Abstract
Gene signatures have been developed for estrogen receptor-positive breast cancer to complement pathological factors in providing prognostic information. The 70-gene and the 21-gene signatures identify patients who may not require adjuvant chemotherapy. Gene signatures in triple-negative disease and HER2-positive disease have not been fully developed yet, although studies demonstrate heterogeneity within these subgroups. Further research is needed before genotyping will help in making clinical decisions in triple-negative and HER2-positive disease. Molecular subtyping of breast cancer led to define luminal, basal, and HER2-enriched subtypes, which have specific clinical behavior. This approach may lead to identify new subgroups requiring specific therapies. Standardization of techniques will be required to translate investigations to the clinic.
Introduction
Breast cancer is a major health issue in developed countries. Early diagnosis and the use of adjuvant therapies have contributed to improve survival, but still 87,000 women died of breast cancer in the European Union in 2011 [1]. Further improvement in outcome could be achieved through a more adequate use of existing therapies. In this context, gene signatures have been incorporated into clinical practice to complement traditional pathology and guide clinical decisions.
The information provided by prognostic or predictive factors is important if it helps in making clinical decisions. Regarding the management of early breast cancer, the indication for adjuvant radiation therapy depends on classic pathological factors, whereas the need for hormonal therapy and adjuvant trastuzumab depends on the results of immunohistochemistry. However, the indication for adjuvant chemotherapy is more complex, because no single biomarker predicts that the patient will benefit from this kind of treatment. Gene signatures are useful because they may identify patients who will require adjuvant chemotherapy.
This paper will deal with the accepted indications of gene signatures in the management of breast cancer, with an emphasis on strengths and limitations of available tools. It will also offer a glimpse into future applications.
Gene Signatures in ER+ Disease
In patients with ER+ disease, adjuvant chemotherapy is recommended in the presence of risk factors (young age, positive lymph nodes, tumor size of more than 1 cm, or poorly differentiated histology/high proliferation index). However, if the patient presents with just one of these factors, the decision to administer chemotherapy is not obvious. The widespread use of adjuvant chemotherapy, even in women with an intermediate risk of relapse, has greatly contributed to improve prognosis in the last decades but at the cost of over-treatment in many patients [2]. Classic parameters have been integrated in software applications—such as Adjuvant! Online [3]—to help doctors in calculating risk of relapse and benefit from adjuvant therapy. However, uncertainty remains in many cases even with the use of this software.
Gene signatures can complement classic prognostic factors to obtain more accurate prognostic information. The 70-gene signature (MammaPrint; Agendia, Amsterdam, The Netherlands) and the 21-gene signature (OncoType; Genomic Health, Redwood City, CA) are being used in selected patients with early ER+ disease to identify those women that will be cured even if they do not receive adjuvant chemotherapy [4,5]. These signatures have been extensively studied and are widely used in Europe and in USA [6–8]. The National Cancer Comprehensive Network guidelines indicate that the 21-gene signature can be considered in women with tumors >0.5 cm, HER2-negative disease, and either N0 or N1 mi (micrometastasis) disease [9]. The National Cancer Comprehensive Network recommendations do not mean that all tumors with such characteristics should be studied with a multigene product: Doctors should select cases within these groups if they doubt about the need for adjuvant chemotherapy.
Both the 70-gene and the 21-gene signatures have predictive, in addition to prognostic, value. This means that, in women allocated to high-risk groups, adjuvant chemotherapy significantly improves disease-free survival, as opposed to those in low-risk groups [10,11]. Analyses confirming the additional predictive value of gene signatures were based on clinical trials including anthracyclines but not taxanes, although it is reasonable to think that similar conclusions can be drawn with regard to current chemotherapy schemes.
Limitations of the 70-gene and the 21-gene signatures include price, the requirement to send samples to a reference center, and, in the case of OncoType, the existence of an intermediate group for which the value of adjuvant chemotherapy is not clear. The clinical trial TailorX, which has completed accrual, will define if patients in the intermediate-risk group do benefit from chemotherapy. Finally, in the case of very small tumors, it may be difficult to isolate enough material to perform the test.
There are other tools designed for ER+ tumors. MapQuant Dx (Ipsogen, Marseille, France) is a 97-gene genomic grade index that splits grade 2 tumors into two categories: one with an outcome similar to that of grade 1 tumors and the other similar to grade 3 [12]. Patients in the favorable group could be treated without chemotherapy. It is microarray based and requires freshly prepared tissue. The Breast Cancer Index (BioTheranostics, San Diego, CA) is a seven-gene assay that identifies a low-risk group of patients that can be safely treated with adjuvant tamoxifen and no chemotherapy [13]. It is based on quantitative reverse transcription-polymerase chain reaction (PCR) and can be used in paraffin-embedded tissue. Other signatures have been described [14]. Experience with these products is more limited than with the 21-gene and the 70-gene signatures.
There are some unmet needs in the field of ER+ breast cancer. Gene signatures have been developed for ductal carcinoma but not for lobular carcinoma. Some studies suggest that, although lobular and ER+ ductal invasive carcinomas share common genomic alterations, the former has some specific molecular features [15–17]. On the other hand, gene signatures predict relapses in the first 5 to 10 years of follow-up but less reliably in the long term, maybe because tumors with such a long natural history have distinct, hitherto unknown genetic features.
Gene Signatures in Triple-Negative Disease
Patients with triple-negative breast cancer do not have the option to receive therapy with hormonally directed agents, so chemotherapy remains the standard of care in this setting. Two uncommon varieties of breast cancer—medullary and adenoid cystic carcinoma—are triple-negative tumors associated with an excellent prognosis [18,19]; their diagnosis relies on morphological criteria. With these two exceptions, triple-negative tumors usually have a high proliferation rate and poor prognosis, so it is unlikely that gene signatures may identify a favorable group of patients who could forgo adjuvant chemotherapy. Indeed, no signatures have been validated that demonstrate this. Predictors developed in ER+ disease may not function in the triple-negative context because the predominant processes linked to outcome differ between subtypes. For instance, proliferation is an important feature in ER+ disease, whereas immune responses may be more determinant in estrogen receptor-negative disease [20].
The possibility to predict response to chemotherapy has attracted considerable attention in the field of triple-negative disease. Some studies have focused on one drug or combination of drugs in the neoadjuvant setting, where the main parameter of response is the rate of pathological complete response to chemotherapy [21–23]. However, defining sensitivity in terms of complete response has been an elusive goal. One possible explanation is that there may be multiple mechanisms of drug resistance in a series of tumors, so one single signature will not identify all of them [24].
Gene profiling has also been used to subdivide triple-negative disease into distinct entities. For instance, a ratio of high B-cell and low interleukin-8 (IL-8) metagenes has been associated with good prognosis in this population [25]. In another study, a group of 14 genes identified patients with a good outcome in the absence of adjuvant therapy [26]. In a supervised analysis of publicly available data sets or triple-negative disease, a group of genes related to inflammation and angiogenesis provided prognostic information; the combination of a prognostic profile and a B-cell metagene also predicted response to neo-adjuvant chemotherapy [27]. Other studies have demonstrated that triple-negative disease is a heterogeneous group, which may encompass different molecular entities [26,28–31]. For instance, one study analyzed gene expression profiles from 21 breast cancer data sets and identified six triple-negative subtypes. Cell line models representative of these models were also identified, which allowed targeting driver pathways with specific drugs. This experiment served as proof of concept that gene expression analysis can inform therapy selection.
Subclassifications of triple-negative breast cancer have not yielded therapeutic advantages so far but could do so in the future. Some triple-negative tumors have an aberrant BRCA1 pathway, which could allow therapy with poly (adenosine diphosphate [ADP]-ribose) polymerase inhibitors. However, clinical studies with poly(ADP-ribose) polymerase inhibitors have yielded disappointing results in breast cancer so far [32,33]. These poor results suggest that further investigation is required to find a role for these drugs. Platinum-based therapy could also be useful in triple-negative disease, particularly in cases with concomitant BRCA1 mutation [34]. On the other hand, as epidermal growth factor receptor (EGFR) is part of the basal cluster, it could be a target in this subgroup of breast cancer. One clinical trial of chemotherapy with or without cetuximab (an anti-EGFR antibody) suggested that inhibition of EGFR yields a benefit when the pathway is deactivated [35]. Finally, the melanoma antigen family A could define a very aggressive subgroup of triple-negative breast cancer, particularly in the absence of immune infiltration in the tumor microenvironment [36]. This finding suggests that triple-negative tumors with melanoma antigen family A expression might benefit from drugs that enhance the immune response.
Any gene signature for triple-negative disease should be prospectively tested in the clinic before widespread use, in the same way that the 70-gene and the 21-gene signatures were studied in ER+ disease. Once a gene tool would demonstrate utility to answer a clinically relevant question, it could be incorporated into daily practice.
Gene Signatures in HER2-Positive Disease
Virtually all patients with HER2-positive disease receive chemotherapy plus anti-HER2 therapy, either in the adjuvant, the neoadjuvant, or the metastatic setting. The combination of chemotherapy with two anti-HER2 drugs (for instance, trastuzumab + lapatinib or trastuzumab + pertuzumab) increases response rates in neoadjuvant therapy. This strategy, however, increases costs, so it should be limited to those patients who are less likely to benefit from single-agent therapy. Considerable effort has been devoted to identify mechanisms of resistance to trastuzumab [37–39]. A few studies have dealt with gene profiling to detect resistance to the drug [40,41], but validated signatures that can be taken to the clinic have not been described.
HER2-positive breast cancer is a heterogeneous disease. In general, response to systemic therapy is poorer in ER+ disease than in ER-[42], although prognosis is better in the former. In a retrospective study, the 70-gene signature identified a subgroup of HER2+ early breast cancer with a favorable long-term outcome, which suggests genetic heterogeneity within the HER2+ population [43]. In another study, a 158-gene predictor identified patients with better and worse outcome across multiple data sets [44]. Further insight into the molecular biology of these tumors might lead to the description of new therapeutic targets in specific subtypes of HER2-positive tumors. On the other hand, some patients with metastatic disease survive for prolonged periods of time after treatment with trastuzumab; identification of these patients could have interest not only in advanced disease but also in the adjuvant setting.
Molecular Subtyping
The molecular classification of breast cancer created in the year 2000 established several subgroups of tumors, such as luminal A, luminal B, basal-like, and HER2 enriched [45]. This classification has greatly influenced the way in which breast cancer is currently regarded by clinicians and investigators alike. It is important to understand the difference between subtypes defined by immunohistochemistry and gene data sets: for instance, although many ER+ tumors correspond to the luminal A subtype, there is overlapping with other subtypes. The same happens with triple-negative and basal-like disease or the HER2 subgroup, so terminology should be accurate in scientific communication.
The molecular subgroups harbor prognostic value, because patients with luminal A tumors have better prognosis than patients with other tumor subtypes (particularly luminal B and basal-like). On the contrary, poor prognosis subtypes respond better to chemotherapy compared to luminal A tumors, so the classification also offers predictive information [46]. Recent evidence indicates that the HER2-enriched subtype has a special sensitivity to anthracyclines, whereas luminal and basal-like tumors obtain a similar benefit from an anthracycline- and a non-anthracycline-based adjuvant chemotherapy [47]. Further studies are required to corroborate these results.
The molecular classification of breast cancer has not contributed to modify standard therapy in any subgroup of patients. For instance, the administration of trastuzumab or lapatinib is determined by the results of immunohistochemistry or fluorescence in situ hybridization (FISH) for HER2 and no study has demonstrated that limiting the use of these drugs to patients with the HER2-enriched subtype would yield better results. Moreover, a basal-like or luminal B tumor with HER2 overexpression should also receive anti-HER2 therapy. Likewise, a claudin-low subtype that is associated with a poor outcome has recently been described [46], but optimal therapy for this subtype has not been defined. Studies comparing classic and molecular criteria to select treatment would be required to know if new technologies would allow a more rational use of current therapeutic options.
Two commercial applications offer molecular subtyping at this time (PAM50, ARUP, Salt Lake City, UT and BluePrint, Agendia). Attempts have been made with PAM50 in patients with ER+ disease to identify those who do well without adjuvant chemotherapy, in a similar way to the 70-gene and the 21-gene signatures. A combined prognostic marker that includes the proliferation genes of PAM50 and tumor size identifies a subpopulation with an excellent outcome if treated with hormonal therapy alone [48].
New Technologies
Currently available tools are based on microarrays or quantitative reverse transcription-PCR platforms. The development of massive parallel sequencing can provide more extensive information about genomic abnormalities in breast cancer. As the cost of sequencing is decreasing rapidly, tumors can be sequenced in real time at the clinic, if and when the information is considered clinically useful [49]. However, further refinement of this technology is needed before its results can be used in the clinic. For the moment, massive sequencing may be more suitable for investigational purposes than for daily practice.
Proteomics and metabolomics are promising technologies that are already providing interesting results in the biomarker discovery arena. Again, these “omics“ will require further development in terms of standardization and reproducibility before they are incorporated into the clinic.
The use of all new technologies will also depend on bioinformatics and statistic tools to analyze massive amount of data. Likewise, large sample collections and cooperative/multidisciplinary networks will be required to draw valid conclusions. The recent METABRIC results are a good example in this direction [50]. This consortium analyzed 2000 tumors by integrating views of the genome and transcriptome: 10 breast cancer subtypes were described and validated, including a high-risk estrogen receptor-positive 11q13/14 cis-acting subgroup and a favorable prognosis subgroup devoid of copy number aberrations. This study provides an excellent example of how recent technical breakthroughs bring new opportunities to clinical research, allowing the analysis of cancer samples from multiple perspectives, from genomics to proteomics.
Need for Standardization
Reproducibility of classic markers—basically hormonal receptors and HER2—is a major issue in daily practice and in clinical investigation [51–53]. Lack of reproducibility is accepted only because immunohistochemistry is cheap and widely available, but it is hard to believe that genomic assays were approved for clinical use with a 20% discordance rate. The 70-gene and the 21-gene signatures have shown excellent concordance when repeated on different days by different technicians [54,55], which is probably due to the fact that these gene tests are performed in centralized laboratories. If high-throughput techniques were to be performed locally, reproducibility should be ensured both in large hospitals processing a lot of samples and in small centers.
A Glimpse to the Future
High-throughput technologies have a huge potential to help in the management of breast cancer, but so far their use has been limited. While these tools are not meant to replace the standard pathological workup, they could provide important complementary information in many cases.
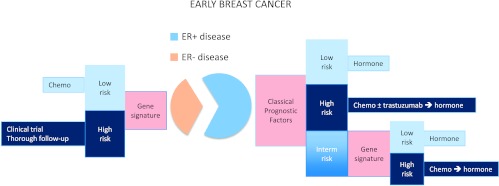
In triple-negative breast cancer, gene profiling is unlikely to find patients who do not need adjuvant chemotherapy, but it could serve a different purpose. Our group performed a discovery PCR study in samples from patients with ER-negative disease who had received adjuvant chemotherapy [56]. A five-gene score splits this population into two groups, one with excellent metastasis-free survival and the other with a 50% chance of distant relapse. This was a small study that is focused in a different target population, but it demonstrates that gene profiling can be applied to patients treated with adjuvant chemotherapy and opens new possibilities. A similar approach could be used in patients with triple-negative disease treated with chemotherapy or in those with HER2+ disease receiving chemotherapy plus trastuzumab. Patients in favorable groups could be safely treated with standard chemotherapy ± trastuzumab, but patients in the poor prognosis group would be ideal candidates for clinical trials with new drugs (Figure 1). By excluding low-risk patients, such trials would be enriched in a population that would be more likely to benefit from the new drug, and as a consequence, fewer patients should be included. In addition, the poor prognosis population could undergo a more thorough follow-up schedule than the general population with breast cancer. Follow-up schedules have become simpler in recent decades but could be modified for the early detection of relapse in selected patients.
Figure 1.
Current clinical use of gene signatures in ER+ disease. Classic prognostic factors include tumor size, lymph node infiltration, grade of differentiation, and age.
Molecular subtyping of breast cancer has changed our view of this disease but has not been used to make clinical decisions so far. The greatest interest of the molecular classification is the possibility to subdivide breast cancer into entities that require specific treatments. The thorough analysis of subtypes may lead to the detection of targetable molecular defects. If a new drug effectively targets a gene marker, the detection of such marker should be incorporated to the standard pathological study. As a consequence, the molecular classification will evolve over time: Indeed, the definition of subtypes has slightly changed since the first description and will continue to do so. Additionally, the molecular classification of breast cancer could be used to tailor follow-up strategies, if it identifies tumors with higher chance of dissemination. In this regard, genes involved in lung and brain dissemination of breast cancer have been described [57,58].
Considering these possibilities, it would be possible to build a two-step process to make decisions in the management in early stage breast cancer (Figure 1). Standard pathological procedures could be complemented with gene signatures—different for ER+ and ER- disease—to identify patients likely to be cured with standard systemic treatment. Patients unlikely to be cured with that approach would require additional treatment. The use of predictive signatures would be fully justified in unfavorable subgroups.
Conclusions
Gene signatures will be incorporated into clinical practice if they help in making clinical decisions. For the moment, some of these tools are being used in ER+ early stage disease to decide whether the patient will forgo adjuvant chemotherapy. In the case of triple-negative or HER2+ tumors, gene signatures could provide information about the best way to administer available drugs, or they could also identify subgroups of patients to be treated with new specific therapies. Clinical studies will have to be done to demonstrate that these hypothetical uses will improve patients' outcome.
References
- 1.Malvezzi M, Arfe A, Bertuccio P, Levi F, La Vecchia C, Negri E. European cancer mortality predictions for the year 2011. Ann Oncol. 2011;22:947–956. doi: 10.1093/annonc/mdq774. [DOI] [PubMed] [Google Scholar]
- 2.Gnant M, Steger GG. Fighting overtreatment in adjuvant breast cancer therapy. Lancet. 2009;374:2029–2030. doi: 10.1016/S0140-6736(09)62097-3. [DOI] [PubMed] [Google Scholar]
- 3.Olivotto IA, Bajdik CD, Ravdin PM, Speers CH, Coldman AJ, Norris BD, Davis GJ, Chia SK, Gelmon KA. Population-based validation of the prognostic model ADJUVANT! for early breast cancer. J Clin Oncol. 2005;23:2716–2725. doi: 10.1200/JCO.2005.06.178. [DOI] [PubMed] [Google Scholar]
- 4.van de Vijver MJ,, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 5.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 6.Albain KS, Barlow WE, Shak S, Hortobagyi GN, Livingston RB, Yeh IT, Ravdin P, Bugarini R, Baehner FL, Davidson NE, et al. Prognostic and predictive value of the 21-gene recurrence score assay in postmenopausal women with node-positive, oestrogen-receptor-positive breast cancer on chemotherapy: a retrospective analysis of a randomised trial. Lancet Oncol. 2010;11:55–65. doi: 10.1016/S1470-2045(09)70314-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mook S, Schmidt MK, Viale G, Pruneri G, Eekhout I, Floore A, Glas AM, Bogaerts J, Cardoso F, Piccart-Gebhart MJ, et al. The 70-gene prognosis-signature predicts disease outcome in breast cancer patients with 1–3 positive lymph nodes in an independent validation study. Breast Cancer Res Treat. 2009;116:295–302. doi: 10.1007/s10549-008-0130-2. [DOI] [PubMed] [Google Scholar]
- 8.Mittempergher L, de Ronde JJ, Nieuwland M, Kerkhoven RM, Simon I, Rutgers EJ, Wessels LF, Van't Veer LJ. Gene expression profiles from formalin fixed paraffin embedded breast cancer tissue are largely comparable to fresh frozen matched tissue. PLoS One. 2011;6:e17163. doi: 10.1371/journal.pone.0017163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.National Cancer Comprehensive Network, author. Breast Cancer. www.nccn.org.
- 10.Knauer M, Mook S, Rutgers EJ, Bender RA, Hauptmann M, van de Vijver MJ, Koornstra RH, Bueno-de-Mesquita JM, Linn SC, van't Veer LJ. The predictive value of the 70-gene signature for adjuvant chemotherapy in early breast cancer. Breast Cancer Res Treat. 2010;120:655–661. doi: 10.1007/s10549-010-0814-2. [DOI] [PubMed] [Google Scholar]
- 11.Paik S, Tang G, Shak S, Kim C, Baker J, Kim W, Cronin M, Baehner FL, Watson D, Bryant J, et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J Clin Oncol. 2006;24:3726–3734. doi: 10.1200/JCO.2005.04.7985. [DOI] [PubMed] [Google Scholar]
- 12.Loi S, Haibe-Kains B, Desmedt C, Lallemand F, Tutt AM, Gillet C, Ellis P, Harris A, Bergh J, Foekens JA, et al. Definition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. J Clin Oncol. 2007;25:1239–1246. doi: 10.1200/JCO.2006.07.1522. [DOI] [PubMed] [Google Scholar]
- 13.Jerevall PL, Ma XJ, Li H, Salunga R, Kesty NC, Erlander MG, Sgroi DC, Holmlund B, Skoog L, Fornander T, et al. Prognostic utility of HOXB13:IL17BR and molecular grade index in early-stage breast cancer patients from the Stockholm trial. Br J Cancer. 2011;104:1762–1769. doi: 10.1038/bjc.2011.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanchez-Navarro I, Gamez-Pozo A, Pinto A, Hardisson D, Madero R, Lopez R, San Jose B, Zamora P, Redondo A, Feliu J, et al. An 8-gene qRT-PCR-based gene expression score that has prognostic value in early breast cancer. BMC Cancer. 2010;10:336. doi: 10.1186/1471-2407-10-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gruel N, Lucchesi C, Raynal V, Rodrigues MJ, Pierron G, Goudefroye R, Cottu P, Reyal F, Sastre-Garau X, Fourquet A, et al. Lobular invasive carcinoma of the breast is a molecular entity distinct from luminal invasive ductal carcinoma. Eur J Cancer. 2010;46:2399–2407. doi: 10.1016/j.ejca.2010.05.013. [DOI] [PubMed] [Google Scholar]
- 16.Zhao H, Langerod A, Ji Y, Nowels KW, Nesland JM, Tibshirani R, Bukholm IK, Karesen R, Botstein D, Borresen-Dale AL, et al. Different gene expression patterns in invasive lobular and ductal carcinomas of the breast. Mol Biol Cell. 2004;15:2523–2536. doi: 10.1091/mbc.E03-11-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bertucci F, Orsetti B, Negre V, Finetti P, Rouge C, Ahomadegbe JC, Bibeau F, Mathieu MC, Treilleux I, Jacquemier J, et al. Lobular and ductal carcinomas of the breast have distinct genomic and expression profiles. Oncogene. 2008;27:5359–5372. doi: 10.1038/onc.2008.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martinez SR, Beal SH, Canter RJ, Chen SL, Khatri VP, Bold RJ. Medullary carcinoma of the breast: a population-based perspective. Med Oncol. 2011;28:738–744. doi: 10.1007/s12032-010-9526-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boujelbene N, Khabir A, Jeanneret Sozzi W, Mirimanoff RO, Khanfir K. Clinical review—breast adenoid cystic carcinoma. Breast. 2012;21:124–127. doi: 10.1016/j.breast.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 20.Desmedt C, Haibe-Kains B, Wirapati P, Buyse M, Larsimont D, Bontempi G, Delorenzi M, Piccart M, Sotiriou C. Biological processes associated with breast cancer clinical outcome depend on the molecular subtypes. Clin Cancer Res. 2008;14:5158–5165. doi: 10.1158/1078-0432.CCR-07-4756. [DOI] [PubMed] [Google Scholar]
- 21.Farmer P, Bonnefoi H, Anderle P, Cameron D, Wirapati P, Becette V, Andre S, Piccart M, Campone M, Brain E, et al. A stroma-related gene signature predicts resistance to neoadjuvant chemotherapy in breast cancer. Nat Med. 2009;15:68–74. doi: 10.1038/nm.1908. [DOI] [PubMed] [Google Scholar]
- 22.Desmedt C, Di Leo A, de Azambuja E, Larsimont D, Haibe-Kains B, Selleslags J, Delaloge S, Duhem C, Kains JP, Carly B, et al. Multifactorial approach to predicting resistance to anthracyclines. J Clin Oncol. 2011;29:1578–1586. doi: 10.1200/JCO.2010.31.2231. [DOI] [PubMed] [Google Scholar]
- 23.Martin M, Romero A, Cheang MC, Lopez Garcia-Asenjo JA, Garcia-Saenz JA, Oliva B, Roman JM, He X, Casado A, de la Torre J, et al. Genomic predictors of response to doxorubicin versus docetaxel in primary breast cancer. Breast Cancer Res Treat. 2011;128:127–136. doi: 10.1007/s10549-011-1461-y. [DOI] [PubMed] [Google Scholar]
- 24.Borst P, Wessels L. Do predictive signatures really predict response to cancer chemotherapy? Cell Cycle. 2010;9:4836–4840. doi: 10.4161/cc.9.24.14326. [DOI] [PubMed] [Google Scholar]
- 25.Rody A, Karn T, Liedtke C, Pusztai L, Ruckhaeberle E, Hanker L, Gaetje R, Solbach C, Ahr A, Metzler D, et al. A clinically relevant gene signature in triple negative and basal-like breast cancer. Breast Cancer Res. 2011;13:R97. doi: 10.1186/bcr3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yau C, Esserman L, Moore DH, Waldman F, Sninsky J, Benz CC. A multigene predictor of metastatic outcome in early stage hormone receptor-negative and triple-negative breast cancer. Breast Cancer Res. 2010;12:R85. doi: 10.1186/bcr2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karn T, Pusztai L, Holtrich U, Iwamoto T, Shiang CY, Schmidt M, Muller V, Solbach C, Gaetje R, Hanker L, et al. Homogeneous datasets of triple negative breast cancers enable the identification of novel prognostic and predictive signatures. PLoS One. 2011;6:e28403. doi: 10.1371/journal.pone.0028403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kreike B, van Kouwenhove M, Horlings H, Weigelt B, Peterse H, Bartelink H, van de Vijver MJ. Gene expression profiling and histopathological characterization of triple-negative/basal-like breast carcinomas. Breast Cancer Res. 2007;9:R65. doi: 10.1186/bcr1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turner N, Lambros MB, Horlings HM, Pearson A, Sharpe R, Natrajan R, Geyer FC, van Kouwenhove M, Kreike B, Mackay A, et al. Integrative molecular profiling of triple negative breast cancers identifies amplicon drivers and potential therapeutic targets. Oncogene. 2010;29:2013–2023. doi: 10.1038/onc.2009.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Teschendorff AE, Caldas C. A robust classifier of high predictive value to identify good prognosis patients in ER-negative breast cancer. Breast Cancer Res. 2008;10:R73. doi: 10.1186/bcr2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Shaughnessy J, Schwartzberg LS, Danso MA, Rugo HS, Miller K, Yardley DA, Carlson RW, Finn RS, Charpentier E, Freese M, et al. A randomized phase III study of iniparib in combination with gemcitabine/carboplatin in metastatic triple-negative breast cancer; 47th Annual Meeting of the American Society of Clinical Oncology; 2011. abs 1007. [Google Scholar]
- 33.Gelmon KA, Tischkowitz M, Mackay H, Swenerton K, Robidoux A, Tonkin K, Hirte H, Huntsman D, Clemons M, Gilks B, et al. Olaparib in patients with recurrent high-grade serous or poorly differentiated ovarian carcinoma or triple-negative breast cancer: a phase 2, multicentre, open-label, non-randomised study. Lancet Oncol. 2011;12:852–861. doi: 10.1016/S1470-2045(11)70214-5. [DOI] [PubMed] [Google Scholar]
- 34.Silver DP, Richardson AL, Eklund AC, Wang ZC, Szallasi Z, Li Q, Juul N, Leong CO, Calogrias D, Buraimoh A, et al. Efficacy of neoadjuvant cisplatin in triple-negative breast cancer. J Clin Oncol. 2010;28:1145–1153. doi: 10.1200/JCO.2009.22.4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Shaughnessy J, Weckstein DJ, Vukelja SJ, McIntyre K, Krekow L, Holmes FA, Asmar L, Blum JL. Preliminary results of a randomized phase II study of weekly irinotecan/carboplatin with or without cetuximab in patients with metastatic breast cancer; San Antonio Breast Cancer Symposium; 2007. abs 308. [Google Scholar]
- 36.Karn T, Pusztai L, Ruckhaberle E, Liedtke C, Muller V, Schmidt M, Metzler D, Wang J, Coombes KR, Gatje R, et al. Melanoma antigen family A identified by the bimodality index defines a subset of triple negative breast cancers as candidates for immune response augmentation. Eur J Cancer. 2012;48:12–23. doi: 10.1016/j.ejca.2011.06.025. [DOI] [PubMed] [Google Scholar]
- 37.Minuti G, Duchnowska R, Jassem J, Roncalli M, O'Brien T, Fabi A, Landi L, Di Marsico R, Biernat W, Czartoryska-Arlukowicz B, et al. MET and hepatocyte growth factor increased gene copy number is associated to trastuzumab failure in HER2 positive metastatic breast cancer; San Antonio Breast Cancer Symposium; 2011. P5-13-07. [Google Scholar]
- 38.Garrett JT, Arteaga CL. Resistance to HER2-directed antibodies and tyrosine kinase inhibitors: mechanisms and clinical implications. Cancer Biol Ther. 2011;11:793–800. doi: 10.4161/cbt.11.9.15045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gajria D, Chandarlapaty S. HER2-amplified breast cancer: mechanisms of trastuzumab resistance and novel targeted therapies. Expert Rev Anticancer Ther. 2011;11:263–275. doi: 10.1586/era.10.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khoury T, Kanehira K, Wang D, Ademuyiwa F, Mojica W, Cheney R, Morrison C, Conroy J, Nowak N, Liu S. Breast carcinoma with amplified HER2: a gene expression signature specific for trastuzumab resistance and poor prognosis. Mod Pathol. 2010;23:1364–1378. doi: 10.1038/modpathol.2010.125. [DOI] [PubMed] [Google Scholar]
- 41.Vegran F, Boidot R, Coudert B, Fumoleau P, Arnould L, Garnier J, Causeret S, Fraise J, Dembele D, Lizard-Nacol S. Gene expression profile and response to trastuzumab-docetaxel-based treatment in breast carcinoma. Br J Cancer. 2009;101:1357–1364. doi: 10.1038/sj.bjc.6605310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gianni L, Eiermann W, Semiglazov V, Manikhas A, Lluch A, Tjulandin S, Zambetti M, Vazquez F, Byakhow M, Lichinitser M, et al. Neoadjuvant chemotherapy with trastuzumab followed by adjuvant trastuzumab versus neoadjuvant chemotherapy alone, in patients with HER2-positive locally advanced breast cancer (the NOAH trial): a randomised controlled superiority trial with a parallel HER2-negative cohort. Lancet. 2010;375:377–384. doi: 10.1016/S0140-6736(09)61964-4. [DOI] [PubMed] [Google Scholar]
- 43.Knauer M, Cardoso F, Wesseling J, Bedard PL, Linn SC, Rutgers EJ, van't Veer LJ. Identification of a low-risk subgroup of HER-2-positivebreast cancer by the 70-gene prognosis signature. Br J Cancer. 2010;103:1788–1793. doi: 10.1038/sj.bjc.6605916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Staaf J, Ringner M, Vallon-Christersson J, Jonsson G, Bendahl PO, Holm K, Arason A, Gunnarsson H, Hegardt C, Agnarsson BA, et al. Identification of subtypes in human epidermal growth factor receptor 2-positive breast cancer reveals a gene signature prognostic of outcome. J Clin Oncol. 2010;28:1813–1820. doi: 10.1200/JCO.2009.22.8775. [DOI] [PubMed] [Google Scholar]
- 45.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 46.Prat A, Perou CM. Deconstructing the molecular portraits of breast cancer. Mol Oncol. 2011;5:5–23. doi: 10.1016/j.molonc.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheang MC, Voduc KD, Tu D, Jiang S, Leung S, Chia SK, Shepherd L, Levine MN, Pritchard KI, Davies SR, et al. Responsiveness of intrinsic subtypes to adjuvant anthracycline substitution in the NCIC-CTG MA.5 randomized trial. Clin Cancer Res. 2012;18:2402–2412. doi: 10.1158/1078-0432.CCR-11-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nielsen TO, Parker JS, Leung S, Voduc D, Ebbert M, Vickery T, Davies SR, Snider J, Stijleman IJ, Reed J, et al. A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor-positive breast cancer. Clin Cancer Res. 2010;16:5222–5232. doi: 10.1158/1078-0432.CCR-10-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weigelt B, Pusztai L, Ashworth A, Reis-Filho JS. Challenges translating breast cancer gene signatures into the clinic. Nat Rev Clin Oncol. 2012;9:58–64. doi: 10.1038/nrclinonc.2011.125. [DOI] [PubMed] [Google Scholar]
- 50.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, Fitzgibbons PL, Francis G, Goldstein NS, Hayes M, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer (unabridged version) Arch Pathol Lab Med. 2010;134:e48–e72. doi: 10.5858/134.7.e48. [DOI] [PubMed] [Google Scholar]
- 52.Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, Fitzgibbons PL, Francis G, Goldstein NS, Hayes M, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol. 2010;28:2784–2795. doi: 10.1200/JCO.2009.25.6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rhodes A, Jasani B, Barnes DM, Bobrow LG, Miller KD. Reliability of immunohistochemical demonstration of oestrogen receptors in routine practice: interlaboratory variance in the sensitivity of detection and evaluation of scoring systems. J Clin Pathol. 2000;53:125–130. doi: 10.1136/jcp.53.2.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Glas AM, Floore A, Delahaye LJ, Witteveen AT, Pover RC, Bakx N, Lahti-Domenici JS, Bruinsma TJ, Warmoes MO, Bernards R, et al. Converting a breast cancer microarray signature into a high-throughput diagnostic test. BMC Genomics. 2006;7:278. doi: 10.1186/1471-2164-7-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cronin M, Sangli C, Liu ML, Pho M, Dutta D, Nguyen A, Jeong J, Wu J, Langone KC, Watson D. Analytical validation of the Oncotype DX genomic diagnostic test for recurrence prognosis and therapeutic response prediction in node-negative, estrogen receptor-positive breast cancer. Clin Chem. 2007;53:1084–1091. doi: 10.1373/clinchem.2006.076497. [DOI] [PubMed] [Google Scholar]
- 56.Espinosa E, Sánchez Navarro I, Gámez Pozo A, Lamarca A, Pinto A, Ciruelos E, Fresno Vara JA. Desarrollo de un perfil de 5 genes con valor pronóstico en cáncer de mama con receptores hormonales negativos (A prognostic 5-gene score in hormonal receptor-negative breast cancer); XII Congreso de la Sociedad Española de Oncología Médica, Málaga; 2011. abs 37. [Google Scholar]
- 57.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, Massague J. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bos PD, Zhang XH, Nadal C, Shu W, Gomis RR, Nguyen DX, Minn AJ, van de Vijver MJ, Gerald WL, Foekens JA, et al. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459:1005–1009. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]