Engineered kinetochores reveal distinct functions of the CCAN in recruiting CENP-A to the centromere and acting as a structural core to directly recruit kinetochore proteins.
Abstract
CENP-A acts as an important epigenetic marker for kinetochore specification. However, the mechanisms by which CENP-A is incorporated into centromeres and the structural basis for kinetochore formation downstream of CENP-A remain unclear. Here, we used a unique chromosome-engineering system in which kinetochore proteins are targeted to a noncentromeric site after the endogenous centromere is conditionally removed. Using this system, we created two distinct types of engineered kinetochores, both of which were stably maintained in chicken DT40 cells. Ectopic targeting of full-length HJURP, CENP-C, CENP-I, or the CENP-C C terminus generated engineered kinetochores containing major kinetochore components, including CENP-A. In contrast, ectopic targeting of the CENP-T or CENP-C N terminus generated functional kinetochores that recruit the microtubule-binding Ndc80 complex and chromosome passenger complex (CPC), but lack CENP-A and most constitutive centromere-associated network (CCAN) proteins. Based on the analysis of these different engineered kinetochores, we conclude that the CCAN has two distinct roles: recruiting CENP-A to establish the kinetochore and serving as a structural core to directly recruit kinetochore proteins.
Introduction
The kinetochore provides the critical chromosomal interface with microtubules from the mitotic spindle for faithful chromosome segregation during mitosis. Recent studies have revealed that the kinetochore is specified on centromeric chromatin by sequence-independent epigenetic mechanisms (Black and Cleveland, 2011; Perpelescu and Fukagawa, 2011). The centromere-specific histone H3 variant CENP-A is a key epigenetic marker because all active centromeres, including natural or experimentally induced neocentromeres, contain CENP-A (Ishii et al., 2008; Marshall et al., 2008; Shang et al., 2010; Perpelescu and Fukagawa, 2011; Black and Cleveland, 2011; Burrack and Berman, 2012). In addition, ectopic localization of CENP-A to noncentromeric loci induces kinetochore-like structures in vertebrate or Drosophila melanogaster cells (Barnhart et al., 2011; Guse et al., 2011; Mendiburo et al., 2011), which suggests that CENP-A is an upstream factor for kinetochore assembly. Several molecules including Mis16/RbAp46, the Mis18 complex, and HJURP are involved in CENP-A recruitment to centromeres (Hayashi et al., 2004; Fujita et al., 2007; Dunleavy et al., 2009; Foltz et al., 2009). However, it is still unknown how centromeres are specified by CENP-A through these molecules.
In addition to CENP-A, a group of 16 chromatin-proximal proteins termed the constitutive centromere-associated network (CCAN) also associate with centromeric chromatin. The CENP-T-W-S-X complex, a CCAN component, directly binds to centromeric DNA using a histone-like structure (Hori et al., 2008; Nishino et al., 2012). CENP-T has its histone-like domain at the C terminus and also has an extended N-terminal region, which is critical for its association with outer kinetochore proteins (Gascoigne et al., 2011; Suzuki et al., 2011). Thus, CENP-T interacts with both centromeric chromatin and microtubule-binding kinetochore complexes. To support this model, we previously transiently expressed a CENP-T–LacI fusion to target it to a noncentromere LacO locus and observed formation of a kinetochore-like structure at this site (Gascoigne et al., 2011). The CCAN component CENP-C also has DNA-binding activity (Saitoh et al., 1992; Yang et al., 1996) and connects with the outer kinetochore Mis12 complex (Przewloka et al., 2011; Screpanti et al., 2011). Transient targeting of CENP-C to a noncentromere LacO locus induces the recruitment of some outer kinetochore proteins, similar to CENP-T (Gascoigne et al., 2011). In each case, these ectopic kinetochore-like structures lacked CENP-A. Such experiments using ectopic localization of kinetochore proteins provide useful information to understand kinetochore assembly. However, as transient expression of proteins was used previously, there were questions regarding whether CENP-T or CENP-C are able to induce formation of a “stable” kinetochore structure in the absence of CENP-A. Thus, it is critical to define how the CCAN contributes to centromere specification and how the CCAN functions as a structural core for outer kinetochore assembly to establish functional kinetochores.
To address these questions, we developed a unique experimental system in which an endogenous centromere is conditionally removed (Shang et al., 2010; Gascoigne et al., 2011) after the ectopic localization of kinetochore components to a noncentromere LacO locus of the chicken chromosome Z. After growth and selection, we isolated stable cell lines in which the endogenous centromere of chromosome Z was completely removed, but a functional kinetochore is formed at the noncentromeric LacO locus by the ectopic targeting of LacI fusion proteins. Using this experimental system, we isolated two distinct types of engineered kinetochores. The first type of ectopic kinetochore, formed by the targeting of full-length of HJURP, CENP-C, CENP-I, or the C-terminus of CENP-C, contains the full complement of kinetochore components including CENP-A. The second type of engineered kinetochore, formed by ectopic targeting of the CENP-T N terminus (1–530 aa) or CENP-C N terminus (1–643 aa), lacks CENP-A and most CCAN proteins, but recruits the microtubule-binding Ndc80 complex and the chromosome passenger complex (CPC). Based on these results, we conclude that the CCAN has two distinct roles: recruiting CENP-A to establish the kinetochore and serving as a structural core to recruit the Ndc80 complex and the CPC independently of CENP-A. CENP-C possesses dual roles, with its N terminus acting as a structural platform and its C terminus directing the recruitment of CENP-A.
Results
Ectopic localization of inner kinetochore proteins causes efficient kinetochore formation at a noncentromere locus
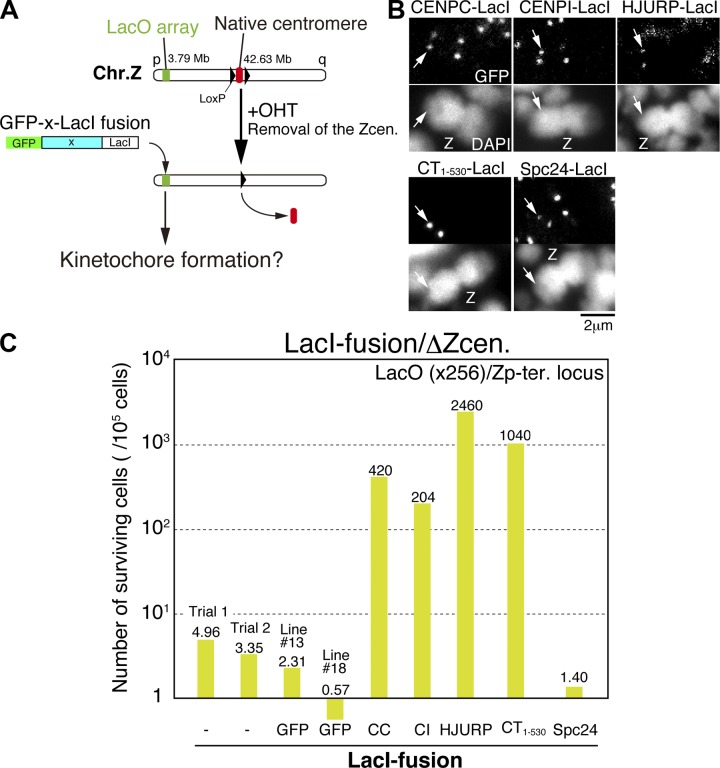
We previously demonstrated that the chicken Z chromosome contains a centromere of 30–50 kb lacking the tandem repetitive sequences typically found in other vertebrates (Shang et al., 2010). We further established an experimental system to conditionally excise the Z chromosome centromere using chromosome engineering (Shang et al., 2010). Here, we combined centromere excision with ectopic localization of each kinetochore protein to a noncentromere locus using the LacO–LacI system (Fig. 1 A). We generated GFP (N-terminal)/LacI (C-terminal) double fusion constructs with multiple different kinetochore proteins, including the CENP-T N terminus (1–530 aa), CENP-C, CENP-I, Spc24, and the CENP-A–specific chaperone HJURP (Dunleavy et al., 2009; Foltz et al., 2009). We confirmed that the CENP-C, CENP-I, and Spc24 fusion proteins were stably expressed and localized to both the LacO locus and the endogenous centromere (Fig. 1 B). We detected the HJURP-LacI fusion at the LacO locus (Fig. 1 B), but not endogenous centromeres, because HJURP localized to centromeres only in G1 (Dunleavy et al., 2009; Foltz et al., 2009). The CENP-T N terminus also localized only to the LacO locus because of the absence of its C-terminal centromere-targeting domain (Fig. 1 B). After expression of the LacI fusion protein, we removed the endogenous centromere from chromosome Z by activation of Cre recombinase using hydroxytamoxifen (OHT) addition and isolated the surviving clones (see timeline in Fig. S1 A). After removal of the endogenous centromere in cells expressing GFP-LacI, we isolated surviving clones at rate of 1–2 × 10−5, similar to the rate observed in cells lacking the LacI fusion protein (Fig. 1 C). Analysis of the surviving clones demonstrated that chromosome Z lacking the centromere had fused to another chromosome or acquired a neocentromere to prevent loss of the chromosome. By analyzing the position of the neocentromeres of these surviving clones, we did not detect neocentromeres at the LacO locus (unpublished data). In contrast to cells expressing GFP-LacI, the survival rate was increased 200–2,000 times in cells expressing LacI fusions to the CENP-T N terminus, CENP-C, CENP-I, or HJURP (Fig. 1 C). In the surviving clones for each of these diverse LacI fusions, an ectopic kinetochore was formed at the LacO locus. We will refer to these stable selected cell lines with the ectopic kinetochore at the LacO locus after removal of endogenous Z centromere as CT1–530-LacI/ΔZcen (for CENP-T 1–530), CC-LacI/ΔZcen (for CENP-C), CI-LacI/ΔZcen (for CENP-I), or HJ-LacI/ΔZcen (for HJURP) cell lines. We tested cells with various copy numbers of LacO repeats and found that 256 copies displayed the highest efficiency for isolating surviving clones (Fig. S1 B). Thus, we used 256 LacO repeats for all further experiments. We also used an independent cell line in which the LacO sequence was integrated at a different locus and found that ectopic kinetochores were similarly formed at the LacO locus after expression of LacI fusions with CENP-T N terminus or HJURP (unpublished data).
Figure 1.
Ectopic localization of CCAN proteins causes efficient kinetochore formation at a noncentromere locus. (A) An experimental design to examine kinetochore formation at a noncentromeric locus after removal of endogenous centromere of chromosome Z. The 256 copies of LacO repeats are integrated on the 3.79-Mb locus of chromosome Z in DT40 cells (LacO locus). GFP/LacI double fusion constructs with multiple different proteins (X) were expressed in cells with the LacO locus. The endogenous centromere of chromosome Z was removed by activation of Cre-recombinase (OHT addition). After removal of endogenous centromere, surviving clones were isolated. Surviving cells are expected to have an ectopic kinetochore at the LacO locus on chromosome Z. Experimental timeline is shown in Fig. S1. (B) Localization of GFP/LacI double fusion with multiple different proteins at the LacO locus before removal of endogenous centromere. Arrows show the LacO locus. All tested fusion proteins localize into the LacO locus. Z, Z chromosome. (C) The survival rate of cells after removal of endogenous centromere of chromosome Z in each assay. The survival rate was increased in cells expressing LacI fusion to CENP-C (CC), CENP-I (CI), HJURP, or CENP-T N terminus (CT1–530), compared with that of cells expressing GFP-LacI or lacking LacI fusion protein. The survival rate was not increased in cells expressing LacI-Spc24. The assay was completed once for each experiment except for CENP-C fusion. In the case of CENP-C, the assay was done twice.
In contrast to the formation of stable ectopic kinetochores for CENP-C, CENP-I, CENP-T, and HJURP, expression of a GFP-Spc24-LacI fusion did not increase the survival rate after excision of the endogenous centromere. This suggests that the Spc24-LacI fusion did not induce kinetochore formation at the LacO locus, even though Ndc80 was recruited to the LacO locus (Fig. S1 C). Spc24 is a component of the Ndc80 complex, which interacts directly with microtubules (Cheeseman et al., 2006; DeLuca et al., 2006; Alushin et al., 2010; DeLuca and Musacchio, 2012). This data suggests that the presence of a microtubule-binding complex at the LacO locus is not sufficient for kinetochore function and that other factors are required for formation of functional kinetochores in vertebrate cells. Together, these results demonstrate that it is possible to generate engineered ectopic kinetochores that are stably propagated after removal of the endogenous centromere.
CENP-T–derived ectopic kinetochores are IPTG-sensitive, whereas HJURP-, CENP-C–, or CENP-I–induced kinetochores are IPTG-independent
To analyze the features of the kinetochores induced by ectopic localization of the selected kinetochore proteins to the LacO locus, we next added IPTG into each surviving cell line. Addition of IPTG disrupts the interaction of LacI fusion proteins with the LacO sequences. Thus, after IPTG addition, we tested the requirement of each LacI fusion protein to maintain kinetochore function at the LacO locus. Cell growth and the fidelity of chromosome segregation in clones with ectopic kinetochores induced by LacI fusions to full-length CENP-C (CC-LacI/ΔZcen cells), CENP-I (CI-LacI/ΔZcen cells), or HJURP (HJ-LacI/ΔZcen cells) were indistinguishable in the absence or presence of IPTG (Fig. 2 A). Consistent with this, we did not detect loss of chromosome Z in CC-LacI/ΔZcen, CI-LacI/ΔZcen, or HJ-LacI/ΔZcen cells in the absence or presence of IPTG (Figs. 2 B and S2 A). These data indicate that these LacI fusion proteins are not necessary to maintain kinetochore function at the LacO locus after the kinetochore has been formed, which suggests that other kinetochore components that were recruited to the LacO locus are able to stably propagate a kinetochore structure at this site. Indeed, CENP-A, CENP-C, CENP-T, and Ndc80 all localized to the CENP-C–, CENP-I–, and HJURP-derived kinetochores (Fig. 2 C). As CENP-A has been proposed to act as an epigenetic mark for functional kinetochores, we also performed chromatin immunoprecipitation (ChIP) experiments with anti-CENP-A antibodies to test CENP-A incorporation into the LacO locus. Indeed, we detected LacO sequences in CENP-A immunoprecipitates in cells with the CENP-C–, CENP-I–, or HJURP-derived kinetochores (Fig. 2 D).
Figure 2.
Two distinct types of engineered kinetochores at the LacO locus were created. (A) Growth curve of the stable selected cell lines with the ectopic kinetochore at the LacO locus after removal of endogenous centromere of chromosome Z in the presence (red line) or absence of IPTG (black line). Each cell line was referred to as CC-LacI/ΔZcen (for CENP-C–LacI expression), CI-LacI/ΔZcen (for CENP-I–LacI expression), or HJ-LacI/ΔZcen (for HJURP-LacI expression). This measurement was completed once in each cell line. (B) Representative images of chromosome Z (Z) stained by the Z-specific probe (red) during anaphase of CC-LacI/ΔZcen, CI-LacI/ΔZcen, or HJ-LacI/ΔZcen cells after addition of IPTG. Arrows indicate FISH signals by a Z chromosome–specific satellite probe. (C) Localization of CENP-A, CENP-C, CENP-T, and Ndc80 at the LacO locus in CC-LacI/ΔZcen, CI-LacI/ΔZcen, or HJ-LacI/ΔZcen cells. These proteins were detected at the LacO locus as red signals. To define the chromosome Z, the Z-specific satellite probes are hybridized (green) on the q arm of chromosome Z (Zq). These experiments were performed in the absence of IPTG. Arrows indicate red signals stained with each antibody. (D) CENP-A–associated DNAs were isolated by ChIP with anti–CENP-A antibodies from CC-LacI/ΔZcen, CI-LacI/ΔZcen, or HJ-LacI/ΔZcen cells. Cells expressing GFP-LacI were also used as a control. The chromatin was isolated as a mononucleosome fraction digested with MNase. The CENP-A–associated DNAs were hybridized with a probe containing the LacO sequence or a centromere DNA from chromosome 5. LacO sequences were detected in CENP-A immunoprecipitates in CC-LacI/ΔZcen, CI-LacI/ΔZcen, or HJ-LacI/ΔZcen cells, but not in GFP-LacI cells. (E) Growth curve of CT1–530-LacI/ΔZcen cells in the presence or absence of IPTG. After addition of IPTG, these cells rapidly died (red solid line). This measurement was completed once in each condition. (F) Representative images of lagging chromosome Z stained by the Z-specific probe (red) during anaphase of CT1–530-LacI/ΔZcen cells after addition of IPTG. (G) Examination of loss or gain of chromosome Z after addition of IPTG to CT1–530-LacI/ΔZcen cells. As a control, numbers of chromosome 1 were also scored. Chromosome Z was lost in >80% of cells at 24 h and 48 h after the addition of IPTG (top). Loss or gain of chromosome 1 was not observed after addition of IPTG to CT1–530-LacI/ΔZcen cells (bottom). This experiment was completed once in each condition (CT1–530-LacI/+Zcen +IPTG, n = 223; CT1–530-LacI/ΔZcen 0 h, n = 140; 24 h, n = 201; 48 h, n = 94). (H) CENP-A–associated DNAs isolated by ChIP with anti–CENP-A antibodies from CT1–530-LacI/ΔZcen cells were hybridized with a probe containing the LacO sequence or a centromere DNA from chromosome 5. LacO sequences were not detected in CENP-A immunoprecipitates in CT1–530-LacI/ΔZcen cells.
In contrast, cells with ectopic kinetochores induced by LacI fusion to the CENP-T N terminus (CT1–530-LacI/ΔZcen cells) stopped growing after addition of IPTG and died rapidly (Figs. 2 E and S2 B). We frequently observed lagging chromosomes in these cells after addition of IPTG (Fig. 2 F). Indeed, based on FISH analysis, chromosome Z was lost in >80% of cells after the addition of IPTG (Fig. 2 G). We note that ∼20% cells did not express the LacI–CENP-T fusion, which caused chromosome loss even in the absence of IPTG (Fig. 2 G). We observed similar IPTG sensitivity for ectopic kinetochores induced by a LacI fusion with full-length CENP-T (Fig. S2, C and D). These results indicate that the CENP-T–LacI fusion is essential to maintain the function of the induced kinetochore at the LacO locus. Consistent with the lack of a stable kinetochore structure in the absence of the LacI fusion, we found that LacO sequences are not present in CENP-A immunoprecipitates from CENP-T–derived kinetochores (Figs. 2 H and S2 E), which indicates that CENP-A is not recruited to these sites.
In total, the ectopic kinetochores induced by CENP-T are different from those induced by CENP-C, CENP-I, or HJURP with the latter proteins required to create, but not to maintain a functional kinetochore structure. In contrast, CENP-T–LacI functions as a structural platform that is necessary to maintain kinetochore function at the LacO locus.
The CENP-C N and C termini play distinct roles in kinetochore assembly
The LacI fusion to full-length CENP-C induced formation of a complete kinetochore structure, including CENP-A, at the LacO locus (Fig. 2, A–D). We next sought to define the domains of CENP-C that are responsible for this activity (Fig. 3 A). We found that expression of LacI fusions to the CENP-C N terminus (1–643 aa) or C terminus (601–864 aa) both induced efficient kinetochore formation at the LacO locus (Fig. 3 A). We note that the CENP-C C terminus, including the Mif2 homology domain, forms a homodimer based on previous analyses of CENP-C/Mif2 (Cohen et al., 2008). We will refer to these stable selected cell lines with the ectopic kinetochores induced by the CENP-C N terminus (1–643 aa) or C terminus (601–864 aa) at the LacO locus as CC1–643-LacI/ΔZcen or CC601–864-LacI/ΔZcen cell lines, respectively (Fig. 3 A). However, although stable cell lines were obtained for each fusion, these cell lines displayed differing sensitivity to IPTG. CC601–864-LacI/ΔZcen cells recruited a full range of kinetochore proteins including CENP-A, and showed growth that was insensitive to IPTG (Fig. 3, B–D). In contrast, after the addition of IPTG, CC1–643-LacI/ΔZcen cells stopped growing, displayed lagging chromosomes and chromosome loss, and rapidly died (Fig. 3, E–G; and Fig. S3), similar to cell lines with CENP-T–derived kinetochores. In addition, CENP-A was absent from LacO sequences in CC1–643-LacI/ΔZcen cell lines based on ChIP analysis (Fig. 3 H).
Figure 3.
Characterization of the CENP-C–derived kinetochores. (A) Diagram showing the chicken CENP-C sequence and the tested deletion constructs for the LacO–LacI–based kinetochore induction assay shown in Fig. 1. The yellow box shows a putative binding domain with the Mis12 complex. Mif2 II and Mif2 III show homology regions with yeast Mif2 (red). The survival rate of cells after removal of native centromere of chromosome Z in cells expressing each LacI fusion construct with multiple deletions is shown in the diagram (right). The survival rate was increased in cells expressing LacI fusion with CENP-C (1–643) or CENP-C (601–864). The assay was completed twice for CC full-length, once for CC1–643, once for CC74–643, once for CC1–165, twice for CC601–761, and three times for CC601–864. (B) Growth curve of CC601–864-LacI/ΔZcen cells in the presence (red line) or absence of IPTG (black line). This measurement was completed once in each condition. (C) Localization of CENP-A, CENP-C, CENP-T, and Ndc80 at the LacO locus in CC601–864-LacI/ΔZcen cells. These experiments were performed in the absence of IPTG. Arrows indicate red signals stained with each antibody. Zq, Z chromosome. (D) CENP-A–associated DNAs isolated by ChIP with anti–CENP-A antibodies from CC601–864-LacI/ΔZcen cells were hybridized with a probe containing the LacO sequence or a centromere DNA from chromosome 5. (E) Growth curve of CC1–643-LacI/ΔZcen cells in the presence or absence of IPTG. This measurement was completed once in each condition. (F) Representative images of lagging chromosome Z stained by the Z-specific probe (red) during anaphase of CC1–643-LacI/ΔZcen cells after addition of IPTG. (G) Examination of loss or gain of chromosome Z after addition of IPTG to CC1–643-LacI/ΔZcen cells (top). As a control, numbers of chromosome 1 were also scored (bottom). Chromosome Z was lost in >80% of cells at 48 h after the addition of IPTG. Loss or gain of chromosome 1 was not observed after addition of IPTG to CC1–643-LacI/ΔZcen cells. This experiment was completed once in each condition (CC1–643-LacI/+Zcen +IPTG, n = 229; CC1–643-LacI/ΔZcen 0 h, n = 281; 24 h, n = 235; 48 h, n = 204). (H) CENP-A–associated DNAs isolated by ChIP with anti-CENP-A antibodies from CC1–643-LacI/ΔZcen cells were hybridized with a probe containing the LacO sequence or a centromere DNA from chromosome 5. LacO sequences were not detected in CENP-A immunoprecipitates in CC1–643-LacI/ΔZcen cells. (I) Localization of Mis12 at the LacO locus in cells expressing LacI fusions with CENP-C (1–643), CENP-C (74–643), or CENP-C (1–165). Mis12 signals were detected in cells expressing LacI fusion with CENP-C (1–643), but not detected in cells expressing LacI fusions with CENP-C (74–643) or CENP-C (1–165). Arrows indicate the position of the LacO sequence. Z, Z chromosome.
The CENP-C N terminus has been shown to interact directly with the Mis12 complex (Gascoigne et al., 2011; Przewloka et al., 2011; Screpanti et al., 2011). To test whether the recruitment of the Mis12 complex by the CENP-C N terminus is essential for ectopic kinetochore formation, we analyzed Mis12 localization to the LacO locus in cells expressing CENP-C (1–643 aa), CENP-C (74–643 aa), or CENP-C (1–165 aa) before removal of the endogenous centromere (Fig. 3 I). We detected Mis12 localization in cells expressing CENP-C (1–643 aa) at the LacO locus in which ectopic kinetochores efficiently formed (Fig. 3, A and I), but not in cells expressing CENP-C (74–643 aa) or CENP-C (1–165 aa) at the LacO locus, in which ectopic kinetochores were not formed (Fig. 3 A). In total, we conclude that CENP-C has dual roles for kinetochore assembly, with the N terminus (1–643 aa) acting as a structural core to assemble outer kinetochore through recruitment of the Mis12 complex and the C terminus (601–864 aa) functioning to form full kinetochores through the recruitment of CENP-A.
The CENP-T and CENP-C N termini–derived kinetochores lack CENP-A and CCAN proteins
Although CT1–530-LacI/ΔZcen and CC1–643-LacI/ΔZcen cells stopped growing after IPTG addition, both cell lines grew well in the absence of IPTG (Fig. 3). To confirm that chromosome segregation in these cells is normal, we examined the behavior of chromosome Z with CENP-T N terminus– or CENP-C N terminus–derived kinetochores by a live-cell imaging. The duration of mitosis (from NEBD to anaphase onset) in both cell lines was ∼30 min in the absence of IPTG, similar to wild-type cells (Fig. 4 A). In addition, lagging chromosomes were rarely detected in these cell lines in the absence of IPTG (Fig. 4 B), although we did observe lagging chromosomes in the presence of IPTG (Fig. S4). These observations confirm that both CENP-T N terminus– and CENP-C N terminus–derived kinetochores are functional in the absence of IPTG. We next examined the localization of other proteins to these ectopically derived kinetochores. Based on immunofluorescence analysis, we did not detect CENP-A, CENP-C, CENP-H, or CENP-O at the LacO locus in CT1–530-LacI/ΔZcen cells (Fig. 4 C, top) or CENP-A, CENP-T, CENP-H, and CENP-O at the LacO locus in CC1–643-LacI/ΔZcen cells (Fig. 4 D, top). As both CT1–530-LacI/ΔZcen and CC1–643-LacI/ΔZcen cells grew well, these localization data strongly suggest that CENP-A and most CCAN proteins are dispensable for kinetochore function if CENP-T N terminus or CENP-C N terminus is constitutively targeted to chromosomes using an independent means.
Figure 4.
The CENP-T and CENP-C N terminus–derived kinetochores are functional. (A) Mitotic progression of CT1–530-LacI/ΔZcen, CC1–643-LacI/ΔZcen, or wild-type DT40 cells observed by live-cell imaging under a microscope in the absence of IPTG. Selected images are shown. (B) Numbers of anaphase cells with normal or abnormal segregation of chromosome Z in CT1–530-LacI/ΔZcen or CC1–643-LacI/ΔZcen cells (in the absence of IPTG). It is rare to detect cells with abnormal segregation of chromosome Z in the absence of IPTG. GFP-LacI/Zp-ter LacO cells were used as a control. This measurement was completed once (GFP-LacI/Zp-ter LacO, n = 139; CT1–530-LacI/ΔZcen, n = 122; CC1–643-LacI/ΔZcen, n = 229). (C) Localization of CCAN (top), outer kinetochore (middle), and checkpoint proteins (bottom) at the LacO locus in CT1–530-LacI/ΔZcen cells. Each tested protein is shown in red and the LacI fusion with CENP-T (1–530) is shown in green. All tested CCAN proteins were not detected at the LacO locus, but outer kinetochore and checkpoint proteins were detected. Data were also summarized in F. (D) Localization of CCAN (top), outer kinetochore (middle), and checkpoint proteins (bottom) at the LacO locus in CC1–643-LacI/ΔZcen cells. Each tested protein is shown in red and the LacI fusion with CENP-C (1–643) is shown in green. Arrows indicate the position of the LacO sequence. Z, Z chromosome. (E) Relative signal intensities of Ndc80 or Mis12 at the LacO locus compared with those at endogenous kinetochores in CT1–530-LacI/ΔZcen or CC1–643-LacI/ΔZcen cells. Error bars indicate mean ± SD. (F) Summary of the localization results of various kinetochore proteins at the LacO locus in CT1–530-LacI/ΔZcen or CC1–643-LacI/ΔZcen cells. +, positive localization at the LacO locus; −, negative localization at the LacO locus.
We next tested whether the microtubule-binding kinetochore proteins are recruited to CENP-T N terminus– or CENP-C N terminus–derived kinetochores in CT1–530-LacI/ΔZcen or CC1–643-LacI/ΔZcen cells. We detected Ndc80 and KNL1 at ectopic kinetochores formed by the CENP-T N terminus or CENP-C N terminus (Fig. 4, C and D). In addition, most checkpoint proteins were loaded at these ectopic kinetochores when CT1–530-LacI/ΔZcen or CC1–643-LacI/ΔZcen cells were treated with the microtubule-depolymerizing drug nocodazole (Fig. 4, C and D). Although outer kinetochore proteins were detected at both CENP-T N terminus– and CENP-C N terminus–derived kinetochores, the localization of these proteins based on their relative signal intensities were different between CENP-T N terminus– and CENP-C N terminus–derived kinetochores (Fig. 4, E and F). Ndc80 localization to CENP-T N terminus–derived kinetochores was stronger than to CENP-C N terminus kinetochores. In contrast, Mis12 localization at CENP-T N terminus kinetochores was weaker than that observed at CENP-C N terminus kinetochores. These differences are consistent with the distinct localization requirements observed in CENP-C– or CENP-T–deficient cells (Kwon et al., 2007; Cheeseman et al., 2008; Hori et al., 2008; Gascoigne et al., 2011), and with the stoichiometries that we have previously observed after transient expression of the CENP-T and CENP-C N termini to ectopic loci (Gascoigne et al., 2011). Overall, all tested inner kinetochore (CCAN) proteins failed to localize to CENP-T N terminus– and CENP-C N terminus–derived ectopic kinetochores, whereas the tested outer kinetochore proteins were effectively targeted, indicating that the CENP-T or CENP-C N termini bypass the function of the CCAN proteins to recruit outer kinetochore proteins and form a functional kinetochore.
Creation of the CENP-T–derived kinetochores on plasmid DNA
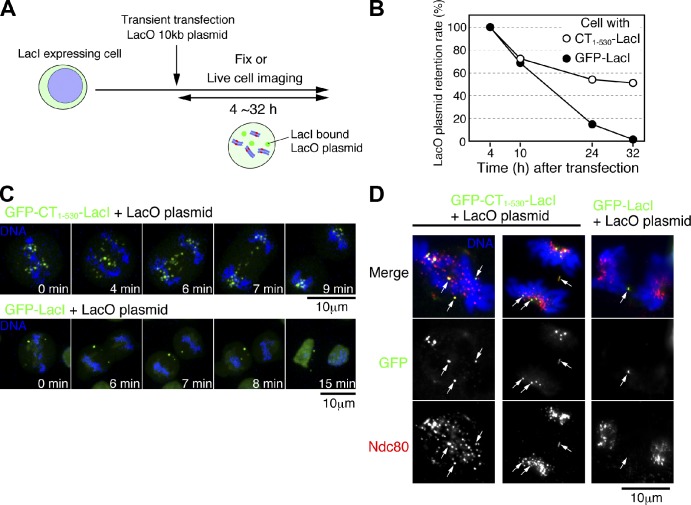
In the previous section, we generated CENP-T or CENP-C N terminus–derived kinetochores on chromosome Z, which bypass the requirement for most CCAN components and recruit outer kinetochore components to create a functional kinetochore-like structure. Therefore, targeting of CENP-T to any unit of DNA should similarly generate a kinetochore-like structure on that DNA. To test this, we transiently introduced a LacO-containing plasmid DNA into cells expressing LacI-fused CENP-T (1–530) (Fig. 5 A). This plasmid was retained in only 15.1% of control cells expressing GFP-LacI 24 h after transfection and was completely lost by 32 h. In contrast, the plasmid was maintained in 51.1% of CENP-T (1–530)–LacI expressing cells at 32 h (Fig. 5 B). Live-cell imaging analysis demonstrated that many plasmids were distributed to daughter cells during chromosome segregation in CENP-T–LacI–expressing cells, although the plasmid was easily lost in GFP-LacI–expressing cells (Fig. 5 C). Importantly, we found that Ndc80 localized to the LacO-containing plasmid in cells expressing CENP-T–LacI (Fig. 5 D). We conclude that the CENP-T N terminus is sufficient to recruit the Ndc80 complex to generate the kinetochore-like structure on plasmid DNA.
Figure 5.
The kinetochore-like structure is generated on the LacO-containing plasmid DNA in cells expressing LacI fusion with CENP-T N terminus. (A) The LacO-containing plasmids were transiently introduced into cells expressing LacI fusion with CENP-T N terminus (1–530). After introduction of the plasmids, cells were characterized at each time point. (B) Percentage of cells retaining the LacO plasmids at each time point after introduction of the plasmids into cells expressing LacI fusion with CENP-T N terminus (1–530). The plasmids were also introduced into cells expressing GFP-LacI fusion. The plasmids were retained in 51.1% of cells expressing LacI fusion with CENP-T N terminus (1–530) at 32 h after transfection, whereas all plasmids were lost in all cells expressing GFP-LacI fusion at 32 h after transfection. This experiment was completed once (cell with CT1–530, n > 340; cell with GFP-LacI, n > 160). (C) Behavior of the LacO-containing plasmids (green) during mitosis of cell expressing LacI fusion with CENP-T N terminus (1–530) (top) or GFP-LacI fusion (bottom). Cells were observed by live-cell imaging under a microscope. Selected images are shown. Many plasmids were segregated into daughter cells expressing LacI fusion with CENP-T N terminus (1–530) (see 9 min of top panel), whereas many plasmids were lost in cells expressing GFP-LacI fusion. (D) Localization of Ndc80 on the LacO-containing plasmids. Colocalization of CENP-T–LacI with Ndc80 was observed (left and middle), whereas colocalization of GFP-LacI with Ndc80 was not observed (right).
The CPC is recruited to the inner centromere at ectopic CENP-T– or CENP-C–derived kinetochores
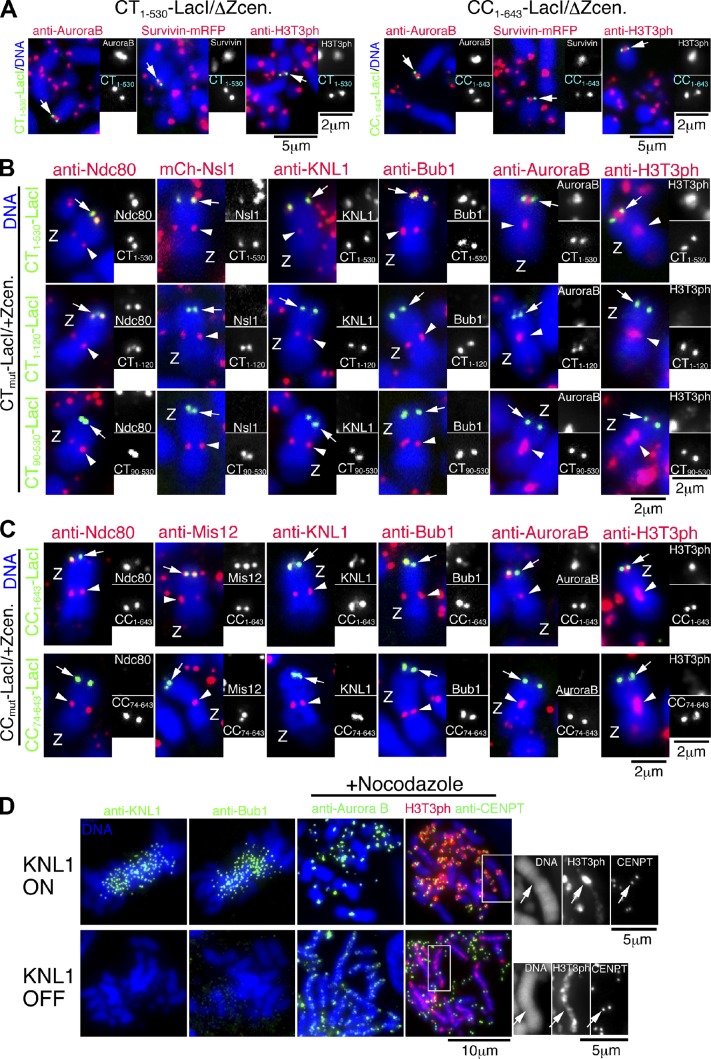
In addition to outer kinetochore components, kinetochores require the function of inner centromere proteins that contribute to the spindle checkpoint and sister chromatid cohesion (Ruchaud et al., 2007; Carmena et al., 2009). Threonine 3 of histone H3 is phosphorylated at the inner centromere coupled with the CPC recruitment (Kelly et al., 2010; Wang et al., 2010; Yamagishi et al., 2010). Therefore, we tested whether the CPC and T3 phosphorylation of histone H3 were present at the inner centromere region of ectopic induced kinetochores. Indeed, we detected the CPC proteins Aurora B and Survivin and phosphorylated histone H3 between sister ectopic kinetochores induced by either the CENP-T or CENP-C N terminus at the LacO locus (Fig. 6 A), which indicates that inner centromere proteins are recruited to these ectopic kinetochores. In addition, we observed that the distance between sister kinetochores induced by the CENP-T or CENP-C N terminus on the LacO locus was increased, but that sisters were not completely separated by the presence of tension from spindle microtubules (Fig. S5, A–C), which suggests that sister chromatid cohesion was established and protected at these ectopic kinetochore regions.
Figure 6.
The CPC and KNL1 are recruited into the CENP-T or CENP-C N terminus–derived kinetochores. (A) Localization of Aurora B, Survivin, and phosphorylated T3 of histone H3 (H3T3ph) at the LacO locus in CT1–530-LacI/ΔZcen or CC1–643-LacI/ΔZcen cells. Each protein is shown in red and CENP-T–LacI or CENP-C–LacI signals are shown in green. Aurora B, Survivin, and H3T3ph are detected in the inner centromere between sister kinetochores induced by both CENP-T and CENP-C N termini at the LacO locus. An antibody against H3T3ph was provided by H. Kimura (Osaka University, Osaka, Japan). (B) Localization of Ndc80, Nsl1, KNL1, Bub1, Aurora B, and H3T3ph at the LacO locus in cells expressing LacI fusion with CENP-T (1–530) (top), CENP-T (1–120) (middle), or CENP-T (90–530) (bottom). Each tested protein is shown in red. CENP-T derivatives are shown in green at the LacO locus. Data were summarized in Fig. S5 E. Z, Z chromosome. (C) Localization of Ndc80, Mis12, KNL1, Bub1, Aurora B, and H3T3ph at the LacO locus in cells expressing LacI fusion with CENP-C (1–643) (top) or CENP-C (74–643) (bottom). Each tested protein is shown in red. CENP-C derivatives are shown in green at the LacO locus. Arrows indicate the position of the LacO sequence. Data are summarized in Fig. S5 F. (D) Localization of Bub1, Aurora B, or H3T3ph in KNL1-deficient DT40 cells (bottom, KNL1 OFF). Top panels show localization of Bub1, Aurora B, or H3T3ph in DT40 cells expressing KNL1 (KNL1 ON). Bub1 localization was abolished in KNL1 OFF cells (bottom). Whereas Aurora B and H3T3ph signals were concentrated on inner centromere in KNL1 ON cells, these signals were diffused along chromosome axis in KNL1 OFF cells. Arrows indicate the position of the centromere.
The Ndc80 complex is necessary but not sufficient for functional kinetochore formation
Ectopic targeting of the CENP-T N terminus or CENP-C N terminus is sufficient to induce the recruitment of outer kinetochore and inner centromere proteins to form a functional kinetochore (Figs. 4 and 6 A). We next sought to determine which region of CENP-T or CENP-C is responsible for directing kinetochore assembly. We have previously shown that the N-terminal 100 aa of CENP-T is necessary for the localization of the Ndc80 complex to kinetochores (Gascoigne et al., 2011). Consistent with this, we observed that Ndc80 was recruited to the LacO locus in cells expressing a LacI fusion to the extreme CENP-T N terminus (1–120), whereas Ndc80 was not detected on the LacO locus in cells expressing an N-terminal deletion of CENP-T (90–530)–LacI (Fig. 6 B). The inability to recruit Ndc80 to the LacO locus in the CENP-T (90–530)–LacI fusion prevented kinetochore formation (Fig. S5 D). Although Ndc80 was recruited to the LacO locus in cells expressing the CENP-T–LacI N terminus (1–120), functional kinetochore formation was not induced in these cells after removal of endogenous centromere (Fig. S5 D). Similarly, recruitment of the Ndc80 complex by targeting Spc24 to the LacO locus did not induce kinetochore formation (Figs. 1 C and S1 C). These results indicate that the Ndc80 complex is necessary but not sufficient for functional kinetochore formation. Consistent with this, cells expressing a LacI fusion to the extreme CENP-T N terminus (1–120) lacked detectable KNL1, Bub1, or the CPC at the LacO locus (Fig. 6 B, middle). In addition, deletion of the CENP-T N terminus caused mislocalization of KNL1, Bub1, and CPC (Fig. 6 B, bottom; and Fig. S5 E). We also focused on the CENP-C N terminus, which is responsible for binding of the Mis12 complex (Gascoigne et al., 2011; Przewloka et al., 2011; Screpanti et al., 2011). Although the CENP-C N terminus (1–643 aa) induced assembly of IPTG-sensitive kinetochores, an N-terminal deletion of this region (74–643 aa) did not induce kinetochore assembly (Fig. 3). Consistent with this, we did not detect KNL1, Bub1, or the CPC at the LacO locus in cells expressing the CENP-C–LacI N terminus deletion (74–643) (Fig. 6 C, bottom; and Fig. S5 F).
The localization of the CPC to centromeres is mediated by Bub1 (Yamagishi et al., 2010), and Bub1 associates with KNL1 (Kiyomitsu et al., 2007). Thus, we hypothesized that KNL1 recruitment is essential for functional kinetochore formation for the CENP-T or CENP-C N termini–derived kinetochores. Consistent with this, we found that the centromere localization of Bub1 and the CPC were abolished in KNL1-deficient DT40 cells (Fig. 6 D).
Theses results demonstrate that the N-terminal region of CENP-T (1–89 aa) is required for recruiting the Ndc80 complex and that both the CENP-T N terminus and the middle region of CENP-T (121–530) are required for the localization of KNL1, Bub1, and CPC to centromeres. The CENP-C N terminus (1–73) is also essential to recruit KNL1 through an interaction with the Mis12 complex. In total, we conclude that the presence of both the Ndc80 complex at outer kinetochores and the CPC recruited by the KNL1–Bub1 pathway to inner centromeres is necessary to form a functional kinetochore in the CENP-T or CENP-C N terminus–derived kinetochores.
Discussion
In vertebrates, the kinetochore is specified on centromeric chromatin by sequence-independent epigenetic mechanisms. Recent work suggests that CENP-A is an important epigenetic marker for the kinetochore specification (Black and Cleveland, 2011; Perpelescu and Fukagawa, 2011). In support of this idea, several groups demonstrated that ectopic localization of CENP-A to noncentromere loci induces a kinetochore-like structure in vertebrate or Drosophila cells (Barnhart et al., 2011; Guse et al., 2011; Mendiburo et al., 2011). This suggests that CENP-A functions as a mark for kinetochore assembly. However, it was unclear how CENP-A is targeted and maintained at centromeric chromatin and how functional kinetochores are formed after the specification of this site by CENP-A. Here, we developed a unique experimental system to directly test the functional role of kinetochore proteins during kinetochore assembly. Our work demonstrates that the CCAN has two distinct roles: recruiting CENP-A to establish a kinetochore and providing a structural core to recruit outer kinetochore and inner centromere proteins (Fig. 7).
Figure 7.
Model of kinetochore assembly in the engineered kinetochores. (A) Molecular architecture of the CENP-T or CENP-C N terminus–derived kinetochores. CENP-A and CCAN proteins are not detected in both the CENP-T or CENP-C N terminus–derived kinetochores. Function of these kinetochores is sensitive to IPTG addition. The CENP-T N terminus directly binds to the Ndc80 complex and the CENP-C N terminus binds to the Mis12 complex, which associates with the Ndc80 complex. Although both kinetochores do not contain CENP-A and CCAN, the CPC is recruited. If CENP-T or CENP-C N terminus were steadily supplied to the LacO locus, functional kinetochores would be formed. This indicates that CENP-T or CENP-C N terminus is sufficient for formation of functional kinetochores in the absence of CENP-A and most CCAN proteins. (B) Kinetochore formation at the LacO locus induced by LacI fusion with HJURP, CENP-I, full-length CENP-C, or CENP-C C terminus (601–864). These kinetochores are resistant to addition of IPTG, and full kinetochore components are recruited. HJURP is known as a CENP-A–specific chaperone. CENP-I and CENP-C C terminus function as a mark for CENP-A incorporation. Once a full kinetochore is formed at the LacO locus, Lac-I fusions with HJURP, CENP-I, or CENP-C proteins are dispensable. Characterization of this type of kinetochore suggests that a major function of CENP-C or CENP-I is recruitment of CENP-A.
The CCAN functions to recruit CENP-A
The ectopic localization of HJURP, CENP-C, or CENP-I LacI fusion proteins induced kinetochore formation at the noncentromeric LacO locus. However, in these cells, once a kinetochore was formed, the LacI fusion protein became dispensable for kinetochore function. This suggests that a major function of these proteins is to mark centromere position and recruit CENP-A (Fig. 7 B). We and others have previously demonstrated that the localization of CENP-C and CENP-I occurs downstream of CENP-A at endogenous kinetochores (Nishihashi et al., 2002; Goshima et al., 2003; Régnier et al., 2005; Liu et al., 2006; Okada et al., 2006), which suggests that CENP-C or CENP-I recognizes and is targeted to the CENP-A containing chromatin. We also have demonstrated that deposition of newly synthesized CENP-A at centromeres depends on CENP-H/I–associated proteins (Okada et al., 2006). Considering these previous results together with the experiments presented here, we propose the following model. CENP-A is initially deposited at centromeres through the CENP-A–specific chaperone HJURP, and the CCAN is subsequently targeted to CENP-A–containing chromatin. After mitosis, additional CENP-A must be supplied to the centromeres (Jansen et al., 2007). At this stage, newly synthesized CENP-A is targeted into centromeres using the CCAN as a mark (Fig. 7 B). Once CENP-A is recruited to this site, the pathways that act to recruit additional outer kinetochore proteins act to assemble a full functional kinetochore at this locus.
The CCAN serves a structural platform for outer kinetochore assembly
In contrast to kinetochores induced by full-length HJURP, CENP-C, or CENP-I, kinetochores induced by the CENP-T or CENP-C N terminus require the LacI fusion protein for both the establishment and maintenance of these synthetic kinetochores (Fig. 7 A). This is because CENP-A is not recruited to the CENP-T or CENP-C N termini–derived kinetochores and thus lacks the ability to epigenetically propagate this structure in the absence of the DNA-binding LacI protein. Instead, the LacO sequences are essential for the LacI fusion protein with the CENP-T N terminus or CENP-C N terminus proteins to generate a kinetochore structure. Even if CENP-A is not targeted to these sites, chromosome segregation in cells with a CENP-T or CENP-C N terminus–derived kinetochore is normal, indicating that these fusions are sufficient to serve as a structural platform for outer kinetochore assembly. Most CCAN proteins were absent from these kinetochores, but are required at endogenous kinetochores. We propose that the CCAN functions in two important ways: (1) The CCAN recruits CENP-A to mark centromere position; and (2) the CCAN functions to target and maintain CENP-T and CENP-C at kinetochores. CENP-T and CENP-C in turn function as a structural platform for outer kinetochore assembly.
CENP-C has dual roles for kinetochore formation
Our molecular dissection experiments revealed that the CENP-C C terminus functions to recruit CENP-A, whereas the CENP-C N terminus serves as a structural platform for outer kinetochore assembly through its interaction with the Mis12 complex. CENP-C is a conserved inner kinetochore protein, which shows sequence similarity with budding yeast Mif2 (Saitoh et al., 1992; Brown, 1995). The Mif2 homology region is restricted to the C-terminal region of CENP-C, which possesses the CENP-A recruiting activity in our assay, suggesting that this activity may be conserved. Recent work has proposed that the conserved C-terminal region of CENP-C binds to M18BP1/KNL2 (Moree et al., 2011; Dambacher et al., 2012). Therefore, it is possible that the CENP-C C terminus recruits CENP-A through its interaction with M18BP1/KNL2. We observed that the CENP-C C terminus (601–864) could localize endogenous kinetochores (unpublished data), which suggests that this domain may be critical for kinetochore specification. Carroll et al. (2010) demonstrated that the middle region of human CENP-C (426–537 aa) directly interacts with the CENP-A nucleosome. We did not observe centromere formation at the LacO locus induced by localization of chicken CENP-C (74–643 aa), which contains the potential CENP-A–binding region, suggesting that the direct binding of CENP-A nucleosome for CENP-C is not sufficient to recruit CENP-A. CENP-C forms a dimer (Cohen et al., 2008) and CENP-C (74–643 aa) is likely to lack dimerization activity. Dimerization may be important for the recruitment of CENP-A by CENP-C. Consistent with this, CENP-C (601–761 aa), which lacks the dimerization domain, did not induce kinetochore formation at the LacO locus, whereas CENP-C (601–864 aa), which has the CENP-A recruiting activity, forms a dimer.
The CENP-C N terminus serves as a structural platform for outer kinetochore assembly. Although the extreme 70-aa region of the CENP-C N terminus is essential for the Mis12 binding and is required for kinetochore assembly, CENP-C (1–165) did not induce outer kinetochore assembly in our assay. However, as we observed formation of IPTG-sensitive kinetochores by expression of LacI fusions with CENP-C (1–324) (unpublished data), multiple regions within the N-terminal region (1–324 aa) of CENP-C may be required for recruitment of Mis12 complex and outer kinetochore assembly.
In total, the CENP-C C-terminal 200-aa region including the conserved dimerization domain is essential for CENP-A recruitment, and the N-terminal region of CENP-C is essential for outer kinetochore assembly.
Both the Ndc80 complex and KNL1 are necessary to form a functional kinetochore
An important aspect of kinetochore function is the generation of an interface with microtubules from the mitotic spindle. As the Ndc80 complex localizes to outer kinetochores and binds to microtubules directly, localization of the Ndc80 complex to kinetochores is an essential step in the formation of a functional kinetochore. The CENP-T or CENP-C N-terminus either directly or indirectly recruits the Ndc80 complex to the LacO locus, which is necessary for functional kinetochore formation. However, the Ndc80 complex is not sufficient for kinetochore formation, as ectopic localization of the Ndc80 complex (using Spc24 or CENP-T[1–120] LacI fusions) did not induce kinetochore formation (Figs. 1 and 6). We propose that, in addition to recruiting the Ndc80 complex to kinetochores, recruitment of the CPC is also essential to form a functional kinetochore and that the CENP-T or CENP-C N terminus is responsible for CPC recruitment through the KNL1–Bub1 pathway (Fig. 6). We demonstrated that the middle region of CENP-T is required for the localization of KNL1 and Bub1. The CENP-C N terminus also recruits KNL1 to kinetochores through the Mis12 complex–KNL1 interaction, which facilitates CPC recruitment (Fig. 6).
In addition to recruiting the CPC to centromeres, the middle region of CENP-T contributes to kinetochore stretching (Suzuki et al., 2011). CENP-T (1–120)–LacI recruits the Ndc80 complex to the LacO locus, but does not induce kinetochore formation (Fig. 6 B). This may be caused by defects in kinetochore flexibility or the way in which Ndc80 is positioned with respect to the DNA. Conformational changes of CENP-T governed by the CENP-T middle region may create an organization and dynamic rearrangement of the microtubule interface that is necessary for the correct attachment of kinetochores to microtubules. We conclude that multifunctionality of CENP-T and CENP-C allows the creation of artificial kinetochores at noncentromeric regions.
Toward the creation of a stable artificial chromosome
In yeast, ectopic localization of the microtubule-binding Dam1 complex to a noncentromeric region induced formation of functional synthetic kinetochores on a plasmid-based minichromosome (Kiermaier et al., 2009; Lacefield et al., 2009). In vertebrate cells, it has been complicated to use plasmid systems because plasmid DNA is not correctly replicated or stably maintained. Therefore, we have developed a unique chromosome-engineering system and demonstrated that creation of an artificial kinetochore can maintain an artificial chromosome. “First-generation” artificial chromosomes were created using α-satellite DNA, which spans the human centromere region (Harrington et al., 1997; Ikeno et al., 1998). However, establishing stable cell lines with these artificial chromosomes occurs with very low efficiency, and the structure of these artificial chromosomes is typically rearranged after introduction of the chromosome vector into cells. In addition, the α-satellite DNA-derived artificial chromosomes cannot be easily controlled and manipulated because it is unclear how kinetochore proteins are assembled on the long α-satellite arrays. Artificial kinetochores generated using targeting of kinetochore proteins in cells provide a much more applicable approach. Indeed, we demonstrated that ectopic targeting can generate CENP-T–derived kinetochore-like structures on plasmid DNA (Fig. 5). Our experimental approach has provided important insights into the basic mechanisms of kinetochore assembly and will also be useful for the creation of artificial chromosomes in vertebrate cells for therapeutic or industrial purposes.
Materials and methods
Deletion of centromere DNA and ectopic targeting of LacI fusion proteins
Target constructs containing LoxP sequence or LacO sequence and several LacI fusion constructs were created by a standard method. LacO and LacI plasmids were provided by A. Belmont (University of Illinois at Urbana-Champaign, Champaign, IL). DNA constructs were transfected with an electroporator (Gene Pulser II; Bio-Rad Laboratories) into DT40 cells. All DT40 cells were cultured at 38°C in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum, 1% chicken serum, 2-mercaptoethanol, penicillin, and streptomycin. To activate the Cre-recombinase (Mer-Cre-Mer), 4-hydroxytamoxifen (OHT; Sigma-Aldrich) was added to the culture medium to a final concentration of 100 nM (Shang et al., 2010). A Mer-Cre-Mer plasmid was provided by M. Reth (Max Planck Institute of Immunobiology and Epigenetics, Freiburg, Germany). KNL1 conditional knockout cell lines were previously created (Cheeseman et al., 2008). In these cells, exon 8 and 9 of KNL1 were disrupted and expression of KNL1 was sustained by KNL1 cDNA under the tetracycline-responsive promoter. If tetracycline were added to these cells, KNL1 expression was suppressed.
FISH and immunofluorescence
DT40 metaphase spreads were prepared by the cytospin method and fixed in 4% paraformaldehyde for 15 min at room temperature. After a brief treatment with 0.5% NP-40 in PBS, glass slides were dehydrated in ethanol. Chromosome DNAs were heat-denatured at 95°C for 5 min. DNA probes were labeled with Biotin-16-dUTP (Roche) by nick translation procedure. Probe DNAs were denatured at 75°C for 10 min, and hybridization was performed at 37°C in a humidity chamber overnight. Slides were washed and Z chromosome detected by Z-specific macrosatellite DNA (Hori et al., 1996) was used as a probe. Immunofluorescent staining of DT40 cells was performed a using anti–CENP-A (a rabbit antibody against a synthetic peptide for chicken CENP-A 2–15 aa), anti–CENP-C (a rabbit antibody against recombinant chicken CENP-C 1–330 aa), anti–CENP-O (a rabbit antibody against recombinant chicken full-length CENP-O), anti–CENP-T (a rabbit antibody against recombinant chicken full-length CENP-T), anti-Mis12 (a rabbit antibody against recombinant chicken full-length Mis12), anti-Ndc80 (a rabbit antibody against recombinant chicken Ndc80 465–640 aa), anti-KNL1 (a rabbit antibody against recombinant chicken KNL1 1,242–1,667 aa), anti-Bub1 (a rabbit antibody against recombinant chicken Bub1 903–1,087 aa), anti-BubR1 (a rabbit antibody against chicken recombinant full-length BubR1), anti-Mad2 (a rabbit antibody against recombinant chicken full-lengthMad2), or anti–Aurora B antibodies (a rabbit antibody against recombinant chicken full-length Aurora B). Some of these antibodies were used in previous studies (Kline et al., 2006; Okada et al., 2006; Hori et al., 2008). Several GFP or mCherry fusion constructs were also used to examine protein localization. Immunofluorescence and FISH images were collected with a cooled charge-coupled device camera (Cool Snap HQ; Roper Scientific) mounted on an inverted microscope (IX71; Olympus) with a 100× objective lens together with a filter wheel. We also used the high-speed spinning disc confocal system (Yokogawa) to collect images. Z-sections (n = 15–25) were acquired at 0.3-µm steps. All subsequent analysis and processing of images were performed using MetaMorph software (Molecular Devices).
ChIP
For immunoprecipitation of DT40 cells with anti–CENP-A antibodies, a nuclear fraction of cells was collected and digested with excess MNase (60 U/ml; Takara Bio Inc.) at 37°C for 20 min. The samples were solubilized, incubated with protein G Sepharose bead (GE Healthcare)–conjugated anti–CENP-A antibodies for 2–4 h at 4°C, and washed 4 times with 1 ml of buffer B (20 mM Tris-HCl, pH 8.0, 5 mM EDTA, 500 mM NaCl, and 0.2% Tween 20). DNA was extracted from immunoprecipitates and Southern hybridization was performed using a LacO sequence or centromere sequence from chromosome 5 as a probe.
Live-cell imaging
Cells were stained with Hoechst 33342 for 10–15 min at a final concentration of 100 ng/ml and washed with culture medium. Fluorescently stained cells expressing GFP-fused CENP-T (1–530)– or CENP-C (1–643)–LacI were observed with a confocal scanner box (Cell Voyager CV1000; Yokogawa) with an oil immersion objective lens (Plan-Apochromat 60×, NA 1.40). Time-lapse images of living cells were recorded at 5-min intervals with an exposure time of 0.2 s. Z sections (n = 15–25) for GFP signals were acquired at 0.3-µm steps for each time point. Hoechst 33342 images were collected with a single image at each time point to avoid cell damage.
Online supplemental material
Fig. S1 shows characterization of the CENP-T–derived kinetochores. Fig. S2 demonstrates characterization of both ITPG-sensitive and -insensitive kinetochores. Fig. S3 shows cell-cycle analysis for the CENP-C N terminus–derived kinetochores. Fig. S4 shows abnormal mitotic progression of CT1–530-LacI/ΔZcen or CC1–643-LacI/ΔZcen cells in the presence of IPTG. Fig. S5 demonstrates that centromere cohesion is normal in the CENP-T or CENP-C N terminus–derived kinetochores. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201210106/DC1.
Supplementary Material
Acknowledgments
The authors are very grateful to K. Suzuki, K. Nakaguchi, M. Takahashi, and K. Kita for technical assistance. We also thank I.M. Cheeseman for useful comments and for critical reading of the manuscript, M. Reth for providing us a Mer-Cre-Mer plasmid, and A. Belmont for providing us LacO and LacI plasmids.
This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan and the Cabinet Office of the Government of Japan through its Funding Program for Next Generation World-Leading Researchers to T. Fukagawa. T. Hori is supported by Precursory Research for Embryonic Science and Technology of the Japan Science and Technology Agency and by Grants-in-Aid for Scientific Research from MEXT.
Footnotes
Abbreviations used in this paper:
- CCAN
- constitutive centromere-associated network
- ChIP
- chromatin immunoprecipitation
- CPC
- chromosome passenger complex
References
- Alushin G.M., Ramey V.H., Pasqualato S., Ball D.A., Grigorieff N., Musacchio A., Nogales E. 2010. The Ndc80 kinetochore complex forms oligomeric arrays along microtubules. Nature. 467:805–810 10.1038/nature09423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart M.C., Kuich P.H., Stellfox M.E., Ward J.A., Bassett E.A., Black B.E., Foltz D.R. 2011. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J. Cell Biol. 194:229–243 10.1083/jcb.201012017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black B.E., Cleveland D.W. 2011. Epigenetic centromere propagation and the nature of CENP-a nucleosomes. Cell. 144:471–479 10.1016/j.cell.2011.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M.T. 1995. Sequence similarities between the yeast chromosome segregation protein Mif2 and the mammalian centromere protein CENP-C. Gene. 160:111–116 10.1016/0378-1119(95)00163-Z [DOI] [PubMed] [Google Scholar]
- Burrack L.S., Berman J. 2012. Flexibility of centromere and kinetochore structures. Trends Genet. 28:204–212 10.1016/j.tig.2012.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena M., Ruchaud S., Earnshaw W.C. 2009. Making the Auroras glow: regulation of Aurora A and B kinase function by interacting proteins. Curr. Opin. Cell Biol. 21:796–805 10.1016/j.ceb.2009.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll C.W., Milks K.J., Straight A.F. 2010. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J. Cell Biol. 189:1143–1155 10.1083/jcb.201001013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., Chappie J.S., Wilson-Kubalek E.M., Desai A. 2006. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 127:983–997 10.1016/j.cell.2006.09.039 [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Hori T., Fukagawa T., Desai A. 2008. KNL1 and the CENP-H/I/K complex coordinately direct kinetochore assembly in vertebrates. Mol. Biol. Cell. 19:587–594 10.1091/mbc.E07-10-1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen R.L., Espelin C.W., De Wulf P., Sorger P.K., Harrison S.C., Simons K.T. 2008. Structural and functional dissection of Mif2p, a conserved DNA-binding kinetochore protein. Mol. Biol. Cell. 19:4480–4491 10.1091/mbc.E08-03-0297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dambacher S., Deng W., Hahn M., Sadic D., Fröhlich J., Nuber A., Hoischen C., Diekmann S., Leonhardt H., Schotta G. 2012. CENP-C facilitates the recruitment of M18BP1 to centromeric chromatin. Nucleus. 3:101–110 10.4161/nucl.18955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca J.G., Musacchio A. 2012. Structural organization of the kinetochore-microtubule interface. Curr. Opin. Cell Biol. 24:48–56 10.1016/j.ceb.2011.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca J.G., Gall W.E., Ciferri C., Cimini D., Musacchio A., Salmon E.D. 2006. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 127:969–982 10.1016/j.cell.2006.09.047 [DOI] [PubMed] [Google Scholar]
- Dunleavy E.M., Roche D., Tagami H., Lacoste N., Ray-Gallet D., Nakamura Y., Daigo Y., Nakatani Y., Almouzni-Pettinotti G. 2009. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 137:485–497 10.1016/j.cell.2009.02.040 [DOI] [PubMed] [Google Scholar]
- Foltz D.R., Jansen L.E., Bailey A.O., Yates J.R., III, Bassett E.A., Wood S., Black B.E., Cleveland D.W. 2009. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 137:472–484 10.1016/j.cell.2009.02.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Hayashi T., Kiyomitsu T., Toyoda Y., Kokubu A., Obuse C., Yanagida M. 2007. Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev. Cell. 12:17–30 10.1016/j.devcel.2006.11.002 [DOI] [PubMed] [Google Scholar]
- Gascoigne K.E., Takeuchi K., Suzuki A., Hori T., Fukagawa T., Cheeseman I.M. 2011. Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell. 145:410–422 10.1016/j.cell.2011.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Kiyomitsu T., Yoda K., Yanagida M. 2003. Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway. J. Cell Biol. 160:25–39 10.1083/jcb.200210005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guse A., Carroll C.W., Moree B., Fuller C.J., Straight A.F. 2011. In vitro centromere and kinetochore assembly on defined chromatin templates. Nature. 477:354–358 10.1038/nature10379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington J.J., Van Bokkelen G., Mays R.W., Gustashaw K., Willard H.F. 1997. Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat. Genet. 15:345–355 10.1038/ng0497-345 [DOI] [PubMed] [Google Scholar]
- Hayashi T., Fujita Y., Iwasaki O., Adachi Y., Takahashi K., Yanagida M. 2004. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell. 118:715–729 10.1016/j.cell.2004.09.002 [DOI] [PubMed] [Google Scholar]
- Hori T., Suzuki Y., Solovei I., Saitoh Y., Hutchison N., Ikeda J.E., Macgregor H., Mizuno S. 1996. Characterization of DNA sequences constituting the terminal heterochromatin of the chicken Z chromosome. Chromosome Res. 4:411–426 10.1007/BF02265048 [DOI] [PubMed] [Google Scholar]
- Hori T., Amano M., Suzuki A., Backer C.B., Welburn J.P., Dong Y., McEwen B.F., Shang W.H., Suzuki E., Okawa K., et al. 2008. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell. 135:1039–1052 10.1016/j.cell.2008.10.019 [DOI] [PubMed] [Google Scholar]
- Ikeno M., Grimes B., Okazaki T., Nakano M., Saitoh K., Hoshino H., McGill N.I., Cooke H., Masumoto H. 1998. Construction of YAC-based mammalian artificial chromosomes. Nat. Biotechnol. 16:431–439 10.1038/nbt0598-431 [DOI] [PubMed] [Google Scholar]
- Ishii K., Ogiyama Y., Chikashige Y., Soejima S., Masuda F., Kakuma T., Hiraoka Y., Takahashi K. 2008. Heterochromatin integrity affects chromosome reorganization after centromere dysfunction. Science. 321:1088–1091 10.1126/science.1158699 [DOI] [PubMed] [Google Scholar]
- Jansen L.E., Black B.E., Foltz D.R., Cleveland D.W. 2007. Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 176:795–805 10.1083/jcb.200701066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly A.E., Ghenoiu C., Xue J.Z., Zierhut C., Kimura H., Funabiki H. 2010. Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science. 330:235–239 10.1126/science.1189505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiermaier E., Woehrer S., Peng Y., Mechtler K., Westermann S. 2009. A Dam1-based artificial kinetochore is sufficient to promote chromosome segregation in budding yeast. Nat. Cell Biol. 11:1109–1115 10.1038/ncb1924 [DOI] [PubMed] [Google Scholar]
- Kiyomitsu T., Obuse C., Yanagida M. 2007. Human Blinkin/AF15q14 is required for chromosome alignment and the mitotic checkpoint through direct interaction with Bub1 and BubR1. Dev. Cell. 13:663–676 10.1016/j.devcel.2007.09.005 [DOI] [PubMed] [Google Scholar]
- Kline S.L., Cheeseman I.M., Hori T., Fukagawa T., Desai A. 2006. The human Mis12 complex is required for kinetochore assembly and proper chromosome segregation. J. Cell Biol. 173:9–17 10.1083/jcb.200509158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon M.S., Hori T., Okada M., Fukagawa T. 2007. CENP-C is involved in chromosome segregation, mitotic checkpoint function, and kinetochore assembly. Mol. Biol. Cell. 18:2155–2168 10.1091/mbc.E07-01-0045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacefield S., Lau D.T., Murray A.W. 2009. Recruiting a microtubule-binding complex to DNA directs chromosome segregation in budding yeast. Nat. Cell Biol. 11:1116–1120 10.1038/ncb1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.T., Rattner J.B., Jablonski S.A., Yen T.J. 2006. Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J. Cell Biol. 175:41–53 10.1083/jcb.200606020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall O.J., Chueh A.C., Wong L.H., Choo K.H. 2008. Neocentromeres: new insights into centromere structure, disease development, and karyotype evolution. Am. J. Hum. Genet. 82:261–282 10.1016/j.ajhg.2007.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendiburo M.J., Padeken J., Fülöp S., Schepers A., Heun P. 2011. Drosophila CENH3 is sufficient for centromere formation. Science. 334:686–690 10.1126/science.1206880 [DOI] [PubMed] [Google Scholar]
- Moree B., Meyer C.B., Fuller C.J., Straight A.F. 2011. CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly. J. Cell Biol. 194:855–871 10.1083/jcb.201106079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihashi A., Haraguchi T., Hiraoka Y., Ikemura T., Regnier V., Dodson H., Earnshaw W.C., Fukagawa T. 2002. CENP-I is essential for centromere function in vertebrate cells. Dev. Cell. 2:463–476 10.1016/S1534-5807(02)00144-2 [DOI] [PubMed] [Google Scholar]
- Nishino T., Takeuchi K., Gascoigne K.E., Suzuki A., Hori T., Oyama T., Morikawa K., Cheeseman I.M., Fukagawa T. 2012. CENP-T-W-S-X forms a unique centromeric chromatin structure with a histone-like fold. Cell. 148:487–501 10.1016/j.cell.2011.11.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada M., Cheeseman I.M., Hori T., Okawa K., McLeod I.X., Yates J.R., III, Desai A., Fukagawa T. 2006. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat. Cell Biol. 8:446–457 10.1038/ncb1396 [DOI] [PubMed] [Google Scholar]
- Perpelescu M., Fukagawa T. 2011. The ABCs of CENPs. Chromosoma. 120:425–446 10.1007/s00412-011-0330-0 [DOI] [PubMed] [Google Scholar]
- Przewloka M.R., Venkei Z., Bolanos-Garcia V.M., Debski J., Dadlez M., Glover D.M. 2011. CENP-C is a structural platform for kinetochore assembly. Curr. Biol. 21:399–405 10.1016/j.cub.2011.02.005 [DOI] [PubMed] [Google Scholar]
- Régnier V., Vagnarelli P., Fukagawa T., Zerjal T., Burns E., Trouche D., Earnshaw W., Brown W. 2005. CENP-A is required for accurate chromosome segregation and sustained kinetochore association of BubR1. Mol. Cell. Biol. 25:3967–3981 10.1128/MCB.25.10.3967-3981.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruchaud S., Carmena M., Earnshaw W.C. 2007. Chromosomal passengers: conducting cell division. Nat. Rev. Mol. Cell Biol. 8:798–812 10.1038/nrm2257 [DOI] [PubMed] [Google Scholar]
- Saitoh H., Tomkiel J., Cooke C.A., Ratrie H., III, Maurer M., Rothfield N.F., Earnshaw W.C. 1992. CENP-C, an autoantigen in scleroderma, is a component of the human inner kinetochore plate. Cell. 70:115–125 10.1016/0092-8674(92)90538-N [DOI] [PubMed] [Google Scholar]
- Screpanti E., De Antoni A., Alushin G.M., Petrovic A., Melis T., Nogales E., Musacchio A. 2011. Direct binding of Cenp-C to the Mis12 complex joins the inner and outer kinetochore. Curr. Biol. 21:391–398 10.1016/j.cub.2010.12.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang W.H., Hori T., Toyoda A., Kato J., Popendorf K., Sakakibara Y., Fujiyama A., Fukagawa T. 2010. Chickens possess centromeres with both extended tandem repeats and short non-tandem-repetitive sequences. Genome Res. 20:1219–1228 10.1101/gr.106245.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A., Hori T., Nishino T., Usukura J., Miyagi A., Morikawa K., Fukagawa T. 2011. Spindle microtubules generate tension-dependent changes in the distribution of inner kinetochore proteins. J. Cell Biol. 193:125–140 10.1083/jcb.201012050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Dai J., Daum J.R., Niedzialkowska E., Banerjee B., Stukenberg P.T., Gorbsky G.J., Higgins J.M. 2010. Histone H3 Thr-3 phosphorylation by Haspin positions Aurora B at centromeres in mitosis. Science. 330:231–235 10.1126/science.1189435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi Y., Honda T., Tanno Y., Watanabe Y. 2010. Two histone marks establish the inner centromere and chromosome bi-orientation. Science. 330:239–243 10.1126/science.1194498 [DOI] [PubMed] [Google Scholar]
- Yang C.H., Tomkiel J., Saitoh H., Johnson D.H., Earnshaw W.C. 1996. Identification of overlapping DNA-binding and centromere-targeting domains in the human kinetochore protein CENP-C. Mol. Cell. Biol. 16:3576–3586 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.