Abstract
Macrodactyly is a discrete congenital anomaly consisting of enlargement of all tissues localized to the terminal portions of a limb, typically within a ‘nerve territory’. The classic terminology for this condition is ‘lipofibromatous hamartoma of nerve’ or Type I macrodactyly. The peripheral nerve, itself, is enlarged both in circumference and in length. It is not related to neurofibromatosis (NF1), nor is it associated with vascular malformations, such as in the recently reported CLOVES syndrome. The specific nerve pathophysiology in this form of macrodactyly has not been well described and a genetic etiology for this specific form of enlargement is unknown. To identify the genetic cause of macrodactyly, we used whole-exome sequencing to identify somatic mutations present in the affected nerve of a single patient. We confirmed a novel mutation in PIK3CA (R115P) present in the patient's affected nerve tissue but not in blood DNA. Sequencing PIK3CA exons identified gain-of-function mutations (E542K, H1047L or H1047R) in the affected tissue of five additional unrelated patients; mutations were absent in blood DNA available from three patients. Immunocytochemistry confirmed AKT activation in cultured cells from the nerve of a macrodactyly patient. Additionally, we found that the most abnormal structure within the involved nerve in a macrodactylous digit is the perineurium, with additional secondary effects on the axon number and size. Thus, isolated congenital macrodactyly is caused by somatic activation of the PI3K/AKT cell-signaling pathway and is genetically and biochemically related to other overgrowth syndromes.
INTRODUCTION
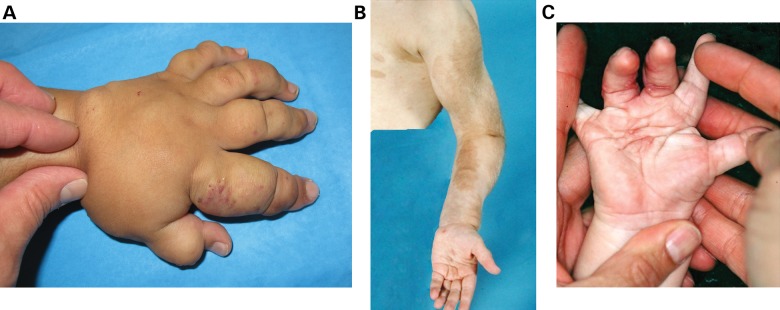
Congenital-isolated limb enlargement or overgrowth is rare, occurring in approximately 1 in 100 000 live births (1). The specific form studied in this report is the most common, or type I macrodactyly (Fig. 1A). It is a specific condition characterized by fibrofatty tissue enlargement and bony overgrowth with resultant loss of function due to overgrowth of a part of a limb. It is a benign condition, with no cases of malignant degeneration reported. Phenotypic heterogeneity has been observed in the anatomic extent of involvement (e.g. partial digit to entire hand or foot) and the rate of growth. Some involved digits may be abnormally large yet grow in proportion to the remainder of the limb. Other macrodactylous digits may demonstrate rapid growth with increasing disproportion to the uninvolved parts of the body. Type I macrodactyly classically affects a ‘nerve territory’ and the individual peripheral nerve is both enlarged and elongated (Fig. 1B). The treatment of macrodactyly to date has been ablative, consisting of debulking by excision of a portion of the involved digit, epiphysiodesis to stop longitudinal skeletal growth and amputation when the enlarged parts are no longer functional. Macrodactyly has been described as an unsolved condition by the International Federation of Societies for Surgery of the Hand.
Figure 1.
Macrodactyly causes disproportionate overgrowth of affected digits. (A) Patient with type I macrodactyly affecting the index and middle fingers shown at 5 months of age. (B) Macrodactyly affects a ‘nerve territory’, invariably affecting the median nerve of the affected hand.
In some cases, progression may extend proximally and include the entire limb, a condition known as macrodystrophia lipomatosa (2). Macrodystrophia lipomatosa, as clinically described by Ho et al. (2), has recently been linked to PIK3CA mosaicism (3). Unlike other syndromes with limb hypertrophy, such as Klippel–Trenaunay (Fig. 2A) and Parks–Webber syndromes, macrodactyly is not caused by increased vascular supply during development (2). In type I macrodactyly, the vessels are usually disproportionately small in the involved tissues. Compared with neurofibromatosis (NF1), congenital pseudoarthrosis and malignant degeneration do not occur in type I macrodactyly. In NF1, the enlargement is typically related to more proximal sensory nerves (rather than the distal nerves) and overgrowth of the digits is rare (Fig. 2B). Another peculiar condition that includes mildly enlarged syndactylized digits, though not in a nerve territory, and a predictable propensity for keloid scarring is clinically different from type I macrodactyly (4) (Fig. 2C).
Figure 2.
Overgrowth is apparent in disorders distinct from type I macrodactyly. (A) The left hand of an infant with Klippel–Trenaunay Syndrome (KT). KT is caused by increased vascularity causing severe overgrowth. Vascular markings are evident with soft tissue overgrowth characteristic of KT. (B) Overgrowth of the left arm of a patient with Neurofibromatosis type I (NF1). Overgrowth of the hand or macrodactyly is rarely seen in NF1 patients. (C) Infant with a keloid-forming overgrowth. This keloid-forming overgrowth is different from the overgrowth present in type I macrodactyly, which does not form keloid scars.
Next-generation sequencing provides a cost-effective method to generate a genome-wide sequence in modest patient populations. In particular, whole-exome capture and sequencing have proved to be a powerful method to identify new genes causing Mendelian disease. For rare and recessive disorders, this approach can identify causal mutations in a very few unrelated patients (5,6). Consequently, exome sequencing has enabled rapid identification of disease genes for many Mendelian disorders (5). Exome sequencing strategies are also proving effective for revealing causal mutations for non-Mendelian diseases such as schizophrenia (7,8), autism (9,10) and mental retardation (11) likely arising from de novo mutations (12). Similarly, comparing exome sequences of matched lesional tissue and blood samples has yielded somatic disease mutations causing neoplastic conditions, both malignant and benign. Somatic mosaic mutations in the AKT1 gene encoding the v-AKT protooncogene (AKT) were recently identified in patients with Proteus syndrome (13). We, therefore, elected to apply next-generation sequencing strategies to identify causal mutations in lesional tissues of type I macrodactyly patients. In the course of our study, similar investigations of other overgrowth syndromes identified somatic activating mutations that cluster in the PI3K-AKT signaling pathway (3,14–16).
We find that somatic mutations in PIK3CA in the affected nerve cause macrodactylous overgrowth. We also describe the pathology of lesional nerve tissue in macrodactyly. Our results support the concept that a highly localized overgrowth condition can arise from the same genetic underpinnings as generalized mosaic overgrowth syndromes, raising questions about the timing and specific tissue localization of these initial genetic events during development.
RESULTS
The proband (Patient 1) was a healthy 5-year-old girl who presented with enlarged index and middle fingers on one hand, with other digits being normal in size compared with the uninvolved hand. The enlarged digits were noticed at birth; however, the patient's medical history was otherwise unremarkable and no trauma was reported. No malformations caused by vascular abnormalities were noted. The patient was diagnosed with macrodactyly of the index and middle fingers and the growth was monitored during subsequent visits. She maintained the use of the digits; however, at ∼8 years of age, the affected digits were disproportionately larger and required surgical debulking, epiphysiodesis and osteotomy to correct the angular deformity of the digit.
We hypothesized that macrodactyly was caused by somatic mutations promoting cell growth. DNA from the nerve tissue of a macrodactylous digit was analyzed by whole-exome sequencing and compared with exome sequence of whole-blood DNA from the same patient. The exome sequence of nerve and blood samples provided a median 42- and 56-fold coverage across RefSeq genes, respectively (Supplementary Material, Figs. S1 and S2). Candidate gene analysis was performed with the hypothesis that macrodactyly is caused by rare nonsynonymous (NS) somatic mutations present in the affected peripheral nerve tissues and not in the germline. Exome sequencing identified 11 338 and 9495 NS single nucleotide variants (SNVs) in the affected nerve and germline samples, respectively.
NS sequence variants were excluded as disease candidates by their (a) presence in 28 control samples sequenced by our group using a similar method, (b) presence in the exome sequence of the germline sample and (c) presence in dbSNP 135. In order to fully exclude the variants present in the germline sample which were not identified during sequence analysis, we excluded the variants present in at least one read of the germline sample. Our analysis identified two candidate variants. The first was a SNV in UBXN11 (C392G). Computational analyses using PhastCons (17) and genomic evolutionary rate profiling (GERP) (18) produced weak scores (0.012 and −2.83, respectively) for this SNV, suggesting that UBXN11 was an unlikely candidate. The exome sequence depth (d) at this SNV was low in both the nerve (d = 12) and blood samples (d = 8), this variant was clustered with four additional SNVs within 14bp and all reads at this position were unidirectional in both the blood and tissue samples. As expected, Sanger sequencing both blood and nerve samples confirmed that the UBXN11 SNV was false positive in the nerve exome sequence alignment (data not shown). The second candidate mutation was a missense (R115P) SNV in PIK3CA. The mutation was present in 13 of 46 (28%) reads in the nerve exome and absent from all 45 reads in the germline exome (Supplementary Material, Figs. S3 and S4). Both the PhastCons (1.0) and GERP (5.52) scores suggested that this mutation was potentially deleterious. The PIK3CA mutation was not present in ∼6500 exomes available in the Exome Variant Server (http://evs.gs.washington.edu/EVS/). Sanger sequencing confirmed that the R115P mutation in PIK3CA was present in DNA from lesional tissue but not in blood of Patient 1 (Table 1 and Supplementary Material, Table S1).
Table 1.
PI3K mutations identified in type I macrodactyly patients
| Patient | PIK3CA | Somatic |
|---|---|---|
| Patient 1 | R115P | Y |
| Patient 2 | H1047L | N/A |
| Patient 3 | H1047R | Y |
| Patient 4 | E542K | Y |
| Patient 5 | E542K | Y |
| Patient 6 | H1047R | N/A |
The mutation in Patient 1 was identified by whole-exome sequencing; others were identified by Sanger sequencing exons 2, 10 and 21. DNA from whole blood was unavailable for Patients 2 and 6. N/A, not available.
Additional evidence suggested that the R115P mutation in PIK3CA (PI3K) was a likely candidate for macrodactyly. First, somatic activation of AKT, a downstream target of PI3K, was recently described in patients with Proteus syndrome (13). Second, another mutation of the same amino acid position (R115L) was annotated in the catalog of somatic mutations in cancer database from a squamous cell carcinoma (19,20), suggesting that mutations at p.Arg115 may be pathogenic.
To provide additional evidence for pathogenic PI3K somatic mutations in macrodactyly, exons 2, 10 and 21 were amplified from DNA extracted from the affected tissue of seven additional unrelated macrodactyly patients as well as nerve from an amputated polydactyly control digit and sequenced by the Sanger method. PIK3CA exons 10 and 21 encode the helical and kinase domains, respectively, and have been shown to be frequently mutated in cancer (21). Exon 2 was mutated in the proband. Of the seven additional macrodactyly patients, three known gain-of-function mutations were identified in five patients; no mutation was identified in the polydactyly control (Table 1 and Supplementary Material, Table S1). Complete PIK3CA sequencing in the two negative patients failed to identify mutations, possibly due to the limitations of Sanger sequencing in mixed cell populations. Alternatively, activating mutation in other genes of the PI3K-AKT-mTOR pathway may cause type I macrodactyly; genetic heterogeneity was observed among patients with megalencephaly–polymicrogyria–polydactyly–hydrocephalus syndrome resulting from the activation of the same pathway (16). Two macrodactyly patients shared somatic mutations (E542K) in the helical domain. Three patients had two different mutations at amino acid p.His1047 (H1047R & H1047L) in the kinase domain. The H1047R mutation was somatic in one patient; blood samples for comparison were unavailable for the remaining two patients. PI3K amino acids p.Glu542 and p.His1047 are located in the helical and kinase domains, respectively, and are known hotspots for somatic gain-of-function mutations in cancer (22,23). The R115P mutation identified by exome sequencing lies in a linker sequence between the adaptor-binding and the Ras-binding domains (24).
PIK3CA encodes the p110α catalytic subunit of the phosphoinositide-3-kinase heterodimer. Upon activation, PI3K phosphorylates phosphatidylinositol-4,5-bisphosphate (PIP2) at the third position, generating PIP3 (25). PIP3 molecules activate downstream signaling proteins, such as AKT and the pyruvate dehydrogenase kinase, isozyme 1 (PDK1) (26). Specifically, PIP3 binds the pleckstrin homology domain of AKT, resulting in a conformational change that allows PDK1 to phosphorylate Thr308 of AKT (27). AKT is also phosphorylated by the mammalian target of rapamycin complex 2 (mTORC2) at Ser473, resulting in greater AKT activation (28). PI3K mutations at amino acids p.Glu542 and p.His1047 increase intracellular AKT phosphorylation, which promotes numerous cellular processes including cell survival and proliferation (29). To test AKT activation in macrodactylous tissue, we performed immunocytochemistry using cultured cells derived from the affected nerve of Patient 6 who harbored a H1047R mutation in affected tissue. Compared with control, macrodactyly cells clearly showed increased staining, indicating greater levels of Ser473-phosphorylated AKT (Fig. 3).
Figure 3.
Immunocytochemistry reveals increased AKT activation in cultured cells from the macrodactylous nerve of Patient 6. (A and B) Negative (no antibody) immunocytochemistry of cells cultured from the nerves of patients with polydactyly (A) and macrodactyly (B). (C and D) Representative immunocytochemistry showing no significant increase of Ser473-phosphorylated AKT in cells from the nerve of a polydactylic digit (C) compared with a clear increase in cells from the macrodactylous nerve (D). Scale bars, 200 μm.
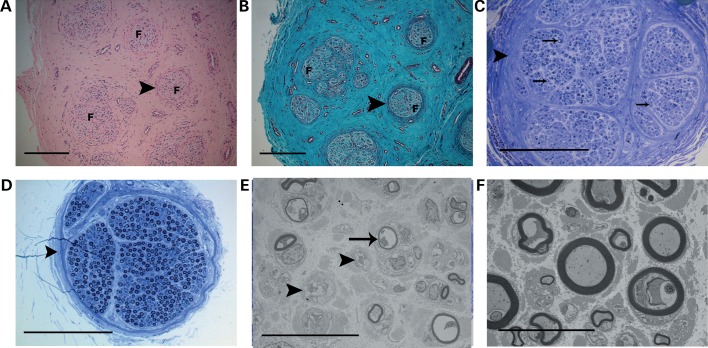
To further define the morphological changes in macrodactylous digits, resected nerve segments from Patient 6 were evaluated at the light microscopic and ultrastructural levels. Peripheral nerves from the macrodactylous digit showed separation of individual nerve fascicles by dense endoneurial connective tissue (Fig. 4A and B). Additionally, macrodactylous nerve fascicles had thickened perineurial sheaths. Nerve axons were also markedly different in the macrodactylous nerve fascicle compared with polydactyly control. The number of myelinated axons was markedly decreased in the macrodactylous nerve (Fig. 4C) compared with the nerve from a polydactyly specimen (Fig. 4D). Ultrastructural evaluation of the nerves showed a decrease in the number of myelinated axons; the remaining myelinated axons were generally smaller than those in the polydactyly control and were invested by disproportionately thinner myelin sheaths (Fig. 4E and F). The macrodactylous nerve also contained increased numbers of swollen, dissociated Schwann cell processes compared with control nerve, further indicative of axonal loss (Fig. 4E and F). It is possible that the axonal changes in the macrodactyly nerve reflect injury imposed by the dramatically increased amount of endoneurial connective tissue and interstitial fat, leading to changes similar to those encountered in chronic entrapment neuropathies (30).
Figure 4.
Perineurial overgrowth of the peripheral nerve from the macrodactylous digit of Patient 6. (A and B) Hematoxylin and eosin (A) and Gomori trichrome (B) stains of the paraffin section of peripheral nerve. Nerve fascicles (F) are separated by endoneurial connective tissue and have thickened, concentrically arrayed perineurium (arrowhead). Scale bars, 250 μm. (C and D) Toluidine blue-stained resin sections from the macrodactylous digit of Patient 6 (C) and a polydactylous control digit (D). Macrodactylous nerve has fewer myelinated axons (arrows) and thicker perineurial sheath (arrowhead). Scale bar, 250 μm. (E and F) Uranyl acetate-lead citrate-stained sections from the macrodactylous digit (E) and a polydactyly control (F). Macrodactylous nerve has fewer myelinated axons with disproportionately thinner myelin sheaths (arrow). The macrodactylous nerve also has an increased number of swollen Schwann cell processes (arrowhead). Scale bar, 20 μm.
DISCUSSION
Here, we report somatic activating mutations in PI3K in multiple patients with type I macrodactyly, a congenital overgrowth of peripheral nerve-supporting tissues of the digits. PI3K is an upstream regulator of the AKT-mTOR cell-signaling pathway. Upon activation by receptor tyrosine kinases, G-protein-coupled receptors or by Ras, PI3K phosphorylates PIP2 and the resultant product (PIP3) promotes the activation of AKT (31). The lipid kinase activity of PI3K is balanced by the phosphatase activity of PTEN, which dephosphorylates PIP3. Both activating mutations in PI3K and AKT and loss-of-function mutations in PTEN promote cell proliferation and are frequent in cancers. We identified somatic mutations at p.Glu542 and p.His1047 of PI3K in the lesional tissue of macrodactyly patients. Mutation at these positions increase PI3K activity and are frequent in cancer (21). Mutation of p.Glu542 disrupts binding at the N-SH2 domain of the p85 regulatory subunit, while mutation of p.His1047 likely causes a conformational change resulting in Ras-independent activation (32–34).
Activation of the PI3K-AKT cell-signaling axis has been recently linked to other overgrowth syndromes. Mosaic activating mutations have been described for Proteus syndrome (13), CLOVES syndrome (14), megalencephaly-capillary malformation (15,16) and fibroadipose hyperplasia (3). These overgrowth disorders are variably accompanied with other malformations, including musculoskeletal abnormalities (scoliosis, bone overgrowth) and vascular anomalies. However, unlike these other somatic overgrowth syndromes, macrodactyly is a localized gigantism without associated vasculopathy. Our patients presented with unilateral macrodactyly of the fingers and, to date, no other comorbidities have been observed. The extent to which somatic PI3K-AKT activation causes different congenital overgrowth syndromes is unclear.
The phenotypic variability of PI3K-AKT-associated overgrowth syndromes suggests that the timing of the mutational event during development as well as tissue specificity leads to distinctly different disease syndromes. Indeed, we noted that one patient (Patient 4) who harbored an E542K somatic mutation was diagnosed with overlap of both macrodactyly of the index and middle fingers as well as true muscular hemihypertrophy, which was not caused by pseudohypertrophy from adipose infiltration. Muscular hemihypertrophy is a congenital, typically unilateral, progressive overgrowth of muscle resulting in ectopic muscle formation in the affected limb and dramatically hypertrophic muscle tissue. Similar to the macrodactylous nerve tissue, we identified the patient's somatic PI3K mutation (E542K) in muscle tissue, suggesting that PI3K-AKT activation in this patient is the cause of both the nerve macrodactyly and muscle hemihypertrophy (Supplementary Material, Table S1).
Characterizing the degree of tissue involvement for each diagnosis will be important in defining any therapeutic potential of PI3K-AKT inhibitors, which are currently in clinical trials for the treatment of various cancers (35). Patients with localized overgrowth, such as in macrodactyly, may benefit from targeted monotherapy directed at PI3K specifically or the PI3K-AKT signaling pathway generally. It is worth noting that small molecule inhibitors that limit AKT activation are currently being investigated as a potential treatment for neuroblastoma (36). Rapamycin indirectly targets PI3K and also may be useful in treating macrodactyly. Oral administration of Rapamycin was effective in reducing the tissue overgrowth in patients with inactivating PTEN mutations (37,38).
We add type I macrodactyly to a growing list of overgrowth syndromes caused by hotspot mutations in PI3K which were previously thought to maintain oncogenic potential (39). Whether macrodactyly patients, as well as others with somatic PI3K-AKT-associated overgrowth disorders, are at greater risk of developing malignant neoplasms is at present unknown.
MATERIALS AND METHODS
Patient recruitment
This study was approved by the Research Advisory Panel of Texas Scottish Rite Hospital for Children and the Institutional Review Board of the University of Texas Southwestern Medical Center. A written informed consent was provided by all subjects. Blood and tissue samples were collected during surgical debulking procedures performed as standard of care for treatment of macrodactyly.
Whole-exome sequencing
DNA was extracted from whole blood and surgically removed ‘affected’ tissue using the QIAamp DNA Mini kit (Qiagen, Valencia, CA, USA). Whole-exome capture and sequencing were performed by the McDermott Center next-generation Sequencing Core facility at the University of Texas Southwestern Medical Center. Briefly, 1 μg of DNA was prepared using the TruSeq DNA sample preparation kit v2 (Illumina, San Diego, CA, USA), including sonication using a Covaris S2. Samples were ligated with barcoded adapters and subsequently purified and size selected with Ampure XP beads to an average size of ∼450 bp. Four barcoded samples were pooled and exome capture performed using the TruSeq Exome Enrichment kit (Illumina). The Truseq Exome kit targets ∼62 Mb of protein-coding and regulatory untranslated regions of the genome. Four samples were sequenced together on a single lane of an Illumina HiSeq 2000 using the paired-end sequencing protocol for 100 bp sequence fragments.
Sequence reads were aligned to the reference genome (b37) using the Burrows–Wheeler aligner (40). Alignments were processed as follows: (a) poorly mapped and low-quality reads were removed using Samtools (41), (b) duplicate reads were removed using Picard, (c) base-quality score recalibration and local re-alignment near indels were performed using the genome analysis toolkit (GATK) (42). Variants were identified using the GATK and Samtools and annotated using SeattleSeq Annotation 134 software (http://snp.gs.washington.edu/SeattleSeqAnnotation134/index.jsp).
Sanger sequencing
The individual coding sequence and flanking introns of PIK3CA (NM_006218.2) exons 2, 10 and 21 and UBXN11 (NM_183008.2) exon 16 were amplified by PCR from DNA extracted from the affected tissue and whole blood (if available). PCR primer sequences are provided in Supplementary Material, Table S2. Amplification products were treated with Exo-Sap-IT (Affymetrix, Santa Clara, CA, USA) and sequenced using Sanger sequencing. Individual chromatograms were manually inspected using FinchTV. The sequence peak for the H1047R mutation in nerve tissue from Patient 3 was small (Supplementary Material, Table S1); therefore, the amplification product was subcloned by conventional TA-cloning into vector pcDNA3.1/V5-His (Life Technologies). Sequencing individual clones confirmed the heterozygous H1047R mutation (data not shown).
Cell culture
Cells from the affected nerve tissue of Patient 6 were cultured in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum and 1% antibiotic (penicillin/streptomycin). The cultured cells from the nerve tissue rapidly reached confluence with spindle-shaped morphology, consistent with fibroblasts. DNA was extracted from the cultured cells using the QIAamp DNA Mini kit (Qiagen) and sequenced to confirm the presence of the PI3K H1047R mutation.
Immunocytochemistry
The cells were cultured into Collagen I 4-well Culture Slides (BD Biosciences, San Jose, CA, USA). The cells were fixed with 4% paraformaldehyde in PBS for 20 min at room temperature and washed with PBS. Universal IHC blocking solution was added for 30 min. Slides were incubated with a rabbit monoclonal antibody specific for Ser473-phosphorylated AKT (D9E) XP (1 : 50 dilution; Cell Signaling Technology, Danvers, MA, USA) overnight at 4°C and washed with PBS. Slides were blocked with a polymer penetration enhancer for 30 min, then washed with PBS. The cells were incubated with polyclonal HRP anti-mouse/rabbit IgG and washed with PBS. DAB solutions were applied to the slide and the cells counterstained with Mayer's hematoxylin for 2 min and washed in PBS. The cells were imaged using a DP70 Olympus microscope at ×200 magnification.
Paraffin histology
The segments of peripheral nerve designated for paraffin histology were fixed overnight in 10% neutral buffered formalin. Following fixation, the nerve segments were dehydrated in sequentially graded ethyl alcohol and xylene, embedded in paraffin and sectioned at 4 μm intervals in the transverse plane with a Leitz® rotary microtome. The sections were rehydrated through sequentially graded xylene and ethyl alcohol in water. The sequential sections were stained with hematoxylin and eosin stain, Gomori trichrome, Masson trichrome, Verhoeff van Giesen and periodic acid Schiff stains and evaluated by light microscopy.
Resin histology and electron microscopy
The segments of peripheral nerve were fixed overnight in 3% buffered glutaraldehyde. Following fixation, the nerve segments were washed in phosphate buffer, fixed for 1 h in 1% osmium tetroxide, dehydrated in graded ethyl alcohol and placed in propylene oxide for 10 min. The segments were taken through graded incubations of propylene oxide to pure resin and polymerized overnight at 65°C. Following resin embedding, the segments were sectioned in the transverse plane at 1.5 μm intervals on a Leica® Ultracut ultramicrotome, stained with toluidine blue and evaluated by light microscopy. For ultrastructural evaluation, resin blocks were sectioned in the transverse plane at 100 nm intervals, stained sequentially with uranyl acetate and lead citrate and examined with a Hitachi® 7500 transmission electron microscope at 100 kV. In addition to digital nerves harvested from macrodactylous specimens, peripheral nerve from digits of patients with polydactyly were prepared as controls.
SUPPLEMENTARY MATERIAL
FUNDING
This study was supported by Texas Scottish Rite Hospital for Children. Funding to pay the Open Access publication charges for this article was provided by Texas Scottish Rite Hospital for Children.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank L. Wheeler, J. Mills, V. Laws, K. Mauldin, J. Brandon, S. Shafer, D. Zhang, N. Kamiya, M. Romero-Ortega and the Texas Scottish Rite Tissue Bank Core Facility for technical assistance. The authors also thank H. Hobbs and J. Cohen for helpful discussion.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Flatt A. E. The Care of Congenital Hand Anomalies. St. Louis, Missouri: Quality Medical Publishing, Inc; 1994. [Google Scholar]
- 2.Ho C.A., Herring J.A., Ezaki M. Long-term follow-up of progressive macrodystrophia lipomatosa. A report of two cases. J. Bone Joint Surg. Am. 2007;89:1097–1102. doi: 10.2106/JBJS.F.01378. doi:10.2106/JBJS.F.01378. [DOI] [PubMed] [Google Scholar]
- 3.Lindhurst M.J., Parker V.E., Payne F., Sapp J.C., Rudge S., Harris J., Witkowski A.M., Zhang Q., Groeneveld M.P., Scott C.E., et al. Mosaic overgrowth with fibroadipose hyperplasia is caused by somatic activating mutations in PIK3CA. Nat. Genet. 2012;44:928–933. doi: 10.1038/ng.2332. doi:10.1038/ng.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muzaffar A.R., Rafols F., Masson J., Ezaki M., Carter P.R. Keloid formation after syndactyly reconstruction: associated conditions, prevalence, and preliminary report of a treatment method. J. Hand Surg. [Am.] 2004;29:201–208. doi: 10.1016/j.jhsa.2003.10.017. doi:10.1016/j.jhsa.2003.10.017. [DOI] [PubMed] [Google Scholar]
- 5.Bamshad M.J., Ng S.B., Bigham A.W., Tabor H.K., Emond M.J., Nickerson D.A., Shendure J. Exome sequencing as a tool for Mendelian disease gene discovery. Nat. Rev. Genet. 2011;12:745–755. doi: 10.1038/nrg3031. doi:10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- 6.Ng S.B., Buckingham K.J., Lee C., Bigham A.W., Tabor H.K., Dent K.M., Huff C.D., Shannon P.T., Jabs E.W., Nickerson D.A., et al. Exome sequencing identifies the cause of a Mendelian disorder. Nat. Genet. 2010;42:30–35. doi: 10.1038/ng.499. doi:10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu B., Roos J.L., Dexheimer P., Boone B., Plummer B., Levy S., Gogos J.A., Karayiorgou M. Exome sequencing supports a de novo mutational paradigm for schizophrenia. Nat. Genet. 2011;43:864–868. doi: 10.1038/ng.902. doi:10.1038/ng.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Girard S.L., Gauthier J., Noreau A., Xiong L., Zhou S., Jouan L., Dionne-Laporte A., Spiegelman D., Henrion E., Diallo O., et al. Increased exonic de novo mutation rate in individuals with schizophrenia. Nat. Genet. 2011;43:860–863. doi: 10.1038/ng.886. doi:10.1038/ng.886. [DOI] [PubMed] [Google Scholar]
- 9.Neale B.M., Kou Y., Liu L., Ma'ayan A., Samocha K.E., Sabo A., Lin C.F., Stevens C., Wang L.S., Makarov V., et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. doi:10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Roak B.J., Deriziotis P., Lee C., Vives L., Schwartz J.J., Girirajan S., Karakoc E., Mackenzie A.P., Ng S.B., Baker C., et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat. Genet. 2011;43:585–589. doi: 10.1038/ng.835. doi:10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vissers L.E., de Ligt J., Gilissen C., Janssen I., Steehouwer M., de Vries P., van Lier B., Arts P., Wieskamp N., del Rosario M., et al. A de novo paradigm for mental retardation. Nat. Genet. 2010;42:1109–1112. doi: 10.1038/ng.712. doi:10.1038/ng.712. [DOI] [PubMed] [Google Scholar]
- 12.Veltman J.A., Brunner H.G. De novo mutations in human genetic disease. Nat. Rev. Genet. 2012;13:565–575. doi: 10.1038/nrg3241. doi:10.1038/nrg3241. [DOI] [PubMed] [Google Scholar]
- 13.Lindhurst M.J., Sapp J.C., Teer J.K., Johnston J.J., Finn E.M., Peters K., Turner J., Cannons J.L., Bick D., Blakemore L., et al. A mosaic activating mutation in AKT1 associated with the Proteus syndrome. New Engl. J. Med. 2011;365:611–619. doi: 10.1056/NEJMoa1104017. doi:10.1056/NEJMoa1104017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kurek K.C., Luks V.L., Ayturk U.M., Alomari A.I., Fishman S.J., Spencer S.A., Mulliken J.B., Bowen M.E., Yamamoto G.L., Kozakewich H.P., et al. Somatic mosaic activating mutations in PIK3CA cause CLOVES syndrome. Am. J. Hum. Genet. 2012;90:1108–1115. doi: 10.1016/j.ajhg.2012.05.006. doi:10.1016/j.ajhg.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee J.H., Huynh M., Silhavy J.L., Kim S., Dixon-Salazar T., Heiberg A., Scott E., Bafna V., Hill K.J., Collazo A., et al. De novo somatic mutations in components of the PI3K-AKT3-mTOR pathway cause hemimegalencephaly. Nat. Genet. 2012;44:941–945. doi: 10.1038/ng.2329. doi:10.1038/ng.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Riviere J.B., Mirzaa G.M., O'Roak B.J., Beddaoui M., Alcantara D., Conway R.L., St-Onge J., Schwartzentruber J.A., Gripp K.W., Nikkel S.M., et al. De novo germline and postzygotic mutations in AKT3, PIK3R2 and PIK3CA cause a spectrum of related megalencephaly syndromes. Nat. Genet. 2012;44:934–940. doi: 10.1038/ng.2331. doi:10.1038/ng.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. doi:10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cooper G.M., Stone E.A., Asimenos G., Green E.D., Batzoglou S., Sidow A. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. doi:10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bamford S., Dawson E., Forbes S., Clements J., Pettett R., Dogan A., Flanagan A., Teague J., Futreal P.A., Stratton M.R., et al. The COSMIC (catalogue of somatic mutations in cancer) database and website. Brit. J. Cancer. 2004;91:355–358. doi: 10.1038/sj.bjc.6601894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stransky N., Egloff A.M., Tward A.D., Kostic A.D., Cibulskis K., Sivachenko A., Kryukov G.V., Lawrence M.S., Sougnez C., McKenna A., et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. doi:10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bader A.G., Kang S., Zhao L., Vogt P.K. Oncogenic PI3K deregulates transcription and translation. Nat. Rev. Cancer. 2005;5:921–929. doi: 10.1038/nrc1753. doi:10.1038/nrc1753. [DOI] [PubMed] [Google Scholar]
- 22.Gymnopoulos M., Elsliger M.A., Vogt P.K. Rare cancer-specific mutations in PIK3CA show gain of function. Proc. Natl Acad. Sci. USA. 2007;104:5569–5574. doi: 10.1073/pnas.0701005104. doi:10.1073/pnas.0701005104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Samuels Y., Wang Z., Bardelli A., Silliman N., Ptak J., Szabo S., Yan H., Gazdar A., Powell S.M., Riggins G.J., et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. doi:10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 24.Huang C.H., Mandelker D., Gabelli S.B., Amzel L.M. Insights into the oncogenic effects of PIK3CA mutations from the structure of p110alpha/p85alpha. Cell Cycle. 2008;7:1151–1156. doi: 10.4161/cc.7.9.5817. doi:10.4161/cc.7.9.5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vanhaesebroeck B., Leevers S.J., Ahmadi K., Timms J., Katso R., Driscoll P.C., Woscholski R., Parker P.J., Waterfield M.D. Synthesis and function of 3-phosphorylated inositol lipids. Annu. Rev. Biochem. 2001;70:535–602. doi: 10.1146/annurev.biochem.70.1.535. doi:10.1146/annurev.biochem.70.1.535. [DOI] [PubMed] [Google Scholar]
- 26.Hers I., Vincent E.E., Tavare J.M. Akt signalling in health and disease. Cell Signal. 2011;23:1515–1527. doi: 10.1016/j.cellsig.2011.05.004. doi:10.1016/j.cellsig.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 27.Calleja V., Alcor D., Laguerre M., Park J., Vojnovic B., Hemmings B.A., Downward J., Parker P.J., Larijani B. Intramolecular and intermolecular interactions of protein kinase B define its activation in vivo. PLOS Biol. 2007;5:e95. doi: 10.1371/journal.pbio.0050095. doi:10.1371/journal.pbio.0050095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alessi D.R., Andjelkovic M., Caudwell B., Cron P., Morrice N., Cohen P., Hemmings B.A. Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 1996;15:6541–6551. [PMC free article] [PubMed] [Google Scholar]
- 29.Ikenoue T., Kanai F., Hikiba Y., Obata T., Tanaka Y., Imamura J., Ohta M., Jazag A., Guleng B., Tateishi K., et al. Functional analysis of PIK3CA gene mutations in human colorectal cancer. Cancer Res. 2005;65:4562–4567. doi: 10.1158/0008-5472.CAN-04-4114. doi:10.1158/0008-5472.CAN-04-4114. [DOI] [PubMed] [Google Scholar]
- 30.Rempel D.M., Diao E. Entrapment neuropathies: pathophysiology and pathogenesis. J. Electromyogr. Kines. 2004;14:71–75. doi: 10.1016/j.jelekin.2003.09.009. doi:10.1016/j.jelekin.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 31.Zhao L., Vogt P.K. Class I PI3K in oncogenic cellular transformation. Oncogene. 2008;27:5486–5496. doi: 10.1038/onc.2008.244. doi:10.1038/onc.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang C.H., Mandelker D., Schmidt-Kittler O., Samuels Y., Velculescu V.E., Kinzler K.W., Vogelstein B., Gabelli S.B., Amzel L.M. The structure of a human p110alpha/p85alpha complex elucidates the effects of oncogenic PI3Kalpha mutations. Science. 2007;318:1744–1748. doi: 10.1126/science.1150799. doi:10.1126/science.1150799. [DOI] [PubMed] [Google Scholar]
- 33.Miled N., Yan Y., Hon W.C., Perisic O., Zvelebil M., Inbar Y., Schneidman-Duhovny D., Wolfson H.J., Backer J.M., Williams R.L. Mechanism of two classes of cancer mutations in the phosphoinositide-3-kinase catalytic subunit. Science. 2007;317:239–242. doi: 10.1126/science.1135394. doi:10.1126/science.1135394. [DOI] [PubMed] [Google Scholar]
- 34.Zhao L., Vogt P.K. Helical domain and kinase domain mutations in p110alpha of phosphatidylinositol 3-kinase induce gain of function by different mechanisms. Proc. Natl Acad. Sci. USA. 2008;105:2652–2657. doi: 10.1073/pnas.0712169105. doi:10.1073/pnas.0712169105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holmes D. PI3K pathway inhibitors approach junction. Nat. Rev. Drug Discov. 2011;10:563–564. doi: 10.1038/nrd3527. doi:10.1038/nrd3527. [DOI] [PubMed] [Google Scholar]
- 36.Segerstrom L., Baryawno N., Sveinbjornsson B., Wickstrom M., Elfman L., Kogner P., Johnsen J.I. Effects of small molecule inhibitors of PI3K/Akt/mTOR signaling on neuroblastoma growth in vitro and in vivo. Int. J. Cancer. 2011;129:2958–2965. doi: 10.1002/ijc.26268. doi:10.1002/ijc.26268. [DOI] [PubMed] [Google Scholar]
- 37.Iacobas I., Burrows P.E., Adams D.M., Sutton V.R., Hollier L.H., Chintagumpala M.M. Oral rapamycin in the treatment of patients with hamartoma syndromes and PTEN mutation. Pediatr. Blood Cancer. 2011;57:321–323. doi: 10.1002/pbc.23098. doi:10.1002/pbc.23098. [DOI] [PubMed] [Google Scholar]
- 38.Marsh D.J., Trahair T.N., Martin J.L., Chee W.Y., Walker J., Kirk E.P., Baxter R.C., Marshall G.M. Rapamycin treatment for a child with germline PTEN mutation. Nat. Clin. Pract. Oncol. 2008;5:357–361. doi: 10.1038/ncponc1112. [DOI] [PubMed] [Google Scholar]
- 39.Bader A.G., Kang S., Vogt P.K. Cancer-specific mutations in PIK3CA are oncogenic in vivo. Proc. Natl Acad. Sci. USA. 2006;103:1475–1479. doi: 10.1073/pnas.0510857103. doi:10.1073/pnas.0510857103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H., Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. doi:10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. doi:10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. doi:10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.