Abstract
The classical ‘lock-and-key’ and ‘induced-fit’ mechanisms for binding both originated in attempts to explain features of enzyme catalysis. For both of these mechanisms and for their recent refinements, enzyme catalysis requires exquisite spatial and electronic complementarity between the substrate and the catalyst. Thus, binding models derived from models originally based on catalysis will be highly biased towards mechanisms that utilize structural complementarity. If mere binding without catalysis is the endpoint, then the structural requirements for the interaction become much more relaxed. Recent observations on specific examples suggest that this relaxation can reach an extreme lack of specific 3D structure, leading to molecular recognition with biological consequences that depend not only upon structural and electrostatic complementarity between the binding partners but also upon kinetic, entropic, and generalized electrostatic effects. In addition to this discussion of binding without fixed structure, examples in which unstructured regions carry out important biological functions not involving molecular recognition will also be discussed. Finally, we discuss whether ‘intrinsically disordered protein’ (IDP) represents a useful new concept.
Introduction
Preparations of the enzyme emulsin cleave β-glycosides, but not α-glycosides, while preparations of invertase cleave α-glycosides, but not β-glycosides. From these observations, Emil Fischer suggested in 1894 that the enzyme and substrate exert a mutual effect on each other like a lock and key [1-2]. Thus, the lock-and-key hypothesis was originally derived from studies on catalysis, not molecular recognition.
A short time later, in 1897, Paul Ehrlich applied the lock-and-key hypothesis to the problem of how an antibody binds specifically to a particular antigen [3-4]. In this example, the interaction results in specific binding, not catalysis. Thus, Ehrlich converted the lock-and-key hypothesis from explaining enzyme specificity to explaining protein-based molecular recognition.
However, the lock-and-key model gives rise to some interesting questions. For enzymatic transfer of a phosphate from ATP to an –OH acceptor via a lock-and-key mechanism, why doesn't an –OH from water simply outcompete the –OH from the acceptor, thus leading to ATP hydrolysis instead of phosphate transfer? Daniel Koshland suggested that sequestering the reactants from water via small conformational changes that he called “induced fit” would solve the problem, especially since water itself would be too small to induce the needed conformational changes [5-6]. Induced fit is the formation of an encounter complex between molecules in conformations distinct from their final conformations, followed by mutual structural adjustment until the intimate fit between the two partners is realized. Subsequent structural studies on a number of enzymes revealed substantial conformational changes upon substrate binding, which are consistent with induced fit but could also be explained by other mechanisms [7].
Besides induced fit, an alternative model called ‘conformational selection’ has been proposed to explain the association between a macromolecule and a flexible ligand [8]. In conformational selection, the ligand assumes an ensemble of conformations, and the protein binds to the conformation that gives the best fit to the binding site. Conformational selection and induced fit are often discussed as the two extreme possible mechanisms for the binding to a flexible ligand such as an IDP to a structured partner.
For enzymes, the lock-and-key and the induced-fit mechanisms lead to different kinetic equations, which can be considered as parts of a unified reaction cycle [9]. Analysis of enzyme kinetic data in terms of this unified reaction cycle suggests that substrate and enzyme concentrations determine whether conformational selection, induced fit, or a mixture of the two underlies the reaction [10]. The key point here is that not only is the structure of the final complex important but also the mechanism used to achieve the final complex.
As for non-catalytic molecular recognition by flexible proteins, many DNA-binding proteins contain regions that undergo disorder-to-order transitions upon binding to their DNA partners [11]. Georg Schulz collected several of these examples and suggested in 1979 that such a disorder-to-order binding mechanism could be helpful for some biological interactions by enabling the combination of relatively high specificity and low affinity [12]. Thermodynamic studies of several protein-DNA interactions gave further support for such disorder-to-order transitions, in which the authors, Ruth Spolar and Tom Record, described the binding process as “coupled-binding and folding,” or in one place “extreme induced fit” [13]. Just as for enzyme catalysis, it has been suggested that conformational selection and coupled binding and folding (sometimes called induced fit) represent the extreme possibilities for binding, and just as for enzymes, it has been suggested that either mechanism or even mixtures of the two mechanisms may be involved for any particular interaction [14].
As with the lock-and-key hypothesis, induced fit was originally developed to explain specific features of enzyme catalyzed reactions, and this concept was later converted to a molecular recognition mechanism when the underlying ideas were applied to non-catalytic binding interactions.
While enzyme catalysis very often occurs without significant disorder, for several enzymes disorder-to-order backbone rearrangements is a feature. In these cases, the substrate binds to part of the active site that is structured and then a disordered region folds onto the substrate typically including residues involved in the catalysis (and often to exclude water) and then the same region unfolds again to release the product [15-16].
Recent studies on enzyme reaction mechanisms using faster methods than previously available have revealed multiple steps in which conformational changes in the substrates are complementary to multiple conformational changes in the enzymes. These multiple substeps involve small amounts of energy and only slight conformational changes, resulting in an overall better fit and larger energy decrease in the transition state energy than could likely be achieved by one large conformational change [7].
The importance of this background for the current discussion is that binding for the catalytic event is necessarily highly structured: the enzyme has to bind more tightly to a molecular intermediate form than to the undistorted ground state or to the product(s) [7,9], and so the physiochemical requirements for catalysis depend on interactions with exquisite steric and electrostatic complementarity between the enzyme and the ligand. However, we argue that if the catalytic requirement for the interaction is dropped and mere binding is the biological function, then the molecular recognition event itself ought to become more relaxed. The question here is whether molecular recognition in the absence of catalysis can become so relaxed that fixed 3D structure is no longer a requirement for binding.
Effects on binding without specific structure formation
Suppose an unstructured protein binds to a partner without forming structure. How would one evaluate such a complex in the absence of structure? How does one determine the biological significance of such an interaction if one were found? To address these issues, several examples are presented below in a progression from the most to the least amount of structure.
In the first example, a region of intrinsic protein disorder binds to a partner via a disorder-to-order transition with part of the structure remaining unstructured. Such events are very common. In one study of short segments that bind to protein partners, out of 372 binding segments containing 10,434 residues, 13% of the residues remained unstructured after binding as determined from the lack of electron density in the crystal structures. This is substantially higher than the 7% figure for disordered residues observed for a set of 848 structured protein monomers [17]. Are these disordered regions merely present or do they contribute to the binding free energy?
In a few of the interactions containing IDP regions, studies have been carried out on the effects of removing all, or fractions, of the structurally unobserved regions (reviewed in [18]). Removal of such flanking disordered regions has been shown to result in both positive and negative changes in the free energy of binding (Fuxreiter, personal communication).
In one case, the disordered splicing factor 1 (SF1) binds to the large subunit of the U2 small nuclear RNA auxiliary actor (U2AF), the SF1 segment binds by a motif of 10 residues with Kd of 23.8 nM, and removal of residues not in physical contact with U2AF reduces the Kd to 55.6 nM. On the other hand, the full-length SF1 binding with U2AF has a Kd of 11.8 nM, indicating that the flanking but unstructured regions contribute significantly to binding free energy [19]. These data demonstrate binding energy without specific structure formation. Evidently, the steric complementarity required for enzyme catalysis has become considerably relaxed for mere association.
In the case above, the disordered region exhibits a measurable free energy of association with the remainder of the protein without the formation of specific structure. There are at least three alternative mechanisms as discussed below.
One possibility is that the unobserved residues alter the polypeptide in the unbound state. The removal of these residues would then affect the binding constant in the bound state. For example, if the binding mechanism were to depend on the conformational selection mechanism, then the unobserved residues could reduce the amount of time spent in a binding-competent conformation by a variety of mechanisms, thus reducing the on-rate, and lowering the overall affinity [8].
A second possibility is that there are several different conformations that enable the disordered protein to bind to the surface via short-range contacts. Having several such binding structures could result in missing structure in X-ray experiments due to incoherent scattering or to missing data in NMR experiments due to exchange broadening. Indeed, several examples of the same disordered region changing structure to bind to different surfaces have been observed [20-21]. All that would be needed is to have the alternative binding modes close to each other on the same binding surface. It could be argued that such a mechanism still uses structure for binding, just that there are multiple structures of similar energy that can interconvert over timescales required to collect either the X-ray or NMR data. We would give the counter argument that adopting multiple, different, interconverting structures is distinctly different from becoming structured upon binding.
A third possibility is that electrostatic interactions could occur over long range without the formation of any specific structure at all. Such a result would truly be interaction without a specific complementary structure. Consideration of electrostatic interactions leads us to the second example.
The second example involves the interaction between Sic1 and CDC4. In this example, the Sic1 protein is unstructured but contains nine similar motifs that are well separated along the sequence. Each of these motifs contains a central serine or threonine that can become phosphorylated [22]. Each phosphorylated Sic1 motif is recognized by the CDC4 protein, which recruits Sic1 for ubiquitination, thereby targeting it for degradation in late G1 phase, an event necessary for the onset of DNA replication [23]. Replacing different numbers of serines or threonines with alanines leads to loss of viability if fewer than six phosphorylation sites remain [22]. In vitro studies show that individual phosphorylated motifs bind weakly, but the affinity and the steepness of the binding isotherm increases as more sites are phosphorylated. When any six sites are phosphorylated, the affinity sharply increases and the binding isotherm is so sharp it resembles an on-off switch [22].
The CDC4 molecule contains just one site (!) that associates with the phosphorylated motif. If there is only one binding site, how do the additional sites increase the affinity and sharpen the binding isotherm? The main effect seems to be electrostatic interactions that keep the phosphorylated motifs near to the single binding site, so that, as soon as one hops off, another is nearby to hop on [24]. A mathematical description of these effects has been developed, and the resulting mean field statistical mechanical model for the electrostatic interactions gives reasonably good agreement with the experimental data, including both estimates of the threshold number of phosphorylation sites for binding, and also including experimental affinities of CDC4 for Sic1 fragments with different total charges [25].
The Sic1-CDC4 and the SF1-U2AF examples discussed above demonstrate that non-structured protein can contribute very significantly to binding free energy. As reviewed by Tompa and Fuxreiter [18] many additional examples have been observed in which unstructured regions contribute both positively and negatively to the binding free energy between an IDP and its protein partner. We anticipate that similar results will be found for IDPs binding to nucleic acids if such examples have not been found already. These observations clearly make the point that unstructured regions of protein can contribute to binding free energy without becoming structured in the final complex.
While the CDC4 has a single site for binding one phosphate group and the flanking amino acids on Sic1, which has multiple sites of phosphorylation along with sequence-similar motifs for the flanking amino acids in order to fit onto the binding site on CDC4, the mathematical model explaining the increased affinity arising from the remainder of the molecule does not require specific structure, merely electrostatic interactions between a dynamic IDP and its partner. The question then arises whether such non-specific interactions could lead to specific protein association without the formation of any long lasting complementary interfaces between an IDP and its partner. In other words, is there a mechanism by which an IDP could bind to a partner without itself forming specific structure?
Binding without structure formation: is it possible?
The various experimental observations above could be combined to yield a model with an overall electrostatic attraction between an IDP and its partner, coupled with several local docking interactions that rapidly convert from one to another. Such an interaction mechanism could lead to a specific association between an IDP and its partner without the formation of stable structure.
Let's consider this possibility from a more traditional view. Protein complex formation typically involves at least two steps. Upon meeting, an encounter complex is formed, either proceeding towards the final complex or towards dissociation. Evidence suggests that encounter complexes are dominated by electrostatic forces, but hydrophobic interactions can also play a role [26]. An interesting variant is to consider the subsequent events in terms of game theory, according to which the interacting partners continually affect the conformational landscapes of each other in such a way that consecutive steps depend on prior steps until the final complex is formed [27,28]. What if the folding funnel for the overall complex were rather flat, with no energy minimum corresponding to one specific structure? In this case, the moves and counter-moves would continue endlessly, leading to a long-lived, dynamically fluctuating encounter complex that, of course, could dissociate at any moment. Since there are data supporting the existence of encounter complexes, the question then becomes whether it is possible for encounter complexes to be long-lived.
The above discussion has an interesting parallel to the molten globule concept. The capability of a polypeptide chain to adopt partially folded intermediates was first proposed as a part of the frame-work model of protein folding, with a collapsed, but internally dynamic, short-lived transient intermediate protein form in protein folding [29]. In later studies it became apparent the intermediate with a collapsed, but internally dynamic, structure was shown to be a stable form for some proteins, following slight structural destabilization using a variety of treatments [30]. Next, this intermediate was shown to be transiently populated at the early stages of the globular protein folding [31]. Finally, certain proteins were suggested to form molten globules in their functional states [32]. So, will encounter complexes turn out to exhibit a similar progression, being first recognized as transients, then as stable forms under some particular conditions, then as stable forms under physiological conditions for some protein sequences? Time will tell.
Has any protein-protein interaction without any structure formation ever been observed?
A number of IDPs interact with each other to form dimers that sometimes exhibit very simple folds such as leucine zippers [33], and that at other times exhibit more complex folds such as helical bundles [34,35]. For a few examples, the IDP dimers are highly dynamic with only localized regions that show evidence of structure formation [36,18]. All of these examples, more or less, fit the standard view of coupled binding and folding or induced fit.
However, Sigalov and co-workers have reported homodimerization of the cytoplasmic region of the T cell receptor zeta subunit that is not accompanied by measurable shifts in the CD spectra [37] nor by measurable chemical shift changes in the NMR spectra possibly suggesting association without structure formation [38]. Other explanations for this observation include the possibility that the CD spectra might not be sensitive enough to pick up formation of highly localized structure or technical problems such as exchange broadening for the same protein regions before and after dimerization could obscure the formation of structure during the interaction, so further work needs to be carried out to prove that the molecular association is truly occurring without the formation of protein structure. Furthermore, the initial report was in 2004 and other laboratories have not yet reported similar findings, and this absence of confirmatory data adds uncertainty to Sigalov's interpretation of his observations. However, given the results for a number of complexes in which IDP regions have been shown to contribute to the overall free energy of the protein-protein interaction [24,25,39,40] and given evidence for the existence of encounter complexes along with the various models to explain their interaction without structure [26-28], it is our opinion that Sigalov's results cannot be dismissed out of hand. While our own view is that specific protein-protein interactions likely require at least some localized structure at a key contact point such as observed for the Sic1-CDC4 interaction, formation of protein complexes with highly transient structure formation ought to be taken as a possibility until ruled out by further studies.
Flexible linkers, flexibility, and molecular recognition
Direct involvement in molecular recognition is not the only type of biological function carried out by proteins. In addition, disorder can affect molecular recognition without direct involvement in the binding interface. Two aspects will be discussed here: flexible linkers and free energy in the unbound state. These examples emphasize that the final 3D structure of the complex is not the only biologically important aspect of molecular recognition, and that the on-rates, off-rates, and conformational changes enabling association and dissociation can also contribute to biological function.
Flexible linkers can enable combinations of interactions that would not be sterically allowed by completely rigid structures. For example, calmodulin uses a flexible linker between two domains to wrap around its target helix [41]. Formation and dissociation of the calmodulin-target complex would simply not be possible without the flexibility of the linker. Multiple zinc fingers connected by flexible linkers enable certain transcription factors to wrap up their target DNA molecules [42]. Again such complexes could not form or dissociate without the flexibility of the linkers.
Flexible linkers can also affect rates of association and affinities. A particularly interesting example is provided by voltage gated ion channels. Such channels exhibit three states: closed (voltage sensitive), open, and inactive (voltage insensitive). While in the open state, ion flow through the open channel collapses the cell membrane potential. Thus, the amount of time spent in the open state is important for the biological function of the channel. The mechanism of closing is via a ‘ball and chain’, where the terminus of a disordered region closes the open channel by a binding event [43-44]. Lengthening of the ‘chain’ slows the closure, shortening the chain speeds it up [45,46], suggesting the possibility that the ball undergoes a random-walk search for binding site, but more detailed studies are needed to confirm this model [47]. Comparing the orthologous voltage channels in sperm from different mammals shows that the exon corresponding to the chain region has significant length variability that arises from insertions and deletions (indels). The number of indel substitutions is 5 to 8 times higher than is generally observed in genomic studies and indels within the disordered regions are considerably longer than average indels, suggesting that positive selection is occurring [48]. The authors suggest that the observed chain-region length variability, which can affect sperm motility, may be an important determinant in sperm competition, thus accounting for the positive selection.
Another very interesting example is provided by the entirely unstructured ~200 residue kinase inhibitor p27kip1 [49], which plays a key role in the control of eukaryotic cell division by the inhibition of several different cyclin-dependent protein kinases (Cdks) [50,51]. For one such complex, ~70 residues of p27kip1 wrap around the outside of a dimer of a cyclin and its cognate Cdk [52]. The flexibility of p27kip1 allows it to associate and dissociate segmentally [50,53], thereby providing opportunities for regulation and control. More specifically, segmental dissociation enables phosphorylation of p27kip1's Y88 by a non-receptor tyrosine kinase, leading to the exposure of the Cdk's active site, which then phosphorylates p27kip1's T187. This second phosphorylation provides a signal for ubiquitination, which then leads to digestion of p27kip1 via the proteasome, which in turn promotes cell cycle progression [50,51].
This pathway of phosphorylation leading to ubiquitination, in turn leading to degradation, is very commonly observed in eukaryotic cells as a means to remove or deplete the levels of key regulatory proteins [54]. Both phosphorylation and ubiquitination commonly occur in regions of disorder [55-57], and having a sufficiently long region of disorder appears to be important for entry into the proteasome [58].
As a final example of the consequences of flexible linkers on molecular recognition, Kuriyan and Eisenberg [59] argue that the proximity brought about by flexible linkers brings about an amplification of the effects on one domain of random mutations in the colocalized domain. Through natural selection, this amplification of effects by proximity leads to specific interactions and to a startling variety of complex allosteric controls.
As for effects arising from the free energy of the unbound state, the unbound, flexible state is the starting point for the many examples involving disorder-to-order transitions upon binding to their partners, which in turn might be structured or also disordered. While structure accounts for the final recognition, the rate of association or dissociation might also be important for biological function: mutations that do not involve any of the interacting residues but that affect the free energy in the unbound state would affect the final binding constant [60]. Such an alteration in free energy by mutation could also be viewed as an alteration in the underlying protein ensemble. This ensemble view was recently used to explain mutational effects on binding events that lead to allostery [61,62]. In this view, rather than affecting a Rube-Goldberg type pathway underlying allostery, the mutation could be affecting the conformational ensemble and hence the allostery. In a recent study, the binding constants of associations between structured proteins were shown to have a correlation with the measured off-rates and to be rather independent of on-rates. On the other hand, binding constants of associations involving an initially disordered protein were shown to have a correlation with the on-rates [63]. A possible mechanism here is that a reduced free energy in the unbound state (as compared to the bound state) would be expected to both reduce the final affinity and to slow the on-rate. These disorder-dependent effects on binding kinetics and affinity values don't directly involve the molecular details of the bound state, but are nevertheless likely to lead to important biological consequences.
One-to-many binders and multifacial complexes
IDPs are known to participate in one-to-many and many-to-one interactions, where one IDP or one intrinsically disordered region (IDR) binds to multiple partners potentially gaining very different structures in the bound state, or where multiple unrelated IDPs/IDRs bind to one partner, potentially gaining similar structures in the bound state [20,21,60].
The one-to-many binding mechanism is especially interesting since it might generate multifacial complexes, where the same region of an IDP can be engaged in interaction with multiple unrelated partners and be able to fold into very different conformations in the bound state. One of the illustrative examples of such one-to-many binders is p53, a single C-terminal region, which is known to interact with at least four different partners [20]. The amino acids involved in each interaction show a significant overlap and no two of these interactions could exist simultaneously. Furthermore, the same residues adopt helix, sheet, and two different irregular structures when associated with the different partners. Finally, the same amino acids are buried to very different extent in each of the molecular associations [20]. These results show that one of the functional advantages of IDPs/IDRs over ordered proteins and domains is the ability of one disordered segment to bind to multiple partners due to its ability to adopt different conformations in the bound state.
Recent analysis revealed that the C-terminal recognition domain of p53 is not a unique entity and several other IDPs can be engaged in the formation of multifacial complexes [21]. These examples highlight the transient nature of the intrinsic disorder-based interactions and emphasize the extreme adaptability of IDPs. In general, complexes involving disordered proteins are drastically different from the complexes formed by ordered proteins.
The case of the elastomeric proteins
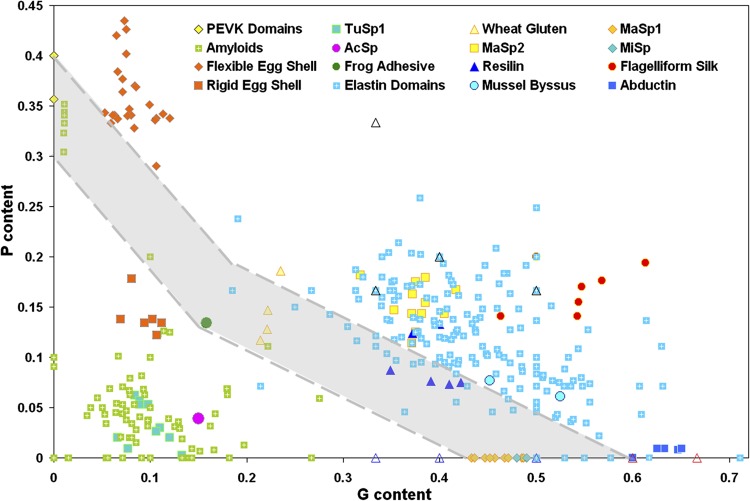
An important example of biologically important yet disordered complexes is that of the elastomeric proteins, which have a wide range of crucial functions and are involved in various unique mechanisms where they provide the high efficiency elastic recoil necessary to undergo reversible deformation [64]. These proteins are found in the human arterial wall, the capture spiral of spider webs, the hinge of scallop shells, and are involved in the jumping mechanism of fleas. Since it has been suggested that the elastic recoil of proteins is due to a combination of internal energy and entropy, and since the dominant driving force in this recoil process is the increased entropy of the relaxed state relative to the stretched state, it was pointed out that intrinsic disorder plays a crucial role in the function of these rubber-like elastomeric proteins. In agreement with this hypothesis, the functional aggregates of these proteins were described as intrinsically disordered or fuzzy complexes with high polypeptide chain entropy [64]. Although these disordered elastomers possess a broad range of sequence motifs, mechanical properties and biological functions, all of them are dramatically enriched in proline and glycine residues [65]. This P and G enrichment plays a central role in defining the elastin-like properties of disordered elastomers that form disordered functional aggregates (including disordered fibers), and clearly separates all elastomeric proteins from the amyloidogenic proteins/peptides that form insoluble amyloid-like fibrils characterized by the cross-β-sheet structure with the β-strands perpendicular to the fiber axis. Figure 1 illustrates this observation by showing a two-dimensional diagram that relates the P and G contents of natural elastomeric protein domains and proteins that were experimentally shown to form amyloid fibrils [65]. In this plot, a clear separation is seen between elastomeric and amyloidogenic sequences. These data provide a clear explanation of why elastomeric proteins are expected to form disordered fibers: their amino acid sequences are enriched in the structure-breaking G and P residues and therefore are naturally selected not to form lengthy ordered segments.
Figure 1. A two-dimensional plot correlating proline and glycine content for a wide variety of elastomeric and amyloidogenic peptides.
Elastomeric proteins are characterized by high PG content and are located in the upper-right part of this plot. Contrarily, amyloidogenic peptides are characterized by low PG content and therefore are located in the left bottom corner of the plot. The coexistence region (shaded in gray) contains P and G compositions consistent with both amyloidogenic and elastomeric properties. Elastomeric proteins, including the domains of elastin, major ampullate spindroin (MaSp) 2, flagelliform silk, the elastic domains of mussel byssus thread, and abductin, appear above a composition threshold (upper dashed line). Amyloidogenic sequences are primarily found below the PG-threshold, along with rigid lizard egg shells, tubulliform silk (TuSp1), a protective silk for spider eggs, and aciniform silk (AcSp), used for wrapping prey. The coexistence region contains amyloid-like peptides as well as the elastomeric adhesive produced by the frog Notaden bennetti, the PEVK domains of titin, wheat glutenin protein, and the strongest spider silks, namely MaSp1 and minor ampullate spindroin (MiSp). Figure reproduced from [65]. Abbreviations: AcSp, aciniform silk; MaSp, major ampullate spindroin; MiSp, minor ampullate spindroin; TuSp1, tubulliform silk.
The ‘IDP’ concept: necessary or not?
To our knowledge, the first report of a significant-sized region of missing electron density was in the structure of the extracellular nuclease from Staphylococcus aureus, as described in 1971 [66]. Two such regions were observed and the authors suggested that these were “disordered” and both highly solvated and highly mobile. The authors also reported the extreme trypsin sensitivity of these regions. Even earlier, optical rotatory dispersion was used to identify a few proteins that appeared to be fully unstructured under apparently physiological conditions, and from such studies one author suggested that there is a category of “disordered proteins” [67].
Subsequently many regions of proteins have been found to lack 3D structure under apparently physiological conditions in the absence of a binding partner, and NMR has revealed many additional proteins that appeared to be entirely unstructured. Release 6.0 of DISPROT [68] lists 667 proteins with 1,467 disordered regions that are associated with biological function; this set includes 112 wholly disordered proteins.
When such disordered proteins and regions were first discovered, the standard view was that they were somehow likely to be structured, except that they were denatured during isolation or lacked a critical partner that got lost during isolation. Indeed, some of the scientists who were involved in carrying out early, key work on these proteins [69,70] tell us that when they were graduate students doing the work in the citations just given, they repeated protein purification multiple times using different protocols because neither they nor their advisors could believe that unstructured proteins could be carrying out the biological functions being observed (Daughdrill, Kriwacki, personal communications).
A key development in the study of these proteins in our view has been the development of disorder predictors that used amino acid sequence or composition as inputs [71,72]. These predictors give results much better than expected by chance, leading to the conclusion that, to a considerable degree, lack of structure is encoded by the amino acid sequence. In other words, disordered proteins and regions have amino acid compositions that are distinctly different from the compositions of structured proteins. Thus, this observation links disorder to the DNA sequence, leading to an extension of the standard Central Dogma. That is, the standard Central Dogma is given by the following steps: (1) DNA sequence, (2) RNA sequence, (3) Protein sequence, (4) structure, and (5) function. The extension, however, is given by the following steps: (1) DNA sequence, (2) RNA sequence, (3) Protein sequence, (4) intrinsically disordered ensemble, and (5) function.
In our view, a portion of ‘folding code’ (and sometimes a significant part of it) that defines the ability of ordered proteins to spontaneously gain a unique biologically active structure is missing for IDPs. This missing portion of the ‘folding code’ (or a part of it) can be supplemented by binding partner(s). As a result, a key difference between structured and disordered proteins is that the former fold first and then bind to their partners while the latter remain unfolded until they bind their partners. Other researchers suggest that this distinction makes no difference. To emphasize that this is a distinction without a difference, recently the term “proteins waiting for partners” (PWPs) was proposed as an alternative to the term “disordered” [73]. Above we describe many examples in which disordered proteins have functions other than partner binding, so this term cannot be used for all types of disordered protein. Also, it is not clear how the PWPs concept would apply to examples such as the C-terminus of p53 described above, in which the same disordered region assumes four different conformations when binding to four different partners. This example suggests the possibility that the same disordered region can switch from one partner to another, with the disordered region changing its shape as it changes its partner. This sort of behavior seems to be much more dynamic than just ‘waiting for a partner’.
‘Flexibility’ is often proposed to describe motions in proteins covering both folded and unfolded forms. When we started studying these proteins, one of us chose the descriptor ‘natively unfolded protein’ [72,74] and the other one of us chose ‘disordered protein’ [71]. Both of us considered but rejected ‘flexibility’ as a descriptor. Our views regarding ‘flexibility’ were that this term is applied to both structured and unfolded proteins but describes entirely different processes for the two protein forms. That is, for structured proteins, flexibility refers to periodic or slightly aperiodic motions as atoms oscillate about their equilibrium positions, with higher flexibility referring to larger amplitudes for the oscillations. During these oscillations, the overall shape of the molecule changes very little. On the other hand, for unfolded proteins, flexibility refers to massive changes in backbone and side chain dihedral angles, leading to large-scale changes in overall shape. Given that flexibility has entirely different meanings for structured and unstructured proteins, using the same term for both protein forms tends to blur the very large differences in behavior.
With regard to replacing disorder with either flexibility or PWPs, our view on these suggestions can be summarized by a well-known phrase: “What's in a name? That which we call a rose by any other name would smell as sweet”. [75].
The fundamental distinction between folding first and then binding as compared to concomitant binding and folding is reflected by the marked differences in the amino acid composition between the two types of proteins. Indeed, early theoretical studies on protein folding suggested that whether a protein folds or not depends on its amino acid composition, and if it has a composition commensurate with folding, then the sequence patterns determine which fold is favored [76]. We [72,77,78] and others [79] have pointed out that the amino acid compositions of IDPs are entirely consistent with their lack of folding. The high polarity of these sequences is very much along the lines of the suggestions from the early theoretical studies for sequences that would fail to fold [76].
Researchers who question the existence of IDPs in vivo often point out that cells have elaborate mechanisms to deal with misfolded proteins and that disordered proteins would be cleared by these mechanisms. Thus, they conclude that disordered proteins cannot exist in cells except transiently. In our opinion, such suggestions are misguided for three reasons. First, the misfolded protein response is confined to the endoplasmic reticulum, so it is unclear to what extent other parts of the cell are under surveillance for protein misfolding. Second, such suggestions are based on the assumption that an amino acid sequence that is commensurate with folding (e.g. a high level of hydrophobic groups and aromatics) but that is unfolded or misfolded and that a highly polar sequence that has evolved to be unfolded will both be readily recognized by the ‘unfolded protein response systems’ and cleared by the cell. We think that it is equally likely that IDPs and regions have evolved to avoid the unfolded protein response by having sequences not recognized by those systems. Where is the proof that these systems recognize all types of sequences? Indeed, the mechanism by which the unfolded protein response recognizes its substrate proteins is currently ambiguous, and whether, or which, IDPs are cleared by this system is not yet understood (Ron Wek, personal communication). Also, in-cell NMR data demonstrates the existence and stability of IDPs even when inside both prokaryotic and eukaryotic cells [80-90]; why doesn't the misfolded protein response rapidly remove these proteins? Third, to use humans as an example organism, some of our proteins turn over with half-lives of less than a minute while others exist for the life of the human, giving almost eight orders of magnitude difference in protein stability. The stability of each protein is an important aspect of its biology. Studies on the relationship between protein lifetimes and protein disorder suggest that some disordered proteins have long lifetimes, but, on the other hand, the short-lifetime proteins are rich in disorder [91]. Perhaps both a disordered region and a particular signal, such as the PEST motif [92,93], are needed for a protein to exhibit a short half-life. The important point here is that disordered regions likely help to promote short protein life-times as an important aspect of this biology or to put it another way, life-time modulation is an important biological function of disordered proteins.
Is there any evidence that disorder is a product of evolution? Studies on the evolution of structured proteins suggest that regions of proteins with a high packing density show fewer amino acid changes over evolutionary time as compared to regions with lower packing densities. That is, if positional mutation rates (expressed as Shannon's entropy) are plotted versus tightness of packing (expressed as 1/density), virtually a straight line is observed with lower packing densities showing higher sequence variability (see Figure 3 in [94]). Of course many years earlier it was pointed out that the residues in the core of a protein family exhibit fewer mutations over evolutionary time as compared to residues on the surface of the same proteins [95]. Thus, mutation rates are strongly correlated with structural features of proteins. If the mutation rates of the structured and disordered regions of proteins are compared, in general (but not always), the mutation rates of the disordered regions are much higher than the mutation rates of the structured parts of the same proteins [96-98]. In our view, these observations are simply explained assuming that disordered proteins evolve differently from ordered proteins to maintain their disordered structure under physiological conditions, which is necessary for their functions.
Until recently, disordered proteins or regions were largely ignored. However, each week there are now about 17 publications (estimated by Caron Morales from the last 10 weeks using DisProt's standard keyword search of PubMed) that focus on the characterization and functions of these proteins. It could be reasonably argued that there is nothing new in the IDP concept, that all of the current views of these proteins follow naturally from long-held views of protein structure and function. From a chemistry and physics point of view, that is certainly true. However, the fact that these proteins were largely ignored previously and now they are being actively studied suggests that developing the IDP concepts has served the useful purpose of bringing attention to these proteins and to understanding the biological functions with which they are involved. The reader is invited to make up his or her own mind regarding the utility, or lack thereof, of the IDP concept.
Acknowledgments
This work was supported in part by the grant EF 0849803 from the National Science Foundation (to A.K.D and V.N.U.) and the Program of the Russian Academy of Sciences for the “Molecular and Cellular Biology” (to V.N.U.). Caron Morales is thanked for estimating the number of publications appearing in PubMed each week on intrinsically disordered proteins.
Abbreviations
- Cdk
cyclin-dependent protein kinase
- IDP
intrinsically disordered protein
- IDR
intrinsically disordered region
- PWPs
proteins waiting for partners
- SF1
splicing factor 1
- U2AF
U2 small nuclear RNA auxiliary actor
Disclosure
The authors declare that they have no disclosures
The electronic version of this article is the complete one and can be found at: http://f1000.com/prime/reports/b/5/1
References
- 1.Fischer E. Einfluss der configuration aurf die wirkung der enzyme. Ber Dt Chem Ges. 1894;27:2985–93. doi: 10.1002/cber.18940270364. [DOI] [Google Scholar]
- 2.Lemieux RU, Spohr U. How Emil Fischer was led to the lock and key concept for enzyme specificity. Adv Carbohydr Chem Biochem. 1994;50:1–20. doi: 10.1016/S0065-2318(08)60149-3. [DOI] [PubMed] [Google Scholar]
- 3.Ehrlich P. Die werthbemessugn des diphtherieheilserums und deren theoretische grundlagen. Klimishes Jahrbuch. 1897;6:299–326. [Google Scholar]
- 4.Tanford C, Reynolds J. Oxford: Oxford University Press; 2001. Nature's Robots; pp. 176–87. [Google Scholar]
- 5.Koshland DE., Jr Enzyme flexibility and enzyme action. J Cell Comp Physiol. 1959;54:245–58. doi: 10.1002/jcp.1030540420. [DOI] [PubMed] [Google Scholar]; www.f1000.com/prime/12818962
- 6.Koshland DE., Jr Crazy, but correct. Nature. 2004;432:447. doi: 10.1038/432447a. [DOI] [PubMed] [Google Scholar]
- 7.Hammes GG, Benkovic SJ, Hammes-Schiffer S. Flexibility, diversity, and cooperativity: pillars of enzyme catalysis. Biochemistry. 2011;50:10422–30. doi: 10.1021/bi201486f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burgen AS, Roberts GC, Feeney J. Binding of flexible ligands to macromolecules. Nature. 1975;253:753–5. doi: 10.1038/253753a0. [DOI] [PubMed] [Google Scholar]
- 9.Hammes GG. New York: Academic Press; 1982. Enzyme catalysis and regulation. [Google Scholar]
- 10.Hammes GG, Chang YC, Oas TG. Conformational selection or induced fit: a flux description of reaction mechanism. Proc Natl Acad Sci U S A. 2009;106:13737–41. doi: 10.1073/pnas.0907195106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nadassy K, Wodak SJ, Janin J. Structural features of protein-nucleic acid recognition sites. Biochemistry. 1999;38:1999–2017. doi: 10.1021/bi982362d. [DOI] [PubMed] [Google Scholar]
- 12.Schulz GE. Nucleotide Binding Proteins. In: Balaban M, editor. Molecular Mechanisms of Biological Recognition. Amsterdam: Elsevier/North-Holland Biomedical Press; 1979. pp. 79–94. [Google Scholar]
- 13.Spolar RS, Record MT., Jr Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994;263:777–84. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]
- 14.Espinoza-Fonseca LM. Reconciling binding mechanisms of intrinsically disordered proteins. Biochem Biophys Res Commun. 2009;382:479–82. doi: 10.1016/j.bbrc.2009.02.151. [DOI] [PubMed] [Google Scholar]; www.f1000.com/prime/717968034
- 15.Fersht AR, Knill-Jones JW, Bedouelle H, Winter G. Reconstruction by site-directed mutagenesis of the transition state for the activation of tyrosine by the tyrosyl-tRNA synthetase: a mobile loop envelopes the transition state in an induced-fit mechanism. Biochemistry. 1988;27:1581–7. doi: 10.1021/bi00405a028. [DOI] [PubMed] [Google Scholar]; www.f1000.com/prime/717968047
- 16.Schnell JR, Dyson HJ, Wright PE. Effect of cofactor binding and loop conformation on side chain methyl dynamics in dihydrofolate reductase. Biochemistry. 2004;43:374–83. doi: 10.1021/bi035464z. [DOI] [PubMed] [Google Scholar]; www.f1000.com/prime/717968035
- 17.Mohan A, Oldfield CJ, Radivojac P, Vacic V, Cortese MS, Dunker AK, Uversky VN. Analysis of molecular recognition features (MoRFs) J Mol Biol. 2006;362:1043–59. doi: 10.1016/j.jmb.2006.07.087. [DOI] [PubMed] [Google Scholar]
- 18.Tompa P, Fuxreiter M. Fuzzy complexes: polymorphism and structural disorder in protein-protein interactions. Trends Biochem Sci. 2008;33(1):2–8. doi: 10.1016/j.tibs.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 19.Selenko P, Gregorovic G, Sprangers R, Stier G, Rhani Z, Kramer A, Sattler M. Structural basis for the molecular recognition between human splicing factors U2AF65 and SF1/mBBP. Mol Cell. 2003;11:965–76. doi: 10.1016/S1097-2765(03)00115-1. [DOI] [PubMed] [Google Scholar]; www.f1000.com/prime/717968048
- 20.Oldfield CJ, Meng J, Yang JY, Yang MQ, Uversky VN, Dunker AK. Flexible nets: disorder and induced fit in the associations of p53 and 14-3-3 with their partners. BMC Genomics. 2008;9(Suppl 1):S1. doi: 10.1186/1471-2164-9-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsu WL, Oldfield C, Meng J, Huang F, Xue B, Uversky VN, Romero P, Dunker AK. Intrinsic protein disorder and protein-protein interactions. Pac Symp Biocomput. 2012:116–127. [PubMed] [Google Scholar]
- 22.Nash P, Tang X, Orlicky S, Chen Q, Gertler FB, Mendenhall MD, Sicheri F, Pawson T, Tyers M. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414:514–21. doi: 10.1038/35107009. [DOI] [PubMed] [Google Scholar]; www.f1000.com/prime/1002406
- 23.Verma R, Annan RS, Huddleston MJ, Carr SA, Reynard G, Deshaies RJ. Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science. 1997;278:455–60. doi: 10.1126/science.278.5337.455. [DOI] [PubMed] [Google Scholar]
- 24.Mittag T, Orlicky S, Choy WY, Tang X, Lin H, Sicheri F, Kay LE, Tyers M, Forman-Kay JD. Dynamic equilibrium engagement of a polyvalent ligand with a single-site receptor. Proc Natl Acad Sci U S A. 2008;105:17772–7. doi: 10.1073/pnas.0809222105. [DOI] [PMC free article] [PubMed] [Google Scholar]; www.f1000.com/prime/1127153
- 25.Borg M, Mittag T, Pawson T, Tyers M, Forman-Kay JD, Chan HS. Polyelectrostatic interactions of disordered ligands suggest a physical basis for ultrasensitivity. Proc Natl Acad Sci U S A. 2007;104:9650–5. doi: 10.1073/pnas.0702580104. [DOI] [PMC free article] [PubMed] [Google Scholar]; www.f1000.com/prime/717968036
- 26.Ubbink M. The courtship of proteins: understanding the encounter complex. FEBS Lett. 2009;583:1060–6. doi: 10.1016/j.febslet.2009.02.046. [DOI] [PubMed] [Google Scholar]; www.f1000.com/prime/717968049
- 27.Antal MA, Bode C, Csermely P. Perturbation waves in proteins and protein networks: applications of percolation and game theories in signaling and drug design. Curr Protein Pept Sci. 2009;10:161–72. doi: 10.2174/138920309787847617. [DOI] [PubMed] [Google Scholar]
- 28.Kovacs IA, Szalay MS, Csermely P. Water and molecular chaperones act as weak links of protein folding networks: energy landscape and punctuated equilibrium changes point towards a game theory of proteins. FEBS Lett. 2005;579:2254–60. doi: 10.1016/j.febslet.2005.03.056. [DOI] [PubMed] [Google Scholar]; www.f1000.com/prime/717968037
- 29.Ptitsyn OB. Stages in the mechanism of self-organization of protein molecules. Dokl Akad Nauk SSSR. 1973;210:1213–5. [PubMed] [Google Scholar]; www.f1000.com/prime/717968050
- 30.Dolgikh DA, Gilmanshin RI, Brazhnikov EV, Bychkova VE, Semisotnov GV, Venyaminov S, Ptitsyn OB. Alpha-Lactalbumin: compact state with fluctuating tertiary structure? FEBS Lett. 1981;136:311–5. doi: 10.1016/0014-5793(81)80642-41. [DOI] [PubMed] [Google Scholar]
- 31.Gilmanshin RI, Ptitsyn OB. An early intermediate of refolding alpha-lactalbumin forms within 20 ms. FEBS Lett 223. 1987;223:327–9. doi: 10.1016/0014-5793(87)80313-7. [DOI] [PubMed] [Google Scholar]
- 32.Bychkova VE, Pain RH, Ptitsyn OB. The 'molten globule' state is involved in the translocation of proteins across membranes? FEBS Lett. 1988;238:231–4. doi: 10.1016/0014-5793(88)80485-X. [DOI] [PubMed] [Google Scholar]; www.f1000.com/prime/717968038
- 33.Bracken C, Carr PA, Cavanagh J, Palmer AG., 3rd Temperature dependence of intramolecular dynamics of the basic leucine zipper of GCN4: implications for the entropy of association with DNA. J Mol Biol. 1999;285:2133–46. doi: 10.1006/jmbi.1998.2429. [DOI] [PubMed] [Google Scholar]; www.f1000.com/prime/717968051
- 34.Xu D, Tsai CJ, Nussinov R. Mechanism and evolution of protein dimerization. Protein Sci. 1998;7:533–44. doi: 10.1002/pro.5560070301. [DOI] [PMC free article] [PubMed] [Google Scholar]; www.f1000.com/prime/717968039
- 35.Gunasekaran K, Tsai CJ, Nussinov R. Analysis of ordered and disordered protein complexes reveals structural features discriminating between stable and unstable monomers. J Mol Biol. 2004;341:1327–41. doi: 10.1016/j.jmb.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 36.Fuxreiter M, Tompa P. Fuzzy complexes: a more stochastic view of protein function. Adv Exp Med Biol. 2012;725:1–14. doi: 10.1007/978-1-4614-0659-4_1. [DOI] [PubMed] [Google Scholar]
- 37.Sigalov A, Aivazian D, Stern L. Homooligomerization of the cytoplasmic domain of the T cell receptor zeta chain and of other proteins containing the immunoreceptor tyrosine-based activation motif. Biochemistry. 2004;43:2049–61. doi: 10.1021/bi035900h. [DOI] [PubMed] [Google Scholar]; www.f1000.com/prime/717968052
- 38.Sigalov AB, Zhuravleva AV, Orekhov VY. Binding of intrinsically disordered proteins is not necessarily accompanied by a structural transition to a folded form. Biochimie. 2007;89:419–21. doi: 10.1016/j.biochi.2006.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu J, Faeder JR, Camacho CJ. Toward a quantitative theory of intrinsically disordered proteins and their function. Proc Natl Acad Sci U S A. 2009;106:19819–23. doi: 10.1073/pnas.0907710106. [DOI] [PMC free article] [PubMed] [Google Scholar]; www.f1000.com/prime/1671956
- 40.Awile O, Krisko A, Sbalzarini IF, Zagrovic B. Intrinsically disordered regions may lower the hydration free energy in proteins: a case study of nudix hydrolase in the bacterium Deinococcus radiodurans. PLoS Comput Biol. 2010;6:e1000854. doi: 10.1371/journal.pcbi.1000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meador WE, Means AR, Quiocho FA. Modulation of calmodulin plasticity in molecular recognition on the basis of x-ray structures. Science. 1993;262:1718–21. doi: 10.1126/science.8259515. [DOI] [PubMed] [Google Scholar]; www.f1000.com/prime/717968040
- 42.Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252:809–17. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]; www.f1000.com/prime/717968053
- 43.Armstrong CM, Bezanilla F. Inactivation of the sodium channel. II. Gating current experiments. J Gen Physiol. 1977;70:567–90. doi: 10.1085/jgp.70.5.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gomez-Lagunas F, Armstrong CM. The relation between ion permeation and recovery from inactivation of shakerB K+ channels. Biophys J. 1994;67:1806–15. doi: 10.1016/S0006-3495(94)80662-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoshi T, Zagotta WN, Aldrich RW. Two types of inactivation in Shaker K+ channels: effects of alterations in the carboxy-terminal region. Neuron. 1991;7:547–56. doi: 10.1016/0896-6273(91)90367-9. [DOI] [PubMed] [Google Scholar]
- 46.Zagotta WN, Hoshi T, Aldrich RW. Restoration of inactivation in mutants of Shaker potassium channels by a peptide derived from ShB. Science. 1990;250:568–71. doi: 10.1126/science.2122520. [DOI] [PubMed] [Google Scholar]
- 47.Liebovitch LS, Selector LY, Kline RP. Statistical properties predicted by the ball and chain model of channel inactivation. Biophys J. 1992;63:1579–85. doi: 10.1016/S0006-3495(92)81732-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Podlaha O, Zhang J. Positive selection on protein-length in the evolution of a primate sperm ion channel. Proc Natl Acad Sci U S A. 2003;100:12241–6. doi: 10.1073/pnas.2033555100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bienkiewicz EA, Adkins JN, Lumb KJ. Functional consequences of preorganized helical structure in the intrinsically disordered cell-cycle inhibitor p27(Kip1) Biochemistry. 2002;41:752–9. doi: 10.1021/bi015763t. [DOI] [PubMed] [Google Scholar]; www.f1000.com/prime/1004689
- 50.Galea CA, Nourse A, Wang Y, Sivakolundu SG, Heller WT, Kriwacki RW. Role of intrinsic flexibility in signal transduction mediated by the cell cycle regulator, p27 Kip1. J Mol Biol. 2008;376:827–38. doi: 10.1016/j.jmb.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]; www.f1000.com/prime/717968041
- 51.Dunker AK, Uversky VN. Signal transduction via unstructured protein conduits. Nat Chem Biol. 2008;4:229–30. doi: 10.1038/nchembio0408-229. [DOI] [PubMed] [Google Scholar]
- 52.Russo AA, Jeffrey PD, Patten AK, Massague J, Pavletich NP. Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A-Cdk2 complex. Nature. 1996;382:325–31. doi: 10.1038/382325a0. [DOI] [PubMed] [Google Scholar]; www.f1000.com/prime/717968027
- 53.Lacy ER, Wang Y, Post J, Nourse A, Webb W, Mapelli M, Musacchio A, Siuzdak G, Kriwacki RW. Molecular basis for the specificity of p27 toward cyclin-dependent kinases that regulate cell division. J Mol Biol. 2005;349:764–73. doi: 10.1016/j.jmb.2005.04.019. [DOI] [PubMed] [Google Scholar]
- 54.Xue B, Dunker AK, Uversky VN. The roles of intrinsic disorder in orchestrating the wnt-pathway. J Biomol Struct Dyn. 2012;29:843–61. doi: 10.1080/073911012010525024. [DOI] [PubMed] [Google Scholar]
- 55.Iakoucheva LM, Radivojac P, Brown CJ, O'Connor TR, Sikes JG, Obradovic Z, Dunker AK. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32:1037–49. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gao J, Thelen JJ, Dunker AK, Xu D. Musite, a tool for global prediction of general and kinase-specific phosphorylation sites. Mol Cell Proteomics. 2010;9:2586–600. doi: 10.1074/mcp.M110.001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Radivojac P, Vacic V, Haynes C, Cocklin RR, Mohan A, Heyen JW, Goebl MG, Iakoucheva LM. Identification, analysis, and prediction of protein ubiquitination sites. Proteins. 2010;78:365–80. doi: 10.1002/prot.22555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Inobe T, Fishbain S, Prakash S, Matouschek A. Defining the geometry of the two-component proteasome degron. Nat Chem Biol. 2011;7:161–7. doi: 10.1038/nchembio.521. [DOI] [PMC free article] [PubMed] [Google Scholar]; www.f1000.com/prime/8880963
- 59.Kuriyan J, Eisenberg D. The origin of protein interactions and allostery in colocalization. Nature. 2007;450:983–90. doi: 10.1038/nature06524. [DOI] [PubMed] [Google Scholar]
- 60.Dunker AK, Garner E, Guilliot S, Romero P, Albrecht K, Hart J, Obradovic Z, Kissinger C, Villafranca JE. Protein disorder and the evolution of molecular recognition: theory, predictions and observations. Pac Symp Biocomput. 1998:473–484. [PubMed] [Google Scholar]
- 61.Manson A, Whitten ST, Ferreon JC, Fox RO, Hilser VJ. Characterizing the role of ensemble modulation in mutation-induced changes in binding affinity. J Am Chem Soc. 2009;131:6785–93. doi: 10.1021/ja809133u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hilser VJ. Biochemistry. An ensemble view of allostery. Science. 2010;327:653–4. doi: 10.1126/science.1186121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Prakash MK. Insights on the role of (dis)order from protein-protein interaction linear free-energy relationships. J Am Chem Soc. 2011;133:9976–9. doi: 10.1021/ja201500z. [DOI] [PubMed] [Google Scholar]; www.f1000.com/prime/11660957
- 64.Rauscher S, Pomes R. Structural disorder and protein elasticity. Adv Exp Med Biol. 2012;725:159–83. doi: 10.1007/978-1-4614-0659-4_10. [DOI] [PubMed] [Google Scholar]; www.f1000.com/prime/717968042
- 65.Rauscher S, Baud S, Miao M, Keeley FW, Pomes R. Proline and glycine control protein self-organization into elastomeric or amyloid fibrils. Structure. 2006;14:1667–76. doi: 10.1016/j.str.2006.09.008. [DOI] [PubMed] [Google Scholar]; www.f1000.com/prime/1086043
- 66.Arnone A, Bier CJ, Cotton FA, Day VW, Hazen EE, Jr., Richardson DC, Yonath A, Richardson JS. A high resolution structure of an inhibitor complex of the extracellular nuclease of Staphylococcus aureus. I. Experimental procedures and chain tracing. J Biol Chem. 1971;246:2302–16. [PubMed] [Google Scholar]; www.f1000.com/prime/717968054
- 67.Jirgensons B. Classification of proteins according to conformation. Die Macromolekulare Chemie. 1966;91:74–86. doi: 10.1002/macp.1966.020910105. [DOI] [Google Scholar]
- 68.Sickmeier M, Hamilton JA, LeGall T, Vacic V, Cortese MS, Tantos A, Szabo B, Tompa P, Chen J, Uversky VN, Obradovic Z, Dunker AK. DisProt: the Database of Disordered Proteins. Nucleic Acids Res. 2007;35(Database issue):D786–93. doi: 10.1093/nar/gkl893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kriwacki RW, Hengst L, Tennant L, Reed SI, Wright PE. Structural studies of p21Waf1/Cip1/Sdi1 in the free and Cdk2-bound state: conformational disorder mediates binding diversity. Proc Natl Acad Sci U S A. 1996;93:11504–9. doi: 10.1073/pnas.93.21.11504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Daughdrill GW, Chadsey MS, Karlinsey JE, Hughes KT, Dahlquist FW. The C-terminal half of the anti-sigma factor, FlgM, becomes structured when bound to its target, sigma 28. Nat Struct Biol. 1997;4:285–91. doi: 10.1038/nsb0497-285. [DOI] [PubMed] [Google Scholar]; www.f1000.com/prime/717968029
- 71.Romero P, Obradovic Z, Kissinger K, Villafranca JE, Dunker AK. Identifying disordered regions in proteins from amino acid sequence. IEEE International Conference on Neural Networks; Huston, TX: 1997. [Google Scholar]
- 72.Uversky VN, Gillespie JR, Fink AL. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins. 2000;41:415–27. doi: 10.1002/1097-0134(20001115)41:3<415::AID-PROT130>3.3.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 73.Janin J, Sternberg MJE. Protein flexibility, not disorder, is intrinsic to molecular recognition. F1000 Biol Rep. 2013;5:2. doi: 10.3410/B5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Uversky VN, Gillespie JR, Millett IS, Khodyakova AV, Vasiliev AM, Chernovskaya TV, Vasilenko RN, Kozlovskaya GD, Dolgikh DA, Fink AL, Doniach S, Abramov VM. Natively unfolded human prothymosin alpha adopts partially folded collapsed conformation at acidic pH. Biochemistry. 1999;38:15009–16. doi: 10.1021/bi990752+. [DOI] [PubMed] [Google Scholar]
- 75.Shakespeare W. The Most Excellent and Lamentable Tragedie of Romeo and Juliet. London: Cuthbert Burby; 1599. [Google Scholar]
- 76.Shakhnovich EI, Gutin AM. Engineering of stable and fast-folding sequences of model proteins. Proc Natl Acad Sci U S A. 1993;90:7195–9. doi: 10.1073/pnas.90.15.7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xie Q, Arnold GE, Romero P, Obradovic Z, Garner E, Dunker AK. Sequence Attribute Method for Determining Relationships Between Sequence and Protein Disorder. Genome Inform Ser Workshop Genome Inform. 1998;9:193–200. [PubMed] [Google Scholar]
- 78.Dunker AK, Brown CJ, Obradovic Z. Identification and functions of usefully disordered proteins. Adv Protein Chem. 2002;62:25–49. doi: 10.1016/S0065-3233(02)62004-2. [DOI] [PubMed] [Google Scholar]
- 79.Dosztanyi Z, Csizmok V, Tompa P, Simon I. The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J Mol Biol. 2005;347:827–39. doi: 10.1016/j.jmb.2005.01.071. [DOI] [PubMed] [Google Scholar]; www.f1000.com/prime/717968043
- 80.McNulty BC, Young GB, Pielak GJ. Macromolecular crowding in the Escherichia coli periplasm maintains alpha-synuclein disorder. J Mol Biol. 2006;355:893–7. doi: 10.1016/j.jmb.2005.11.033. [DOI] [PubMed] [Google Scholar]; www.f1000.com/prime/1029874
- 81.Li C, Charlton LM, Lakkavaram A, Seagle C, Wang G, Young GB, Macdonald JM, Pielak GJ. Differential dynamical effects of macromolecular crowding on an intrinsically disordered protein and a globular protein: implications for in-cell NMR spectroscopy. J Am Chem Soc. 2008;130:6310–1. doi: 10.1021/ja801020z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li C, Wang GF, Wang Y, Creager-Allen R, Lutz EA, Scronce H, Slade KM, Ruf RA, Mehl RA, Pielak GJ. Protein (19)F NMR in Escherichia coli. J Am Chem Soc. 2010;132:321–7. doi: 10.1021/ja907966n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schlesinger AP, Wang Y, Tadeo X, Millet O, Pielak GJ. Macromolecular crowding fails to fold a globular protein in cells. J Am Chem Soc. 2011;133:8082–5. doi: 10.1021/ja201206t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fauvet B, Fares MB, Samuel F, Dikiy I, Tandon A, Eliezer D, Lashuel HA. Characterization of Semisynthetic and Naturally N alpha-Acetylated alpha-Synuclein in Vitro and in Intact Cells: IMPLICATIONS FOR AGGREGATION AND CELLULAR PROPERTIES OF alpha-SYNUCLEIN. J Biol Chem. 2012;287:28243–62. doi: 10.1074/jbc.M112.383711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Binolfi A, Theillet FX, Selenko P. Bacterial in-cell NMR of human alpha-synuclein: a disordered monomer by nature? Biochem Soc Trans. 2012;40:950–4. doi: 10.1042/BST20120096. [DOI] [PubMed] [Google Scholar]
- 86.Thongwichian R, Selenko P. In-cell NMR in Xenopus laevis oocytes. Methods Mol Biol. 2012;895:33–41. doi: 10.1007/978-1-61779-927-3_3. [DOI] [PubMed] [Google Scholar]; www.f1000.com/prime/717968055
- 87.Bekei B, Rose HM, Herzig M, Dose A, Schwarzer D, Selenko P. In-cell NMR in mammalian cells: part 1. Methods Mol Biol. 2012;895:43–54. doi: 10.1007/978-1-61779-927-3_4. [DOI] [PubMed] [Google Scholar]; www.f1000.com/prime/717968044
- 88.Bekei B, Rose HM, Herzig M, Selenko P. In-cell NMR in mammalian cells: part 2. Methods Mol Biol. 2012;895:55–66. doi: 10.1007/978-1-61779-927-3_5. [DOI] [PubMed] [Google Scholar]; www.f1000.com/prime/717968056
- 89.Bekei B, Rose HM, Herzig M, Stephanowitz H, Krause E, Selenko P. In-cell NMR in mammalian cells: part 3. Methods Mol Biol. 2012;895:67–83. doi: 10.1007/978-1-61779-927-3_6. [DOI] [PubMed] [Google Scholar]; www.f1000.com/prime/717968045
- 90.Ito Y, Mikawa T, Smith BO. In-cell NMR of intrinsically disordered proteins in prokaryotic cells. Methods Mol Biol. 2012;895:19–31. doi: 10.1007/978-1-61779-927-3_2. [DOI] [PubMed] [Google Scholar]; www.f1000.com/prime/717968057
- 91.Tompa P, Prilusky J, Silman I, Sussman JL. Structural disorder serves as a weak signal for intracellular protein degradation. Proteins. 2008;71:903–09. doi: 10.1002/prot.21773. [DOI] [PubMed] [Google Scholar]; www.f1000.com/prime/717968046
- 92.Rogers SW, Rechsteiner MC. Microinjection studies on selective protein degradation: relationships between stability, structure, and location. Biomed Biochim Acta. 1986;45:1611–8. [PubMed] [Google Scholar]
- 93.Singh GP, Ganapathi M, Sandhu KS, Dash D. Intrinsic unstructuredness and abundance of PEST motifs in eukaryotic proteomes. Proteins. 2006;62:309–15. doi: 10.1002/prot.20746. [DOI] [PubMed] [Google Scholar]
- 94.Jernigan RL, Kloczkowski A. Packing regularities in biological structures relate to their dynamics. Methods Mol Biol. 2007;350:251–76. doi: 10.1385/1-59745-189-4:251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bordo D, Argos P. Evolution of protein cores. Constraints in point mutations as observed in globin tertiary structures. J Mol Biol. 1990;211:975–88. doi: 10.1016/0022-2836(90)90087-3. [DOI] [PubMed] [Google Scholar]
- 96.Brown CJ, Takayama S, Campen AM, Vise P, Marshall TW, Oldfield CJ, Williams CJ, Dunker AK. Evolutionary rate heterogeneity in proteins with long disordered regions. J Mol Evol. 2002;55:104–10. doi: 10.1007/s00239-001-2309-6. [DOI] [PubMed] [Google Scholar]
- 97.Daughdrill GW, Narayanaswami P, Gilmore SH, Belczyk A, Brown CJ. Dynamic behavior of an intrinsically unstructured linker domain is conserved in the face of negligible amino acid sequence conservation. J Mol Evol. 2007;65:277–88. doi: 10.1007/s00239-007-9011-2. [DOI] [PubMed] [Google Scholar]; www.f1000.com/prime/717968058
- 98.Brown CJ, Johnson AK, Daughdrill GW. Comparing models of evolution for ordered and disordered proteins. Mol Biol Evol. 2010;27:609–21. doi: 10.1093/molbev/msp277. Erratum in: Mol Biol Evol. 2012, 29:443. [DOI] [PMC free article] [PubMed] [Google Scholar]; www.f1000.com/prime/3231956