Abstract
The eukaryotic basic leucine zipper (bZIP) transcription factors play critical roles in the organismal response to the environment. Recently, a novel YAP-like bZIP, restorer of secondary metabolism A (RsmA), was found in a suppressor screen of an Aspergillus nidulans secondary metabolism (SM) mutant in which overexpression of rsmA was found to partially remediate loss of SM in Velvet Complex mutants. The Velvet Complex is a conserved fungal transcriptional heteromer that couples SM with sexual development in fungi. Here we characterized and contrasted SM in mutants of RsmA and four other A. nidulans bZIP proteins (NapA, ZipA, ZipB and ZipC) with predicted DNA binding motifs similar to RsmA. Only two overexpression mutants exhibited both SM and sexual abnormalities that were noteworthy: OE : : rsmA resulted in a 100-fold increase in sterigmatocystin and a near loss of meiotic spore production. OE : : napA displayed decreased production of sterigmatocystin, emericellin, asperthecin, shamixanthone and epishamixanthone, coupled with a shift from sexual to asexual development. Quantification of bZIP homodimer and heterodimer formation using fluorescence resonance energy transfer (FRET) suggested that these proteins preferentially self-associate.
Introduction
Dimeric basic leucine zipper (bZIP) proteins are conserved transcriptional enhancers found in all eukaryotes. These proteins play critical roles, often species-specific, in many aspects of organismal development. A distinct subset of proteins within the bZIP family is referred to as yeast activator protein (Yap) transcription factors, which have been well defined in yeast (Rodrigues-Pousada et al., 2010). Members of the Yap family, eight proteins in yeast, contain a conserved set of residues in their basic region conferring distinct DNA-binding properties that overlap and differ from those of the canonical Gcn4 factor. The Yap family binding site was characterized as TTAC/GTAA and the terms ‘Yap site’ or ‘Yap response element’ (YRE) were coined. Yap proteins frequently mediate stress responses, and are often associated with resistance to reactive oxygen species (ROS), osmotic imbalances, drugs or heavy metals.
Since the finding of the first Yap protein in Saccharomyces cerevisiae (Moye-Rowley et al., 1989), several orthologues have been characterized in other fungi. Similar to the reported roles in yeast, these proteins typically are associated with resistance to ROS or antifungals. The Yap1 orthologues Afyap1, Aoyap1, Apyap1 and MoAP1 in the human pathogen Aspergillus fumigatus, the mycotoxic pathogens Aspergillus ochraceus and Aspergillus parasiticus, and the rice pathogen Magnaporthe oryzae, respectively, all mediate the oxidative stress response, and in the latter pathogen, MoAP1 is critical for pathogenicity (Guo et al., 2011; Lessing et al., 2007; Qiao et al., 2008; Reverberi et al., 2007, 2012). Yap-like proteins in Aspergillus nidulans and Neurospora crassa have also been characterized in ROS and heavy metal resistance (Asano et al., 2007; Tian et al., 2011).
A recently reported and novel function for bZIPs is association of these proteins with secondary metabolite production in filamentous fungi. In a mutagenesis screen for secondary metabolism (SM) activation in the filamentous fungus A. nidulans, a Yap-like bZIP, termed restorer of secondary metabolism A (RsmA), has been identified (Shaaban et al., 2010). Overexpression of rsmA was able to partially compensate for loss of LaeA and VeA, two members of the fungal-specific Velvet Complex required for global regulation of SM and sexual development in A. nidulans (Bayram et al., 2008). Yap-like bZIPs have also been associated with negative regulation of secondary metabolites. For example, deletion of Aoyap1 and Apyap1 in A. ochraceus and A. parasiticus resulted in increases of ochratoxin and aflatoxin, respectively (Reverberi et al., 2007, 2012). These latter two studies led to the hypothesis that fungal SM can be triggered by stress-response pathways.
To further explore this linkage of bZIP response pathways to SM in the model aspergillus A. nidulans, we assessed the effects of deletion and overexpression of five Yap-like proteins, including RsmA. We found that both RsmA and NapA, previously identified as having a major role in the fungal stress response, have significant and distinct effects on SM and sexual development, a linkage previously characterized by the Velvet Complex (Bayram & Braus, 2012).
Methods
Fungal strains and culture conditions.
The fungal strains and plasmids used in this study are listed in Table 1. All strains were grown at 37 °C on glucose minimum medium (GMM) (Shimizu & Keller, 2001) with appropriate supplements corresponding to the auxotrophic markers, and were maintained as glycerol stocks at −80 °C. Escherichia coli strain DH5α was propagated in Luria–Bertani (LB) medium with appropriate antibiotics for plasmid DNA.
Table 1. Plasmids and fungal strains used in this study.
pXX, Plasmid; RXX, ascospore recombinant; TXX, original transformant.
| Strain or plasmid | Description | Reference or source |
| Strains | ||
| RDIT9.32 | veA | Tsitsigiannis et al. (2004b) |
| RDIT55.37 | pyroA4, veA | Tsitsigiannis et al. (2004a) |
| RWY2.12 | gpdA(p) : : rsmA : : A. fumigatus pyrG, veA | Yin et al. (2012) |
| RJMP1.49 | pyrG89, pyroA4, ΔnkuA : : argB, veA | Shaaban et al. (2010) |
| HZS189 | paba1, ribo2B, ΔatfA : : riboB, veA1 | Balázs et al. (2010) |
| TMS6.30 | pyrG89 : : ΔrsmA : : A. parasiticus pyrG, pyroA4, veA | Shaaban et al. (2010) |
| TWY6.2 | pyrG89; ΔzipA : : pyroA A. fumigatus, pyroA4, ΔnkuA : : argB; veA | This study |
| TWY7.3 | pyrG89; ΔnapA : : pyroA A. fumigatus, pyroA4, ΔnkuA : : argB; veA | This study |
| TWY8.3 | pyrG89; ΔzipB : : pyroA A. fumigatus, pyroA4, ΔnkuA : : argB; veA | This study |
| TWY13.15 | pyrG89; A. fumigatus pyroA : : gpdA(p) : : napA, pyroA4, ΔnkuA : : argB; veA | This study |
| TWY14.3 | pyrG89; A. fumigatus pyroA : : gpdA(p) : : zipA, pyroA4, ΔnkuA : : argB; veA | This study |
| TWY15.5 | pyrG89; A. fumigatus pyroA : : gpdA(p) : : zipB, pyroA4, ΔnkuA : : argB; veA | This study |
| TWY17.10 | pyrG89; A. fumigatus pyroA : : gpdA(p) : : zipC, pyroA4, ΔnkuA : : argB; veA | This study |
| RWY8.5 | ΔrsmA : : pyrG A. parasiticus, veA | This study |
| RWY9.2 | ΔzipA : : pyroA A. fumigatus, veA | This study |
| RWY10.3 | ΔnapA : : pyroA A. fumigatus, veA | This study |
| RWY11.1 | ΔzipB : : pyroA A. fumigatus, veA | This study |
| RWY17.3 | A. fumigatus pyroA : : gpdA(p) : : napA, pyroA4, veA | This study |
| RWY18.4 | A. fumigatus pyroA : : gpdA(p) : : zipA, pyroA4, veA | This study |
| RWY19.1 | A. fumigatus pyroA : : gpdA(p) : : zipB, pyroA4, veA | This study |
| RWY22.1 | A. fumigatus pyroA : : gpdA(p) : : zipC, pyroA4, veA | This study |
| Plasmids | ||
| pTlex3 | Yeast two-hybrid bait vector | Cho et al. (2003) |
| pWY25.16 | A. fumigatus pyroA : : gpdA in pGEM-T Easy vector | Yin et al. (2012) |
| pWY16 | zipA cDNA in pTlex3 | This study |
| pWY17 | napA cDNA in pTlex3 | This study |
| pWY18 | zipB cDNA in pTlex3 | This study |
| pWY19 | zipC cDNA in pTlex3 | This study |
Creation of fungal strains.
The oligonucleotide sequences for PCR primers are given in Table S1 available with the online version of this paper. PCR amplification was carried out on a C1000 Thermal Cycler from Bio-Rad. For creation of overexpression strains of napA (AN7513), zipA (AN11891), zipB (AN8772) and zipC (AN10378) at the native locus, double joint PCR procedures (Yu et al., 2004) were carried out. Briefly, the marker gene cassette (A. fumigatus pyroA and a constitutive A. nidulans gpdA promoter) was amplified by using pWY25.16 (Table 1) as template and used as marker gene for all overexpression bZIP mutants. Approximately 1 kb fragments upstream and downstream of the target genes were amplified from genomic DNA of A. nidulans using the designated primers (Table S1), respectively. These three amplified PCR fragments were then purified with a QIAquick gel extraction kit (Qiagen), quantified, and fused using double-joint PCR procedures. The final PCR product was amplified with the primer pairs NEST_for and _rev or the end primers of each flanking region, confirmed with endonuclease digestion and purified for fungal transformation. Except for the first-round PCR with Pfu Ultra II DNA polymerases (Agilent), all PCR steps were performed by using an Expand Long Template PCR system (Roche) according to the manufacturer’s instructions. Using the same double-joint PCR strategy (Yu et al., 2004), napA, zipA and zipB deletion strains were created with A. fumigatus pyroA as the selectable maker. Fungal protoplast preparation and transformation were carried out as described by Bok & Keller (2004). Samples (5 µg) of the double-joint cassette were used to overexpress/delete bZIPs by using A. nidulans strain RJMP1.49 (pyrG89, pyroA4, ΔnkuA : : argB, veA) as the recipient host. Overexpression and deletion strains were verified by PCR and Southern blot analysis.
Prototrophic strains, other than the bZIP mutation, were generated by sexual crossing for physiological studies, according to standard methods (Pontecorvo et al., 1953). Briefly, crossing TMS6.30 with HZS189 yielded RWY8.5 (ΔrsmA). Crossing RDIT55.37 with TWY6.2, TWY7.3, TWY8.3, TWY13.15, TWY14.3, TWY15.5 and TWY17.10 created RWY9.2 (ΔzipA), RWY10.3 (ΔnapA), RWY11.1 (ΔzipB), RWY17.3 (OE : : napA), RWY18.4 (OE : : zipA), RWY19.1 (OE : : zipB) and RWY22.1 (OE : : zipC), respectively. The genotypes of the progeny were determined by growth on selection media and PCR confirmation with designated primers (Table S1). Possession of the veA or veA1 allele was determined by using the primers veA+For and veA+Rev and veA1For and veA+Rev, respectively (Table S1).
Northern analysis.
Extractions were made from mycelia of cultures where 106 spores ml−1 were grown in 50 ml liquid GMM at 37 °C for 48 h with shaking at 250 r.p.m. Mycelia were harvested and lyophilized. RNA was extracted by using Isol-RNA Lysis Reagent according to the manufacturer’s instructions (5 Prime). About 30 µg of total RNA was used for RNA blot analysis. RNA blots were hybridized with designated DNA fragments, which were generated by PCR using gene-specific primers as shown in Table S1. All experiments were performed in duplicate.
Sterigmatocystin examination.
Samples (5 µl) of 105 spores of A. nidulans strains were point- or overlay-inoculated onto YAG medium (5 g yeast extract l−1, 15 g agar l−1 and 20 g d-glucose l−1, supplemented with 1 ml of a trace element solution l−1 and 200 µg pyridoxine l−1) or GMM (Shimizu & Keller, 2001) with pyridoxine at a concentration of 200 µg l−1, and incubated for 4 or 7 days at 37 °C under both light and dark conditions. An equal-size agar plug, 7 mm in diameter, was removed from the centre of each plate culture, homogenized in 3 ml nanopure water and extracted with an equal amount of chloroform by agitation for 30 min at room temperature. The chloroform extracts were then dried completely at room temperature and resuspended in 100 µl chloroform. Metabolites were separated in the developing solvent toluene : ethyl acetate : acetic acid (TEA; 8 : 1 : 1) on silica-coated TLC plates (Shwab et al., 2007), and photographs were taken following exposure to UV radiation at 254 and 366 nm wavelengths.
LC–MS analysis.
The same cultures from 7-day YAG plates were used for secondary metabolite analysis by LC-MS. Briefly, three 7 mm diameter agar plugs were taken from each strain and transferred to a 10 ml vial. The plugs were extracted with 2 ml methanol followed by 2 ml CH2Cl2/methanol (1 : 1) each with 1 h sonication. The organic extract was transferred to a fresh 7 ml vial, in which the organic solvents were evaporated by TurboVap LV (Caliper Life Sciences) to dryness. The crude extract was then redissolved in 0.2 ml DMSO/methanol (1 : 4). After filtration, 10 µl of DMSO/methanol extract was injected for HPLC-photodiode array detection-MS (HPLC-DAD-MS) analysis as described previously (Bok et al., 2009).
For determining fold differences, negative ion electrospray ionization (ESI) at m/z 317 was used for the detection of asperthecin by using an extracted ion chromatogram (EIC). Positive mode at m/z 325, 409, 389 and 389 was used for the detection of sterigmatoycstin, emericellin, shamixanthone and epishamixanthone, respectively. The fold differences were calculated according to the following formula: (area(sample)−area(blank))/(area(WT)−area(blank)).
Analysis of spore production.
Before adding chloroform for secondary metabolite extractions of YAG overlay samples, 100 µl of spore-agar suspension described above was collected for each strain to use for ascospore (sexual spore) and conidia (asexual spore) quantification. Serial dilutions were carried out in double-distilled water, and spores were quantified with a haemocytometer.
Stress sensitivity assays on nutrient agar plates.
To estimate the stress sensitivity of the mutants, 105 freshly grown (6 days) conidia washed and suspended in 5 µl 0.9 % NaCl, 0.01 % Tween 80 (Eigentler et al., 2012) were spotted on minimal-nitrate medium agar plates (Balázs et al., 2010; Hagiwara et al., 2007, 2008), which were also supplemented with one of the following stress-generating agents (concentrations and mechanisms of actions are given in parentheses): diamide (2 mmol l−1, triggers glutathione redox imbalance), menadione sodium bisulphite (MSB; 0.12 mmol l−1, increases intracellular superoxide concentrations), tert-butylhydroperoxide (tBOOH; 0.8 mmol l−1, accelerates lipid peroxidation) and H2O2 (6.0 mmol l−1, increases intracellular peroxide concentrations). The surface cultures of RWY17.3 (OE : : napA), RWY18.4 (OE : : zipA) and RWY19.1 (OE : : zipB) were always supplemented with pyridoxine at a concentration of 200 µg l−1. All stress plates were incubated at 37 °C for 5 days (Balázs et al., 2010), the colony diameters were measured and the percentage growth inhibition was calculated. The growth inhibition of the mutant strains was always compared with that of the RDIT9.32 reference strain.
Determination of physiological parameters in submerged cultures of A. nidulans.
A. nidulans strains (RDIT9.32: wild-type, RWY10.3 ΔnapA, RWY17.3 OE : : napA) were pre-grown in Erlenmeyer flasks (500 ml) containing 100 ml minimal-nitrate medium (pH 6.5) also supplemented with 200 µg pyridoxine l−1 (wild-type, OE : : napA) as required. Culture media were inoculated with 5×107 spores and incubated at 37 °C and at 3.7 Hz shaking frequency. Oxidative stress was induced by the addition of 0.8 mmol tBOOH l−1 to exponential growth phase (18 h) cultures, and samples were taken at 28 h (10 h stress exposure) and 42 h (24 h stress exposure) incubation times for the determination of selected physiological parameters.
The intracellular reactive species (RS) levels were characterized by the formation of 2′,7′-dichlorofluorescein (DCF) from 2′,7′-dichlorofluorescin diacetate (Halliwell & Gutteridge, 2007). RS includes all ROS and reactive nitrogen species, which oxidize 2′,7′-dichlorofluorescin to DCF (Halliwell & Gutteridge, 2007). At the incubation times tested, 10 µmol 2′,7′-dichlorofluorescin diacetate ml−1 was added to 20 ml aliquots of the cultures, and after incubating further for 1 h in 100 ml culture flasks, the mycelia were harvested by centrifugation. The production of DCF was determined spectrofluorimetrically (Emri et al., 1997, 1999).
Changes in the specific activities of certain antioxidant enzymes were also recorded in separate experiments. In these cases, cell-free extracts were prepared by X-pressing and centrifugation (Emri et al., 1997). Specific catalase (Roggenkamp et al., 1974) and glutathione peroxidase (GPx; Chiu et al., 1976) activities were measured. Briefly, catalase and GPx activities were determined spectrophotometrically, measuring H2O2 decomposition and NADPH diminution rates, respectively (Roggenkamp et al., 1974; Chiu et al., 1976). In the GPx assay, cumene hydroperoxide was used as substrate and the glutathione disulphide formed was reduced by glutathione reductase auxiliary enzyme, which oxidizes NADPH cofactor (Chiu et al., 1976).
In sterigmatocystin determinations, mycelia from 24 h cultures were filtered and washed. After lyophilization, sterigmatocystin was extracted by 70 % (v/v) acetone from 20 mg quantities of the freeze-dried mycelial powder. The sterigmatocystin content of the solutions was quantified on silica gel according to Klich et al. (2001).
Plasmid construction.
bZIP ORFs were amplified from an A. nidulans cDNA library with designated primers (Table S1). Then, each bZIP fragment was cloned into pTlex3 using the Quick-change method (Bok & Keller, 2012) to obtain pWY16 to pWY19. All plasmids were sequenced for confirmation before use.
Plasmid preparation, digestion with restriction enzyme, gel electrophoresis, blotting, hybridization and probe preparation were performed by standard methods (Sambrook et al., 1989). Aspergillus DNA for diagnostic PCR was isolated as described previously (Shaaban et al., 2010). Sequence data were analysed in the SeqBuilder (v. 7.0) of the Lasergene software package from dnastar.
Cloning, expression, purification and labelling of proteins for FRET.
bZIP domains were cloned from plasmids pWY16 to pWY19 (Table 1) or A. nidulans genomic DNA into a modified pTXB1 (NEB) plasmid using the sequence and ligation-independent cloning (SLIC) method (Li & Elledge, 2007), or were restriction-digested with XhoI and NsiI. All constructs were verified by sequencing. Sequences for the proteins used are listed in Table 2. Proteins were expressed as intein–chitin binding domain fusions in RP3098 E. coli by growing 1 l LB cultures at 37 °C to OD600 0.4–0.8, at which point expression was induced with 0.5 mM IPTG. Cultures were then incubated for 3–4 h and cells pelleted. Cells were resuspended in buffer (20 mM HEPES, pH 8.0, 500 mM NaCl, 2 mM EDTA, 1 M guanidine-HCl, 0.2 mM PMSF and 0.1 % Triton X-100) and lysed using sonication. The lysate was then split and each half was poured over a column containing 1 ml chitin beads (NEB). The column was washed and then equilibrated with EPL buffer [50 mM HEPES, pH 8.0, 500 mM NaCl, 200 mM 2-mercaptoethane sulfonate (MESNA), 1 M guanidine-HCl]. To cleave the intein and label the proteins with a fluorescent dye, the columns were incubated with EPL buffer containing 1 mg cysteine-lysine-dye ml−1, where the dye was either fluorescein or carboxytetramethylrhodamine (TAMRA; Celtek). Columns were capped and incubated for 16 h. Cleaved and labelled proteins were eluted and diluted fivefold into denaturing buffer (6 M guanidine-HCl, 5 mM imidazole, 0.5 M NaCl, 20 mM Tris, 1 mM DTT, pH 7.9). This solution was flowed over columns containing 1 ml nickel-nitrilotriacetic acid (Ni-NTA) resin. After washing, proteins were eluted with 60 % acetonitrile/0.1 % trifluoroacetic acid. Labelled proteins were lyophilized, resuspended and desalted using spin-columns (Bio-Rad). Proteins were stored in 10 mM potassium phosphate, pH 4.5, at −80 °C. Peptide concentrations were measured in 6 M guanidine-HCl/100 mM sodium phosphate, pH 7.4, using the absorbance of the dye with an absorption coefficient of 68 000 M−1 cm−1 at 499 nm for fluorescein and 86 000 M−1 cm−1 at 560 nm for TAMRA. Molecular masses of A. nidulans fluorescein-labelled proteins were determined by MS to be within 0.1 % of the expected mass.
Table 2. Protein sequences utilized for FRET.
| Protein | Sequence* |
| NapA | KKPGRKPLTSEPTSKRKAQNRAAQRAFRERKEKHLKDLEAKVEELQKASDSANQENGLLKAQVERLQVELREYRKRLSWVT |
| RsmA | EKDKDGIGITPAQSKRKAQNRAAQRAFRERKERHVRDLEEKVSNLQQESSNLLADNERLKREIARYSTENEILR |
| ZipA | LLASEEGKKLSSKERRQLRNKVSARAFRSRRKEYIGQLENEVAQKTNEAHELRQQNRALCDENARLTDLVRQLL |
| ZipB | STKENASEPTDPGLRRKEQVRRAQQTYRLRKESYIKSLEREILHLRTAKSDLTGETRKLRAEVRRLRQVIEQHG |
| ZipC | ETPKTYGKRPLSTSKRAAQNRAAQRAFRQRKESYIRKLEEQVKEYEVMSQEYKALQAENYQLREYVINLQSRLL |
| JUN | SPIDMESQERIKAERKRMRNRIAASKCRKRKLERIARLEEKVKTLKAQNSELASTANMLREQVAQLKQKVMNHV |
| FOS | KVEQLSPEEEEKRRIRRERNKMAAAKCRNRRRELTDTLQAETDQLEDEKSALQTEIANLLKEKEKLEFILAAHR |
All sequences contained the linkers SHHHHHHDWKGSS at the N terminus and GA at the C terminus.
FRET measurements.
TAMRA-labelled proteins diluted in 1 mM tris (2-carboxyethyl)phosphine (TCEP) were serially diluted twofold in black 384-well non-binding surface plates (Corning). This resulted in 12 concentrations of each TAMRA protein from 1000 to 0 nM with each well containing 30 µl. Fluorescein-labelled proteins were diluted to 80 nM in 1 mM TCEP and 10 µl of each was transferred to the 384-well plate. A 40 µl volume of 2× binding buffer (100 mM potassium phosphate, pH 7.4, 300 mM KCl, 0.2 % BSA, 0.2 % Tween-20) was then added to each well and mixed. Binding reactions were set up using a Tecan Freedom EVO liquid-handling robot. Plates were incubated for 105 min at 37 °C. Following incubation, plates were read using a fluorescence plate reader (Molecular Devices) with excitation at 480 nm and emission at 525 nm. Plates were then transferred to 21 °C and incubated for 60 min and measured again. Plates were then transferred to 4 °C and incubated for 60 min and measured again.
To fit equilibrium disassociation constants, a system of ordinary differential equations (ODEs) was integrated in MATLAB as described previously (Ashenberg et al., 2011). The Kd of each homodimer along with an upper and lower baseline was fitted. Homodimer Kds were used subsequently to determine the Kds of each heterodimer. When fitting heterodimers, the ODEs describing donor or acceptor homodimer formation were only included in the system when their respective Kds were less than 5000 nM (otherwise, the system was modelled with no homodimer species). Each curve was manually inspected. For several curves with bad upper or lower baselines, one or more points were removed to allow for an improved fit. All interactions identified were also observed in a replicate experiment.
Statistical analysis.
For statistical analyses, data were analysed with the GraphPad Instate software package, version 5.01 using the Tukey–Kramer multiple comparison test at P≤0.05.
Results
Selection of bZIPs
RsmA, identified as a suppressor in a ΔlaeA screen, was found to activate sterigmatocystin through binding to the Yap-like site TTAGTAA (Yin et al., 2012). In yeast, Yap proteins often exhibit overlapping roles, so we reasoned that other A. nidulans Yap-like bZIPs would also affect SM. We examined this possibility by characterizing four other bZIP proteins that bore residues conserved with respect to RsmA/Yap proteins (as assessed by clustal w analysis) in the DNA binding motif and also showed differential expression in microarray profiles of laeA mutants (Bok et al., 2006; data not shown). These proteins included the already characterized NapA (AN7513) (Asano et al., 2007), as well as three uncharacterized proteins (AN11891, AN8772 and AN10378, hereinafter called ZipA, ZipB and ZipC). Fig. 1 shows the alignment of the DNA binding and leucine zipper motifs of these five proteins in comparison with Yap-family proteins and the human Yap-like bZIPs JUN and FOS (Ferguson & Goodrich, 2001).
Fig. 1.
Comparison of the Yap bZIP domains with A. nidulans bZIPs. Sequences of the eight S. cerevisiae Yap bZIP domains (Yap1 to Yap8) compared with corresponding regions from five A. nidulans bZIPs (RsmA, NapA, ZipA, ZipB and ZipC) and two human bZIPs (JUN and FOS). In the basic region, residues that directly interact with base pairs and Yap-specific residues are indicated in bold type; in the leucine zipper region, the conserved leucines (or other residues) are also indicated in bold type.
Deletion and overexpression of RsmA has already been reported (Shaaban et al., 2010), and similar procedures were conducted to create the other bZIP deletion and overexpression mutants (Table 1). The only mutant that we failed to create was the zipC deletion, perhaps due to a near-lethal effect of loss of this gene on fungal growth. Genomic DNA was extracted from all transformants and analysed by PCR followed by Southern blot analysis (Figs S1 and S2). Overexpression strains were also confirmed by Northern analysis (Fig. S3). Prototrophic strains were created according to standard sexual crossing methods (Pontecorvo et al., 1953) and were always PCR-confirmed with designated primers (Table S1). We found that several of the overexpression mutants displayed a pyridoxine marker gene effect, and thus added pyridoxine to the media of all strains for secondary metabolite and sexual spore assessment, and for selected stress test assessments as described below.
Secondary metabolite assessment
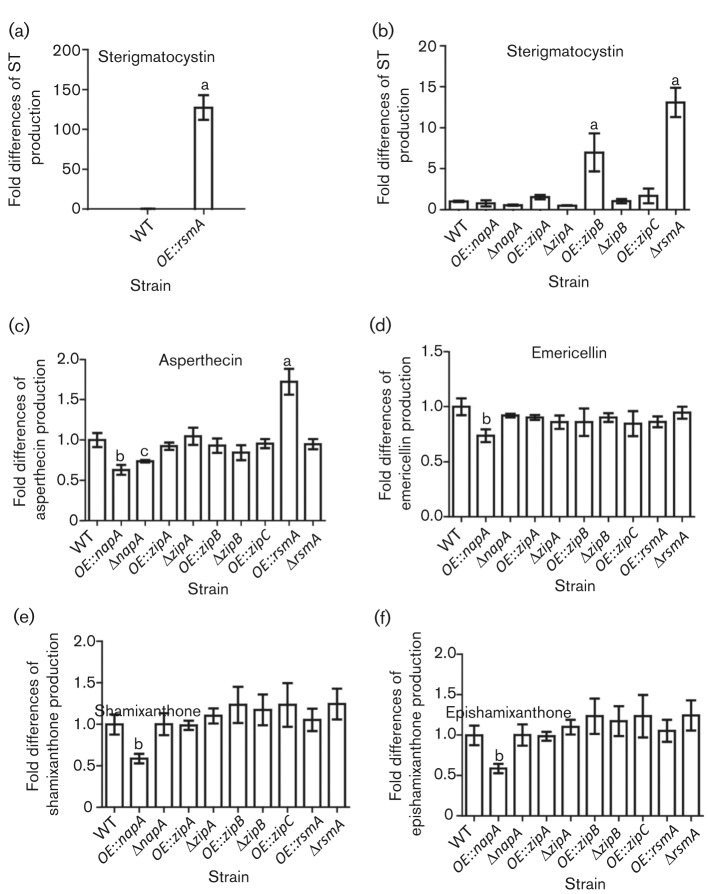
As production of secondary metabolites is greatly dependent on culture conditions, secondary metabolite production of the bZIP mutants was assessed under a variety of conditions. We first assessed sterigmatocystin production, as this metabolite is produced in all growth media. The effect of overexpression or deletion of the different bZIP genes on sterigmatocystin production was dependent on growth medium and light conditions. As reported earlier, sterigmatocystin was always increased in the OE : : rsmA (RMY2.12) strain, but its levels varied depending on the growth medium in the other bZIP mutant backgrounds (Fig. S4). For comparison’s sake, we quantified production of five secondary metabolites, asperthecin, sterigmatocystin, emericellin, shamixanthone and epishamixanthone, under the same solid-state culture conditions (Fig. 2).
Fig. 2.
LC-MS analysis of secondary metabolite production of bZIP strains. Strains were grown on YAG medium with overlay inoculation at 37 °C for 7 days under dark conditions before extraction. Sterigmatocystin (ST) (a, b), asperthecin (c), emericellin (d), shamixanthone (e) and epishamixanthone (f) were analysed by LC-MS. Data were analysed with the GraphPad Instate software package, version 5.01, using the Tukey–Kramer multiple comparison test at P≤0.05. Mean values with different letters show significant differences.
The most dramatic effect on secondary metabolite production was that of OE : : rsmA, which increased sterigmatocystin production by 100-fold (Fig. 2a). Although several other mutants showed increased sterigmatocystin production under these conditions (Fig. 2b), the increases were modest compared with that of the OE : : rsmA strain. OE : : rsmA also showed a significant increase in asperthecin, as reported previously (Yin et al., 2012). Conversely, OE : : napA presented a phenotype of significant decreases in emericellin, asperthecin, shamixanthone and epishamixanthone production (Fig. 2c–f), and, dependent on growth conditions, decreased sterigmatocystin production (Fig. S4).
Sexual development
Because secondary metabolite synthesis has been genetically linked with sexual development, we also assessed production of the meiotic spore, the ascospore, in all mutants. Ascospore production was significantly reduced in three overexpression strains: OE : : napA, OE : : rsmA and OE : : zipA (Fig. 3a). Macroscopic examination of these three strains showed that decreased sexual spore production was correlated with increases in asexual spore production in OE : : napA and OE : : zipA (Fig. 3b); this was verified upon quantification of the asexual conidia (Fig. 3c). In contrast, OE : : rsmA developed only sexually, but the sexual fruiting bodies, the cleistothecia, contained few ascospores (Fig. 3b, c).
Fig. 3.
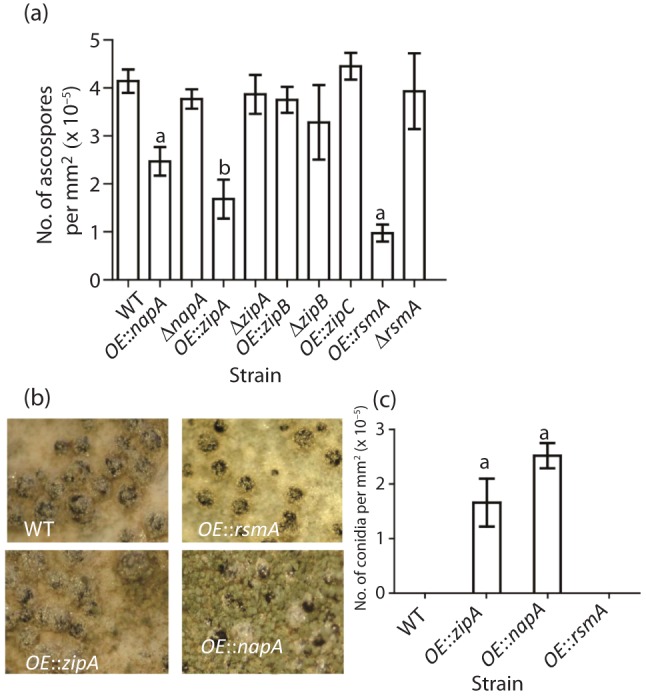
Assessment of asexual and sexual spore production by bZIP strains. (a) Ascospore numbers of bZIP strains which were grown on YAG plates for 7 days. (b) Cleistothecia are shown for four bZIP strains which have decreased ascospore counts in (a). (c) Conidia numbers of selected bZIP strains from (a). Statistical differences were analysed in each strain group with Prism 5 software, version 5.01 (GraphPad software), using the Tukey–Kramer multiple comparison tests at P≤0.05. Mean values with different letters are significantly different.
Stress challenge
The oxidative stress tolerances of all bZIP mutants were tested on nutrient agar stress plates (Fig. 4 and Fig. S5). As expected, the deletion of napA resulted in increased oxidative stress sensitivity in the presence of a series of well-characterized oxidants (diamide, MSB, tBOOH, H2O2), which is in accordance with previous reports (Asano et al., 2007). The overexpression of napA increased moderately the diamide and tBOOH tolerance of the fungus. In general, the other bZIP proteins had little effect on stress sensitivity as determined by growth of mutants on common stress media; however, OE : : zipA and OE : : zipC were more sensitive to MSB and diamide, respectively, and ΔzipA showed an increased tolerance to hydrogen peroxide (Fig. S5).
Fig. 4.
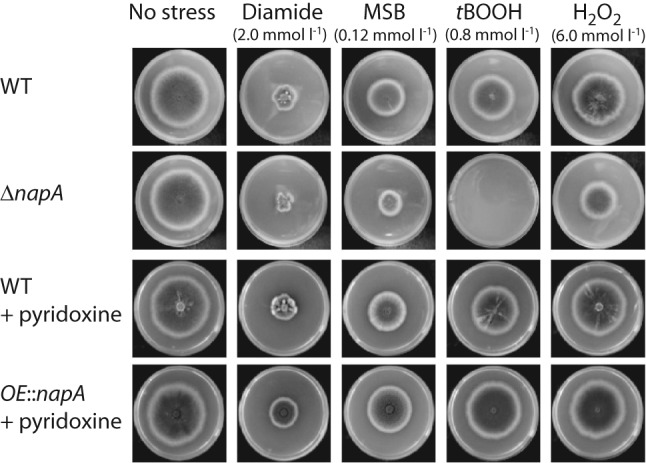
Comparison of the oxidative stress tolerances of napA mutants. RDIT9.32 (wild-type), RWY10.3 (ΔnapA) and RWY17.3 (OE : : napA) strains were incubated at 37 °C for 5 days, and selected culture media were supplemented with 200 µg pyridoxine l−1, as required. Stress-initiated percentage changes (ΔA) in growth (colony diameters) were determined for each strain and stress-generating agent, and differences between the percentage changes recorded for the mutants and the RDIT9.32 control strain (ΔΔA = ΔAmutant−ΔARDIT9.32) were calculated. Significantly decreased stress tolerances were found in oxidative stress (MSB, tBOOH and H2O2)-exposed RWY10.3 (ΔnapA) cultures (−80 % <ΔΔA <−20 %); meanwhile, growth stimulation caused by the overexpression of napA (RWY17.3) was ΔΔA ≤15 % under different oxidative stress treatments. Pyridoxine supplementation increased slightly the oxidative stress resistance of RDIT9.32 (ΔΔA ≤5 %), which was similar to previous findings (Balázs et al., 2010).
NapA involvement in SM is stress-related
The above studies clearly demonstrated that NapA had a unique role in the fungal response to stress, SM and sexual development that, in many respects, was opposite to that of RsmA. Whereas RsmA regulation of sterigmatocystin is positive via DNA binding of aflR (Yin et al., 2012), studies of NapA deletants in A. ochraceus and A. parasiticus (Aoyap1 and Apyap1, respectively) suggested that the effect of this bZIP factor on SM was indirect. Specifically, a hypothesis was proposed that the loss of Aoyap1/Apyap1 disabled the redox balance in fungi, leading to activation of secondary metabolite synthesis (Reverberi et al., 2007, 2008, 2012). We therefore set out to examine the relationship of oxidative stress regulation and sterigmatocystin production more exhaustively in the napA mutants. To do this, biomass, sterigmatocystin and RS (Halliwell & Gutteridge, 2007) production as well as changes in the specific catalase and glutathione peroxidase activities were measured in exponential growth phase cultures of wild-type, ΔnapA and OE : : napA exposed to tBOOH, a lipid peroxidation-inducing agent (Table 3).
Table 3. Biomass, sterigmatocystin, RS and antioxidant enzyme production in A. nidulans ΔnapA and OE : : napA mutants.
WT, wild-type.
| Strain and treatment | Physiological parameter* | |||||||||
| Biomass (DCM†) production (g l−1) | Sterigmatocystin production [mg (g DCM)−1] | RS‡ production [pmol DCF (g DCM)−1] | Specific antioxidant enzyme activities | |||||||
| Catalase [kat (kg protein−1)] | Glutathione peroxidase (GPx) [mkat (kg protein)−1] | |||||||||
| 0 h | 10 h | 24 h | 24 h | 10 h | 24 h | 10 h | 24 h | 10 h | 24 h | |
| WT | 2.0±0.3 | 4.5±0.6 | 4.2±0.5 | 1.5±0.5 | 0.25±0.05 | 0.19±0.05 | 0.21±0.02 | 0.28±0.03§ | 0.54±0.05 | 0.31±0.03§ |
| WT+tBOOH | 2.1±0.3 | 2.2±0.6¶ | 4.4±0.5§ | 3.1±1¶ | 27±5¶ | 0.52±0.06§¶ | 0.56±0.05¶ | 0.31±0.04§ | 0.68±0.05¶ | 0.28±0.03§ |
| ΔnapA | 2.1±0.3 | 4.4±0.3 | 3.6±0.4§ | 1.4±0.5 | 0.41±0.06# | 0.47±0.07# | 0.12±0.02# | 0.20±0.03§# | 0.68±0.07# | 0.33±0.03§ |
| ΔnapA+tBOOH | 2.1±0.2 | 1.8±0.3¶ | 3.4±0.4§# | 3.3±1¶ | 32±7¶ | 1.8±0.5§¶# | 0.31±0.03¶# | 0.28±0.03¶ | 0.76±0.08 | 0.65±0.07¶# |
| Pyridoxine supplementation | ||||||||||
| WT | 2.2±0.3 | 4.5±0.6 | 4.2±0.5 | 1.6±0.5 | 0.24±0.05 | 0.17±0.05 | 0.22±0.02 | 0.27±0.04 | 0.51±0.05 | 0.21±0.03§|| |
| WT+tBOOH | 2.1±0.2 | 4.6±0.6|| | 4.0±0.5 | 3.0±1¶ | 19±4||¶ | 0.44±0.05§¶ | 0.39±0.02||¶ | 0.28±0.03§ | 0.63±0.05¶ | 0.23±0.03§ |
| OE : : napA | 2.3±0.3 | 4.8±0.5 | 4.0±0.5 | 0.7±0.3** | 0.3±0.05 | 0.19±0.04§ | 0.19±0.02 | 0.26±0.03§ | 0.62±0.06** | 0.53±0.07** |
| OE : : napA+tBOOH | 2.2±0.3 | 4.8±0.5 | 4.2±0.5 | 1.0±0.3¶** | 1.1±0.2¶** | 0.18±0.05§** | 0.21±0.02** | 0.25±0.03 | 0.68±0.06 | 0.47±0.07§** |
Mean±sd values calculated from three independent experiments are presented.
DCM, dry cell mass.
RS includes all intracellular oxidants which react to give rise to 2′,7′-dichlorofluorescein from from 2′,7′-dichlorofluorescin diacetate (Halliwell & Gutteridge, 2007).
||¶#**Significant differences (P<0.05, where P values were calculated using Student’s t test) between 10 and 24 h incubations (§), caused by pyridoxine (||) supplementation or tBOOH treatment (¶) or initiated by ΔnapA (#) or OE : : napA (**), respectively.
First, we compared wild-type with ΔnapA. There were no statistical differences in biomass accumulation in these two strains, regardless of tBOOH treatment, and in both strains the addition of tBOOH elicited sterigmatocystin overproduction. Differences were observed in RS production and antioxidant enzyme activity in the ΔnapA strain as compared with wild-type with or without tBOOH addition. In general, RS production was higher in the deletion mutants, with decreases in catalase activity at early time points and higher glutathione peroxidase activity at 24 h for the tBOOH treatment.
The comparison of wild-type with OE : : napA required pyridoxine supplementation as described above. As expected, the addition of pyridoxine ameliorated the slow growth normally associated with tBOOH treatment. However, this compound still had the effect of inducing sterigmatocystin overproduction in the wild-type. Notably, sterigmatocystin levels in both treatments were statistically decreased in the OE : : napA strain compared with wild-type. Also, unlike in the wild-type, addition of tBOOH did not increase sterigmatocystin in this strain (0.7±0.3 versus 1.0±0.3). Most striking was the large decrease of RS production in the tBOOH-treated OE : : napA strain. This appeared largely independent of catalase and glutathione peroxidase activity, which showed little change compared with the wild-type.
FRET analysis of A. nidulans bZIP proteins
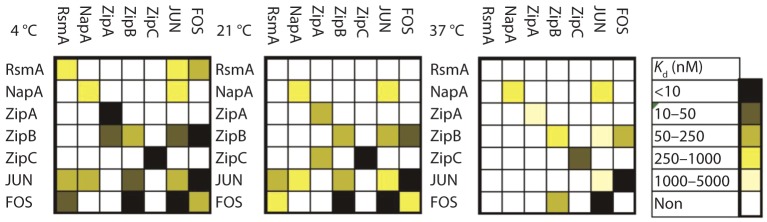
bZIP proteins function as either homo- or heterodimers, and the same bZIP can exist as a homodimer in some instances and a heterodimer in others (Vinson et al., 2006). To examine the likelihood of homodimer and heterodimer interactions of seven A. nidulans bZIP proteins, we quantified the interactions of 15 pairs of leucine-zipper dimerization domains using a solution FRET assay. We also included the human bZIP proteins FOS and JUN as controls, because they are known to form very stable heterodimers, with weaker homo-association. The assay measured protein–protein association in the absence of DNA. FRET analysis at three temperatures supported preferential homo-association of all five A. nidulans bZIP proteins, with strong self-association of ZipC. Heterodimer formation was also observed between ZipA and ZipB (at 4 °C) and ZipA and ZipC (at 21 °C, Fig. 5). Although binding to DNA is expected to stabilize all complexes, the differential protein–protein affinities that we observed may be important for competitive binding to transcriptional regulatory elements. Interestingly, several A. nidulans bZIPs were able to bind to human FOS and JUN leucine-zipper domains, emphasizing the conserved sequence and structure of this ancient protein family.
Fig. 5.
FRET analysis of A. nidulans bZIP proteins. Dissociation constants at 4, 21 and 37 °C for selected A. nidulans bZIPs and human JUN and FOS are presented as a heat map, with the strength of each interaction indicated by the scale at the right. Fluorescein-labelled proteins are in rows and TAMRA-labelled proteins are in columns.
Discussion
This study was motivated by a recent finding that a newly discovered A. nidulans Yap-like bZIP called RsmA was instrumental in regulating the production of the anti-predation metabolite sterigmatocystin (Shaaban et al., 2010; Yin et al., 2012), a compound known to show insecticidal activities towards several organisms (Gunst et al., 1982; Matasyoh et al., 2011). However, no obvious role for this protein was observed in canonical stress responses, in contrast to those reported for other Yap-like bZIPs including those of Aspergillus species (Asano et al., 2007; Hagiwara et al., 2008; Reverberi et al., 2012; Sakamoto et al., 2009). We hypothesized that Yap-like bZIPs might preferentially govern different defensive responses in A. nidulans, and thus created a series of deletion and overexpression strains of a subset of Yap-like bZIPs to examine this hypothesis.
We found that only two of the five bZIPs examined had a large effect on SM. As reported previously, OE : : rsmA increased production of sterigmatocystin and asperthecin (Yin et al., 2012). In contrast, OE : : napA produced lower amounts of shamixanthone, asperthecin, emericellin, epishamixanthone and sterigmatocystin (dependent on culture conditions, Table 3, Fig. S4) than the wild-type. NapA orthologues in A. ochraceus and A. parasiticus have been reported to be negative regulators of ochratoxin and aflatoxin in these two species, thus fitting with our observations (Reverberi et al., 2007, 2012). In solid-state cultivation, OE : : zipA also showed some increase in sterigmatocystin (Fig. 2), but the effect of this allele on sterigmatocystin was minimal under other culture conditions (Fig. S4). As noted previously (Shaaban et al., 2010), loss of rsmA resulted in some increase in sterigmatocystin as well (Fig. 2). In the previous study, it was suggested that rsmA loss may allow for another bZIP protein to activate sterigmatocystin; perhaps ZipA fulfils this role.
Several studies have reported a linkage between secondary metabolite production and sexual development. Overexpression of both rsmA and napA negatively affected sexual development but through different mechanisms. Although increases in SM are generally associated with sexual development (Bayram et al., 2008; Lee et al., 2012), the data do not always distinguish between fruiting body formation (cleistothecia in A. nidulans) and ascospore production. Here the OE : : rsmA strains produced cleistothecia but few ascospores were generated in the cleistothecia. Possibly the extreme increase in sterigmatocystin production could come at a cost to sexual spore production, and initial data from an OE : : rsmA ΔaflR strain suggest that this could be the case (our unpublished data). On the other hand, the decrease of ascospore numbers in the OE : : napA strain was associated with a shift to increased asexual growth. Whereas the cleistothecia produced in this strain produced abundant ascospores, there were simply fewer cleistothecia. The one other bZIP to show a sexual developmental phenotype, OE : : zipA, was similar to the OE : : napA strain. Considered together, our data here support the noted linkage of SM and sexual development.
An assessment of stress sensitivities in the bZIP mutants confirmed that NapA plays a major role in resistance to oxidative stress, with napA loss yielding a more sensitive oxidative stress phenotype and OE : : napA a more resistant phenotype. The effect of NapA loss has been examined in several fungi, including A. nidulans, with all studies showing this protein to be important in protection from oxidative and other stressors (Asano et al., 2007; Thön et al., 2010). However, to our knowledge, this is the first examination of OE : : napA in A. nidulans.
bZIP-type transcription factors may regulate secondary metabolite production, either directly by binding at promoters of genes encoding key elements of the biosynthetic machinery, as shown for RsmA and other bZIPs (Roze et al., 2011; Yin et al., 2012), or via modulating intracellular ROS levels through fine-tuning the activity of the antioxidative defence system, as demonstrated for NapA orthologues in A. ochraceus and A. parasiticus (Reverberi et al., 2007, 2008, 2012). This theory has also been supported by observations that the inhibition of oxidation processes including lipid peroxidation hinders biosynthesis of mycotoxins, including aflatoxin (Jayashree & Subramanyam, 1999, 2000), and the addition of lipid peroxidation-stimulating agents to fungal cultures increases the yields of certain metabolites (e.g. ochratoxin A; Reverberi et al., 2012).
To further assess this possible relationship of oxidative stress and SM, the effect of NapA on sterigmatocystin and antioxidative defence was examined in greater depth. This bZIP seems to be important in combating tBOOH-induced stress, and also in the regulation of cellular recovery processes from this stressor (Table 3). For instance, both ΔnapA and OE : : napA mutants showed aberrancies in either growth recovery or RS concentrations with the tBOOH treatment as compared with the wild-type strain; these were in part associated with changes in catalase and glutathione peroxidase activities. Thus it would seem that like ApYapA and AoYap1, A. nidulans NapA is an important element in modulating intracellular RS levels through the regulation of antioxidative enzyme activities. Although the deletion of napA did not increase sterigmatocystin yields under the conditions tested here (unlike in the cases of ApyapA and Aoyap1 loss and its effect on aflatoxin and ochratoxin), overexpression of this gene resulted in a strain unable to increase sterigmatocystin in response to tBOOH, thus supporting a ROS involvement in SM in this Aspergillus species.
Finally, it is unclear what role(s) ZipA, ZipB and ZipC play in fungal biology. As seen in Fig. 5, the FRET data did not support a high probability of heterodimer formation among these five proteins, with the possible exception of ZipC/ZipA and ZipA/ZipB. One of our future goals will be to examine double mutants (e.g. ΔzipAΔzipB) and the use of RNA interference technology to downregulate zipC for a further understanding of any effect of these proteins on Aspergillus development.
Acknowledgements
This work was supported by National Institutes for Health (NIH) National Institute of General Medical Sciences grant PO1GM084077 to N. P. K. and C. C. W., and NIH grant R01GM067681 to A. E. K. The authors are indebted to Mrs Lászlóné Gábor Tóth and Mr Imre Pócsi (University of Debrecen) for their valuable technical assistance. One of us (I. P.) was financially supported by the European Union and European Social Fund co-financed TÁMOP 4.2.1./B-09/1/KONV-2010-0007 and TÁMOP-4.2.2/B-10/1-2010-002 projects. We thank Scott Klasek for helping with physiological experiments.
Abbreviations:
- DCF
dichlorofluorescein
- FRET
fluorescence resonance energy transfer
- MSB
menadione sodium bisulphite
- ROS
reactive oxygen species
- RS
reactive species
- SM
secondary metabolism
- TAMRA
carboxytetramethylrhodamine
- tBOOH
tert-butylhydroperoxide
Footnotes
Two supplementary tables and five supplementary figures are available with the online version of this paper.
References
- Asano Y., Hagiwara D., Yamashino T., Mizuno T. (2007). Characterization of the bZip-type transcription factor NapA with reference to oxidative stress response in Aspergillus nidulans. Biosci Biotechnol Biochem 71, 1800–1803. 10.1271/bbb.70133 [DOI] [PubMed] [Google Scholar]
- Ashenberg O., Rozen-Gagnon K., Laub M. T., Keating A. E. (2011). Determinants of homodimerization specificity in histidine kinases. J Mol Biol 413, 222–235. 10.1016/j.jmb.2011.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balázs A., Pócsi I., Hamari Z., Leiter E., Emri T., Miskei M., Oláh J., Tóth V., Hegedűs N. & other authors (2010). AtfA bZIP-type transcription factor regulates oxidative and osmotic stress responses in Aspergillus nidulans. Mol Genet Genomics 283, 289–303. 10.1007/s00438-010-0513-z [DOI] [PubMed] [Google Scholar]
- Bayram O., Braus G. H. (2012). Coordination of secondary metabolism and development in fungi: the velvet family of regulatory proteins. FEMS Microbiol Rev 36, 1–24. 10.1111/j.1574-6976.2011.00285.x [DOI] [PubMed] [Google Scholar]
- Bayram O., Krappmann S., Ni M., Bok J. W., Helmstaedt K., Valerius O., Braus-Stromeyer S., Kwon N. J., Keller N. P. & other authors (2008). VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 320, 1504–1506. 10.1126/science.1155888 [DOI] [PubMed] [Google Scholar]
- Bok J. W., Keller N. P. (2004). LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot Cell 3, 527–535. 10.1128/EC.3.2.527-535.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok J. W., Keller N. P. (2012). Fast and Easy Method for Construction of Plasmid Vectors Using Modified Quick-Change Mutagenesis. Humana Press; 10.1007/978-1-62703-122-6_11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok J. W., Hoffmeister D., Maggio-Hall L. A., Murillo R., Glasner J. D., Keller N. P. (2006). Genomic mining for Aspergillus natural products. Chem Biol 13, 31–37. 10.1016/j.chembiol.2005.10.008 [DOI] [PubMed] [Google Scholar]
- Bok J. W., Chiang Y.-M., Szewczyk E., Reyes-Dominguez Y., Davidson A. D., Sanchez J. F., Lo H.-C., Watanabe K., Strauss J. & other authors (2009). Chromatin-level regulation of biosynthetic gene clusters. Nat Chem Biol 5, 462–464. 10.1038/nchembio.177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu D. T., Stults F. H., Tappel A. L. (1976). Purification and properties of rat lung soluble glutathione peroxidase. Biochim Biophys Acta 445, 558–566. 10.1016/0005-2744(76)90110-8 [DOI] [PubMed] [Google Scholar]
- Cho J. H., Yun S. S., Jang Y. K., Cha M. J., Kwon N. J., Chae S. K. (2003). Identification and cloning of jipA encoding a polypeptide that interacts with a homolog of yeast Rad6, UVSJ in Aspergillus nidulans. J Microbiol 41, 46–51. [Google Scholar]
- Eigentler A., Pócsi I., Marx F. (2012). The anisin1 gene encodes a defensin-like protein and supports the fitness of Aspergillus nidulans. Arch Microbiol 194, 427–437. 10.1016/S0891-5849(97)00065-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emri T., Pócsi I., Szentirmai A. (1997). Glutathione metabolism and protection against oxidative stress caused by peroxides in Penicillium chrysogenum. Free Radic Biol Med 23, 809–814. 10.1016/S0891-5849(97)00065-8 [DOI] [PubMed] [Google Scholar]
- Emri T., Pócsi I., Szentirmai A. (1999). Analysis of the oxidative stress response of Penicillium chrysogenum to menadione. Free Radic Res 30, 125–132. 10.1080/10715769900300131 [DOI] [PubMed] [Google Scholar]
- Ferguson H. A., Goodrich J. A. (2001). Expression and purification of recombinant human c-Fos/c-Jun that is highly active in DNA binding and transcriptional activation in vitro. Nucleic Acids Res 29, e98. 10.1093/nar/29.20.e98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunst K., Chinnici J. P., Llewellyn G. C. (1982). Effects of aflatoxin B, aflatoxin B, aflatoxin G and sterigmatocystin on viability, rates of development, and body length in two strains of Drosophila melanogaster (Diptera). J Invertebr Pathol 39, 388–394. 10.1016/0022-2011(82)90064-7 [DOI] [PubMed] [Google Scholar]
- Guo M., Chen Y., Du Y., Dong Y., Guo W., Zhai S., Zhang H., Dong S., Zhang Z. & other authors (2011). The bZIP transcription factor MoAP1 mediates the oxidative stress response and is critical for pathogenicity of the rice blast fungus Magnaporthe oryzae. PLoS Pathog 7, e1001302. 10.1371/journal.ppat.1001302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara D., Asano Y., Marui J., Furukawa K., Kanamaru K., Kato M., Abe K., Kobayashi T., Yamashino T., Mizuno T. (2007). The SskA and SrrA response regulators are implicated in oxidative stress responses of hyphae and asexual spores in the phosphorelay signaling network of Aspergillus nidulans. Biosci Biotechnol Biochem 71, 1003–1014. 10.1271/bbb.60665 [DOI] [PubMed] [Google Scholar]
- Hagiwara D., Asano Y., Yamashino T., Mizuno T. (2008). Characterization of bZip-type transcription factor AtfA with reference to stress responses of conidia of Aspergillus nidulans. Biosci Biotechnol Biochem 72, 2756–2760. 10.1271/bbb.80001 [DOI] [PubMed] [Google Scholar]
- Halliwell B., Gutteridge J. M. C. (2007). Chapter 5. Measurement of Reactive Species. In Free Radicals in Biology and Medicine, 4th edn, pp. 268–330. Oxford: Oxford University Press. [Google Scholar]
- Jayashree T., Subramanyam C. (1999). Antiaflatoxigenic activity of eugenol is due to inhibition of lipid peroxidation. Lett Appl Microbiol 28, 179–183. 10.1046/j.1365-2672.1999.00512.x [DOI] [PubMed] [Google Scholar]
- Jayashree T., Subramanyam C. (2000). Oxidative stress as a prerequisite for aflatoxin production by Aspergillus parasiticus. Free Radic Biol Med 29, 981–985. 10.1016/S0891-5849(00)00398-1 [DOI] [PubMed] [Google Scholar]
- Klich M., Mendoza C., Mullaney E., Keller N., Bennett J. W. (2001). A new sterigmatocystin-producing Emericella variant from agricultural desert soils. Syst Appl Microbiol 24, 131–138. 10.1078/0723-2020-00007 [DOI] [PubMed] [Google Scholar]
- Lee J., Myong K., Kim J. E., Kim H. K., Yun S. H., Lee Y. W. (2012). FgVelB globally regulates sexual reproduction, mycotoxin production and pathogenicity in the cereal pathogen Fusarium graminearum. Microbiology 158, 1723–1733. 10.1099/mic.0.059188-0 [DOI] [PubMed] [Google Scholar]
- Lessing F., Kniemeyer O., Wozniok I., Loeffler J., Kurzai O., Haertl A., Brakhage A. A. (2007). The Aspergillus fumigatus transcriptional regulator AfYap1 represents the major regulator for defense against reactive oxygen intermediates but is dispensable for pathogenicity in an intranasal mouse infection model. Eukaryot Cell 6, 2290–2302. 10.1128/EC.00267-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M. Z., Elledge S. J. (2007). Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat Methods 4, 251–256. 10.1038/nmeth1010 [DOI] [PubMed] [Google Scholar]
- Matasyoh J. C., Dittrich B., Schueffler A., Laatsch H. (2011). Larvicidal activity of metabolites from the endophytic Podospora sp. against the malaria vector Anopheles gambiae. Parasitol Res 108, 561–566. 10.1007/s00436-010-2098-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moye-Rowley W. S., Harshman K. D., Parker C. S. (1989). Yeast YAP1 encodes a novel form of the jun family of transcriptional activator proteins. Genes Dev 3, 283–292. 10.1101/gad.3.3.283 [DOI] [PubMed] [Google Scholar]
- Pontecorvo G., Roper J. A., Chemmons L. M., MacDonald K. D., Bufton A. W. (1953). The genetics of Aspergillus nidulans. Adv Genet 5, 141–238. 10.1016/S0065-2660(08)60408-3 [DOI] [PubMed] [Google Scholar]
- Qiao J., Kontoyiannis D. P., Calderone R., Li D., Ma Y., Wan Z., Li R., Liu W. (2008). Afyap1, encoding a bZip transcriptional factor of Aspergillus fumigatus, contributes to oxidative stress response but is not essential to the virulence of this pathogen in mice immunosuppressed by cyclopthosphamide and triamcinolone. Med Mycol 46, 773–782. 10.1080/13693780802054215 [DOI] [PubMed] [Google Scholar]
- Reverberi M., Zjalic S., Punelli F., Ricelli A., Fabbri A. A., Fanelli C. (2007). Apyap1 affects aflatoxin biosynthesis during Aspergillus parasiticus growth in maize seeds. Food Addit Contam 24, 1070–1075. 10.1080/02652030701553244 [DOI] [PubMed] [Google Scholar]
- Reverberi M., Zjalic S., Ricelli A., Punelli F., Camera E., Fabbri C., Picardo M., Fanelli C., Fabbri A. A. (2008). Modulation of antioxidant defense in Aspergillus parasiticus is involved in aflatoxin biosynthesis: a role for the ApyapA gene. Eukaryot Cell 7, 988–1000. 10.1128/EC.00228-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverberi M., Gazzetti K., Punelli F., Scarpari M., Zjalic S., Ricelli A., Fabbri A. A., Fanelli C. (2012). Aoyap1 regulates OTA synthesis by controlling cell redox balance in Aspergillus ochraceus. Appl Microbiol Biotechnol 95, 1293–1304. 10.1007/s00253-012-3985-4 [DOI] [PubMed] [Google Scholar]
- Rodrigues-Pousada C., Menezes R. A., Pimentel C. (2010). The Yap family and its role in stress response. Yeast 27, 245–258. 10.1002/yea.1752 [DOI] [PubMed] [Google Scholar]
- Roggenkamp R., Sahm H., Wagner F. (1974). Microbial assimilation of methanol induction and function of catalase in Candida boidinii. FEBS Lett 41, 283–286. 10.1016/0014-5793(74)81230-5 [DOI] [PubMed] [Google Scholar]
- Roze L. V., Chanda A., Wee J., Awad D., Linz J. E. (2011). Stress-related transcription factor AtfB integrates secondary metabolism with oxidative stress response in aspergilli. J Biol Chem 286, 35137–35148. 10.1074/jbc.M111.253468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K., Iwashita K., Yamada O., Kobayashi K., Mizuno A., Akita O., Mikami S., Shimoi H., Gomi K. (2009). Aspergillus oryzae atfA controls conidial germination and stress tolerance. Fungal Genet Biol 46, 887–897. 10.1016/j.fgb.2009.09.004 [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T. (1989). Molecular Cloning: a Laboratory Manual, 2nd edn Cold Spring Harbor, NY: Cold Spring Harbor Laboratory. [Google Scholar]
- Shaaban M. I., Bok J. W., Lauer C., Keller N. P. (2010). Suppressor mutagenesis identifies a velvet complex remediator of Aspergillus nidulans secondary metabolism. Eukaryot Cell 9, 1816–1824. 10.1128/EC.00189-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K., Keller N. P. (2001). Genetic involvement of a cAMP-dependent protein kinase in a G protein signaling pathway regulating morphological and chemical transitions in Aspergillus nidulans. Genetics 157, 591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shwab E. K., Bok J. W., Tribus M., Galehr J., Graessle S., Keller N. P. (2007). Histone deacetylase activity regulates chemical diversity in Aspergillus. Eukaryot Cell 6, 1656–1664. 10.1128/EC.00186-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thön M., Al Abdallah Q., Hortschansky P., Scharf D. H., Eisendle M., Haas H., Brakhage A. A. (2010). The CCAAT-binding complex coordinates the oxidative stress response in eukaryotes. Nucleic Acids Res 38, 1098–1113. 10.1093/nar/gkp1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C., Li J., Glass N. L. (2011). Exploring the bZIP transcription factor regulatory network in Neurospora crassa. Microbiology 157, 747–759. 10.1099/mic.0.045468-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsitsigiannis D. I., Kowieski T. M., Zarnowski R., Keller N. P. (2004a). Endogenous lipogenic regulators of spore balance in Aspergillus nidulans. Eukaryot Cell 3, 1398–1411. 10.1128/EC.3.6.1398-1411.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsitsigiannis D. I., Zarnowski R., Keller N. P. (2004b). The lipid body protein, PpoA, coordinates sexual and asexual sporulation in Aspergillus nidulans. J Biol Chem 279, 11344–11353. 10.1074/jbc.M310840200 [DOI] [PubMed] [Google Scholar]
- Vinson C., Acharya A., Taparowsky E. J. (2006). Deciphering B-ZIP transcription factor interactions in vitro and in vivo. Biochim Biophys Acta 1759, 4–12. 10.1016/j.bbaexp.2005.12.005 [DOI] [PubMed] [Google Scholar]
- Yin W. B., Amaike S., Wohlbach D. J., Gasch A. P., Chiang Y. M., Wang C. C., Bok J. W., Rohlfs M., Keller N. P. (2012). An Aspergillus nidulans bZIP response pathway hardwired for defensive secondary metabolism operates through aflR. Mol Microbiol 83, 1024–1034. 10.1111/j.1365-2958.2012.07986.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J. H., Hamari Z., Han K. H., Seo J. A., Reyes-Domínguez Y., Scazzocchio C. (2004). Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet Biol 41, 973–981. 10.1016/j.fgb.2004.08.001 [DOI] [PubMed] [Google Scholar]