Abstract
Viral protein synthesis is completely dependent upon the translational machinery of the host cell. However, many RNA virus transcripts have marked structural differences from cellular mRNAs that preclude canonical translation initiation, such as the absence of a 5′ cap structure or the presence of highly structured 5′UTRs containing replication and/or packaging signals. Furthermore, whilst the great majority of cellular mRNAs are apparently monocistronic, RNA viruses must often express multiple proteins from their mRNAs. In addition, RNA viruses have very compact genomes and are under intense selective pressure to optimize usage of the available sequence space. Together, these features have driven the evolution of a plethora of non-canonical translational mechanisms in RNA viruses that help them to meet these challenges. Here, we review the mechanisms utilized by RNA viruses of eukaryotes, focusing on internal ribosome entry, leaky scanning, non-AUG initiation, ribosome shunting, reinitiation, ribosomal frameshifting and stop-codon readthrough. The review will highlight recently discovered examples of unusual translational strategies, besides revisiting some classical cases.
Introduction
No virus encodes its own ribosome. Indeed, it has been proposed that the distinction between cellular life and the virus world could be based simply on whether an entity encodes ribosomes or a capsid (Raoult & Forterre, 2008). Nevertheless, whilst viruses seem to be almost entirely dependent upon their hosts for the provision of components of the translational machinery, they have evolved a profusion of non-canonical mechanisms to allow translation to be customized to their specific needs. Indeed, in RNA viruses in particular, non-canonical translation seems to be more the rule than the exception, with some individual viruses employing several different mechanisms. Here, we review the different types of non-canonical translational mechanisms utilized by viruses of eukaryotes, focusing on RNA viruses, but including also examples from retro-transcribing viruses. The focus is on the different translational strategies that RNA viruses employ for accessing multiple ORFs in mRNAs. Such strategies include internal ribosome entry, leaky scanning, non-AUG initiation, ribosome shunting, reinitiation, ribosomal frameshifting and stop-codon readthrough (summarized in Fig. 1). Although several excellent reviews have been written previously on similar topics, the recent explosion in the pace of sequencing has seen many interesting new examples of non-canonical translation come to light in just the past few years. This review will discuss some of these recently discovered examples, besides revisiting some classical cases.
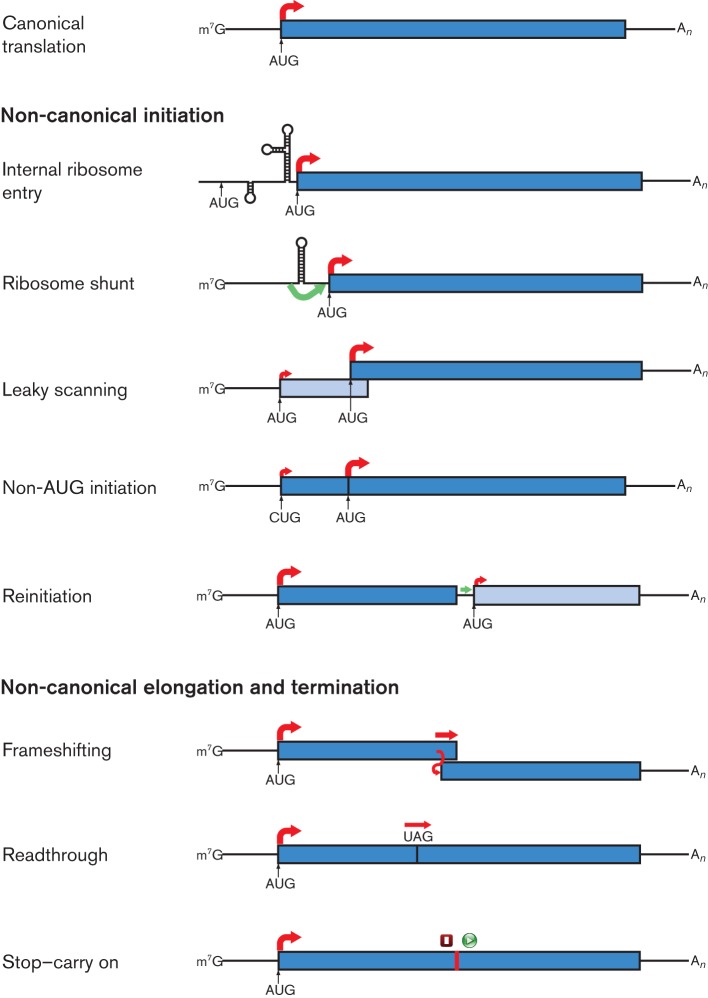
Fig. 1.
Examples of non-canonical translational mechanisms utilized by RNA viruses. Canonical eukaryotic mRNA translation is shown in the top panel. Red arrows indicate initiation of protein synthesis (at the start of an ORF) or continuation of translation by 80S ribosomes, with thicker arrows indicating the predominant path taken by ribosomes (not to scale). Green arrows indicate the probable movement of 40S subunits in a non-canonical manner. Where two distinct polypeptides are synthesized, the ORFs are shown in different shades of blue; where a recoding event during elongation leads to C-terminal extension of a polypeptide, the two ORFs are shown in the same colour. In the stop–carry on mechanism, both termination and initiation steps are non-canonical, as indicated by the red square and green circle.
Canonical translation in eukaryotes
In order to appreciate alternative translational mechanisms, it is instructive to review first the standard course of events in translation of eukaryotic cellular mRNAs, the vast majority of which bear a 5′ cap structure (m7G) and a 3′ poly(A) tail. Translation can be divided into four stages: initiation, elongation, termination and ribosome recycling. The initiation step (reviewed by Jackson et al., 2010) begins upon recognition of the 5′ cap structure by initiation factor (eIF) 4E, which recruits eIF4G, a scaffold protein, which in turn engages eIF4A and eIF4B and also the poly(A)-binding protein (PABP). PABP is actually bound at the poly(A) tail at the 3′ end of the mRNA, and its interaction with eIF4G leads to mRNA circularization (see below). The resulting complex of initiation factors recruits the 43S preinitiation complex, comprising the 40S subunit of the ribosome together with eIF3, eIF1, eIF1A, eIF5 and the ternary complex eIF2–Met–tRNAi–GTP. This is thought to be largely via interaction between eIF4G and eIF3. Following recruitment to a 5′-proximal position, the preinitiation complex scans along the mRNA until it encounters the first AUG codon. Scanning is assisted by the helicase eIF4A and its cofactor eIF4B, which unwind RNA secondary structures in the 5′UTR of the mRNA. The initiation factors eIF1 and eIF1A are key to the recognition of the AUG codon and its surrounding nucleotide context. Once an AUG codon is recognized, eIF5 triggers hydrolysis of eIF2-bound GTP, the 40S subunit locks into a closed conformation with Met–tRNAi in the ribosomal P-site, and eIF1, eIF2–GDP and eIF5 are released. Then, eIF5B and GTP catalyse joining of the 60S subunit of the ribosome to form the 80S ribosome and release of eIF5B and eIF1A. Translation proceeds to the elongation stage, and eIF3 and its associated eIF4G are thought to be released shortly thereafter.
During elongation, consecutive triplet codons are recognized in the ribosomal A-site by cognate aminoacylated tRNAs delivered to the A-site by eukaryotic elongation factor eEF1A. As each codon is recognized in the A-site, a peptide bond is formed, transferring the nascent polypeptide chain from the P-site tRNA to the A-site tRNA. Translocation, catalysed by eEF2, passes the P-site deacylated tRNA to the E-site (where it is released from the ribosome) and the A-site peptidyl–tRNA to the P-site, thus also moving the mRNA through the ribosome and opening the A-site for the next round of elongation. Elongation continues until a termination codon (UAG, UGA or UAA in the standard genetic code) enters the A-site. These codons are recognized by eukaryotic release factor eRF1, which, together with eRF3 and GTP, mediates termination of translation and release of the newly synthesized protein via hydrolysis of the P-site peptidyl–tRNA (reviewed by Jackson et al., 2012). Post-termination ribosomes are separated into component 40S and 60S subunits by ATP and ABCE1 [a member of the ATP-binding cassette (ABC) family of proteins]. Release of the P-site deacylated tRNA and of the 40S subunit from the mRNA is mediated by eIF3, eIF1 and eIF1A, which are then thought to remain associated with the 40S subunit. The mRNA circularization described earlier is believed to enhance translation, possibly by aiding ribosome recycling and/or by tethering initiation factors to the message (Wells et al., 1998; Jackson et al., 2010).
Why is non-canonical translation so abundant in viruses?
One of the major challenges facing RNA viruses is the 5′-end dependence of canonical eukaryotic translation initiation, which generally permits the synthesis of only a single protein from a given mRNA. RNA viruses must generally express multiple structural and enzymic proteins to complete their replicative cycle, and they have evolved a variety of strategies to meet this requirement. Some are compatible with 5′-end-dependent translation; for example, the production of functionally monocistronic subgenomic RNAs (sgRNAs; e.g. coronaviruses and closteroviruses) or the use of segmented genomes where most segments are monocistronic (e.g. reoviruses and orthomyxoviruses). Another common strategy is to encode long polyproteins that are subsequently processed by virus-encoded or host proteases to generate the viral proteome (e.g. picornaviruses and flaviviruses). However, the use of these mechanisms has consequences. Viruses with segmented genomes have to ensure the correct packaging of the different segments, or must be able to tolerate the reduction in specific infectivity if segments are packaged randomly or individually into separate virions. Polyprotein expression can be considered to be an inefficient way of exploiting the available resources of the host cell as the mature virus proteins are produced in equal amounts, even though the enzymic proteins are often required in much smaller quantities than the structural proteins. Moreover, while some viral proteins may be expressed from sgRNAs, in probably all positive-strand RNA viruses the components of the replication complex must still be translated from the genomic RNA. Non-canonical translational mechanisms provide a number of alternative ways to express multiple proteins from a single mRNA.
RNA viruses also have very compact genomes, with the largest around 30 kb (e.g. some members of the families Coronaviridae and Reoviridae). Thus there is strong selective pressure to optimize their coding capacity, for example via the utilization of overlapping ORFs. Non-canonical translational strategies such as leaky scanning, ribosomal frameshifting and alternative initiation are essential in facilitating access to such ORFs. Non-canonical translational mechanisms may also help to overcome the challenges imposed by the marked structural differences present in many viral transcripts in comparison with typical cellular mRNAs. For example, packaging and/or replication signals within the 5′UTR of the genomic RNA or RNA segments can inhibit scanning-dependent translation initiation. Ribosome shunting or internal ribosome entry sites (IRESes) can be employed to circumvent such impediments. Non-canonical translational mechanisms may also play roles in regulating the expression level and/or timing of expression of various proteins.
It should be noted that many RNA viruses lack the machinery to add a 5′ cap and/or poly(A) tail to their transcripts and have evolved alternative mechanisms for ribosome recruitment and/or mRNA circularization. Similarly, several viruses have evolved proteins and or RNA structural elements that further enhance translation of the viral mRNAs. Due to space limitations, such mechanisms will mostly not be discussed here. Many excellent reviews covering these topics, besides some cautionary critiques, are available (Kozak, 2004; Dreher & Miller, 2006; Kneller et al., 2006; Edgil & Harris, 2006; Miller & White, 2006; Kozak, 2007; Miller et al., 2007; Thiébeauld et al., 2007; Nicholson & White, 2011; Walsh & Mohr, 2011).
Non-canonical initiation
Internal ribosome entry
IRESes are highly organized, complex RNA structures that recruit ribosomes to internal positions on mRNAs (reviewed by Kieft, 2008; Balvay et al., 2009). In viruses, they are often employed as a way to facilitate translation initiation whilst allowing replication elements and/or packaging signals to be accommodated within the 5′UTR. They may also function to allow translation of viral mRNAs to continue, even when host-cell translation is inhibited, for example by viral protease cleavage of initiation factors required for 5′-cap-dependent translation. IRESes can also be used to access internal ORFs that would otherwise be inaccessible. Viral IRESes vary in both the degree of dependence on initiation factors and the precision with which the initiation site is selected. In picornaviruses, where IRESes were first described (Jang et al., 1988; Pelletier & Sonenberg, 1988), two major classes (types I and II) have been identified that are distinct in structure and sequence, but typically require most of the canonical initiation factors for activity, including eIF3, eIF4A and the C-terminal domain of eIF4G, besides the eIF2–Met–tRNAi–GTP ternary complex (reviewed by Belsham, 2009; other types of picornavirus IRES – such as those found in Aichi virus and hepatitis A virus – will not be discussed here). In those picornaviruses harbouring type I IRESes (poliovirus and other enteroviruses), the initiator AUG for translation of the viral polyprotein is located some distance downstream of the site of recruitment of the 40S subunit to the IRES, and some form of scanning is required to locate it. In type II IRESes, found in cardioviruses such as Theiler’s murine encephalomyelitis virus and aphthoviruses such as foot-and-mouth disease virus, the initiator AUG is close to the ribosome entry point and little, if any, scanning is required. In contrast, the IRES of hepatitis C hepacivirus (family Flaviviridae) requires fewer initiation factors (eIF3, eIF5 and the eIF2–Met–tRNAi–GTP ternary complex), recruits 40S subunits directly, and places the initiator AUG into the ribosomal P-site without any requirement for scanning (reviewed by Lukavsky, 2009). Similar IRESes are also found in some pestiviruses and teschoviruses. Yet other types of IRESes have been described in human immunodeficiency virus (HIV)-1, HIV-2 and other retroviruses (reviewed by Chamond et al., 2010), dicistroviruses (see below) and various other viruses. On a related note, under conditions of eIF2 phosphorylation (which results in global inhibition of translation, often in response to virus infection), initiation may proceed via an eIF2-independent route that involves the cellular protein ligatin, provided there exist structures within the mRNA that position the initiation codon directly in the ribosomal P-site (Skabkin et al., 2010). This mechanism has been demonstrated in the sgRNA of Sindbis alphavirus, and can also occur on the hepatitis C virus IRES.
As the type I and type II picornavirus IRESes do not necessarily place the ribosome directly onto a specific initiation codon, they have the potential to direct initiation at more than one site on the mRNA. One example where a second initiation site is utilized functionally occurs in Theiler’s murine encephalomyelitis virus, where a 156-codon ORF overlaps the 5′ end of the polyprotein ORF in the +1 reading frame (Kong & Roos, 1991; van Eyll & Michiels, 2002). The ORF, which encodes the L* protein, is translated from an AUG codon positioned 13 nt 3′ of the polyprotein AUG initiation codon. In neurovirulent strains, the L* AUG codon is replaced with an ACG codon, but some level of translation still occurs (van Eyll & Michiels, 2002). It has been suggested that translation of L* may be facilitated by the IRES placing a proportion of scanning-competent ribosomes 3′ of the polyprotein initiation codon or otherwise promoting leaky scanning beyond the polyprotein AUG, although the exact mechanism(s) are unknown (van Eyll & Michiels, 2002). A similarly positioned but otherwise unrelated overlapping coding sequence (with an upstream ACG initiator) appears likely to be present in turdivirus 3 (family Picornaviridae, genus Paraturdivirus). On the other hand, in foot-and-mouth disease virus, two in-frame AUG codons are used as alternative initiation sites to produce different isoforms (Lab and Lb) of the leader protease (reviewed by Belsham, 2005).
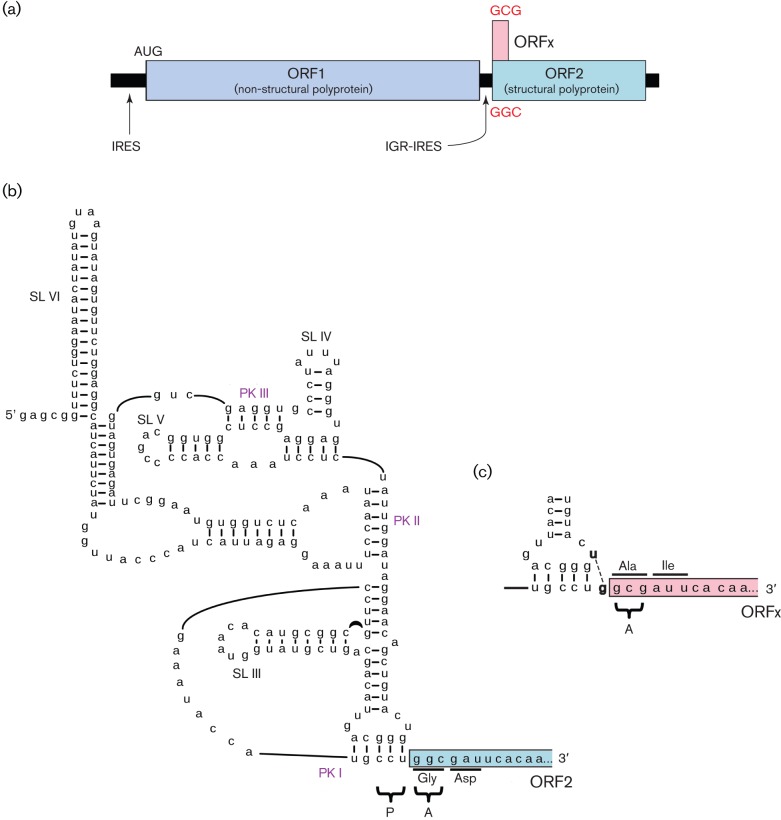
Another type of IRES has been described in the genomes of members of the family Dicistroviridae – a family of positive-stranded monopartite viruses that infect arthropods. Unusually, dicistroviruses have two non-overlapping coding sequences where translation of each is directed by a distinct IRES. The intergenic region IRES (IGR-IRES) that directs translation of the 3′ ORF encoding structural proteins is very unusual. It is short (typically around 180 nt) in comparison to picornavirus IRESes (typically around 450 nt) and compact, but is folded elegantly by virtue of RNA pseudoknotting into a structure that can partly mimic E- and P-site tRNAs, including the P-site codon : anticodon duplex (Fig. 2). The IGR-IRES binds to ribosome subunits and assembles translationally competent 80S ribosomes, which remarkably can initiate translation on a non-AUG codon in the A-site (in contrast to the P-site in conventional initiation) without any requirement for Met–tRNAi or any of the canonical initiation factors (Wilson et al., 2000; Jang et al., 2009). Precise placement of the ribosome on the mRNA and the absence of initiation factors would seem to preclude any form of leaky scanning in this case. Nonetheless, members of at least one group of dicistroviruses (Israeli acute bee paralysis and related viruses) express an additional protein from a short ORF that overlaps the 5′-proximal region of the structural polyprotein ORF in the +1 reading frame (Ren et al., 2012). Translation of this ORF appears to be directed by an extra base-pairing interaction in the P-site anticodon : codon-mimicking duplex of the IGR-IRES that facilitates a proportion of incoming A-site tRNAs to pair not to the structural polyprotein initiation codon, but instead to the codon offset by +1 nt (Fig. 2; Ren et al., 2012). Besides dicistroviruses, a number of other viruses appear to employ the strategy of expressing two polyproteins from separate IRESes. Canine picodicistrovirus appears to have two picornavirus-like IRESes, with the 3′ ORF encoding the RNA-dependent RNA polymerase (RdRp) and other non-structural proteins (Woo et al., 2012), while several other unclassified dicistronic positive-strand RNA viruses may have a dicistrovirus-like IGR-IRES, although the details have not yet been determined and alternative translation strategies have not been definitively ruled out (Boros et al., 2011).
Fig. 2.
The Israeli acute paralysis dicistrovirus IGR-IRES directs translation of two overlapping ORFs. (a) Genome map. Distinct IRESes direct translation of non-structural and structural polyproteins. The IGR-IRES also directs translation of ORFx, which overlaps ORF2 in the +1 reading frame. (b) Schematic of the IGR-IRES, showing pseudoknots (PK) I, II and III and stem–loops (SL) III, IV, V and VI. PK I occupies the ribosomal P-site and translation of ORF2 initiates at the GGC codon in the ribosomal A-site. (c) The formation of an additional base pair in PK I (U-G; bold) leads instead to initiation at the +1 frame GCG codon and translation of ORFx. Modified from a figure kindly provided by E. Jan (Ren et al., 2012).
Hepatitis C virus also has an ORF named core+1 (alternatively F or ARF) that overlaps the 5′-proximal region of the polyprotein ORF (Walewski et al., 2001; reviewed by Vassilaki & Mavromara, 2009). There is little doubt that the core+1 ORF is expressed in vivo, at least at some level, as infected patients develop antibodies against core+1 peptides (Walewski et al., 2001). Many different mechanisms have been proposed to account for expression of the core+1 ORF, including ribosomal frameshifting, transcriptional slippage, and independent initiation at either AUG or non-AUG codons. However, which, if any, of these mechanisms are utilized by the virus, and whether the products provide any functional benefit for the virus, remains unclear despite extensive research.
Leaky scanning
In the scanning model of initiation, 40S ribosomal subunits bind close to the 5′ cap and scan linearly until they encounter the first AUG codon (Kozak, 2002). However, in some circumstances, a significant proportion of scanning ribosomes fail to initiate at the first AUG codon but, instead, continue scanning until they reach an alternative initiation codon further downstream (reviewed by Kozak, 2002). This process is termed leaky scanning and may allow the expression of multiple C-terminally coincident isoforms of a single protein (in-frame alternative initiation sites), distinct proteins encoded by different overlapping ORFs (alternative initiation sites in different reading frames) or even distinct proteins encoded by non-overlapping sequential ORFs. The distance scanned can be considerable – for example, in rice tungro bacilliform tungrovirus, ribosomes scan through a region of around 895 nt to translate the third of three consecutive ORFs (Fütterer et al., 1997). Several other cases have been identified where three or even four distinct proteins are translated from a single transcript, often involving initiation at non-AUG codons (see below), besides AUG codons with poor context (Turina et al., 2000; Castaño et al., 2009). Leaky scanning is perhaps the mechanism most commonly used by RNA viruses to translate functionally multicistronic messages (Table 1). The efficiency of initiation at a potential initiation codon is modulated by its context, i.e. the identity of the nucleotides immediately preceding and immediately following the initiation codon. In mammals, the optimal context includes a G at +4 and a purine at −3 (the initiation codon itself corresponds to nucleotides +1 to +3), where the presence of an A at −3 is the strongest single indicator of efficient initiation (Kozak, 1986). Contexts with an A at −3, or a G at −3 and a G at +4, may be regarded as ‘strong’. Although the optimal context appears to vary between taxa, preference for an A (or G) at the −3 position is widespread in many animals, plants and fungi (Nakagawa et al., 2008). When the context of the first AUG on the message is suboptimal, there is potential for efficient leaky scanning.
Table 1. Examples of known and suspected cases of leaky scanning.
See also Table 2 for cases that involve non-AUG initiation. Full details of references in Tables 1–5 are available from the authors on request.
| Taxon | Products | References |
| Umbravirus | ORF3/ORF4 | Taliansky & Robinson (2003) |
| Luteovirus | CP/MP | Dinesh-Kumar & Miller (1993) |
| Polerovirus | CP/MP | Tacke et al. (1990) |
| Polerovirus | P0/P1 | Mayo et al. (1989) |
| Sobemovirus | P1/P2a | Sivakumaran & Hacker (1998) |
| Tombusvirus | MP/p20 | Johnston & Rochon (1996) |
| Aureusvirus | MP/p14 | Rubino & Russo (1997) |
| Pelarspovirus | MP1/MP2/CP | Castaño et al. (2009) |
| Panicovirus | MP1/MP2/CP/p15 | Turina et al. (2000) |
| Machlomovirus | MP1/MP2/CP | Scheets (2000) |
| Machlomovirus | p32/replicase | Nutter et al. (1989) |
| Tymovirus | p69/replicase | Weiland & Dreher (1989) |
| Potexvirus | TGB2/TGB3 | Verchot et al. (1998) |
| Hordeivirus RNA2 | TGB2/TGB3 | Zhou & Jackson (1996) |
| Pecluvirus RNA2 | CP/p39 | Herzog et al. (1995) |
| Arterivirus | ORF5a/GP5 | Firth et al. (2011) |
| Betacoronavirus (some species) | N/I | Senanayake et al. (1992) |
| Hepevirus | ORF3/CP | Graff et al. (2006) |
| Caliciviridae – murine norovirus, neboviruses, some sapoviruses | VP1/ORF4 | Simmonds et al. (2008); McFadden et al. (2011) |
| Omegatetravirus RNA2 | p17/CP | Hanzlik et al. (1995) |
| Mammalian orthoreovirus, segment S1 | σ1/σ1s | Ernst & Shatkin (1985) |
| Avian orthoreovirus and Nelson Bay orthoreovirus, segment S1 | p10/p17 | Racine et al. (2007) |
| Rotavirus A, segment 11 | NSP5/NSP6 | Mattion et al. (1991) |
| Respirovirus | P/C | Giorgi et al. (1983) |
| Morbillivirus | P/C | Bellini et al. (1985) |
| Henipavirus | P/C | Lo et al. (2009) |
| Orthobunyavirus (some species) | N/NSs | Fuller et al. (1983) |
| Hantavirus (some species) | N/NSs | Vera-Otarola et al. (2012) |
| Orthomyxoviridae – influenza virus B | NB/NA | Williams & Lamb (1989) |
| Orthomyxoviridae – influenza virus A | PB1/PB1-F2/N40 | Chen et al. (2001); Wise et al. (2009) |
| Isavirus, segment 8 | P6/P7 | Rimstad & Mjaaland (2002) |
| Tungrovirus | ORF I/ORF II/ORF III | Fütterer et al. (1997) |
| Badnavirus | ORF I/ORF II/ORF III | Pooggin et al. (1999) |
One of the earliest examples described is found in segment S1 of mammalian orthoreovirus (Ernst & Shatkin, 1985). Here, the coding sequence for a 14 kDa non-structural protein, σ1s, lies entirely within the coding sequence for the 49 kDa attachment protein, σ1. The σ1 AUG initiation codon has a suboptimal context (cggAUGg) and the σ1s AUG codon is 58 nt 3′ in the +1 reading frame. Another early example of leaky scanning occurs in the small segment of the orthobunyaviruses. Here, the NSs protein is translated from an ORF of around 100 codons that overlaps the 5′-terminal region of the N (nucleocapsid) protein ORF (Fuller et al., 1983). A similar N/NSs arrangement is also present in hantaviruses (Vera-Otarola et al., 2012). Such short, overlapping ORFs, often evolved relatively late, have a more limited phylogenetic distribution than the ancestral ORFs that they overlap, and the encoded proteins tend to have ancillary functions (Rancurel et al., 2009).
Besides suboptimal context surrounding the first AUG codon on a message, leaky scanning may also be promoted by a number of other mechanisms. If an AUG codon is very close to the 5′ end of the transcript, then it is often not recognized efficiently, with the efficiency diminishing as the 5′UTR length decreases below 30 nt, and particularly below 12 nt (Sedman et al., 1990; Kozak, 1991). In murine norovirus (family Caliciviridae), translation of an ORF overlapping the capsid coding sequence appears to initiate at the third AUG codon on the sgRNA (underlined in gugaAUGaggAUGagugAUGg; McFadden et al., 2011) despite the presence of two upstream AUG codons, the second of which is in a good context with an A at −3. It seems likely that, in this case, the shortness of the 5′ leader (4 or 10 nt) promotes leaky scanning past the first two AUG codons (which, nonetheless, must also be utilized efficiently for expressing the capsid protein). It should be noted that calicivirus RNAs lack a 5′ cap and instead possess a viral protein (VPg) linked covalently to the 5′ end. The VPg interacts with eIF4E and other initiation factors but, in murine norovirus, the role of these interactions in translation remains uncertain (Daughenbaugh et al., 2006; Chaudhry et al., 2006). It is possible that the presence of VPg facilitates 40S binding on a short leader.
The close proximity of a downstream AUG codon to a preceding AUG codon (e.g. within approx. 10 nt) can also increase the efficiency of leaky scanning. This has been demonstrated in both tymoviruses and in segment 6 of influenzavirus B (4 nt separation; Williams & Lamb, 1989; Matsuda & Dreher, 2006), and may also be relevant to murine norovirus (see above). The data suggest that scanning may involve alternating forward thrusts and backwards relaxations so that downstream AUG codons can sometimes capture a proportion of scanning ribosomes that might otherwise scan back to a slightly upstream AUG codon. [Conversely, initiation at a downstream AUG codon may stimulate initiation at an upstream AUG codon via a mechanism which is thought to depend on a scanning 40S subunit stacking up behind a ribosome initiating at the downstream AUG codon in an appropriate position for initiation at the upstream AUG codon (Dinesh-Kumar & Miller, 1993).]
Leaky scanning can also be promoted by short upstream ORFs. Ribosomes that translate a short ORF have the capacity to resume scanning and reinitiate on a downstream ORF, but it can take time for such ribosomes to reacquire the relevant initiation factors, and some intervening AUG codons may be efficiently bypassed (see also section entitled Reinitiation). Indeed, there are a number of cases of leaky scanning – including the PB1-F2 protein of influenza virus A – where there are one or more intervening AUG codons between the initiation codons of the ORFs that encode functional products (Chen et al., 2001; Wise et al., 2009; Racine & Duncan, 2010). Such AUG codons would be expected to ‘soak up’ a proportion of scanning ribosomes – depending on the strength of their contexts – but, provided the ORFs are short, these ribosomes may still be able to reinitiate on the major downstream ORF(s). Thus, some cases of leaky scanning probably also include an element of reinitiation and possibly also a degree of shunting or non-linear scanning (Racine & Duncan, 2010; see section on ribosome shunting). RNA structure in the scanned region, besides the location, length and amino acid composition of short intervening ORFs, may all influence the proportion of ribosomes that eventually reach the major downstream ORF(s).
Non-AUG initiation
Eukaryotic protein synthesis begins almost exclusively (but see discussion of the dicistrovirus IGR-IRES above, and see also Skabkin et al., 2010) with methionine, brought to the ribosome by Met–tRNAi, a tRNA that differs from the standard (elongation) Met–tRNA. However, initiation does not necessarily have to occur at an AUG codon. Near-cognate codons, such as CUG and ACG, can under certain circumstances also be recognized by Met–tRNAi. Initiation at a non-AUG codon normally requires a strong context (e.g. an A or G at −3 and a G at +4) and is enhanced when an RNA structure (e.g. a stem–loop) is able to form at a distance of approximately 14 nt 3′ of the initiation codon, so that it is positioned at the entrance of the mRNA channel when the potential initiation codon is in the P-site of the ribosome (Kozak, 1990; see also Clyde & Harris, 2006). The codons CUG, GUG, ACG, AUU, AUA, AUC and UUG are known to allow appreciable levels of initiation (e.g. 2–30 %), with CUG being the most efficient non-AUG initiation codon in many systems (reviewed by Touriol et al., 2003). Non-AUG initiation may be widely used as a regulatory mechanism by cellular organisms (Ivanov et al., 2008; Ingolia et al., 2011) but, at present, it is unclear whether such regulatory aspects have relevance to the use of non-AUG initiation by RNA viruses.
As non-AUG initiation is nearly always relatively inefficient, an inevitable consequence is that a large proportion of ribosomes will scan past the non-AUG initiation site and initiate instead at downstream AUG codons or other near-cognate non-AUG codons. Thus, instances of non-AUG initiation in RNA viruses generally form part of a leaky-scanning mechanism for translating multiple N-terminal extension isoforms of a protein, or multiple proteins from alternative reading frames (Table 2). One of the first cases of non-AUG initiation described occurs in Sendai respirovirus (family Paramyxoviridae). An upstream in-frame ACG codon is used to initiate translation of C′, an N-terminally extended version of the C protein (Curran & Kolakofsky, 1988). C′ and C are encoded by an ORF that mostly overlaps the 5′ region of the P (phosphoprotein) coding sequence, with initiation codons in the order C′ (ACG), P (AUG), C (AUG). The AUG initiation codon for P lacks a purine at −3, thus all three proteins can be translated via leaky scanning. In the related virus, human parainfluenza respirovirus 1, C′, P and C proteins are produced, but here C′ translation initiates at a GUG codon with surprisingly high efficiency (Boeck et al., 1992). An N-terminally extended version of the Gag polyprotein of murine leukemia gammaretrovirus has also been shown to initiate on an upstream in-frame non-AUG codon, in this case a CUG (Prats et al., 1989). The N-terminal extension includes a signal peptide that directs the product to the endoplasmic reticulum. While the AUG-initiated Gag is the precursor of the virion structural proteins, the N-terminally extended version is not incorporated into virions, but undergoes glycosylation, is displayed on the surface of cells and plays a role in virus release (Nitta et al., 2010). A homologous extension in feline leukemia gammaretrovirus initiates at an AUG codon with a weak context, thus potentially allowing leaky scanning to produce the N-terminally truncated isoform (Laprevotte et al., 1984).
Table 2. Examples of known and suspected cases of non-AUG initiation.
| Taxon | Product | Initiation codon | References |
| Respirovirus – Sendai virus | C′ (N-term. extension of C) | ACG | Curran & Kolakofsky (1988); Gupta & Patwardhan (1988) |
| Respirovirus – human parainfluenza virus 1 | " | GUG | Boeck et al. (1992) |
| Murine leukemia gammaretrovirus | Gag N-term. extension | CUG | Prats et al. (1989) |
| Soil-borne wheat mosaic furovirus and other furoviruses | CP N-term. extension | CUG | Shirako (1998) |
| Panicovirus – Panicum mosaic virus | Second movement protein of carmovirus-like double gene block | GUG | Turina et al. (2000) |
| Panicovirus – cocksfoot mild mosaic virus | " | CUG | Ziegler et al. (2009) |
| Machlomovirus – maize chlorotic mottle virus | " | CUG? | |
| Pelargonium line pattern virus | " | GUG | Castaño et al. (2009) |
| Pelargonium chlorotic ring pattern virus | " | GUG | |
| Rosa rugosa leaf distortion virus | " | CUG | |
| TGP carmovirus 3 | " | CUG | Scheets et al. (2011) |
| TGP carmovirus 1 | " | CUG/ACG? | |
| Allexiviruses and blackberry virus E | TGB3 | CUG? | Kanyuka et al. (1992); Sabanadzovic et al. (2011) |
| Lily potexvirus X | TGB3 | ACG? | Chen et al. (2005) |
| Strawberry mild yellow edge-associated potexvirus | TGB1 | CUG? | Jelkmann et al. (1992) |
| Hibiscus chlorotic ringspot carmovirus | p27 (N-term. extension of p25) | CUG | Koh et al. (2006) |
| Aquareovirus A, segment 7 | ORF1 (FAST protein) | CUG | Racine et al. (2009) |
| Torovirus | Predicted 30K protein | CUG | Firth & Atkins (2009) |
| Rice tungro bacilliform tungrovirus | ORF I | AUU | Fütterer et al. (1996) |
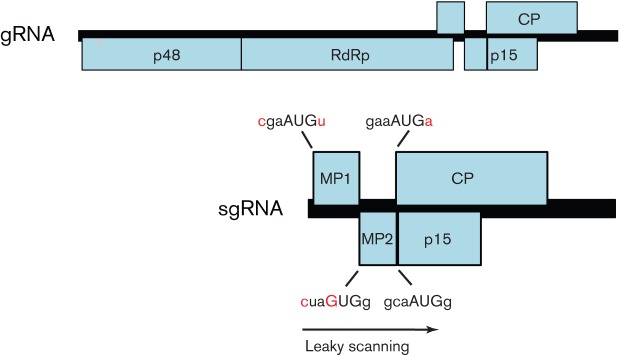
Many examples of non-AUG initiation come from plant viruses. In some cases, non-AUG initiation and leaky scanning are used to express three or even four separate proteins from a single transcript. One classical case, in rice tungro bacilliform tungrovirus (family Caulimoviridae), involves three consecutive ORFs where translation of the first ORF initiates at an AUU codon (facilitated by ribosome shunting; see below), the second ORF initiates at an AUG codon with a poor context, and the first two ORFs contain no other AUG codons, despite spanning around 895 nt, thus allowing leaky scanning also to the third ORF (Fütterer et al., 1996, 1997). A second case that is looking increasingly widespread occurs in some members of the family Tombusviridae. Many viruses in this family produce two coding 3′-co-terminal sgRNAs – one to express the coat protein and another for expressing additional proteins from ORFs either 5′ or 3′ of the coat protein ORF. However, Panicum mosaic panicovirus produces only a single coding sgRNA from which four proteins, including the two carmovirus-like movement proteins, are expressed via a combination of non-AUG initiation and leaky scanning (Turina et al., 2000; Fig. 3). The first movement protein, p8, has an AUG codon in a weak context; the second movement protein, p6.6 (which may be required in lower quantities; Li et al., 1998), is expressed from a GUG initiation codon; and the coat protein and p15 (whose coding sequence overlaps the coat protein coding sequence) are expressed from AUG codons. Unusually, the GUG initiation codon is in a suboptimal context (cuaGUGg; cf. aacCUGg at the corresponding position in cocksfoot mild mosaic panicovirus). Maize chlorotic mottle machlomovirus and Pelargonium line pattern virus (proposed genus Pelarspovirus) also lack a separate sgRNA for coat protein expression and, like the panicoviruses, these and several related viruses also appear to use non-AUG initiation to express the second movement protein (Castaño et al., 2009; Scheets et al., 2011).
Fig. 3.
Genome map of Panicum mosaic panicovirus. MP1, MP2, CP and p15 are all expressed from a single sgRNA via a combination of leaky scanning and non-AUG initiation. Initiation codons are indicated in upper case and nucleotides that differ from a strong initiation context are indicated in red.
Another well-conserved but poorly appreciated case of non-AUG initiation appears to occur in the allexiviruses (family Alphaflexiviridae). Nearly all plant viruses encode one or more movement proteins that facilitate cell-to-cell movement through plasmodesmata. For many plant viruses, this takes the form of the ‘triple gene block’ – an evolutionarily conserved module that comprises three proteins, TGB1, TGB2 and TGB3 (Morozov & Solovyev, 2003). The TGBs are generally expressed from sgRNAs but TGB3 – which is required in much lower quantities than TGB2 – is normally translated inefficiently from the same sgRNA as TGB2 via leaky scanning (Verchot et al., 1998; Morozov & Solovyev, 2003). While the allexiviruses clearly contain a TGB3 coding sequence, it lacks a suitable AUG initiation codon (Kanyuka et al., 1992). Instead, in these viruses the TGB2 AUG initiation codon consistently has a weak context (C or U at −3, A at +4) thus facilitating leaky scanning, and TGB3 translation probably depends on non-AUG initiation. A conserved CUG codon in a strong context (A at −3, G at +4) provides one possible initiation site.
Ribosome shunting
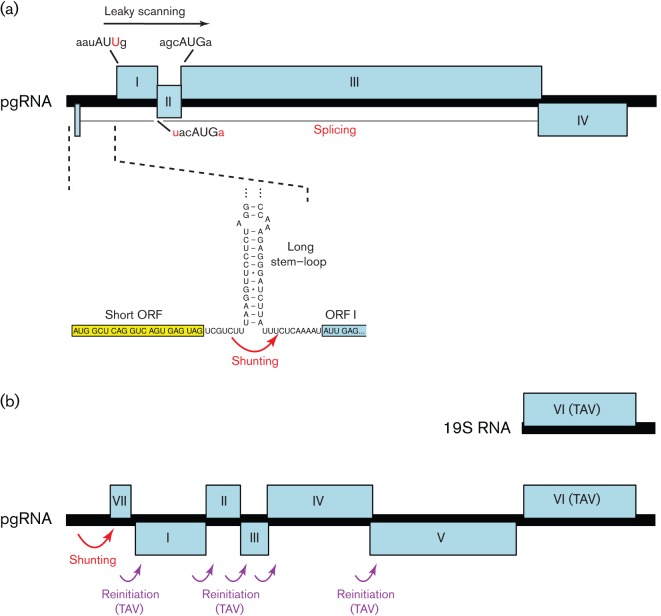
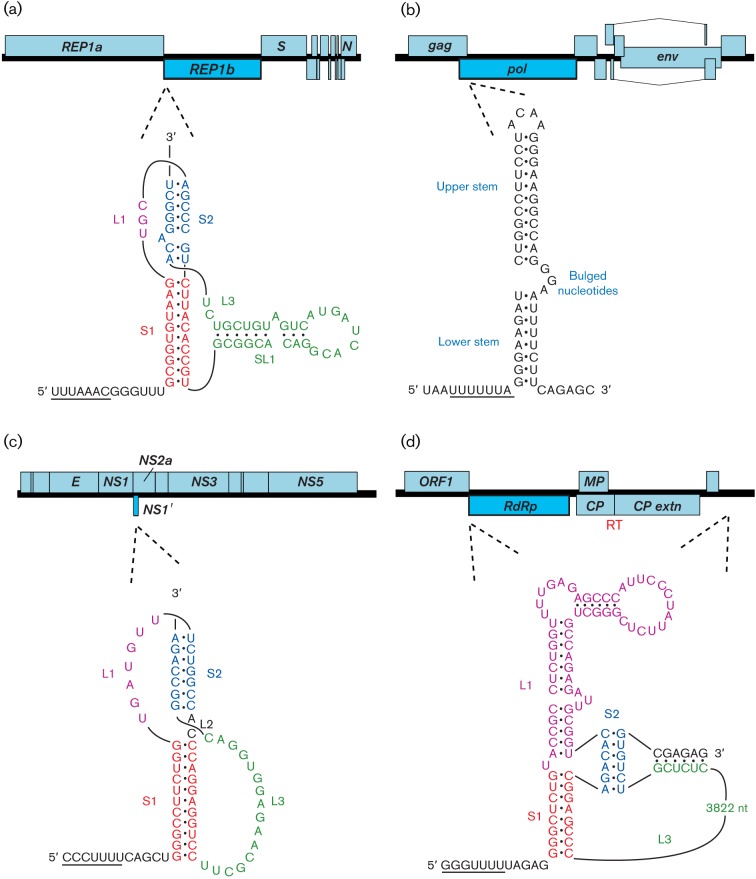
While IRESes allow internal entry of ribosomes on a message in a 5′-end-independent manner, shunting allows ribosomes to access downstream ORFs in a manner that is 5′-end-dependent but, at least partly, scanning-independent. One of the best-studied examples occurs in viruses of the family Caulimoviridae – a family of plant-infecting pararetroviruses (Fütterer et al., 1993; reviewed by Thiébeauld et al., 2007). These viruses produce a longer-than-genome-length pregenomic RNA (pgRNA) from the circular genomic DNA. The pgRNA serves as the template for reverse transcription. Some members have 3′ ORFs that are translated from spliced or subgenomic RNAs. However, the pgRNA is generally polycistronic – containing several consecutive ORFs that are translated either via leaky scanning (in members of the genera Tungrovirus and Badnavirus; see above) or via reinitiation (in members of the genera Caulimovirus and Soymovirus; see below). The pgRNA is capped and has a long 5′UTR, much of which is predicted to fold into a large stem–loop structure. A short ORF terminates just upstream of the stem–loop (Fig. 4). Translation is 5′-cap-dependent and the 40S subunits of ribosomes that scan to and translate the short ORF are, upon termination, able to bypass the stem–loop (comprising e.g. approx. 480 nt including eight AUG codons in cauliflower mosaic caulimovirus) and resume scanning at a landing site just 3′ of the stem–loop (Schmidt-Puchta et al., 1997; Pooggin et al., 2006). It is thought that this ability depends on the small subunit of the ribosome retaining certain initiation factors during translation of the short ORF (see also section entitled Reinitiation), but that the (temporary) loss of other initiation factors promotes discontinuous scanning across the base of the stem–loop. The length and position of the short ORF, but not its sequence, are important for shunting and the short ORF must be translated for efficient shunting to occur (Hemmings-Mieszczak et al., 2000; Pooggin et al., 2006). The large stem–loop and 5′-adjacent short ORF are predicted to be present in most sequenced members of the family, suggesting that the shunt mechanism is a common feature of members of the Caulimoviridae (Pooggin et al., 1999).
Fig. 4.
Ribosome shunting, reinitiation and leaky scanning in members of the family Caulimoviridae. (a) Translation of ORF I of the pgRNA of rice tungro bacilliform tungrovirus is by ribosome shunting. Here, 40S complexes released after translation of the short 5′-most ORF (yellow) are able to shunt past a stable stem–loop in the 5′UTR (red arrow) and continue scanning the mRNA. Reinitiation subsequently takes place at the start codons of either ORF I (a non-AUG codon, AUU), ORF II (weak context AUG) or ORF III (strong context AUG). ORF IV is expressed from a spliced mRNA. (b) In cauliflower mosaic caulimovirus, a similar shunting mechanism is used to access the 5′-most coding ORF, VII. However, downstream ORFs I–V are translated via reinitiation events that are stimulated by a viral reinitiation factor, transactivator viroplasmin (TAV), expressed from an sgRNA (see text).
Shunting and/or partly scanning-independent mechanisms have also been proposed to explain non-canonical translation observed for the Y1 and Y2 proteins (N-terminally truncated versions of C) in Sendai respirovirus (de Breyne et al., 2004), the σC gene on segment S1 of avian orthoreovirus and Nelson Bay orthoreovirus (Racine & Duncan, 2010), the P (polymerase) gene of avihepadnaviruses (Cao & Tavis, 2011), and the gag gene of spumaviruses (Schepetilnikov et al., 2009). Except for the last, these cases of shunting appear not to involve the short ORF and 3′-adjacent stem–loop configuration that occurs in members of the family Caulimoviridae.
Reinitiation
After translation termination, the 40S and 60S subunits of the ribosome dissociate and, generally, both subunits leave the message. However, after translating a very short ORF (e.g. less than 30 codons), the 40S subunit may remain associated with the message, resume scanning, and reinitiate translation at a downstream AUG codon (reviewed by Jackson et al., 2012). The ability of 40S subunits to remain associated with the message after translating a short ORF is thought to depend on certain initiation factors remaining attached to the ribosome during translation of the short ORF. After translation of the short ORF, the 40S subunit of the ribosome is not immediately competent to reinitiate, but becomes competent after scanning for some distance. This is thought to correspond to the time required to reacquire certain other initiation factors, including the eIF2–Met–tRNAi–GTP ternary complex. Such short ORFs are thought to be widely used in cellular genes to regulate expression of downstream protein-encoding ORFs in response to the cellular environment (Morris & Geballe, 2000; Ingolia et al., 2011). In contrast, reinitiation after translation of a long ORF – e.g. one that encodes a functional product – is much rarer and requires special signals within the mRNA or specific trans-acting protein factors (reviewed by Thiébeauld et al., 2007; Powell, 2010; Jackson et al., 2012). A number of distinct mechanisms appear to be used by different viruses (Table 3).
Table 3. Examples of known and suspected cases of reinitiation.
| Taxon | Products | References |
| Caliciviridae – Lagovirus, Vesivirus, Norovirus, Sapovirus, Nebovirus | VP1/VP2 | Meyers (2003); Luttermann & Meyers (2007); Napthine et al. (2009) |
| Caliciviridae – some noroviruses | Replicase/VP1 | McCormick et al. (2008) |
| Orthomyxoviridae – influenza virus B, segment 7 | M1/BM2 | Horvath et al. (1990); Powell et al. (2008); Powell et al. (2011) |
| Pneumovirinae – Pneumovirus, Metapneumovirus | M2-1/M2-2 | Ahmadian et al. (2000); Gould & Easton (2005); Gould & Easton (2007) |
| Totiviridae – Victorivirus | Gag/Pol | Huang & Ghabrial (1996); Soldevila & Ghabrial (2000); Ghabrial & Nibert (2009); Li et al. (2011) |
| Hypovirus – Cryphonectria hypoviruses 1 and 2 | ORFA/ORFB | Shapira et al. (1991); Hillman et al. (1994); Guo et al. (2009) |
| Caulimoviridae – Caulimovirus, Soymovirus | Multiple consecutive ORFs | Fütterer & Hohn (1991); Scholthof et al. (1992); Maiti et al. (1998) |
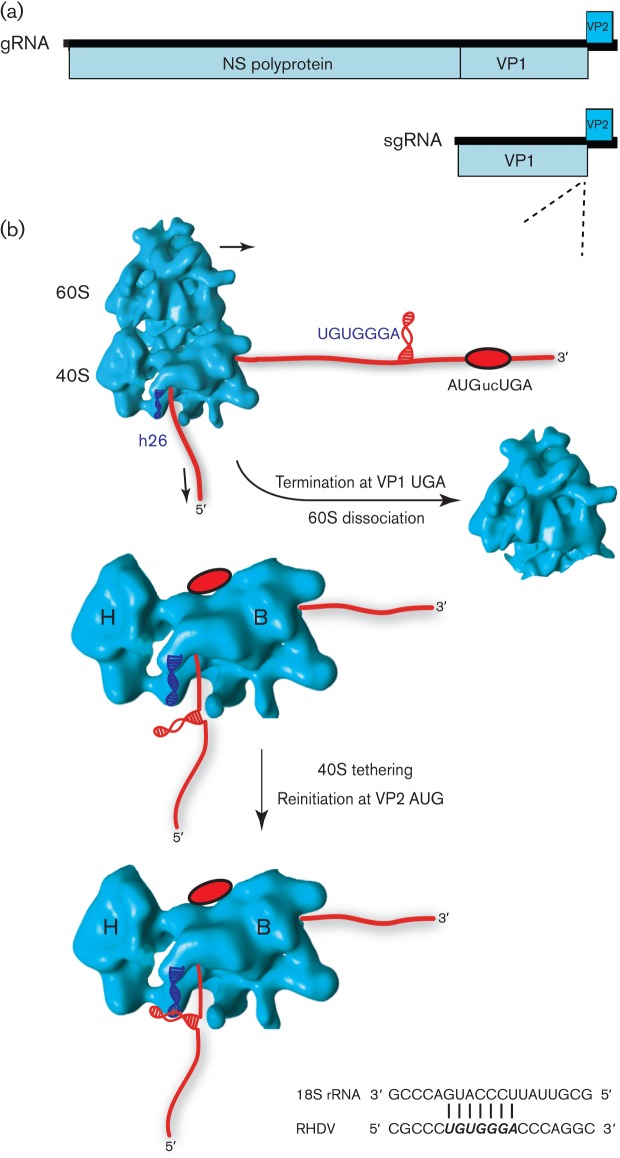
Calicivirus genomes generally contain at least three protein-encoding ORFs: ORF1 is translated from the genomic RNA and encodes the non-structural proteins, whilst ORF2 and ORF3 are translated from a single sgRNA and encode, respectively, the major capsid protein and a small basic protein that is a minor component of the virion (Herbert et al., 1996). In some genera (e.g. Lagovirus and Sapovirus), ORF2 is contiguous with ORF1 so that some capsid protein is also translated from the genomic RNA as a fusion with the non-structural polyprotein, even though the major source of capsid protein translation is still the sgRNA (Fig. 5). ORF3 is positioned at the 3′ end of the sgRNA such that its initiation codon is located very close to the ORF2 termination codon (often AUGnnUGA or overlapping as UAAUG or AUGA), and is translated via reinitiation after translation of ORF2 (Meyers, 2003; Luttermann & Meyers, 2007; Pöyry et al., 2007; Napthine et al., 2009). Here, reinitiation is dependent on RNA sequence motifs typically within the 40–90 nucleotides directly upstream of the ORF2 termination codon. This sequence region is termed the TURBS (termination upstream ribosome-binding site) and contains a short sequence motif (motif 1; UGGGA and flanking nucleotides) that is complementary to the loop region of helix 26 of 18S rRNA (the RNA component of the 40S subunit). Interaction between motif 1 and 18S rRNA has been shown to be required for efficient reinitiation in a yeast system in which 18S rRNA could be mutated (Luttermann & Meyers, 2009). The TURBS has also been shown to bind eIF3 (Pöyry et al., 2007). It is thought that a proportion of 40S subunits of ribosomes terminating translation of ORF2 are tethered with eIF3 to the mRNA via interaction with the TURBS and, following recruitment of the eIF2–Met–tRNAi–GTP ternary complex, such subunits may subsequently initiate translation of ORF3 (Fig. 5). Notably, and in contrast to reinitiation after a very short ORF, eIF4G is not required (Pöyry et al., 2007). A similar reinitiation mechanism also appears to be used by influenza B virus for translation of the BM2 protein (Horvath et al., 1990). Here, the M1 and BM2 coding sequences overlap with the sequence UAAUG and an upstream TURBS, comprising around 45 nt upstream of the termination codon and incorporating an appropriately positioned UGGGA motif, is again crucial for reinitiation (Powell et al., 2011). Mutational analyses have demonstrated that increasing the distance between the TURBS and the termination codon reduces reinitiation efficiency, presumably due to reduced tethering of post-termination 40S subunits, but reinitiation still preferentially occurs at the natural position with respect to the TURBS (Pöyry et al., 2007; Powell et al., 2011). In some cases, initiation codons some distance downstream of the natural reinitiation site may be utilized at reduced efficiency if the natural reinitiation site is mutated (Powell et al., 2011). Non-AUG initiation codons may also be utilized, although AUG codons are preferred if available (Luttermann & Meyers, 2007; Pöyry et al., 2007).
Fig. 5.
Proposed model for termination–reinitiation in caliciviruses. (a) Genome map of the calicivirus rabbit hemorrhagic disease lagovirus (RHDV). Expression of VP2 is by termination–reinitiation during translation of the viral sgRNA. (b) As the ribosome approaches the termination–reinitiation site (red oval; AUGucUGA in RHDV), the stretch of RNA containing TURBS motif 1 (UGUGGGA), predicted to be located in an RNA secondary structure, is translated and may be remodelled. During termination, the secondary structure is located in the mRNA exit channel of the ribosome [located between the head (H) and body (B) of the 40S subunit] and in close proximity to the solvent-accessible helix 26 (h26) of 18S rRNA (indicated as a blue helix). Base pairing between complementary residues in motif 1 and h26 occurs (shown at the bottom), with the interaction likely to be stabilized by eIF3 (not shown), also known to contact the TURBS and 18S rRNA. Together, these interactions act to tether the ribosome to the viral RNA, preventing its dissociation, allowing time for the recruitment of initiation factors and subsequent reinitiation on the downstream VP2 ORF.
Interestingly, despite the production of an sgRNA for ORF2 expression, some noroviruses are also capable of translating ORF2 via reinitiation after translation of ORF1 from the genomic RNA (McCormick et al., 2008). Again, this reinitiation is dependent on an upstream TURBS. As certain other caliciviruses are also capable of expressing the capsid protein from the genomic RNA due to ORF2 being contiguous with ORF1 (see above), it has been hypothesized that the capsid protein may play additional roles early in virus infection before sgRNAs are produced (McCormick et al., 2008).
A termination–reinitiation mechanism is also used by pneumoviruses and metapneumoviruses to express their M2-1 and M2-2 proteins from a single transcript (Ahmadian et al., 2000; Gould & Easton, 2007). As with caliciviruses, reinitiation is dependent on sequences upstream of the termination codon, but a much larger sequence region is involved (e.g. approx. 250 nt are required to achieve 80 % of the wild-type reinitiation efficiency in respiratory syncytial pneumovirus) and a TURBS-like sequence has not been identified. Interestingly, the reinitiation AUG codon can be some distance upstream of the termination codon (e.g. up to 29 nt in respiratory syncytial pneumovirus, although two other AUG codons that are closer to the termination codon are also utilized; Ahmadian et al., 2000). In members of the genus Victorivirus of the family Totiviridae, reinitiation after translation of the capsid (Gag) coding sequence is used to translate the polymerase (Pol) – which is required in much lower quantities – from the single genomic RNA (Li et al., 2011). The two ORFs often overlap with AUGA or sometimes AUGnnUAG or UAGnnAUG. Again, upstream sequences – e.g. at least 32 nt in Helminthosporium victoriae victorivirus 190S – have been shown to be important for reinitiation (Li et al., 2011).
Whilst reinitiation in the above viruses is mediated partly or wholly by RNA sequences directly upstream of the termination codon, a very different reinitiation mechanism is used by members of the genera Caulimovirus and Soymovirus in the family Caulimoviridae (see section entitled Ribosome shunting). Here, reinitiation is mediated by a viral protein ‘transactivator viroplasmin’ or TAV (Fütterer & Hohn, 1991; Scholthof et al., 1992; reviewed by Thiébeauld et al., 2007). In cauliflower mosaic caulimovirus, TAV is encoded by ORF VI, which is translated from a subgenomic 19S transcript, while several other consecutive ORFs are translated from the pgRNA via TAV-mediated reinitiation (Fig. 4). TAV-mediated reinitiation does not appear to require any specific sequence elements in the RNA transcript (Fütterer & Hohn, 1991). Through interactions with the host proteins TOR and RISP, TAV is thought to tether eIF3 to the elongating ribosome during translation of the upstream ORF, thus assisting reinitiation on downstream ORFs (Schepetilnikov et al., 2011).
Non-canonical elongation and termination
Non-canonical translation mechanisms that act during elongation, or via altered readings of termination signals, are known as recoding mechanisms (reviewed by Atkins & Gesteland, 2010). These fall into four main categories: ribosomal frameshifting, whereby a proportion of ribosomes are directed into a different reading frame by shifting forwards or backwards 1 or 2 nt; bypassing, in which a proportion of ribosomes skip over a larger number of nucleotides and continue translating; stop-codon redefinition and readthrough, whereby a proportion of ribosomes fail to terminate at a stop codon but instead insert a standard amino acid (readthrough) or a non-standard amino acid such as selenocysteine or pyrrolysine (redefinition); and stop–carry on – a mechanism that directs co-translational separation of the peptide chain by preventing peptide-bond formation at a specific site. Programmed bypassing and selenocysteine/pyrrolysine insertion are not, however, known to be utilized by eukaryote-infecting RNA viruses.
Ribosomal frameshifting
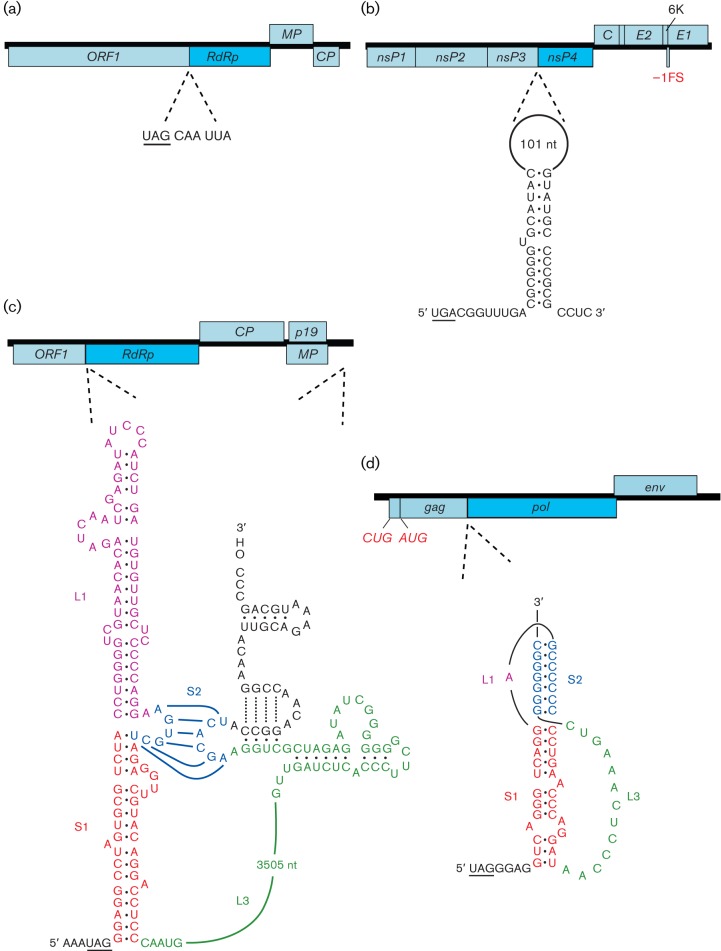
Programmed −1 ribosomal frameshifting was first described as the mechanism by which the Gag–Pol polyprotein of Rous sarcoma alpharetrovirus is expressed from overlapping gag and pol ORFs (Jacks & Varmus, 1985; Jacks et al., 1988). Related signals have since been documented in many other viruses (see Table 4), including the clinically important lentiviruses HIV-1 and HIV-2, human T-cell lymphotrophic deltaretrovirus types 1 and 2, and the coronavirus responsible for severe acute respiratory syndrome (SARS-CoV). Frameshifting has also been increasingly recognized in conventional cellular genes of both prokaryotes and eukaryotes, as well as in other replicating elements, such as insertion sequences and transposons (reviewed by Giedroc & Cornish, 2009; Brierley et al., 2010). The mRNA signal for −1 frameshifting in eukaryotic systems comprises two elements: a slippery sequence with consensus X_XXY_YYZ (underlines separate zero-frame codons; XXX represents any three identical nucleotides, YYY represents AAA or UUU, and Z represents A, C or U) where the ribosome changes frame, and a downstream stimulatory RNA structure (Fig. 6; reviewed by Giedroc & Cornish, 2009; Brierley et al., 2010). Appropriate spacing (typically 5–9 nt) between the slippery sequence and the stimulatory RNA is also required for efficient frameshifting.
Table 4. Examples of known and suspected cases of programmed −1 ribosomal frameshifting.
| Taxon | Product | References |
| Astroviridae – Avastrovirus, Mamastrovirus | Replicase | Marczinke et al. (1994) |
| Arteriviridae – Arterivirus | Replicase | den Boon et al. (1991) |
| Coronaviridae – Alphacoronavirus, Betacoronavirus, Gammacoronavirus, Bafinivirus, Torovirus | Replicase | Brierley et al. (1987); Bredenbeek et al. (1990); Herold & Siddell (1993); Snijder et al. (1990); Thiel et al. (2003) |
| Roniviridae – Okavirus | Replicase | Cowley et al. (2000) |
| Unclassified Nidovirales – Nam Dinh virus, Cavally virus | Replicase | Nga et al. (2011) |
| Sobemovirus | Replicase | Mäkinen et al. (1995) |
| Barnaviridae – Barnavirus | Replicase | Revill et al. (1994) |
| Polemovirus | Replicase | aus dem Siepen et al. (2005) |
| Luteoviridae – Polerovirus, Enamovirus | Replicase | Demler & de Zoeten (1991); Prüfer et al. (1992); Garcia et al. (1993) |
| Luteoviridae – Luteovirus | Replicase | Brault & Miller (1992) |
| Umbravirus | Replicase | Demler et al. (1993); Gibbs et al. (1996) |
| Tombusviridae – Dianthovirus | Replicase | Xiong et al. (1993); Kim & Lommel (1994); Tajima et al. (2011) |
| Totiviridae – Trichomonasvirus (Trichomonas vaginalis viruses 2, 3 and 4) | Replicase | Bessarab et al. (2000); Bessarab et al. (2011) |
| Totiviridae – Totivirus (Saccharomyces cerevisiae viruses L–A and L–BC, Tuber aestivum virus 1, black raspberry virus F) | Replicase | Diamond et al. (1989); Dinman et al. (1991) |
| Totiviridae – Giardiavirus | Replicase | Wang et al. (1993); Li et al. (2001) |
| Totiviridae – ‘Artivirus, (penaeid shrimp infectious myonecrosis virus, Armigeres subalbatus virus, Drosophila melanogaster totivirus, Omono River virus) | Replicase | Nibert (2007); Zhai et al. (2010); Wu et al. (2010); Isawa et al. (2011) |
| Unclassified Totiviridae (Lentinula edodes mycovirus, Phlebiopsis gigantea mycovirus 1, Phlebiopsis gigantea mycovirus 2, Fusarium graminearum mycovirus 3, piscine myocarditis virus, grapevine associated totivirus 2) | Replicase | Ohta et al. (2008); Kozlakidis et al. (2009); Yu et al. (2009); Haugland et al. (2011); Al Rwahnih et al. (2011) |
| Unclassified dsRNA viruses (Rosellinia necatrix megabirnavirus, Spissistilus festinus virus 1, Circulifer tenellus virus 1) | Replicase | Chiba et al. (2009); Spear et al. (2010) |
| Retroviridae – Lentivirus, Alpharetrovirus | Reverse transcriptase (Gag–Pol) | Jacks & Varmus (1985); Jacks et al. (1988a); Jacks et al. (1988b); Morikawa & Bishop (1992) |
| Retroviridae – Betaretrovirus, Deltaretrovirus | Reverse transcriptase (Gag–Pro–Pol) | Moore et al. (1987); Jacks et al. (1987); Nam et al. (1988); Mador et al. (1989); Nam et al. (1993) |
| Unclassified ssRNA+ viruses – Acyrthosiphon pisum virus, rosy apple aphid virus | CP-extension | van der Wilk et al. (1997) |
| Togaviridae – Alphavirus | TF | Firth et al. (2008); Chung et al. (2010) |
| Flaviviridae – Flavivirus (Japanese encephalitis serogroup) | NS1′ | Firth & Atkins (2009); Melian et al. (2010) |
| Flaviviridae – Flavivirus (insect-specific flaviviruses) | FIFO | Firth et al. (2010) |
| Picornaviridae – Cardiovirus | 2B* | Loughran et al. (2011) |
Fig. 6.
Programmed ribosomal frameshifting in viral mRNAs. In each of the four examples, the genome is indicated as an ORF map, with the location of the frameshift site shown by dotted lines. In SARS coronavirus (a), Japanese encephalitis flavivirus (c) and barley yellow dwarf luteovirus (d), frameshifting is stimulated by an RNA pseudoknot (including a long-range interaction in the luteovirus). In HIV-1 (b), frameshifting is stimulated by a two-stem helix, although the upper stem makes the major contribution to frameshifting efficiency. In each case, the slippery shift site sequence is underlined. Note that, in (c), the unprocessed frameshift product generates a truncated polyprotein, unlike the other examples, where the frameshift facilitates extension of the polyprotein. Spliced and subgenomic RNAs are not shown and polyprotein cleavage products are only indicated where specifically relevant. ‘RT’ indicates a stop codon readthrough site.
There is considerable experimental support for the idea that ‘tandem slippage’ of ribosome-bound peptidyl– and aminoacyl–tRNAs on the slippery sequence occurs upon encounter of the stimulatory RNA, with the tRNAs detaching from the zero-frame codons (XXY_YYZ) and re-pairing in the −1 frame (XXX_YYY), thus allowing for perfect re-pairing except at the wobble positions (Jacks et al., 1988). As the codon : anticodon duplex in the P-site is not monitored as strictly as that in the A-site (Ogle et al., 2001), certain deviations from the canonical XXX of the slippery site are tolerated, including GUU in equine arteritis arterivirus, GGA in insect-specific flaviviruses, and GGU in encephalomyocarditis cardiovirus. The stimulatory RNA generally takes the form of a stem–loop or RNA pseudoknot and, in most cases, is a discrete local element. However, some 3′ stimulatory structures have an additional long-range base-pairing component that may provide a regulatory link between translation and replication (e.g. barley yellow dwarf luteovirus; Barry & Miller, 2002). How the stimulatory RNAs function to promote frameshifting is still uncertain, but accumulating evidence implicates the intrinsic unwinding activity of the ribosome (Qu et al., 2011), with the stimulatory RNA exhibiting resistance to unwinding, perhaps by presenting an unusual topology (Plant & Dinman, 2005; Namy et al., 2006). Failure to unwind the stimulatory RNA appropriately has been proposed to induce tension in the mRNA, leading to uncoupling of the codon : anticodon complexes and realignment of the tRNAs in the −1 frame (Plant & Dinman, 2005; Namy et al., 2006).
Many positive-strand RNA viruses, most retroviruses, and some members of the family Totiviridae of dsRNA viruses make use of −1 frameshifting to express their RdRp or reverse transcriptase (Table 4). Indeed, in the case of members of the genera Betaretrovirus and Deltaretrovirus, where the protease gene (pro) is encoded as a separate ORF, two frameshifts are required to express the Gag–Pro–Pol polyprotein. There are numerous potential advantages in using frameshifting as an expression strategy. In retroviruses and totiviruses, for example, it allows the virus to generate a defined ratio of Gag : Gag–Pol that is likely to be optimized for virion assembly and allows facile targeting of the replicative enzymes to the virion core. It also obviates the need to produce a separate mRNA for expression of the viral polymerase. Similarly, in many positive-strand RNA viruses, frameshifting may serve to produce the polymerase at a fixed ratio relative to other components of the replication complex (reviewed by Ahlquist, 2006). Indeed, artificially altering the frameshifting efficiency has proven to be attenuating in several cases (Dulude et al., 2006; Plant et al., 2010), although in HIV-1 there is evidence that a modest stimulation of frameshifting can actually increase infectivity (Miyauchi et al., 2006).
Frameshifting is also utilized for the expression of proteins unrelated to polymerases. In Acyrthosiphon pisum virus, an unclassified virus that is related distantly to viruses of the family Picornaviridae, −1 frameshifting at the end of a long polyprotein-encoding ORF1 provides access to a 3′-terminal ORF2 (van der Wilk et al., 1997). Here, the 3′ end of ORF1 encodes the major virion protein (34K), while the transframe fusion gives rise to a minor virion protein (66K). In alphaviruses, the Japanese encephalitis serogroup of flaviviruses, and the cardioviruses, −1 frameshifting provides access to short ORFs overlapping internal regions of long polyprotein-encoding ORFs to generate transframe proteins that are N-terminally coincident with one of the polyprotein cleavage products (Firth et al., 2008; Melian et al., 2010; Loughran et al., 2011). In the Japanese encephalitis serogroup, frameshifting adds a 52 aa transframe C-terminal extension to the NS1 protein to produce the NS1′ protein. In the alphaviruses, frameshifting produces an 8 kDa protein that shares the N-terminal approximately two-thirds with the 6K protein, but has a hydrophilic instead of a hydrophobic C-terminal region. In encephalomyocarditis cardiovirus, frameshifting produces a 128 or 129 aa protein, 2B*, in which just the N-terminal 11–12 aa are encoded by the zero frame.
In a few taxa, +1 (or −2) frameshifting appears to be utilized to express the viral polymerase. However, in viruses these mechanisms have been far less well-studied than −1 tandem slippage and remain poorly understood, in part because the frameshift efficiencies, where investigated, often seem to be very low. In members of the Closteroviridae – a family of large positive-strand plant-infecting RNA viruses – the RdRp is encoded by ORF2, which is in the +1 frame relative to ORF1, while many 3′ ORFs are translated from sgRNAs. As the RdRp is required to produce sgRNAs, it was proposed that ORF2 is translated via +1 frameshifting at or near the end of ORF1 (Agranovsky et al., 1994). Indeed now, with many highly divergent genome sequences available, it is clear from comparative sequence analysis that mechanisms involving AUG-initiation cannot, in general, mediate ORF2 translation. However the exact frameshifting mechanism remains something of a mystery. In many closteroviruses, frameshifting is thought to occur on a highly conserved GUU_stop_C motif at the ORF1 stop codon and may involve +1 nt P-site slippage from GUU to UUU, with the slow-to-decode stop codon in the A-site. In citrus tristeza closterovirus, however, frameshifting appears to occur upstream of the ORF1 stop codon (as evidenced by a conserved overlap region and high conservation at ORF1-frame synonymous sites for at least 25 codons upstream of the ORF1 stop codon), and frameshifting has been suggested to occur on a GUU_CGG_C sequence that aligns with the GUU_stop_C sequence in other closteroviruses (Karasev et al., 1995; but compare with Çevik, 2001).
Whilst many members of the family Totiviridae utilize −1 tandem slippage to express a Gag–Pol fusion, or reinitiation to express Gag and Pol separately, a few members apparently utilize +1 or −2 frameshifting. Trichomonas vaginalis virus 1 (genus Trichomonasvirus) is particularly interesting. Here, ORF2 is in the +1 frame relative to ORF1 and nucleotide sequence analysis indicates that frameshifting is most likely to occur on a conserved CC_CUU_UUU sequence (Su & Tai, 1996; Goodman et al., 2011). Notably, the 5′ CC is conserved despite the corresponding ORF1-frame xCC codon being GCC, UCC or ACC in different isolates, suggesting that frameshifting is by −2 nt and not +1 nt. In contrast, in Trichomonas vaginalis viruses 2, 3 and 4, ORF2 is in the −1 frame relative to ORF1, and −1 frameshifting is predicted to occur on a G_GGC_CCY heptanucleotide – a sequence that, due to the C_CCY A-site tetranucleotide, is not usually associated with efficient frameshifting, but which may nevertheless be adequate for the low level of frameshifting that is apparently required by these viruses.
The Amalgamaviridae – a recently proposed family of monopartite dsRNA viruses (Martin et al., 2011), besides Leishmania RNA virus 1 (genus Leishmaniavirus, family Totiviridae; Kim et al., 2005), and the unclassified positive-strand RNA viruses chronic bee paralysis virus and Lake Sinai viruses 1 and 2 (Olivier et al., 2008), also appear to require +1 slippage to express their RdRp, although the shift sites have not yet been definitively localized nor have alternative mechanisms been ruled out.
Recently, a short conserved ORF that overlaps the P3-encoding region of the polyprotein ORF in probably all members of the Potyviridae (the largest family of RNA plant viruses) was shown to be translated and essential for virus infectivity (Chung et al., 2008; Wen & Hajimorad, 2010; Wei et al., 2010; for earlier insights see also Gibbs & Keese, 1995; Choi et al., 2001). In turnip mosaic potyvirus, the ORF (known as pipo) is expressed as part of an approximately 25 kDa product that is believed to correspond to a fusion of PIPO with the N-terminal region of P3 (i.e. P3N–PIPO). The frameshifting mechanism has not yet been determined, but current evidence suggests that it occurs at the level of translation rather than transcription, and most likely involves a highly conserved GAA_AAA_A motif at the 5′ end of the pipo ORF (Chung et al., 2008). Notably, the GAA_AAA_A motif is in a different frame relative to the canonical X_XXY_YYZ −1 tandem slippage site, suggesting a +2 rather than a −1 shift.
Stop-codon readthrough
Translation termination is generally a highly efficient process, but is influenced by the nature of the stop codon present (UAA, UAG or UGA) and its flanking nucleotides, especially the immediately adjacent 3′ base (reviewed by Bertram et al., 2001). Some termination codon contexts are noticeably ‘leaky’ (e.g. UGAC; McCaughan et al., 1995), i.e. they allow ‘readthrough’ at frequencies ranging from 0.3 to 5 % (Bertram et al., 2001). In readthrough, the stop codon is decoded by a near-cognate or suppressor tRNA, and translation continues to the next termination codon. Readthrough is exploited in the expression of several viral and cellular genes, where it is referred to as programmed readthrough (reviewed by Beier & Grimm, 2001; Namy & Rousset, 2010). Readthrough allows the production of a C-terminally extended polypeptide at a defined frequency. In viruses, it is often used to express the polymerase, but another common use is to append an extension domain to a proportion of coat proteins. In the luteoviruses, for example, readthrough at the end of the coat protein gene generates a protein required for aphid transmission (Brault et al., 1995). Similarly, in benyviruses and pomoviruses the coat protein extension is required for transmission by their plasmodiophorid vectors (reviewed by Adams et al., 2001).
The efficiency of readthrough can be influenced by elements located both 5′ and 3′ of the suppressed stop codon. Adenosines in the two positions immediately preceding the stop codon have been shown to stimulate readthrough (albeit in a yeast-cell environment) and are a feature common to many viral readthrough cases – notably in the tobamoviruses, poleroviruses and luteoviruses (Beier & Grimm, 2001; Tork et al., 2004). Downstream stimulators generally fall into two classes: the 3′-adjacent nucleotides, which are thought to act at the level of primary sequence, and more distal elements that typically involve RNA secondary structures. The effect of the immediately 3′-adjacent nucleotide(s) may be specific to the identity of the stop codon (Bonetti et al., 1995), and this may relate to competition between the release factor and potential near-cognate tRNAs binding to the stop codon. In contrast, 3′ RNA structures typically beginning around 8 nt 3′ of the stop codon are a common (but not ubiquitous) feature of different types of readthrough. The exact mechanism(s) by which such structures promote readthrough is not currently known, but possibilities include modulation of ribosome activity directly through mRNA–protein or mRNA–rRNA interactions; interference with release factor function through steric hindrance; or, similar to frameshift stimulatory RNAs, by providing a barrier to unwinding by a ribosome-associated helicase (Qu et al., 2011; although at least some readthrough-stimulating structures do not, on their own, cause significant ribosome pausing; Napthine et al., 2012).
The various 3′ motifs that stimulate readthrough in eukaryote-infecting viruses have been divided into three broad classes (Beier & Grimm, 2001; Fig. 7; Table 5). The type I motif is exemplified by tobacco mosaic virus and other tobamoviruses. Here, readthrough of a UAG codon in the replicase gene is stimulated by the six nucleotides immediately following the stop codon, with the consensus motif for efficient readthrough being UAG_CAR_YYA (Skuzeski et al., 1991; R = purine, Y = pyrimidine). The same motif is utilized by a number of other plant viruses; for example in benyviruses and pomoviruses, where readthrough generates extended versions of the viral coat protein. Although natural cases of CARYYA-stimulated readthrough generally involve a UAG codon, CARYYA can also stimulate readthrough of UGA and UAA codons (Skuzeski et al., 1991). The type II motif was originally defined as generally comprising a UGA stop codon followed by a CGG or CUA triplet (Beier & Grimm, 2001). It was later proposed that most instances of readthrough in this class also involve a 3′ RNA structure component – often comprising an extended stem–loop structure beginning around 8 nt 3′ of the stop codon (Firth et al., 2011; Napthine et al., 2012). Type II readthrough occurs in the replicase gene of a number of alphaviruses (although not all alphaviruses have an internal stop codon in their replicase gene), the replicase gene of tobraviruses, pecluviruses, furoviruses and pomoviruses, the coat protein extension gene of furoviruses, and the VP9/VP9′ gene of coltiviruses.
Fig. 7.
Programmed stop codon readthrough in viral mRNAs. In each of the four examples, the genome is indicated as an ORF map, with the location of the readthrough site shown by dotted lines. In tobacco mosaic tobamovirus (a), only a short local sequence context 3′ of the recoded UAG is required for efficient readthrough. In Venezuelan equine encephalitis alphavirus (b), carnation italian ringspot tombusvirus (c) and murine leukemia gammaretrovirus (d), the 3′ stimulator is an RNA secondary structure: an extended stem–loop in (b), an RNA pseudoknot in (d) and long-range base pairing in (c). Spliced and subgenomic RNAs are not shown and polyprotein cleavage products are only indicated where specifically relevant. ‘FS’ indicates a ribosomal frameshift site.
Table 5. Examples of known and suspected cases of stop-codon readthrough.
| Taxon | Product | Type | Stop-codon context | RNA structure | References |
| Alphavirus (species that utilize readthrough) | Replicase | II | UGA-CGG, UGA-CUA | Extended stem–loop | Li & Rice (1993); Firth et al. (2011) |
| Tobravirus RNA1 | Replicase | II | UGA-CGG | Extended stem–loop* | Urban et al. (1996); Firth et al. (2011) |
| Pecluvirus RNA1 | Replicase | II | UGA-CGG | Extended stem–loop? | |
| Furovirus RNA1 | Replicase | II | UGA-CGG | Extended stem–loop? | |
| Pomovirus RNA1 | Replicase | II | UGA-CGG, UAA-CGG etc. | Extended stem–loop* | |
| Tobamovirus | Replicase | I | A-UAG-CAA-UUA | Skuzeski et al. (1991); Zerfass & Beier (1992) | |
| Providence virus | Replicase | I | UAG-CAA-CUA | Walter et al. (2010) | |
| Tombusviridae (except Dianthovirus) | Replicase | III | UAG-GGR | 3′-proximal structure+long-distance 3′ base pairing | Cimino et al. (2011) |
| Gammaretrovirus | Reverse transcriptase | III | UAG-G | Compact pseudoknot | Wills et al. (1991); Alam et al. (1999) |
| Epsilonretrovirus | Reverse transcriptase | III | UAG-R | Extended stem–loop† | |
| Furovirus RNA2 | CP-extension | II | UGA-CGG etc. | Extended stem–loop? | |
| Pomovirus RNA2 | CP-extension | I | UAG-CAA-UYA, UAA-CAA-UUA | ||
| Luteovirus, Polerovirus | CP-extension | III | AAA-UAG-GUA | Long-distance 3′ base pairing? | Brown et al. (1996); Bruyère et al. (1997) |
| Rose spring dwarf-associated luteovirus | CP-extension | II | UGA-CGG | ||
| Enamovirus | CP-extension | UGA-GGG | Demler & de Zoeten (1991) | ||
| Benyvirus | CP-extension | I | UAG-CAA-UUA | Compact stem–loop* | |
| Rice stripe necrosis benyvirus | CP-extension | III | UAG-GGG | Compact stem–loop* | |
| Coltivirus, segment 9 | VP9′ | II | UGA-CGG | Extended stem–loop | Jaafar et al. (2004); Napthine et al. (2012) |
R. Ling & A. E. Firth, unpublished data.
S. Napthine, K. E. Deigan & I. Brierley, unpublished data.
Type III readthrough motifs comprise a more diverse class, but generally involve a UAG stop codon, a 3′-adjacent G or purine-rich octanucleotide, and some form of 3′ RNA structure. In murine leukemia virus and other gammaretroviruses, efficient readthrough of a UAG codon in the replicase gene requires a compact 3′-adjacent pseudoknot structure, with the identity of certain nucleotides in the 8 nt ‘spacer’ region between the stop codon and the pseudoknot also being important (Alam et al., 1999; Houck-Loomis et al., 2011). In luteoviruses and poleroviruses, efficient readthrough of a UAG codon is dependent on 3′-adjacent sequences, but an element located approximately 700–750 nt 3′ of the stop codon is also important, and long-distance RNA base pairing between the 3′-proximal and 3′-distal elements has been suggested as a possible mechanism (Brown et al., 1996). In members of the family Tombusviridae, such as carnation italian ringspot tombusvirus and turnip crinkle carmovirus, efficient readthrough is dependent on a large 3′-proximal RNA stem–loop structure, besides long-distance RNA base pairing between this structure and an element near the 3′ end of the genome, 3.5 kb away (Cimino et al., 2011). Similar long-distance interactions have been predicted for other genera in the family Tombusviridae, including Necrovirus, Aureusvirus and Panicovirus, and it has been proposed that the long-distance interaction may play a regulatory role by linking translation (of the RdRp) with replication (Cimino et al., 2011).
Stop–carry on
Stop–carry on is mediated by the amino acid motif D(V/I)ExNPGP, which, together with less-conserved but nonetheless functionally important upstream amino acids also within the ribosome exit tunnel (approx. 30 aa in total), prevents formation of a peptide bond between glycine and the final proline (Donnelly et al., 2001; Doronina et al., 2008; reviewed by Brown & Ryan, 2010). Nonetheless, translation can continue (with proline as the N-terminal amino acid of the downstream product) with up to near-100 % efficiency. It is thought that the structure of the nascent peptide within the ribosome prevents Pro–tRNA from binding in the A-site, but instead allows eRF1 to bind and, with eRF3, release the nascent peptide, following which Pro–tRNA is able to bind and translation proceeds. As such, stop–carry on provides an alternative mechanism to proteolytic cleavage for producing multiple protein products from a single ORF. Stop–carry on cassettes are present in diverse RNA viruses and have probably evolved (or been acquired) independently on more than one occasion. One of the most well-known occurrences is at the junction between the 2A and 2B proteins of members of several genera of the family Picornaviridae, including the aphthoviruses, cardioviruses, erboviruses and teschoviruses. Stop–carry on cassettes also occur in (some) members of the taxa Iflavirus, Dicistroviridae, Tetraviridae, Rotavirus, Cypovirus and Totiviridae, with some viruses having multiple stop–carry on cassettes (Luke et al., 2008).
Alternatives to non-canonical translation
Although this review has focused on the unusual translational mechanisms that viruses employ to cope with the unique constraints imposed by their compact genomes and atypical mRNAs, a number of RNA viruses have evolved various non-translational mechanisms that in some ways achieve similar results. As discussed previously, many viruses make use of sgRNAs, segmented genomes and post-translationally cleaved polyproteins in order to express the multiple proteins necessary for their replicative cycle. In addition, retro-transcribing viruses, besides a small proportion of RNA viruses, including orthomyxoviruses (e.g. influenzavirus) and bornaviruses, enter the host-cell nucleus where they make use of the host-cell splicing machinery for producing alternative transcripts.
An interesting parallel may be drawn between ribosomal frameshifting and a completely different mechanism – transcriptional slippage – that is utilized by several monopartite negative-strand RNA viruses (order Mononegavirales), notably viruses in the subfamily Paramyxovirinae, which includes measles virus (genus Morbillivirus), mumps virus (genus Rubulavirus) and parainfluenzaviruses (genus Respirovirus). In these viruses, programmed polymerase slippage or stuttering occurs at a specific site (3′-UnCm-5′, transcribed to AnGm in the mRNA; n+m≥8) during transcription of the phosphoprotein mRNAs, leading to the efficient insertion of one or more extra Gs in a proportion of transcripts (reviewed by Kolakofsky et al., 2005). Thus, translation of the mRNAs leads to the production of different N-terminally coincident, C-terminally distinct proteins, with the relative proportion determined by species-specific details of the stuttering site. Transcriptional slippage is also used in the glycoprotein gene of ebolaviruses (family Filoviridae, order Mononegavirales; Sanchez et al., 1996; Volchkova et al., 2011).
A similar parallel may be made between translational codon redefinition (including stop-codon readthrough) and programmed RNA editing. The most common form of RNA editing in vertebrates is editing of adenosine to inosine by ADARs (adenosine deaminases acting on RNA; reviewed by Wulff & Nishikura, 2010). Inosine is read by the translational apparatus as guanosine; thus, for example, a templated UAG stop codon may subsequently be edited to UIG in a proportion of mRNA transcripts and translated as tryptophan. A slight variation of this mechanism is utilized by hepatitis delta virus – a subviral RNA satellite that replicates nuclearly and is dependent on hepatitis B virus for envelope proteins and, unusually, on cellular (normally DNA-dependent) RNA polymerase II for replication and transcription. The hepatitis delta virus genome is only known to encode one protein, known as δAg (delta antigen), which is translated in two forms of 24 and 27 kDa, both of which are essential (reviewed by Taylor, 2006). The larger form, which is required for virion assembly, has a 19–20 aa C-terminal extension and is produced late in infection upon ADAR editing of the coding-sense antigenome. The edit is copied to new genome-sense RNA and leads to the replacement of a UAG stop codon with a UGG tryptophan codon in transcribed mRNAs (Polson et al., 1996).
Concluding remarks
Together, it is clear that RNA viruses provide a fascinating plethora of examples of non-standard mechanisms for gene expression. Traditionally, virus research has focused on those species that are most relevant to humankind – that is, human viruses and the viruses of commercially important plants and animals, besides viruses of human parasites such as Leishmania and Trichomonas, and viruses of a few model organisms such as yeast and Drosophila. Recent years, however, have seen an explosion in the rate of acquisition of new sequencing data and, as sequencing turns more to environmental samples, there is the opportunity to sample viruses from a much larger diversity of hosts (e.g. diverse fungi, insects and protists). Some such viruses are highly divergent from known viruses and are likely to provide examples of new translational mechanisms, besides many new variations of previously identified mechanisms. This is particularly so for viruses of organisms (or organelles) with unusual components of the translational apparatus, such as unusual ribosomes or unusual tRNA types and abundances. Other newly discovered viruses are related more closely to known viruses but can provide a broader phylogenetic baseline for computational comparative analyses that can be used to detect undiscovered elements in economically and medically important species.
The discovery of novel translational elements is increasingly driven by bioinformatic analysis of sequence databases, but new data resources (e.g. whole-proteome mass spectrometry and whole-transcriptome ribosome profiling; Ingolia et al., 2011) will play an increasingly important role. Although non-canonical translation appears to be less significant in DNA viruses and cellular genes, there are still many such examples ranging from the rather common presence of short ORFs in 5′ leaders to the highly conserved and functionally critical frameshift sites in release factor 2 and antizyme genes. The identification and characterization of novel types of non-canonical translation in RNA viruses will aid in cellular genome annotation by building a catalogue of biologically feasible mechanisms, and generating ‘search patterns’ that can be used as part of automated annotation pipelines for both cellular and viral genomes. In this review, we have included some examples where experimental details remain uncertain (e.g. hypothesized −2 frameshifting in a trichomonasvirus and +2 frameshifting in potyviruses) because we believe that it is useful for annotators of viral genomes to be aware of such possibilities.
Exceptions to the canonical translational rules can be programmed or incidental. The latter may be thought of as translational noise and occur in probably all genes for a small proportion of translating ribosomes. However, they are not subject to strong purifying selection so are generally not phylogenetically preserved over significant evolutionary distances. On the other hand, programmed exceptions generally (though not always) involve a significant proportion of translating ribosomes and tend to be subject to strong purifying selection and phylogenetic conservation. Not surprisingly, there are continua in the dimensions of efficiency, functionality and evolutionary conservation, and it is not always obvious whether a given case of non-canonical translation is programmed or incidental.
The study of non-canonical translation can lead to the development of extremely valuable tools for molecular biological research and biotechnology. A case in point is the use of stop–carry on cassettes for equimolar co-expression of multiple proteins from a single transcript. Non-canonical translational mechanisms (in particular frameshifting in HIV) have also been proposed as potential targets for antiviral drugs. This is particularly attractive if it can be demonstrated that a given mechanism is not utilized for host gene expression. Finally, by acquiring a greater understanding of the extent to which the translational machinery can be subverted from canonical cap-dependent scanning initiation and triplet decoding, and the mechanisms for achieving such subversions, one will also develop a greater understanding of the canonical mechanisms of eukaryotic translation – arguably (together with counterparts in bacteria and archaea) the most important process in the modern biosphere.
Acknowledgements
The authors would like to thank T. D. K. Brown and W. A. Miller for critical review of the manuscript and, especially, J. F. Atkins for unceasing support, encouragement, and countless fruitful discussions. The authors apologize to those whose work has not been cited or discussed due to space limitations. A. E. F. is supported by a grant from the Wellcome Trust (088789). I. B. is funded by grants from the Biotechnology and Biological Sciences Research Council, UK (BB/G008205/1, BB/G020272/1).
References
- Adams M. J., Antoniw J. F., Mullins J. G. (2001). Plant virus transmission by plasmodiophorid fungi is associated with distinctive transmembrane regions of virus-encoded proteins. Arch Virol 146, 1139–1153 10.1007/s007050170111 [DOI] [PubMed] [Google Scholar]
- Agranovsky A. A., Koonin E. V., Boyko V. P., Maiss E., Frötschl R., Lunina N. A., Atabekov J. G. (1994). Beet yellows closterovirus: complete genome structure and identification of a leader papain-like thiol protease. Virology 198, 311–324 10.1006/viro.1994.1034 [DOI] [PubMed] [Google Scholar]
- Ahlquist P. (2006). Parallels among positive-strand RNA viruses, reverse-transcribing viruses and double-stranded RNA viruses. Nat Rev Microbiol 4, 371–382 10.1038/nrmicro1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmadian G., Randhawa J. S., Easton A. J. (2000). Expression of the ORF-2 protein of the human respiratory syncytial virus M2 gene is initiated by a ribosomal termination-dependent reinitiation mechanism. EMBO J 19, 2681–2689 10.1093/emboj/19.11.2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam S. L., Wills N. M., Ingram J. A., Atkins J. F., Gesteland R. F. (1999). Structural studies of the RNA pseudoknot required for readthrough of the gag-termination codon of murine leukemia virus. J Mol Biol 288, 837–852 10.1006/jmbi.1999.2713 [DOI] [PubMed] [Google Scholar]
- Atkins J. F., Gesteland R. F. (2010). Recoding: Expansion of Decoding Rules Enriches Gene Expression. Heidelberg: Springer [Google Scholar]
- Balvay L., Soto Rifo R., Ricci E. P., Decimo D., Ohlmann T. (2009). Structural and functional diversity of viral IRESes. Biochim Biophys Acta 1789, 542–557 [DOI] [PubMed] [Google Scholar]
- Barry J. K., Miller W. A. (2002). A −1 ribosomal frameshift element that requires base pairing across four kilobases suggests a mechanism of regulating ribosome and replicase traffic on a viral RNA. Proc Natl Acad Sci U S A 99, 11133–11138 10.1073/pnas.162223099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier H., Grimm M. (2001). Misreading of termination codons in eukaryotes by natural nonsense suppressor tRNAs. Nucleic Acids Res 29, 4767–4782 10.1093/nar/29.23.4767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsham G. J. (2005). Translation and replication of FMDV RNA. Curr Top Microbiol Immunol 288, 43–70 10.1007/3-540-27109-0_3 [DOI] [PubMed] [Google Scholar]
- Belsham G. J. (2009). Divergent picornavirus IRES elements. Virus Res 139, 183–192 10.1016/j.virusres.2008.07.001 [DOI] [PubMed] [Google Scholar]
- Bertram G., Innes S., Minella O., Richardson J., Stansfield I. (2001). Endless possibilities: translation termination and stop codon recognition. Microbiology 147, 255–269 [DOI] [PubMed] [Google Scholar]
- Boeck R., Curran J., Matsuoka Y., Compans R., Kolakofsky D. (1992). The parainfluenza virus type 1 P/C gene uses a very efficient GUG codon to start its C′ protein. J Virol 66, 1765–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonetti B., Fu L., Moon J., Bedwell D. M. (1995). The efficiency of translation termination is determined by a synergistic interplay between upstream and downstream sequences in Saccharomyces cerevisiae. J Mol Biol 251, 334–345 10.1006/jmbi.1995.0438 [DOI] [PubMed] [Google Scholar]
- Boros Á., Pankovics P., Simmonds P., Reuter G. (2011). Novel positive-sense, single-stranded RNA (+ssRNA) virus with di-cistronic genome from intestinal content of freshwater carp (Cyprinus carpio). PLoS One 6, e29145 10.1371/journal.pone.0029145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brault V., van den Heuvel J. F., Verbeek M., Ziegler-Graff V., Reutenauer A., Herrbach E., Garaud J. C., Guilley H., Richards K., Jonard G. (1995). Aphid transmission of beet western yellows luteovirus requires the minor capsid read-through protein P74. EMBO J 14, 650–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Gilbert R. J. C., Pennell S. (2010). Pseudoknot-dependent programmed −1 ribosomal frameshifting: structures, mechanisms and models. In Recoding: Expansion of Decoding Rules Enriches Gene Expression, pp. 149–174 Edited by Atkins J. F., Gesteland R. F. Heidelberg: Springer; 10.1007/978-0-387-89382-2_7 [DOI] [Google Scholar]
- Brown J. D., Ryan M. D. (2010). Ribosome “skipping”: “stop-carry on” or “StopGo” translation. In Recoding: Expansion of Decoding Rules Enriches Gene Expression, pp. 101–121 Edited by Atkins J. F., Gesteland R. F. Heidelberg: Springer; 10.1007/978-0-387-89382-2_5 [DOI] [Google Scholar]
- Brown C. M., Dinesh-Kumar S. P., Miller W. A. (1996). Local and distant sequences are required for efficient readthrough of the barley yellow dwarf virus PAV coat protein gene stop codon. J Virol 70, 5884–5892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao F., Tavis J. E. (2011). RNA elements directing translation of the duck hepatitis B virus polymerase via ribosomal shunting. J Virol 85, 6343–6352 10.1128/JVI.00101-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaño A., Ruiz L., Hernández C. (2009). Insights into the translational regulation of biologically active open reading frames of Pelargonium line pattern virus. Virology 386, 417–426 10.1016/j.virol.2009.01.017 [DOI] [PubMed] [Google Scholar]
- Çevik B. (2001). Characterization of the RNA-dependent RNA polymerase gene of citrus tristeza closterovirus PhD dissertation, University of Florida. [Google Scholar]
- Chamond N., Locker N., Sargueil B. (2010). The different pathways of HIV genomic RNA translation. Biochem Soc Trans 38, 1548–1552 10.1042/BST0381548 [DOI] [PubMed] [Google Scholar]
- Chaudhry Y., Nayak A., Bordeleau M. E., Tanaka J., Pelletier J., Belsham G. J., Roberts L. O., Goodfellow I. G. (2006). Caliciviruses differ in their functional requirements for eIF4F components. J Biol Chem 281, 25315–25325 10.1074/jbc.M602230200 [DOI] [PubMed] [Google Scholar]
- Chen W., Calvo P. A., Malide D., Gibbs J., Schubert U., Bacik I., Basta S., O’Neill R., Schickli J. & other authors (2001). A novel influenza A virus mitochondrial protein that induces cell death. Nat Med 7, 1306–1312 10.1038/nm1201-1306 [DOI] [PubMed] [Google Scholar]
- Choi I. R., Hall J. S., Henry M., Zhang L., Hein G. L., French R., Stenger D. C. (2001). Contributions of genetic drift and negative selection on the evolution of three strains of wheat streak mosaic tritimovirus. Arch Virol 146, 619–628 10.1007/s007050170167 [DOI] [PubMed] [Google Scholar]
- Chung B. Y., Miller W. A., Atkins J. F., Firth A. E. (2008). An overlapping essential gene in the Potyviridae. Proc Natl Acad Sci U S A 105, 5897–5902 10.1073/pnas.0800468105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimino P. A., Nicholson B. L., Wu B., Xu W., White K. A. (2011). Multifaceted regulation of translational readthrough by RNA replication elements in a tombusvirus. PLoS Pathog 7, e1002423 10.1371/journal.ppat.1002423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyde K., Harris E. (2006). RNA secondary structure in the coding region of dengue virus type 2 directs translation start codon selection and is required for viral replication. J Virol 80, 2170–2182 10.1128/JVI.80.5.2170-2182.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran J., Kolakofsky D. (1988). Ribosomal initiation from an ACG codon in the Sendai virus P/C mRNA. EMBO J 7, 245–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughenbaugh K. F., Wobus C. E., Hardy M. E. (2006). VPg of murine norovirus binds translation initiation factors in infected cells. Virol J 3, 33 10.1186/1743-422X-3-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Breyne S., Monney R. S., Curran J. (2004). Proteolytic processing and translation initiation: two independent mechanisms for the expression of the Sendai virus Y proteins. J Biol Chem 279, 16571–16580 10.1074/jbc.M312391200 [DOI] [PubMed] [Google Scholar]
- Dinesh-Kumar S. P., Miller W. A. (1993). Control of start codon choice on a plant viral RNA encoding overlapping genes. Plant Cell 5, 679–692 10.1105/tpc.5.6.679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly M. L., Luke G., Mehrotra A., Li X., Hughes L. E., Gani D., Ryan M. D. (2001). Analysis of the aphthovirus 2A/2B polyprotein ‘cleavage’ mechanism indicates not a proteolytic reaction, but a novel translational effect: a putative ribosomal ‘skip’. J Gen Virol 82, 1013–1025 [DOI] [PubMed] [Google Scholar]
- Doronina V. A., Wu C., de Felipe P., Sachs M. S., Ryan M. D., Brown J. D. (2008). Site-specific release of nascent chains from ribosomes at a sense codon. Mol Cell Biol 28, 4227–4239 10.1128/MCB.00421-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher T. W., Miller W. A. (2006). Translational control in positive strand RNA plant viruses. Virology 344, 185–197 10.1016/j.virol.2005.09.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulude D., Berchiche Y. A., Gendron K., Brakier-Gingras L., Heveker N. (2006). Decreasing the frameshift efficiency translates into an equivalent reduction of the replication of the human immunodeficiency virus type 1. Virology 345, 127–136 10.1016/j.virol.2005.08.048 [DOI] [PubMed] [Google Scholar]
- Edgil D., Harris E. (2006). End-to-end communication in the modulation of translation by mammalian RNA viruses. Virus Res 119, 43–51 10.1016/j.virusres.2005.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst H., Shatkin A. J. (1985). Reovirus hemagglutinin mRNA codes for two polypeptides in overlapping reading frames. Proc Natl Acad Sci U S A 82, 48–52 10.1073/pnas.82.1.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth A. E., Chung B. Y., Fleeton M. N., Atkins J. F. (2008). Discovery of frameshifting in alphavirus 6K resolves a 20-year enigma. Virol J 5, 108 10.1186/1743-422X-5-108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth A. E., Wills N. M., Gesteland R. F., Atkins J. F. (2011). Stimulation of stop codon readthrough: frequent presence of an extended 3′ RNA structural element. Nucleic Acids Res 39, 6679–6691 10.1093/nar/gkr224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller F., Bhown A. S., Bishop D. H. (1983). Bunyavirus nucleoprotein, N, and a non-structural protein, NSs, are coded by overlapping reading frames in the S RNA. J Gen Virol 64, 1705–1714 10.1099/0022-1317-64-8-1705 [DOI] [PubMed] [Google Scholar]
- Fütterer J., Hohn T. (1991). Translation of a polycistronic mRNA in the presence of the cauliflower mosaic virus transactivator protein. EMBO J 10, 3887–3896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fütterer J., Kiss-László Z., Hohn T. (1993). Nonlinear ribosome migration on cauliflower mosaic virus 35S RNA. Cell 73, 789–802 10.1016/0092-8674(93)90257-Q [DOI] [PubMed] [Google Scholar]
- Fütterer J., Potrykus I., Bao Y., Li L., Burns T. M., Hull R., Hohn T. (1996). Position-dependent ATT initiation during plant pararetrovirus rice tungro bacilliform virus translation. J Virol 70, 2999–3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fütterer J., Rothnie H. M., Hohn T., Potrykus I. (1997). Rice tungro bacilliform virus open reading frames II and III are translated from polycistronic pregenomic RNA by leaky scanning. J Virol 71, 7984–7989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs A. J., Keese P. K. (1995). In search of the origins of viral genes. In Molecular Basis of Virus Evolution, pp. 76–90 Edited by Gibbs A. J., Calisher C. H., Garcia-Arenal F. Cambridge: Cambridge University Press; 10.1017/CBO9780511661686.008 [DOI] [Google Scholar]
- Giedroc D. P., Cornish P. V. (2009). Frameshifting RNA pseudoknots: structure and mechanism. Virus Res 139, 193–208 10.1016/j.virusres.2008.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman R. P., Freret T. S., Kula T., Geller A. M., Talkington M. W., Tang-Fernandez V., Suciu O., Demidenko A. A., Ghabrial S. A. & other authors (2011). Clinical isolates of Trichomonas vaginalis concurrently infected by strains of up to four Trichomonasvirus species (family Totiviridae). J Virol 85, 4258–4270 10.1128/JVI.00220-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould P. S., Easton A. J. (2007). Coupled translation of the second open reading frame of M2 mRNA is sequence dependent and differs significantly within the subfamily Pneumovirinae. J Virol 81, 8488–8496 10.1128/JVI.00457-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmings-Mieszczak M., Hohn T., Preiss T. (2000). Termination and peptide release at the upstream open reading frame are required for downstream translation on synthetic shunt-competent mRNA leaders. Mol Cell Biol 20, 6212–6223 10.1128/MCB.20.17.6212-6223.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert T. P., Brierley I., Brown T. D. (1996). Detection of the ORF3 polypeptide of feline calicivirus in infected cells and evidence for its expression from a single, functionally bicistronic, subgenomic mRNA. J Gen Virol 77, 123–127 10.1099/0022-1317-77-1-123 [DOI] [PubMed] [Google Scholar]
- Horvath C. M., Williams M. A., Lamb R. A. (1990). Eukaryotic coupled translation of tandem cistrons: identification of the influenza B virus BM2 polypeptide. EMBO J 9, 2639–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houck-Loomis B., Durney M. A., Salguero C., Shankar N., Nagle J. M., Goff S. P., D’Souza V. M. (2011). An equilibrium-dependent retroviral mRNA switch regulates translational recoding. Nature 480, 561–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia N. T., Lareau L. F., Weissman J. S. (2011). Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147, 789–802 10.1016/j.cell.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I. P., Loughran G., Atkins J. F. (2008). uORFs with unusual translational start codons autoregulate expression of eukaryotic ornithine decarboxylase homologs. Proc Natl Acad Sci U S A 105, 10079–10084 10.1073/pnas.0801590105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T., Varmus H. E. (1985). Expression of the Rous sarcoma virus pol gene by ribosomal frameshifting. Science 230, 1237–1242 10.1126/science.2416054 [DOI] [PubMed] [Google Scholar]
- Jacks T., Madhani H. D., Masiarz F. R., Varmus H. E. (1988). Signals for ribosomal frameshifting in the Rous sarcoma virus gag–pol region. Cell 55, 447–458 10.1016/0092-8674(88)90031-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson R. J., Hellen C. U., Pestova T. V. (2010). The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11, 113–127 10.1038/nrm2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson R. J., Hellen C. U., Pestova T. V. (2012). Termination and post-termination events in eukaryotic translation. Adv Protein Chem Struct Biol 86, 45–93 10.1016/B978-0-12-386497-0.00002-5 [DOI] [PubMed] [Google Scholar]
- Jang S. K., Kräusslich H. G., Nicklin M. J., Duke G. M., Palmenberg A. C., Wimmer E. (1988). A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol 62, 2636–2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang C. J., Lo M. C., Jan E. (2009). Conserved element of the dicistrovirus IGR IRES that mimics an E-site tRNA/ribosome interaction mediates multiple functions. J Mol Biol 387, 42–58 10.1016/j.jmb.2009.01.042 [DOI] [PubMed] [Google Scholar]
- Kanyuka K. V., Vishnichenko V. K., Levay K. E., Kondrikov D. Y., Ryabov E. V., Zavriev S. K. (1992). Nucleotide sequence of shallot virus X RNA reveals a 5′-proximal cistron closely related to those of potexviruses and a unique arrangement of the 3′-proximal cistrons. J Gen Virol 73, 2553–2560 10.1099/0022-1317-73-10-2553 [DOI] [PubMed] [Google Scholar]
- Karasev A. V., Boyko V. P., Gowda S., Nikolaeva O. V., Hilf M. E., Koonin E. V., Niblett C. L., Cline K., Gumpf D. J. & other authors (1995). Complete sequence of the citrus tristeza virus RNA genome. Virology 208, 511–520 10.1006/viro.1995.1182 [DOI] [PubMed] [Google Scholar]
- Kieft J. S. (2008). Viral IRES RNA structures and ribosome interactions. Trends Biochem Sci 33, 274–283 10.1016/j.tibs.2008.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. N., Choi J. H., Park M. W., Jeong S. J., Han K. S., Kim H. J. (2005). Identification of the +1 ribosomal frameshifting site of LRV1-4 by mutational analysis. Arch Pharm Res 28, 956–962 10.1007/BF02973883 [DOI] [PubMed] [Google Scholar]
- Kneller E. L., Rakotondrafara A. M., Miller W. A. (2006). Cap-independent translation of plant viral RNAs. Virus Res 119, 63–75 10.1016/j.virusres.2005.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolakofsky D., Roux L., Garcin D., Ruigrok R. W. (2005). Paramyxovirus mRNA editing, the “rule of six” and error catastrophe: a hypothesis. J Gen Virol 86, 1869–1877 10.1099/vir.0.80986-0 [DOI] [PubMed] [Google Scholar]
- Kong W. P., Roos R. P. (1991). Alternative translation initiation site in the DA strain of Theiler’s murine encephalomyelitis virus. J Virol 65, 3395–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. (1986). Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 44, 283–292 10.1016/0092-8674(86)90762-2 [DOI] [PubMed] [Google Scholar]
- Kozak M. (1990). Downstream secondary structure facilitates recognition of initiator codons by eukaryotic ribosomes. Proc Natl Acad Sci U S A 87, 8301–8305 10.1073/pnas.87.21.8301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. (1991). A short leader sequence impairs the fidelity of initiation by eukaryotic ribosomes. Gene Expr 1, 111–115 [PMC free article] [PubMed] [Google Scholar]
- Kozak M. (2002). Pushing the limits of the scanning mechanism for initiation of translation. Gene 299, 1–34 10.1016/S0378-1119(02)01056-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. (2004). How strong is the case for regulation of the initiation step of translation by elements at the 3′ end of eukaryotic mRNAs? Gene 343, 41–54 10.1016/j.gene.2004.08.011 [DOI] [PubMed] [Google Scholar]
- Kozak M. (2007). Lessons (not) learned from mistakes about translation. Gene 403, 194–203 10.1016/j.gene.2007.08.017 [DOI] [PubMed] [Google Scholar]
- Laprevotte I., Hampe A., Sherr C. J., Galibert F. (1984). Nucleotide sequence of the gag gene and gag–pol junction of feline leukemia virus. J Virol 50, 884–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. Z., Qu F., Morris T. J. (1998). Cell-to-cell movement of turnip crinkle virus is controlled by two small open reading frames that function in trans. Virology 244, 405–416 10.1006/viro.1998.9125 [DOI] [PubMed] [Google Scholar]
- Li H., Havens W. M., Nibert M. L., Ghabrial S. A. (2011). RNA sequence determinants of a coupled termination-reinitiation strategy for downstream open reading frame translation in Helminthosporium victoriae virus 190S and other victoriviruses (family Totiviridae). J Virol 85, 7343–7352 10.1128/JVI.00364-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughran G., Firth A. E., Atkins J. F. (2011). Ribosomal frameshifting into an overlapping gene in the 2B-encoding region of the cardiovirus genome. Proc Natl Acad Sci U S A 108, E1111–E1119 10.1073/pnas.1102932108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukavsky P. J. (2009). Structure and function of HCV IRES domains. Virus Res 139, 166–171 10.1016/j.virusres.2008.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke G. A., de Felipe P., Lukashev A., Kallioinen S. E., Bruno E. A., Ryan M. D. (2008). Occurrence, function and evolutionary origins of ‘2A-like’ sequences in virus genomes. J Gen Virol 89, 1036–1042 10.1099/vir.0.83428-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttermann C., Meyers G. (2007). A bipartite sequence motif induces translation reinitiation in feline calicivirus RNA. J Biol Chem 282, 7056–7065 10.1074/jbc.M608948200 [DOI] [PubMed] [Google Scholar]
- Luttermann C., Meyers G. (2009). The importance of inter- and intramolecular base pairing for translation reinitiation on a eukaryotic bicistronic mRNA. Genes Dev 23, 331–344 10.1101/gad.507609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R. R., Zhou J., Tzanetakis I. E. (2011). Blueberry latent virus: an amalgam of the Partitiviridae and Totiviridae. Virus Res 155, 175–180 10.1016/j.virusres.2010.09.020 [DOI] [PubMed] [Google Scholar]
- Matsuda D., Dreher T. W. (2006). Close spacing of AUG initiation codons confers dicistronic character on a eukaryotic mRNA. RNA 12, 1338–1349 10.1261/rna.67906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaughan K. K., Brown C. M., Dalphin M. E., Berry M. J., Tate W. P. (1995). Translational termination efficiency in mammals is influenced by the base following the stop codon. Proc Natl Acad Sci U S A 92, 5431–5435 10.1073/pnas.92.12.5431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick C. J., Salim O., Lambden P. R., Clarke I. N. (2008). Translation termination reinitiation between open reading frame 1 (ORF1) and ORF2 enables capsid expression in a bovine norovirus without the need for production of viral subgenomic RNA. J Virol 82, 8917–8921 10.1128/JVI.02362-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden N., Bailey D., Carrara G., Benson A., Chaudhry Y., Shortland A., Heeney J., Yarovinsky F., Simmonds P. & other authors (2011). Norovirus regulation of the innate immune response and apoptosis occurs via the product of the alternative open reading frame 4. PLoS Pathog 7, e1002413 10.1371/journal.ppat.1002413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melian E. B., Hinzman E., Nagasaki T., Firth A. E., Wills N. M., Nouwens A. S., Blitvich B. J., Leung J., Funk A. & other authors (2010). NS1′ of flaviviruses in the Japanese encephalitis virus serogroup is a product of ribosomal frameshifting and plays a role in viral neuroinvasiveness. J Virol 84, 1641–1647 10.1128/JVI.01979-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers G. (2003). Translation of the minor capsid protein of a calicivirus is initiated by a novel termination-dependent reinitiation mechanism. J Biol Chem 278, 34051–34060 10.1074/jbc.M304874200 [DOI] [PubMed] [Google Scholar]
- Miller W. A., White K. A. (2006). Long-distance RNA–RNA interactions in plant virus gene expression and replication. Annu Rev Phytopathol 44, 447–467 10.1146/annurev.phyto.44.070505.143353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller W. A., Wang Z., Treder K. (2007). The amazing diversity of cap-independent translation elements in the 3′-untranslated regions of plant viral RNAs. Biochem Soc Trans 35, 1629–1633 10.1042/BST0351629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyauchi K., Komano J., Myint L., Futahashi Y., Urano E., Matsuda Z., Chiba T., Miura H., Sugiura W., Yamamoto N. (2006). Rapid propagation of low-fitness drug-resistant mutants of human immunodeficiency virus type 1 by a streptococcal metabolite sparsomycin. Antivir Chem Chemother 17, 167–174 [DOI] [PubMed] [Google Scholar]
- Morozov S. Y., Solovyev A. G. (2003). Triple gene block: modular design of a multifunctional machine for plant virus movement. J Gen Virol 84, 1351–1366 10.1099/vir.0.18922-0 [DOI] [PubMed] [Google Scholar]
- Morris D. R., Geballe A. P. (2000). Upstream open reading frames as regulators of mRNA translation. Mol Cell Biol 20, 8635–8642 10.1128/MCB.20.23.8635-8642.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S., Niimura Y., Gojobori T., Tanaka H., Miura K. (2008). Diversity of preferred nucleotide sequences around the translation initiation codon in eukaryote genomes. Nucleic Acids Res 36, 861–871 10.1093/nar/gkm1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namy O., Rousset J. P. (2010). Specification of standard amino acids by stop codons. In Recoding: Expansion of Decoding Rules Enriches Gene Expression, pp. 79–100 Edited by Atkins J. F., Gesteland R. F. Heidelberg: Springer; 10.1007/978-0-387-89382-2_4 [DOI] [Google Scholar]
- Namy O., Moran S. J., Stuart D. I., Gilbert R. J., Brierley I. (2006). A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. Nature 441, 244–247 10.1038/nature04735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napthine S., Lever R. A., Powell M. L., Jackson R. J., Brown T. D., Brierley I. (2009). Expression of the VP2 protein of murine norovirus by a translation termination–reinitiation strategy. PLoS One 4, e8390 10.1371/journal.pone.0008390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napthine S., Yek C., Powell M. L., Brown T. D., Brierley I. (2012). Characterization of the stop codon readthrough signal of Colorado tick fever virus segment 9 RNA. RNA 18, 241–252 10.1261/rna.030338.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson B. L., White K. A. (2011). 3′ Cap-independent translation enhancers of positive-strand RNA plant viruses. Curr Opin Virol 1, 373–380 10.1016/j.coviro.2011.10.002 [DOI] [PubMed] [Google Scholar]
- Nitta T., Kuznetsov Y., McPherson A., Fan H. (2010). Murine leukemia virus glycosylated Gag (gPr80gag) facilitates interferon-sensitive virus release through lipid rafts. Proc Natl Acad Sci U S A 107, 1190–1195 10.1073/pnas.0908660107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle J. M., Brodersen D. E., Clemons W. M., Jr, Tarry M. J., Carter A. P., Ramakrishnan V. (2001). Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292, 897–902 10.1126/science.1060612 [DOI] [PubMed] [Google Scholar]
- Olivier V., Blanchard P., Chaouch S., Lallemand P., Schurr F., Celle O., Dubois E., Tordo N., Thiéry R. & other authors (2008). Molecular characterisation and phylogenetic analysis of chronic bee paralysis virus, a honey bee virus. Virus Res 132, 59–68 10.1016/j.virusres.2007.10.014 [DOI] [PubMed] [Google Scholar]
- Pelletier J., Sonenberg N. (1988). Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334, 320–325 10.1038/334320a0 [DOI] [PubMed] [Google Scholar]
- Plant E. P., Dinman J. D. (2005). Torsional restraint: a new twist on frameshifting pseudoknots. Nucleic Acids Res 33, 1825–1833 10.1093/nar/gki329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plant E. P., Rakauskaite R., Taylor D. R., Dinman J. D. (2010). Achieving a golden mean: mechanisms by which coronaviruses ensure synthesis of the correct stoichiometric ratios of viral proteins. J Virol 84, 4330–4340 10.1128/JVI.02480-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polson A. G., Bass B. L., Casey J. L. (1996). RNA editing of hepatitis delta virus antigenome by dsRNA–adenosine deaminase. Nature 380, 454–456 10.1038/380454a0 [DOI] [PubMed] [Google Scholar]
- Pooggin M. M., Fütterer J., Skryabin K. G., Hohn T. (1999). A short open reading frame terminating in front of a stable hairpin is the conserved feature in pregenomic RNA leaders of plant pararetroviruses. J Gen Virol 80, 2217–2228 [DOI] [PubMed] [Google Scholar]
- Pooggin M. M., Ryabova L. A., He X., Fütterer J., Hohn T. (2006). Mechanism of ribosome shunting in rice tungro bacilliform pararetrovirus. RNA 12, 841–850 10.1261/rna.2285806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell M. L. (2010). Translational termination-reinitiation in RNA viruses. Biochem Soc Trans 38, 1558–1564 10.1042/BST0381558 [DOI] [PubMed] [Google Scholar]
- Powell M. L., Leigh K. E., Pöyry T. A., Jackson R. J., Brown T. D., Brierley I. (2011). Further characterisation of the translational termination-reinitiation signal of the influenza B virus segment 7 RNA. PLoS One 6, e16822 10.1371/journal.pone.0016822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöyry T. A., Kaminski A., Connell E. J., Fraser C. S., Jackson R. J. (2007). The mechanism of an exceptional case of reinitiation after translation of a long ORF reveals why such events do not generally occur in mammalian mRNA translation. Genes Dev 21, 3149–3162 10.1101/gad.439507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prats A. C., De Billy G., Wang P., Darlix J. L. (1989). CUG initiation codon used for the synthesis of a cell surface antigen coded by the murine leukemia virus. J Mol Biol 205, 363–372 10.1016/0022-2836(89)90347-1 [DOI] [PubMed] [Google Scholar]
- Qu X., Wen J. D., Lancaster L., Noller H. F., Bustamante C., Tinoco I., Jr (2011). The ribosome uses two active mechanisms to unwind messenger RNA during translation. Nature 475, 118–121 10.1038/nature10126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racine T., Duncan R. (2010). Facilitated leaky scanning and atypical ribosome shunting direct downstream translation initiation on the tricistronic S1 mRNA of avian reovirus. Nucleic Acids Res 38, 7260–7272 10.1093/nar/gkq611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rancurel C., Khosravi M., Dunker A. K., Romero P. R., Karlin D. (2009). Overlapping genes produce proteins with unusual sequence properties and offer insight into de novo protein creation. J Virol 83, 10719–10736 10.1128/JVI.00595-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raoult D., Forterre P. (2008). Redefining viruses: lessons from mimivirus. Nat Rev Microbiol 6, 315–319 10.1038/nrmicro1858 [DOI] [PubMed] [Google Scholar]
- Ren Q., Wang Q. S., Firth A. E., Chan M. M., Gouw J. W., Guarna M. M., Foster L. J., Atkins J. F., Jan E. (2012). Alternative reading frame selection mediated by a tRNA-like domain of an internal ribosome entry site. Proc Natl Acad Sci U S A 109, E630–E639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez A., Trappier S. G., Mahy B. W., Peters C. J., Nichol S. T. (1996). The virion glycoproteins of Ebola viruses are encoded in two reading frames and are expressed through transcriptional editing. Proc Natl Acad Sci U S A 93, 3602–3607 10.1073/pnas.93.8.3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheets K., Blinkova O., Melcher U., Palmer M. W., Wiley G. B., Ding T., Roe B. A. (2011). Detection of members of the Tombusviridae in the Tallgrass Prairie Preserve, Osage County, Oklahoma, USA. Virus Res 160, 256–263 10.1016/j.virusres.2011.06.023 [DOI] [PubMed] [Google Scholar]
- Schepetilnikov M., Schott G., Katsarou K., Thiébeauld O., Keller M., Ryabova L. A. (2009). Molecular dissection of the prototype foamy virus (PFV) RNA 5′-UTR identifies essential elements of a ribosomal shunt. Nucleic Acids Res 37, 5838–5847 10.1093/nar/gkp609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepetilnikov M., Kobayashi K., Geldreich A., Caranta C., Robaglia C., Keller M., Ryabova L. A. (2011). Viral factor TAV recruits TOR/S6K1 signalling to activate reinitiation after long ORF translation. EMBO J 30, 1343–1356 10.1038/emboj.2011.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Puchta W., Dominguez D., Lewetag D., Hohn T. (1997). Plant ribosome shunting in vitro. Nucleic Acids Res 25, 2854–2860 10.1093/nar/25.14.2854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholthof H. B., Gowda S., Wu F. C., Shepherd R. J. (1992). The full-length transcript of a caulimovirus is a polycistronic mRNA whose genes are trans activated by the product of gene VI. J Virol 66, 3131–3139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedman S. A., Gelembiuk G. W., Mertz J. E. (1990). Translation initiation at a downstream AUG occurs with increased efficiency when the upstream AUG is located very close to the 5′ cap. J Virol 64, 453–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skabkin M. A., Skabkina O. V., Dhote V., Komar A. A., Hellen C. U., Pestova T. V. (2010). Activities of Ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes Dev 24, 1787–1801 10.1101/gad.1957510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skuzeski J. M., Nichols L. M., Gesteland R. F., Atkins J. F. (1991). The signal for a leaky UAG stop codon in several plant viruses includes the two downstream codons. J Mol Biol 218, 365–373 10.1016/0022-2836(91)90718-L [DOI] [PubMed] [Google Scholar]
- Su H. M., Tai J. H. (1996). Genomic organization and sequence conservation in type I Trichomonas vaginalis viruses. Virology 222, 470–473 10.1006/viro.1996.0446 [DOI] [PubMed] [Google Scholar]
- Taylor J. M. (2006). Hepatitis delta virus. Virology 344, 71–76 10.1016/j.virol.2005.09.033 [DOI] [PubMed] [Google Scholar]
- Thiébeauld O., Pooggin M. M., Ryabova L. A. (2007). Alternative translation strategies in plant viruses. Plant Viruses 1, 1–20 [Google Scholar]
- Tork S., Hatin I., Rousset J. P., Fabret C. (2004). The major 5′ determinant in stop codon read-through involves two adjacent adenines. Nucleic Acids Res 32, 415–421 10.1093/nar/gkh201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touriol C., Bornes S., Bonnal S., Audigier S., Prats H., Prats A. C., Vagner S. (2003). Generation of protein isoform diversity by alternative initiation of translation at non-AUG codons. Biol Cell 95, 169–178 10.1016/S0248-4900(03)00033-9 [DOI] [PubMed] [Google Scholar]
- Turina M., Desvoyes B., Scholthof K. B. (2000). A gene cluster encoded by panicum mosaic virus is associated with virus movement. Virology 266, 120–128 10.1006/viro.1999.0069 [DOI] [PubMed] [Google Scholar]
- van der Wilk F., Dullemans A. M., Verbeek M., Van den Heuvel J. F. (1997). Nucleotide sequence and genomic organization of Acyrthosiphon pisum virus. Virology 238, 353–362 10.1006/viro.1997.8835 [DOI] [PubMed] [Google Scholar]
- van Eyll O., Michiels T. (2002). Non-AUG-initiated internal translation of the L* protein of Theiler’s virus and importance of this protein for viral persistence. J Virol 76, 10665–10673 10.1128/JVI.76.21.10665-10673.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilaki N., Mavromara P. (2009). The HCV ARFP/F/core+1 protein: production and functional analysis of an unconventional viral product. IUBMB Life 61, 739–752 10.1002/iub.201 [DOI] [PubMed] [Google Scholar]
- Vera-Otarola J., Solis L., Soto-Rifo R., Ricci E. P., Pino K., Tischler N. D., Ohlmann T., Darlix J. L., López-Lastra M. (2012). The Andes hantavirus NSs protein is expressed from the viral small mRNA by a leaky scanning mechanism. J Virol 86, 2176–2187 10.1128/JVI.06223-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verchot J., Angell S. M., Baulcombe D. C. (1998). In vivo translation of the triple gene block of potato virus X requires two subgenomic mRNAs. J Virol 72, 8316–8320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volchkova V. A., Dolnik O., Martinez M. J., Reynard O., Volchkov V. E. (2011). Genomic RNA editing and its impact on Ebola virus adaptation during serial passages in cell culture and infection of guinea pigs. J Infect Dis 204 (Suppl 3), S941–S946 10.1093/infdis/jir321 [DOI] [PubMed] [Google Scholar]
- Walewski J. L., Keller T. R., Stump D. D., Branch A. D. (2001). Evidence for a new hepatitis C virus antigen encoded in an overlapping reading frame. RNA 7, 710–721 10.1017/S1355838201010111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D., Mohr I. (2011). Viral subversion of the host protein synthesis machinery. Nat Rev Microbiol 9, 860–875 10.1038/nrmicro2655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei T., Zhang C., Hong J., Xiong R., Kasschau K. D., Zhou X., Carrington J. C., Wang A. (2010). Formation of complexes at plasmodesmata for potyvirus intercellular movement is mediated by the viral protein P3N-PIPO. PLoS Pathog 6, e1000962 10.1371/journal.ppat.1000962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells S. E., Hillner P. E., Vale R. D., Sachs A. B. (1998). Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell 2, 135–140 10.1016/S1097-2765(00)80122-7 [DOI] [PubMed] [Google Scholar]
- Wen R. H., Hajimorad M. R. (2010). Mutational analysis of the putative pipo of soybean mosaic virus suggests disruption of PIPO protein impedes movement. Virology 400, 1–7 10.1016/j.virol.2010.01.022 [DOI] [PubMed] [Google Scholar]
- Williams M. A., Lamb R. A. (1989). Effect of mutations and deletions in a bicistronic mRNA on the synthesis of influenza B virus NB and NA glycoproteins. J Virol 63, 28–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson J. E., Pestova T. V., Hellen C. U., Sarnow P. (2000). Initiation of protein synthesis from the A site of the ribosome. Cell 102, 511–520 10.1016/S0092-8674(00)00055-6 [DOI] [PubMed] [Google Scholar]
- Wise H. M., Foeglein A., Sun J., Dalton R. M., Patel S., Howard W., Anderson E. C., Barclay W. S., Digard P. (2009). A complicated message: Identification of a novel PB1-related protein translated from influenza A virus segment 2 mRNA. J Virol 83, 8021–8031 10.1128/JVI.00826-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P. C., Lau S. K., Choi G. K., Huang Y., Teng J. L., Tsoi H. W., Tse H., Yeung M. L., Chan K. H. & other authors (2012). Natural occurrence and characterization of two internal ribosome entry site elements in a novel virus, canine picodicistrovirus, in the picornavirus-like superfamily. J Virol 86, 2797–2808 10.1128/JVI.05481-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulff B. E., Nishikura K. (2010). Substitutional A-to-I RNA editing. Wiley Interdiscip Rev RNA 1, 90–101 [DOI] [PMC free article] [PubMed] [Google Scholar]