml Steel factor (Stemcell Technologies; referred to as GF-SFM). Viable cell counts were determined by hemocytometer counts of trypan blue-excluding cells and the cells diluted in fresh medium to maintain the cell concentration below 106 cells/ml.
Virus production
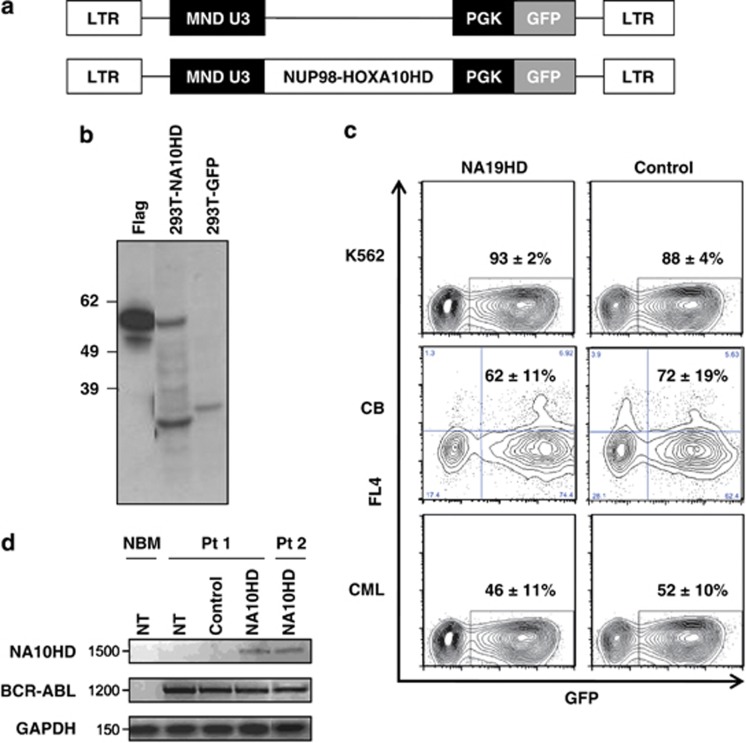
The NA10HD fusion gene cDNA3 was cloned into the (MND-PGK-green fluorescent protein (GFP) lenti-viral vector19 (Figure 1a) and the construct sequence was verified. Virus was produced by transient transfection of 293T cells. Protein production in these cells was verified by western blotting (Figure 1b) and virus-containing supernatants were concentrated.20 K562 cells exposed to NA10HD or control virus preparations for 6 h contained 93±2% and 88±4% GFP+ cells, respectively, 2 days later (n=4, Figure 1c), indicative of equivalent titers.
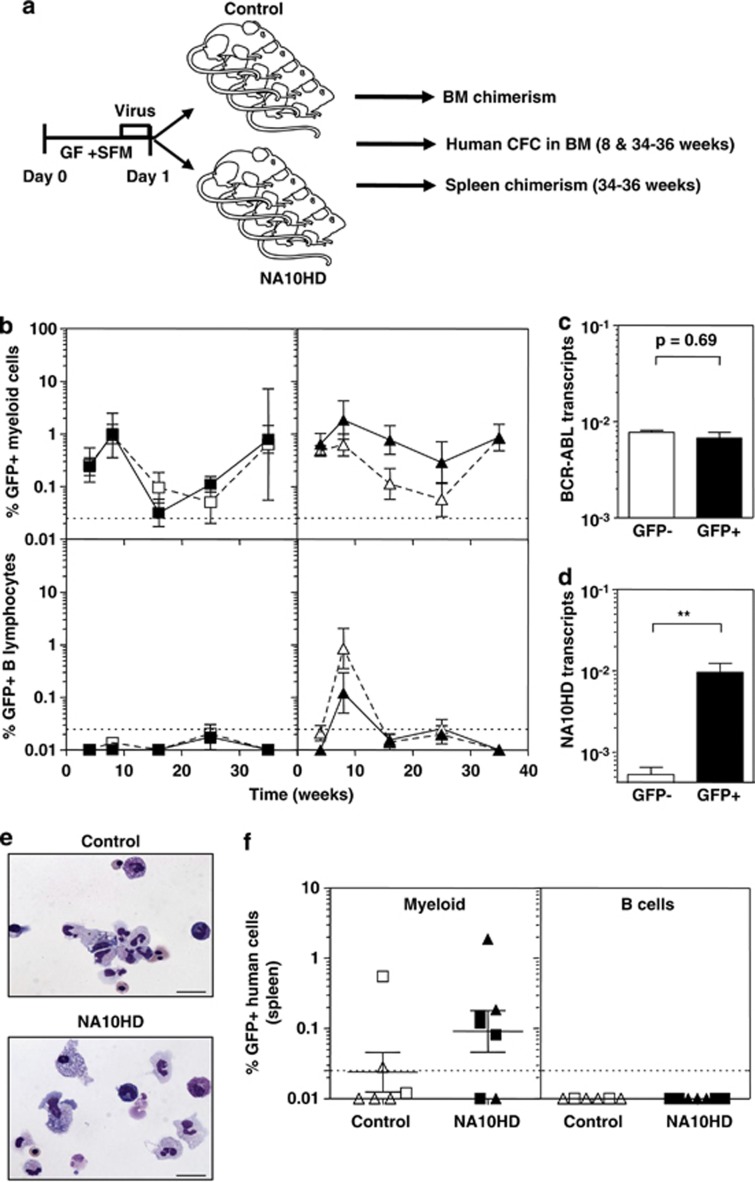
Figure 1.
Vector validation. (a) Lenti-viruses used. The promoter region is shown in black. (b) Western blot showing flag-tagged NA10HD protein in transfected 293T cells. The expected 56 kDa size protein is present only in NA10HD-transfected 293T cells. Bands smaller than 39 kDa probably reflect cross reaction of the monoclonal ANTI-FLAG M2 antibody with naturally occurring FLAG-like epitopes in the cell lysates. Molecular weight markers (kDa) are indicated on the left. (c) Representative FACS profiles showing the gene transfer efficiency to: K562 cells measured 2 days after exposure to virus (upper panels), CD34+ CB cells assessed after their maintenance in suspension culture with GFs for 7 days after exposure to virus (middle panels) and CD34+ CP-CML cells (bottom) assessed 2 days after exposure to virus (lower panels). Values shown are mean±s.e.m. for the % CD34+ (CB) or total (K562 and CML) GFP+ cells for both constructs (n=4 for each cell type). (d) NA10HD, BCR-ABL and GAPDH transcript detection by quantitative reverse transcriptase-PCR. Values shown are mean±s.d. (n=3). GFP, green fluorescent protein; LTR, long terminal repeat; NBM, normal bone marrow CD34+ cells that had not been transduced (NT); NUP98-HOXA10HD, N-terminal region of NUCLEOPROTEIN-98 fused to the HOXA10 homeodomain (NA10HD); Pt1, transcripts in CD34+ cells from patient 1 that were NT or transduced with empty control vector (control) or transduced with NA10HD-virus and then cultured for 7 days in GF-SFM; Pt 2, transcripts in single 12-week-LTC-derived colonies initiated with NA10HD-transduced CD34+ cells from patient 2. Values shown are the mean±s.d. (n=3).
Transduction protocol
Primary cells were prestimulated overnight (∼16 h) in GF-SFM at a concentration of ⩽5 × 105 viable CD34+ cells/ml, then transferred in virus-containing supernatant plus GF and SFM into tissue culture wells that had been precoated with 10 μg/cm2 human fibronectin (Sigma) and preincubated with concentrated control or NA10HD lenti-viral supernatant for 2 h at 4 °C (⩽105 cells per well in 96-well plates and ⩽106 cells per well in 24-well plates). After exposing the cells to virus at 37 °C for 24 h (CB cells) or 6 h (CML cells), they were removed, washed with PBS and used as described.
Xenotransplantation
Eight to ten-week-old NOD/scid-IL-2 receptor γ-chain null (NSG) mice were irradiated with 3.15–3.25 Gy of 137Cs γ-rays and then injected intravenously with the cells indicated. To analyze the human cells present at later times, BM aspirates, or harvested and suspended BM or spleen cells were then incubated in ammonium chloride to lyse the RBCs before being washed and stained with the following anti-human-specific antibodies: GlyA-PE (10F7MN labeled by contract), CD45-APC (2D3), CD34-APC (8G12), CD19-PE (4G7), CD14-PE (M0P9), CD15-PE and CD3-APC (SK7) from Becton Dickinson, and CD20-PE (L27) and CD33-PE (P67.6) from Stemcell Technologies, after incubation with a blocking reagent consisting of PBS with 2% fetal bovine serum, 5% human AB serum, anti-human CD32 blocking antibody (clone IV.3 from STEMCELL Technologies), as well as an anti CD16/CD32 antibody (2.4G2). To detect and genotype human CFCs in samples obtained from repopulated mouse tissues, human CD45+ cells were first isolated by FACS before plating the cells in methylcellulose assays. All procedures involving these mice were carried out in accordance with the Canadian Council on Animal Care guidelines with approval from the University of British Columbia Animal Care Committee.
Statistical analysis
Results are shown as the arithmetic or geometric mean±s.e.m. or s.d. as indicated. Differences between groups were assessed using the Student's t-test or the Whitney test. A Spearman ranked Wilcoxon test was used for the correlation analyses. P values <0.05 were assigned statistical significance.
Results
NA10HD transduction of CD34+ CB cells produces a rapid and sustained increase in LTC-ICs without affecting their differentiation
To determine whether forced expression of NA10HD would promote expansion of normal human HSCs in vitro, we used conditions that would be biologically similar to those used in previous mouse experiments (Figure 2a). This involved transducing CD34+ CB cells and then measuring the total number of transduced (GFP+) HSCs (here assayed as LTC-ICs) present both immediately after transduction and later, after culturing the cells for another 7 days in GF-SFM (that is, after a total of 2 and 9 days in vitro under conditions previously shown to maintain unmanipulated CB LTC-ICs over this period21). The efficiency of transduction achieved using this protocol was 62±11 (NA10HD virus) and 72±19% (control virus), as determined from the frequency of GFP+ cells within the CD34+ population present in the GF-SFM cultures 7 days post-transduction. These 7-day suspension culture measurements confirmed the high and similar titers of both virus stocks obtained using K562 cells (Figure 1) and justified direct comparisons of the effects of the two different virus preparations.
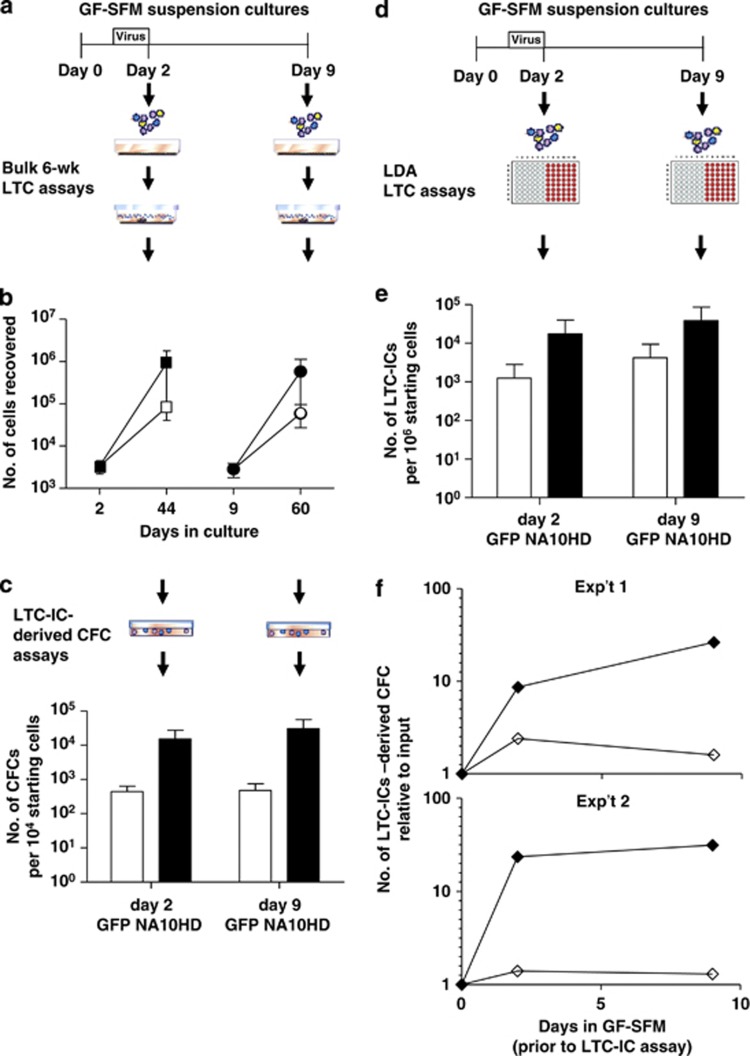
Figure 2.
Forced expression of NA10HD in CD34+ CB cells produces a rapid and sustained increase in LTC-ICs in vitro. (a, e) Experimental designs used to assess effects of NA10HD on LTC-IC activity in cultures of transduced CD34+ CB cells using bulk (a) and limiting dilution (e) LTC-IC assays. Cells were assayed both at the end of the period of exposure to virus (day 2) and after another 7 days (day 9) in GF-SFM suspension cultures. Data for control-transduced cells are shown as open symbols/bars and data for NA10HD-transduced cells are shown as black symbols/bars. (b) Comparison of total cell numbers in 6-week bulk LTCs initiated with control or NA10HD-transduced cells. Values shown are mean±s.d. (n=3). (c) Comparison of the LTC-IC activity of control or NA10HD-transduced cells as measured by their 6-week CFC content in bulk LTC-IC assays. Values shown are mean±s.e.m. (n=3) of CFC outputs expressed per 104 starting (day 0, pretransduction) cells. (d) Same data as in panel (c) but normalized to corresponding values for the same number of unmanipulated CD34+ cells in the two respective CB samples for which this initial value was determined and shown as a time course plot. (f) Comparison of the frequency of control and NA10HD-transduced LTC-ICs detected by limiting dilution analysis. Values shown are based on the combined data from two experiments and are expressed per 106 starting (pretransduction) cells.
The total number of cells (Figure 2b) and CFCs (Figure 2c) present in bulk 6-week LTCs set up with NA10HD-transduced CD34+ CB cells immediately post-transduction were both significantly higher than corresponding values for control virus-transduced cells (10 and 30-fold higher, respectively). Bulk LTC-IC assays performed on transduced cells cultured in GF-SFM for another 7 days (day 9 in GF-SFM) showed a further, albeit modest (twofold) increase in the differential yield of CFCs between the LTC-IC assays of the NA10HD- and control-transduced cells (Figures 2b and c). Relative to pretransduction values, the final increase in NA10HD-transduced LTC-IC-derived CFCs was 60-fold, whereas the number of control-transduced LTC-IC-derived CFCs was <2-fold higher than the pretransduction number (Figure 2f). LTC-IC measurements using limiting dilution analysis protocol (Figure 2d) detected ∼14-fold more LTC-ICs in the assays of the NA10HD-transduced cells assayed either immediately post-transduction or after the cells had been maintained for another 7 days in GF-SFM by comparison to control-transduced cells (Figure 2e). Six-week CFC outputs per LTC-IC calculated in these limiting dilution analysis experiments both immediately and 7 days post-transduction yielded values for the NA10HD-transduced LTC-ICs that appeared slightly reduced (7 and 4 CFCs/LTC-IC) relative to the controls (16 and 12 CFCs/LTC-IC), which were similar to previously published values for CB LTC-ICs assayed similarly.17 Thus, it seems likely that NA10HD expression results in an expansion of CB LTC-ICs that may have slightly less proliferative potential. More than 90% of the colonies obtained in all of these cultures were granulopoietic, and indistinguishable in size and appearance between the groups (data not shown). Thus, the major impact of forced NA10HD expression in primitive normal human hematopoietic cells is a rapid and sustained increase in the number of cells that display LTC-IC activity without altering their differentiation potential.
NA10HD transduction of primitive CP-CML cells produces a rapid increase in Ph+/BCR-ABL + CML LTC-ICs without affecting their differentiation in vitro
The effects of forced NA10HD expression on Ph+/BCR-ABL+ and on co-existing normal adult LTC-ICs from CP-CML patients were examined using a similar experimental design (Figure 3a), as devised for the CB experiments. For the CML experiments, we selected four CP patient samples whose LTC-IC compartments we had previously found to range from predominantly Ph+/BCR-ABL+ (>93%) to predominantly normal (<6% Ph+/BCR-ABL+, Supplementary Table S1). The CD34+ cells from these samples were transduced using the same overnight prestimulation protocol as for the CD34+ CB cells, but a shorter period of exposure to virus to enable later in vivo experiments to be performed using the same transduction protocol without incurring a significant loss of CP-CML cells with repopulating activity. Assessment of the efficiency of gene transfer to CD34+ CML cells achieved using the shorter period of exposure to virus was similar (46±11% for the NA10HD virus and 52±10% for the control virus, Figures 1c and d) to that obtained with CD34+ CB cells.
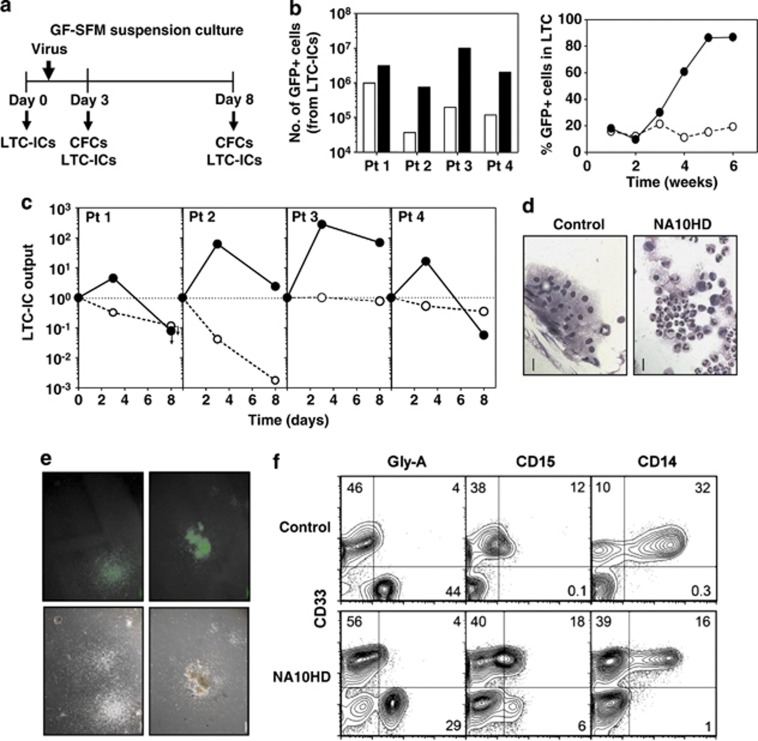
Figure 3.
Forced expression of NA10HD in CD34+ cells from CML patients promotes a rapid and sustained increase in LTC-ICs without effects on their ability to differentiate. (a) Experimental design used to assess the effect of NA10HD on LTC-IC activity in cultures of CD34+ cells isolated from four CP-CML patient's samples that initially contained variable numbers of Ph+/BCR-ABL+ LTC-IC assays were performed 2 and 7 days post-transduction (day 3 and 8 in GF-SFM suspension cultures). (b) Data for control-transduced cells are shown as open symbols/bars and data for NA10HD-transduced cells are shown as black symbols/bars. Comparison of total GFP+ cell numbers in 6-week-old LTCs initiated with NA10HD- or control-transduced cells (left panel) and time course data from one experiment (patient 2). (c) Comparison of the number of GFP+ CFCs in the 6-week-old LTCs initiated with NA10HD- or control-transduced cells in the suspension cultures assessed 2 and 7 days after exposure to virus. All values were normalized to the day 0 value for each patient. Arrows indicate values below the detection limit corresponding to the data point. (d) Giemsa staining of cells (mainly macrophages) found in the non-adherent fraction of 5-week-old LTCs initiated with control-transduced cells from patient 2 as compared with the non-adherent fraction of corresponding LTCs initiated with NA10HD-transduced cells found to contain mainly granulocytes. Scale bar: 50 μm. (e) Photomicrographs of colonies produced in methylcellulose assays of LTC-derived CFCs derived from both control- and NA10HD-transduced CML cells illustrating their normal appearance. Scale bar: 500 μm. (f) Representative FACS profiles of immuno-stained cells harvested from CFC assays of 6-week LTCs initiated with control- or NA10HD-transduced cells showing their similar content of mature erythroid (GlyA+CD33−), immature (CD33+CD15+) and mature (CD33−CD15+) granulocytes, and monocytes/macrophages (CD33+CD14+).
Bulk LTC-IC assays performed 2 days post-transduction (day 3 in GF-SFM) showed a consistently marked and selectively enhanced production of GFP+ cells (Figure 3b) and CFCs (Figure 3c) from the NA10HD-transduced cells after 6 weeks. For two of the CML samples (patients 2 and 3), this differential was well sustained for another 5 days (7 days post-transduction and day 8 in GF-SFM), but was lost in the other two cases (Figure 3c). Cells released in both types of LTCs were mainly differentiating granulopoietic cells with very rare blasts (Figure 3d). The CFCs present at the end of the 6 weeks of LTC in both cases also generated colonies of cells with a normal morphology (Figure 3e) and phenotype (Figure 3f). Taken together, these findings show that forced expression of NA10HD enhances LTC-IC production in primitive cells from CP-CML patients without affecting their ability to differentiate.
To discriminate effects of NA10HD transduction on primitive Ph+/BCR-ABL+ and residual normal cells in the same samples, the CFCs produced in the LTC-IC assays were genotyped. In addition, serial LTC-IC assay (replating) experiments were undertaken and the colonies again genotyped (Figure 4a). NA10HD-transduced cells showed the same consistently marked increase in LTC-IC numbers (bulk assays) when assessed 2 days post-transduction (Supplementary Table S2). The genotyping analyses showed a rapid and particularly marked increase in Ph+/BCR-ABL+ LTC-ICs (Figure 4b). This effect of NA10HD on Ph+/BCR-ABL+ LTC-ICs was sufficient to preclude the detection of any accompanying effect on primitive normal LTC-ICs in three patients' samples. However, for patient 4, where only normal LTC-ICs were detectable in the input cells (Figure 4c and Supplementary Table S2), a significant enhancing effect of NA10HD on normal adult LTC-ICs was clearly evident. The results of the secondary bulk LTC-IC assays of NA10HD-transduced cells further demonstrated a continuing marked enhancement of normal and Ph+/BCR-ABL+ LTC-IC production in the primary LTCs (Figures 4b and c). The daughter CFCs generated in these secondary LTC-IC assays again were morphologically normal (data not shown). In addition, G-banded metaphases of NA10HD-transduced cells from both normal and Ph+/BCR-ABL+ LTC-derived colonies showed no new chromosomal abnormalities (data not shown).
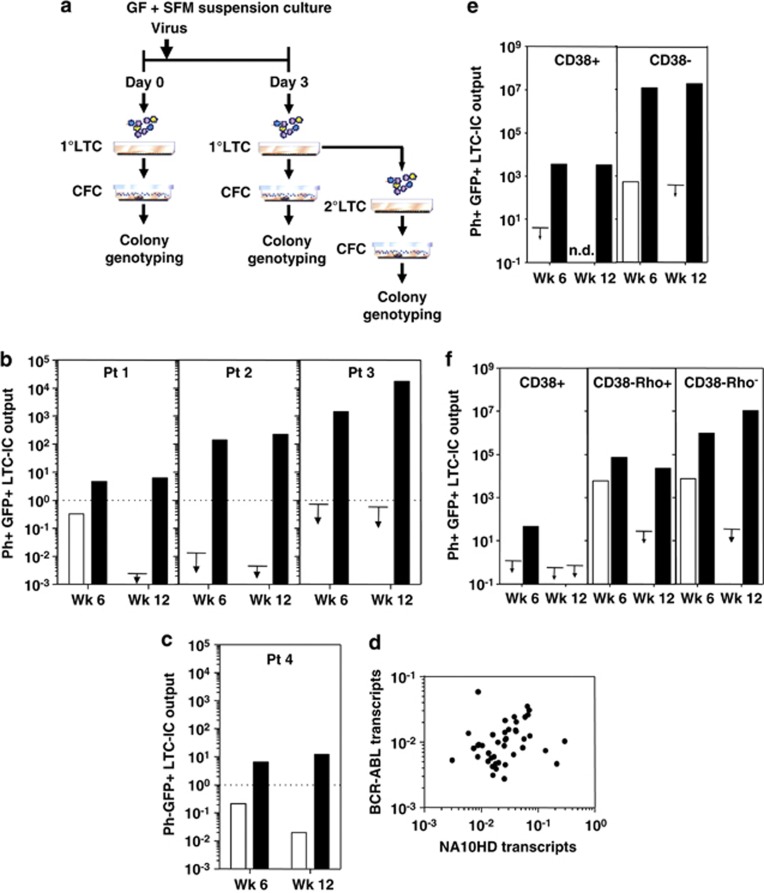
Figure 4.
NA10HD markedly enhances the expansion of normal and Ph+/BCR-ABL+ LTC-ICs in vitro in LTCs and has the greatest effect on the more primitive CD34+CD38− cells. (a) Experimental design (using cells from the same four CP-CML patients studied in Figure 3). Assays were performed on cells that had been maintained in GF-SFM suspension cultures for 3 days after exposure to virus. (b) Data for control-transduced cells are shown in open symbols/bars and for NA10HD-transduced cells are shown in black symbols/bars. Comparison of the absolute number of LTC-derived GFP+ BCR-ABL+ CFCs found in primary (wk 6) and secondary (wk 12) LTC-IC assays normalized to day 0 values (pretransduction values, dotted lines) obtained from the same number of starting CD34+ cells from the same patient. Arrows indicate values below the detection limit corresponding to the data point. (c) The same data for the LTC-IC-derived GFP+ normal (BCR-ABL−) CFCs are shown separately for patient 4 (as no BCR-ABL+ CFCs were found in LTCs initiated with these cells). (d) Comparison of BCR-ABL transcript levels as a function of NA10HD transcript levels (quantified relative to GAPDH transcripts) in extracts of individual colonies produced from CFCs generated in 6-week LTCs initiated with transduced cells from patients 1, 2 and 3. Results are expressed as 2−(Ct BCR−ABL−Ct GAPDH) for BCR-ABL transcripts and 2−(Ct NA10HD−Ct GAPDH) for each colony (R2=0.0007; Pearson test showing no significant correlation, P=0.87). (e) Comparison of the effect of NA10HD on the CD38+ and CD38− subsets of CD34+ CML cells (patient 2). Data shown are the absolute number of LTC-derived GFP+ BCR-ABL+ CFCs produced in primary (wk 6) and secondary (wk 12) LTC-IC assays expressed per 105 starting CD34+CD38+ cells or CD34+CD38− cells. (f) The same experimental design was followed as in panel (e) but the CD34+CD38− cells were additionally subdivided into a Rho+ and Rho− fraction (cells from patient 1). Arrows show detection limits. ND, not done.
As predicted from the predominance of GFP+ and Ph+/BCR-ABL+ colonies in the LTC-IC assays of the NA10HD-transduced cells from patients 1–3, NA10HD transcripts were also readily detectable in the same colonies (Figure 4d). These levels were highly variable between individual colonies, but showed no correlation with BCR-ABL transcript levels in the same colonies (R2=0.0007, P=0.87; Pearson test). A similarly variable expression of NA10HD was seen in transduced K562 cell clones (Supplementary Figure S1A), where BCR-ABL transcript levels were more similar to one another (and to the levels measured in control-transduced K562 cells, P=0.92 unpaired t-test, data not shown). NA10HD-transduced K562 cells also showed no alteration of their sensitivity to imatinib mesylate (Supplementary Figure S1B), indicating that the forced expression of NA10HD has no effect on BCR-ABL kinase activity.
The ability of NA10HD to enhance CML LTC-IC numbers in vitro is restricted to a primitive subset of CD34+ cells
To characterize the cells that NA10HD targets to produce increased numbers of Ph+/BCR-ABL+ LTC-ICs in vitro, we undertook additional experiments using the same design as shown in Figure 4a, but starting with various FACS-purified subsets of CD34+ cells (isolated from patients 1 and 2). These included starting populations of CD34+CD38+ and CD34+CD38− cells (Figure 4e) or the CD34+CD38+ cells plus the CD34+CD38− cells further subdivided according to their ability to efflux Rho (Figure 4f). Forced expression of NA10HD in the CD34+CD38+ cells enhanced CFC outputs in both the primary and secondary LTC-IC assays of the transduced cells. In contrast, LTC-ICs in the corresponding assays of control-transduced cells were below detectable levels, as expected from experiments with unmanipulated cells.10 However, the LTC-IC numbers obtained from the NA10HD-transduced CD34+CD38+ cells was still much lower (∼3000-fold) than those obtained from the NA10HD-transduced CD34+CD38− cells, or the Rho− subset of these. Thus, although the latter two subsets represent small fractions of the total CD34+ populations (∼10% and ∼0.1%, respectively), their contribution to the NA10HD-stimulated production of LTC-ICs accounts for the bulk of the increases measured from the total CD34+ cells.
NA10HD has little effect on the long-term growth or differentiation of primary CML cells in NSG mice
To evaluate the effects of NA10HD transduction of primitive CP-CML cell growth and differentiation in an in vivo setting, we transplanted sublethally irradiated NSG mice intravenously with 106 CD34+ cells each from patients 1 or 2 (with predominantly Ph+/BCR-ABL+ LTC-ICs) immediately post-transduction. We then analyzed the human cell content of BM aspirates obtained from these mice up to 36 weeks later (Figure 5a). The results showed that, independent of the vector used to transduce the initial cells, a population of human myeloid cells was maintained for the entire period that the mice were followed with no (patient 1) or very few (and only at early times, patient 2) human lymphoid cells detected, as expected for transplants of Ph+/BCR-ABL+ CP-CML stem cells. Approximately, 15 to 30% of the human cells regenerated were GFP+ in both the myeloid (CD33+) and lymphoid (CD19/20+) lineages (Figure 5b). GFP+ CD34+ cells were also detected in the BM aspirates obtained 8 weeks after transplantation (0.02±0.01% and 0.01±0.01% in recipients of NA10HD- and control-transduced cells, respectively, from patient 1, and 0.35±0.13% and 0.02±0.01%, respectively, for patient 2). Assessment of the frequency (per 104 human CD45+ cells present) and genotype of the human CFCs present in the 8-week BM aspirates showed that both Ph+/BCR-ABL+ and normal human CFCs were also readily detectable in the same BM aspirates from the mice (Table 1). Nine months post-transplantation, Ph+/BCR-ABL+ CML cells were still readily detectable in the BM of the NSG mice, as demonstrated by the levels of BCR-ABL in both the GFP+ and GFP− regenerated human cells (Figure 5c) and by the restricted presence of NA10HD transcripts in the GFP+ human cells (Figure 5d). Ph+/BCR-ABL+ CFCs were also detected in assays of the human CD45+ cells harvested at this time and were possibly more prevalent in the NA10HD-transduced (GFP+) cells (Table 2). However, the levels of BCR-ABL transcripts found in NA10HD-expressing colonies and control colonies were not statistically different (data not shown). In addition, both the morphology (Figure 5e) and Lin markers (Supplementary Figure S2) expressed on the cells they produced in vitro were indistinguishable from the colonies produced by the CFCs in the original primary CP-CML samples from these patients. Examination of the spleens removed from the mice 9 months post-transplant showed that all human cells present were myeloid, consistent with the lymphoid-deficient chimerism seen in the BM of these same mice (Figure 5f).
Figure 5.
NA10HD-transduced CML cells show unperturbed repopulating activity in NSG mice. (a) Experimental design. The harvested progeny of 106 initial CD34+ cells from patients 1 and 2 were injected immediately after transduction into each of eight sublethally irradiated NSG mice that were then monitored as indicated. (b) Data for control-transduced cells are shown in open symbols/bars and for NA10HD-transduced cells are shown in black symbols/bars. Comparison of the percentage of total human myeloid (CD45+CD33+CD14+CD15+GlyA+, top panel) and mature B-lymphoid (CD45+CD19+CD34−, bottom panel) GFP+ cells detected in the BM of the NSG mice at different times post-transplantation. Data for patient 1 is shown in the left panel and for patient 2 in the right panel. (c) Comparison of BCR-ABL transcript levels in FACS-purified human CD45+ cells obtained from the BM of mice transplanted with NA10HD- or control-transduced CP-CML cells 35 weeks before. Results are expressed as 2−(Ct BCR−ABL−Ct GAPDH). (d) Comparison of NA10HD transcript levels in human CD45+GFP− (white bar) and human CD45+GFP+ (black bar) cells isolated from the BM of five (of the eight original) mice transplanted with NA10HD-transduced CML cells 35 weeks before. Results are expressed as 2−(Ct BCR−ABL−Ct GAPDH). The difference between the two values is statistically significant (**P=0.008, Mann–Whitney). (e) Wright-Giemsa-stained cytospins of colonies produced from CFCs harvested from mice that received transduced cells from patient 1 showing normal myeloid differentiation in both cases. (f) Presence of human myeloid cells and absence of human B-lymphoid cells in the spleens of mice transplanted 35 weeks before with control- or NA10HD-transduced cells.
Table 1. Human myeloid progenitor quantification in mouse BM at 8 weeks post-transplantation.
| Cells infused | Mouse No | GFP | Patient No |
CFCs/104
CD45+
cells in mouse BM |
B/A+ CFCs/total CFCs | |||
|---|---|---|---|---|---|---|---|---|
| BFU-E | CFU-GM | CFU-GEMM | Total | |||||
| Control GFP | 17 | − | 1 | 28 | 40 | <1 | 68 | 13/13 |
| + | 1 | 6 | <1 | 7 | ND | |||
| 18 | − | 2 | 1 | 4 | <1 | 6 | ND | |
| + | <1 | 3 | <1 | 3 | ND | |||
| NA10HD | 12 | − | 2 | 2 | 19 | <1 | 21 | 0/14 |
| + | 125 | 84 | 1 | 210 | 6/16 | |||
| 15 | − | 1 | 46 | 79 | <1 | 125 | ND | |
| + | <1 | 2 | <1 | 2 | ND | |||
| 20 | − | 2 | 5 | 10 | <1 | 15 | 0/14 | |
| + | 139 | 117 | <1 | 256 | 12/13 | |||
| 24 | − | 2 | 1 | 2 | <1 | 3 | ND | |
| + | 192 | 137 | 3 | 333 | 8/8 | |||
Abbreviations: B/A+, BCR-ABL+; BM, bone marrow; CFC, colony-forming cell; GEMM, granulopoietic, erythroid and multi-potent myeloid; GFP, green fluroscent protein; GM, granulopoietic; ND, not done.
Table 2. Human myeloid progenitor quantification in mouse BM 34 to 36 weeks post-transplantation.
| Cells infused | Mouse No | GFP | Patient No |
CFCs/104
CD45+
cells in mouse BM |
Ph+CFCs/total CFCs | |||
|---|---|---|---|---|---|---|---|---|
| BFU-E | CFU-GM | CFU-GEMM | Total | |||||
| Control GFP | 10 | − | 2 | <0.3 | 1 | <0.3 | 1 | ND |
| 17 | − | 1 | 20 | 183 | <1 | 203 | 13/13 | |
| + | 29 | 230 | 1 | 260 | 14/14 | |||
| NA10HD | 11 | − | 1 | 95 | 141 | <1 | 236 | 12/13 |
| + | 48 | 92 | <1 | 140 | 13/13 | |||
| 12 | − | 2 | <0.3 | 1 | <0.3 | 1 | ND | |
| + | <0.5 | 0.5 | <0.5 | 0.5 | ND | |||
| 19 | − | 1 | 38 | 270 | <1 | 308 | 12/12 | |
| + | 22 | 166 | <1 | 188 | 11/11 | |||
Abbreviations: BM, bone marrow; CFC, colony-forming cell; GEMM, granulopoietic, erythroid and multi-potent myeloid; GFP, green fluroscent protein; GM, granulopoietic; ND, not done.
Effects of NA10HD on primitive human hematopoietic cells are not mediated by detectable changes in their proliferation or survival
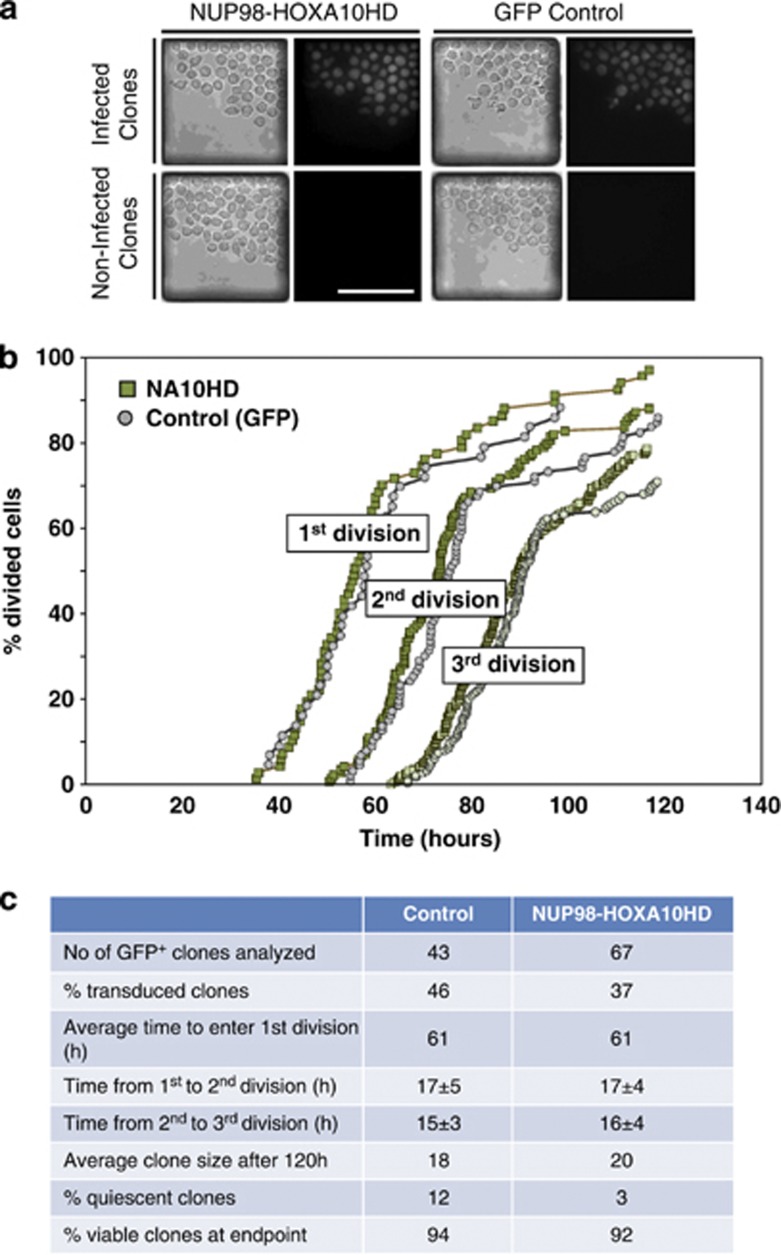
We next asked whether forced expression of NA10HD might affect the cell cycle entry or cell cycle transit time of primitive human hematopoietic cells. Accordingly, single CD34+CD38− cells were isolated by FACS from CB, transduced and then, 24 h later, introduced into a microfluidic cell culture array containing a large number of nanoliter-volume wells preloaded with GF-SFM.22 The loaded chips were kept at 37 °C and cells imaged every 5 min for 96 h to determine the frequency and rate of clone formation of the GFP+ cells. The results revealed no differences between NA10HD-transduced and control (GFP+) CB cells (Figure 6). Thus, by day 5 post-transduction (after 2 days in the microfluidic array), the average size of the clones produced by the NA10HD- and control-transduced cells was the same (Figure 6c). Similarly, K562 cells transduced with NA10HD also did not show any difference in their clonogenic efficiency (Supplementary Figure S3A), nor in their doubling time in bulk suspension cultures (Supplementary Figure S3B), and this latter result held for at least 10 passages (data not shown).
Figure 6.
NUP98-HOXA10HD does not alter the initial cell division kinetics of transduced CD34+CD38− CB cells. (a) Bright field and fluorescent photomicrographs of clones generated from single NA10HD and control-transduced CD34+CD38− CB cells after 4 days in microfluidic cultures in GF-SFM. Single cells that were successfully infected could easily be distinguished from non-expressing cells by their GFP fluorescence. Scale bar, 100 μm. (b) Comparison of the rates of cell division of individual transduced cells (clones that contained 100% GFP+ cells after 96 h of culture) when imaged every 5 min. Data are plotted as the cumulative rates of completion of one, two and three divisions over the period monitored, expressed as a percent of the number of cells/clones monitored (n=43 for control-transduced cells (grey symbols) and n=67 for NA10HD-transduced cells (green symbols). (c) Control- and NA10HD-transduced populations were loaded sequentially into the microfluidic cell culture array. A set of fluorescent images was taken at the end of the experiment to identify GFP+ clones. The % transduced clones was calculated by dividing the number of GFP+ clones by the initial number of single cells for each population. Cells were imaged every 5 min and the time of first, second and third division identified by manual inspection of the videos. The number of viable cells in each clone at the end of the experiment was counted manually. Clones were labeled as quiescent if they contained a single viable cell that had not divided during the course of the experiment. The % viability was measured by counting the number of clones, transduced or not, that contained at least one viable cell at the end of the experiment divided by the initial number of single cells for each population. Results are shown as mean±s.d.
Discussion
Many genes have now been identified as having a role in regulating HSC expansion, but few of these have thus far translated into clinically useful strategies for expanding normal or leukemic human HSCs ex vivo. Likewise, these findings have not yet led to improved methods for treating human acute leukemia. Nevertheless, the HOX gene family and related regulators have remained of paramount interest, based on evidence of the effects of their perturbed (elevated) expression in primitive normal and leukemic cells.2 Such studies support the widely held thesis that a full-blown leukemia involves the acquisition of a deregulated self-renewal mechanism that shares features with that operative in normal HSCs.
Interestingly, there are now several examples where altered Hox gene functionality has produced marked enhancing effects on various HSC subsets without causing or contributing to a leukemogenic process. The first example was the demonstration that forced overexpression of HOXB4 in adult mouse BM cells can produce a marked (40-fold) expansion of HSCs in short-term cultures containing early acting GFs with resumption of a normal growth and differentiation behavior following their transplantation into an in vivo environment.23 Subsequent studies showed a more modest effect of forced overexpression of HOXB4 on cultured human hematopoietic cells.24 More recently, we described a similar but greatly magnified effect (>1000-fold expansion of HSCs) in short-term cultures of primitive mouse BM cells transduced with NA10HD, again without perturbing the subsequent normal functionality of the expanded HSC when they were transplanted in vivo.3 These findings were then followed by a similar study in nonhuman primate cells where a marked but transient effect on neutrophil recovery was obtained using NA10HD-transduced bone marrow CD34+ cells.25 Thus, it appears that enhancement/acquisition of self-renewal and contributions to leukemogenicity can be molecularly dissociated.
We now demonstrate that forced overexpression of NA10HD in primitive human cells enables the number of LTC-ICs present after short periods of culture to be markedly increased (>10-fold above starting numbers). This finding thus fulfills expectations from previous studies including the observation that the effects on human cells are less than those reported for mice. The possible expansion of a more differentiated subset of human LTC-ICs (characterized by a reduced CFC output) observed here is reminiscent of the results obtained when nonhuman primate transplants were transduced with NA10HD.25 However, differences in the vectors and cell manipulation protocols need also to be considered as potential different contributors to similar outcomes.
We also obtained evidence here that the effects on primitive normal human cells include HSCs in CB and adult BM (CP-CML patient 4 whose HSCs were predominantly normal), suggesting a potential broad applicability of this finding in the future. As a first test of a possible leukemogenic activity of NA10HD in transduced human cells, we examined its effect on the growth and differentiation of primitive Ph+/BCR-ABL+ CP-CML cells with LTC-IC (in vitro) and NSG mouse repopulating activity (in vivo). However, within the time frames of these experiments (2 and up to 8 months, respectively), the growth of NA10HD-transduced normal or Ph+/BCR-ABL+ cells was not significantly altered. There was also no apparent effect on their cell cycle kinetics or subsequent ability to differentiate in vitro (or in vivo) and no evidence of progression to leukemia. Taken together, these findings indicate that sufficiently elevated levels of NA10HD can selectively upregulate the self-renewal probability of normal and Ph+/BCR-ABL+ human HSCs and/or their immediate progeny.
Further evidence that NA10HD affects the self-renewal machinery of primitive human and mouse cells in a similar fashion can be inferred from the results of experiments designed to characterize the major subset of human CD34+ cells affected. These showed that the primary targets are the Rho−CD38− subset of CD34+ cells, a population previously shown to be in G0. Interestingly, during the initial 2-day period, when a marked increase in LTC-IC activity was detected, we found that fewer than 20% of the engineered CD34+CD38− CB cells had completed a first division. These results suggest that NA10HD confers LTC-IC activity on normal CD34+CD38−Rho− cells that would otherwise not possess this property and may do so even before the cells exit G0. A precedent for such an effect is provided by previous observations of the opposite effect obtained on highly purified, quiescent mouse HSCs where these were shown to undergo co-ordinated changes in transcription factor expression and loss of long-term repopulating activity long before undergoing a first division.26 It is also important to note that some CD34+CD38− cells do not produce CFCs within 6 weeks under LTC conditions but do so after 12 weeks.27, 28 We also found that the transduced NA10HD cDNA increased the output of secondary LTC-ICs from normal, as well as CML, cells seeded into primary LTCs. This suggests either that NA10HD enhances the self-renewal of cells that normally display 6-week LTC-IC activity or that NA10HD recruits deeply quiescent cells with greater self-renewal potential into the detectable LTC-IC pool.
An increased chimerism in vivo but unaffected differentiation and lineage output has also been seen in autologous nonhuman primate recipients of transduced cells,25 as well as in congenic3, 4 or allogeneic recipients29 of NA10HD-transduced cells. More recently, AML1α has been found to have a similar effect on mouse HSCs.30 BMI1, another key regulator of normal and leukemic stem cells, was found to enhance human CML LTC-IC activity and also the replating ability of clonogenic CML cells, but their differentiation remained unaffected. However, when CB CD34+ cells were co-engineered to express both BCR-ABL and BMI1, doubly transduced cells initiated a lymphoid leukemia in recipient mice.31 The NA10HD fusion gene thus appears as a potentially more selective and hence promising tool for expanding normal HSCs and CP-CML stem cells. However, any translation of such a strategy into clinical practice will clearly require additional efforts to avoid risk of undesirable vector integration sites and more prolonged assessment of NA10HD in nonhuman primate models.
It is interesting to note that a very marked effect of NA10HD on both primitive normal and Ph+/BCR-ABL+ in the samples obtained from the CP-CML patients was seen when the cells were maintained in vitro under LTC conditions, whereas for the transduced CB cells maintained in shorter term cultures containing only GFs, there was only minimal evidence of a continued HSC expansion. Thus, it seems likely that the effect of NA10HD on human targets may be dependent on additional extrinsic cues provided by stromal cells. This possibility is consistent with previous mouse HSC experiments that have shown a critical effect of the cytokines added to enable HOX gene overexpression to salvage the self-renewal mechanism of mouse HSCs maintained in vitro. In these, IL-3, IL-6 and SF have all been shown to be positive effectors in contrast to TPO which, in combination with other cytokines, was found to be inhibitory.4 Therefore, NA10HD-engineered primitive CML cells may also offer an interesting model to probe the ability of extrinsic factors to affect CP-CML stem cell self-renewal responses.
This study also documents for the first time, the long-term and largely myeloid-restricted regenerative capacity that CP-CML stem cells can display in sublethally irradiated NSG mice when they are transplanted with sufficient CP-CML stem cells. In addition, we show that a relatively short transduction protocol can achieve a transduction efficiency of these cells to allow follow-up of their behavior for periods of more than 8 months. The lack of a significant effect of NA10HD on these cells or their progeny over several months argues against an ability of NA10HD on its own to cause progression of CP-CML cells to blast crisis even though a marked enhancing effect on their self-renewal is obtained. Recent development of humanized mouse models such as NSG mice transgenic for human IL-3, GM-CSF and SCF will be of interest to further evaluate the leukemogenic potential of NA10HD on human HSCs.32
In summary, forced expression of NA10HD in primitive normal human hematopoietic cells and their CP-CML counterparts selectively, rapidly, sustainably and markedly enhances their numbers in vitro in the presence of known and unknown GFs produced by stromal cells. This effect is not accompanied by any immediate or overt effect on their initial cell cycle regulation or ability to differentiate in vitro or in vivo or transformation even in CP-CML cells. Thus, NA10HD should prove a useful tool to rapidly increase primitive normal or CP-CML stem cell numbers for their further characterization or use for testing new therapeutic drugs.
Acknowledgments
We thank the staff of the Stem Cell Assay Laboratory, Glen Edin and Margaret Hale for their technical assistance, as well as Claudia Benz and Alice Cheung for their fruitful scientific discussions. We also thank the Flow Cytometry Core Facility of the Terry Fox Laboratory and the Animal Resource Center of the BC Cancer Agency. We also acknowledge the importance of the Hematology Cell Bank of BC and its many contributors, supporters and staff for enabling access to many of the patients' cell samples used in this study. This work was supported by grants from the National Cancer Institute of Canada with funds from the Canadian Cancer Society and the Terry Fox Foundation, and grants from the Canadian Institutes of Health and the Natural Sciences and Engineering Research Council of Canada. Ivan Sloma held a fellowship from La Fondation pour la Recherche Médicale and from La Fondation de France. Philip Beer held a Kay Kendall Leukemia Fund Intermediate Fellowship from the UK.
AUTHOR CONTRIBUTIONS
Experiments were designed by IS, SI and CJE with input from YZ, RKH, JP and CH. CD34+ cell isolation and feeder line preparation were performed by KL. The NA10HD lenti-virus vector was cloned by SI. Primary cell engineering, CFC and LTC-IC assays were performed by IS (CML) and SI (CB). Colony genotyping was performed by IS, DL, KR and CB. Transplantations, and flow cytometry sorting and analysis were carried out primarily by IS with assistance from PB and DL. Microfluidic experiments were designed by VL and IS, and performed by VL with assistance from IS for cell preparation. Statistical assessment was done by IS and CJE. Manuscript was written by IS, SI and CJE with input from VL, RKH, JP and CH.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Leukemia website (http://www.nature.com/leu)
Supplementary Material
References
- Seita J, Weissman IL. Hematopoietic stem cell: self-renewal versus differentiation. Wiley Interdiscip Rev Syst Biol Med. 2010;2:640–653. doi: 10.1002/wsbm.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argiropoulos B, Humphries RK. Hox genes in hematopoiesis and leukemogenesis. Oncogene. 2007;26:6766–6776. doi: 10.1038/sj.onc.1210760. [DOI] [PubMed] [Google Scholar]
- Ohta H, Sekulovicac S, Bakovic S, Eaves CJ, Pineault N, Gasparetto M, et al. Near-maximal expansions of hematopoietic stem cells in culture using NUP98-HOX fusions. Exp Hematol. 2007;35:817–830. doi: 10.1016/j.exphem.2007.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekulovic S, Gasparetto M, Lecault V, Hoesli CA, Kent DG, Rosten P, et al. Ontogeny stage-independent and high-level clonal expansion in vitro of mouse hematopoietic stem cells stimulated by an engineered NUP98-HOX fusion transcription factor. Blood. 2011;118:4366–4376. doi: 10.1182/blood-2011-04-350066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis RE, Boice JD, Stovall M, Bernstein L, Holowaty E, Karjalainen S, et al. Relationship of leukemia risk to radiation dose following cancer of the uterine corpus. J Natl Cancer Inst. 1994;86:1315–1324. doi: 10.1093/jnci/86.17.1315. [DOI] [PubMed] [Google Scholar]
- Weiss HA, Darby SC, Fearn T, Doll R. Leukemia mortality after X-ray treatment for ankylosing spondylitis. Radiat Res. 1995;142:1–11. [PubMed] [Google Scholar]
- Ichimaru M, Ishimaru T, Mikami M, Yamada Y, Ohkita T. Technical Report RERF TR 13-81. Radiation Effects Research Foundation: Hiroshima; 1981. Incidence of leukemia in a fixed cohort of atomic bomb survivors and controls, Hiroshima and Nagasaki, October 1950–December 1978. [Google Scholar]
- Goto T, Nishikori M, Arlin Z, Gee T, Kempin S, Burchenal J, et al. Growth characteristics of leukemic and normal hematopoietic cells in Ph' + chronic myelogenous leukemia and effects of intensive treatment. Blood. 1982;59:793–808. [PubMed] [Google Scholar]
- Udomsakdi C, Eaves CJ, Swolin B, Reid DS, Barnett MJ, Eaves AC. Rapid decline of chronic myeloid leukemic cells in long-term culture due to a defect at the leukemic stem cell level. Proc Natl Acad Sci USA. 1992;89:6192–6196. doi: 10.1073/pnas.89.13.6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petzer AL, Eaves CJ, Barnett MJ, Eaves AC. Selective expansion of primitive normal hematopoietic cells in cytokine-supplemented cultures of purified cells from patients with chronic myeloid leukemia. Blood. 1997;90:64–69. [PubMed] [Google Scholar]
- Daley GQ, Van Etten RA, Baltimore D. Induction of chronic myelogenous leukemia in mice by the P210bcr/abl gene of the Philadelphia chromosome. Science. 1990;247:824–830. doi: 10.1126/science.2406902. [DOI] [PubMed] [Google Scholar]
- Li S, Ilaria RL, Million RP, Daley GQ, Van Etten RA. The P190, P210, and P230 forms of the BCR/ABL oncogene induce a similar chronic myeloid leukemia-like syndrome in mice but have different lymphoid leukemogenic activity. J Exp Med. 1999;189:1399–1412. doi: 10.1084/jem.189.9.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Ng E, Yip C, Eisterer W, Chalandon Y, Stuible M, et al. Primitive interleukin 3 null hematopoietic cells transduced with BCR-ABL show accelerated loss after culture of factor-independence in vitro and leukemogenic activity in vivo. Blood. 2002;100:3731–3740. doi: 10.1182/blood-2002-05-1324. [DOI] [PubMed] [Google Scholar]
- Schemionek M, Elling C, Steidl U, Baumer N, Hamilton A, Spieker T, et al. BCR-ABL enhances differentiation of long-term repopulating hematopoietic stem cells. Blood. 2010;115:3185–3195. doi: 10.1182/blood-2009-04-215376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udomsakdi C, Eaves CJ, Lansdorp PM, Eaves AC. Phenotypic heterogeneity of primitive leukemic hematopoietic cells in patients with chronic myeloid leukemia. Blood. 1992;80:2522–2530. [PubMed] [Google Scholar]
- McKenzie JL, Takenaka K, Gan OI, Doedens M, Dick JE. Low rhodamine 123 retention identifies long-term human hematopoietic stem cells within the Lin-CD34+CD38− population. Blood. 2007;109:543–545. doi: 10.1182/blood-2006-06-030270. [DOI] [PubMed] [Google Scholar]
- Hogge DE, Lansdorp PM, Reid D, Gerhard B, Eaves CJ. Enhanced detection, maintenance, and differentiation of primitive human hematopoietic cells in cultures containing murine fibroblasts engineered to produce human steel factor, interleukin-3, and granulocyte colony-stimulating factor. Blood. 1996;88:3765–3773. [PubMed] [Google Scholar]
- Fraser C, Eaves CJ, Kalousek DK. Fluorodeoxyuridine synchronization of hemopoietic colonies. Cancer Genet Cytogenet. 1987;24:1–6. doi: 10.1016/0165-4608(87)90077-x. [DOI] [PubMed] [Google Scholar]
- Logan AC, Nightingale SJ, Haas DL, Cho GJ, Pepper KA, Kohn DB. Factors influencing the titer and infectivity of lentiviral vectors. Hum Gene Ther. 2004;15:976–988. doi: 10.1089/hum.2004.15.976. [DOI] [PubMed] [Google Scholar]
- Imren S, Payen E, Westerman KA, Pawliuk R, Fabry ME, Eaves CJ, et al. Permanent and panerythroid correction of murine beta thalassemia by multiple lentiviral integration in hematopoietic stem cells. Proc Natl Acad Sci USA. 2002;99:14380–14385. doi: 10.1073/pnas.212507099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conneally E, Cashman J, Petzer A, Eaves C. Expansion in vitro of transplantable human cord blood stem cells demonstrated using a quantitative assay of their lympho-myeloid repopulating activity in nonobese diabetic-scid/scid mice. Proc Natl Acad Sci USA. 1997;94:9836–9841. doi: 10.1073/pnas.94.18.9836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecault V, Vaninsberghe M, Sekulovic S, Knapp DJ, Wohrer S, Bowden W, et al. High-throughput analysis of single hematopoietic stem cell proliferation in microfluidic cell culture arrays. Nat Methods. 2011;8:581–586. doi: 10.1038/nmeth.1614. [DOI] [PubMed] [Google Scholar]
- Antonchuk J, Sauvageau G, Humphries RK. HOXB4-induced expansion of adult hematopoietic stem cells ex vivo. Cell. 2002;109:39–45. doi: 10.1016/s0092-8674(02)00697-9. [DOI] [PubMed] [Google Scholar]
- Buske C, Feuring-Buske M, Abramovich C, Spiekermann K, Eaves CJ, Coulombel L, et al. Deregulated expression of HOXB4 enhances the primitive growth activity of human hematopoietic cells. Blood. 2002;100:862–868. doi: 10.1182/blood-2002-01-0220. [DOI] [PubMed] [Google Scholar]
- Watts KL, Zhang X, Beard BC, Chiu SY, Trobridge GD, Humphries RK, et al. Differential effects of HOXB4 and NUP98-HOXA10hd on hematopoietic repopulating cells in a nonhuman primate model. Hum Gene Ther. 2011;22:1475–1482. doi: 10.1089/hum.2011.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent DG, Dykstra BJ, Cheyne J, Ma E, Eaves CJ. Steel factor coordinately regulates the molecular signature and biologic function of hematopoietic stem cells. Blood. 2008;112:560–567. doi: 10.1182/blood-2007-10-117820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao QL, Thiemann FT, Petersen D, Smogorzewska EM, Crooks GM. Extended long-term culture reveals a highly quiescent and primitive human hematopoietic progenitor population. Blood. 1996;88:3306–3313. [PubMed] [Google Scholar]
- Petzer AL, Hogge DE, Landsdorp PM, Reid DS, Eaves CJ. Self-renewal of primitive human hematopoietic cells (long-term-culture-initiating cells) in vitro and their expansion in defined medium. Proc Natl Acad Sci USA. 1996;93:1470–1474. doi: 10.1073/pnas.93.4.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Even Y, Bennett JL, Sekulovic S, So L, Yi L, McNagny K, et al. NUP98-HOXA10hd-expanded hematopoietic stem cells efficiently reconstitute bone marrow of mismatched recipients and induce tolerance. Cell Transplant. 2011;20:1099–1108. doi: 10.3727/096368910X545068. [DOI] [PubMed] [Google Scholar]
- Tsuzuki S, Seto M. Expansion of functionally defined mouse hematopoietic stem and progenitor cells by a short isoform of RUNX1/AML1. Blood. 2012;119:727–735. doi: 10.1182/blood-2011-06-362277. [DOI] [PubMed] [Google Scholar]
- Rizo A, Horton SJ, Olthof S, Dontje B, Ausema A, van Os R, et al. BMI1 collaborates with BCR-ABL in leukemic transformation of human CD34+ cells. Blood. 2010;116:4621–4630. doi: 10.1182/blood-2010-02-270660. [DOI] [PubMed] [Google Scholar]
- Wunderlich M, Chou FS, Link KA, Mizukawa B, Perry RL, Carroll M, et al. AML xenograft efficiency is significantly improved in NOD/SCID-IL2RG mice constitutively expressing human SCF, GM-CSF and IL-3. Leukemia. 2010;24:1785–1788. doi: 10.1038/leu.2010.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.