Abstract
The Vacuole Membrane Protein 1 -VMP1- is a pancreatitis-associated transmembrane protein whose expression triggers autophagy in several human diseases. In the current study, we unveil the mechanism through which this protein induces autophagosome formation in mammalian cells. We show that VMP1 autophagy-related function requires its 20-aminoacid C-terminus hydrophilic domain (VMP1-AtgD). This is achieved through its direct binding to the BH3 motif of Beclin 1 leading to the formation of a complex with the Class III phosphatidylinositol-3 kinase (PI3K) hVps34, a key positive regulator of autophagy, at the site where autophagosomes are generated. This interaction also concomitantly promotes the dissociation of Bcl-2, an autophagy inhibitor, from Beclin 1. Moreover, we show that the VMP1-Beclin 1-hVps34 complex favors the association of Atg16L1 and LC3 with the autophagosomal membranes. Collectively, these findings reveal that VMP1 expression recruits and activates the Class III PI3K complex at the site of autophagosome formation during mammalian autophagy.
Autophagy is an evolutionarily conserved transport pathway that involves the sequestration and delivery of cytoplasmic material into the lysosome, where it is degraded and recycled1. This catabolic process is involved in the turnover of long-lived proteins and other cellular macromolecules, and it might play a protective role in tumor development, aging, cell death, and intracellular pathogen invasions2,3.
Macroautophagy (hereafter autophagy) involves the formation of double-membrane autophagosomes around the targeted cargoes, which include large structures such as organelles and protein aggregates. Autophagosomes then fuse with lysosomes exposing their cargoes to the hydrolytic content of this organelles4. This cellular process essential to maintain cellular homeostasis is regulated in an analogous manner to secretion and endocytosis, where related molecules on distinct organelle membranes mediate the flux of vesicular transport by protein-protein interactions. Since the discovery of yeast autophagy-related (Atg) proteins5, autophagosome formation has been dissected at the molecular level but a lot of questions about the molecular mechanism underlying this process remain unanswered. Autophagosomes can be considered unique organelles because they do not contain marker proteins of other subcellular compartments6. In mammalian cells, the sequential association of at least a subset of the Atg proteins leads to the assembly of the pre-autophagosomal structures (PAS), which is believed to be the site where the precursor structure of the autophagosomes, the phagophores, are generated7. The PAS and phagophore formation also requires phosphatidylinositol 3-phosphate (PI3P)8 and it is believed to be associated to specific subdomains of the endoplasmic reticulum (ER) termed omegasomes9,10. Among the key mediators initiating autophagosome formation, there is a set of evolutionarily conserved Atg. gene products; the kinase-containing Ulk1/2 complex (Atg1 in yeast), the Class III phosphatidylinositol 3-kinase (Class III PI3K) complex (composed by Beclin 1/Atg6-hVps34, hVps15 and Atg14L), the ubiquitin-like conjugation systems leading to the formation of the Atg5–Atg12–Atg16L1 complex and the LC3/Atg8 phosphatidylethanolamine- conjugate (e.g. LC3-II)11. A second group of Atg proteins, which does not have orthologous in yeast, has also recently emerged and appear to play a key role in regulating autophagy in high eukaryotes. One of these proteins is the transmembrane Vacuole-Membrane-Protein-1 (VMP1), whose expression triggers autophagy in mammalian cells even under nutrient-rich conditions12,13,14. Conversely, autophagy is completely blocked in absence of VMP112.
Crucially, VMP1 is expressed early during the onset of several pathologies, including diabetes mellitus, pancreatitis and pancreatic cancer12,15,16. Recently, we have reported that VMP1 expression is induced by hyperstimulation of Gq-coupled CCK receptor in pancreatic acinar cells during acute pancreatitis14 and by mutated K-Ras in pancreatic cancer cells17. Its tissue-specific transgenic-expression in vivo prevents pancreatic cell death induced by acute pancreatitis14. Besides having role in triggering autophagy as a cell response in pathological situations, VMP1 is required for autophagosome formation in mammalian cells in all conditions7,18. VMP1, along with Ulk1 and Atg14, localizes to the sites where autophagosome are formed independently of the other Atg proteins7 highlighting an upstream function of VMP1 in this process. We have previously shown that VMP1 is a transmembrane protein localizing the ER19, where the autophagosome are thought to be generated10. Therefore, we hypothesized that VMP1 may serve as a platform that promotes the optimal organization of the Atg machinery leading to the formation of the PAS and subsequently of autophagosomes.
Beclin 1 is a haploinsufficient tumor suppressor and an important effector of autophagy. Beclin 1 is a subunit of the Class III PI3K complex, the action of which is antagonized by Bcl-220,21. Beclin 1 contains a BH3 domain that mediates its interaction with Bcl-2, but the avidity of this interaction is quite weak compared to BH3 domains present in the proteins involved in apoptosis regulation22,23. The interaction between Bcl-2 and Beclin 1 leads to inhibition of autophagy by interfering with the formation and activity of the autophagy promoter complex, Beclin 1-PI3K21. In view of the multiples pathways relying on Bcl-2 at the ER and the relatively weak binding between Bcl-2 and Beclin 1, a mechanism to ensure adequate partitioning of Beclin 1 to the autophagy pathway must exist. Mammalian Beclin 1 is present in distinct Class III PI3K complexes24. Each complex seems to have a core consisting of Beclin 1, hVps34, and hVps15 and specific interactors, such as Atg14L, UVRAG, or Rubicon, conferring them distinct functions in membrane trafficking25. The Atg14L-containing PI3K complex is directly involved in autophagosome formation26,27,28. The specific molecular mechanism determining the association of the Class III PI3K complex to the PAS, however, remains to be fully understood.
In this study, we have elucidated the mechanism through which Beclin 1 and hVps34 are recruited to the PAS. This event involves the binding between the 20-aminoacid of the C-terminal hydrophilic domain of VMP1, which we have called the VMP1 autophagy-related domain (VMP1-AtgD)12,29, with the BH3 domain of Beclin 1. This interaction leads the formation of a VMP1-Beclin 1-hVps34 complex and the subsequent association of Atg16L1 to the autophagosomal membranes, providing a model describing one of the key steps in the PAS formation and autophagy regulation in mammalian cells.
Results
VMP1 interacts with the BH3 domain of Beclin 1
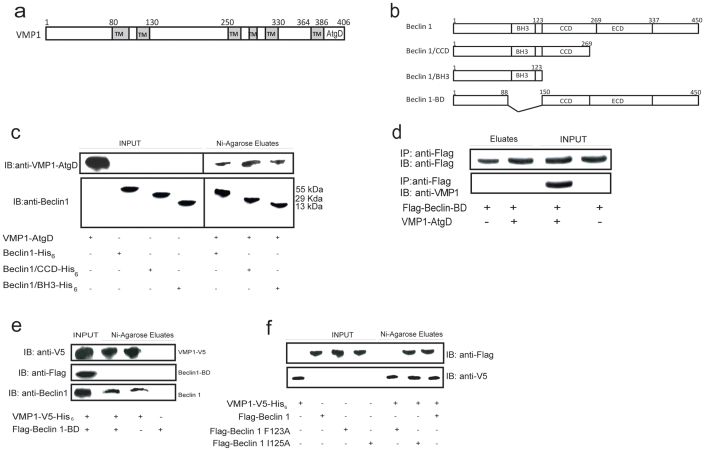
We have previously demonstrated that the C-terminal domain of VMP1, VMP1-AtgD (Fig. 1a), is essential for autophagy and for the interaction with Beclin 112. Therefore, to further delineate the interaction mechanism between VMP1 and Beclin 1, we have set out an in vitro assay to determine the domain of Beclin 1 that binds to VMP1. In a first series of experiments, we performed in vitro pull-down assays using three different recombinant Beclin 1 peptides (Fig. 1b) with the VMP1-AtgD synthetic peptide (hydrophilic C-terminal residues 386–406). We found that all the three recombinant Beclin 1 peptides (Beclin 11-450, Beclin 1/CCD1-269 and Beclin 1/BH31-123) were able to bind the VMP1-AtgD synthetic peptide (Fig. 1c). Taking into account that the BH3 domain is present in all the three Beclin 1 peptides that bound to VMP1-AtgD peptide, these results suggested that the BH3 domain is the one implicated in VMP1-Beclin 1 interaction. We refined our search by using an expression plasmid encoding the Beclin 1-defective mutant, i.e. Beclin 1-BDΔ88-150 (lacking the BH3 domain). First we performed in vitro pull-down assays with anti-Flag magnetic beads. Lysates obtained from HeLa cells transfected with the plasmid expressing Flag-tagged Beclin 1-BDΔ88-150 were incubated with the VMP1-AtgD synthetic peptide. Figure 1d shows that Beclin 1-BD was not able to precipitate the VMP1-AtgD. Then, homogenates obtained from HeLa cells transfected with the plasmids expressing Flag-tagged Beclin 1-BD and VMP1-V5-His6-tagged were subjected to coimmunoprecipitation assays using nickel-agarose beads. As shown in Figure 1e, VMP1 was not able to bind with the mutant Beclin 1-BD. Interestingly; VMP1 was able to interact to the endogenous Beclin 1, which contains the BH3 domain. We performed additional pull-down assays using a mutant with a single amino acid substitution in the BH3 domain, the Flag-Beclin 1 F123A, which has been previously shown to disrupt the interaction of Beclin 1 with Bcl-221. Thus, we evaluate if this mutant is able to interact with VMP1. Also we evaluated Flag-Beclin 1 I125A, which is a control mutant. We performed pull-down assays in HeLa cells transfected with VMP1-V5 and Flag-Beclin 1, Flag-Beclin 1 F123A or Flag-Beclin 1 I125A expression plasmids. Figure 1f shows that while Beclin 1 and Beclin 1 I125A are able to interact with VMP1, Beclin 1 F123A does not co-isolate with VMP1; confirming that the BH3 domain of Beclin 1 is required for VMP1-Beclin 1 interaction. Collectively, these results demonstrate that VMP1 is able to interact with the BH3 domain of Beclin 1.
Figure 1. VMP1-AtgD interacts with the BH3 domain of Beclin 1.
(a) Schematic representation of VMP1 protein primary structure with the position of six hydrophilic domains. TM, transmembrane domain; AtgD Autophagy related domain. (b) Schematic diagram of Beclin 1 recombinant peptides and Beclin 1-BD. The predicted functional domains of human Beclin 1 are indicated: BH3, Bcl-2 binding domain; CCD, coiled coil domain; ECD, evolutionarily conserved domain. (c) Pull-down assays. His6-fused recombinant peptides of Beclin 1 (Beclin 11-450; Beclin1/CCD1-269; Beclin1/BH31-123) bound to nickel-agarose beads were incubated with the VMP1-AtgD purified peptide. Eluates were separated and subjected to Beclin 1 and VMP1-AtgD immunoblotting. Data are representative of three independent experiments. (d) Pull-down assay using anti-Flag antibody of lysates from cells transfected with a plasmid encoding for Flag-Beclin-BD incubated with a synthetic VMP1-AtgD peptide. Eluates were separated and subjected to anti-Flag and anti-VMP1-AtgD immunoblotting. (e) Co-precipitation assays. Lysates from HeLa cells transfected with pCR3.1-Flag-Beclin 1-BD and pcDNA4-VMP1 expression plasmids were incubated with nickel-agarose beads. After washes, eluted proteins were separated and subjected to Flag, V5 and Beclin 1 immunoblotting (to detect endogenous Beclin 1). (f) Pull-down assays. Lysates from HeLa cells transfected with VMP1-V5 and Flag-Beclin 1, Flag-Beclin 1 F123A or Flag-Beclin1 I125A were incubated with nickel-agarose beads. Eluates were subjected to SDS-PAGE and immunoblotting using anti-V5 or anti-Flag antibodies. Data are representative of three independent experiments.
VMP1 interacts with Beclin 1 in vivo in human cells
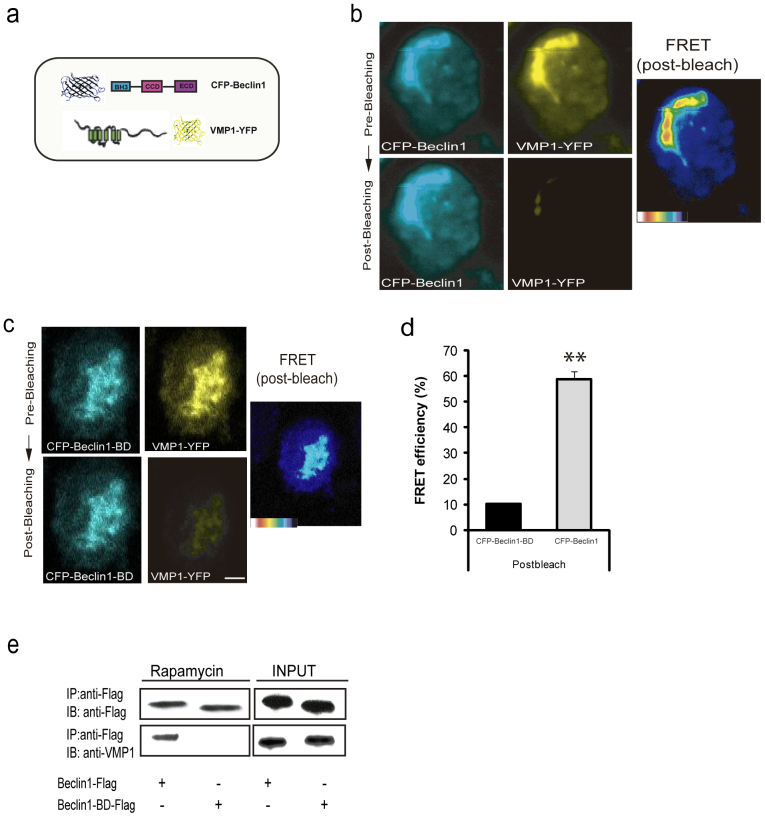
In order to confirm VMP1-Beclin 1 in vivo interaction in human cells, we used the fluorescence resonance energy transfer (FRET) analysis by using acceptor photo-bleaching method30, an established technique that relies on close spatial proximity of fluorescent molecules (see Materials and Methods). Evidence of increased donor fluorescence after photo-bleaching, indicates energy transfer between the donor and the acceptor, which can only occur when the donor-acceptor distance is less than 10 nm, a distance that is too short to be occupied by another protein. The FRET constructs were made tagging YFP to the VMP1 C-terminus and CFP to Beclin 1 N-terminus (Fig. 2a). Plasmids encoding CFP-Beclin 1 and VMP1-YFP were transiently transfected into HeLa cells, and the CFP and YFP fluorescent signals were acquired before (Fig. 2b; top panel) and after photo-bleaching (Fig. 2b; bottom panel). Confocal images of CFP-Beclin 1 are typically and noticeable brighter in fluorescence intensity after photo-bleaching the YFP fused to VMP1 (Fig. 2b). The pseudo-color images represent the increase of pixel density before and after bleaching. The color in the bar represents the pixel density of the image and thus the intensity of interaction, indicating an in vivo interaction between VMP1 and Beclin 1 (Fig. 2b). As a negative control, we employed the CFP-Beclin 1-BD fusion, which lacks the BH3 domain. Figure 2c shows that when VMP1-YFP was bleached, there was no increase in CFP-Beclin 1-BD signaling (bottom panel). Accurate quantification of the signals Fig. 2d showed that the average energy transfer efficiency for the cells expressing CFP-Beclin 1-BD with photo-bleached acceptors was 12.7% compared with 57% in the ones carrying CFP-Beclin 1. Taken together, our results confirm that VMP1 interacts with Beclin 1 in vivo.
Figure 2. FRET analysis of VMP1-Beclin 1 in vivo interaction.
(a) Schematic representation of the FRET pair used: CFP tagged N-terminal of Beclin 1 and YFP tagged c-terminal of VMP1. (b) In vivo detection of interaction of VMP1-YFP and CFP-Beclin 1 by acceptor photo-bleaching FRET. VMP1-YFP and CFP-Beclin 1 were transiently transfected into the HeLa cells. Images were acquired before (top panels: CFP-Beclin 1 and VMP1-YFP) and after (bottom panels: CFP-Beclin 1 and VMP1-YFP) photo-bleaching the VMP1-YFP. (c) CFP-Beclin 1-BD was used as a negative control. Images were acquired before (top panels: CFP-Beclin 1-BD and VMP1-YFP) and after (bottom panels: CFP-Beclin1-BD and VMP1-YFP) photo-bleaching of VMP1-YFP. Pseudo-colored images are shown in both cases indicating the intensity of interaction. (d) The bar graphs represent FRET efficiency. All data are represented as mean ± SD. Asterisks indicate a significant difference in the Student's t-test (**p< 0.01). (e) Coimmunoprecipitation assays. Lysates from HeLa cells treated with rapamycin (2 h) and transfected with pSG5-Flag-Beclin 1 or pCR3.1-Flag-Beclin 1-BD expression plasmids were incubated with anti-Flag-magnetics beads. Eluted proteins were separated and subjected to anti-Flag and anti-VMP1 immunoblotting. Bar, 10 μm. Data are representative of three independent experiments.
Previous studies have shown that rapamycin induces autophagy via VMP1 expression12. Therefore, we explored whether endogenous VMP1 interacts with the BH3 domain of Beclin 1 during rapamycin-induced autophagy. HeLa cells were transfected with a vector expressing Flag-tagged Beclin 1 or Beclin 1-BD, and coimmunoprecipitation assays were performed using anti-Flag antibodies bound to magnetic beads. In cells treated with rapamycin and expressing Flag-Beclin 1, VMP1 was coimmunoprecipitated with Beclin 1 (Fig. 2e). In contrast, Flag-Beclin 1-BD was not able to immunoprecipitate endogenous VMP1 under the same conditions (Fig. 2e). Together, these results demonstrate that the interaction between endogenous VMP1 and the BH3 domain of Beclin 1 occurs during autophagy in mammalian cells.
VMP1 releases Bcl-2 from Beclin 1, driving Beclin 1 into the autophagy pathway
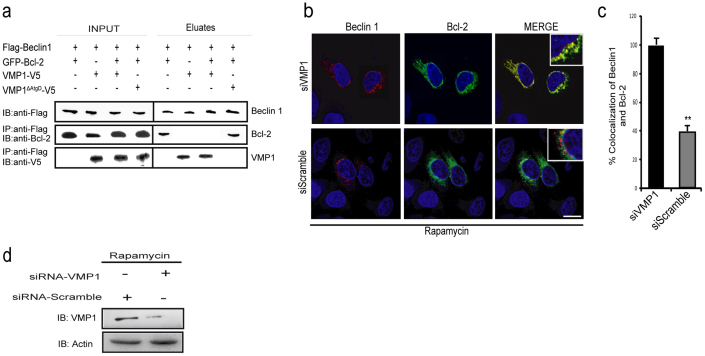
It has been demonstrated that Beclin 1 interacts with Bcl-2 through its BH3 domain21,23,31. This interaction is reduced when autophagy is stimulated by nutrient deprivation and the dissociation between Beclin 1-Bcl-2 is crucial for autophagy induction21,32. To evaluate the relevance of the VMP1-Beclin 1 interaction in this regulatory context we studied whether VMP1 promotes the dissociation of Beclin 1-Bcl-2 complex upon autophagy induction. We thus performed coimmunoprecipitation experiments in cells co-expressing Flag-Beclin 1, GFP-Bcl-2 and VMP1-V5 or VMP1ΔAtgD-V5. Cell lysates were incubated with anti-Flag antibodies bound to magnetic beads and the eluates were subjected to immunoblot analyses. In cells expressing Beclin 1 and Bcl-2, Bcl-2 was coimmunoprecipitated with Beclin 1 (Fig. 3a) while in cells expressing VMP1 and Beclin 1, VMP1 was co-isolated with Beclin 1 (Fig. 3a). Importantly, in lysates of cells simultaneously expressing VMP1, Beclin 1 and Bcl-2; Beclin 1 was found associated with VMP1 but not with Bcl-2. As expected, in cells expressing VMP1ΔAtgD (which lack the Atg domain and is not able to interact with Beclin 1)12, Bcl-2 co-immunoprecipitated with Beclin 1 (Fig. 3a). To confirm that VMP1 regulates the disruption of the Bcl-2-Beclin 1 complex, we evaluate the role of endogenous VMP1 during rapamycin-induced autophagy using a specific siRNA to knock down VMP1 expression in HeLa cells. We found that the levels of Bcl-2-Beclin 1 colocalization was increased in cells transfected with VMP1 siRNA, but not in cells transfected with scramble siRNA (Fig. 3b and 3c). Collectively, these results indicate that under autophagy VMP1 expression promotes the dissociation between Beclin 1 and Bcl-2 probably through binding competition.
Figure 3. VMP1 displaces Bcl-2 from the BH3 domain of Beclin 1.
(a) Coimmunoprecipitation assays. Lysates from HeLa cells transfected with pSG5-Flag-Beclin 1, pEGFP-Bcl-2 and pcDNA4-VMP1 or pcDNA4-VMP1ΔAtgD expression plasmids incubated with anti-Flag-magnetics beads. Eluted proteins were separated and subjected to Beclin 1, VMP1 and Bcl-2 immunoblotting. (b) Representative confocal microscopy images of HeLa cells co-transfected with Beclin 1-RFP, GFP-Bcl-2 and treated with rapamycin under/without VMP1 knockdown expression. (c) Percentage of Bcl-2 spots colocalizing with Beclin 1. The panels on the right show an enlarged view of the boxed regions on the left. Bar, 10 μm. (d) The VMP1 knocking down was validated by western blot analysis. Results represent mean ± SD for combined data from four independent experiments.
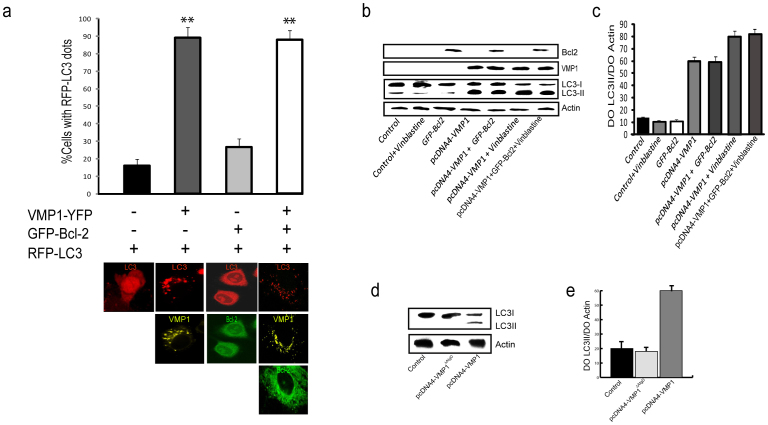
In order to evaluate the ability of VMP1 to drive Beclin 1 function into autophagy, HeLa, PANC-1 and AR42J cells cultured in presence of nutrients and growth factors, were co-transfected with plasmids expressing RFP-LC3, a marker protein for autophagosomal membranes, and VMP1-YFP and/or GFP-Bcl-2 before determining the percentage of cells with RFP-LC3 puncta. As expected, the number of RFP-LC3-positive puncta per cell was significantly higher in cells expressing VMP1 than in those expressing Bcl-2. Interestingly, the distribution pattern of RFP-LC3 in cells that co-expressed VMP1 and Bcl-2 was identical to the one in cells expressing VMP1 alone (Fig. 4a). Representative confocal images are shown below bars in Fig 4a. During autophagy, the cytosolic form of LC3, e.g. LC3-I undergoes first a C-terminus proteolytic cleavage and then a lipid modification on autophagosomal membrane to become LC3-II31,33. Thus to measure autophagy induction in cells expressing VMP1 and/or Bcl-2, we analyzed LC3-I conversion into LC3-II by western blot. As expected and in accordance with the literature21, we found increased LC3-II levels in cells transfected with the VMP1 construct. Similarly, cells co-expressing VMP1 and Bcl-2 were more prone to autophagy induction than Bcl-2 expressing cells (Fig. 4b, lanes 4 and 5, and 4c). Then we evaluated LC3 processing in VMP1ΔAtgD and Bcl-2 expressing cells. As expected, VMP1ΔAtgD was not able to increase LC3-II levels (Fig. 4d and 4e, lane 2). Analogous results were obtained in pancreatic PANC-1 and AR42J cells (data not shown). We concluded that under autophagy-inducing condition, VMP1 is involved in the dissolution of Bcl-2-Beclin 1 complex, necessary for the Beclin 1-mediated induction of autophagy.
Figure 4. VMP1-Beclin 1 interaction favors autophagosome formation.
(a) HeLa cells co-transfected with RFP-LC3, VMP1-YFP and GFP-Bcl-2. Fluorescence microscopic quantitation of autophagy (% cells with RFP-LC3 dots) was determined as described under “Materials and methods”. Representative images of HeLa cells co-transfected with RFP-LC3, VMP1-YFP and/or GFP-Bcl-2 are shown below graphic bar. (b) Lysates of pcDNA4-VMP1 and GFP-Bcl-2 plasmid-transfected HeLa cells with or without vinblastine treatment were analyzed by western blot. LC3-I (apparent mobility, 18kDa) and LC3-II (16kDa), VMP1, Bcl-2 and Actin proteins were determinated. (c) Quantification of protein signal intensities from western blots showing LC3-II levels after normalization to the control protein Actin. (d) Lysates of pcDNA4-VMP1 and pcDNA4-VMP1ΔAtgD plasmid-transfected HeLa cells were analyzed by western blot. LC3-I (apparent mobility, 18kDa) and LC3-II (16kDa) were determinated. (e) Quantification of protein signal intensities from western blots showing LC3-II levels after normalization to the control protein Actin. Results represent mean ± SD for combined data from four independent experiments. Asterisks indicate a significant difference versus control in two-way ANOVA (**p< 0.01).
Identification of the VMP1-Beclin 1-hVps34 complex acting in autophagy
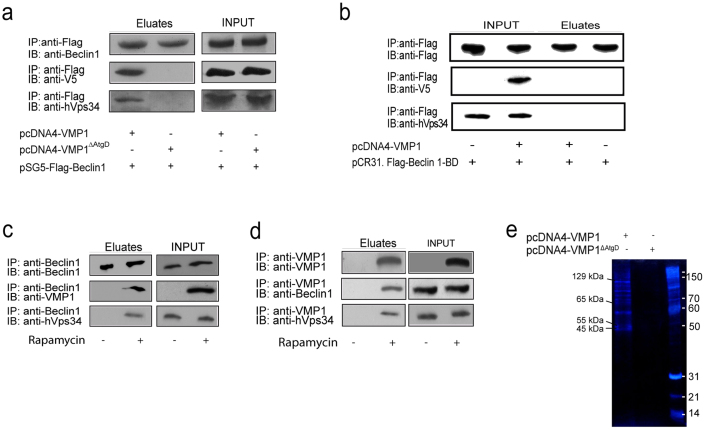
VMP1 expression triggers autophagosome formation12 and it has been shown that this protein localizes at the PAS together with Ulk1 and Atg14L7. Beclin 1 participates in autophagy as part of the Atg14L-containing Class III PI3K complex34,35,36. Generation of PI3P by this complex is crucial for the recruitment of several other Atg proteins onto autophagosomal membranes36. To further delineate the VMP1-Beclin 1 interaction during autophagy, we carried out coimmunoprecipitation assays to test whether the Class III PI3K complex is associated with VMP1. First, lysates from HeLa cells transfected with plasmid expressing Flag-Beclin 1 and VMP1-V5-His6 or VMP1ΔAtgD-V5-His6 were incubated with anti-Flag antibody bound to magnetics beads. As shown in Figure 5a, VMP1 and endogenous hVps34 co-immunoprecipitate with Beclin 1 in cells expressing VMP1. On the contrary, in cells expressing VMP1ΔAtgD, neither VMP1 nor hVps34 were coisolated with Beclin 1 (Fig. 5a). These results indicate that VMP1 is part of the Class III PI3K complex during autophagy. Next, HeLa cells transfected with Flag-Beclin 1-BD and VMP1-V5 were incubated with anti-Flag antibody bound to magnetics beads. Surprisingly, neither VMP1 nor hVps34 were able to co-immunoisolate with Beclin 1-BD (Fig. 5b), suggesting that VMP1-Beclin 1 interaction may be involved in the mammalian PI3K complex formation during autophagy. Then, we treated HeLa cells with rapamycin and using anti-Beclin 1 or anti-VMP1 antibodies bound to magnetic beads we evaluated the formation of the VMP1-Beclin 1-hVps34 complex. We found that during rapamycin-induced autophagy Beclin 1 is able to coimmunoprecipitate with endogenous VMP1 and hVps34 (Fig. 5c), and VMP1 coimmunoprecipitates with endogenous Beclin 1 and hVps34 (Fig. 5d). Unfortunately, our system failed to detect endogenous Atg14L in the whole homogenates as well as in VMP1 or Beclin 1 immunoprecipitates from rapamycin treated cells (data not shown). Finally, HeLa cells were transfected with the VMP1-V5-His6 construct and lysates were precipitated using nickel-agarose beads. Resulting precipitates were subjected to SDS-PAGE and silver nitrate staining. Bands corresponding to molecular weights suggesting the co-isolation of hVps34 (129kDa), Atg14L (65kDa), and Beclin 1 (55kDa) were present in the VMP1 precipitate (Fig. 5e). Collectively, results described above, allow us to propose that VMP1 is associated to the autophagy-specific PI3K complex during mammalian autophagy.
Figure 5. Identification of the VMP1-Beclin 1-hVps34 complex.
(a) Coimmunoprecipitation assays. Lysates from HeLa cells transfected with pSG5-Flag-Beclin 1 and pcDNA4-VMP1ΔAtgD or pcDNA4-VMP1 expression plasmids incubated with anti-Flag covalently bound to magnetic beads. After washes, eluted proteins were separated and subjected to anti-Beclin 1, V5 and hVps34 immunoblotting. (b) Coimmunoprecipitation assays. Lysates from HeLa cells transfected with pCR3.1-Flag-Beclin 1-BD and pcDNA4-VMP1 expression plasmids incubated with anti-Flag covalently bound to magnetic beads. After washes, eluted proteins were separated and subjected to anti-Flag, V5 and hVps34 immunoblotting. (c) Coimmunoprecipitation assays. Lysates from HeLa cells treated with rapamycin were incubated with anti-Beclin 1 antibody. Eluted proteins were subjected to anti-Beclin 1, anti-VMP1 or anti-hVps34 antibody. (d) Coimmunoprecipitation assays. Lysates from HeLa cells treated with rapamycin were incubated with anti-VMP1 antibody. Eluted proteins were subjected to anti-VMP1, anti-Beclin 1 or anti-hVps34 antibody. (e) HeLa cells transfected with a plasmid harboring pcDNA4-VMP1-V5 were coprecipitated using nickel-agarose beads. pcDNA4-VMP1ΔAtg expressing cells were used as control. The precipitates were subjected to SDS-PAGE and silver nitrate staining. Data is representative of three independent experiments.
The VMP1-Beclin 1 complex promotes PI3P generation on autophagosomal membranes
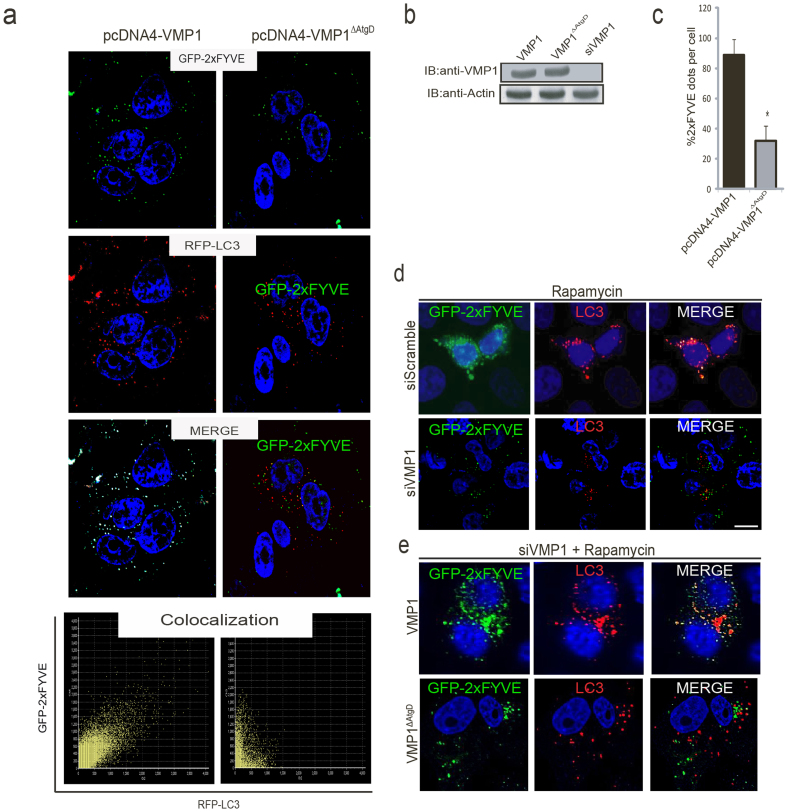
To test whether the VMP1-Beclin 1 interaction regulates the generation of PI3P in the autophagosomal membranes; we analyzed PI3K activity in cells expressing either VMP1 or VMP1ΔAtgD. The production of PI3P by Class III PI3K can be visualized and quantified using the GFP-tagged double FYVE domain of the Hrs protein36,37, which specifically binds to PI3P. We thus used the GFP-2xFYVE probe to detect PI3P synthesis on autophagosomal membranes identified with RFP-LC3. HeLa cells were co-transfected with plasmids expressing GFP-2xFYVE, RFP-LC3 and VMP1-V5 or VMP1ΔAtgD-V5. We found a remarkable colocalization between 2xFYVE and LC3 in cells undergoing autophagy triggered by VMP1 expression (Fig. 6a left panel), revealing that VMP1 is able to recruit the activity of the PI3K complex to the autophagosomal membranes. On the contrary, the VMP1ΔAtgD mutant failed to recruit the PI3K complex onto autophagosomal membranes, since almost no colocalization between 2xFYVE and RFP-LC3 was found (Fig. 6a right panel). Moreover, quantification of GFP-2xFYVE puncta per cell revealed that PI3P production was significantly diminished in VMP1ΔAtgD-expressing cells compared to VMP1-expressing cells (Fig. 6b). These data indicate that the Atg domain of VMP1 is required to recruit PI3K activity to the autophagosomal membrane. Subsequently, we evaluated PI3K activity when VMP1 endogenous expression is activated under rapamycin-induced autophagy before and after downregulation of VMP1 expression12. Figure 6d upper panel shows that GFP-2xFYVE positive puncta colocalizes with LC3 in rapamycin-treated cells. On the contrary, almost no colocalization between 2xFYVE and LC3 was found after downregulation of VMP1 expression in rapamycin-treated cells (Fig. 6d bottom panel). We then transfected VMP1-knocked-down cells with pcDNA4-VMP1 or pcDNA4-VMP1ΔAtgD under rapamycin treatment. Figure 6e shows that VMP1 expression is able to rescue the generation of PI3P in autophagosomal membranes. Remarkably, VMP1ΔAtgD expression was not able to rescue VMP1 mediated recruitment of PI3K activity to the autophagosomal membrane. These results demonstrate that VMP1 is essential for the localization of PI3K activity to the PAS in mammalian autophagy and confirm that the interaction between the Atg domain of VMP1 and Beclin 1 is required for bringing the mammalian Beclin 1 –PI3K complex activity to the site of autophagosome formation.
Figure 6. The VMP1-Beclin 1 complex promotes PI3P generation in the autophagosomal membranes.
(a) Representative confocal microscopy images of HeLa cells co-transfected with plasmids encoding for GFP-2xFYVE, RFP-LC3 and VMP1-V5, VMP1ΔAtgD-V5. Colocalization images were evaluated using the Colocalization Scatterplot. (b) The presence of VMP1 or VMP1ΔAtgD in lysates corresponding to the experiments in “a” was evaluated by immunoblotting using VMP1 antibody. (c) The percentage of cells with GFP-2xFYVE puncta was determined in cells expressing VMP1 or in VMP1ΔAtgD. Asterisks indicate a significant difference versus VMP1 in two-way ANOVA (*p< 0.05). (d) Representative confocal images of HeLa cells under rapamycin treatment after downregulation of VMP1 expression using a specific VMP1 siRNA. (e) VMP1 expression was downregulated using the specific siRNA in HeLa cells treated with rapamycin. After 2 hours of treatment cells were transfected during 24 h with pcDNA4-VMP1 or pcDNA4-VMP1ΔAtgD. Bar, 10 μm. Data is representative of three independent experiments.
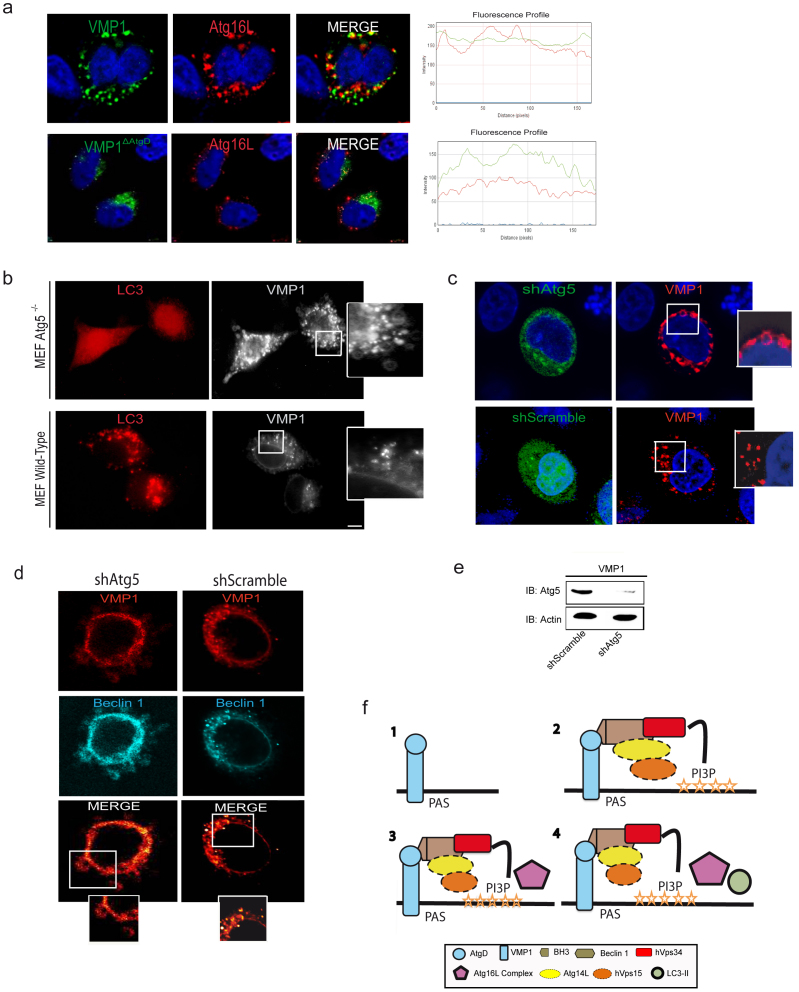
The generation of PI3P by the Class III PI3K complex is crucial for the recruitment of several other Atg proteins onto autophagosomal membranes36,37,38. To monitor this event, we decided to analyze the localization of Atg16L1 to the PAS. Therefore, HeLa cells were transfected with a plasmid expressing VMP1-GFP or VMP1ΔAtgD-GFP before investigating endogenous Atg16L1 localization by immunofluorescence. Figure 7a shows that VMP1 partially colocalizes with Atg16L1 during VMP1-induced autophagy. On the contrary VMP1ΔAtgD fails to colocalize with Atg16L1 (Fig. 7a), suggesting that VMP1-Beclin 1 complex formation is required for Atg16L1 localization to the PAS. It has been reported that Atg16L1 form a complex with Atg12-Atg5 that is involved in determining the site of Atg8/LC3 lipidation32,38. We then investigated whether VMP1-induced LC3 recruitment onto autophagosomal membranes depends on the Atg12-Atg5-Atg16L1. Accordingly, we simultaneously transfected Atg5−/− and wild-type, mouse embryonic fibroblasts (MEFs) with plasmid expressing VMP1-GFP and RFP-LC3. Surprisingly, we found a remarkable accumulation of large VMP1 labeled membranes in Atg5−/− MEFs, most of which were negative for the RFP-LC3 PUNCTA. (Fig. 7b). Then, HeLa cells were transfected with VMP1 after downregulation of Atg5 expression using a specific shRNA. Figure 7c shows accumulation of VMP1 in large membrane structures in Atg5 silenced cells. Further, we transfected HeLa cells with VMP1 and Beclin 1 expression plasmids after downregulation of Atg5 expression to determine whether Beclin 1 was still recruited to the PAS by VMP1 in absence of Atg5. We found that Atg5-depleted cells displayed VMP1-Beclin 1 complex accumulation in puncta also of large size (Fig. 7d). These findings indicate that VMP1-Beclin 1 complex acts upstream of the Atg12-Atg5-Atg16L1 complex during autophagosome formation. Collectively, the above-described results demonstrate that VMP1 expression, through the interaction with Beclin 1, regulates the generation of PI3P on the autophagosomal membranes promoting the localization of other Atg proteins to the PAS, including Atg16L1 and LC3.
Figure 7. VMP1-Beclin 1 complex is upstream of the Atg12-Atg5-Atg16L1complex.
(a) HeLa cells cultured under nutrient and grown factor-replete conditions and transfected with the VMP1-GFP or the VMP1ΔAtgD-GFP plasmids. Endogenous Atg16L1 was analyzed by immunofluorescence by confocal microscopy. Merge image and two hot fluorescence spots profile are shown to prove specific colocalization. (b) Representative images of MEF Atg5−/− and wild-type cells concomitantly transfected with VMP1-GFP and RFP-LC3 expressing plasmids and observed in a fluorescence microscope. (c) HeLa cells were transfected with RFP-VMP1 after downregulation of Atg5 using a specific shRNA. (d) HeLa cells concomitantly transfected with RFP-VMP1, CFP-Beclin 1 and shRNA-Atg5 or shRNA-Scramble. The panels on the right and bottom show an enlarged view of the boxed regions. (e) Western blot shows downregulation of Atg5 in HeLa cells transfected with shAtg5. Bar, 10 μm. Data in a-e are representative of four independent experiments. (f) This panel illustrates the proposed model of VMP1-Beclin 1 autophagy induction: (1) VMP1 expression is induced. (2) The autophagy-specific PI3K complex is recruited to the PAS and generates PI3P. (3) The Atg12-Atg5-Atg16L1 complex is recruited. (4) Lipidation and localization of LC3 at the PAS.
Discussion
In previous works we have demonstrated that VMP1 expression triggers autophagy and that this transmembrane protein is essential for autophagosome formation in mammalian cells12. In the current study, we report that autophagy induction by VMP1 expression is mediated through the interaction between the VMP1-AtgD and the Beclin 1-BH3 domains. This event allows the localization of the Class III PI3K activity on the autophagosome formation site, i.e. the PAS. We have also demonstrated that the binding of the VMP1-AtgD domain to the Beclin 1 BH3 motif promotes the displacement of Bcl-2, a negative regulator of autophagy, driving Beclin 1 to the autophagic pathway. VMP1-AtgD is required for both forming the VMP1-Beclin 1-hVps34 complex and inducing the formation of PI3P on the autophagosomal membranes. Finally, VMP1-Beclin 1-hVps34 complex, favors the localization of Atg16L1 and LC3 to the PAS. These findings collectively reveal that VMP1 is part of the Beclin 1-Class III PI3K complex that regulates autophagy in mammalian cells at least under certain conditions.
To the best of our knowledge VMP1 is the only transmembrane Atg protein with no homologue in yeast and other low eukaryotes. Our data have revealed that the interaction between VMP1 and Beclin 1 requires Beclin 1 BH3 domain. Interestingly, Atg6/Vps30, the yeast homologue of Beclin 1, does not possess a BH3 motif. Another protein that associates with the BH3 domain of Beclin 1 is Bcl-2. Binding of Bcl-2 to Beclin1 inhibits autophagy and accordingly the dissociation of Bcl-2 from Beclin 1 is an important regulatory event to induce this pathway39. During normal growth conditions Bcl-2 binding to Beclin 1 is maximal, and when autophagy is induced this interaction is strongly reduced21. Here, we found that VMP1 expression leads to the dissolution of the Beclin 1-Bcl-2 complex, indicating that VMP1 is involved in driving Beclin 1 into the autophagic process by removing the negative regulator Bcl-2. Thus, we propose that VMP1, through the interaction with the BH3 domain of Beclin 1, regulates the initial steps of autophagy in mammalian cells.
Recent findings provide biochemical evidence that Beclin 1 is present in distinct Class III PI3K complexes25. These Beclin 1 complexes are involved at different stages of autophagy and/or of the endocytic trafficking. Each complex has a core consisting of Beclin 1, hVps34, and hVps15. Our results identify VMP1 as a new interactor of one or more Beclin 1-hVps34 complex, and provide a possible idea about how the Beclin 1-hVps34 complex could control autophagosome formation. Our results suggest that VMP1-Beclin 1 interaction is required for the formation of the VMP1-Beclin 1-hVps34 complex during mammalian autophagy. VMP1 can be immunoprecipitated with both Beclin 1 and hVps34, but Beclin 1 fails to coprecipitate with hVps34 when VMP1 lacks its Atg domain (Fig. 5a). These data may appear inconsistent with previously published data proposing a direct association between Beclin 1 and hVps3434,43. However, those results were obtained in cells undergoing autophagy25, and during autophagy is when the VMP1-Beclin 1 interaction occurs12. Indeed, Beclin 1 is not able to coisolate with hVps34 when its BH3 domain is interacting with Bcl-221. These data are consistent with our results, because when autophagy is not induced (Fig. 5c, 5d) or Beclin 1 is not able to interact with VMP1 (Fig.5b), Beclin1-hVps34 complex is not detectable.
Atg14L is another component specifically associated with the Beclin 1-containing PI3K complex involved in the autophagosome biogenesis24,26. The antibodies in our possession unfortunately failed to detect endogenous Atg14L in cell extracts from either rapamycin- or VMP1-treated cells. However, a band of 65kDa was found in the eluates when we tried to overcome this problem by performing large scale pull-down done using VMP1 as bait, suggesting that Atg14L could be part of the VMP1-Beclin 1-hVps34 complex. Our results are consistent with the findings of Itakura and co-workers7, which reported that VMP1 co-localizes with Atg14L at the PAS. To our knowledge, this is the first study analyzing the endogenous proteins involved in the formation of the Class III PI3K complex.
We found that the VMP1-Beclin 1 complex promotes PI3P generation on autophagosomal membranes. Our data show partial but remarkable co-localization between LC3 and Hrs 2XFYVE upon rapamycin treatment and VMP1 expression. These findings seem to be different from data published by Axe, et al.9, which suggest that multiple pools of PI3P, apart from the endoplasmic reticulum, are labeled by 2xFYVE during amino acid starvation. Nevertheless, rapamycin treatment or VMP1 overexpression may favor the PI3P generation in the autophagosome formation site. In fact, we demonstrated that endogenous VMP1 expression is required for the docking of the PI3K complex to autophagosomal membranes since 2xFYVE did not colocalize with these intermediates when VMP1 expression is downregulated under rapamycin treatment. Moreover, VMP1ΔAtgD mutant failed to rescue rapamycin treated cells from VMP1 down-regulation (Fig. 5e), supporting that the VMP1-Beclin 1 interaction through the VMP1-AtgD is required for the proper localization of PI3K activity during mammalian autophagy.
In mammalian cells, Atg16L1 associated with the Atg12-Atg5 conjugated is involved in determining the site of Atg8/LC3 lipidation32,38. The two conjugation systems are closely related, as Atg12–Atg5-Atg16L1 is necessary for the efficient LC3 lipidation in vivo and in vitro40,41. Our findings suggest that VMP1-Beclin 1 interaction regulates membrane association of the Atg12-Atg5-Atg16L1 complex during autophagy in agreement with data obtained in yeast42. We demonstrate that VMP1 promotes the generation of PI3P favoring the localization of Atg16L1 and LC3 to the PAS during autophagosome formation. Consistently, we also found that PI3K activity does not localize to the autophagosomal membranes when VMP1 is not able to interact with Beclin 1. Our data also indicate that the VMP1-Beclin 1 complex functions upstream of Atg16L1 complex. This notion is also supported by the observation that the VMP1-Beclin 1 complex accumulates in membrane structures, probably an autophagosomal precursor, when Atg5 is downregulated. Altogether, our findings suggest that VMP1 allows the PI3P-enrichement at the PAS and trigger autophagosome biogenesis by recruiting, forming and possibly activating the VMP1-Beclin 1-hVps34 complex. The formation of the VMP1-Beclin 1-hVps34 complex favors the recruitment of Atg16L1 and LC3 to promote proper autophagosome formation (Fig. 7f).
Remarkably, VMP1 expression triggers autophagosome formation in mammalian cells even under nutrient-replete conditions12. Pathological processes such as pancreatitis12 and diabetes mellitus16 as well as tumor-cell transformation activate VMP1-mediated autophagy in human tissues and tumor-cell lines15,17. In particular, while human normal pancreas does not express detectable VMP1 levels, pancreatic diseases activate VMP1-mediated autophagy. Data presented here show VMP1 being a key regulator of the early steps of autophagosome formation in mammalian cells possibly acting as a scaffold protein. These characteristics collectively point at VMP1 as a unique Atg protein that regulates autophagy induction in numerous human diseases. Our findings thus set the bases for future studies on the regulation of autophagy in pathological situations and highlight VMP1 as a positive target for therapeutic interventions.
Methods
Mammalian cell lines, transfections and treatments
Human HeLa cell line, pancreatic cancer cell lines PANC-1, rat pancreatic AR42J and Mouse Embryonic Fibroblast, (MEF) Atg5−/− or MEF wild-type cells were used. Cells were transfected using FuGENE-6 reagent (Promega) as indicated by the manufacturer. Cells were incubated for 24 h in DMEM supplemented with 10% FBS in plates at 80% confluence and then transfected. Cells were treated with 55 μM rapamycin (Calbiochem) for 2 h. For inhibition of autophagosome-lysosomal fusion, cells were incubated with 0.05 nM vinblastine (Sigma).
VMP1-AtgD
The synthetic 20-aminoacid-carboxiterminal hydrophilic peptide (residues 386–406) was used for pull-down assays and to develop the Rabbit anti-VMP1-AtgD.
Plasmids
Plasmids pRFP-LC3, pSG5-Flag-epitope-tagged human Beclin 1, pGFP-Bcl-2, pCR3.1-Flag-Beclin 1–BD21 and pGFP-2xFYVE (Fab1, YOTB, Vac1, and pEEA1-domain) were kindly provided by Dr. Maria I. Colombo (Universidad Nacional de Cuyo, Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), Argentina). pCR3.1-Flag-Beclin1 F123A and pCR3.1-Flag-Beclin1 I125A were kindly provided by Dr. Beth Levine (UT Southwestern Medical Center, Texas, United States). pcDNA4-V5-His6 (Invitrogen) containing full-length rat VMP1 cDNA (NM_138839) or VMP1ΔAtgD, deficient mutant, in which the VMP1 C-terminus peptide (aa 378–406) was deleted as was described in Ropolo et al.12 pEGFP-VMP1, pEGFP-VMP1ΔAtgD, pYFP-VMP1, pRFP-VMP1, pRFP-Beclin 1, pCFP-Beclin 1 and pECFP-Beclin1-BD were designed and constructed by our laboratory. Atg5 down-regulation was made using the plasmid pCMS3-H1p-EGFP, which was kindly provided by Dr D. Billadeau (Department of Immunology, College of Medicine, Mayo Clinic, United States). The plasmid contains an Atg5 short hairpin RNA construction (sense 5′-GCAACUCUGGAUGGGAUUG-3′, and antisense 5′- CAAUCCCAUCCAGAGUUGC-3′) or a scramble sequence, and a separate transcriptional cassette for EGFP to identify transfected cells. Cells were also transfected using Oligofectamine (Invitrogen) with siRNA pre-designed for human VMP1 mRNA using 5′-GGCAGAAUAUUGUCCUGUGtt-3′, as sense, and 5′-CACAGGACAAUAUUCUCUGCCtt-3′, as antisense (Ambion ID32935).
Antibodies
Polyclonal goat anti-LC3, goat anti-Beclin 1 (Santa Cruz Biotechnology, Inc.); rabbit anti-Flag, rabbit anti-Bcl-2 (Sigma-Aldrich); monoclonal mouse anti-V5 and rabbit anti-hVps34 (Invitrogen); and rabbit anti-Atg16L1 (ABR), were used according manufacturer. Rabbit anti-VMP1-AtgD (residues 386–406) was developed in our laboratory. Anti-mouse Alexa Fluor 594 (Molecular Probes) antibody was used for immunofluorescence. Peroxidase-labeled anti-rabbit, anti-mouse, and anti- goat IgG antibodies were used for western blot according Amersham Biosciences.
Acceptor photo-bleaching förster resonance energy transfer (fret)
The acceptor photo-bleaching method of FRET was used to study protein-protein interaction. We used CFP-Beclin 1 and VMP1-YFP constructs. Conditions establishing that the Beclin 1 and VMP1 constructs behaved similarly to wild-type non-fluorescent tagged proteins were previously reported12. In FRET analysis, an acceptor fluorescent molecule can be quenched and causes fluorescence of a donor molecule if the respective proteins of interested tagged with each fluorescent molecule are closed enough. FRET between CFP-Beclin 1 (donor) and VMP1-YFP (acceptor) was performed using the acceptor photo-bleaching method on an Olympus FV1000 confocal microscope outlined in the FV10 Olympus software. HeLa cells were transiently transfected with CFP-Beclin 1 or CFP-Beclin 1-BD and VMP1-YFP using FuGENE-6 (Promega). 24 h after transfection, cells were fixed with 4% paraformaldehyde and mounted with DABCO. A UPLSAPO 60X O NA: 1.35 objective was used. Regions of interest (ROI) were selected and images collected before and after bleaching with the 514 nm laser line at maximum intensity (100%). Excitation wavelength was 458 nm for CFP and 514 nm for YFP. Emission wavelength was BP 465–510 nm for CFP and 518–561 nm for YFP. The change in the fluorescence intensity between pre- and post-bleach donor values (efficiency, E) was calculated using the formula E = Fpre/Fpost, and was shown as a percentage.
Beclin 1 constructions
Beclin 1 coding sequence was cloned in the expression plasmid pET22b. The pET22b vector contains a poly-His sequence that is added to the carboxy terminal end of the peptide to facilitate its purification. We generated the following constructions: pET22-Beclin1/BH3 (Beclin1 amino terminal domain; aa 1-123), pET22-Beclin1/CCD (BH3 domain and CCD domain; aa 1-269) and pET22/Beclin1 (complete protein; aa 1-450). The constructions were used in induction assays performed in the bacterial strain BL21 (DE3) Codon plus RIL. This strain contains an extra plasmid that codifies for rare tRNA codons of Arg, Ile and Leu, which work to express proteins in mammals. For the induction assays, bacteria's were transformed by the heat shock method, and then grown overnight (ON) in LB medium. Clones were selected for miniexpression assays. Bacteria culture of each clone was grown and expression was induced by the addition of the lactose analogue IPTG (2 mM). Cultures were grown for 4 h. The bacterial pellets were then lysed with lysozime 1 mg/ml and a buffer containing NaH2PO4 50 mM, NaCl 300 mM, histidine 10 mM pH8. Finally, we obtained bacterial lysates containing the recombinant proteins Beclin 1, Beclin1/CCD and Beclin1/BH3. In each case an aliquot of the lysate was used to test the interaction with the VMP1-AtgD peptide (aa 386–406), in a pull-down assay.
Immunofluorescence
After treatments, HeLa cells were fixed during 15 minutes with 4% p-formaldehyde in PBS and immediately washed several times with PBS. Cells were incubated with primary antibodies overnight at 4°C, according to manufacturer. Rabbit anti-mouse Alexa Fluor 594 antibody was used for immunofluorescence. Samples were mounted in DABCO (Sigma-Aldrich) with DAPI (Sigma-Aldrich) as nuclear marker and observed in an inverted LSM Olympus FV1000 using an UPLSAPO 60X O NA: 1.35 objectives.
Fluorescence microscopy
After transfection, cells were fixed during 15 minutes with 4% p-formaldehyde in PBS and immediately washed several times with PBS. Samples were mounted in DABCO (Sigma-Aldrich) with DAPI (Sigma-Aldrich) as nuclear marker and observed in an inverted LSM Olympus FV1000 using an UPLSAPO 60X O NA: 1.35 objective and in a fluorescence microscope Nikon Eclypse 200 (Plan100). To evaluate colocalization, we used the Olympus software Colocalization Scatterplot coordinate system.
Coprecipitation assays
Cells were lysed (Lysis buffer: 50 mM Na2HPO4, 300 mM NaCl, pH 8.0, 0.5% Tween-20, 0.5% Triton X-100, 10 mM imidazole) and cell lysates were incubated with 50 ml of supermagnetics beads (Sigma-Aldrich) ON at 4°C; afterwards the supernatant was removed using a magnetic device (MPC, Dynal Inc.) and saved as unbound fraction. The beads were intensively washed with lysis buffer and 20 mM imidazole. We eluted the bounded proteins with lysis buffer supplemented with 200 mM imidazole. Samples were resolved on SDS-PAGE, and detected by the appropriate antibodies. Transfected cells were lysed (Lysis buffer: 50 mM Na2HPO4, 300 mM NaCl, pH 8.0, 0.5% Tween-20, 0.5% Triton X-100, 10 mM imidazole) and cell lysates were incubated for 20-min at 4°C, followed by a 30-min centrifugation at 14000 rpm. The supernatants containing His-tagged proteins were saved as unbound fraction. Ni-NTA beads were intensively washed with washed buffer (50 mM tris pH 8.0 and 120 mM NaCl). The bound proteins were eluted with lysis buffer supplemented with 200 mM imidazole. Samples were resolved on SDS-PAGE, and detected by immunoblotting. Protein expression levels were quantified using the ImageJ densitometry; gel analyzer command. For silver nitrate staining after SDS-PAGE gels were fixed in 30% ethanol, 10% acetic acid. Then, gels were impregnate with 12 mM silver nitrate. When the adequate degree of staining has been achieved, transfer the gel to the Tris stop solution.
Pull-down assays
Lysates were incubated with a nickel-agarose matrix (Qiagen) during 24 h. The matrix was then washed with a buffer (NaH2PO4 50 mM and NaCl 300 mM pH 8.0). Then the VMP1 recombinant peptide (VMP1-AtgD) was added when corresponding and the system was incubated for 24 h. Next, supernatant was separated (L), and after three successive washes, we proceeded to the elution step of the retained complex by adding a buffer with an excess of histidine. Finally, we evaluated the presence of the VMP1-AtgD peptide and the recombinant Beclin 1 fragment by western Blot assays.
Statistical analysis
Data are expressed as mean ± SD. Student's t test was used for comparisons between 2 groups and ANOVA test to assess more than 2 groups. Differences were considered significant when p <0.05.
Ethics statement
The Ethics Committee of the School of Pharmacy and Biochemistry, University of Buenos Aires, CD/2011 approved the experiments.
Author Contributions
M.I.M., performed experiments, analyzed data, prepared figures and wrote the manuscript; A.R., designed experiments and wrote the manuscript; A.L.R., performed experiments; V.B., developed analytical tools; and M.I.V., developed the hypothesis, designed experiments, analyzed the output data and wrote the main manuscript text. All authors reviewed the manuscript.
Acknowledgments
Authors are grateful to Dr. Fulvio Reggiori for his insightful comments and expert helping in the drafting of the manuscript. This work was supported by grants from the Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT-PICT01627-MIV and ANPCyT-PICT0411-AR), the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET-PIP2527-MIV) and the University of Buenos Aires (UBA-UBACyT M072-MIV). MIM and ALR are CONICET fellows.
References
- Levine B. & Klionsky D. J. Development by self-digestion: Molecular mechanisms and biological functions of autophagy. Dev Cell 6, 463–477 (2010). [DOI] [PubMed] [Google Scholar]
- Hara T. et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441, 885–889 (2006). [DOI] [PubMed] [Google Scholar]
- Qu X. et al. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell 128, 931–946 (2007). [DOI] [PubMed] [Google Scholar]
- Yang Z. & Klionsky D. J. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 22(2), 124–131 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D. J. et al. A unified nomenclature for yeast autophagy-related genes. Dev Cell 5, 539–45 (2003). [DOI] [PubMed] [Google Scholar]
- Mari M., Tooze S. A. & Reggiori F. . The puzzling origin of the autophagosomal membrane. F1000 Biology Reports 3, 25 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E. & Mizushima N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy 6, 764–776 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z. & Klionsky D. J. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 9(10), 1102–9 (2007). [DOI] [PubMed] [Google Scholar]
- Axe E. L. et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 182(4), 685–701 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi-Nishino M. et al. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol. 11, 1433–7 (2007). [DOI] [PubMed] [Google Scholar]
- Madeo F., Tavernarakis N. & Kroemer G. Can autophagy promote longevity? Nature Cell Biology. 12, 842–846 (2010). [DOI] [PubMed] [Google Scholar]
- Ropolo A. et al. The pancreatitis-induced vacuole membrane protein 1 triggers autophagy in mammalian cells. J Biol Chem. 282, 124–133 (2007). [DOI] [PubMed] [Google Scholar]
- Vaccaro M. I., Ropolo A., Grasso D. & Iovanna J. L. A novel mammalian trans-membrane protein reveals an alternative initiation pathway for autophagy. Autophagy 4(3), 388–90 (2008). [DOI] [PubMed] [Google Scholar]
- Grasso D. et al. Zymophagy, a novel selective autophagy pathway mediated by VMP1-USP9x-p62, prevents pancreatic cell death. J Biol Chem. 286(10), 8308–24 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo R. P. et al. Gemcitabine induces the VMP1-mediated autophagy pathway to promote apoptotic death in human pancreatic cancer cells. Pancreatology 10(1), 19–26 (2010). [DOI] [PubMed] [Google Scholar]
- Grasso D. et al. Autophagy and VMP1 expression are early cellular events in experimental diabetes. Pancreatology 9(1–2), 81–8 (2009). [DOI] [PubMed] [Google Scholar]
- Lo Re A. E. et al. A novel AKT1-GLI3-VMP1 pathway mediates KRAS-induced autophagy in cancer cells. J Biol Chem. 287(30), 25325–34 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y. et al. C. elegans screen identifies autophagy genes specific to multicellular organisms. Cell 141, 1042–1055 (2010). [DOI] [PubMed] [Google Scholar]
- Dusetti N. J. et al. Cloning and expression of the rat vacuole membrane protein 1 (VMP1), a new gene activated in pancreas with acute pancreatitis, which promotes vacuole formation. Biochem Biophys Res Commun 18, 290(2), 641–9 (2002). [DOI] [PubMed] [Google Scholar]
- Liang X. H. et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 402, 672–6 (1999). [DOI] [PubMed] [Google Scholar]
- Pattingre S. et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 122, 927–39 (2005). [DOI] [PubMed] [Google Scholar]
- Feng W., Huang S., Wu H. & Zhang M. Molecular basis of Bcl-xL's target recognition versatility revealed by the structure of Bcl-xL in complex with the BH3 domain of Beclin-1. J Mol Biol. 372(1), 223-35 (2007). [DOI] [PubMed] [Google Scholar]
- Oberstein A., Jeffrey P. D. & Shi Y. Crystal structure of the Bcl-XL-Beclin 1 peptide complex: Beclin 1 is a novel BH3-only protein. J Biol Chem. 282, 13123–13132 (2007). [DOI] [PubMed] [Google Scholar]
- Itakura E., Kishi C., Inoue K., Mizushima N. Beclin 1 form two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell 19, 5360–5372 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funderburk S. F., Wang Q. J. & Yue Z. The Beclin 1-VPS34 complex – at the crossroads of autophagy and beyond. Trends in Cell Biology 20, 355–362 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga K. et al. Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J Cell Biol. 190(4), 511–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y. et al. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 11(4), 468–76 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Yoshimori T. & Ohsumi Y. The role of atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 27, 107–32 (2011). [DOI] [PubMed] [Google Scholar]
- Klionsky D. Look people, "Atg" is an abbreviation for "autophagy-related." That's it. Autophagy 8–9, 1–2 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piston D. W. & Kremers G. J. Fluorescent protein FRET: the good, the bad and the ugly. Trends Biochem Sci. 32, 407–414 (2007). [DOI] [PubMed]
- Klionsky D. J., Cuervo A. M. & Seglen P. O. Methods for monitoring autophagy from yeast to human. Autophagy 3, 181–206 (2007). [DOI] [PubMed] [Google Scholar]
- Wei Y., Bassik M., Levine B. JNK1-Mediated Phosphorylation of Bcl-2 Regulates Starvation-Induced Autophagy. Mol Cell 30(6), 678–688 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y. et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19, 5720–28 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara A., Kabeya Y., Ohsumi Y. & Yoshimori T. Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep. 2, 330–335 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y. & Klionsky D. J. Physiological functions of Atg6/Beclin 1: a unique autophagy-related protein. Cell Research. 17, 839–849 (2007). [DOI] [PubMed]
- Sun Q. et al. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. PNAS. 105(49), 19211–16 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillooly D. J. et al. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 19, 4577–4588 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N. et al. The Atg16L1 complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell 5, 2092–100 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C. et al. Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature 18, 481(7382), 511–5 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. & Klionsky D. J. Eaten alive: a history of macroautophagy. Nat Cell Biol. 12(9), 814–22 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita M. et al. Structure of Atg5-Atg16, a complex essential for autophagy. J Biol Chem 282(9), 6763–72 (2007). [DOI] [PubMed] [Google Scholar]
- Suzuki K., Kubota Y., Sekito T. & Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells 12(2), 209–18 (2007). [DOI] [PubMed] [Google Scholar]
- Furuya N., Yu J., Byfield M., Pattingre S. & Levine B. The evolutionary domain of Beclin 1 is required for Vps34 binding, Autophagy and tumor suppression fuction. Autophagy 1(1), 46–52 (2005). [DOI] [PubMed] [Google Scholar]