Abstract
Marrow stromal cells are adult stem cells from bone marrow that can differentiate into multiple nonhematopoietic cell lineages. Previous reports demonstrated that single-cell-derived colonies of marrow stromal cells contained two morphologically distinct cell types: spindle-shaped cells and large flat cells. Here we found that early colonies also contain a third kind of cell: very small round cells that rapidly self-renew. Samples enriched for the small cells had a greater potential for multipotential differentiation than samples enriched for the large cells. Also, the small cells expressed a series of surface epitopes and other proteins that potentially can be used to distinguish the small cells from the large cells. The results suggested it will be important to distinguish the major subpopulations of marrow stromal cells in defining their biology and their potential for cell and gene therapy.
Bone marrow contains at least two kinds of stem cells, hematopoietic stem cells and stem cells for nonhematopoietic tissues (1–26), variously referred to as mesenchymal stem cells or marrow stromal cells (MSCs). MSCs are of interest because they are easily isolated from a small aspirate of bone marrow and readily generate single-cell-derived colonies (1–5, 25, 27); the single-cell-derived colonies can be expanded through as many as 50 population doublings in about 10 weeks (25), and they can differentiate into osteoblasts, adipocytes, chondrocytes (1–13), myocytes (9), astrocytes, oligodendrocytes, and neurons (17, 23, 26, 27). For these reasons, the cells are currently being tested for their potential use in cell and gene therapy for a number of human diseases (22, 24). Previous reports (3, 11, 12) demonstrated that single-cell-derived colonies of human MSCs are heterogeneous in that they contain at least two morphologically distinct kinds of cells: spindle-shaped cells and large cuboidal or flattened cells. Here we have extended our previous observations (25) to demonstrate that the colonies also contain extremely small cells that are rapidly self-renewing (RS cells). The RS cells appear to be the earliest progenitors in the cultures and have the greatest potential for multilineage differentiation. They differ from more mature cells in the same cultures by a series of surface epitopes and expressed proteins.
Methods
Isolation and Growth of MSCs.
To isolate human MSCs, bone marrow aspirates of 10–20 ml were taken from the iliac crest of normal donors ranging in age from 19 to 49 years old under an Institutional Review Board approved protocol. Nucleated cells were isolated with a density gradient [Ficoll/Paque (Pharmacia)] and resuspended in complete culture medium [α MEM, GIBCO/BRL; 20% FBS, lot-selected for rapid growth of MSCs (Atlanta Biologicals); 100 units/ml of penicillin/100 μg/ml of streptomycin/2 mM l-glutamine (GIBCO/BRL)]. All of the cells were plated in 25 ml of medium in a 175-cm2 culture dish (Falcon) and incubated at 37°C with 5% humidified CO2. After 24 h, nonadherent cells were discarded, and adherent cells were thoroughly washed twice with PBS. The cells were incubated for 5–7 days, harvested with 0.25% trypsin and 1 mM EDTA for 5 min at 37°C, and then replated at about three cells/cm2 in an intercommunicating system of culture flasks (6,320 cm2; Cell Factory; Nunc). After 14 days, the cells (from passage 1) were harvested with trypsin/EDTA, suspended at 1≈2 × 106 cells/ml in 5% DMSO and 30% FBS, and frozen at 1-ml aliquots in liquid nitrogen. To expand a culture, a frozen stock of MSCs was thawed, plated at 5,000 cells/cm2, and incubated for 5–7 days. The cells were harvested and diluted for further expanding by plating at initial densities of about three cells/cm2 and culturing in the complete culture medium. The cells were lifted by incubation with 0.25% trypsin and 1 mM EDTA for 5 min at 37°C, and cell numbers were counted with a hemocytometer. The relative numbers of small and agranular cells (RS-1 cells), small and granular cells (RS-2) cells, and large and more mature cells (mMSCs) were assayed by forward and side light scattering (FACS Vantage SE; Becton Dickinson). For colony-forming unit assays, cultures were incubated for 10–14 days, washed with PBS, and stained with 0.5% Crystal violet in methanol for 5–10 min at room temperature. Alkaline phosphatase activity was assayed in cell lysates with a p-nitrophenol phosphate disodium solution (Sigma 104).
Electron Microscopy.
RS cells were prepared by lifting 14-day cultures of MSCs and passing the cells through a 10-μm filter (Millipore). FACS assays (25) indicated that about 95% of the cells in the filtrate were RS cells. Cells on the filter were used as a source of mMSCs. FACS assays indicated that about 90% of the cells were mMSCs. The cells were pelleted by centrifugation, fixed in 2% glutaraldehyde, and cut into 3-mm3 blocks. The samples were stained with 2% osmium and 2% aqueous uranyl acetate. After dehydration, the blocks were cut into 70-μm sections. The sections were stained with alcoholic uranyl acetate and counterstained with bismuth subnitrite. Images were obtained with an electron microscope (JOEL JEM 1010) and a charge-coupled device camera (Hamamatsu, Middlesex, NJ). Electron micrographs were prepared by the Biomedical Imaging Core Facility of the University of Pennsylvania Medical Center.
Assays for Differentiation.
For osteogenic differentiation (21), cells were plated at 1,000 cells/cm2 in 2-cm2 wells and grown to 50–70% confluency in 5 days. They were then incubated in osteogenic medium (10−8 M dexamethasone/0.2 mM ascorbic acid/10 mM β-glycerolphosphate; Sigma). The medium was replaced every 3–4 days for 21 days. Cultures were washed with PBS, fixed in a solution of ice-cold 70% ethanol for 1 h, and stained for 10 min with 1 ml of 40 mM Alizarin red (pH 4.1; Sigma) (21). Because of the mineral deposited over the cells, the extent of mineralization was estimated by dividing the area covered with mineral by the total area of mineral plus unmineralized cells. For adipogenic differentiation (21), 50–70% confluent cultures were incubated in complete medium supplemented with 0.5 μM hydrocortisone/0.5 mM isobutylmethylxanthine/60 μM indomethacin. The medium was replaced every 3–4 days for 21 days. Cells were washed with PBS, fixed in 10% formalin for 10 min, and stained for 15 min with fresh Oil Red-O solution (Fisher Scientific). The extent of adipogenic differentiation was estimated as percent of cells containing well-stained oil droplets. For chondrogenic differentiation, we modified (I. Sekiya and D.J.P., unpublished work) the pellet culture system by Johnstone et al. (15). After 200,000 MSCs were centrifuged in a 15-ml polypropylene tube, the pellets were cultured in chondrogenic media that consisted of high-glucose DMEM (Cellgro, Mediatech, Herndon, VA) supplemented with 500 ng/ml of BMP-6 (R & D Systems) as well as 10 ng/ml of transforming growth factor-β3/0.1 μM dexamethasone/50 μg/ml of ascorbate-2-phosphate/40 μg/ml of proline/100 μg/ml of pyruvate/50 mg/ml of ITS+ Premix (Becton Dickinson; 6.25 μg/ml of insulin/6.25 μg/ml of transferrin/6.25 ng/ml of selenious acid/1.24 mg/ml of BSA/5.35 mg/ml of linoleic acid). The medium was replaced every 2–3 days for 21 days. The pellets were embedded in paraffin, cut into 5-mm sections, and stained with Safranin O (Richard–Allen Scientific, Kalamazoo, MI).
Analysis of Surface Epitope by FACS.
The cells were suspended in PBS at a concentration of about 100,000 cells/ml, fixed in 1% methanol or acetone at 4°C for 10 min, and washed with PBS. Nonspecific antigens were blocked by incubating the cells at room temperature for 1 h in 1% BSA and 0.1% goat serum. The cells were washed by centrifugation in three volumes of PBS, and the cell pellet was suspended in 0.5 ml of a primary antibody solution containing 20 μg/ml of antibody, 1% BSA, and 0.1% goat serum. After incubation for 40 min at 4°C, the cells were washed in PBS. The primary antibodies were mouse or rabbit anti-human, obtained from Chemicon, IgM Hybridoma Bank, University of Iowa; PharMingen; Biomedia; and Santa Cruz Biotechnology. For an isotype control, nonspecific mouse or rabbit IgG (Dako, PharMingen, Chemicon, or Santa Cruz) was substituted for the primary antibody. For antibodies that required a second antibody for detection, the cell pellet was incubated under the same conditions for 20 min with anti-mouse or anti-rabbit IgG labeled with FITC or phycoerythrin. The cells were then washed in PBS and suspended in 1 ml of PBS for analysis on a cell sorter.
Proteomic Analyses.
For protein analysis, cell pellets of about 3 million cells were vortexed in 1 ml of lysis buffer (3 mg/ml SDS and 30 mg/ml DTT in 30 mM Tris⋅HCl buffer, pH 7.8) and heated to 80–90°C for 5 min. Ten microliters of RNase (Boehringer Mannheim) was added, and the sample was incubated at 37°C for 5–10 min. The protein was precipitated with 5 ml of acetone at 4°C, recovered by centrifugation, air dried, and dissolved at a concentration of 10 mg/ml of solubilization buffer (2.1 g of urea, 0.8 g of thiourea, 200 mg of 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate, and 40 mg of DTT in 5 ml of water). The samples were separated first by isoelectric focusing (Immobilized pH Gradient Strip, pH 3–10; Amersham Pharmacia) and then on 10–12% polyacrylamide gels in SDS. The gels were stained (Silver Plus; Bio-Rad), and protein spots distinguishing RS cells from mMSCs were identified either by matrix-assisted laser desorption ionization/time-of-flight before and after trypic digestion or by liquid chromotography/MS/MS. The preparation of the two-dimensional gels and analysis of proteins by MS was performed by the W. M. Keck Biomedical Mass Spectrometry Laboratory, Biomedical Research Facility, University of Virginia, Charlottesville, VA.
Results
Appearance of RS Cells and mMSCs in Culture.
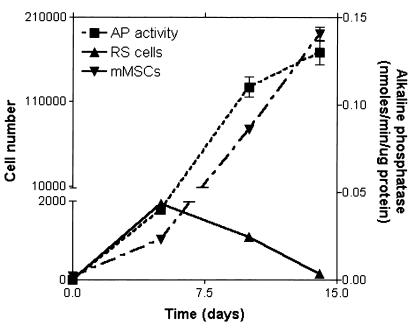
To prepare cultures of human MSCs, nucleated cells were isolated from bone marrow aspirates from normal volunteers and the plastic adherent cells isolated (21, 25). Thereafter, the cells were expanded by plating at an initial density of about three cells/cm2. As reported by previous investigators (1, 2, 5), the cultures underwent an initial lag phase of about 5 days, during which the colonies were seen to arise from single cells. As reported by previous investigators (see ref. 3), the colonies contained both spindle-shaped cells and large flat cells. In addition, we noted that the colonies also contained very small round cells that were primarily detected as they became highly reflective during replication (doublets of RS cells in Fig. 1). The colonies that were formed during the lag phase arose from either RS cells or small spindle-shaped cells. Large adherent cells in the cultures replicated slowly. After about 5 days, each of the colonies contained 20–100 cells (Figs. 1 and 2). Analysis by FACS demonstrated that in the 5-day cultures, RS cells accounted for over half of the cells (Fig. 3). Their rapid replication was apparent by time-lapse photography (Fig. 2 A–C). To prepare purified fractions of RS cells, cells from 14-day cultures were passed through a 10-μm filter (Millipore). The RS cells in the filtrate were about 7 μm in diameter and had a high nucleus-to-cytoplasm ratio (Fig. 2D). In contrast, mMSCs retained on the filter were 15–50 μm in diameter and contained a large number of unidentified vacuoles (Fig. 2E). During the logarithmic growth phase from about day 6 to day 12, the proportion of RS cells decreased, and the proportion of mMSCs greatly increased (Fig. 3). The increase in mMSCs was accompanied by an increase in alkaline phosphatase (Fig. 3), an observation suggesting that some of the cells were differentiating into osteoblast precursors.
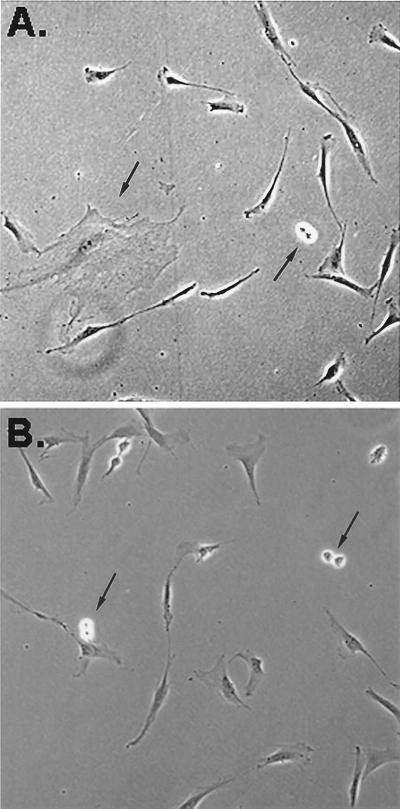
Figure 1.
Phase-contrast photomicrograph of two different colonies of human MSCs 6 days after the cells were plated at three cells/cm2. Arrows indicate one large mMSC and three doublets of recently replicated RS cells. (×200.)
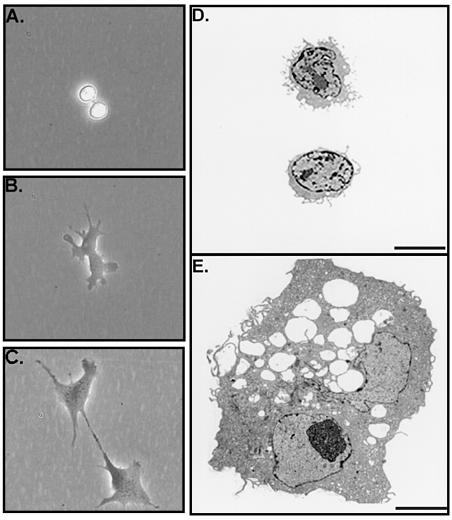
Figure 2.
(A–C) Phase-contrast photomicrographs of a dividing RS cell taken at intervals of 20 min. (D) Electron micrographs of two RS cells. (E) Electron micrograph of an mMSC. As indicated, the mMSCs were vacuolated and frequently binucleate. (Bar = 5 μm.)
Figure 3.
Time course for the expansion of cells and alkaline phosphatase activity in cultures of MSCs. MSCs were plated at an initial density of about 3 cells/cm2 in 75-cm2 plates.
Differentiation of RS Cells and mMSCs.
The ultrafiltration procedure provided only a small yield of purified RS cells because the mMSCs rapidly obstructed the filter pores. Therefore, to test the multipotentiality for differentiation of the two types of cells, cultures that were incubated for 5 days containing about 60% RS cells were compared with cultures that were incubated for 12 days containing about 90% mMSCs (Fig. 3). After the cells were replated at about 1,000 cells/cm2 and incubated in osteogenic medium for 21 days (21), the preparations enriched of RS cells more extensively differentiated into osteoblasts than the preparations enriched for mMSCs (Fig. 4 A and B). The area of mineralization (21) was 1.4–1.7 times greater with the RS cells. After incubation in adipogenic medium under similar conditions, the preparations enriched for RS cells differentiated more extensively into adipocytes than the preparations enriched for mMSCs (Fig. 4 C and D). The number of adipocytes was 3.5- to 6-fold greater with the RS cells. Also, after the cells were aggregated into micropellets and incubated in chondrogenic medium for 21 days (21), the cultures enriched for RS cells differentiated more extensively into cartilage (Fig. 4 E and F). The cartilage pellets obtained with the RS-enriched cultures were larger and stained more extensively for proteoglycans (Fig. 4 E and F). They also contained 1.6-fold higher levels of mRNA for type II collagen, as assayed by reverse transcription–PCR (not shown).
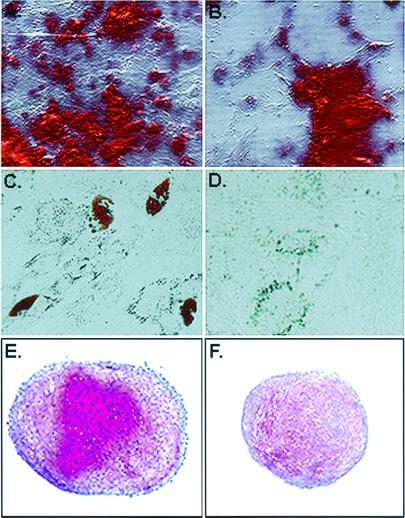
Figure 4.
Assays for differentiation. (A) Cell preparations enriched for RS cells incubated in osteogenic medium for 21 days (21). The mineral in the cultures was detected by staining with Alizarin red. (B) Cell fraction enriched for mMSCs incubated in osteogenic medium for 21 days. (C) Cell fraction enriched for RS cells incubated in adipogenic medium for 21 days (21). Fat droplets in the cells were stained with Oil Red O. (D) Cell fraction enriched for mMSCs incubated in adipogenic medium for 21 days. (E) Cell fraction for RS cells incubated under chondrogenic conditions for 21 days (21). Proteoglycans in paraffin sections of the pellet were stained with Safranin O. (F) Cell fraction enriched for mMSCS incubated under chondrogenic conditions for 21 days. Paraffin sections of the pellet were stained with Safranin O.
Surface Epitopes.
To identify surface epitopes, cultures enriched for RS cells and mMSCs were assayed by using a series of commercially available antibodies. As noted previously (25), FACS analyses distinguished two subtypes of RS cells: small and agranular cells (RS-1 cells) seen in stationary and late-logarithm phase cultures and small granular cells (RS-2 cells) that were seen primarily at the end of the lag phase and that were probably mitotic RS-1 cells. The RS-1 cells and some of the RS-2 cells contained four epitopes not found on mMSCs (Table 1): the vascular endothelial growth factor receptor-2 (FLK-1), TRK (an NFG receptor), transferrin receptor, and annexin II (lipocortin 2). Some but not all of the RS cells contained several other distinguishing epitopes. These epitopes included c-Kit (CD117), the stem cell factor receptor. Also, some but not all of the RS cells contained the epitope for the multidrug resistance gene that is a distinguishing feature of the “side population” of small cells from both muscle and marrow (28, 29). SP cells are positive for c-Kit, negative for CD45, and positive for ScaI, but they are precursors of hematopoietic cells in addition to muscle cells. However, all of the cells in the culture were negative for the hematopoietic stem cell marker CD34 and a series of other markers for hematopoietic precursors. The only exception was that some of the RS-1 and RS-2 cells were positive for CD4, which is expressed on thymocytes and peripheral blood lymphocytes, including T cells. Also of interest was that both the RS-1 and RS-2 cells were negative for STRO-1, an epitope originally suggested as a marker for MSCs (30, 31). However, some of the mMSCs contained the STRO-1 epitope, an observation consistent with their ability to differentiate into osteoblasts (21). Some of the mMSCs contained several other epitopes not found on RS cells. These included receptors for platelet-derived growth factor and epidermal growth factor, an observation suggesting that the previously reported stimulatory effects of these two cytokines in cultures of MSCs primarily expanded the subpopulation of mMSCs (11, 32).
Table 1.
Surface epitopes for RS cells and mMSCs
| Epitopes | RS-1 cells | RS-2 cells | mMSCs |
|---|---|---|---|
| Selective for RS cells | |||
| Vascular endothelial growth factor receptor (FLK-1) | (+) | (+/−) | (−) |
| TRK (C-14; a nerve growth factor receptor) | (+) | (+/−) | (−) |
| Transferrin receptor | (+) | NA | (−) |
| Annexin II (Lipocortin 2) | (+) | NA | (−) |
| Multidrug resistance | (+/−) | (+/−) | (−) |
| Epithelial membrane antigen | (+/−) | (+/−) | (−) |
| CD4 | (+/−) | (+/−) | (−) |
| CD104 | (+/−) | (+/−) | (−) |
| CD117 (c-Kit; stem cell factor receptor) | (+/−) | (+/−) | (−) |
| Selective for mMSCs | |||
| STRO-1 | (−) | (−) | (+/−) |
| PDGF-R | (−) | (−) | (+/−) |
| EGF-R | (−) | (−) | (+/−) |
| CD10 | (−) | (−) | (+) |
| CD147 (Neuroregulin) | (−) | (−) | (+) |
| Nonselective | |||
| Annexin V (Lipocortin 5) | (+/−) | NA | (+/−) |
| HLA-1 | (+/−) | (+/−) | (+/−) |
| Basic FGF receptor | (+/−) | (+/−) | (+/−) |
| CD31 | (+/−) | (+/−) | (+/−) |
| CD38 | (+/−) | (+/−) | (+/−) |
| CD44 (Hyaluronic acid receptor) | (+) | (−) | (+) |
| CD49e (integrin alpha 5) | (+) | (+) | (+) |
| CD59 | (+) | (−) | (+) |
| CD81 | (+/−) | (−) | (+) |
| CD90 (Thy-1) | (+/−) | (−) | (+) |
(+), most cells positive; (+/−), some cells positive; (−), negative. All three subpopulations of MSCs are negative for the hematopoietic markers: CD1a, CD11B (Mac-1), CD14, CD27, CD34, CD43, CD45, and CD133. They were also negative for a series of other markers: CD50 (I-CAM 3), CD53, CD109, CD114 (G-CSFR), HLA-2, CCR5 (chemokine receptor-5), and human L1 (neurite adhesion molecule).
Proteomic Analysis.
To further characterize the subpopulations, proteins differentially expressed in preparations enriched for RS cells and mMSCs were assayed by preparing two-dimensional gels and identifying the proteins by MS. Over 30 proteins were identified in fractions enriched for RS cells that were not detected in fractions enriched for mMSCs (Table 2). Conversely, over 10 proteins were identified in fractions enriched for mMSCs that were not detected in fractions enriched for RS cells.
Table 2.
Proteins differentially expressed in RS cells and mMSCs
| Classification | RS cells | mMSCs | |
|---|---|---|---|
| Stress proteins | Heat shock 27 Tumor rejection antigen (gp 96) | Glutathione-S transferase Peroxiredoxin 1 | T-complex protein, 1-alpha |
| Ion channel and transport | Voltage-dependent-anion channel-2 | ||
| Signal transduction | Protein kinase C substrate Phosphatase 2A inhibitor | ||
| DNA synthesis/repair/ recombination | Esterase D RNase A | ||
| Protein synthesis | Initiation factor 5a Elongation factor 1-alpha Ribosomal protein S12 Ribosomal protein, large, P1 | Ribosomal protein, large, P2 | Initiation factor 2G Ribosomal protein, large, P0 |
| Transcription | Transcription factor BTF 3a | ||
| Surface | Annexin I (lipocortin I) Annexin II (lipocortin II) | Annexin V | |
| Cytoskeleton | Destrin Myosin light chain | Actin β chain | |
| Metabolic | Lactate dehydrogenase A Glycerolaldehyde-3-P dehydrogenase Citrate synthetase Transketolase P-glycerolmutase | Aldo-keto reductase 7(A2) Alpha-amylase inhibitor CM3 Enoyl-CoA hydratase Proteosome subunit, alpha-4 | Lactate dehydrogenase B Phosphoglycerate kinase-1 Enolase-1 Protein disulfide isomerase ER60 precursor |
Discussion
Although cultures of MSCs have been studied extensively for over 30 years (1), rigorous criteria for characterizing the cells have not been developed. Therefore, it is difficult to compare the data from different laboratories. The issue has become particularly pressing since several trials have been initiated in which cultures of MSCs are being used in patients (22, 24). Several groups of investigators developed protocols for preparation of human MSCs by using the criteria of morphologic homogeneity of the cultures and uniform staining with several antibodies (see ref. 18). However, several reports have demonstrated (see refs. 3, 11, 12, and 21) that cultures of human MSCs become morphologically homogeneous only after they were passed several times at high density and lost some of their potential for multilineage differentiation. In contrast, early passage cells and cultures passed at very low plating densities to generate single-cell-derived colonies contained both the small spindle-shaped cells and the large cells originally described by Mets and Verdonk (3). The results here demonstrate that after the cells are plated at very low densities (25), a third morphologically distinct type of cell, RS cells, can readily be detected. RS cells are characterized by their extremely small size, rapid rate of replication, and enhanced potential for multilineage differentiation. Moreover, they can be distinguished from more mature cells in the same cultures by a series of surface epitopes and expressed proteins. Therefore, the results raise the possibility that RS cells may have the greatest potential for long-term engraftment and differentiation in vivo. As the results here emphasized, however, even the subpopulation defined as RS cells were heterogeneous, because they did not stain uniformly for several surface epitopes. Therefore it will be of interest to further subfractionate the RS cell population and determine the potentials of the subpopulations for multilineage differentiation and engraftment to specific tissues.
Acknowledgments
The work was supported in part by grants from the National Institutes of Health (AR47161 and 42210), the Oberkotter Foundation, the HCA–Healthcare Company, and the Louisiana Gene Therapy Research Consortium.
Abbreviations
- MSC
marrow stromal cell
- RS cells
rapidly self-renewing cells
- RS-1 cells
small and agranular RS cells
- RS-2 cells
small and granular RS cells
- mMSC
mature MSC
References
- 1.Friedenstein A J, Chailakhjan R K, Lalykina K S. Cell Tissue Kinet. 1970;3:393–403. doi: 10.1111/j.1365-2184.1970.tb00347.x. [DOI] [PubMed] [Google Scholar]
- 2.Castro-Malaspina H, Gay R E, Resnick G, Kapoor N, Meyers P, Chiarieri D, McKenzie S, Broxmeyer H E, Moore M A. Blood. 1980;56:289–301. [PubMed] [Google Scholar]
- 3.Mets T, Verdonk G. Mech Ageing Dev. 1981;16:81–89. doi: 10.1016/0047-6374(81)90035-x. [DOI] [PubMed] [Google Scholar]
- 4.Piersma A H, Brockbank K G, Ploemacher R E, van Vliet E, Brakel-van Peer K M, Visser P J. Exp Hematol. 1985;13:237–243. [PubMed] [Google Scholar]
- 5.Owen M E, Friedenstein A J. Cell and Molecular Biology of Vertebrate Hard Tissues. Chichester, UK: Ciba Foundation Symposium; 1988. pp. 42–60. [PubMed] [Google Scholar]
- 6.Caplan A I. J Ortho Res. 1991;9:641–650. doi: 10.1002/jor.1100090504. [DOI] [PubMed] [Google Scholar]
- 7.Clark B R, Keating A. Ann NY Acad Sci. 1995;770:70–78. doi: 10.1111/j.1749-6632.1995.tb31044.x. [DOI] [PubMed] [Google Scholar]
- 8.Beresford N N, Bennett J H, Devlin C, Leboy P S, Owen M E. J Cell Sci. 1992;102:341–351. doi: 10.1242/jcs.102.2.341. [DOI] [PubMed] [Google Scholar]
- 9.Wakitani S, Saito T, Caplan A I. Muscle Nerve. 1995;18:1417–1426. doi: 10.1002/mus.880181212. [DOI] [PubMed] [Google Scholar]
- 10.Pereira R F, Halford K W, O'Hara M D, Leeper D B, Sokolov B P, Pollard M D, Bagasra O, Prockop D J. Proc Natl Acad Sci USA. 1995;92:4857–4861. doi: 10.1073/pnas.92.11.4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuznetsov S A, Krebsbach P H, Satomura K, Kerr J, Riminucci M, Benayahu D, Robey P G. J Bone Miner Res. 1997;12:1335–1347. doi: 10.1359/jbmr.1997.12.9.1335. [DOI] [PubMed] [Google Scholar]
- 12.Bruder S P, Jaiswal N, Haynesworth S E. J Cell Biochem. 1997;64:278–294. doi: 10.1002/(sici)1097-4644(199702)64:2<278::aid-jcb11>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 13.Prockop D J. Science. 1997;276:711–774. doi: 10.1126/science.276.5309.71. [DOI] [PubMed] [Google Scholar]
- 14.Pereira R F, O'Hara M D, Laptev A V, Halford K W, Pollard M D, Class R, Simon D, Livezey K, Prockop D J. Proc Natl Acad Sci USA. 1998;95:1142–1147. doi: 10.1073/pnas.95.3.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnstone B, Hering T M, Caplan A I, Goldberg V M, Yoo J U. Exp Cell Res. 1998;238:265–272. doi: 10.1006/excr.1997.3858. [DOI] [PubMed] [Google Scholar]
- 16.Ferrari G, Cusella-De Angelis G, Coletta M, Paolucci E, Stornaiuolo A, Cossu G, Mavilio F. Science. 1998;279:1528–1530. doi: 10.1126/science.279.5356.1528. [DOI] [PubMed] [Google Scholar]
- 17.Azizi S A, Stokes D G, Augelli B J, DiGirolamo C M, Prockop D J. Proc Natl Acad Sci USA. 1998;95:3908–3913. doi: 10.1073/pnas.95.7.3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pittenger M F, Mackay A M, Beck S C, Jaiswal R K, Douglas R, Mosca J D, Morman M A, Simonetti D W, Craig S, Marshak D R. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 19.Nilsson S K, Dooner M S, Weier H U, Frenkel B, Lian J B, Stein G S, Quesenberry P J. J Exp Med. 1999;189:729–734. doi: 10.1084/jem.189.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hou Z, Nguyen Q, Frenkel B, Nilsson S K, Milne M, van Wijnen A J, Stein J L, Quesenberry P, Lian J B, Stein G S. Proc Natl Acad Sci USA. 1999;96:7294–7299. doi: 10.1073/pnas.96.13.7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DiGirolamo C M, Stokes D, Colter D, Phinney D G, Class R, Prockop D J. Br J Haematol. 1999;107:275–281. doi: 10.1046/j.1365-2141.1999.01715.x. [DOI] [PubMed] [Google Scholar]
- 22.Horwitz E M, Prockop D J, Fitzpatrick L A, Koo W W, Gordon P L, Neel M, Sussman M, Orchard P, Marx J C, Pyeritz R E, Brenner M K. Nat Med. 1999;5:309–313. doi: 10.1038/6529. [DOI] [PubMed] [Google Scholar]
- 23.Kopen G C, Prockop D J, Phinney D G. Proc Natl Acad Sci USA. 1999;96:10711–10716. doi: 10.1073/pnas.96.19.10711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caplan A I. Clin Orthop Relat Res. 2000;379:567–570. doi: 10.1097/00003086-200010001-00010. [DOI] [PubMed] [Google Scholar]
- 25.Colter D, Class R, DiGirolamo C M, Prockop D J. Proc Natl Acad Sci USA. 2000;97:3213–3218. doi: 10.1073/pnas.070034097. . (First Published March 21, 2000; 10.1073/pnas.070034097) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chopp M, Zhang X H, Li Y, Wang L, Chen J, Lu D, Lu M, Rosenblum M. NeuroReport. 2000;11:3001–3005. doi: 10.1097/00001756-200009110-00035. [DOI] [PubMed] [Google Scholar]
- 27.Woodbury D, Schwarz E J, Prockop D J, Black I B. J Neurosci Res. 2000;61:364–370. doi: 10.1002/1097-4547(20000815)61:4<364::AID-JNR2>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 28.Goodell M A, Rosenzweig M, Kim H, Marks D F, DeMaria M, Paradis G, Grupp S A, Sieff C A, Mulligan R C, Johnson R P. Nat Med. 1997;3:1337–1345. doi: 10.1038/nm1297-1337. [DOI] [PubMed] [Google Scholar]
- 29.Jackson K A, Mi T, Goodell M A. Proc Natl Acad Sci USA. 1999;96:14482–14486. doi: 10.1073/pnas.96.25.14482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gronthos S, Zannettino A C, Graves S E, Ohta S, Hay S J, Simmons P J. J Bone Miner Res. 1999;14:47–56. doi: 10.1359/jbmr.1999.14.1.47. [DOI] [PubMed] [Google Scholar]
- 31.Walsh S, Jefferiss C, Stewart K, Jordan G R, Screen J, Beresford J N. Bone. 2000;27:185–195. doi: 10.1016/s8756-3282(00)00319-7. [DOI] [PubMed] [Google Scholar]
- 32.Kuznetsov S A, Friedenstein A J, Robey P G. Br J Haematol. 1997;97:561–570. doi: 10.1046/j.1365-2141.1997.902904.x. [DOI] [PubMed] [Google Scholar]