Abstract
We report the discovery of two enteroviruses detected in nasopharyngeal samples obtained from subjects with respiratory disease in Peru. Phylogenetic analysis indicated that both viruses belong to a clade within the species Human enterovirus C, which includes the recently characterized human enteroviruses 109 and 104. Members of this clade have undergone significant genomic rearrangement, as indicated by deletions in the hypervariable region of the 5′ UTR and the VP1 protein, as well as recombination within the non-structural genes. Our findings and review of published sequences suggests that several novel human enterovirus C serotypes are currently circulating worldwide.
Introduction
Enteroviruses (EVs) are non-enveloped, positive-sense, ssRNA viruses in the family Picornaviridae. They are transmitted via multiple routes (faecal–oral, oral–oral, respiratory droplets and fomites) and are amongst the most commonly diagnosed viral infections in humans. Although most infections are asymptomatic or associated with only mild disease, EVs can cause pneumonia, myocarditis, aseptic meningitis, encephalitis or acute flaccid paralysis (Pallansch & Roos, 2007). A typical human enterovirus (HEV) genome is approximately 7.5 kb in length and encodes a polyprotein ORF flanked by 5′ and 3′ UTRs. The polyprotein is cleaved post-translationally to yield four structural viral proteins (VP4, VP2, VP3 and VP1) and seven non-structural proteins (2A, 2B, 2C, 3A, 3B, 3C and 3D) (Racaniello, 2007). New HEV types have been classified by sequence divergence within the VP1 capsid gene, with novel serotypes possessing <75 % nucleotide identity and <85 % amino acid identity relative to known HEVs (Knowles et al., 2012; Oberste et al., 1999a, b). Based on genetic characteristics, all HEV serotypes are grouped into four species: HEV-A, -B, -C and -D (Knowles et al., 2012).
Over 100 HEV types are recognized; 23 belong to the species Human enterovirus C (HEV-C) (http://www.picornaviridae.com). Two of these, EV-C104 and EV-C109, were detected in recent large-scale cohort studies of respiratory disease in Switzerland and Nicaragua, respectively, and represent a novel monophyletic clade within HEV-C (Tapparel et al., 2009; Yozwiak et al., 2010). The closest serotypes to EV-C104 and EV-C109 are CV-A1, A19 and A22 (Brown et al., 2003). Since their initial characterizations, both EV-C104 and EV-C109 have been detected in respiratory-disease samples in other countries (Kaida et al., 2012; Pankovics et al., 2012; Piralla et al., 2010).
During a MassTag PCR cohort study of respiratory disease in Peru, we identified sequences of two novel viruses that clustered within the same HEV-C lineage as EV-C104 and EV-C109. Both genotypes were similar to viruses detected recently in Europe and Asia. In this report, we describe the full genomes of both viruses and present phylogenetic comparisons to related serotypes.
Results
Samples (nasal and oropharyngeal swabs and sputum; n = 244) collected in Peru were examined for viruses associated with respiratory disease by MassTag PCR (Briese et al., 2005). A 350 bp 5′ UTR sequence amplified from two EV-positive samples (numbers PER153 and PER161) exhibited low similarity (<85 %) to the 5′ UTRs of EV-C104 and EV-C109. Both samples were collected in November 2010 in two separate locations; PER153 from a 2-year-old boy in Lima, and PER161 in Tumbes from a 5-year-old girl. The complete genome sequences of PER153 and PER161 were obtained by PCR, using available EV-C104 and EV-C109 genome sequences (GenBank accession numbers AB686524 and GQ865517, respectively) as reference.
Complete genome analysis
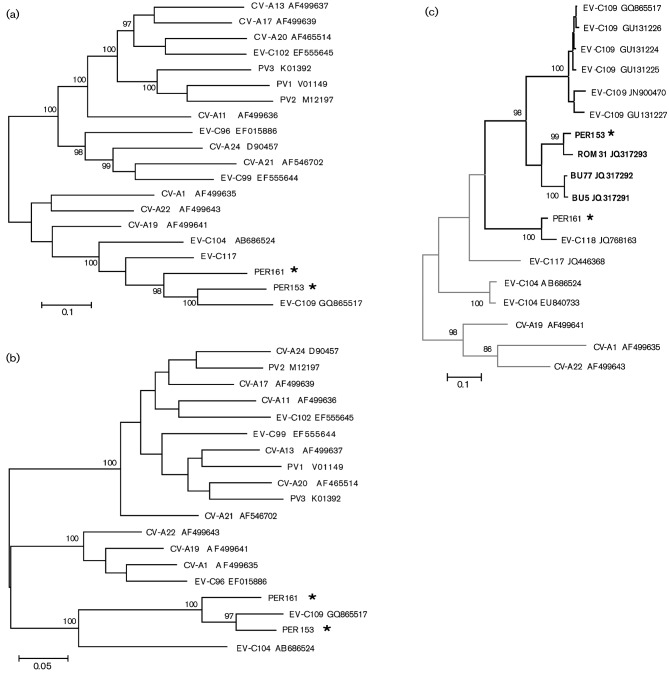
Sequence alignment and genetic distance comparison of the full genomes indicated that the ORFs for PER153 and PER161 viruses were 75.7 % identical at the nucleotide level and 89.3 % identical at the amino acid level. When compared with other complete HEV genomes, PER153 and PER161 were related most closely to EV-C104 and EV-C109 throughout the polyprotein ORF (Fig 1a, b, Table 1). PER153 is also related closely (>95 %) to novel partial sequences of an enterovirus strain detected in Romania (GenBank accession numbers JQ317293 and JN967687; 2010); PER161 is >92 % similar to strains ISR10 and ISR38, detected in Israel (GenBank accession numbers JN967685 and JN967686, respectively; 2011). Based on partial sequences, ISR10 has been designated EV-C118 (http://www.picornaviridae.com).
Fig. 1.
Phylogenetic relationship of PER153 and PER161 to other HEV-C serotypes. (a, b) Maximum-likelihood phylogenetic trees constructed from available complete (a) HEV-C P1 sequences and (b) HEV-C 3D sequences. (c) Maximum-likelihood phylogenetic tree based on a 326 nt fragment of the VP1 gene. Analysis is based on sequences in GenBank related most closely to PER153 and PER161. The branches of genotypes with deletions within the 5′ UTR are shown in black; other branches are shown in grey. For strains ROM31, BU5, BU77 and ISR10, the 5′ UTR sequence is not yet available. Strains with a 3 bp deletion are indicated in bold. In all trees, GenBank accession numbers are indicated next to the strain designation and * indicates the two genotypes identified in this study.
Table 1. Percentage difference of nucleotide and amino acid sequences among the polyprotein of related HEV-C serotypes.
Amino acid difference is shown in parentheses.
| PER153 | PER161 | EV-C109 | EV-C104 | CV-A1 | CV-A19 | |
| PER153 | – | – | – | – | – | – |
| PER161 | 24.3 (10.7) | – | – | – | – | – |
| EV-C109 | 17.9 (5.6) | 24.7 (10.1) | – | – | – | – |
| EV-C104 | 35.0 (17.9) | 35.5 (17.5) | 34.5 (17.4) | – | – | – |
| CV-A1 | 40.8 (25.2) | 40.0 (25.0) | 39.7 (24.2) | 37.5 (21.1) | – | – |
| CV-A19 | 39.5 (23.8) | 38.8 (22.9) | 38.2 (22.5) | 34.8 (17.9) | 23.3 (9.6) | – |
| CV-A22 | 39.6 (24.4) | 39.5 (23.8) | 39.2 (23.7) | 35.6 (19.5) | 22.7 (10.7) | 22.3 (8.2) |
Structural genes
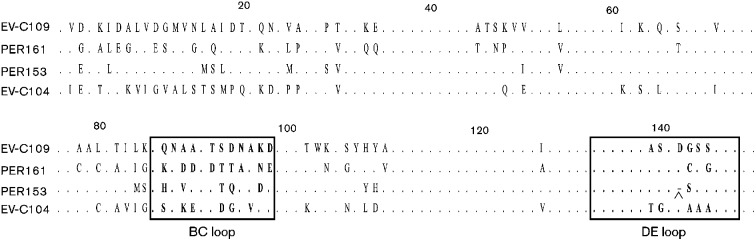
Comparison of full-length VP1 sequences from PER153 and PER161 indicated that each virus fulfilled the criteria for designation as a novel serotype, with <75 % nucleotide and <85 % amino acid identity to the closest serotype (Table 2). A maximum-likelihood phylogenetic tree generated with 326 nt of VP1 sequence from closely related EVs showed that PER161 is ancestral to PER153 and EV-C109, and that EV-C104 is a descendant of an ancestor to all three genotypes (Fig. 1c). Although PER153 has close similarity to EV-C109, the VP1 gene of PER153 has a 3 bp deletion that results in the loss of an aspartic acid residue at position 142 (Fig. 2). Alignment of the VP1 amino acid sequence indicated that the deletion is localized in the DE loop of VP1. This deletion is also present in the ROM31 strain and in related Bulgarian genotypes BU5 and BU77 (GenBank accession numbers JQ317291 and JQ317292, respectively), located within the same phylogenetic branch, but not in the recognized EV-C109 sequences or the ancestral EV-C104 and EV-C117 sequences. Analysis of the full-length VP1 protein indicates that the majority of amino acid differences among these viruses occur within the N-terminal portion and the BC loop of the VP1 gene (Fig. 2). Overall, high sequence variation was observed throughout the capsid genes (VP4–VP1), with PER161 and PER153 showing only 66.5 and 73.1 % nucleotide identity, respectively, to the next closest serotype, EV-C109.
Table 2. Nucleotide and amino acid percentage difference within individual genes of PER153, PER161, EV-C109, EV-C104 and EV-C117.
Amino acid difference is shown in parentheses.
| Gene | Serotype | PER153 | PER161 | EV-C109 | EV-C104 |
| VP4 | PER153 | – | – | – | – |
| PER161 | 20.6 (4.0) | – | – | – | |
| EV-C109 | 18.9 (2.6) | 23.9 (6.7) | – | – | |
| EV-C104 | 23.8 (4.1) | 29.3 (8.2) | 27.2 (6.8) | – | |
| EV-C117* | 24.1 (4.1) | 30.4 (8.2) | 28.9 (6.8) | 20.6 (0) | |
| VP2 | PER153 | – | – | – | – |
| PER161 | 30.3 (10.6) | – | – | – | |
| EV-C109 | 26.0 (6.2) | 32.1 (11.3) | – | – | |
| EV-C104 | 34.9 (18.7) | 36.1 (20.6) | 34.6 (18.6) | – | |
| EV-C117* | 30.5 (14.0) | 31.8 (13.0) | 33.7 (13.9) | 30.2 (13.5) | |
| VP3 | PER153 | – | – | – | – |
| PER161 | 35.9 (21.4) | – | – | – | |
| EV-C109 | 31.4 (14.2) | 34.6 (16.0) | – | – | |
| EV-C104 | 43.2 (27.7) | 40.5 (23.9) | 39.1 (22.1) | – | |
| EV-C117* | 38.6 (25.0) | 34.6 (21.7) | 38.3 (19.7) | 32.0 (12.1) | |
| VP1 | PER153 | – | – | – | – |
| PER161 | 41.5 (30.8) | – | – | – | |
| EV-C109 | 30.8 (15.7) | 38.8 (27.1) | – | – | |
| EV-C104 | 45.1 (36.5) | 46.3 (35.8) | 47.3 (34.8) | – | |
| EV-C117* | 37.9 (25.1) | 40.5 (27.7) | 40.4 (26.3) | 38.8 (23.2) | |
| 2A | PER153 | – | – | – | – |
| PER161 | 22.0 (8.6) | – | – | – | |
| EV-C109 | 14.0 (2.1) | 24.6 (9.3) | – | – | |
| EV-C104 | 31.8 (10.0) | 24.0 (4.9) | 30.2 (9.3) | – | |
| 2B | PER153 | – | – | – | – |
| PER161 | 24.8 (6.2) | – | – | – | |
| EV-C109 | 10.7 (4.1) | 22.5 (6.2) | – | – | |
| EV-C104 | 30.7 (14.1) | 36.5 (13.1) | 31.3 (14.0) | – | |
| 2C | PER153 | – | – | – | – |
| PER161 | 20.6 (4.3) | – | – | – | |
| EV-C109 | 13.1 (0.9) | 21.7 (4.3) | – | – | |
| EV-C104 | 30.8 (12.3) | 33.7 (12.2) | 31.0 (13.0) | – | |
| 3A | PER153 | – | – | – | – |
| PER161 | 22.4 (9.2) | – | – | – | |
| EV-C109 | 17.3 (4.5) | 23.3 (9.2) | – | – | |
| EV-C104 | 37.2 (21.0) | 37.9 (19.9) | 37.7 (20.9) | – | |
| 3B | PER153 | – | – | – | – |
| PER161 | 27.2 (14.8) | – | – | – | |
| EV-C109 | 16.8 (4.8) | 22.7 (9.8) | – | – | |
| EV-C104 | 45.5 (9.6) | 34.7 (14.7) | 34.4 (4.8) | – | |
| 3C | PER153 | – | – | – | – |
| PER161 | 18.0 (5.4) | – | – | – | |
| EV-C109 | 12.2 (2.7) | 20.2 (6.5) | – | – | |
| EV-C104 | 36.7 (15.6) | 38.1 (16.8) | 33.1 (16.3) | – | |
| 3D | PER153 | – | – | – | – |
| PER161 | 12.9 (3.8) | – | – | – | |
| EV-C109 | 8.8 (1.8) | 14.1 (4.3) | – | – | |
| EV-C104 | 33.0 (13.0) | 33.8 (12.5) | 33.0 (13.0) | – |
For EV-C117, only VP4–VP1 sequences are available.
Fig. 2.
Alignment of the first 150 aa of VP1 from EV-C109, PER153, PER161 and EV-C104. Amino acids in the BC and DE loops are indicated in bold and boxed; ∧ indicates the deletion site in PER153. Variant residues are indicated by their letter designation.
Non-structural proteins
The non-structural part of the EV genome encodes proteins involved in viral protein processing and genome replication. Both PER153 and PER161 contain conserved sequence motifs important for the function of picornaviral non-structural proteins. Catalytic triads H20-D38-C109 in 2A and H40-E71-C147 in 3C were present, as well as the cis-acting replication element (cre) and cysteine-rich motif CX2CX8CX4C in 2C and an RNA-binding domain KFRDI in 3C. The NTP-binding motif (S/T)KVEQGKS in the 3D protein was present, although the sequence was SKIKTGKS in PER153 and SKIKAGKS in PER161. All RNA polymerase signature motifs were also conserved.
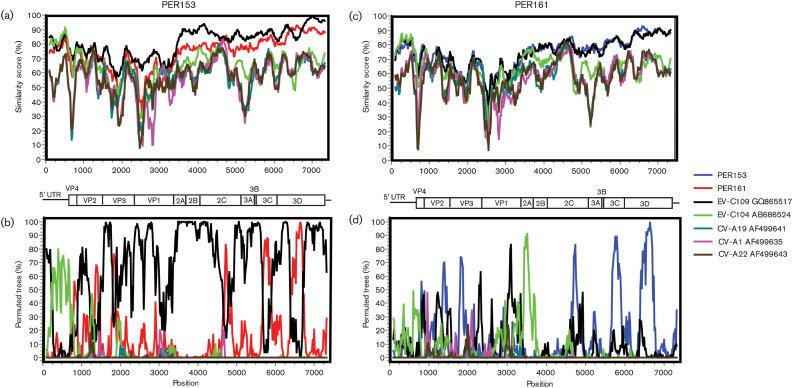
Similarity-plot analysis of the full genomes showed that both viruses were similar to EV-C109 throughout the non-structural portions of the genome (Fig. 3a, c). For every gene in the non-structural region, PER153 and PER161 had >95 and >90 % amino acid similarity to EV-C109, respectively (Table 2). Similarity was highest in the 3D gene; comparison with the only other full-length polyprotein EV-C109 sequence in GenBank, Hungarian strain L87/HUN/2007 (accession no. JN900470; 2007), revealed that, in the 3D gene, EV-C109 from Nicaragua clustered more closely with PER153 than L87/HUN/2007 (data not shown).
Fig. 3.
Similarity plots and bootscanning analysis comparing PER153 (a, b) and PER161 (c, d) with EV-C104, EV-C109, CV-A1, CV-A19 and CV-A22. The EV genetic map is shown as a reference. Colour legend is shown on the right.
Recombination within the non-structural regions is a frequent finding in EVs (Bessaud et al., 2011; Brown et al., 2003; Lukashev, 2005; Lukashev et al., 2005; Oberste et al., 2004). Bootscan analyses performed to identify possible recombination among viruses within this clade indicated several potential recombination events. In PER153, multiple fragments within the non-structural genome parts clustered with PER161, indicating likely ancestral recombination events between these viruses (Fig. 3b, d).
UTR
A typical EV 5′ UTR is approximately 700–750 nt in length (Knowles et al., 2012). The upstream region (approx. 600 nt) is highly conserved and includes the internal ribosomal entry site (IRES). Both EV-C109 and EV-C104 contain a HEV-A-like 5′ UTR that is thought to be the product of an ancestral interspecies recombination event (Tapparel et al., 2009; Yozwiak et al., 2010). Analysis of a region encompassing nt 42–602 of the 5′ UTR showed that PER153 and PER161 clustered with EV-C104 and EV-C109. Bootscanning analysis demonstrated that the majority of the PER153 5′ UTR sequence was EV-C104-like, although the flanking regions were more similar to EV-C109 (Fig. 3b). The 5′ UTR hypervariable region (100–150 bp in length) lies between the end of the IRES and the beginning of the polyprotein ORF, and usually shows little sequence conservation, even within EV serotypes. Relative to the ancestral EV-C104 strain, viruses PER153 and PER161 had a truncated hypervariable region, similar to the truncation described in EV-C109. In comparison to EV-C104, the length of the hypervariable region was 52 and 50 nt shorter in PER153 and PER161, respectively, which mirrored the 50 nt truncation found in EV-C109.
Discussion
In this report, we describe the discovery and genome characterization of two novel HEV-C viruses detected in samples collected in Peru from children with respiratory disease. Full-genome analyses demonstrated that both viruses were related most closely to EV-C109 and EV-C104, and belong to a distinct clade within HEV-C that is genetically different from the other serotypes within this species. The receptor for the viruses in this clade is still unknown, and none of the serotypes have been cultured. Both viruses exhibited comparatively low similarity to any HEV-C serotype within the structural genes and significantly higher similarity to EV-C109 than to EV-C104 within the non-structural genes, with evidence of recombination within this latter region.
The capsid genes of both PER153 and PER161 viruses have diverged significantly from those of their closest serotypes, and fulfil the molecular criteria for designation as novel serotypes. However, sequence differences between PER153 and EV-C109 are very close to the proposed cut-off (15.7 % amino acid difference in VP1) and serological data for this novel clade of viruses may be needed to demarcate their types unequivocally. Both PER153 and PER161 are very similar to EV-C109 within the non-structural part of the genome, and the overall genome sequence of PER153 is similar to that of EV-C109 (<6 % amino acid diversity). Thus, PER153 may represent the result of an ancestral recombination between a virus with an EV-C109-like non-capsid region, and a capsid region of another virus (Fig. 3a). Furthermore, it cannot be excluded that PER153 may represent only a distant genotype of EV-C109, or may be identical to HEV types for which no sequence is currently available.
In EVs, neutralizing epitopes are mainly located in the exposed loops connecting the β-strands of the capsid proteins (Minor et al., 1986; Page et al., 1988). The loops are under host immune pressure and undergo rapid evolutionary change compared with the rest of the EV genomes. Differences in serotype are often associated with mutations within the loops, which alter the antigenicity of the viral epitopes. Within VP1, the BC and DE loops are among the main targets for neutralizing antibodies. Sequences of PER153 and PER161 both show high amino acid variation amongst each other and related HEV-C serotypes within the BC loop. Although the changes within the DE loop are not as extensive, PER153 has a single amino acid deletion within this region; this may affect virus–host interactions.
Both PER153 and PER161 feature a truncated hypervariable region of the 5′ UTR, similar to one found in EV-C109. With the exception of EV-C104, all novel viruses in this clade with available full genome sequences contain a truncated hypervariable region. Such deletions are not common and it is noteworthy that current circulating strains of EV-D68, a virus associated with respiratory disease, also exhibit various truncations within the hypervariable regions of their 5′ UTRs, which is probably a recent event based on comparisons with ancestral strains (Tokarz et al., 2012). The specific role that such a 5′ UTR alteration might serve, and whether it increases fitness within the viruses responsible for respiratory-tract infections, remain to be determined.
Initially described in 2009 and 2010, respectively, EV-C104 and EV-C109 appear to be distributed worldwide. EV-C109 has been reported in Hungary, Nicaragua and the Democratic Republic of Congo. EV-C104 has been detected in Switzerland, Italy and Japan (Grard et al., 2010; Kaida et al., 2012; Pankovics et al., 2012; Piralla et al., 2010, 2012; Tapparel et al., 2009; Yozwiak et al., 2010). The viruses reported here have also been detected in samples from Europe and the Middle East, and were all collected within a period of 5 months. This finding suggests a global distribution, although the overall incidence appears to be low. We did not detect either virus in additional samples, possibly due to cohort size. For comparison, the incidence of EV-C109 in Nicaragua was calculated to be 1.6 % within 310 samples (Yozwiak et al., 2010). It is likely that a larger cohort would reveal more information on prevalence and identify additional strains of these viruses. Judging from its distribution on other continents, we anticipate that future studies will identify these viruses in more patients with respiratory disease.
Methods
Samples (nasal swabs, oropharyngeal swabs and sputum) were collected as part of a cohort study on influenza-like illness in Peru (Razuri et al., 2012). Samples (n = 244) were examined for respiratory-disease agents by MassTag PCR using a panel specific for viruses associated with respiratory disease, including influenza A and B viruses, human rhinovirus, human enterovirus, human metapneumovirus, human parainfluenza viruses 1–4, human coronaviruses 229E and OC43, and respiratory syncytial viruses A and B (Briese et al., 2005). Samples positive by MassTag PCR for picornaviruses were confirmed by a single-plex PCR assay targeting a conserved region within the 5′ UTR with primers 5′-fwd, GGTCAAGCACTTCTGTTTCCC, and 5′-rev, GAAACACGGWCACCCAAAGTASTCG. Full-genome sequencing of PER153 and PER161 was done by consensus PCR, using EV-C104 and EV-C109 genome sequences as references. Multiple sequence alignments, phylogenetic trees and genetic distance matrices were generated in mega 5.05 (Tamura et al., 2007). Phylogenetic trees were generated using the maximum-likelihood method with 1000 bootstrap replicates. Similarity plots and recombination analyses were performed using SimPlot 3.5 and Recombination Detection Program (rdp) v.3.44, with manual bootscanning using the Kimura distance model with a 200 nt window and a step size of 20 nt. VP1 secondary structure prediction was generated using swiss-model (Arnold et al., 2006). PER153 and PER161 sequences were deposited in GenBank (accession numbers JX393301 and JX393302).
The study protocols in which the human samples were taken were approved by the US Naval Medical Research Center Institutional Review Board in compliance with all applicable Federal regulations governing the protection of human subjects.
Acknowledgements
We thank Meera Bhat and Thomas Briese for assistance with the manuscript. The authors thank Claudia Guezala, Hugo Razuri, Candice Romero, Yeny Tinoco and Gabriela Salmon-Mulanovich for assistance with the cohort study. This work was supported by National Institutes of Health grant AI57158 (North-east Biodefence Center-Lipkin) and by grants from the Defense Threat Reduction Agency and the Armed Forces Health Science Center Global Emerging Infections Surveillance Program. The views expressed in this article are those of the authors and do not necessarily reflect the official policy or position of the Department of the Navy, Department of Defense, nor the US Government.
References
- Arnold K., Bordoli L., Kopp J., Schwede T. (2006). The swiss-model workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 10.1093/bioinformatics/bti770 [DOI] [PubMed] [Google Scholar]
- Bessaud M., Joffret M. L., Holmblat B., Razafindratsimandresy R., Delpeyroux F. (2011). Genetic relationship between cocirculating human enteroviruses species C. PLoS ONE 6, e24823. 10.1371/journal.pone.0024823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briese T., Palacios G., Kokoris M., Jabado O., Liu Z., Renwick N., Kapoor V., Casas I., Pozo F. & other authors (2005). Diagnostic system for rapid and sensitive differential detection of pathogens. Emerg Infect Dis 11, 310–313 10.3201/eid1102.040492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown B., Oberste M. S., Maher K., Pallansch M. A. (2003). Complete genomic sequencing shows that polioviruses and members of human enterovirus species C are closely related in the noncapsid coding region. J Virol 77, 8973–8984 10.1128/JVI.77.16.8973-8984.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grard G., Drexler J. F., Lekana-Douki S., Caron M., Lukashev A., Nkoghe D., Gonzalez J. P., Drosten C., Leroy E. (2010). Type 1 wild poliovirus and putative enterovirus 109 in an outbreak of acute flaccid paralysis in Congo, October–November 2010. Euro Surveill 15, 19723. [DOI] [PubMed] [Google Scholar]
- Kaida A., Kubo H., Sekiguchi J., Hase A., Iritani N. (2012). Enterovirus 104 infection in adult, Japan, 2011. Emerg Infect Dis 18, 882–883 10.3201/eid1805.111890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles N. J., Hovi T., Hyypiä T., King A. M. Q., Lindberg A. M., Pallansch M. A., Palmenberg A. C., Simmonds P., Skern T. & other authors (2012). Picornaviridae. In Virus Taxonomy: Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses, pp. 855–880 Edited by King A. M. Q., Adams M. J., Carstens E. B., Lefkowitz E. J. San Diego, CA: Elsevier [Google Scholar]
- Lukashev A. N. (2005). Role of recombination in evolution of enteroviruses. Rev Med Virol 15, 157–167 10.1002/rmv.457 [DOI] [PubMed] [Google Scholar]
- Lukashev A. N., Lashkevich V. A., Ivanova O. E., Koroleva G. A., Hinkkanen A. E., Ilonen J. (2005). Recombination in circulating human enterovirus B: independent evolution of structural and non-structural genome regions. J Gen Virol 86, 3281–3290 10.1099/vir.0.81264-0 [DOI] [PubMed] [Google Scholar]
- Minor P. D., Ferguson M., Evans D. M., Almond J. W., Icenogle J. P. (1986). Antigenic structure of polioviruses of serotypes 1, 2 and 3. J Gen Virol 67, 1283–1291 10.1099/0022-1317-67-7-1283 [DOI] [PubMed] [Google Scholar]
- Oberste M. S., Maher K., Kilpatrick D. R., Flemister M. R., Brown B. A., Pallansch M. A. (1999a). Typing of human enteroviruses by partial sequencing of VP1. J Clin Microbiol 37, 1288–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberste M. S., Maher K., Kilpatrick D. R., Pallansch M. A. (1999b). Molecular evolution of the human enteroviruses: correlation of serotype with VP1 sequence and application to picornavirus classification. J Virol 73, 1941–1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberste M. S., Peñaranda S., Maher K., Pallansch M. A. (2004). Complete genome sequences of all members of the species human enterovirus A. J Gen Virol 85, 1597–1607 10.1099/vir.0.79789-0 [DOI] [PubMed] [Google Scholar]
- Page G. S., Mosser A. G., Hogle J. M., Filman D. J., Rueckert R. R., Chow M. (1988). Three-dimensional structure of poliovirus serotype 1 neutralizing determinants. J Virol 62, 1781–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallansch M. R., Roos R. (2007). Enteroviruses. In Fields Virology, 5th edn, pp. 839–893 Edited by Knipe D. M., Howley P. M. Philadelphia, PA: Lippincott Williams & Wilkins [Google Scholar]
- Pankovics P., Boros A., Szabó H., Székely G., Gyurkovits K., Reuter G. (2012). Human enterovirus 109 (EV109) in acute paediatric respiratory disease in Hungary. Acta Microbiol Immunol Hung 59, 285–290 10.1556/AMicr.59.2012.2.13 [DOI] [PubMed] [Google Scholar]
- Piralla A., Rovida F., Baldanti F., Gerna G. (2010). Enterovirus genotype EV-104 in humans, Italy, 2008–2009. Emerg Infect Dis 16, 1018–1021 10.3201/eid1606.091533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piralla A., Lilleri D., Sarasini A., Marchi A., Zecca M., Stronati M., Baldanti F., Gerna G. (2012). Human rhinovirus and human respiratory enterovirus (EV68 and EV104) infections in hospitalized patients in Italy, 2008–2009. Diagn Microbiol Infect Dis 73, 162–167 10.1016/j.diagmicrobio.2012.02.019 [DOI] [PubMed] [Google Scholar]
- Racaniello V. (2007). Picornaviridae: the viruses and their replication. In Fields Virology, 5th edn, pp. 795–839 Edited by Knipe D. M., Howley P. M. Philadelphia, PA: Lippincott Williams & Wilkins [Google Scholar]
- Razuri H., Romero C., Tinoco Y., Guezala M. C., Ortiz E., Silva M., Reaves E., Williams M., Laguna-Torres V. A. & other authors (2012). Population-based active surveillance cohort studies for influenza: lessons from Peru. Bull World Health Organ 90, 318–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K. D. J., Dudley J., Nei M., Kumar S. (2007). mega4: Molecular Evolutionary Genetics Analysis (mega) software version 4.0. Mol Biol Evol 24, 1596–1599 10.1093/molbev/msm092 [DOI] [PubMed] [Google Scholar]
- Tapparel C., Junier T., Gerlach D., Van-Belle S., Turin L., Cordey S., Mühlemann K., Regamey N., Aubert J. D. & other authors (2009). New respiratory enterovirus and recombinant rhinoviruses among circulating picornaviruses. Emerg Infect Dis 15, 719–726 10.3201/eid1505.081286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokarz R., Firth C., Madhi S. A., Howie S. R., Wu W. Y., Sall A. A., Haq S., Briese T., Lipkin W. I. (2012). Worldwide emergence of multiple clades of enterovirus 68. J Gen Virol 93, 1952–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yozwiak N. L., Skewes-Cox P., Gordon A., Saborio S., Kuan G., Balmaseda A., Ganem D., Harris E., DeRisi J. L. (2010). Human enterovirus 109: a novel interspecies recombinant enterovirus isolated from a case of acute pediatric respiratory illness in Nicaragua. J Virol 84, 9047–9058 10.1128/JVI.00698-10 [DOI] [PMC free article] [PubMed] [Google Scholar]