Abstract
An ‘intrinsically disordered protein’ (IDP) is assumed to be unfolded in the cell and perform its biological function in that state. We contend that most intrinsically disordered proteins are in fact proteins waiting for a partner (PWPs), parts of a multi-component complex that do not fold correctly in the absence of other components. Flexibility, not disorder, is an intrinsic property of proteins, exemplified by X-ray structures of many enzymes and protein-protein complexes. Disorder is often observed with purified proteins in vitro and sometimes also in crystals, where it is difficult to distinguish from flexibility. In the crowded environment of the cell, disorder is not compatible with the known mechanisms of protein-protein recognition, and, foremost, with its specificity. The self-assembly of multi-component complexes may, nevertheless, involve the specific recognition of nascent polypeptide chains that are incompletely folded, but then disorder is transient, and it must remain under the control of molecular chaperones and of the quality control apparatus that obviates the toxic effects it can have on the cell.
Introduction
Flexibility and disorder are two different concepts. When it applies to a polypeptide chain that has hundreds of internal degrees of freedom, flexibility describes concerted changes that affect a few degrees of freedom, modifying the overall structure without destroying it. Disorder implies a lack of constraints on many or all the degrees of freedom of the chain and no permanent structure, but the flexibility of proteins is intrinsic, part of their function, and an essential feature of molecular recognition. Many X-ray structures, some going back to the early 1970s, illustrate how a protein can adjust its conformation while making specific interactions with a ligand. Disorder does occur in the test tube, as purified polypeptides are seen to lack a permanent structure. The concept of “intrinsically disordered proteins” (IDPs) assumes that the lack of structure also occurs in the cell, and that a disordered polypeptide is capable of specific molecular recognition and performs a viable biological function [1-7]. The evidence is currently scant for both assumptions. In vivo, most proteins are part of oligomeric assemblies and multi-component complexes, and the disorder observed with purified polypeptides in vitro may result from the absence of other components. On the other hand, disorder-order transitions are sometimes observed both in the crystal and in solution when two proteins form a complex. In such cases, accepted mechanisms of protein-protein recognition may account for observed kinetics of the association reaction, but they do not explain its specificity in the crowded environment of the cell. Nevertheless, disorder must occur in vivo when polypeptide chains are being synthesized, and it may represent a serious obstacle to the self-assembly of multi-component complexes. The concept of IDP provides no plausible model for that process, and we suggest that most, if not all, IDPs are in fact PWPs (proteins waiting for a partner) protected from promiscuous interactions by chaperones and subject to the quality control apparatus of the cell until they meet their cognate partners.
IDPs are (mostly) artifacts of current methods of protein production
In the last twenty years, the great majority of proteins used in biophysical and structural studies have been over-expressed from cloned DNA fragments in Escherichia coli or another expression host. The procedure, standard in structural genomics, has obvious limitations in spite of its success. The target protein may be part of a hetero-complex or a multi-component assembly in the source organism, where it interacts with other polypeptide chains, nucleic acids, or prosthetic groups. These components are absent, or at least not over-expressed, in the expression host, and the target may not fold properly without making these interactions. The long tail segments present in many ribosomal proteins illustrate the case: they are disordered in the purified protein but fully ordered in the ribosome, where interactions with the RNA determine their conformation [8-9].
Genome-wide studies of protein-protein interactions by genetic (yeast two-hybrid) and analytical (tandem-affinity purification coupled to mass spectrometry) methods indicate that a majority of eukaryotic proteins are part of hetero-complexes coded by more than one gene. In the yeast Saccharomyces cerevisiae, at least 70% of the proteins involved in transcription and translation are known to be part of assemblies that contain an average of 4.7 components [10], and the list is still far from complete. When we launched the Orsay Yeast Structural Genomics pilot-program in 2001, we knew hetero-complexes to be a problem, though not to what extent. In fact, of the 208 S. cerevisiae open reading frames (ORFs) that we selected as targets, 75% were expressed at a satisfactory level in E. coli, but only 25% could be purified in a soluble form. Nearly half of those gave crystals of some sort, but few were suitable for structure determination [11-12]. The low yield of the purification procedure and the poor quality of the crystals suggested that many of our targets did not fold properly, so we deleted terminal segments that sequence-based procedures predicted to be disordered. The new constructs were often expressed at a higher level, but only one in four showed better solubility, and only seven yielded better crystals [13]. When the pilot-program was completed in 2005, it had produced a structure for 12 novel proteins, only 6% of the initial set. Yet, the ORF-by-ORF approach proved fruitful. The procedure developed for the pilot-program yielded many other X-ray structures in Orsay, and it could easily be adapted to prepare yeast hetero-complexes in the frame of the 3D-Repertoire and SPINE2-Complexes European programs [14-15].
Other structural genomics programs have had a similar experience on a much larger scale [16]. Expressing individual eubacterial or archaeal ORFs in E. coli often yields more soluble proteins (up to 50%) than we got with yeast. The great majority are homo-oligomers, and so were most of the yeast proteins we solved. Mammalian proteins, including human, do far worse: less than 10% express as soluble material. Moreover, very few of the mammalian structures determined by structural genomics programs (or in other labs for that matter) are of full-length proteins. Most are fragments, often single domains cut out of ORFs that are too large for expression in E. coli or in vitro. Splitting a mammalian ORF into putative domains yields many constructs that do not express into soluble proteins, and when some remain unfolded, it may just be due to all the intra- and inter-chain contacts that cannot be made.
Flexibility versus disorder in crystals and in macromolecular recognition
Although a crystal is definitely not the best place to find disorder, the first sequence-based methods to identify IDPs relied on features observed in crystal structures [17-18]. In a Protein Data Bank (PDB) entry, residues that are present in the sequence, but not the coordinate set, count as disordered, but to a crystallographer ‘disorder’ only means that the electron density is low, and its atomic interpretation uncertain. The corresponding atoms either have a high B-factor or are reported as ‘missing’. The B-factor measures the mean-square fluctuation of the atomic position: an atom that moves by 1.25 Å has B≈120 Å2 and a weak electron density. In the PDB, only 3% of the protein atoms have such high B-factors, because when a side chain or a chain segment has a weak density it usually counts as ‘missing’, even though the amplitude of its movement may be less than the length of a covalent bond. A low electron density can also mean that the atom occupies several discrete positions, but this is rarely reported: in the PDB, alternate positions concern only 0.8% of all protein atoms, almost all of them side chain atoms. A chain segment with two conformations is likely to be ‘missing’, albeit far from a state of intrinsic disorder. Even when a whole domain is ‘missing’, there may be no actual disorder. An early example is Kol, an immunoglobulin that forms crystals in which only the antigen-binding Fab moieties are in contact. The Fc moieties are free to move in the empty space in between, and they lack electron density even though they are fully structured [19]. The linker peptide, a short polyproline II helix, is flexible but not disordered either [20].
Kol illustrates how flexibility has been part of protein crystallography almost from the beginning, and its functional importance was soon recognized [21]. Whereas this is now commonplace, other cases dating from the same early period are still worth citing: hemoglobin, where flexibility is required for the allosteric transition, and the NAD-dependent dehydrogenases. X-ray structures determined in the 1970s show how the dehydrogenases change from an open conformation in the absence of the coenzyme, to a closed one in its presence [22-24]. The transition, which involves movements of flexible loops and/or hinge rotations of domains and subunits, remodels the active site and allows the coenzyme to enter and leave. Thus, it must play an essential part in the catalytic cycle.
A decade before any 3D structure was known, Koshland [25-26] had predicted enzymes to be flexible and offered substrate-induced conformation changes as the answer to the question: how does hexokinase manage to transfer the gamma-phosphate of ATP to a sugar hydroxyl and not to water, equally reactive and much more abundant? X-ray structures have shown the prediction to be correct for hexokinase [27], and for many other enzymes. Lysozyme, ribonuclease A, and chymotrypsin, initially seemed to prove Koshland wrong, but we now know that their apparent rigidity is the exception, not the rule (and the requirement for excluding water does not hold for hydrolases). Moreover, the flexibility of chymotrypsin was soon established by a structure of its precursor chymotrypsinogen [28], from which it differs by the cleavage of a single peptide bond. The cleavage induces main chain movements throughout the molecule, including the active site and the substrate binding pocket. The related trypsin/trypsinogen system also displays a large change in conformation, and, interestingly, two competing sets of X-ray structures describe it in different ways, possibly as alternative interpretations of a weak density: Felhammer et al. [29] see disordered loops in trypsinogen becoming ordered in trypsin, where Kossiakoff et al. [30] describe movements between defined positions.
Trypsinogen also illustrates the role of flexibility (or disorder-order transitions) in macromolecular recognition: it becomes fully ordered and trypsin-like, when it binds the pancreatic trypsin inhibitor [31]. In general, flexibility shows up as conformation changes when comparing two X-ray structures obtained with and without a ligand; the ligand can be anything from H+ or a metal ion to DNA or another protein. Disorder-order transitions are less common, and the disorder may only be apparent. An early example concerns DNA recognition by the lactose operon repressor (LacR), a 154 kDa tetramer. One-dimensional proton nuclear magnetic resonance (NMR) spectra, albeit unresolved as expected for a protein this size, contained narrow lines that could be attributed to the DNA-binding ‘headpiece’ (residues 1-61) [32-33]. The headpiece is folded, but flexibly connected to the protein body. It is also mobile in crystals, and it takes a fixed position only in the presence of the cognate DNA [34]. As a result, the PDB reports it as ‘missing’ in the free repressor (entry 1LBI) but present in the DNA complex (entry 1EFA). Here again, flexibility implies no disorder, and its functional role is obvious: the headpiece can orient itself relative to the DNA double helix much faster than the rotational diffusion of the whole tetramer would allow. This would be useless if it was not properly folded.
Modeling rigid and flexible recognition
Rigid body macromolecular recognition accounts for the high stability and specificity of antigen-antibody, enzyme-inhibitor, and many other types of protein-protein complexes. Its mechanism is relatively well understood: two complementary protein surfaces come into contact to form an interface that typically involves 24 residues and buries 800 Å2 of protein surface on each component [35-36]. In such systems, docking algorithms that simulate the association of the free components generally yield good quality models of the assembly [37-39]. These algorithms take into account a number of properties, including electrostatics, but shape recognition is their essential criterion. Their performance degrades quickly when the molecules change conformation, and then flexibility must be simulated in order to generate acceptable solutions [40-41].
The kinetics of association are rather simple in the absence of conformation changes. A single bimolecular step is usually observed, and the rate constant (kon) is in the range 5.104-5.108 M-1s-1 [42], compatible with a simple diffusion-collision mechanism. The lower bound of the range corresponds to random collisions that yield a stable complex if, and only if, the proper regions of the two protein surfaces happen to face each other. The lock-and-key model requires in principle the two binding patches to be perfectly positioned and oriented. It effectively predicts kon=0 but with more reasonable assumptions on the geometry of the transition state, kon evaluates to 105-106 M-1s-1 [43-44], and most enzyme-inhibitor and antigen-antibody complexes have binding rates in this range. LacR binds DNA much faster than this, but it undergoes facilitated one-dimensional diffusion along the double helix, a mechanism applicable only to DNA recognition [45-46]. Long-range electrostatic interactions modulate binding rates in a way that can be modeled from the charge distribution on the protein surfaces [44,47-49], and that quantitatively explains most of the larger kon values reported in [42].
Flexibility adds a level of complexity to the binding process. Conformation changes and disorder-to-order transitions are expected to make association slower, but in a way that is difficult to model. A plausible mechanism of flexible recognition is conformer selection: a fraction of the receptors pre-exist in the correct conformation, and only those can bind the ligand (‘receptor’ and ‘ligand’ are here for convenience only). An alternative is induced fit: most, if not all, of the receptor molecules are able to form a low affinity complex with the ligand, and the interaction promotes the conformation changes that yield the stable assembly. Both mechanisms predict the binding kinetics to be biphasic, but the first order step (the conformation change) is often fast on the time scale of the experiment, and only one phase is detected. Its rate should be proportional to the fraction of the receptors that have the correct conformation, if conformer selection is the dominant mechanism, and to the probability that the intermediate evolves into the product before it dissociates, if induced fit applies.
Kinetic data are available on many systems that involve conformation changes and a few that display disorder-to-order transitions. Conformer selection can often be excluded. For instance, the NAD-dependent dehydrogenases must be in an open conformation when they bind the coenzyme, which they do at nearly diffusion-limited rates. On the other hand, conformer selection certainly contributes to protein-protein recognition when the conformation changes are of limited amplitude [49-51]. However, the observed binding rates either imply that native-like conformations are highly populated to start with, or that induced fit coexists with conformer selection. Thus, induced fit must be the dominant mechanism when trypsinogen binds pancreatic trypsin inhibitor. The affinity of the precursor for pancreatic trypsin inhibitor is eight orders of magnitude less than for trypsin, due to koff increasing by six orders while kon decreases by only two [52-53]. Conformer selection would require 1% of the trypsinogen molecules to preexist in a trypsin-like conformation, whereas the actual fraction is estimated to be less than one in a million.
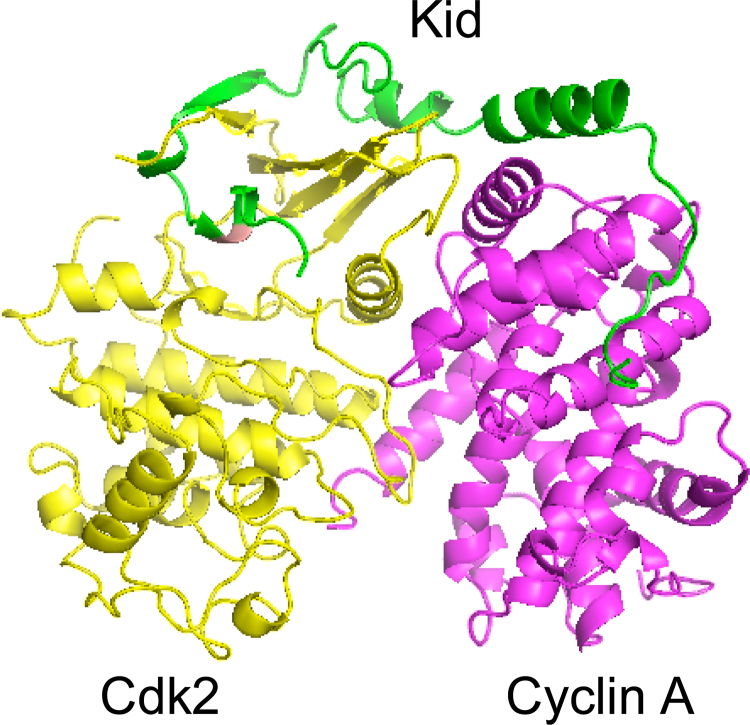
Conformer selection may also involve (partly) disordered proteins. An example is the kinase inhibitory domain (KID) of the p27Kip1 cyclin-dependent inhibitor. In solution, KID contains a significant amount of α-helix detected by NMR and circular dichroism [54]. In crystals of the ternary complex with Cdk2 and cyclin A (Figure 1), its N-terminal half is partly helical and interacts with the cyclin, whereas the C-terminal half forms an open loop in contact with the kinase [55]. KID has nanomolar affinity for either Cdk2 alone or cyclin A alone and remarkable binding kinetics: kon is low (5.103 M-1s-1) for Cdk2 alone, and high (1.6 to 3.106 M-1s-1) for cyclin A alone. It is also high with the Cdk2-cyclin complex, but then the reaction is biphasic and the second step as slow as for Cdk2 alone [54]. Albeit compatible with induced folding, the kinetics suggest a conformer selection mechanism by which the cyclin quickly associates with the many KID molecules that contain a helical fragment, while the C-terminal conformation recognized by the kinase is very rare.
Figure 1. The p27Kip1-Cdk2-cyclin A ternary complex.
In the crystal structure [55] (Protein Data Bank entry 1JSU), the kinase inhibitory domain of p27Kip1 (KID, in green) is observed to be partly helical and interact with both the kinase and the cyclin. When it is free in solution, KID is mostly disordered, but some of the helical structure is already present [54], making conformer selection a plausible alternative to induced fit as an explanation of the rapid association with the cyclin.
Disorder in vivo: does it exist, and how does the cell deal with it?
But what is the actual state of KID in the living cell? There, the overall protein concentration reaches hundreds of grams per liter, orders of magnitude above the concentrations of the purified proteins in test tube experiments [56]. As a result, proteins disordered in vitro may become partially ordered, and this can be tested in the test tube by adding molecular crowding agents. These agents have little effect on KID [57], but FlgM, a 97-residue polypeptide that binds the transcription factor σ28, gains structure in their presence. In dilute solution, FlgM is disordered except for transient α-helices in its C-terminal half. This half becomes fully ordered upon binding to σ28, while the N-terminal half remains disordered [58-59]. Adding a high concentration of other proteins (bovine serum albumin or ovalbumin), or glucose, induces structure in the C-terminal, but not the N-terminal half. Remarkably, in-cell NMR shows that the polypeptide over-expressed in E. coli also has an ordered C-terminal and a disordered N-terminal half. As E. coli σ28 is not over-expressed, an interaction with it cannot explain that transition, and it may be induced by the crowded environment in the cell [60].
If the disorder seen in vitro for KID, FlgM and other putative IDPs effectively occurs in the cell, it must affect the stability, the kinetics and the specificity of the interactions that mediate the function of these polypeptides. A conformation change or a disorder-order transition costs free energy, and it should make the assembly less stable. It does in trypsinogen/pancreatic trypsin inhibitor relative to trypsin/pancreatic trypsin inhibitor, but in general flexible recognition is associated with the formation of large interfaces [35], and the additional interactions must offset that cost. Thus, KID loses over 2800 Å2 of accessible surface area in contact with Cdk2-cyclin A, four times as much as an antibody in contact with the cognate antigen. The kinetic constraints could be of more consequence: the low kon of KID for binding Cdk2 (as opposed to Cdk2-cyclin A) predicts the binary complex to form in about an hour at a KID concentration of 10-7 M, and such a long lag is probably not compatible with its inhibitory function.
However, the most significant constraint in recognition is specificity. A disordered polypeptide chain has no defined shape, and it contains the same chemical groups as all other proteins, positioned more or less at random in space. How can it recognize, or be recognized by, another biomolecule? In a test tube experiment, the cognate interactions have no (or very few) competitors; in vivo they have thousands or millions. A linear sequence motif, or the presence of modified residues (phosphorylation, for instance), can serve as identification in some cases, but in general disorder must imply promiscuity, and be incompatible with all but a few cellular functions.
Another argument against this is that promiscuous interactions can be toxic [61], and cells have efficient quality control mechanisms designed to prevent them and to degrade or sequester misfolded polypeptides. Artificial conditions, such as over-expression in E. coli, may allow putative IDPs to escape quality control, but the problem of handling disorder in the cell is more general. It concerns all nascent polypeptide chains, and, most of all, those that will form oligomers. Nascent polypeptides are protected by chaperone proteins as they exit the ribosome, or sequestered by chaperonins, such as GroES/GroEL in bacteria, until their folding is completed. How they assemble to form oligomers is not understood at present. Their subunits are often unstable in vitro, and, whether folded or partially unfolded, they carry large hydrophobic surface patches that are prone to non-specific interactions. In the cell, their concentration must be kept low, and their assembly cannot be fast because it is a second or higher order reaction. With hetero-complexes, stoichiometry raises an additional question. It cannot be exact when subunits are independently synthesized on the ribosome, and the component in excess is a source of promiscuous interactions. Here again, the cell protects itself by using chaperones and protein degradation. Hemoglobin is an example: in beta-thalassemia, the alpha-chains are produced in excess of the beta-chains. They cannot form homo-tetramers (the beta-chains do) and are unstable, but a specialized chaperone prevents them from releasing heme and damaging the red blood cells [62]; in precursor cells, excess alpha-chains are polyubiquitinated and degraded by the proteasome [63].
Conclusion
The hemoglobin alpha-chain is not an IDP, it is a PWP, and, as such, it represents a very common situation. We contend that most, if not all, putative IDPs are in fact PWPs. They are unfolded in the test tube, but in vivo they are folded and part of a multi-component assembly (possibly of more than one). In general, molecular disorder is not compatible with function. A partly disordered polypeptide may be capable of specific recognition through a conformer selection mechanism, but then it is the ordered population that reacts, and the disorder is neither intrinsic nor functional. While disorder cannot be entirely avoided in the cell, it remains transient, and it is kept to a minimum by sophisticated mechanisms of biosynthesis and quality control. The mechanism that limits the damage an improper assembly of hemoglobin can cause in beta-thalassemia is probably one of many. Research in the field is very active and highly relevant to human health, and we may expect more to be discovered in coming years.
Acknowledgments
We thank Dr. Sameer Velankar (Hinxton, UK) for the statistics on atomic B-factors and occupancies in the Protein Data Bank.
Abbreviations
- IDP
intrinsically disordered protein
- KID
kinase inhibitory domain
- LacR
lactose operon repressor
- NMR
nuclear magnetic resonance
- ORF
open reading frame
- PDB
Protein Data Bank
- PWP
proteins waiting for a partner
Disclosure
MJES is Director and Shareholder of Equinox Pharma Ltd, which is involved in the commercialization and exploitation of chemoinformatics and bioinformatics software.
The electronic version of this article is the complete one and can be found at: http://f1000.com/prime/reports/b/5/2
References
- 1.Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol. 1999;293:321–31. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 2.Uversky VN, Oldfield CJ, Dunker AK. Showing your ID: intrinsic disorder as an ID for recognition, regulation and cell signaling. J Mol Recognit. 2005;18:343–84. doi: 10.1002/jmr.747. [DOI] [PubMed] [Google Scholar]
- 3.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 4.Dunker AK, Silman I, Uversky VN, Sussman JL. Function and structure of inherently disordered proteins. Curr Opin Struct Biol. 2008;18:756–64. doi: 10.1016/j.sbi.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Boehr DD, Nussinov R, Wright PE. The role of dynamic conformational ensembles in biomolecular recognition. Nat Chem Biol. 2009;5:789–96. doi: 10.1038/nchembio1209-954d. Erratum in: Nat Chem Biol.5:954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dyson HJ. Expanding the proteome: disordered and alternatively folded proteins. Q Rev Biophys. 2011;44:467–518. doi: 10.1017/S0033583511000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunker AK, Uversky VN. The case for intrinsically disordered proteins (IDPs) playing contributory roles in molecular recognition without a stable 3D structure. F1000 Biol Rep. 2013;5:1. doi: 10.3410/B5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chandra Sanyal S, Liljas A. The end of the beginning: structural studies of ribosomal proteins. Curr Opin Struct Biol. 2000;10:633–6. doi: 10.1016/S0959-440X(00)00143-3. [DOI] [PubMed] [Google Scholar]
- 9.Klein DJ, Moore PB, Steitz TA. The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J Mol Biol. 2004;340:141–77. doi: 10.1016/j.jmb.2004.03.076. [DOI] [PubMed] [Google Scholar]; www.f1000.com/prime/1019572
- 10.Pu S, Wong J, Turner B, Cho E, Wodak SJ. Up-to-date catalogues of yeast protein complexes. Nucleic Acids Res. 2009;37:825–31. doi: 10.1093/nar/gkn1005. [DOI] [PMC free article] [PubMed] [Google Scholar]; www.f1000.com/prime/717968016
- 11.Quevillon-Cheruel S, Collinet B, Trésaugues L, Minard P, Henckes G, Aufrère R, Blondeau K, Zhou CZ, Liger D, Bettache N, Poupon A, Aboulfath I, Leulliot N, Janin J, van Tilbeurgh H. Cloning, production, and purification of proteins for a medium-scale structural genomics project. Methods Mol Biol. 2007;363:21–37. doi: 10.1007/978-1-59745-209-0_2. [DOI] [PubMed] [Google Scholar]
- 12.Leulliot N, Trésaugues L, Bremang M, Sorel I, Ulryck N, Graille M, Aboulfath I, Poupon A, Liger D, Quevillon-Cheruel S, Janin J, van Tilbeurgh H. High-throughput crystal-optimization strategies in the South Paris Yeast Structural Genomics Project: one size fits all? Acta Crystallogr D Biol Crystallogr. 2007;61:664–70. doi: 10.1107/S0907444905000028. [DOI] [PubMed] [Google Scholar]
- 13.Quevillon-Cheruel S, Leulliot N, Gentils L, van Tilbeurgh H, Poupon A. Production and crystallization of protein domains: how useful are disorder predictions? Curr Protein Pept Sci. 2007;8:151–60. doi: 10.2174/138920307780363433. [DOI] [PubMed] [Google Scholar]; www.f1000.com/prime/717968017
- 14.Brooks MA, Gewartowski K, Mitsiki E, Létoquart J, Pache RA, Billier Y, Bertero M, Corréa M, Czarnocki-Cieciura M, Dadlez M, Henriot V, Lazar N, Delbos L, Lebert D, Piwowarski J, Rochaix P, Böttcher B, Serrano L, Séraphin B, van Tilbeurgh H, Aloy P, Perrakis A, Dziembowski A. Systematic bioinformatics and experimental validation of yeast complexes reduces the rate of attrition during structural investigations. Structure. 2010;18:1075–82. doi: 10.1016/j.str.2010.08.001. [DOI] [PubMed] [Google Scholar]; www.f1000.com/prime/717968018
- 15.Collinet B, Friberg A, Brooks MA, van den Elzen T, Henriot V, Dziembowski A, Graille M, Durand D, Leulliot N, Saint André C, Lazar N, Sattler M, Séraphin B, van Tilbeurgh H. Strategies for the structural analysis of multi-protein complexes: lessons from the 3D-Repertoire project. J Struct Biol. 2011;175:147–58. doi: 10.1016/j.jsb.2011.03.018. [DOI] [PubMed] [Google Scholar]
- 16.Structural Genomics Consortium. China Structural Genomics Consortium. Northeast Structural Genomics Consortium. Gräslund S, Nordlund P, Weigelt J, Hallberg BM, Bray J, Gileadi O, Knapp S, Oppermann U, Arrowsmith C, Hui R, Ming J, dhe-Paganon S, Park HW, Savchenko A, Yee A, Edwards A, Vincentelli R, Cambillau C, Kim R, Kim SH, Rao Z, Shi Y, Terwilliger TC, Kim CY, Hung LW, Waldo GS, Peleg Y, Albeck S, Unger T, Dym O, Prilusky J, Sussman JL, Stevens RC, Lesley SA, Wilson IA, Joachimiak A, Collart F, Dementieva I, Donnelly MI, Eschenfeldt WH, Kim Y, Stols L, Wu R, Zhou M, Burley SK, Emtage JS, Sauder JM, Thompson D, Bain K, Luz J, Gheyi T, Zhang F, Atwell S, Almo SC, Bonanno JB, Fiser A, Swaminathan S, Studier FW, Chance MR, Sali A, Acton TB, Xiao R, Zhao L, Ma LC, Hunt JF, Tong L, Cunningham K, Inouye M, Anderson S, Janjua H, Shastry R, Ho CK, Wang D, Wang H, Jiang M, Montelione GT, Stuart DI, Owens RJ, Daenke S, Schütz A, Heinemann U, Yokoyama S, Büssow K, Gunsalus KC. Protein production and purification. Nat Methods. 2008;25:135–46. doi: 10.1038/nmeth.f.202. [DOI] [PMC free article] [PubMed] [Google Scholar]; www.f1000.com/prime/1120082
- 17.Romero P, Obradovic Z, Kissinger CR, Villafranca JE, Garner E, Guilliot S, Dunker AK. Thousands of proteins likely to have long disordered regions. Pac Symp Biocomput. 1998:437–48. [PubMed] [Google Scholar]
- 18.Romero P, Obradovic Z, Kissinger CR, Villafranca JE, Dunker AK. Identifying Disordered Regions in Proteins from Amino Acid Sequences. Proc. I.E.E.E. International Conference on Neural Networks. 1997:90–5. [Google Scholar]
- 19.Huber R, Deisenhofer J, Colman PM, Matsushima M, Palm W. Crystallographic structure studies of an IgG molecule and an Fc fragment. Nature. 1976;264:415–20. doi: 10.1038/264415a0. [DOI] [PubMed] [Google Scholar]
- 20.Kessler H, Mronga S, Müller G, Moroder L, Huber R. Conformational analysis of a IgG1 hinge peptide derivative in solution determined by NMR spectroscopy and refined by restrained molecular dynamics simulations. Biopolymers. 1991;31:1189–204. doi: 10.1002/bip.360311007. [DOI] [PubMed] [Google Scholar]
- 21.Huber R, Bennett WS., Jr Functional significance of flexibility in proteins. Biopolymers. 1983;22:261–79. doi: 10.1002/bip.360220136. [DOI] [PubMed] [Google Scholar]
- 22.Adams MJ, Buehner M, Chandrasekhar K, Ford GC, Hackert ML, Liljas A, Rossmann MG, Smiley IE, Allison WS, Everse J, Kaplan NO, Taylor SS. Structure-function relationships in lactate dehydrogenase. Proc Natl Acad Sci U S A. 1973;70:1968–72. doi: 10.1073/pnas.70.7.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.White JL, Hackert ML, Buehner M, Adams MJ, Ford GC, Lentz PJ, Jr, Smiley IE, Steindel SJ, Rossmann MG. A comparison of the structures of apo dogfish M4 lactate dehydrogenase and its ternary complexes. J Mol Biol. 1976;102:759–79. doi: 10.1016/0022-2836(76)90290-4. [DOI] [PubMed] [Google Scholar]
- 24.Brändén CI. Coenzyme-induced conformational changes and subunit interactions of liver alcohol dehydrogenase. Biochem Soc Trans. 1977;5:612–5. doi: 10.1042/bst0050612. [DOI] [PubMed] [Google Scholar]; www.f1000.com/prime/717968021
- 25.Koshland DE., Jr Enzyme flexibility and enzyme action. J Cell Comp Physiol. 1959;54:245–58. doi: 10.1002/jcp.1030540420. [DOI] [PubMed] [Google Scholar]; www.f1000.com/prime/12818962
- 26.Koshland DE., Jr Correlation of structure and function in enzyme action. Science. 1963;142:1533–41. doi: 10.1126/science.142.3599.1533. [DOI] [PubMed] [Google Scholar]
- 27.Bennett WS, Jr, Steitz TA. Glucose-induced conformational change in yeast hexokinase. Proc Natl Acad Sci USA. 1979;75:4848–52. doi: 10.1073/pnas.75.10.4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Freer ST, Kraut J, Robertus JD, Wright HT, Xuong NH. Chymotrypsinogen: 2.5-angstrom crystal structure, comparison with alpha-chymotrypsin, and implications for zymogen activation. Biochemistry. 1970;9:1997–2009. doi: 10.1021/bi00811a022. [DOI] [PubMed] [Google Scholar]; www.f1000.com/prime/717968022
- 29.Fehlhammer H, Bode W, Huber R. Crystal structure of bovine trypsinogen at 1-8 A resolution. II. Crystallographic refinement, refined crystal structure and comparison with bovine trypsin. J Mol Biol. 1977;111:415–38. doi: 10.1016/S0022-2836(77)80062-4. [DOI] [PubMed] [Google Scholar]
- 30.Kossiakoff AA, Chambers JL, Kay LM, Stroud RM. Structure of bovine trypsinogen at 1.9 A resolution. Biochemistry. 1977;16:654–64. doi: 10.1021/bi00623a016. [DOI] [PubMed] [Google Scholar]
- 31.Bode W, Schwager P, Huber R. The transition of bovine trypsinogen to a trypsin-like state upon strong ligand binding. The refined crystal structures of the bovine trypsinogen-pancreatic trypsin inhibitor complex and of its ternary complex with Ile-Val at 1.9 A resolution. J. Mol. Biol. 1978;118:99–112. doi: 10.1016/0022-2836(78)90246-2. [DOI] [PubMed] [Google Scholar]; www.f1000.com/prime/717968023
- 32.Buck F, Rüterjans H, Beyreuther K. 1H NMR study of the lactose repressor from Escherichia coli. FEBS Lett. 1978;96:335–8. doi: 10.1016/0014-5793(78)80430-X. [DOI] [PubMed] [Google Scholar]
- 33.Wade-Jardetzky N, Bray RP, Conover WW, Jardetzky O, Geisler N, Weber K. Differential mobility of the N-terminal headpiece in the lac-repressor protein. J Mol Biol. 1979;128:259–64. doi: 10.1016/0022-2836(79)90129-3. [DOI] [PubMed] [Google Scholar]
- 34.Lewis M, Chang G, Horton NC, Kercher MA, Pace HC, Schumacher MA, Brennan RG, Lu P. Crystal structure of the lactose operon repressor and its complexes with DNA and inducer. Science. 1996;271:1247–54. doi: 10.1126/science.271.5253.1247. [DOI] [PubMed] [Google Scholar]; www.f1000.com/prime/717968024
- 35.Lo Conte L, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. J Mol Biol. 1999;285:2177–98. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 36.Chakrabarti P, Janin J. Dissecting protein-protein recognition sites. Proteins. 2002;47:334–43. doi: 10.1002/prot.10085. [DOI] [PubMed] [Google Scholar]
- 37.Smith GR, Sternberg MJ. Prediction of protein-protein interactions by docking methods. Curr Opin Struct Biol. 2002;12:28–35. doi: 10.1016/S0959-440X(02)00285-3. [DOI] [PubMed] [Google Scholar]
- 38.Lensink MF, Wodak SJ. Docking and scoring protein interactions: CAPRI 2009. Proteins. 2010;78:3073–84. doi: 10.1002/prot.22818. [DOI] [PubMed] [Google Scholar]
- 39.Janin J. Protein-protein docking tested in blind predictions: the CAPRI experiment. Mol. Biosystems. 2010;6:2351–62. doi: 10.1039/c005060c. [DOI] [PubMed] [Google Scholar]
- 40.Smith GR, Sternberg MJ, Bates PA. The relationship between the flexibility of proteins and their conformational states on forming protein-protein complexes with an application to protein-protein docking. J Mol Biol. 2005;347:1077–101. doi: 10.1016/j.jmb.2005.01.058. [DOI] [PubMed] [Google Scholar]
- 41.Bonvin AM. Flexible protein-protein docking. Curr Opin Struct Biol. 2006;16:194–200. doi: 10.1016/j.sbi.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 42.Qin S, Pang X, Zhou HX. Automated prediction of protein association rate constants. Structure. 2011;19:1744–51. doi: 10.1016/j.str.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Janin J. The kinetics of protein-protein recognition. Proteins. 1997;28:153–61. doi: 10.1002/(SICI)1097-0134(199706)28:2<153::AID-PROT4>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 44.Vijayakumar M, Wong KY, Schreiber G, Fersht AR, Szabo A, Zhou HX. Electrostatic enhancement of diffusion-controlled protein-protein association: comparison of theory and experiment on barnase and barstar. J Mol Biol. 1998;278:1015–24. doi: 10.1006/jmbi.1998.1747. [DOI] [PubMed] [Google Scholar]
- 45.Berg OG, Winter RB, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981;20:6929–48. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]; www.f1000.com/prime/717968025
- 46.Hammar P, Leroy P, Mahmutovic A, Marklund EG, Berg OG, Elf J. The lac repressor displays facilitated diffusion in living cells. Science. 2012;336:1595–8. doi: 10.1126/science.1221648. [DOI] [PubMed] [Google Scholar]; www.f1000.com/prime/717952254
- 47.Alsallaq R, Zhou HX. Prediction of protein-protein association rates from transition-state theory. Structure. 2007;15:215–24. doi: 10.1016/j.str.2007.01.005. [DOI] [PubMed] [Google Scholar]; www.f1000.com/prime/717968026
- 48.Schreiber G, Shaul Y, Gottschalk KE. Electrostatic design of protein-protein association rates. Methods Mol Biol. 2006;340:235–49. doi: 10.1385/1-59745-116-9:235. [DOI] [PubMed] [Google Scholar]
- 49.Schreiber G, Haran G, Zhou HX. Fundamental aspects of protein-protein association kinetics. Chem Rev. 2009;109:839–60. doi: 10.1021/cr800373w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou HX. Rate theories for biologists. Q Rev Biophys. 2010;43:219–93. doi: 10.1017/S0033583510000120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moal IH, Bates PA. Kinetic rate constant prediction supports the conformational selection mechanism of protein binding. PLoS Comput Biol. 2012;8:e1002351. doi: 10.1371/journal.pcbi.1002351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vincent JP, Lazdunski M. Pre-existence of the active site in zymogens, the interaction of trypsinogen with the basic pancreatic trypsin inhibitor (Kunitz) FEBS Lett. 1976;63:240–4. doi: 10.1016/0014-5793(76)80103-2. [DOI] [PubMed] [Google Scholar]
- 53.Pasternak A, Liu X, Lin TY, Hedstrom L. Activating a zymogen without proteolytic processing: mutation of Lys15 and Asn194 activates trypsinogen. Biochemistry. 1998;37:16201–10. doi: 10.1021/bi980951d. [DOI] [PubMed] [Google Scholar]
- 54.Lacy ER, Filippov I, Lewis WS, Otieno S, Xiao L, Weiss S, Hengst L, Kriwacki RW. p27 binds cyclin-CDK complexes through a sequential mechanism involving binding-induced protein folding. Nat Struct Mol Biol. 2004;11:358–64. doi: 10.1038/nsmb746. [DOI] [PubMed] [Google Scholar]; www.f1000.com/prime/1019529
- 55.Russo AA, Jeffrey PD, Patten AK, Massagué J, Pavletich NP. Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A-Cdk2 complex. Nature. 1996;382:325–31. doi: 10.1038/382325a0. [DOI] [PubMed] [Google Scholar]; www.f1000.com/prime/717968027
- 56.Ellis RJ. Macromolecular crowding: obvious but underappreciated. Trends Biochem Sci. 2001;26:597–604. doi: 10.1016/S0968-0004(01)01938-7. [DOI] [PubMed] [Google Scholar]
- 57.Flaugh SL, Lumb KJ. Effects of macromolecular crowding on the intrinsically disordered proteins c-Fos and p27(Kip1) Biomacromolecules. 2001;2:538–40. doi: 10.1021/bm015502z. [DOI] [PubMed] [Google Scholar]
- 58.Daughdrill GW, Chadsey MS, Karlinsey JE, Hughes KT, Dahlquist FW. The C-terminal half of the anti-sigma factor, FlgM, becomes structured when bound to its target, sigma 28. Nat Struct Biol. 1997;4:285–91. doi: 10.1038/nsb0497-285. [DOI] [PubMed] [Google Scholar]; www.f1000.com/prime/717968029
- 59.Daughdrill GW, Hanely LJ, Dahlquist FW. The C-Terminal Half of the Anti-Sigma Factor FlgM Contains a Dynamic Equilibrium Solution Structure Favoring Helical Conformations. Biochemistry. 1998;37:1076–82. doi: 10.1021/bi971952t. [DOI] [PubMed] [Google Scholar]
- 60.Dedmon MM, Patel CN, Young GB, Pielak GJ. FlgM gains structure in living cells. Proc Natl Acad Sci USA. 2002;99:12681–4. doi: 10.1073/pnas.202331299. [DOI] [PMC free article] [PubMed] [Google Scholar]; www.f1000.com/prime/717968030
- 61.Vavouri T, Semple JI, Garcia-Verdugo R, Lehner B. Intrinsic protein disorder and interaction promiscuity are widely associated with dosage sensitivity. Cell. 2009;138:198–208. doi: 10.1016/j.cell.2009.04.029. [DOI] [PubMed] [Google Scholar]; www.f1000.com/prime/1162626
- 62.Turbpaiboon C, Wilairat P. Alpha-hemoglobin stabilizing protein: molecular function and clinical correlation. Front Biosci. 2010;15:1–11. doi: 10.2741/3601. [DOI] [PubMed] [Google Scholar]; www.f1000.com/prime/717968031
- 63.Khandros E, Thom CS, D'Souza J, Weiss MJ. Integrated protein quality control pathways regulate free α globin in murine β-thalassemia. Blood. 2012;119:5265–75. doi: 10.1182/blood-2011-12-397729. [DOI] [PMC free article] [PubMed] [Google Scholar]; www.f1000.com/prime/717968033