Abstract
The presence of antibiotic-resistance (AR) genes in foodborne bacteria of enteric origin represents a relevant threat to human health in the case of opportunistic pathogens, which can reach the human gut through the food chain. Streptococcus bovis is a human opportunistic pathogen often associated with infections in immune-compromised or cancer patients, and it can also be detected in the environment, including fermented foods. We have focused on the molecular characterization of a tetracycline (Tet)-resistance gene present in 39 foodborne isolates of S. bovis phenotypically resistant to this drug. The gene was identified as a novel tet(S/M) fusion, encoding a mosaic protein composed of the N-terminal 33 amino acids of Tet(S), in-frame with the Tet(M) coding sequence. Heterologous expression of the mosaic gene was found to confer Tet resistance upon Escherichia coli recipients. Moreover, the tet(S/M) gene was found to be transcriptionally inducible by Tet under the endogenous tet(S) promoter in both S. bovis and E. coli. Nucleotide sequencing of the surrounding genomic region of 16.2 kb revealed large blocks of homology with the genomes of Streptococcus infantarius and Lactococcus lactis. A subregion of about 4 kb containing mosaic tet(S/M) was flanked by two copies of the IS1216 mobile element. PCR amplification with primers directed outwards from the tet(S/M) gene identified the presence of a 4.3 kb circular form corresponding to the intervening chromosomal region between the two IS1216 elements, but lacking a replication origin. The circular element shared extensive overall homology with a region of the multidrug-resistance plasmid pK214 from Lc. lactis, containing tet(S), as well as the IS1216 transposase-containing element and intervening non-coding sequences. Linear reconstruction of the insertion events likely to have occurred within this genomic region, inferred from sequence homology, provides further evidence of the chromosomal rearrangements that drive genomic evolution in complex bacterial communities such as the gut and food microbiota.
Introduction
Streptococcus bovis is an indigenous resident of the gastrointestinal (GI) tract in humans and animals. It is classified as a member of the group D streptococci, which includes the highly related species Streptococcus equinus, Streptococcus caprinus, Streptococcus gallolyticus, Streptococcus galactolyticus, Streptococcus infantarius, Streptococcus macedonius and Streptococcus waius (Herrera et al., 2009). Several Streptococcus species represent serious invasive pathogens, often associated with wound infections, sepsis, abscesses and dental caries in immunocompromised or cancer patients (Moet et al., 2007; Fernández-Ruiz et al., 2010; Al-Jashamy et al., 2010). The association of S. bovis with endocarditis (Gupta et al., 2010), colon cancer (Boleij et al., 2009) and colon adenoma (Kahveci et al., 2010) has also recently been reported. This species is therefore considered a potential pathogen.
The widespread use of antibiotics has applied strong selective pressure in the environment, favouring the survival and spread of antibiotic-resistant (AR) bacterial species. Such environmental selection is reflected in the increasing presence of AR commensal bacteria in the gut microbiota of livestock, which in turn leads to an increased frequency of AR species within the microbiota of fermented foods of animal origin, such as dairy and meat products. AR commensal bacteria do not by themselves represent a threat to human health, but their presence in the gut microbiota of humans and animals is increasingly viewed as a reservoir with the potential of being transmitted to pathogens through genomic exchange (Ammor et al., 2007). This is especially relevant for opportunistic pathogens, which are also capable of acquiring virulence genes and are reported to be important agents of AR nosocomial infection. The key role played by mobile elements in AR gene transfer was further emphasized by the publication of the first sequenced genome of a vancomycin-resistant clinical isolate of Enterococcus faecalis, revealing the presence of up to 38 insertion sequences (ISs) (Paulsen et al., 2003). Transposon-mediated inter-species transfer of AR genes has now been further characterized as one of the main mechanisms contributing to AR spread in bacteria (Wozniak & Waldor, 2010).
In the present work we report the molecular characterization of a novel functional mosaic tetracycline (Tet)-resistance gene that was identified in foodborne isolates of S. bovis. We also studied the genomic context surrounding the mosaic gene, whose sequence shares long stretches of homology with the genomes of other streptococcal species.
Methods
Bacterial strains and growth conditions.
S. bovis strains were cultured in de Man, Rogosa and Sharpe (MRS) medium. Growth conditions and antibiotic concentrations were as previously described (Devirgiliis et al., 2010).
Cloning procedures.
A 2603 bp DNA fragment containing full-length mosaic tet(S/M) and the 5′ and 3′ untranslated regions was amplified using the TetS/M-F-SalI and TetS/M-R-PstI primers (Table 1). The purified PCR product was digested with SalI and PstI and cloned into pBluescript KS+ (Stratagene), linearized with the same enzymes. The resulting construct was used to transform electro-competent Escherichia coli DH5α.
Table 1. Primers used in PCR amplifications.
Nucleotides corresponding to restriction site sequences are in italic type. Primers marked with ‘×’ in the Walking primer column were also used for gene walking.
| Primer | Sequence (5′–3′) | Target region | Annealing temperature (°C) | Walking primer | Source or reference |
| TetS/M-F-SalI | gggcccGTCGACTAGCCATTCTGAAGAGTTATCTTC | −379ATG tet(S) | 64 | This work | |
| TetS/M-R-PstI | gggcccCTGCAGCGAATTATCGGCTCTGCGTCTTTGC | Orf6 stop codon | 64 | This work | |
| STRINF2-REV | GGCATTCTCCAATATGTGGAGC | STRINF_00828 | 62 | × | This work |
| IS1068-FW | CGTCGTCCTTCAAAACAACAAGTGG | llkf_p0022 | 62 | × | This work |
| KUP-FW | CGTTTAATAAGGCCACGAGTGCAGG | llkf_p0020 | 62 | × | This work |
| TetS-1-FW | GGCTATCAGTGTGTAGTAGTATACAGC | tet(S) promoter | 65 | This work | |
| TetS-2-REV | GCTGTATACTACACACTGATAGCCACAGATGC | tet(S) promoter | 68 | × | This work |
| Rev-Ext | GCTTTCCTCTTGTTCGAGTTCCAATGC | tet(M) | 68 | × | Devirgiliis et al. (2009) |
| For-Ext | CATTCACATCGAAGTGCCGCCAAATCC | tet(M) | 68 | Devirgiliis et al. (2009) | |
| Tet-int-FW | CGGATAGATAAAGTACGATA | tet(M) | 54 | × | Devirgiliis et al. (2009) |
| Orf6-FW | GTGGATATTGTGTCCTGTATGTGG | Orf6 | 58 | × | This work |
| STRINF1-REV | GGAGTTTTGCTTCTCATACTTGGGG | STRINF_00831 | 62 | × | This work |
DNA extraction and molecular analysis.
Genomic DNA was extracted using the MagPrep Bacterial Genomic DNA kit (Merck), according to manufacturer’s instructions. PCR amplifications were performed as previously described (Devirgiliis et al., 2008), with the primers listed in Table 1 (Primm, Italy). PCR products were purified with the NucleoSpin Extract II purification kit (Macherey-Nagel), and sequenced by the M-Medical sequencing service (Italy). Southern hybridizations were carried out using standard protocols, with probes labelled with digoxigenin-11-dUTP (Roche Diagnostics). Restriction endonucleases were purchased from Promega.
RT-PCR.
Bacterial RNA was isolated using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions and subsequently treated with DNase I (RNase-free). One microgram of total RNA was reverse-transcribed with M-MLV reverse transcriptase (Invitrogen) in a final volume of 20 µl, using random hexamers as primers. Two microlitres of the reaction mix was then used as template in PCRs with primers amplifying either the 16S rDNA (control) or the tet(S/M) mosaic gene (primers TetS-1-FW and Rev-Ext; Table 1).
Gene walking.
The two-step gene walking method consisted of a walking-PCR (step 1) followed by direct sequencing of the PCR product (step 2), as described in (Pilhofer et al., 2007). Walking-PCRs were performed in a final volume of 50 µl containing 0.5 µM of the specific primer (Table 1), 2.5 U AccuTaq (Sigma), 1× AccuTaq buffer, 2.5 mM of each dNTP and 50 ng template DNA. The cycling program was the same as that reported in (Pilhofer et al., 2007), except for the annealing temperature of specific primers (see Table 1) and the AccuTaq extension temperature (68 °C). Using this method, overlapping DNA fragments ranging from 3 to 4 kb were obtained with walking primers directed outwards, upstream of tet(S/M) (Rev-Ext, TetS-2-REV, KUP-FW, IS1068-FW, STRINF2-REV in Table 1). The sequence of the region downstream of tet(S/M) was obtained following three rounds of walking with primers Tet-int-FW, Orf6-FW and STRINF1-REV (Table 1). Walking-PCR products were purified using a NucleoSpin Extract II kit according to the manufacturer’s instructions and were fully sequenced. Computer-assisted joining of the sequenced fragments to obtain the complete sequence was performed using the web resource CLC Sequence Viewer 6.0.2 (www.clcbio.com).
Results
Identification of a novel tet(S/M) mosaic gene
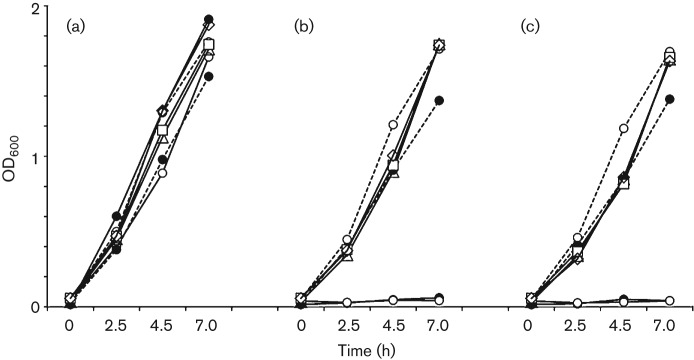
The Tet-resistance determinant in phenotypically resistant foodborne strains of S. bovis isolated in our laboratory was previously reported as tet(M) on the basis of PCR amplification with tet(M)-specific primers (Devirgiliis et al., 2010). To further characterize the resistance gene, three representative isolates belonging to different repetitive extragenic palindromic (rep) groups were analysed at the molecular level. Since tet(M) is most commonly found within mobile elements in Gram-positive bacteria (Flórez et al., 2008), the PCR strategy chosen used primers directed outwards from the tet(M) gene (Table 1, For-Ext and Rev-Ext). These primers could yield an amplicon only in the case of a circular template. This approach yielded a 3.7 kb amplicon (Fig. 1a), which was sequenced fully, revealing a novel Tet-resistance gene represented by a tet(S)–tet(M) fusion. The DNA sequence of the relevant portion of the mosaic tet gene is shown in Fig. 1(b) with the corresponding primary sequence of the protein, consisting of the amino-terminal 33 amino acids of Tet(S), fused to Tet(M) residues 28–639. To ensure that the mosaic gene was indeed responsible for conferring phenotypic resistance to Tet upon the S. bovis isolates, we first tested for the possible presence of other Tet-resistance determinants by PCR and/or by Southern blotting. With these approaches, we could exclude the presence of the most commonly occurring tet resistance genes in Gram-positive bacteria: tet(O), tet(S), tet(L), tet(K) and tet(W) (data not shown). We then tested the ability of the mosaic tet(S/M) gene to confer Tet resistance by heterologous expression in E. coli. A 2.6 kb fragment containing the entire tet(S/M) ORF, with long stretches of 5′ and 3′ untranslated sequences, was cloned into the plasmid vector pBluescript KS+, and the resulting construct was transformed into the Tet-sensitive E. coli DH5α strain. Three independent recombinant colonies were tested for their ability to grow in the presence of Tet in liquid culture, using the MIC for E. coli cells in Tet as a reference concentration (5 mg l−1). As shown in Fig. 2, E. coli transformants expressing S. bovis tet(S/M) were able to survive at the maximum Tet concentration tested (12 mg l−1), with growth curves displaying the same slope as the Tet-resistant E. coli strain XL1Blue, which harbours the Tn10 transposon and is therefore intrinsically resistant to Tet.
Fig. 1.
Amplification and sequence analysis of the mosaic tet(S/M) gene. (a) PCR amplification with primers directed outwards from tet(S/M). Lanes: 1, no DNA control; 2, size marker (1 kb ladder); 3–5, template DNA from S. bovis independent isolates (1315, 1357 and 1400; Devirgiliis et al., 2010). (b) Sequence alignment of mosaic tet(S/M) with tet(S) (accession no. X92946) and tet(M) (U09422), including the portion of coding sequence where fusion occurred between the two tet genes and the untranslated regulatory region upstream of the tet(S) ATG. Transcriptional and translational regulatory sequences (annotated in X92946) are boxed. Mosaic tet(S/M) and tet(M) ATG start codons are underlined. Amino acid residues contributed by Tet(S) in the mosaic protein are shown in bold type.
Fig. 2.
Heterologous expression of Tet(S/M) in E. coli confers Tet resistance. Growth curves of three independent recombinant colonies of E. coli strain DH5α expressing S. bovis tet(S/M) in the absence (a) or presence of Tet at 6 mg l−1 (b) and 12 mg l−1 (c). The three transformants are labelled with □, ▵ and ◊. Negative controls: E. coli strain DH5α (•), strain DH5α with empty vector (○); positive controls: E. coli strain XL1-Blue with empty vector (○, dashed line) or with S. bovis tet(S/M) (•, dashed line).
Genomic localization of tet(S/M)
To begin analysing the genomic context of the newly identified resistance gene, we extracted genomic DNA from the three representative S. bovis strains, and subjected it to Southern blot analysis. Results in Fig. 3 show that tet(S/M) is present in single copy within a genomic EcoRI fragment larger than 10 kb and a HindIII fragment of 3 kb, specifically hybridizing with the tet(M) probe (Fig. 3a). A tnpA probe was used as an internal control, because sequencing of the 3.7 kb amplicon had shown the presence of an IS belonging to the IS1216 family (Mahillon & Chandler, 1998). This probe hybridized with the same genomic EcoRI and HindIII fragments recognized by the tet(M) probe, but also with other fragments, indicating the presence of additional ISs in the genome of S. bovis (Fig. 3b).
Fig. 3.
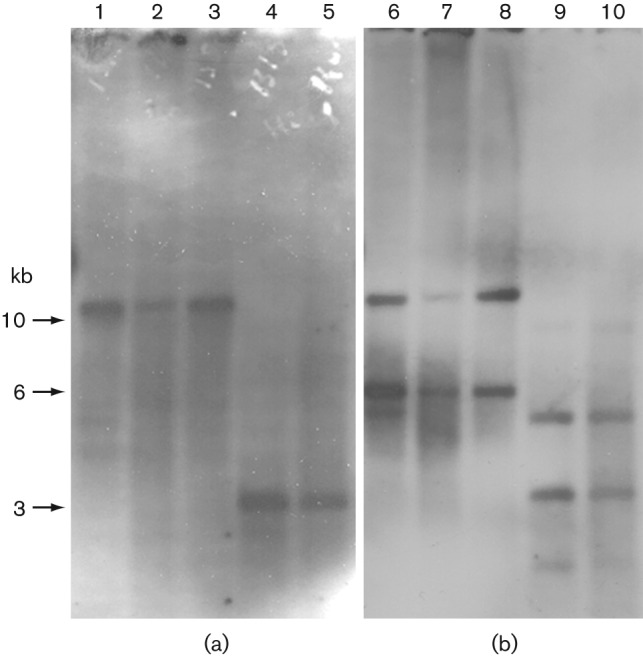
Genomic localization of the tet(S/M) gene. Southern blot analysis of EcoRI- and HindIII-digested genomic DNA from independent isolates of S. bovis probed with tet(M) (a) and tnpA (b) gene fragments. Lanes 1–3 and 6–8, EcoRI-digested DNA from isolates 1315, 1357 and 1400; lanes 4–5 and 9–10, HindIII-digested DNA from isolates 1315 and 1357.
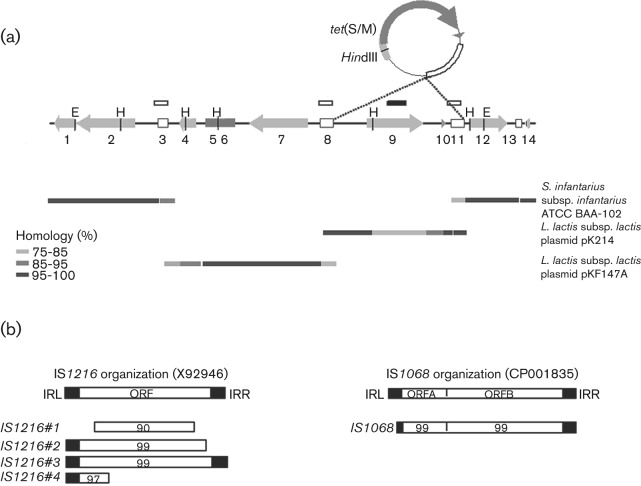
Since the S. bovis genome has not been fully sequenced, we sought to extend sequence information in the chromosomal region containing tet(S/M) using the two-step gene-walking method of Pilhofer et al. (2007) (see Methods). Oligonucleotide primers corresponding to both termini of tet(S/M) were first synthesized, and then used in PCRs to extend into the surrounding sequence in both directions. As more DNA sequence was determined, the same steps were employed to ‘walk’ further into the genome on both sides. The resulting sequence of a chromosomal region of 16 244 nt is schematically represented in Fig. 4(a). Major observations are as follows: a chromosomal subregion containing the tet(S/M) gene is flanked by IS1216 sites, suggesting that excision events could lead to a 4.3 kb circular form. The results of our previous Southern hybridizations were confirmed at the sequence level, as the position of HindIII restriction sites identified the following digestion products: (1) the 3 kb HindIII fragment detected by both tet(S/M) and tnpA probes (Fig. 3, lanes 4–5, 9–10); and (2) the 5 kb fragment detected by the tnpA probe alone (Fig. 3b, lanes 9–10). The remaining sequence showed the presence of three additional ISs, two of them sharing homology with IS6 family members (IS1216), and one with IS3 family members (IS1068) (Mahillon & Chandler, 1998) (Fig. 4b). All IS transposase-encoding ORFs appeared to be truncated by premature stop codons, with the exception of ORF B within IS1068 (Fig. 4b). Overall, blast searches for sequence similarity showed that this region contains stretches of homology to the pkF147A plasmid of Lactococcus lactis subsp. lactis, strain KF147 (GenBank accession no. CP001835), and with a chromosomal region of S. infantarius subsp. infantarius, strain ATCC BAA-102 (ABJK02000017) (Schlegel et al., 2000) (Fig. 4a, Table 2). Noteworthily, the hybrid nature of this region was further suggested by the presence, immediately downstream of the tet(S/M) gene, of an ORF deriving from the broad-host-range conjugative transposon Tn916 (orf6, sharing 87 % homology with orf27 of Lc. lactis pK214) (Fig. 5).
Fig. 4.
Genomic organization of the S. bovis 16 224 bp region flanking the mosaic tet(S/M) gene. (a) Schematic representation of the entire sequenced region. The ORFs are represented by arrows oriented in the direction of transcription. Open boxes represent IS1216-like transposases, while the dark-grey box corresponds to the IS1068-like transposase. HindIII sites (H) and EcoRI sites (E) giving rise to the fragments detected by Southern hybridization are marked. Sequences hybridizing with the tet(M) and tnpA probes are indicated above the genomic structure by closed and open boxes, respectively. Correspondence between the integrated tet(S/M)-containing subregion and the PCR-amplified circular form is indicated by dotted lines. Bars of different grey tones below the linear reconstruction of the entire region indicate homologies with the genomes of sequenced bacterial strains. Gaps between bars represent non-contiguous tracts of homology. (b) Schematic comparison between the IS1216-like and IS1068-like ISs identified in S. bovis and their corresponding reference sequences (accession nos X92946 and CP001835, respectively). Black rectangles represent left (IRL) and right (IRR) inverted repeat sequences; white rectangles indicate transposase ORFs. Numbers within rectangles indicate the percentage nucleotide sequence homology.
Table 2. List of known S. infantarius and Lc. lactis genes homologous to the ORFs detected in the sequenced region of the S. bovis genome depicted in Fig. 4.
| ORF no. | Gene name or locus tag | Protein | Putative function | DNA homology (%) | Amino acid identity (%) | Accession no. | Reference |
| 1 | STRINF_00827 | Hypothetical protein | ATPase component (ABC-type uncharacterized transport system) | 99 | 99 | ABJK02000017 | |
| 2 | STRINF_00828 | Hypothetical protein | Permease component (ABC-type uncharacterized transport system) | 99 | 99 | ABJK02000017 | |
| 3 | tnpA | IS1216 transposase A portion (77–152 aa) | Transposase | 90 | 96 | X92946 | Perreten et al. (1997) |
| 4 | LLKF_p0016 | Resolvase | Resolvase | 92 | 97 | CP001835 | Siezen et al. (2010) |
| 5 | LLKF_p0022 | IS1068 transposase (C terminus) | Transposase | 99 | 98 | CP001835 | Siezen et al. (2010) |
| 6 | LLKF_p0021 | IS1068 transposase (N terminus) | Transposase | 96 | 88 | CP001835 | Siezen et al. (2010) |
| 7 | LLKF_p0020 | Potassium transport system protein kup 1 | Potassium transporter | 99 | 99 | CP001835 | Siezen et al. (2010) |
| 8 | tnpA | Truncated IS1216 transposase A (1–105 aa) | Transposase | 99 | 100 | X92946 | Perreten et al. (1997) |
| 9 | tet (S/M) | Tet(S/M) | Tet resistance | 79 | 77 | X92946 | Perreten et al. (1997) |
| 10 | orf6 | Hypothetical protein | Unknown function | 87 | 81 | X92946 | Perreten et al. (1997) |
| 11 | tnpA | Truncated IS1216 transposase A (1–105 aa) | Transposase | 99 | 100 | X92946 | Perreten et al. (1997) |
| 12 | STRINF_00831 | Hypothetical protein | Catalytic activity (polysaccharide deacetylase domain) | 99 | 98 | ABJK02000017 | |
| 13 | tnpA | N-terminal fragment IS1216 transposase A (1–34 aa) | Transposase | 94 | 88 | X92946 | Perreten et al. (1997) |
| 14 | STRINF_00836 | Hypothetical protein | Regulator of disulfide bond formation | 99 | 99 | ABJK02000017 |
Fig. 5.
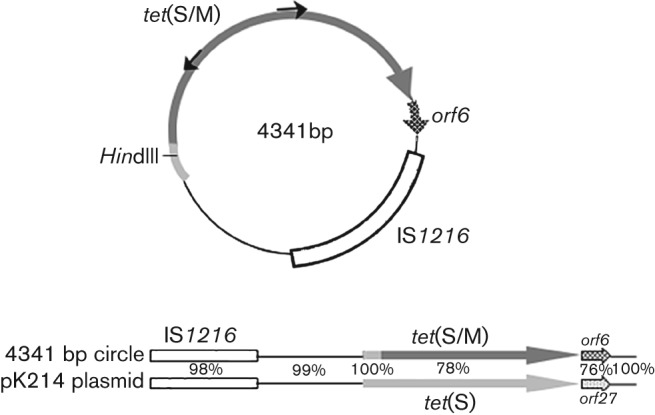
Molecular structure of the 4341 bp circular form containing mosaic tet(S/M). Light grey identifies tet(S) ORF sequences in the mosaic gene, dark grey highlights tet(M) sequences. Sequence comparison between the tet(S/M)-containing circle and nucleotides 25513–29826 of Lc. lactis plasmid pK214 (accession no. X92946) is shown below the circle. Percentage homology is shown for each coding and non-coding subregion.
Tet induction of the mosaic tet(S/M) gene
Tet is well known to induce transcription of the corresponding resistance genes in Gram-negative bacteria (Berens & Hillen, 2003). Tet-dependent regulation of tet gene expression in Gram-positive organisms has been shown to be associated with increased transposition frequency of Tn916 in Ent. faecalis and Bacillus subtilis, by a mechanism involving transcriptional attenuation (Su et al., 1992; Celli & Trieu-Cuot, 1998; Roberts & Mullany, 2009). We have also reported transcriptional induction of tet(M) expression in foodborne strains of Lactobacillus paracasei (Comunian et al., 2010), and we therefore sought to analyse expression of the tet(S/M) mosaic gene in S. bovis strains, where transcription is driven by the tet(S) promoter. To this aim, total RNA was extracted from the Tet-resistant S. bovis strains, grown in the presence or absence of the antibiotic, and subjected to RT-PCR with specific primers mapping on the tet(S) and tet(M) sequences (TetS-1-FW and Rev-Ext in Table 1). As shown in Fig. 6(a), tet(S/M) expression was very low in the absence of Tet in all three strains examined, while it was strongly induced following antibiotic addition to the growth medium at a final concentration of 8 mg l−1. Previously described E. coli transformants expressing an S. bovis tet(S/M) construct (Fig. 2) were also tested for induction by growth in the presence or absence of 12 mg Tet l−1. The results of RT-PCR assays in Fig. 3(b) show that Tet-dependent induction of the mosaic gene, driven by the S. bovis tet(S) promoter, could also be retained in the E. coli heterologous expression system, suggesting that proteins involved in Tet-dependent regulation of gene expression are well conserved in Gram-negative and Gram-positive bacteria.
Fig. 6.
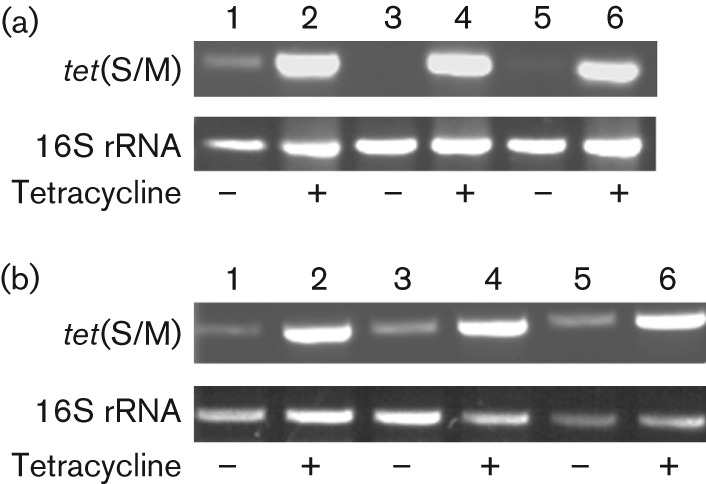
Tet-dependent induction of tet(S/M) expression. RT-PCR analysis of mosaic tet(S/M) gene expression in: (a) three independent S. bovis isolates grown in the presence (lanes 2, 4 and 6) or absence (lanes 1, 3 and 5) of Tet (8 mg l−1); (b) three independent E. coli DH5α transformants expressing S. bovis Tet(S/M), grown in the presence (lanes 2, 4 and 6) or absence (lanes 1, 3 and 5) of Tet (12 mg l−1). Reverse-transcribed 16S rRNA was used as an internal control.
Identification of an intermediate circular form
As described above, amplification of the region containing mosaic tet(S/M) was achieved using diverging primers on the tet(M) sequence that could yield an amplicon only in the presence of a circular template. Furthermore, sequence analysis of the genomic context of the newly identified resistance gene revealed that it localizes within a 4.3 kb chromosomal subregion delimited by ISs, suggestive of potential excision and circularization events. However, no sequence resembling that of a replication origin was identified within the entire 4.3 kb region, suggesting that an excised circle containing tet(S/M) would be unable to sustain autonomous replication. The results of Southern blot hybridization in Fig. 3 seem to confirm the absence of detectable levels of the circular form, as the 4.3 kb HindIII band predicted from the DNA sequence as a linearization product of the circular form could not be detected with either tet(M) or tnpA probes. However, since we were indeed able to amplify a tet(S/M)-containing circle by PCR (Fig. 1a), this result suggests the low abundance or transient nature of the circle. blast sequence similarity searches with the genomic region containing the mosaic tet(S/M) gene revealed homology with a region of Lc. lactis plasmid pK214 (GenBank accession no. X92946, nucleotides 25513–29826), carrying the Tet-resistance gene tet(S) and the IS1216 mobile element (Perreten et al., 1997) (Fig. 5). However, the DNA sequence of the transposase-encoding tnpA gene within the IS1216 element of S. bovis contained a nonsense mutation, leading to a translational STOP codon (UGA) at amino acid position 105.
Discussion
Foodborne bacteria that survive the harsh conditions of the upper GI tract and reach the colon become part of the crowded gut microbiota, thus contributing to its composition, which in turn can affect the health/disease balance of the host (Garrett et al., 2010). Although fermented food products mostly contain probiotic species, they can also contribute a small fraction of bacteria which are not favourable for health, such as opportunistic pathogens capable of acquiring virulence traits in immune-compromised individuals. Among them, enterococci are the most abundant genus found in the human GI tract (Ogier & Serror, 2008), but streptococci are also represented (Herrera et al., 2009). In the latter case, some species have been identified as responsible for common infections (Fernández-Ruiz et al., 2010; Srivastava et al., 2010). Horizontal transfer of AR determinants to pathogens raises growing concern, especially in light of the potential inter-species transferability of AR genes, suggested by their association with mobile elements (Freitas et al., 2011). For this reason it is important not only to identify the occurrence of AR genes in foodborne bacteria but also to characterize their genomic context, with special focus on the regions involved in inter-species and intra-species transfer. Conjugative plasmids containing transposon Tn1546 carrying the van(A) operon seem to have played a relevant role in the recent increase of vancomycin-resistant enterococci (VRE) isolated from poultry and swine sources. Tn1546 variants are associated with IS1216 ISs, which are capable of autonomous transposition and greatly contribute to mobilization of flanking genomic sequences (Novais et al., 2008). Tet(M) and tet(S) have been frequently shown to be in association with the conjugative transposon Tn916 (Roberts & Mullany, 2009). In the present work, we report the association of a novel mosaic Tet-resistance gene [tet(S/M)] with a potentially mobile element closely resembling a region of the Lc. lactis plasmid pK214 (Perreten et al., 1997). The gene was identified in a foodborne strain of the opportunistic pathogen S. bovis formerly isolated from the fermenting microflora of raw buffalo milk (Devirgiliis et al., 2010). Sequencing of the gene and of the surrounding genomic region revealed its molecular organization as that of a novel mosaic Tet-resistance gene consisting of the tet(S) promoter region followed by a small portion of the ORF (encoding 33 N-terminal amino acids) fused in-frame with the tet(M) ORF. Notably, the primary sequences of Tet(S) and Tet(M) share an overall degree of homology of 78 %, aside from their difference in length (646 and 639 amino acids, respectively), which is due to the presence of five additional Tet(S)-specific N-terminal amino acids. We demonstrate that the mosaic tet(S/M) gene is functional, as it was able to confer Tet resistance by heterologous expression in E. coli under the control of the cloned tet(S) promoter. Tet-dependent transcriptional induction of the mosaic gene, which we demonstrated in S. bovis, was retained in the heterologous host. Transcriptional upregulation of tet genes was elegantly described in Gram-positive bacteria as an attenuation mechanism dependent on readthrough of small ORFs downstream of the tet(M) coding region (Su et al., 1992; Celli & Trieu-Cuot, 1998; Roberts & Mullany, 2009). The sequence of the S. bovis mosaic gene does not contain such regulatory ORFs, so it will be extremely interesting to further characterize the mechanism conferring Tet-dependent upregulation upon the tet(S) promoter.
The existence of naturally occurring mosaic Tet-resistance genes has previously been reported in other genera (Stanton et al., 2005; Patterson et al., 2007; van Hoek et al., 2008), but to the best of our knowledge this is the first example of a chimera involving tet(S) and tet(M). Southern blot analysis of total DNA extracted from S. bovis showed that tet(S/M) is present in single copy in the bacterial genome, within a subregion flanked by two IS1216 elements. This region can also be amplified by PCR using primers directed outwards from tet(S/M), indicating the existence of an extra-chromosomal circular form. Complete sequencing of the PCR amplicon revealed the presence of a single copy of IS1216, suggestive of transposase-mediated circularization/excision of the genomic tet(S/M)-containing region, occurring at the two flanking IS1216 sites. However, we could not identify a transfer origin similar to those found in conjugative transposons, and the linearized circle was undetectable in Southern hybridizations. Moreover, in vitro conjugal transfer of Tet resistance to an Ent. faecalis recipient was unsuccessful (Devirgiliis et al., 2010). All this leads us to speculate that the circular form may represent a transposition intermediate rather than a self-replicating extra-chromosomal element, possibly occurring in only a subset of cells and mobilized only within the same cell. Circular elements often represent transient replication intermediates, typical of transposable elements such as Tn916 (Clewell et al., 1995). Replication of such elements is promoted by transposases and generates circular molecules, which can either integrate at different chromosomal locations or be horizontally transferred to other cells (Churchward, 2002). The two IS1216-like repeats flanking the S. bovis tet(S/M) gene might be involved in circularization via homologous recombination. Such non-replicative circular elements harbouring Tet-resistance genes were first described in a Butyrivibrio fibrisolvens transconjugant, where two direct repeats flanking tet(W) in the donor strain were postulated to give rise to a circular intermediate via homologous recombination (Kazimierczak et al., 2006). IS1216-like modules have been reported in association with AR determinants in streptococcal strains. The tet(S) gene in Tet-resistant clinical isolates of Streptococcus dysgalactiae, as well as the erm(T) gene in erythromycin-resistant clinical isolates of S. gallolyticus, are both flanked by two copies of IS1216 in the same orientation (Tsai et al., 2005). In both cases, the structural organization reported for the sequences flanking the AR genes is similar to that of the tet(S)-flanking region in pK214.
Sequencing of a longer, 16 kb genomic region surrounding the mosaic AR gene shows the presence of several ISs, suggestive of a mobilizable region. Although most of the ORFs encoding transposases appear to be truncated, we cannot exclude that some of them might be functional, as evidenced by the presence of the amplifiable tet(S/M)-containing circular form. Notably, it has been reported that C-terminal-truncated transposases play a role in the reduction of transposition activity (Gueguen et al., 2006). Transposase inactivation appears to be a common feature of IS elements, and is recognized as a regulatory mechanism (Mahillon & Chandler, 1998).
The S. bovis genome has not yet been fully sequenced, and very little information is available in databases. The closest species whose entire genome sequence is available is S. infantarius (accession no. ABJK02000000). The taxonomic status of S. bovis strains has been evolving in the past few decades and has progressively changed in parallel with the characterization of new species originally described as S. bovis (Facklam, 2002). Our Tet-resistant foodborne isolates belong to the S. bovis–equinus complex, which includes different species and subspecies isolated from infected humans or animals (Farrow & Collins, 1984; Schlegel et al., 2003). The S. bovis–equinus complex is divided into four DNA clusters, one of which consists of S. infantarius subsp. coli and subsp. infantarius. The latter subspecies has been isolated from foodstuffs and from infected humans, where it was found to be associated with systemic disease in infants (Bouvet et al., 1997; Schlegel et al., 2000, 2004). Due to their association with several human and animal diseases, as well as their occurrence in fermented foods, we believe that it is extremely important to determine the genomic sequences of the most widespread species belonging to the S. bovis–equinus complex, which will serve as a basis to identify the genetic determinants in infectious strains as well as their potential association with AR determinants.
We take these results as strongly indicative of extensive genomic exchanges, possibly through conjugative events, between species that are natural components of the human microbiota, and which should be more thoroughly investigated in light of their potential role as disease-causing agents.
Acknowledgements
The authors wish to thank Kariklia Pascucci for her kind support in daily laboratory work. We acknowledge financial support by a grant from the Italian Ministry of Agriculture, Food & Forestry (MiPAAF) ‘NUME’ (DM 3688/7303/08).
Abbreviations:
- AR
antibiotic resistance (resistant)
- GI
gastrointestinal
- IS
insertion sequence
- Tet
tetracycline
References
- Al-Jashamy K., Murad A., Zeehaida M., Rohaini M., Hasnan J. (2010). Prevalence of colorectal cancer associated with Streptococcus bovis among inflammatory bowel and chronic gastrointestinal tract disease patients. Asian Pac J Cancer Prev 11, 1765–1768. [PubMed] [Google Scholar]
- Ammor M. S., Flórez A. B., Mayo B. (2007). Antibiotic resistance in non-enterococcal lactic acid bacteria and bifidobacteria. Food Microbiol 24, 559–570. 10.1016/j.fm.2006.11.001 [DOI] [PubMed] [Google Scholar]
- Berens C., Hillen W. (2003). Gene regulation by tetracyclines. Constraints of resistance regulation in bacteria shape TetR for application in eukaryotes. Eur J Biochem 270, 3109–3121. 10.1046/j.1432-1033.2003.03694.x [DOI] [PubMed] [Google Scholar]
- Boleij A., Schaeps R. M., Tjalsma H. (2009). Association between Streptococcus bovis and colon cancer. J Clin Microbiol 47, 516. 10.1128/JCM.01755-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet A., Grimont F., Collins M. D., Benaoudia F., Devine C., Regnault B., Grimont P. A. (1997). Streptococcus infantarius sp. nov. related to Streptococcus bovis and Streptococcus equinus. Adv Exp Med Biol 418, 393–395. [DOI] [PubMed] [Google Scholar]
- Celli J., Trieu-Cuot P. (1998). Circularization of Tn916 is required for expression of the transposon-encoded transfer functions: characterization of long tetracycline-inducible transcripts reading through the attachment site. Mol Microbiol 28, 103–117. 10.1046/j.1365-2958.1998.00778.x [DOI] [PubMed] [Google Scholar]
- Churchward G. (2002). Conjugative transposons and related mobile elements. In Mobile DNA II. Edited by Craig N., Craigie R., Gellert M. Washington, DC: American Society for Microbiology. [Google Scholar]
- Clewell D. B., Flannagan S. E., Jaworski D. D., Clewell D. B. (1995). Unconstrained bacterial promiscuity: the Tn916–Tn1545 family of conjugative transposons. Trends Microbiol 3, 229–236. 10.1016/S0966-842X(00)88930-1 [DOI] [PubMed] [Google Scholar]
- Comunian R., Daga E., Dupré I., Paba A., Devirgiliis C., Piccioni V., Perozzi G., Zonenschain D., Rebecchi A. & other authors (2010). Susceptibility to tetracycline and erythromycin of Lactobacillus paracasei strains isolated from traditional Italian fermented foods. Int J Food Microbiol 138, 151–156. 10.1016/j.ijfoodmicro.2009.11.018 [DOI] [PubMed] [Google Scholar]
- Devirgiliis C., Caravelli A., Coppola D., Barile S., Perozzi G. (2008). Antibiotic resistance and microbial composition along the manufacturing process of Mozzarella di Bufala Campana. Int J Food Microbiol 128, 378–384. 10.1016/j.ijfoodmicro.2008.09.021 [DOI] [PubMed] [Google Scholar]
- Devirgiliis C., Coppola D., Barile S., Colonna B., Perozzi G. (2009). Characterization of the Tn916 conjugative transposon in a food-borne strain of Lactobacillus paracasei. Appl Environ Microbiol 75, 3866–3871. 10.1128/AEM.00589-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devirgiliis C., Barile S., Caravelli A., Coppola D., Perozzi G. (2010). Identification of tetracycline- and erythromycin-resistant Gram-positive cocci within the fermenting microflora of an Italian dairy food product. J Appl Microbiol 109, 313–323. [DOI] [PubMed] [Google Scholar]
- Facklam R. (2002). What happened to the streptococci: overview of taxonomic and nomenclature changes. Clin Microbiol Rev 15, 613–630. 10.1128/CMR.15.4.613-630.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrow J. A., Collins M. D. (1984). DNA base composition, DNA–DNA homology and long-chain fatty acid studies on Streptococcus thermophilus and Streptococcus salivarius. J Gen Microbiol 130, 357–362. [DOI] [PubMed] [Google Scholar]
- Fernández-Ruiz M., Villar-Silva J., Llenas-García J., Caurcel-Díaz L., Vila-Santos J., Sanz-Sanz F., Chaves F., Guerra-Vales J. M. (2010). Streptococcus bovis bacteraemia revisited: clinical and microbiological correlates in a contemporary series of 59 patients. J Infect 61, 307–313. 10.1016/j.jinf.2010.07.007 [DOI] [PubMed] [Google Scholar]
- Flórez A. B., Ammor M. S., Mayo B. (2008). Identification of tet(M) in two Lactococcus lactis strains isolated from a Spanish traditional starter-free cheese made of raw milk and conjugative transfer of tetracycline resistance to lactococci and enterococci. Int J Food Microbiol 121, 189–194. 10.1016/j.ijfoodmicro.2007.11.029 [DOI] [PubMed] [Google Scholar]
- Freitas A. R., Coque T. M., Novais C., Hammerum A. M., Lester C. H., Zervos M. J., Donabedian S., Jensen L. B., Francia M. V. & other authors (2011). Human and swine hosts share vancomycin-resistant Enterococcus faecium CC17 and CC5 and Enterococcus faecalis CC2 clonal clusters harboring Tn1546 on indistinguishable plasmids. J Clin Microbiol 49, 925–931. 10.1128/JCM.01750-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett W. S., Gallini C. A., Yatsunenko T., Michaud M., DuBois A., Delaney M. L., Punit S., Karlsson M., Bry L. & other authors (2010). Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe 8, 292–300. 10.1016/j.chom.2010.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueguen E., Rousseau P., Duval-Valentin G., Chandler M. (2006). Truncated forms of IS911 transposase downregulate transposition. Mol Microbiol 62, 1102–1116. 10.1111/j.1365-2958.2006.05424.x [DOI] [PubMed] [Google Scholar]
- Gupta A., Madani R., Mukhtar H. (2010). Streptococcus bovis endocarditis, a silent sign for colonic tumour. Colorectal Dis 12, 164–171. 10.1111/j.1463-1318.2009.01814.x [DOI] [PubMed] [Google Scholar]
- Herrera P., Kwon Y. M., Ricke S. C. (2009). Ecology and pathogenicity of gastrointestinal Streptococcus bovis. Anaerobe 15, 44–54. 10.1016/j.anaerobe.2008.11.003 [DOI] [PubMed] [Google Scholar]
- Kahveci A., Ari E., Arikan H., Koc M., Tuglular S., Ozener C. (2010). Streptococcus bovis bacteremia related to colon adenoma in a chronic hemodialysis patient. Hemodial Int 14, 91–93. 10.1111/j.1542-4758.2009.00400.x [DOI] [PubMed] [Google Scholar]
- Kazimierczak K. A., Flint H. J., Scott K. P. (2006). Comparative analysis of sequences flanking tet(W) resistance genes in multiple species of gut bacteria. Antimicrob Agents Chemother 50, 2632–2639. 10.1128/AAC.01587-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahillon J., Chandler M. (1998). Insertion sequences. Microbiol Mol Biol Rev 62, 725–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moet G. J., Dowzicky M. J., Jones R. N. (2007). Tigecycline (GAR-936) activity against Streptococcus gallolyticus (bovis) and viridans group streptococci. Diagn Microbiol Infect Dis 57, 333–336. 10.1016/j.diagmicrobio.2006.08.001 [DOI] [PubMed] [Google Scholar]
- Novais C., Freitas A. R., Sousa J. C., Baquero F., Coque T. M., Peixe L. V. (2008). Diversity of Tn1546 and its role in the dissemination of vancomycin-resistant enterococci in Portugal. Antimicrob Agents Chemother 52, 1001–1008. 10.1128/AAC.00999-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogier J. C., Serror P. (2008). Safety assessment of dairy microorganisms: the Enterococcus genus. Int J Food Microbiol 126, 291–301. 10.1016/j.ijfoodmicro.2007.08.017 [DOI] [PubMed] [Google Scholar]
- Patterson A. J., Rincon M. T., Flint H. J., Scott K. P. (2007). Mosaic tetracycline resistance genes are widespread in human and animal fecal samples. Antimicrob Agents Chemother 51, 1115–1118. 10.1128/AAC.00725-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen I. T., Banerjei L., Myers G. S., Nelson K. E., Seshadri R., Read T. D., Fouts D. E., Eisen J. A., Gill S. R. & other authors (2003). Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299, 2071–2074. 10.1126/science.1080613 [DOI] [PubMed] [Google Scholar]
- Perreten V., Schwarz F., Cresta L., Boeglin M., Dasen G., Teuber M. (1997). Antibiotic resistance spread in food. Nature 389, 801–802. 10.1038/39767 [DOI] [PubMed] [Google Scholar]
- Pilhofer M., Bauer A. P., Schrallhammer M., Richter L., Ludwig W., Schleifer K. H., Petroni G. (2007). Characterization of bacterial operons consisting of two tubulins and a kinesin-like gene by the novel two-step gene walking method. Nucleic Acids Res 35, e135. 10.1093/nar/gkm836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A. P., Mullany P. (2009). A modular master on the move: the Tn916 family of mobile genetic elements. Trends Microbiol 17, 251–258. 10.1016/j.tim.2009.03.002 [DOI] [PubMed] [Google Scholar]
- Schlegel L., Grimont F., Collins M. D., Régnault B., Grimont P. A., Bouvet A. (2000). Streptococcus infantarius sp. nov., Streptococcus infantarius subsp. infantarius subsp. nov. and Streptococcus infantarius subsp. coli subsp. nov., isolated from humans and food. Int J Syst Evol Microbiol 50, 1425–1434. 10.1099/00207713-50-4-1425 [DOI] [PubMed] [Google Scholar]
- Schlegel L., Grimont F., Ageron E., Grimont P. A., Bouvet A. (2003). Reappraisal of the taxonomy of the Streptococcus bovis/Streptococcus equinus complex and related species: description of Streptococcus gallolyticus subsp. gallolyticus subsp. nov., S. gallolyticus subsp. macedonicus subsp. nov. and S. gallolyticus subsp. pasteurianus subsp. nov. Int J Syst Evol Microbiol 53, 631–645. 10.1099/ijs.0.02361-0 [DOI] [PubMed] [Google Scholar]
- Schlegel L., Grimont F., Grimont P. A., Bouvet A. (2004). New group D streptococcal species. Indian J Med Res 119 (Suppl.), 252–256. [PubMed] [Google Scholar]
- Siezen R. J., Bayjanov J., Renckens B., Wels M., van Hijum S. A., Molenaar D., van Hylckama Vlieg J. E. (2010). Complete genome sequence of Lactococcus lactis subsp. lactis KF147, a plant-associated lactic acid bacterium. J Bacteriol 192, 2649–2650. 10.1128/JB.00276-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A., Walter N., Atkinson P. (2010). Streptococcus bovis infection of total hip arthroplasty in association with carcinoma of colon. J Surg Orthop Adv 19, 125–128. [PubMed] [Google Scholar]
- Stanton T. B., Humphrey S. B., Scott K. P., Flint H. J. (2005). Hybrid tet genes and tet gene nomenclature: request for opinion. Antimicrob Agents Chemother 49, 1265–1266. 10.1128/AAC.49.3.1265-1266.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y. A., He P., Clewell D. B. (1992). Characterization of the tet(M) determinant of Tn916: evidence for regulation by transcription attenuation. Antimicrob Agents Chemother 36, 769–778. 10.1128/AAC.36.4.769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai J. C., Hsueh P. R., Chen H. J., Tseng S. P., Chen P. Y., Teng L. J. (2005). The erm(T) gene is flanked by IS1216V in inducible erythromycin-resistant Streptococcus gallolyticus subsp. pasteurianus. Antimicrob Agents Chemother 49, 4347–4350. 10.1128/AAC.49.10.4347-4350.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoek A. H., Mayrhofer S., Domig K. J., Flórez A. B., Ammor M. S., Mayo B., Aarts H. J. (2008). Mosaic tetracycline resistance genes and their flanking regions in Bifidobacterium thermophilum and Lactobacillus johnsonii. Antimicrob Agents Chemother 52, 248–252. 10.1128/AAC.00714-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak R. A., Waldor M. K. (2010). Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat Rev Microbiol 8, 552–563. 10.1038/nrmicro2382 [DOI] [PubMed] [Google Scholar]