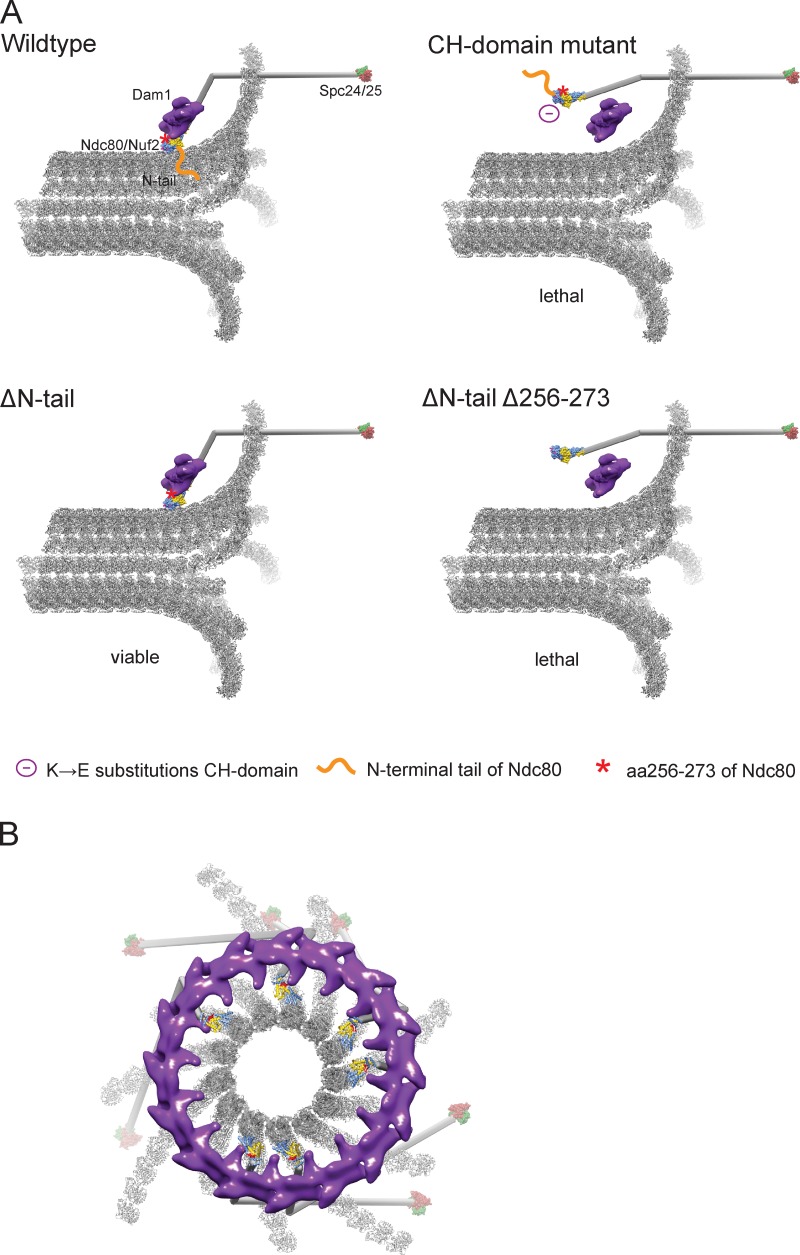
Correct kinetochore–microtubule attachments in budding yeast rely on the conserved CH-domain function of the kinetochore component Ndc80 and its ability to cooperate with the spindle-associated Dam1 complex.
Abstract
Kinetochores are large protein complexes that link sister chromatids to the spindle and transduce microtubule dynamics into chromosome movement. In budding yeast, the kinetochore–microtubule interface is formed by the plus end–associated Dam1 complex and the kinetochore-resident Ndc80 complex, but how they work in combination and whether a physical association between them is critical for chromosome segregation is poorly understood. Here, we define structural elements required for the Ndc80–Dam1 interaction and probe their function in vivo. A novel ndc80 allele, selectively impaired in Dam1 binding, displayed growth and chromosome segregation defects. Its combination with an N-terminal truncation resulted in lethality, demonstrating essential but partially redundant roles for the Ndc80 N-tail and Ndc80–Dam1 interface. In contrast, mutations in the calponin homology domain of Ndc80 abrogated kinetochore function and were not compensated by the presence of Dam1. Our experiments shed light on how microtubule couplers cooperate and impose important constraints on structural models for outer kinetochore assembly.
Introduction
Kinetochores are remarkable molecular machines that link dynamic microtubule plus-ends to centromeres (Santaguida and Musacchio, 2009; Bloom and Joglekar, 2010; Lampert and Westermann, 2011). Ndc80–Hec1 is a heterotetrameric complex that associates with the Mis12 (Mtw1) complex and KNL1 (Spc105; Wei et al., 2005; Cheeseman et al., 2006; Ciferri et al., 2008) to form the KMN network that serves as the central kinetochore–microtubule interface in all eukaryotes. Interference with KMN components is deleterious for the cell, and can lead to a complete lack of microtubule attachments (Wigge and Kilmartin, 2001; Desai et al., 2003; DeLuca et al., 2005). The Ndc80 complex contains conserved CH-domains, which together provide a positively charged interaction surface for the microtubule (Ciferri et al., 2008). Mutations in these two domains partially or completely abrogate microtubule binding in vivo and in vitro (Ciferri et al., 2008; Sundin et al., 2011; Tooley et al., 2011). Additionally, the Ndc80 complex harbors a second microtubule-binding element, the unstructured flexible N-terminal tail of the Ndc80 protein. Although this positively charged domain is a crucial regulatory target of the Aurora B kinase, and might also enhance oligomerization of the Ndc80 complex in humans, (Alushin et al., 2010; DeLuca et al., 2011), it is surprisingly dispensable in budding yeast (Kemmler et al., 2009). Biophysical experiments have demonstrated that multiple Ndc80 complexes can form load-bearing attachments in vitro (Powers et al., 2009), but whether this activity is sufficient to segregate chromosomes in vivo is unclear.
In the single kinetochore–microtubule attachment site of budding yeast the heterodecameric Dam1 complex is additionally crucial for the establishment and regulation of end-on attachments (Cheeseman et al., 2002; Tanaka et al., 2007). In vitro studies have demonstrated that the plus end–tracking Dam1 complex can oligomerize into a ring around the microtubule, providing an enticing explanation for its role as a coupling device (Miranda et al., 2005; Westermann et al., 2005) that is supported by a series of biophysical studies (Asbury et al., 2006; Westermann et al., 2006; Gestaut et al., 2008; Grishchuk et al., 2008).
Recent experiments have provided initial clues about how Ndc80 and Dam1 complexes may work together to couple chromosomes to microtubules. In biochemical reconstitution experiments Dam1 is able to enhance the affinity of Ndc80 for dynamic microtubule plus-tips, suggesting that Dam1 might strengthen Ndc80-mediated microtubule attachments in yeast (Lampert et al., 2010; Tien et al., 2010). The ability to manipulate the kinetochore–microtubule interface has been limited by a lack of knowledge about the mechanism of the Dam1–Ndc80 interaction. Here, we overcome this limitation by identifying residues in the yeast Ndc80 complex that facilitate the interaction with Dam1. This molecular insight allows us to probe the Ndc80–Dam1 interface in vivo and specifically address the differential contributions of these microtubule couplers in the cell.
Results and discussion
Ndc80 CH-domain function is essential in budding yeast and highly conserved
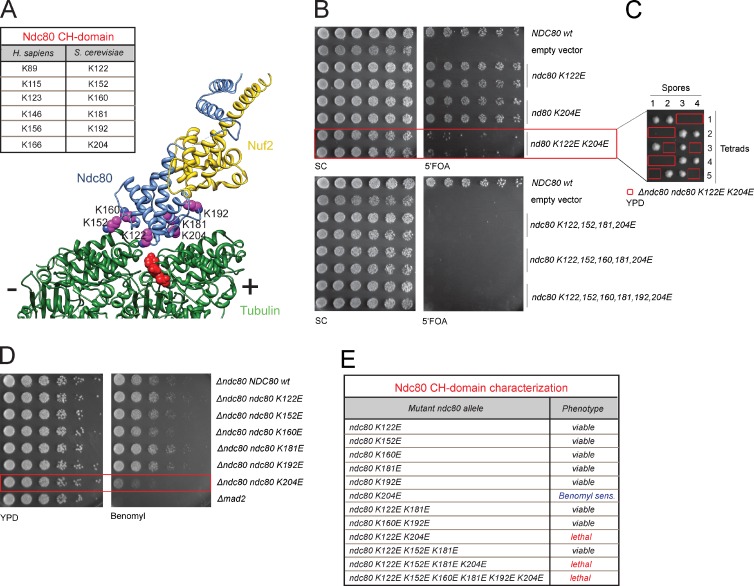
To gain insight into the function of the kinetochore–microtubule interface, we first tested the contribution of the Ndc80 CH-domain based on available structural information (Fig. 1 A; Wilson-Kubalek et al., 2008; Alushin et al., 2010). Although recent functional studies in human cell lines have demonstrated a critical importance of this element for kinetochore–microtubule attachments (Ciferri et al., 2008; Sundin et al., 2011; Tooley et al., 2011), it remains to be shown whether they play a similar role in the attachment configuration of budding yeast. We systematically mutated conserved lysine residues (K122, K152, K160, K181, K192, and K204) previously shown to be critical for binding of the human Ndc80 complex to microtubules in vitro (Ciferri et al., 2008). Viability assays on 5′FOA plates, counter-selecting against wild-type NDC80, demonstrated that all six single lysine mutants could compensate for the loss of the NDC80 wild-type allele. We found, however, that the K122E K204E double mutation was lethal and consistently all other combinations that involved K122E and K204E were inviable (Fig. 1, B, C, and E). In sequence alignments, the position of these residues corresponds precisely to the lysine residues (K89 and K166) that most effectively cripple microtubule binding of the human Ndc80 complex in vitro (Ciferri et al., 2008). In stable haploid integrations a single point mutation, K204E, sensitized cells to the presence of benomyl, suggesting that this residue makes the strongest contribution to the microtubule-binding ability of the Ndc80 complex (Fig. 1, D and E). Intriguingly, a negative charge in the equivalent human residue K166 results in severe chromosome alignment defects in human cells, suggesting that the functional hierarchy among individual lysine residues located in the Ndc80 CH-domain is precisely conserved from yeast to humans (Sundin et al., 2011; Tooley et al., 2011). We conclude that the autonomous microtubule-binding activity of the Ndc80 complex, provided by the CH-domain, is an absolute requirement for a functional kinetochore–microtubule interface, arguing for the existence of an evolutionary conserved binding mode.
Figure 1.
A functional Ndc80 CH-domain is essential in budding yeast. (A) Structure of the human Ndc80 bonsai complex bound to microtubules (Alushin et al., 2010). Critical lysine residues are listed in the box (Ciferri et al., 2008). (B) Plasmid-shuffle assay showing the phenotypes of mutations in the CH-domain of Ndc80. (C) Tetrad dissection of a heterozygous Δndc80 strain with an integrated ndc80 K122E K204E allele. The ndc80 K122E K204E mutant cannot compensate for loss of the wild-type allele. (D) The ndc80 K204E mutant shows increased sensitivity to benomyl. (E) Summary of mutant phenotypes.
Biochemical identification of an Ndc80–Dam1 interaction mutant
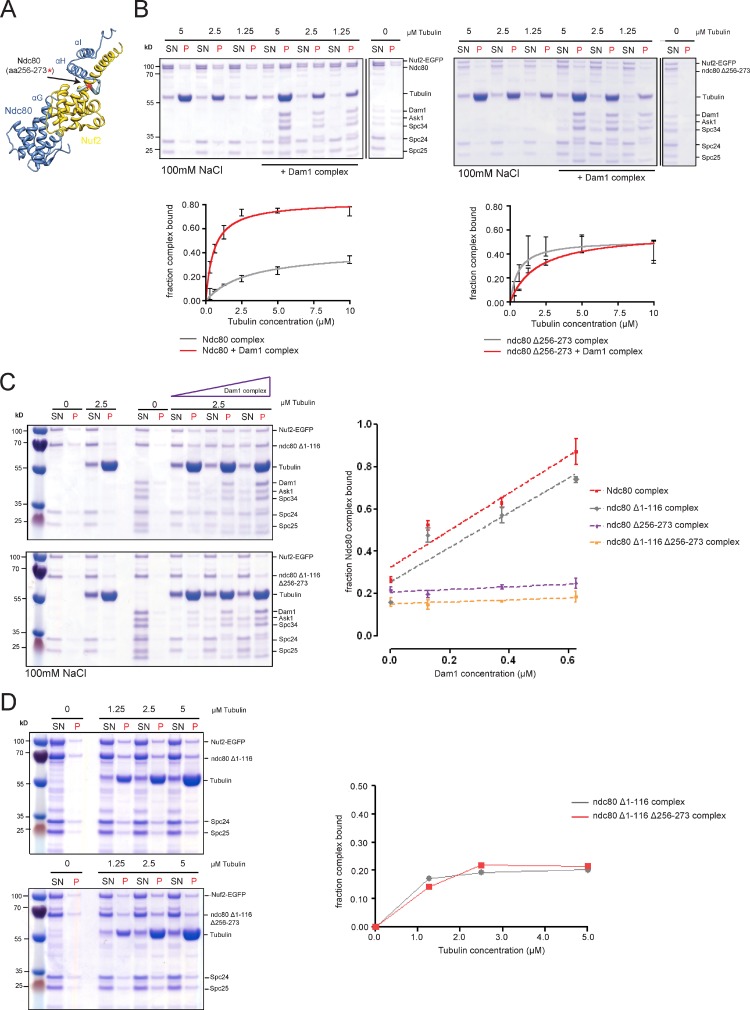
Design of candidate mutations that would interfere with the Ndc80–Dam1 interaction is facilitated by the crystal structure of human Ndc80 bonsai complex (Ciferri et al., 2008), which shows that the Ndc80 CH-domain is connected via a short disordered segment (human residues 203–211, located between helices αG and αH) to the αH-αI helical hairpin preceding the coiled-coil shaft (Fig. 2 A). Two reasons make this part of the molecule an attractive target for structure–function analysis. First, many Saccharomyces species contain large insertions in this region, rich in polar residues that may protrude from the surface, and thus can provide a possible interface for protein–protein interactions. Second, the proximity to the microtubule surface and the location on top of the Nuf2 CH-domain may position it closely to microtubule-bound Dam1 complexes. We tested several candidate mutations for the Dam1–Ndc80 interaction in vitro using microtubule cosedimentation assays. The ndc80 Δ256-273 mutation, further characterized in this study, removes part of the insertion but leaves the CH-domain and the helical αH-αI hairpin unchanged. Bacterially expressed Ndc80 Δ256-273 reconstituted into the full Ndc80 complex (Fig. S1 A) and displayed robust microtubule binding, but in contrast to the wild-type Ndc80, addition of the Dam1 complex was not able to enhance the affinity of Ndc80 Δ256-273 for microtubules (Fig. 2 B).
Figure 2.
Biochemical identification of an Ndc80–Dam1 interaction mutant. (A) Crystal structure of human Ndc80 bonsai complex with secondary structure elements labeled. (B) Gels of microtubule cosedimentation assays and corresponding binding curves of the wild-type and Δ256-273 Ndc80 complexes ± Dam1. Deletion of aa 256–273 abrogates Dam1’s ability to enhance Ndc80’s interaction with microtubules (left). (C) Typical gels of Ndc80 recruitment assays. The Δ1-116 ndc80 complex is cosedimenting with microtubules depending on increasing concentrations of Dam1. In contrast, additional deletion of aa 256–273 abrogates Dam1-mediated recruitment (left). Quantification of recruitment assays (right, n = 3). (D) Gels and corresponding binding curves of cosedimentation assays of the ndc80 N-tail mutant and the Δ1-116 Δ256-273 mutant. Lack of aa 256–273 does not alter the autonomous microtubule-binding activity of the Δ1-116 ndc80 complex under low salt conditions (25 mM; data represent results obtained from three independent experiments).
To further test this effect, we performed cosedimentation assays at a fixed microtubule concentration and titrated the amount of the Dam1 complex. For both wild-type Ndc80 and the Δ1-116 N-tail truncation the addition of Dam1 complex enhanced the association of Ndc80 with microtubules in a dose-dependent manner with the fraction of cosedimenting Ndc80 increasing linearly with the concentration of Dam1. By contrast, deletion of aa 256–273 rendered both Ndc80 and Ndc80 Δ1-116 insensitive to the addition of the Dam1 complex (Fig. 2 C) but left basal autonomous microtubule binding of the complexes unchanged (Fig. 2 D).
Charge-reversing CH-domain mutants (K122E, K204E, or K122E, K152E, K160E, K181E, K192E, K204E) abolished the autonomous microtubule-binding activity of the Ndc80 complex even under low ionic strength. Addition of the Dam1 complex was not able to recruit these mutants to microtubules (Fig. S1 B). These experiments strengthen the notion that the Ndc80–Dam1 interaction occurs on the microtubule and strictly requires the intrinsic microtubule-binding activity of the Ndc80 complex.
Ndc80 mutants display differential effects on cellular fitness
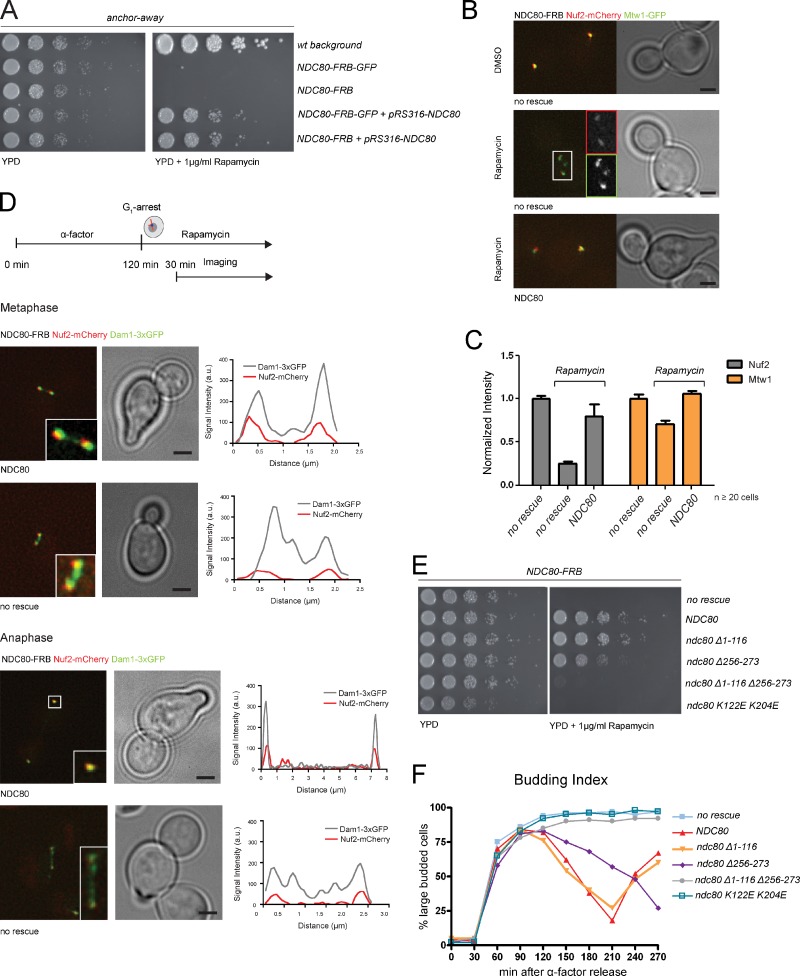
To quickly remove the endogenous Ndc80 protein from the nucleus and test various biochemically defined rescue alleles, we adapted the anchor-away (AA) technique for kinetochore proteins (Fig. S2 A; Haruki et al., 2008). As anticipated, the NDC80-FRB fusion was inviable on rapamycin plates, but could be rescued by expression of wild-type NDC80 from a centromeric plasmid (Fig. 3 A). After 90 min rapamycin exposure the NDC80-FRB-GFP signal became diffuse and was depleted by roughly 90% (Fig. S2 B). This removal of Ndc80 from the kinetochore resulted in a simultaneous reduction of the Nuf2 signal, whereas Mtw1 levels were only slightly affected (Fig. 3, B and C). Consistent with previously reported Ndc80 phenotypes (De Wulf et al., 2003), Mtw1 localization was perturbed in these cells, as it appeared fragmented (Fig. 3 B). Importantly, expression of an Ndc80 wild-type rescue allele restored clustered kinetochores with normal Nuf2 and Mtw1 levels (Fig. 3, B and C).
Figure 3.
Impairing the Dam1–Ndc80 interaction affects cell growth. (A) Ndc80-FRB is lethal on rapamycin plates but rescued by expression of wild-type Ndc80 from a CEN plasmid. (B) Shuttling Ndc80 into the cytoplasm leads to a drop in Nuf2 levels and dispersion of the Mtw1 signal. Expression of wild-type Ndc80 restores Nuf1 and Mtw1 localization. Bars, 2 µm. Insets are magnified 2×. (C) Addition of rapamycin reduced Nuf2 intensity at kinetochores to less than 25%, whereas Mtw1 is moderately affected (75%). Upon expression of wild-type Ndc80, Nuf2 largely remains at kinetochores (n ≥ 20, signal intensity [a.u.] normalized to DMSO-treated cells, error bars represent standard error). (D) Stills of time-lapse movies illustrating Dam1 complex localization in cells depleted for Ndc80. In controls Dam1 is found at kinetochores. A large fraction of Dam1 remains on the spindle in Ndc80-deficient cells (n ≥ 10 from three independent experiments). Bars, 2 µm. Insets are magnified 3×. (E) Spot assay of ndc80 mutants on rapamycin plates. (F) Cell cycle progression of ndc80 mutant strains followed by bud morphology (total percentage of large-budded cells [n = 300] from three independent experiments).
We next addressed the consequences of removing Ndc80 from the nucleus on the subcellular distribution of the Dam1 complex. Cells were arrested in G1, released into rapamycin, and monitored after 30 min. In control cells expressing the wild-type NDC80 rescue allele, we found that the majority of Dam1-3xGFP colocalized with Nuf2 to kinetochores and a minor subpopulation bound to metaphase or anaphase spindles (Fig. 3 D and Videos 1 and 2). In the absence of the rescue allele cells arrested with large buds, short spindles, diminished Nuf2-mCherry signal, and a substantial fraction of Dam1 distributed along nuclear microtubules (Fig. 3 D and Videos 3 and 4). The observed displacement of Dam1 from the kinetochore is in line with previous findings suggesting that kinetochore loading of the Dam1 complex depends on microtubules as well as on Ndc80 (Janke et al., 2002).
We next introduced the ndc80 ΔN1-116, Δ256-273, ΔNΔ256-273, and K122E K204E mutants into the NDC80-FRB background and analyzed their phenotypes on rapamycin plates. Confirming previous reports (Kemmler et al., 2009), the N-tail deletion mutant ndc80 ΔN1-116 grew indistinguishably from a wild-type control strain, whereas the CH-domain mutant (K122E K204E) was lethal on rapamycin (Fig. 3 E). Further, we observed a slow growth phenotype of the ndc80 Δ256-273 mutant that was apparent at all temperatures. Importantly, additional removal of the N-tail from these cells led to lethality in the presence of rapamycin (Fig. 3 E), strongly suggesting that these two structural elements are partially redundant and share an essential function. Analyzing cell cycle progression after synchronization in G1 confirmed the results from the plating assay: the ndc80 Δ256-273 mutant was slow to progress through mitosis, whereas ndc80 ΔNΔ256-273, the K122E K204E mutant, and the strain lacking a rescue allele arrested as large budded cells (Fig. 3 F).
Altering Dam1–Ndc80 cooperation causes chromosome segregation defects
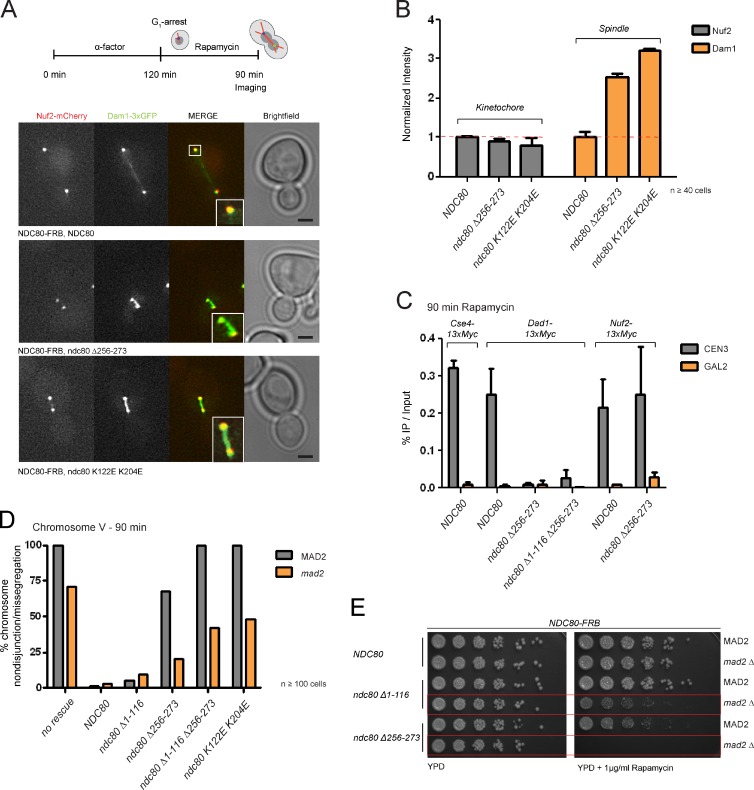
We quantitatively assessed the effects of various Ndc80 mutants on Dam1 localization using live-cell microscopy. We noticed that fusing 3xGFP to Dam1 aggravated the growth defect of ndc80 Δ256-273 on rapamycin plates (Fig. S2 D), possibly due to a direct involvement of the Dam1 C terminus in regulating the interaction between Dam1 and Ndc80 complexes (Cheeseman et al., 2002; Lampert et al., 2010). In support of this notion, microtubule co-pelleting assays revealed a reduced Ndc80 recruitment efficiency for the Dam1 ΔC complex (Fig. S3). Live-cell microscopy showed that 90 min after rapamycin treatment, when wild-type cells had already entered anaphase, both ndc80 Δ256-273 and ndc80 K122E K204E cells exhibited short spindles with high levels of Dam1 decorating the area between the poles. By contrast, the levels of Nuf2 at kinetochores were not significantly affected (Fig. 4, A and B).
Figure 4.
Ndc80 mutants provoke Dam1 complex mislocalization and chromosome segregation defects. (A) Stills of rapamycin-treated cells (90 min) expressing wild-type Ndc80 or ndc80 Δ256-273 and ndc80 K122E K204E mutant complexes. Mutants arrest with Dam1 accumulating on the spindle. Bar, 2 µm. Insets are magnified 2×. (B) Quantification confirms normal Nuf2 localization in the presence of ndc80 mutants but highly elevated levels of Dam1 decorating the spindle (n ≥ 40, signal intensity [a.u.] normalized to Ndc80 wild type, error bars represent standard error). (C) Expression of ndc80 Δ256-273 and Δ1-116 Δ256-273 abolish loading of Dad1 but not Nuf2 to the kinetochore. Percentage of recovered CEN3 was calculated to the WCE input and no AB control was subtracted (n = 3, error bars represent standard error). (D) Percentage of chromosome segregation phenotypes in ndc80 mutants (n ≥ 100 cells from four independent experiments). (E) Growth assay on rapamycin plates comparing wild-type versus Δmad2 background. ndc80 Δ256-273 depends on Mad2 for viability.
As an independent experimental approach to assess the kinetochore association of Dam1 we performed ChIP/qPCR analysis. In an Ndc80 wild-type background, Cse4-13xMyc and Dad1-13xMyc were efficiently bound to CEN3 but not to a control GAL2 locus. In ndc80 Δ256-273 and ndc80 Δ1-116 Δ256-273 strains, however, localization of Dad1-13xmyc to CEN3 was largely abolished (Fig. 4 C), whereas Nuf2 was retained at normal centromeric levels. This supports the conclusion that ndc80 Δ256-273 separates the functions of the Ndc80 complex: complex integrity and recruitment to the centromere remain intact, but promotion of Dam1 localization to the kinetochore and thus assembly of a correct outer kinetochore structure are compromised.
The inability to form a correct kinetochore–microtubule interface might lead to chromosome segregation defects. To test this, we monitored the behavior of GFP-tagged chromosome V in the presence of rapamycin after release from α-factor arrest. Although the control strain and the ndc80 ΔN1-116 mutant segregated chromosome V correctly (98.8% and 95% of large budded cells containing two GFP dots, respectively), this was not the case in ndc80 ΔNΔ256-273 or ndc80 K122E K204E cells. The sister chromatids in these cells remained unresolved and were predominantly localized at the bud-neck. An intermediate phenotype was observed in the ndc80 Δ256-273 strain, which partially distributed chromosome V to the daughter cells (32.1%; Fig. 4 D). Abrogation of the mitotic checkpoint by deleting MAD2 allowed all mutants to enter anaphase and resulted in roughly 40–50% chromosome mis-segregation within a single cell cycle for ndc80 ΔNΔ256-273 and ndc80 K122E K204E, respectively, with the latter sometimes showing a nondisjunction phenotype. This effect was more frequent in the NDC80-FRB strain lacking any rescue allele (70.8%). For the ndc80 ΔN1-116 and Δ256-273 alleles we observed milder mis-segregation defects of 9.5% and 20.8%, respectively.
Given the slow growth phenotype and the high rate of chromosome mis-segregation in the ndc80 Δ256-273 mutant, we asked whether the mitotic checkpoint was essential to keep this strain viable. Growth assays on rapamycin plates demonstrated that the ndc80 Δ256-273 mutation was inviable in a mad2 deletion background (Fig. 4 E). To a lesser extent, elimination of the checkpoint also compromised growth of the ndc80 ΔN1-116 mutant.
In summary, we conclude that interference with the Dam1–Ndc80 interaction in vivo severely compromises chromosome segregation and creates a dependency on the mitotic checkpoint for viability.
Conclusions
Our work provides novel mechanistic insights into the integration of multiple microtubule-binding activities at the kinetochore. We draw three main conclusions from our study.
Kinetochore–microtubule attachments in budding yeast rely critically on the CH-domain of Ndc80.
Our analysis shows that charge-altering mutations in conserved lysine residues abolish the microtubule-binding ability of the yeast complex in vitro and are detrimental for viability (Fig. 5 A). The residues localize to the so-called “toe” region of the Ndc80 CH-domain directly contacting the “toe-print” binding motif formed at the interface between tubulin monomers on the microtubule surface (Alushin et al., 2010).
Figure 5.
Model for kinetochore–microtubule attachments. (A) Current working model of the yeast kinetochore–microtubule interface. The Ndc80 coiled-coil is modeled as a stick with the crystal structures of the Ndc80-Nuf2 CH domains and Spc24-25 on either end. The structure of a Dam1 dimer (two heterodecameric Dam1 complexes) was segmented from the reconstruction of a Dam1 ring (Ramey et al., 2011). Red asterisk denotes the position of Ndc80 residues 256–273. (B) Structural model of the Dam1–Ndc80 interface. The model shows a single Dam1 ring, composed of 16 heterodecamers and six Ndc80 complexes viewed from the minus end of a depolymerizing microtubule attached to the kinetochore. In red are Ndc80 residues 256–273, critical for the Ndc80–Dam1 interaction.
The Ndc80–Dam1 interaction is critical for cell cycle progression and shares an essential function with the N-tail of Ndc80.
Specific interference with Ndc80–Dam1 coupling delays cell cycle progression, creates a dependency on the mitotic checkpoint, and is synthetic-lethal in combination with removal of the Ndc80 N terminus (Fig. 5 A). These results support the notion that, unlike in humans, deletion of the N terminus in yeast Ndc80 is tolerated because its absence is sufficiently balanced by Dam1 complex activity (Lampert et al., 2010; Demirel et al., 2012).
Residues critical for the Ndc80–Dam1 interaction map in close proximity to the CH-domain of Nuf2.
Our biochemical experiments show that residues close to the CH-domains support the Ndc80–Dam1 interaction. This conclusion is independently supported by a recent EM analysis of isolated yeast kinetochores (Gonen et al., 2012). In this study the contact point between the Ndc80 complex and Dam1 rings appeared to be in close proximity to the CH-domains. Combined with our analysis, this challenges the recently proposed role for the evolutionary conserved flexible “loop” (Ndc80 aa 480–520; Maure et al., 2011). Though loop mutants were specifically defective in the establishment of stable end-on attachments, in vitro Dam1-mediated Ndc80 recruitment to microtubules was not affected (Maure et al., 2011). Thus, we speculate that mutations within this domain might indirectly affect Dam1 binding in vivo by lowering the degree of intramolecular Ndc80 flexibility (Maiolica et al., 2007; Ciferri et al., 2008; Wang et al., 2008; Joglekar et al., 2009). The fact that the hinge domain is conserved throughout evolution might suggest a more general role for this element in establishment of attachments (Hsu and Toda, 2011; Schmidt and Cheeseman, 2011).
Combining the available structural data on Ndc80 and Dam1 complexes and the biochemical insights presented in this study allows us to propose one possible model for the Dam1–Ndc80 kinetochore interface (Fig. 5 B). We note that in the future, cryo-EM studies of both complexes simultaneously bound to microtubules have the potential to precisely establish their spatial arrangement and answer whether amino acids 256–273 are directly in contact with Dam1 or whether their deletion impairs Ndc80–Dam1 coupling by a different mechanism. Despite the evolutionary restriction of the Dam1 complex to yeasts, similar concepts for the integration of Ndc80 complex function at the microtubule plus-end are likely also operative in higher eukaryotes.
Materials and methods
Yeast genetics
All strains are derived from strain background W303 unless otherwise stated. Standard procedures were applied to genetically modify strains. Yeast strains used in this study are described in Table S1.
Imaging
Log-phase cells were synchronized in G1 with 10 µg/ml α-factor mating pheromone, washed several times with water, and subsequently released into YPD medium containing 1 µg/ml rapamycin. Stacks and time-lapse movies were collected at ambient temperature (21°C) using live-cell DeltaVision (Applied Precision) microscopy, a UPlanSApo 100× oil immersion objective lens (NA 1.40; Olympus), and a CoolSnap HQ CCD camera (Photometrics). Z-stacks (0.6 µm) were acquired in 20-s intervals, subsequently deconvoluted, projected to two-dimensional images (SoftWoRx software), and further analyzed by MetaMorph (Molecular Devices) or ImageJ (National Institutes of Health) software. For the quantification of kinetochore localization, both kinetochore clusters were manually selected from each cell and the corresponding signal intensities were integrated. Spindle localization signal intensities occurring in-between the sister kinetochores were manually selected and calculated over the whole length. To quantify chromosome segregation, cells were fixed with 4% paraformaldehyde and resuspended in potassium phosphate buffer, pH 7.5, supplemented with sorbitol.
Chromatin immunoprecipitation
50-ml cultures were synchronized in G1 and released into rapamycin-containing medium. Cells were grown to mid-log phase and cross-linked for 30 min with 1% formaldehyde. Chromatin extract preparation and immunoprecipitation were performed as described previously (Mendoza et al., 2009). In brief, cells were resuspended in 50 mM Hepes, pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, and 0.1% sodium-deoxycholate, and lysed using a high-energy cell disrupter (Mini-BeadBeater-16; BioSpec). Subsequently, the chromatin was sonicated to an average length of 500–800 bp, split into two, and incubated with α-Myc (9E10; Covance) coupled or uncoupled Pan Mouse IgG Dynabeads (Invitrogen). Protein–DNA complexes were eluted from the beads with buffer containing 50 mM Tris-HCl, pH 8.0, 10 mM EDTA, and 1% SDS at 65°C. After purification of the DNA, samples were analyzed by quantitative PCR using primer sequences for CEN3 (5′-AAATAGTACAAATAAGTCACAT; 3′-CCATTCAATGAAATATATATTTC) and GAL2 (5′-CGAACTCAGTTCAATGGAGAGT; 3′-CGTCTTCTACCTTATTCACCTC) that have been described previously (Camahort et al., 2009) with a real-time PCR thermocycler (model iQ5), iQ Sybr Green Supermix reagents, and iQ5 software (all from Bio-Rad Laboratories).
Purification of recombinant complexes from bacteria
Expression and purification of the Dam1 complex was performed as described previously (Westermann et al., 2005). Expression, purification, and reconstitution of the full-length Ndc80 complex and mutant derivates was performed as described previously (Lampert et al., 2010). In brief, the two subcomplexes of the Ndc80 complex (Ndc80p-6xHis/Nuf2p-EGFP and Spc24p-6xHis/Spc25p) were separately expressed from the pETDuett or pACYCDuet-1 vectors (EMD Millipore). Bacteria were grown to OD600 = 0.6 at 37°C, induced with 0.2 mM IPTG, and grown for 12–15 h at 18°C. The two subcomplexes were lysed in 25 mM Na2HPO4/NaH2PO4, pH 7.0, 300 mM NaCl, and 20 mM imidazol, and eluted from Ni-NTA beads with 25 mM Na2HPO4/NaH2PO4, pH 7.0, 150 mM NaCl, and 200 mM imidazol. Reconstitution of the full complex was achieved via mixing of the subcomplexes and a second purification step on a Superdex 200 HiLoad 16/60 column (GE Healthcare) in 25 mM Na2HPO4/NaH2PO4, pH 7.0, and 150 mM NaCl. Ndc80 deletion constructs were generated with the Phusion Site-Directed Mutagenesis kit (Thermo Fisher Scientific).
Microtubule-binding assays
For microtubule co-pelleting experiments, purified Ndc80-EGFP and Dam1 complexes were precleared for 5 min at 60 krpm. Taxol-stabilized microtubules were incubated with 0.5 µM of protein complexes for 15 min. The microtubule-bound protein was separated from the unbound fraction via ultracentrifugation (60 krpm, TL100 rotor; Beckman Coulter). Quantification was performed as described previously (Zimniak et al., 2009). For the microtubule recruiting assays the amount of microtubules (2.5 µM), Ndc80 complex (0.5 µM), and the salt concentration (100 mM) were constant in all experiments. The amount of Dam1 complex was varied (0.125–0.625 µM). Binding was quantified using GFP endpoint measurements on a Synergy H1 Hybrid Multi-Mode Microplate Reader (BioTek).
Online supplemental material
Fig. S1 shows purification of recombinant Ndc80 complexes and microtubule cosedimentation assays with CH-domain mutants. Fig. S2 shows a characterization of the anchor-away system, expression analysis of mutant Ndc80 complexes, and growth assays in the Dam1-3xGFP background. Fig. S3 shows Ndc80 recruitment assays with a mutant Dam1ΔC complex. Videos 1–4 show localization of Dam1 and Ndc80 in wild-type and mutant cells. Table S1 describes yeast strains used in this study and is provided as an Excel spreadsheet. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201210091/DC1.
Supplementary Material
Acknowledgments
The authors wish to thank all members of the Westermann laboratory for discussions; René Ladurner, Jan-Michael Peters, and Alexander Schleiffer for critical reading of the manuscript; and Daniel Dyer for experimental help.
Research leading to these results has received funding from the European Research Council under the European Community’s Seventh Framework Program (FP7/2007-2013)/ERC grant agreement (no. 203499), by the Austrian Science Fund FWF (SFB F34-B03), and by the National Institutes of Health (P01 GM051487). E. Nogales is a Howard Hughes Medical Institute Investigator.
References
- Alushin G.M., Ramey V.H., Pasqualato S., Ball D.A., Grigorieff N., Musacchio A., Nogales E. 2010. The Ndc80 kinetochore complex forms oligomeric arrays along microtubules. Nature. 467:805–810 10.1038/nature09423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asbury C.L., Gestaut D.R., Powers A.F., Franck A.D., Davis T.N. 2006. The Dam1 kinetochore complex harnesses microtubule dynamics to produce force and movement. Proc. Natl. Acad. Sci. USA. 103:9873–9878 10.1073/pnas.0602249103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom K., Joglekar A. 2010. Towards building a chromosome segregation machine. Nature. 463:446–456 10.1038/nature08912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camahort R., Shivaraju M., Mattingly M., Li B., Nakanishi S., Zhu D., Shilatifard A., Workman J.L., Gerton J.L. 2009. Cse4 is part of an octameric nucleosome in budding yeast. Mol. Cell. 35:794–805 10.1016/j.molcel.2009.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., Anderson S., Jwa M., Green E.M., Kang Js., Yates J.R., III, Chan C.S., Drubin D.G., Barnes G. 2002. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 111:163–172 10.1016/S0092-8674(02)00973-X [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Chappie J.S., Wilson-Kubalek E.M., Desai A. 2006. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 127:983–997 10.1016/j.cell.2006.09.039 [DOI] [PubMed] [Google Scholar]
- Ciferri C., Pasqualato S., Screpanti E., Varetti G., Santaguida S., Dos Reis G., Maiolica A., Polka J., De Luca J.G., De Wulf P., et al. 2008. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 133:427–439 10.1016/j.cell.2008.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wulf P., McAinsh A.D., Sorger P.K. 2003. Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes Dev. 17:2902–2921 10.1101/gad.1144403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca J.G., Dong Y., Hergert P., Strauss J., Hickey J.M., Salmon E.D., McEwen B.F. 2005. Hec1 and nuf2 are core components of the kinetochore outer plate essential for organizing microtubule attachment sites. Mol. Biol. Cell. 16:519–531 10.1091/mbc.E04-09-0852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca K.F., Lens S.M., DeLuca J.G. 2011. Temporal changes in Hec1 phosphorylation control kinetochore-microtubule attachment stability during mitosis. J. Cell Sci. 124:622–634 10.1242/jcs.072629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirel P.B., Keyes B.E., Chaterjee M., Remington C.E., Burke D.J. 2012. A redundant function for the N-terminal tail of Ndc80 in kinetochore-microtubule interaction in Saccharomyces cerevisiae. Genetics. 192:753–756 10.1534/genetics.112.143818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A., Rybina S., Müller-Reichert T., Shevchenko A., Shevchenko A., Hyman A., Oegema K. 2003. KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes Dev. 17:2421–2435 10.1101/gad.1126303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gestaut D.R., Graczyk B., Cooper J., Widlund P.O., Zelter A., Wordeman L., Asbury C.L., Davis T.N. 2008. Phosphoregulation and depolymerization-driven movement of the Dam1 complex do not require ring formation. Nat. Cell Biol. 10:407–414 10.1038/ncb1702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonen S., Akiyoshi B., Iadanza M.G., Shi D., Duggan N., Biggins S., Gonen T. 2012. The structure of purified kinetochores reveals multiple microtubule-attachment sites. Nat. Struct. Mol. Biol. 19:925–929 10.1038/nsmb.2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk E.L., Efremov A.K., Volkov V.A., Spiridonov I.S., Gudimchuk N., Westermann S., Drubin D., Barnes G., McIntosh J.R., Ataullakhanov F.I. 2008. The Dam1 ring binds microtubules strongly enough to be a processive as well as energy-efficient coupler for chromosome motion. Proc. Natl. Acad. Sci. USA. 105:15423–15428 10.1073/pnas.0807859105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruki H., Nishikawa J., Laemmli U.K. 2008. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol. Cell. 31:925–932 10.1016/j.molcel.2008.07.020 [DOI] [PubMed] [Google Scholar]
- Hsu K.S., Toda T. 2011. Ndc80 internal loop interacts with Dis1/TOG to ensure proper kinetochore-spindle attachment in fission yeast. Curr. Biol. 21:214–220 10.1016/j.cub.2010.12.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C., Ortíz J., Tanaka T.U., Lechner J., Schiebel E. 2002. Four new subunits of the Dam1-Duo1 complex reveal novel functions in sister kinetochore biorientation. EMBO J. 21:181–193 10.1093/emboj/21.1.181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar A.P., Bloom K., Salmon E.D. 2009. In vivo protein architecture of the eukaryotic kinetochore with nanometer scale accuracy. Curr. Biol. 19:694–699 10.1016/j.cub.2009.02.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemmler S., Stach M., Knapp M., Ortiz J., Pfannstiel J., Ruppert T., Lechner J. 2009. Mimicking Ndc80 phosphorylation triggers spindle assembly checkpoint signalling. EMBO J. 28:1099–1110 10.1038/emboj.2009.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampert F., Westermann S. 2011. A blueprint for kinetochores - new insights into the molecular mechanics of cell division. Nat. Rev. Mol. Cell Biol. 12:407–412 10.1038/nrm3133 [DOI] [PubMed] [Google Scholar]
- Lampert F., Hornung P., Westermann S. 2010. The Dam1 complex confers microtubule plus end-tracking activity to the Ndc80 kinetochore complex. J. Cell Biol. 189:641–649 10.1083/jcb.200912021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiolica A., Cittaro D., Borsotti D., Sennels L., Ciferri C., Tarricone C., Musacchio A., Rappsilber J. 2007. Structural analysis of multiprotein complexes by cross-linking, mass spectrometry, and database searching. Mol. Cell. Proteomics. 6:2200–2211 10.1074/mcp.M700274-MCP200 [DOI] [PubMed] [Google Scholar]
- Maure J.F., Komoto S., Oku Y., Mino A., Pasqualato S., Natsume K., Clayton L., Musacchio A., Tanaka T.U. 2011. The Ndc80 loop region facilitates formation of kinetochore attachment to the dynamic microtubule plus end. Curr. Biol. 21:207–213 10.1016/j.cub.2010.12.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza M.A., Panizza S., Klein F. 2009. Analysis of protein-DNA interactions during meiosis by quantitative chromatin immunoprecipitation (qChIP). Methods Mol. Biol. 557:267–283 10.1007/978-1-59745-527-5_17 [DOI] [PubMed] [Google Scholar]
- Miranda J.J., De Wulf P., Sorger P.K., Harrison S.C. 2005. The yeast DASH complex forms closed rings on microtubules. Nat. Struct. Mol. Biol. 12:138–143 10.1038/nsmb896 [DOI] [PubMed] [Google Scholar]
- Powers A.F., Franck A.D., Gestaut D.R., Cooper J., Gracyzk B., Wei R.R., Wordeman L., Davis T.N., Asbury C.L. 2009. The Ndc80 kinetochore complex forms load-bearing attachments to dynamic microtubule tips via biased diffusion. Cell. 136:865–875 10.1016/j.cell.2008.12.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramey V.H., Wong A., Fang J., Howes S., Barnes G., Nogales E. 2011. Subunit organization in the Dam1 kinetochore complex and its ring around microtubules. Mol. Biol. Cell. 22:4335–4342 10.1091/mbc.E11-07-0659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S., Musacchio A. 2009. The life and miracles of kinetochores. EMBO J. 28:2511–2531 10.1038/emboj.2009.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt J.C., Cheeseman I.M. 2011. Chromosome segregation: keeping kinetochores in the loop. Curr. Biol. 21:R110–R112 10.1016/j.cub.2010.12.030 [DOI] [PubMed] [Google Scholar]
- Sundin L.J., Guimaraes G.J., Deluca J.G. 2011. The NDC80 complex proteins Nuf2 and Hec1 make distinct contributions to kinetochore-microtubule attachment in mitosis. Mol. Biol. Cell. 22:759–768 10.1091/mbc.E10-08-0671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Kitamura E., Kitamura Y., Tanaka T.U. 2007. Molecular mechanisms of microtubule-dependent kinetochore transport toward spindle poles. J. Cell Biol. 178:269–281 10.1083/jcb.200702141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tien J.F., Umbreit N.T., Gestaut D.R., Franck A.D., Cooper J., Wordeman L., Gonen T., Asbury C.L., Davis T.N. 2010. Cooperation of the Dam1 and Ndc80 kinetochore complexes enhances microtubule coupling and is regulated by aurora B. J. Cell Biol. 189:713–723 10.1083/jcb.200910142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooley J.G., Miller S.A., Stukenberg P.T. 2011. The Ndc80 complex uses a tripartite attachment point to couple microtubule depolymerization to chromosome movement. Mol. Biol. Cell. 22:1217–1226 10.1091/mbc.E10-07-0626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.W., Long S., Ciferri C., Westermann S., Drubin D., Barnes G., Nogales E. 2008. Architecture and flexibility of the yeast Ndc80 kinetochore complex. J. Mol. Biol. 383:894–903 10.1016/j.jmb.2008.08.077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei R.R., Sorger P.K., Harrison S.C. 2005. Molecular organization of the Ndc80 complex, an essential kinetochore component. Proc. Natl. Acad. Sci. USA. 102:5363–5367 10.1073/pnas.0501168102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S., Avila-Sakar A., Wang H.W., Niederstrasser H., Wong J., Drubin D.G., Nogales E., Barnes G. 2005. Formation of a dynamic kinetochore- microtubule interface through assembly of the Dam1 ring complex. Mol. Cell. 17:277–290 10.1016/j.molcel.2004.12.019 [DOI] [PubMed] [Google Scholar]
- Westermann S., Wang H.W., Avila-Sakar A., Drubin D.G., Nogales E., Barnes G. 2006. The Dam1 kinetochore ring complex moves processively on depolymerizing microtubule ends. Nature. 440:565–569 10.1038/nature04409 [DOI] [PubMed] [Google Scholar]
- Wigge P.A., Kilmartin J.V. 2001. The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J. Cell Biol. 152:349–360 10.1083/jcb.152.2.349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson-Kubalek E.M., Cheeseman I.M., Yoshioka C., Desai A., Milligan R.A. 2008. Orientation and structure of the Ndc80 complex on the microtubule lattice. J. Cell Biol. 182:1055–1061 10.1083/jcb.200804170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimniak T., Stengl K., Mechtler K., Westermann S. 2009. Phosphoregulation of the budding yeast EB1 homologue Bim1p by Aurora/Ipl1p. J. Cell Biol. 186:379–391 10.1083/jcb.200901036 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.