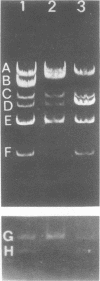
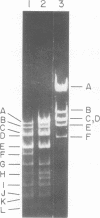
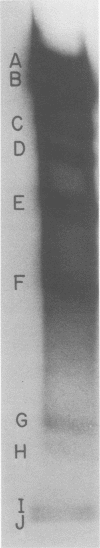
Abstract
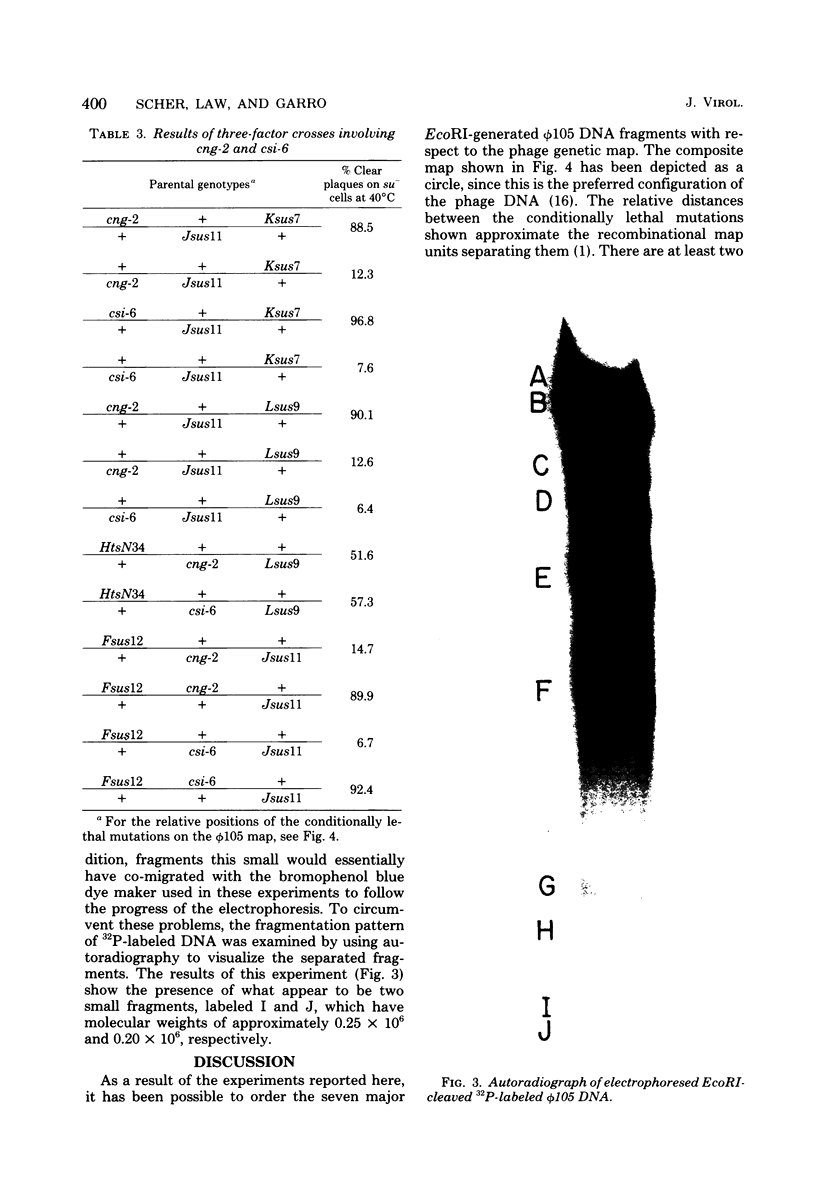
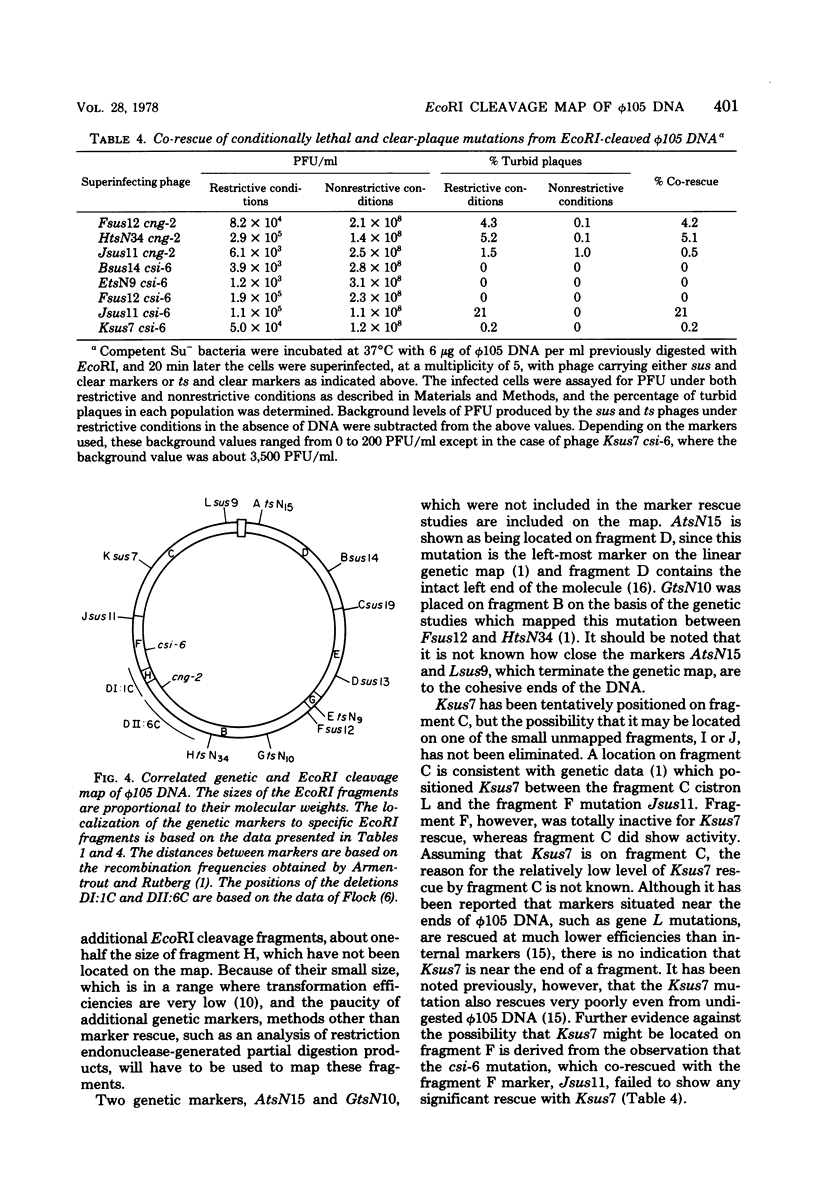
The seven previously identified EcoRI cleavage fragments of phi 105 DNA were ordered with respect to their sites of origin on the phage genome by marker rescue. One fragment, H, did not carry any determinants essential for replication. This fragment was totally missing in a deletion mutant which exhibited a lysogenization-defective phenotype. There is a nonessential region on the phi 105 genome which begins in fragment B, spans fragment H, and ends in fragment F. The size of the nonessential region, as estimated by alterations observed in the fragmentation patterns of deletion mutant DNAs, is approximately 2.7 X 10(6) daltons. Two new EcoRI cleavage fragments with molecular weights of approximately 0.2 X 10(6) were detected by autoradiography of 32P-labeled DNA. These small fragments were not located on the cleavage map.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Armentrout R. W., Rutberg L. Mapping of prophage and mature deoxyribonucleic acid from temperate Bacillus bacteriophage phi 105 by marker rescue. J Virol. 1970 Dec;6(6):760–767. doi: 10.1128/jvi.6.6.760-767.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birdsell D. C., Hathaway G. M., Rutberg L. Characterization of Temperate Bacillus Bacteriophage phi105. J Virol. 1969 Sep;4(3):264–270. doi: 10.1128/jvi.4.3.264-270.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukhari A. I. Bacteriophage mu as a transposition element. Annu Rev Genet. 1976;10:389–412. doi: 10.1146/annurev.ge.10.120176.002133. [DOI] [PubMed] [Google Scholar]
- Chow L. T., Boice L., Davidson N. Map of the partial sequence homology between DNA molecules of Bacillus subtilis bacteriophages SPO2 and phi105. J Mol Biol. 1972 Jul 28;68(3):391–400. doi: 10.1016/0022-2836(72)90093-9. [DOI] [PubMed] [Google Scholar]
- Chow L. T., Davidson N. Electron microscope study of the structures of the Bacillus subtilis prophages, SPO2 and phi105. J Mol Biol. 1973 Apr 5;75(2):257–264. doi: 10.1016/0022-2836(73)90019-3. [DOI] [PubMed] [Google Scholar]
- Flock J. I. Deletion mutants of temperate Bacillus subtilis bacteriophage phi105. Mol Gen Genet. 1977 Oct 24;155(3):241–247. doi: 10.1007/BF00272803. [DOI] [PubMed] [Google Scholar]
- Garro A. J., Law M. F. Relationship between lysogeny, spontaneous induction, and transformation efficiencies in Bacillus subtilis. J Bacteriol. 1974 Dec;120(3):1256–1259. doi: 10.1128/jb.120.3.1256-1259.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAISER A. D. Mutations in a temperate bacteriophage affecting its ability to lysogenize Escherichia coli. Virology. 1957 Feb;3(1):42–61. doi: 10.1016/0042-6822(57)90022-3. [DOI] [PubMed] [Google Scholar]
- Morrison D. A., Guild W. R. Activity of deoxyribonucleic acid fragments of defined size in Bacillus subtilis transformation. J Bacteriol. 1972 Oct;112(1):220–223. doi: 10.1128/jb.112.1.220-223.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins J. B., Zarley C. D., Dean D. H. Restriction endonuclease mapping of bacteriophage phi105 and closely related temperate Bacillus subtilis bacteriophages rho10 and rho14. J Virol. 1978 Oct;28(1):403–407. doi: 10.1128/jvi.28.1.403-407.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polisky B., Greene P., Garfin D. E., McCarthy B. J., Goodman H. M., Boyer H. W. Specificity of substrate recognition by the EcoRI restriction endonuclease. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3310–3314. doi: 10.1073/pnas.72.9.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutberg L., Armentrout R. W. Low-frequency rescue of a genetic marker in deoxyribonucleic acid from Bacillus bacteriophage phi 105 by superinfecting bacteriophage. J Virol. 1970 Dec;6(6):768–771. doi: 10.1128/jvi.6.6.768-771.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutberg L. Heat induction of prophage phi 105 in Bacillus subtilis: bacteriophage-induced bidirectional replication of the bacterial chromosome. J Virol. 1973 Jul;12(1):9–12. doi: 10.1128/jvi.12.1.9-12.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutberg L. Mapping of a temperate bacteriophage active on Bacillus subtilis. J Virol. 1969 Jan;3(1):38–44. doi: 10.1128/jvi.3.1.38-44.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scher B. M., Dean D. H., Garro A. J. Fragmentation of Bacillus bacteriophage phi105 DNA by complementary single-stranded DNA in the cohesive ends of the molecule. J Virol. 1977 Aug;23(2):377–383. doi: 10.1128/jvi.23.2.377-383.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W. Analysis of bacteriophage T7 early RNAs and proteins on slab gels. J Mol Biol. 1973 Sep 15;79(2):237–248. doi: 10.1016/0022-2836(73)90003-x. [DOI] [PubMed] [Google Scholar]
- Thuring R. W., Sanders J. P., Borst P. A freeze-squeeze method for recovering long DNA from agarose gels. Anal Biochem. 1975 May 26;66(1):213–220. doi: 10.1016/0003-2697(75)90739-3. [DOI] [PubMed] [Google Scholar]