Abstract
A major drawback of mechanical and prosthetic heart valves is their inability to permit somatic growth. By contrast, tissue-engineered pulmonary valves potentially have the capacity to remodel and integrate with the patient. For this purpose, adult stem cells may be suitable. Previously, human periodontal ligament cells (PDLs) have been explored as a reliable and robust progenitor cell source for cardiac muscle regeneration (Pelaez, D. Electronic Thesis and Dissertation Database, Coral Gables, FL, May 2011). Here, we investigate the potential of PDLs to support the valve lineage, specifically the concomitant differentiation to both endothelial cell (EC) and smooth muscle cell (SMC) types. We were able to successfully promote PDL differentiation to both SMC and EC phenotypes through a combination of stimulatory approaches using biochemical and mechanical flow conditioning (steady shear stress of 1 dyne/cm2), with flow-based mechanical conditioning having a predominant effect on PDL differentiation, particularly to ECs; in addition, strong expression of the marker FZD2 and an absence of the marker MLC1F point toward a unique manifestation of smooth muscle by PDLs after undergoing steady-flow mechanical conditioning alone, possible by only the heart valve and pericardium phenotypes. It was also determined that steady flow (which was performed using a physiologically relevant [for heart valves] magnitude of ∼5–6 dynes/cm2) augmented the synthesis of the extracellular matrix collagen proteins. We conclude that under steady-flow dynamic culture environments, human PDLs can differentiate to heterogeneous cell populations that are relevant to heart valve tissue engineering. Further exploration of human PDLs for this purpose is thus warranted.
Introduction
Several investigations have shown the potential of adult stem cells for treating vascular injury and disease through tissue engineering and regenerative medicine.1–4 In many cases, progenitor cells are taken from the bone marrow. The Mayer group5 found promising results, creating pulmonary valve leaflets and sections of the main pulmonary artery utilizing bone marrow-derived stem cells (BMSCs). These tissue-engineered heart valves (TEHVs) were nonthrombogenic, promote tissue remodeling, and were found to be durable at the time of explant, 4–8 months after implantation.5 Their sustained functionality during that timeframe provided a positive outlook for TEHV-related research. Yet, the current critical challenges that still remain are to identify a clinically viable autologous cell source, a scaffold with the suitable mechanical and biodegradable properties, an optimized in vitro conditioning system, and a way to track the functional stability of the TEHV implant to successfully move forward from in vitro to in vivo studies, and subsequently to clinical trials.6
While a variety of clinically relevant primary cells and stem cells continue to be investigated in the context of TEHVs, one promising source that has received less attention is periodontal ligament cells (PDLs), which consist of a heterogeneous population of cell types, including cells of mesenchymal origin.7 Clinically, PDLs can be obtained from adult wisdom teeth; developmentally, they derive from the fetal cranial neural crest.8 The primary motivation for using PDLs in tissue engineering is that these cells express a number of important pluripotent stem cell markers (Nanog, Oct4, Sox2, Klf4, SSEA-1, and SSEA-4),8,9 suggestive of enhanced pluripotency over other adult stem cells, and a comparable differentiative capacity to that of embryonic stem cells (ESCs). Moreover, they have demonstrated a strong affinity to differentiate toward cardiovascular cell lineages such as cardiomyocytes.10 In this context, it would seem plausible that another application of PDLs would be in heart valve tissue engineering, in particular owing to their ability to potentially support both the endothelial cell (EC) and smooth muscle cell (SMC) phenotypes,8,11,12 which are innate to heart valves. In addition, it should be pointed out that roughly 30% of the cellular make-up of native heart valves consists of cells that are interstitial in nature, and it is these cells that facilitate continuous remodeling activity to occur throughout adulthood, a critical component to healthy valve functioning.13 From this perspective, PDLs are attractive for heart valve tissue engineering, because they, in theory, possess the necessary embryonic markers to promote continuous valve remodeling, yet are not actually embryonic cells, therefore exempting them from moral, ethical, or legal considerations.
It is known that several growth factors, such as the vascular endothelial growth factor (VEGF), and mechanical stimulation, including fluid-induced shear stresses, support the endothelial phenotype14–17 and result in improvement of the physical integrity of engineered heart valves before implantation.5,6,18–20 Owing to their ESC-like characteristics, availability as a practical, single cell source, and given that these cells can be easily culture expanded, the purpose of this study was to evaluate the utility of PDLs for heart valve tissue engineering under in vitro, dynamic culture environments. Specifically, we combined the use of biochemical stimulants and steady flow-based mechanical conditioning to effectively differentiate PDLs into the EC and SMC phenotypes. Additionally, we evaluated synthesis of the extracellular matrix collagen by the PDL after exposure to steady-flow mechanical conditioning.
Methods
Cell culture
SMCs and ECs
Human vascular cells (pulmonary artery SMCs and ECs; Fisher Scientific) were cultured and expanded under a regular basal medium containing Dulbecco's Modified Eagle Medium (DMEM) high glucose (Invitrogen), 10% fetal bovine serum (FBS; ATCC), and 1% penicillin/streptomycin (ATCC).
Periodontal ligament cells
Periodontal ligament-derived stem cells were isolated as described previously.8 Briefly, human periodontal ligaments were harvested from impacted wisdom teeth that were collected from patients (age <25-years old) at the Dental Clinic of the Miami VA Medical Center with their informed consent, according to our approved institutional review board (IRB) protocols. The periodontal ligaments from different teeth of the same donor were pooled and finely chopped, and cells were released by overnight digestion at 37°C in a high-glucose DMEM supplemented with 10% FBS, 1% antibiotic–antimycotic, 400 U/mL collagenase type I (Worthington Biochemical Corp.), and 1% trypsin/ethylenediaminetetraacetic acid (EDTA) solution (Invitrogen).10 Single-cell suspensions were then obtained by passing the resulting digestion through a 40-μm cell strainer (BD Biosciences). The cells were culture-expanded under a regular basal medium containing DMEM high glucose, 10% FBS, 1% penicillin/streptomycin (ATCC). Note that FBS was incorporated in the digestion protocol due to previously obtained low viability of the cells from overnight digestion without serum. We observed that the inclusion of FBS significantly increased the yield of viable cells recovered from digestion. Since 2009, 8 cell lines with the characteristic phenotypic expression have been isolated using the 10% FBS digestion protocol.8 As to the potency of trypsin, the inactivation of trypsin by serum is specifically caused by the alpha-1-antitrypsin (A1AT) protease inhibitor; however, the FBS that was employed here was heat inactivated, which would denature all enzymatic protease inhibitors, including A1AT. For this reason, we do not anticipate that the FBS utilized for our work would permanently inhibit the trypsin activity. In addition, even if trypsin activity was partially compromised by FBS, the intended use of trypsin/EDTA herein was not intended for the direct digestion of the tissue, but rather for the increase in the yield of single cells in the final suspension. Based on our experience with this digestion, the use of collagenase alone yields sheets or clumps of cells that are extremely difficult to get into adherent cultures. The addition of trypsin/EDTA renders a much higher yield of single, colony-forming, and adherent cells.
Cell differentiation
After PDLs had reached ∼95% confluency, they were separated into four groups, cultured for a period of 1 week, and were categorized as follows at a cell density of 1×106 cells/cm2 per group. Group 1—negative control: PDLs were cultured in a basal medium containing DMEM high glucose, 10% FBS, and 1% penicillin/streptomycin. Group 2–differentiating medium: PDLs were exposed to a cocktail medium formulation (hereafter referred to as the cocktail-differentiating medium) believed to induce SMC and EC differentiation as a similar version of the mixture had been effective in differentiating PDLs into cardiomyocytes.8,10 The cocktail-differentiating medium contains the regular basal medium, as well as 10 ng/mL of vascular endothelial growth factor (VEGF) (BD Biosciences), 25 ng/mL of basic fibroblast growth factor (bFGF) (Invitrogen), 10 ng/mL of insulin-like growth factor (Invitrogen), 10 μM of 5-azacytidine (Sigma-Aldrich), 100 nM ascorbic acid (Sigma-Aldrich), 10 nM oxytocin (Sigma-Aldrich), and 10 nM hydrogen peroxide (Sigma-Aldrich). The cocktail-differentiating medium was changed every day during the 7-day period, with the exception of the hydrogen peroxide, which was added daily. Group 3–steady flow only: PDLs were subjected to a steady shear flow (1 dyne/cm2) while being cultured dynamically (Bioflux200 system; Fluxion Biosciences) (Fig. 1) under a regular basal medium. Group 4–cocktail-differentiating medium and steady flow: this group combined the cocktail medium and the steady shear flow (1 dyne/cm2, Bioflux200 system; Fluxion Biosciences). As a positive control, both human pulmonary artery ECs and SMCs were cultured separately using a regular basal medium.
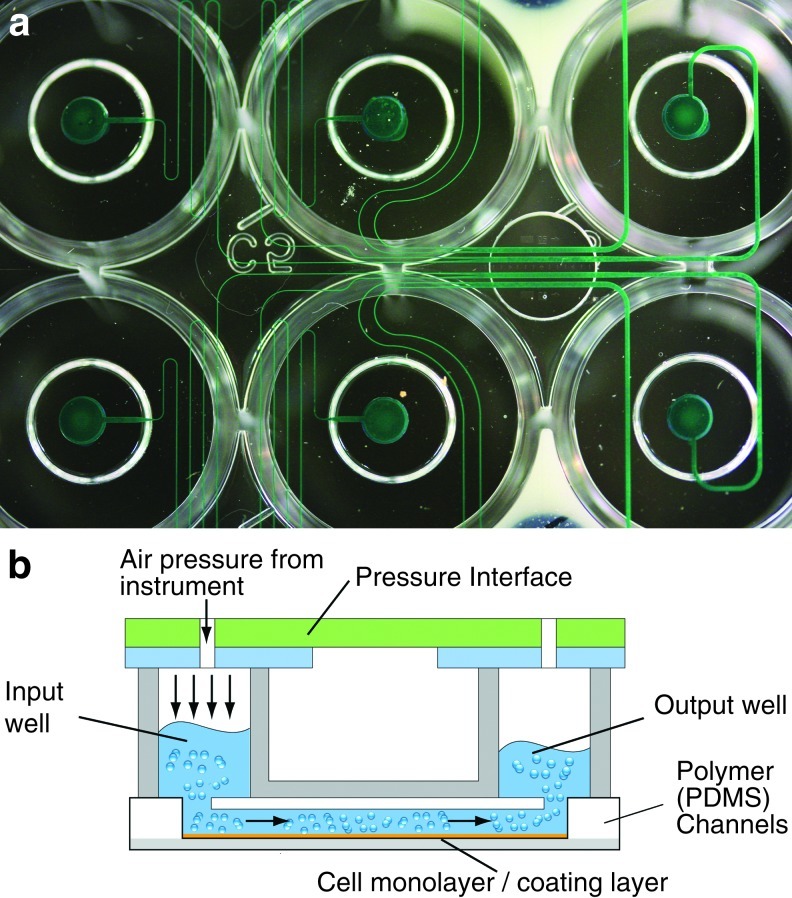
FIG. 1.
(a) Microfluidic component of a shear stress cell assay system (Bioflux200 system; Fluxion Biosciences), which permitted steady-flow conditioning of PDL monolayers. (b) Medium originating from an input well is made to flow continuously over the cell monolayer using a pneumatic pump controller, and which eventually exits to an output well. A precoating of the channel surface with a gel material (rat tail tendon collagen; Roche Applied Science) ensured that the cells were strongly adhered (figures presented here with permission from Fluxion Biosciences). PDL, periodontal ligament. Color images available online at www.liebertpub.com/tea
RNA isolation, reverse transcription, and quantitative real time-polymerase chain reaction
After a 1-week period of culturing cells under the differentiating medium, cells from the four separate groups were subjected to quantitative real time-polymerase chain reaction (qRT-PCR) as previously described21 to determine differentiation to the SMC and EC cell phenotypes. The cells from all groups were rinsed with PBS and detached from the T-75-cm2 flasks (Fisher Scientific) by 0.25% trypsin/EDTA. Cell pellets were collected in a 15-mL conical tube (Fisher Scientific). The pellet was transferred to a 1.5-μL RNase-free tube to perform the RNA isolation procedure. mRNA was purified with the SV Total RNA Isolation System (Promega). A weight of 1 μg of total mRNA was used for reverse transcription reaction with the GoScript™ Reverse Transcription System (Promega). The cDNA was synthesized using an oligo(dT)15 primer, according to the manufacturer's instructions.
Quantitative RT-PCR was performed using the GoTaq® qPCR Master Mix (Promega). Signals were detected with a Step-One Real-Time PCR System (Applied Biosystems). The PCR tube contained two primers and SYBR® green I dye reagent along with the cDNA, a product of reverse transcription mentioned above. The primers (Table 1) were designed to amplify the target sequence, with a majority of sequences obtained using the Basic Local Alignment Search Tool (BLAST) program, National Center for Biotechnology Information (NCBI), except for the markers FZD2 and MLC1F, which were obtained from Brand et al.22 The sample tube was held at 95°C, 2 min before the cycle started to activate the Taq polymerase. The cycling parameters were 95°C, 5 s; 60°C, 45 s; 95°C, 15 s. In this manner, the relative expression of a number of ESC, SMC, EC, osteogenic, chondrogenic, and collagen types 1 and 3 markers were determined for each of the four groups defined earlier (in the Cell differentiation section).
Table 1.
Quantitative RT-Polymerase Chain Reaction Primer Sequences Utilized in This Study
| Gene | Forward primers (5'-3') | Reverse primers (5'-3') |
|---|---|---|
| GAPDH | AATGAAGGGGTCATTGATGG | AAGGTGAAGGTCGGAGTCAA |
| Oct4 | CTCCTGAAGCAGAAGAGGATCAC | CTTCTGGCGCCGGTTACAGAACCA |
| SOX2 | TGCAGTACAACTCCATGACCA | GTGCTGGGACATGTGAAGTCT |
| NANOG | GTCTTCTGCTGAGATGC | AGTTGTTTTTCTGCCACC |
| KLF4 | ACGATCGTGGCCCCGGAAAAGGAC | TGATTGTAGTGCTTTCTGGTGGGCTCC |
| nestin | GCCCTGACCACTCCAGTTTA | GGAGTCCTGGATTTCCTTCC |
| SLUG | TGCTACACAGCAGCCAGATTCC | TTTCTGGGCTGGCCAAACAT |
| p75 | CTGCAAGCAGAACAAGCAAG | GGCCTCATGGGTAAAGGAGT |
| SOX10 | TCTTGTAGTGGGCCTGGATGG | TGAACGCCTTCATGGTGTGG |
| Integrin-β1 | CCTACTTCTGCACGATGTGATG | CCTTTGCTACGGTTGGTTACATT |
| THY1 | TCGCTCTCCTGCTAACAGTCT | CTCGTACTGGATGGGTGAACT |
| PECAM1 | CCAAGGTGGGATCGTGAGG | TCGGAAGGATAAAACGCGGTC |
| FLK1 | GGCCCAATAATCAGAGTGGCA | CCAGTGTCATTTCCGATCACTTT |
| VE-cadherin | GATCAAGTCAAGCGTGAGTCG | AGCCTCTCAATGGCGAACAC |
| TIE-1 | TCGAGCGGCATCTACAGTG | GCACGATGAGCCGAAAGAAG |
| FZD2 | CGGCCCCGCAGCGCCCTGCCC | ACACGAACCCAGGAGGACGCAGGCC |
| MLC1F | GAGTTCTCTAAGGAACAGCAGG | CTGCGTGTCTTTGACAAGGAAGGCAATGG |
| ALP | GTTGCCAAGCTGGGAAGAACAC | CCCACCCCGCTATTCCAAAAC |
| osteocalcin | CACTCCTCGCCCTATTGGC | CCCTCCTGCTTGGACACACAAAG |
| COL1A1 | TACCATGACCGAGACGTGTG | ATAAGACAGCTGGGGAGCAA |
| COL2A1 | AGACTTGCGTCTACCCCAATC | GCAGGCGTAGGAAGGTCATC |
| COL3A1 | GACATCGAGGATTCCCTGGT | CCAATCCCAGCAATGGCAG |
All sequences were obtained using the BLAST program, National Center for Biotechnology Information (NCBI), except for the markers FZD2 and MLC1F, which were obtained from Brand et al.22
RT, real time.
The change in cycle threshold (ΔCt) values were averaged and normalized with GAPDH using the ΔΔCt method.21 Fold changes were calculated as 2−ΔCt, and the gene expression ratio of each of the four groups (negative control, differentiating medium, BioFlux, and differentiating medium and BioFlux) was plotted (Fig. 2).
FIG. 2.
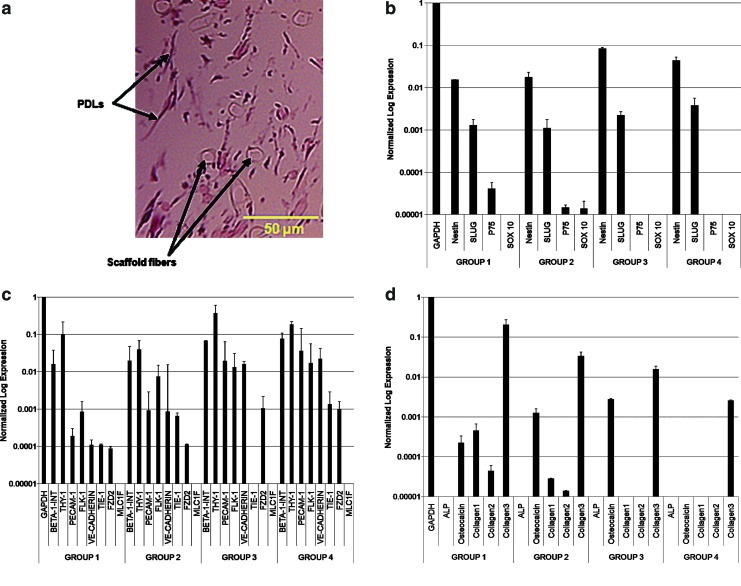
(a) Hematoxylin and eosin (H&E)-stained section of PDLs seeded in a nonwoven 50:50 blend of polyglycolic acid–poly-l-lactic acid scaffold showing the distribution of cells within the three-dimensional culture environment. (b) qRT-PCR-derived expression (average±standard error of the mean [SEM]) of PDLs for assessment of embryonic stem cell markers. Group 1: Cells cultured in regular basal medium. Group 2: Cells cultured in differentiating medium. Group 3: Cell cultured in basal medium and exposed to 1 dyne/cm2. Group 4: Cells cultured in differentiating medium and exposed to 1 dyne/cm2. Each column is normalized by GAPDH. (c) Same analysis as part (b) repeated for smooth muscle cell and endothelial cell markers. (d) Same analysis as part (b) repeated for osteogenic, chondrogenic, and collagen type 1 and 3 markers. The initial sample size was an n=4 wells of standard-sized 8-well plate/group. However, to enable sufficient detection limits for qRT-PCR analysis, specimens were pooled into an n=2–3 samples/group with an r=2–3 measurements/sample. qRT-PCR, quantitative real-time-polymerase chain reaction. Color images available online at www.liebertpub.com/tea
Cell seeding
A nonwoven 50:50 blend of polyglycolic acid (PGA) and poly-l-lactic acid (PLLA) scaffold (Biofelt) was utilized for cell seeding.5 The scaffold has an approximate fiber diameter of 0.012–0.015 mm and a density of 61.75 mg/mL. The PGA:PLLA scaffold was previously utilized in tissue engineering experiments that demonstrated rapid cell growth and an organized three-dimensional (3D) tissue structure.6 Rectangular scaffold samples (n=12) of dimensions (17×6×1 mm) were subjected to ethylene oxide gas sterilization (AN 306, Anprolene; Andersen Products, Inc.) for 12 h. After the sterilization period, each sample was placed in a hybridization tube and seeded with PDLs at a density of 2×106 million cells per cm2. The hybridization tubes (n=12) were then placed on a rotisserie (Fisher Scientific) and rotated at 8 rpm inside a standard cell culture incubator operating at 37°C and 5% CO2 for 1 day with an addition of 7 days (static culture). The basal medium was used identical to the medium described for Group 1 in the cell differentiation studies.
Tissue engineering experiments
The steady-flow mechanical conditioning for the seeded scaffolds was performed in an U-shaped bioreactor.6,23–28 This bioreactor is capable of replicating physiological hemodynamic conditions during in vitro tissue development, which may play an essential role in engineered heart valve tissue formation.24,25 After culturing of the scaffolds (n=6) statically for a total of 8 days, they were separated into two groups: static and flow. Scaffolds (n=3/group) were then cultured for a 2-week period in static and dynamic conditions (22 days of total culture time). Dynamic culture experiments of specimens housed in the U-shaped bioreactor were subjected to a continuous flow rate (850 mL/min), which provided physiologically relevant shear stress magnitudes for aortic heart valves, in the upper limit range of ∼5–6 dynes/cm2.26 All experiments utilized only the basal medium described for Group 1 in the cell differentiation studies.
Histology
After 22 days of tissue culture, the PDL-seeded 50:50 nonwoven blend of PGA:PLLA scaffolds (17×6×1 mm) were fixed in 10% formalin for 24 h. The samples were then dehydrated, paraffin-embedded, and cut into 5-mm-thick serial slices. Sections were subsequently stained with hematoxylin and eosin for morphology as previously described.6,29
DNA quantification
DNA was quantified after 22 days of culture (n=3 specimens/group) as described previously.6 In brief, for each assay, thin samples (8×7×1 mm) were cut from PDL-seeded scaffolds (controls and scaffolds exposed to flow) along their long axis and weighed before extraction. Each sample was then placed in a microcentrifuge tube and extracted in 1 mL of 0.125 mg/mL papain solution for 10 h in a 60°C water bath. The 0.125 mg/mL papain solution must made immediately before use by adding l-cysteine dihydrochloride (Sigma-Aldrich) to a phosphate-buffered EDTA solution (PBE; Sigma-Aldrich) to a concentration of 10 mM, adding papain (minimum 10 units/mg [P4762]; Sigma-Aldrich) to a concentration of 0.125 mg/mL. The PBE solution was made beforehand by adding sodium phosphate dibasic (Sigma-Aldrich) and EDTA (Sigma-Aldrich) to deionized water at concentrations of 100 and 10 mM, respectively. The PBE solution was balanced to pH 6.5 with 0.5 N hydrochloric acid (Sigma-Aldrich) and sterile filtered using a vacuum filtration unit (0.2-mm polyethersulphone (PES) membrane; Nalgene Labware). The extracts were then assayed using the PicoGreen double strand DNA quantitation kit (Invitrogen) as per the manufacturer's instructions and using a Synergy HT Microplate Reader (Biotek).
Extracellular matrix quantification
Collagen and sulfated glycosaminoglycans (S-GAGs) were assayed (n=3 specimens/group) following a similar technique previously used.6 For the collagen assay, thin samples (8×7×1 mm) were cut from PDL-seeded scaffolds (controls and scaffolds exposed to flow) along their long axis and weighed before extraction. Total collagen was extracted from samples using a solution of 0.5 M acetic acid (Sigma-Aldrich) and pepsin (1 mg/mL; Sigma-Aldrich). Each sample was placed in a microcentrifuge tube and incubated in 1 mL of extraction solution on an orbital shaker (Labquake; Thermo Scientific) at 4°C for 16 h. For the S-GAG assay, the same extract used for the DNA assay was utilized. Subsequent to the extraction procedure, the collagen and S-GAG extracts were assayed following the protocols of the manufacturer provided by the Sircol and Blyscan assay kits. (Biocolor Ltd.) using a Synergy HT Microplate Reader (Biotek).
Statistical analysis
Statistical analysis was performed utilizing the statistical package for the social science (SPSS) Software Version 16.0 (IBM, Armonk, NY) for the independent t-tests used to assess significance levels between the different groups in the collagen, S-GAG, and DNA assays (n=3 samples/group). A statistically significant difference between groups was interpreted to occur at a probability value of p<0.05 in all comparisons made.
Results
Cell morphology and phenotype
After 22 days of tissue culture, human PDLs in an elongated morphology (roughly 30 μm in length) were observed to be spatially dispersed (Fig. 2a) among eosinophilic structures and among the presence of the PGA:PLLA scaffold fibers (typical fiber diameter ∼12 μm).
It was determined that the highest level of expression for both SMC (beta-1-integrin and Thy1) and EC (PECAM1, FLK-1, and VE-cadherin) genes was produced from RNA extracted from Groups 3 and 4, that is, when PDLs were exposed to a flow component in the BioFlux system at a steady shear stress of 1 dyne/cm2 (Fig. 2b). Other markers that were strongly expressed after steady flow conditioning (Group 3 versus Groups 1 and 2) was introduced were Sox2, KLF4, FZD2, and COL3A1, while the embryonic marker, Oct4, was only highly expressed in Group 4 (Fig. 2b, c). A number of additional markers (nestin, MLC1F, COL1A1, COL2A1, and ALP) were either negligible or absent in all the groups tested (Fig. 2d).
DNA and extracellular matrix quantification for PDL-seeded scaffolds
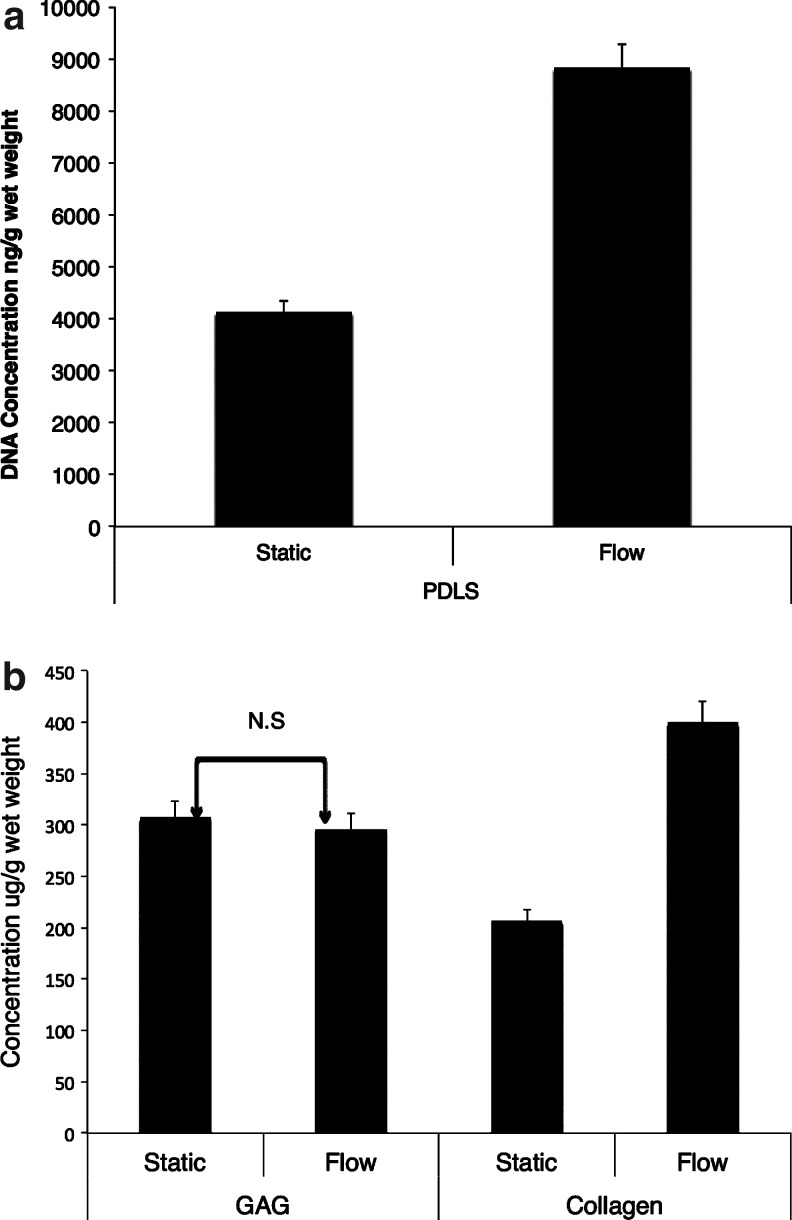
DNA was quantified in the same way as for the SMC-EC-seeded scaffolds. A seeding density of 2×106 cells/scaffold was utilized. Based on a reported value of 7.6 pg of DNA content/cell,6 there was an average initial DNA content of 13.68 μg/scaffold. After 22 days of tissue culture, the DNA concentration decreased to 4130 ng/g (69.8%) wet weight in the static group and to 8830 ng/g wet weight (35.5%) in the group exposed to steady flow (Fig. 3a); these differences were found to be significant (p<0.05). At the conclusion of 22 days, the PDL-seeded scaffolds exposed to flow were found to have a collagen concentration of 400.2±13.2 μg/g wet weight compared with 207.8±10.2 μg/g wet weight in the static group (n=3) (Fig. 3b). This amounted to an ∼93% increase in collagen content in the flow group (p<0.05). On the other hand, the S-GAG concentration was found to be 296.0±15.6 μg/g wet weight in the group exposed to flow, compared to 307.5±15.6 μg/g wet weight in the control group. This represented a 3.7% decrease in the GAG production from the scaffolds exposed to flow in comparison to the static controls, which was not found to be significant (p>0.05).
FIG. 3.
(a) DNA and (b) extracellular matrix (glycosaminoglycan [GAG] and collagen) quantification of PDL-seeded scaffolds after 22 days of incubation in basal medium. Flow group was subjected to ∼5–6 dynes/cm2 over 14 days, following 8 days of static incubation. Extracellular matrix content derived from PDL-seeded scaffolds. Values shown in both parts (a) and (b) are the mean while the error bars represent SEM; (n=3, r=5). Insignificant comparisons are designated by N.S. The group exposed to flow exhibited a 93.3% significant increase in collagen content compared to the static group (p<0.05, n=3, r=5).
Discussion
Over the past decade, the potential of BMSCs to provide regenerative treatment strategies in cardiac disease has been investigated.16,18,23–25 Studies have found that BMSCs differentiate in vitro to cardiomyocytes28,30 and SMCs26,31–33; however, there has not yet been clear evidence of BMSC differentiation to endothelial lineages when seeded on fibrous scaffolds, under biomechanical environments. ESCs, on the other hand, possess the ability to differentiate to cardiac phenotypes, including both SMCs and ECs.11,12 As an alternative to BMSCs and ESCs, in our study, we utilized PDLs, because subpopulations of this cell lineage have been shown to express ESCs markers (Oct4, Sox2, NANOG, KLF4, SSEA-1, and SSEA-4), therefore exhibiting a higher differential potential in comparison to other mesenchymal stem cell lineages.8 Owing to their more primitive state, PDLs offer the possibility to differentiate into neurogenic (ectoderm), cardiomyogenic, osteogenic, and chondrogenic (mesoderm) cell lineages, suggesting extensive pluripotency capacities. To date, the only pluripotent stem cells with the ability to differentiate into the three germ layers (ectoderm, mesoderm, and endoderm) are ESCs and induced pluripotent stem cells (iPS). However, based on recent findings,8 PDLs explicitly are of interest clinically due to their ability to be derived from an adult cell source without the risk of teratoma formation. These properties make PDLs an especially attractive cell source for cardiovascular tissue-engineering therapeutics for use in applications such as severe congenital heart valve disease and anomalies, and potentially in the context of our studies for heart valve tissue engineering.
We found that flow conditioning permits relative strong expression of Sox2 and KLF4, whereas strong Oct4 expression was possible only under combined steady flow and growth factor conditions (Fig. 2b). Nanog expression was negligible in all the groups. However, pluripotency potential could still be maintained through the strong expression of KLF4, that is, to both self-renew and differentiate.34 In addition, we also note the importance of incorporating steady flow in heart valve tissue engineering efforts involving PDLs, since self-renewal will be further regulated by Sox2 expression,35 whereas the sustained use of the cocktail medium in the steady-flow environments would further serve to facilitate periodontal progenitor cell self-renewal by strong expression of Oct4.36
We investigated a number of genetic markers for SMCs namely integrin, beta1, THY1, FZD2, and MLC1F as well as for the EC genes, PECAM1, FLK1, and VE-cadherin. In brief, the function of these essential genes is as follows: integrin, beta1 is involved in cell adhesion and processes such as embryogenesis and tissue repair. THY1 is a regulator of cell–cell and cell–matrix interactions in adhesion, migration, and fibrosis. FZD2 is developmentally regulated and is found to be expressed in heart valve, skin, and pericardium; on the other hand, MLC1F is a smooth muscle marker uniquely expressed only by skin. PECAM1 is involved in leukocyte migration, angiogenesis, and integrin activation. FLK1 functions as a signaling protein for VEGF, whereas TIE1 is a cell surface angiopoietin receptor. Lastly, VE-cadherin is necessary for proper vascular development.
In our study, culturing the cells with the cocktail medium and dynamic conditioning by means of an applied, steady fluid-induced shear stress (1 dyne/cm2) had a notable effect in the PDL gene expression of EC and SMC phenotypes (i.e., Group 4, compared to the no flow Groups, 1 and 2). Interestingly, we found that fluid shear stress had a much more dominant effect in comparison to biochemical stimulants in differentiating PDLs toward the valve phenotype; in particular, EC markers (PECAM1, FLK1 and VE-cadherin) were strongly expressed under steady-flow-only conditions (Group 3) by the PDLs, thereby suggesting that the fluid-induced mechanical stimulation of PDLs alone promotes further EC differentiation and is biomimetic in the context of native ECs that require flow to maintain normostatis. This result again indicates the importance of fluid-induced shear stresses in the regulation of PDLs and as a necessary component to research protocols aiming to regenerate cardiovascular or engineered valve tissues from PDLs. We note that eventually, it appeared that more complete EC differentiation was possible under combined biochemical and flow states. Specifically, strong expression of TIE1 in addition to PECAM1, FLK1, and VE-cadherin was expressed under these coupled states, thus indicating the unique interplay of mechanical and biochemical factors necessary for further transformation of PDLs toward the EC phenotype.
The SMC genes, integrin, beta1 and THY1, were upregulated in PDLs under steady-flow states, without requiring growth factors. It is not entirely surprising that integrin, beta1 and THY1, SMC-related genes, were expressed in the PDLs as they have been observed in undifferentiated PDLs.33 This could be due in part to the association of stem cells in general, to pericytes that display certain SMCs properties.37 These qRT-PCR results are suggestive that, in conjunction with collagen assay results, the application of fluid shear stress to PDLs can enhance differentiation toward a phenotype that warrants interest in heart valve tissue engineering. In addition, our biochemical studies confirmed that flow-based steady-flow conditioning is critical in preserving cellular DNA in growing tissues in vitro and in promoting robust collagen extracellular matrix synthesis by the PDLs. To further reinforce these findings, we found that strong expression of FZD2 was possible only when a steady-flow component was added to the protocol, whereas MLC1F was absent in all the groups, thereby excluding the skin phenotype (Group 3, Fig. 2c). This provides greater confidence that the PDLs were able to express the smooth muscle phenotype more characteristic of valves and cardiovascular tissues rather than generic smooth muscle after exposure to steady flow, a critical observation that, to our knowledge, has never been reported to date. Nonetheless, further analysis of cell markers is still necessary to explicitly demonstrate that the PDLs exhibited unique EC and valve interstitial cell characteristics (e.g., such as examining TnT markers).22
We examined COL1A1 and COL3A1 markers (Fig. 2d) and found a strong presence of isoform 3, but an absence of type 1 collagen expression in the PDL samples that were cultured in two-dimensional (2D) monolayer, in all 4 groups, after 7 days. We note however that 3D scaffold environments will have different cell-to-cell and new cell–scaffold interactions in comparison to 2D culture. In addition, longer culture periods would also alter expression levels. To test this hypothesis, we ran additional experiments in our bioreactor using human PDLs seeded in 50:50 blend of PLLA:PGA nonwoven scaffolds and the 22 day tissue culture duration that was also utilized in the earlier constructs subjected to collagen assays. Our preliminary analysis using qRT-PCR confirmed that an extended culture period beyond 3 weeks in 3D did exhibit equally strong expression of both collagen type 1 and 3 isoforms, the two predominant collagen types in heart valves. Furthermore, the relative strong expression of collagen type 3 during the early stages (i.e., up to 7 days) of in vitro culture is analogous to tissue repair processes that native myxomatous valves undergo in vivo, during which collagen type 3 is, in particular, upregulated.38 Finally, we examined for the presence of additional markers, ALP, osteocalcin, and COL2A1 (Fig. 2d), which would be indicative of diseased valve states, namely along osteogenic and chondrogenic pathways, but these markers were found to be absent in all the groups. This finding suggests that steady-flow mechanical conditioning and the growth factors that were used do not promote undesirable differentiation of the PDLs in the context of heart valve tissue engineering.
Conclusions
In summary we introduced human PDLs as a single, practical, autologous, and potentially allogenic cell source for heart valve tissue engineering. Most importantly, PDLs can produce robust engineered collagen and have reduced loss of cellular DNA under physiologically relevant (∼5–6 dynes/cm2) steady-flow in vitro culturing environments. In addition, PDLs were found to demonstrate early evidence of EC differentiation and robust, possibly valve-associated, SMC phenotypic conversion, when exposed to a fluid-induced shear stress of 1 dyne/cm2 and cultured under differentiating medium conditions. We thus conclude that human PDLs will potentially benefit from an in vitro flow mechanical conditioning component as a part of the heart valve tissue-engineering research protocol.
Acknowledgments
Funding for this work was provided by the American Heart Association Scientist Development Grant, 0830061N. In addition, we thank Dr. Nikolaos Tsoukias and Dr. Yen-Chih Huang for laboratory access and equipment.
Disclosure Statement
No competing financial interests exist.
References
- 1.Kannan R.Y. Salacinski H.J. Sales K. Butler P. Seifalian A.M. The roles of tissue engineering and vascularisation in the development of micro-vascular networks: a review. Biomaterials. 2005;26:1857. doi: 10.1016/j.biomaterials.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 2.Niklason L.E. Gao J. Abbott W.M. Hirschi K.K. Houser S. Marini R. Langer R. Science. Vol. 284. New York, NY: 1999. Functional arteries grown in vitro; p. 489. [DOI] [PubMed] [Google Scholar]
- 3.Zhang G. Drinnan C.T. Geuss L.R. Suggs L.J. Vascular differentiation of bone marrow stem cells is directed by a tunable three-dimensional matrix. Acta Biomater. 2010;6:3395. doi: 10.1016/j.actbio.2010.03.019. [DOI] [PubMed] [Google Scholar]
- 4.Hoerstrup S.P. Sodian R. Daebritz S. Wang J. Bacha E.A. Martin D.P. Moran A.M. Guleserian K.J. Sperling J.S. Kaushal S. Vacanti J.P. Schoen F.J. Mayer J.E., Jr. Functional living trileaflet heart valves grown in vitro. Circulation. 2000;102:III44. doi: 10.1161/01.cir.102.suppl_3.iii-44. [DOI] [PubMed] [Google Scholar]
- 5.Sutherland F.W. Perry T.E. Yu Y. Sherwood M.C. Rabkin E. Masuda Y. Garcia G.A. McLellan D.L. Engelmayr G.C., Jr. Sacks M.S. Schoen F.J. Mayer J.E., Jr. From stem cells to viable autologous semilunar heart valve. Circulation. 2005;111:2783. doi: 10.1161/CIRCULATIONAHA.104.498378. [DOI] [PubMed] [Google Scholar]
- 6.Ramaswamy S. Gottlieb D. Engelmayr G.C., Jr. Aikawa E. Schmidt D.E. Gaitan-Leon D.M. Sales V.L. Mayer J.E., Jr. Sacks M.S. The role of organ level conditioning on the promotion of engineered heart valve tissue development in-vitro using mesenchymal stem cells. Biomaterials. 2010;31:1114. doi: 10.1016/j.biomaterials.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trubiani O. Di Primio R. Traini T. Pizzicannella J. Scarano A. Piattelli A. Caputi S. Morphological and cytofluorimetric analysis of adult mesenchymal stem cells expanded ex vivo from periodontal ligament. Int J Immunopathol Pharmacol. 2005;18:213. doi: 10.1177/039463200501800204. [DOI] [PubMed] [Google Scholar]
- 8.Huang C.Y. Pelaez D. Dominguez-Bendala J. Garcia-Godoy F. Cheung H.S. Plasticity of stem cells derived from adult periodontal ligament. Regen Med. 2009;4:809. doi: 10.2217/rme.09.55. [DOI] [PubMed] [Google Scholar]
- 9.Trubiani O. Zalzal S.F. Paganelli R. Marchisio M. Giancola R. Pizzicannella J. Buhring H.J. Piattelli M. Caputi S. Nanci A. Expression profile of the embryonic markers nanog, OCT-4, SSEA-1, SSEA-4, and frizzled-9 receptor in human periodontal ligament mesenchymal stem cells. J Cell Physiol. 2010;225:123. doi: 10.1002/jcp.22203. [DOI] [PubMed] [Google Scholar]
- 10.Pelaez D. Role of Mechanical Strain on the Cardiomyogenic Differentation of Periodontal Ligament Derived Stem Cells. Coral Gables, FL: University of Miami, Open access dissertations; 2011. p. 573. [Ph.D. Thesis] Paper. [Google Scholar]
- 11.Boyer L.A. Lee T.I. Cole M.F. Johnstone S.E. Levine S.S. Zucker J.P. Guenther M.G. Kumar R.M. Murray H.L. Jenner R.G. Gifford D.K. Melton D.A. Jaenisch R. Young R.A. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.International Stem Cell Initiative. Adewumi O. Aflatoonian B. Ahrlund-Richter L. Amit M. Andrews P.W. Beighton G. Bello P.A. Benvenisty N. Berry L.S. Bevan S. Blum B. Brooking J. Chen K.G. Choo A.B. Churchill G.A. Corbel M. Damjanov I. Draper J.S. Dvorak P. Emanuelsson K. Fleck R.A. Ford A. Gertow K. Gertsenstein M. Gokhale P.J. Hamilton R.S. Hampl A. Healy L.E. Hovatta O. Hyllner J. Imreh M.P. Itskovitz-Eldor J. Jackson J. Johnson J.L. Jones M. Kee K. King B.L. Knowles B.B. Lako M. Lebrin F. Mallon B.S. Manning D. Mayshar Y. McKay R.D. Michalska A.E. Mikkola M. Mileikovsky M. Minger S.L. Moore H.D. Mummery C.L. Nagy A. Nakatsuji N. O'Brien C.M. Oh S.K. Olsson C. Otonkoski T. Park K.Y. Passier R. Patel H. Patel M. Pedersen R. Pera M.F. Piekarczyk M.S. Pera R.A. Reubinoff B.E. Robins A.J. Rossant J. Rugg-Gunn P. Schulz T.C. Semb H. Sherrer E.S. Siemen H. Stacey G.N. Stojkovic M. Suemori H. Szatkiewicz J. Turetsky T. Tuuri T. van den Brink S. Vintersten K. Vuoristo S. Ward D. Weaver T.A. Young L.A. Zhang W. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat Biotechnol. 2007;25:803. doi: 10.1038/nbt1318. [DOI] [PubMed] [Google Scholar]
- 13.Filip D.A. Radu A. Simionescu M. Interstitial cells of the heart valves possess characteristics similar to smooth muscle cells. Circ Res. 1986;59:310. doi: 10.1161/01.res.59.3.310. [DOI] [PubMed] [Google Scholar]
- 14.Hirashima M. Kataoka H. Nishikawa S. Matsuyoshi N. Nishikawa S. Maturation of embryonic stem cells into endothelial cells in an in vitro model of vasculogenesis. Blood. 1999;93:1253. [PubMed] [Google Scholar]
- 15.Yamamoto K. Sokabe T. Watabe T. Miyazono K. Yamashita J.K. Obi S. Ohura N. Matsushita A. Kamiya A. Ando J. Fluid shear stress induces differentiation of Flk-1-positive embryonic stem cells into vascular endothelial cells in vitro. Am J Physiol Heart Circ Physiol. 2005;288:H1915. doi: 10.1152/ajpheart.00956.2004. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y. Miao H. Li S. Chen K.D. Li Y.S. Yuan S. Shyy J.Y. Chien S. Interplay between integrins and FLK-1 in shear stress-induced signaling. Am J Physiol Cell Physiol. 2002;283:C1540. doi: 10.1152/ajpcell.00222.2002. [DOI] [PubMed] [Google Scholar]
- 17.Wu C.C. Chao Y.C. Chen C.N. Chien S. Chen Y.C. Chien C.C. Chiu J.J. Linju Yen B. Synergism of biochemical and mechanical stimuli in the differentiation of human placenta-derived multipotent cells into endothelial cells. J Biomech. 2008;41:813. doi: 10.1016/j.jbiomech.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 18.Hutmacher D.W. Scaffold design and fabrication technologies for engineering tissues—state of the art and future perspectives. J Biomater Sci Polym Ed. 2001;12:107. doi: 10.1163/156856201744489. [DOI] [PubMed] [Google Scholar]
- 19.Hoerstrup S.P. Kadner A. Melnitchouk S. Trojan A. Eid K. Tracy J. Sodian R. Visjager J.F. Kolb S.A. Grunenfelder J. Zund G. Turina M.I. Tissue engineering of functional trileaflet heart valves from human marrow stromal cells. Circulation. 2002;106:I143. [PubMed] [Google Scholar]
- 20.Engelmayr G.C., Jr. Rabkin E. Sutherland F.W. Schoen F.J. Mayer J.E., Jr. Sacks M.S. The independent role of cyclic flexure in the early in vitro development of an engineered heart valve tissue. Biomaterials. 2005;26:175. doi: 10.1016/j.biomaterials.2004.02.035. [DOI] [PubMed] [Google Scholar]
- 21.Vandesompele J. De Preter K. Pattyn F. Poppe B. Van Roy N. De Paepe A. Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brand N.J. Roy A. Hoare G. Chester A. Yacoub M.H. Cultured interstitial cells from human heart valves express both specific skeletal muscle and non-muscle markers. Int J Biochem Cell Biol. 2006;38:30. doi: 10.1016/j.biocel.2005.06.018. [DOI] [PubMed] [Google Scholar]
- 23.Boronyak S.M. El-Kurdi M. Engelmayr G.C. Ramaswamy S. Sacks M.S. Schmidt D.E. Flow-stretch-flexure bioreactor. Sep 1, 2011. (U.S. patent application No. 12/959,906)
- 24.Boronyak S.M. Smelko A. Sacks M.S. Ramaswamy S. Design of a flow-stretch-flexure bioreactor for physiologic conditioning of engineered tissue. Biomedical Engineering Society Annual Fall Meeting; Pittsburgh, PA. Oct 7–10, 2009. [Google Scholar]
- 25.Ramaswamy S., et al. A novel bioreactor for the study of physiological fluid induced stresses for heart valve tissue engineering. (Submitted).
- 26.Engelmayr G.C., Jr. Soletti L. Vigmostad S.C. Budilarto S.G. Federspiel W.J. Chandran K.B. Vorp D.A. Sacks M.S. A novel flex-stretch-flow bioreactor for the study of engineered heart valve tissue mechanobiology. Ann Biomed Eng. 2008;36:700. doi: 10.1007/s10439-008-9447-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramaswamy S. Schmidt D. Boronyak S.M. Sacks M.S. Flow Patterns Under Combined Flexural States for Engineered Heart Valve Tissue Development. Biomedical Engineering Society Annual Fall Meeting; Pittsburgh, PA. 2009. [Google Scholar]
- 28.Ramaswamy S. Boronyak S.M. Schmidt D. Sacks M.S. Design of a Novel Curved Tube Flow-Stretch-Flexure Bioreactor for Mechanistic Studies in Heart Valve Tissue Engineering. Society for Biomaterials, Annual meeting and exposition; San Antonio, TX. 2009. [Google Scholar]
- 29.Ramaswamy S. Uluer M.C. Leen S. Bajaj P. Fishbein K.W. Spencer R.G. Noninvasive assessment of glycosaminoglycan production in injectable tissue-engineered cartilage constructs using magnetic resonance imaging. Tissue Eng Part C Methods. 2008;14:243. doi: 10.1089/ten.tec.2007.0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Engelmayr G.C., Jr. Sales V.L. Mayer J.E., Jr. Sacks M.S. Cyclic flexure and laminar flow synergistically accelerate mesenchymal stem cell-mediumted engineered tissue formation: Implications for engineered heart valve tissues. Biomaterials. 2006;27:6083. doi: 10.1016/j.biomaterials.2006.07.045. [DOI] [PubMed] [Google Scholar]
- 31.Engelmayr G.C., Jr. Sacks M.S. Prediction of extracellular matrix stiffness in engineered heart valve tissues based on nonwoven scaffolds. Biomech Model Mechanobiol. 2008;7:309. doi: 10.1007/s10237-007-0102-1. [DOI] [PubMed] [Google Scholar]
- 32.Schoen F.J. New frontiers in the pathology and therapy of heart valve disease: 2006 Society for Cardiovascular Pathology, Distinguished Achievement Award Lecture, United States-Canadian Academy of Pathology, Atlanta, GA, February 12, 2006. Cardiovasc Pathol. 2006;15:271. doi: 10.1016/j.carpath.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 33.Huang Y. Jia X. Bai K. Gong X. Fan Y. Effect of fluid shear stress on cardiomyogenic differentiation of rat bone marrow mesenchymal stem cells. Arch Med Res. 2010;41:497. doi: 10.1016/j.arcmed.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 34.Geiman D.E. Ton-That H. Johnson J.M. Yang V.W. Transactivation and growth suppression by the gut-enriched Krüppel-like factor (Krüppel-like factor 4) are dependent on acidic amino acid residues and protein-protein interaction. Nucleic Acids Res. 2000;28:1106. doi: 10.1093/nar/28.5.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao R. Daley G.Q. From fibroblasts to iPS cells: induced pluripotency by defined factor. J Cell Biochem. 2008;105:949. doi: 10.1002/jcb.21871. [DOI] [PubMed] [Google Scholar]
- 36.Niwa H. Miyazaki J. Smith A.G. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 37.Tomokiyo A. Maeda H. Fujii S. Monnouchi S. Wada N. Kono K. Yamamoto N. Koori K. Teramatsu Y. Akamine A. A multipotent clonal human periodontal ligament cell line with neural crest cell phenotypes promotes neurocytic differentiation, migration, and survival. J Cell Physiol. 2012;227:2040. doi: 10.1002/jcp.22933. [DOI] [PubMed] [Google Scholar]
- 38.Cole W.G. Chan D. Hickey A.J. Wilcken D.E. Collagen composition of normal and myxomatous human mitral heart valves. Biochem J. 1984;219:451. doi: 10.1042/bj2190451. [DOI] [PMC free article] [PubMed] [Google Scholar]