Abstract
Microdeletions including 5q31 have been reported in only few patients to date. Apart from intellectual disability/developmental delay (ID/DD) of varying degrees, which is common to all reported patients, the clinical spectrum is wide and includes short stature, failure to thrive, congenital heart defects, encephalopathies, and dysmorphic features. We report a patient with a 0.9-Mb de novo deletion in 5q31.2, the smallest microdeletion in 5q31 reported thus far. His clinical presentation includes mild DD, borderline short stature, postnatal microcephaly, and mild dysmorphic signs including microretrognathia. Together with data from 7 reported overlapping microdeletions, analysis of our patient enabled the tentative delineation of a phenotype map for 5q31 deletions. In contrast to the mild phenotype of small microdeletions affecting only 5q31.2, carriers of larger microdeletions which also include subbands 5q31.1 and/or 5q31.3 seem to be more severely affected with congenital malformations, growth anomalies, and severe encephalopathies. A 240-kb smallest region of overlap in 5q31.2 is delineated which contains only 2 genes, CTNNA1 and LRRTM2. We propose LRRTM2 as the most promising candidate gene for ID/DD due to its expression pattern, function as a key regulator of excitatory development, and interaction with Neurexin 1. However, sequence analysis of LRRTM2 in 330 patients with ID/DD revealed no relevant alterations, excluding point mutations in LRRTM2 as a frequent cause of ID/DD in patients without microdeletions.
Key Words: Candidate gene, Developmental delay, Intellectual disability, LRRTM2, Microdeletion, Molecular karyotyping, 5q31.2
Molecular karyotyping has become a routine diagnostic test for intellectual disability/developmental delay (ID/DD), identifying causative copy-number variations (CNVs) with an average detection rate of >12% [Miller et al., 2010]. However, disease associations for many rare CNVs remain unclear, and the dosage-sensitive genes underlying the phenotypes with clear pathogenicity have generally not been identified because often the CNVs are large and encompass many genes [Cooper et al., 2011].
Here, we report a male patient with ID, borderline growth retardation, postnatal microcephaly, a high-arched palate, mild dysmorphisms, and a de novo microdeletion in 5q31.2. Four previously published patients and 3 patients ascertained through the Database of Chromosomal Imbalance and Phenotype in Humans using Ensemble Resources (DECIPHER, https://decipher.sanger.ac.uk/), all with overlapping microdeletions, also showed ID/DD. In this report, we contribute to the clinical characterization of 5q31 microdeletions, define a narrow phenocritical region, and suggest LRRTM2 as a candidate gene for ID/DD.
Clinical Report
Our patient is a 7 years and 3 months old boy and the third-born child of healthy, non-consanguineous German parents. Besides 2 family members with attention-deficit disorder and/or tics, the family history is unremarkable. After an uneventful pregnancy, the patient was born at gestational week 40 with a weight of 2,880 g (3rd percentile), length of 49 cm (3rd–10th percentile), and occipitofrontal circumference (OFC) of 34 cm (3rd–10th percentile). Apgar scores were 10/10. No malformations or other abnormalities were apparent. In the newborn period, feeding difficulties and muscular hypotonia were noted by his parents, as well as longer sleeping periods than observed for his siblings. Microcephaly was diagnosed at 6 weeks (0.5 cm <3rd percentile).
The patient showed mild DD, especially concerning language. His first words were at 18 months; at 7 years, he used only single words. He could sit without support at 12 months and walked independently at 21 months. At 3 years, developmental assessment by MFED (Münchener Funktionelle Entwicklungs-Diagnostik) showed a developmental delay of 1.5 years. At this time, the patient had developed nocturnal awakenings and sometimes showed auto-aggressive and hyperactive behaviors, which were treated with Methylphenidate.
A cranial MRI at 2 years and 3 months showed a bilateral hypoplasia of cerebellar tonsils. At 5 years, mild hyperopia (<1 dpt) and astigmatism were diagnosed, as well as atopic dermatitis.
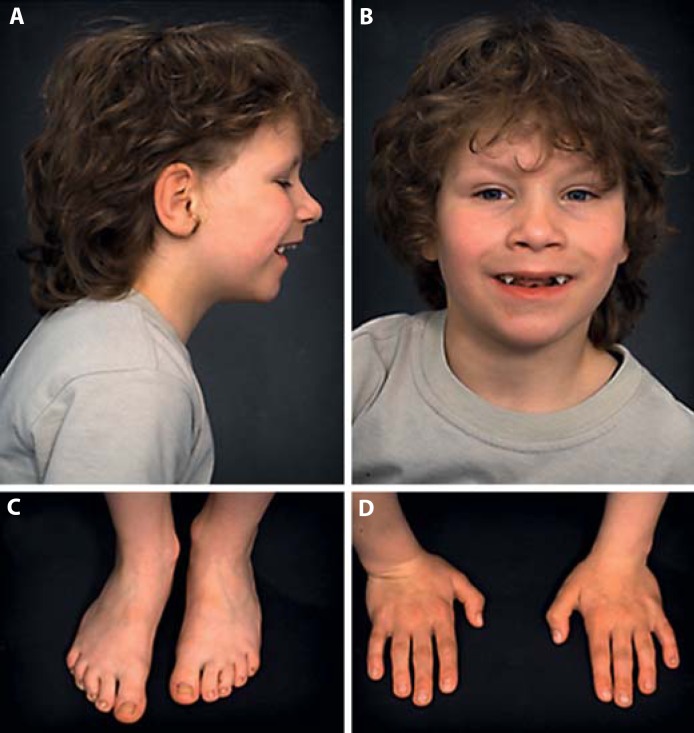
On examination at 7 years and 3 months, his weight was 23 kg (25th–50th percentile), height 116.5 cm (3rd percentile), and OFC 50 cm (<3rd percentile). His mouth was open frequently, but no muscular hypotonia was apparent. He showed mild DD and slight dysmorphisms including deep-set eyes (also present in his parents), a thin upper lip, a high-arched palate, prominent canine teeth (also in the father), and mild microretrognathia (fig. 1). Only mild digital anomalies were present (2nd and 3rd toes: slight syndactyly; 2nd toes: lateral deviation; 3rd toes: medial deviation; 5th toes: hypoplastic nails), as well as 3 café-au-lait spots.
Fig. 1.
Our patient at the age of 7 years and 3 months. A Lateral right: mild microretrognathia, deep-set eyes. B Frontal: thin upper lip, pointed canine teeth. C, D Feet and hands: minor digital anomalies.
Results of conventional karyotyping (400-band level), subtelomeric screening, fragile X testing, and testing for frequent microdeletion syndromes (SALSA MLPA kit P245-A2 Microdeletion Syndromes-1, MRC-Holland, Amsterdam, The Netherlands) were unremarkable.
Materials and Methods
Genomic DNA from our patient was analyzed using an Illumina Human660W-Quad v1 DNA Analysis BeadChip according to the manufacturer's instructions (Illumina, Inc., San Diego, Calif., USA). As published previously, CNV calling and real-time qPCR were performed to confirm the deletion in the patient and determine parental origin [Engels et al., 2009]. In brief, QuantiSNP software version 2.2 was used to determine CNV calls and to calculate log Bayes factors as a measure of confidence for each CNV call. Using Cartagenia BENCH, a software designed for the management and interpretation of CNV data in routine diagnostics and research, all CNV calls with log Bayes factors <7, fewer than 5 consecutive markers, and <20 kb were disregarded, as well as CNVs without known genes or which were covered entirely by benign frequent CNVs according to the Database of Genomic Variants (http://projects.tcag.ca/variation/). Primer sequences are available upon request.
Mutational screening of LRRTM2 was performed on 330 patients (213 male, 117 female) ascertained at the University Medical Genetics Clinics in Bonn (n = 191) and Heidelberg (n = 139), Germany, including 177 with mild ID/DD (IQ >50) and 126 with moderate to severe ID/DD. In 27 patients, the degree of retardation could not be measured, mostly because of their young age (median: 9.5 months). Although the index patient showed only mild DD, slight dysmorphisms and no malformations, the presence of more severe phenotypes cannot yet be excluded for potential point mutation carriers. Thus, the 330 patients were not pre-selected clinically (non-syndromal/isolated ID/DD: n = 95 (29%); syndromal ID/DD: n = 235 (71%), including 70 (21%) with malformations). Standard evaluation consisted of detailed clinical investigation, conventional karyotyping (n = 300 patients, 91%), and exclusion of clinically recognizable syndromes with known etiology. Fragile X syndrome had been tested for and excluded in 176 patients (53%). In 277 individuals (84%), molecular karyotyping had been performed using Affymetrix, Illumina, or Agilent array platforms without detecting any obvious pathogenic aberrations.
LRRTM2 was sequenced in the index patient and in 330 patients with ID/DD using either genomic or whole genome amplified DNA (REPLI-g WGA kit, Qiagen, Hilden, Germany). Segregation of a sequence alteration not listed in dbSNP was analyzed by sequencing parental DNA, if available. Coding exons and exon/intron boundaries of all isoforms were amplified; primer sequences are available upon request. Amplicons were sequenced unidirectionally using Big Dye Terminator v1.1 and a 3500xL Genetic Analyzer (Applied Biosystems, Foster City, Calif., USA). DNA sequences were compared to reference assembly GRCh36. Similarly, the SIL1 gene was sequenced in the index patient.
Results
For our patient, molecular karyotyping revealed a 0.9-Mb heterozygous deletion in chromosome 5q31.2 (chr5:137,750,721–138,687,057; GRCh36) containing 9 known genes: KDM3B, REEP2, EGR1, ETF1, HSPA9, CTNNA1, LRRTM2, SIL1 and MATR3. Quantitative PCR analyses of the patient and his parents demonstrated a de novo occurrence. Although none of the patients with deletions including SIL1 presented with typical clinical signs of Marinesco-Sjögren syndrome, an autosomal recessive disorder characterized by cerebellar ataxia, early-onset cataracts, ID, and muscular hypotonia, mutations in the remaining SIL1 allele were excluded by DNA sequencing in our patient. We sequenced LRRTM2, the most promising candidate gene for ID in the smallest region of overlap (SRO), in 330 patients. Besides one known SNP found in 20 patients (rs11744283), no coding region mutation in LRRTM2 was detected. In 6 patients, a novel heterozygous alteration in intron 1 was detected (c.4 + 74A>G). In silico programs (http://www.fruitfly.org/seqtools/splice.html) predicted no effect on mRNA splicing. Parental DNA from one of the 6 patients was available for sequencing, and the c.4 + 74A>G alteration was also detected in the healthy mother. This suggests no obvious association between the c.4 + 74A>G alteration and the occurrence of ID.
Discussion
Only few patients with de novo microdeletions encompassing 5q31 have been reported. The observed characteristic features include short stature, failure to thrive, ID/DD, hypotonia, club feet, dislocated hips, and characteristic dysmorphic features including prominent forehead, telecanthus/hypertelorism, anteverted nares, cleft or high-arched palate, micro-/retrognathia, and ear abnormalities [Rosenfeld et al., 2011]. The reported microdeletions range from 1.6 to 8.1 Mb and no common SRO has been reported [Mosca et al., 2007; Rosenfeld et al., 2011].
We describe a male patient with the smallest de novo microdeletion (0.9 Mb) involving 5q31.2 reported thus far. His phenotype is relatively mild, with mild DD especially concerning language, microcephaly, borderline short stature with height on the 3rd percentile, muscular hypotonia, and rather mild dysmorphisms including mild microretrognathia and a high palate.
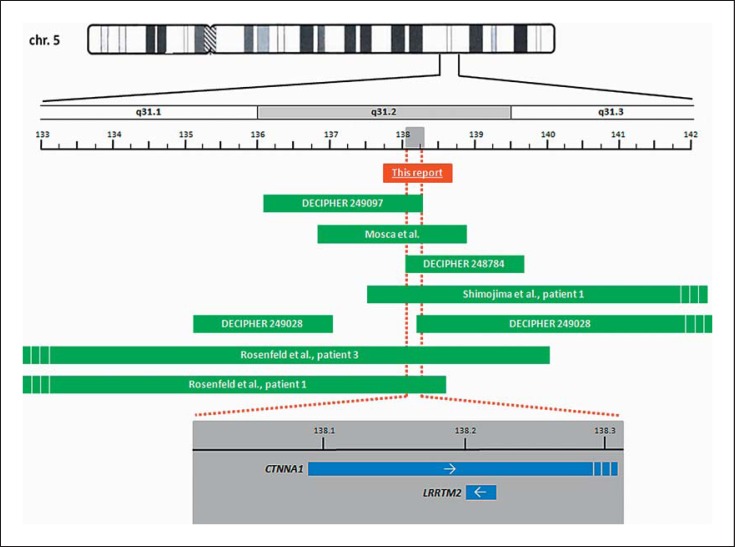
Overlapping microdeletions have been reported in 7 additional patients: 5 are either ‘pure’ deletions (i.e. without other accompanying CNVs) or only accompanied by a most likely benign second aberration [DECIPHER 249097 and 248784; Mosca et al., 2007; Rosenfeld et al., 2011, patient 3; Shimojima et al., 2011], whereas 2 carry relevant additional CNVs [DECIPHER 249028; Rosenfeld et al., 2011, patient 1]. In 4 patients the microdeletions are rather large (5.0–8.1 Mb) and extend either proximally into 5q31.1, distally into 5q31.3, or both. In 3 patients the deletions are confined to 5q31.2, as in our patient (fig. 2, table 1). Interestingly, one of the latter 3 patients showed clinical features similar to our patient, namely mild DD, short stature, microretrognathia, and the absence of malformations [Mosca et al., 2007].
Fig. 2.
Schematic representation of the 5q31.1q31.3 deletions. The dashed red line defines the smallest region of overlap (SRO) of the 6 deletions which are not accompanied by additional relevant imbalances. The grey box represents a magnification of the SRO. Genes within the SRO are depicted by blue bars with the arrows indicating transcription direction. Dashed bars: genes/deletions extend further. All positions are according to assembly GRCh36.
Table 1.
Summary of clinical features observed in individuals with microdeletions of 5q31.1q31.2
| This report | DECIPHER Patient 249097 | Mosca et al. [2007] | DECIPHER Patient 248784 | Shimojima et al. [2011] Patient 1 | DECIPHER Patient 249028 | Rosenfeld et al. [2011] Patient 3 | Rosenfeld et al. [2011] Patient 1 | |
|---|---|---|---|---|---|---|---|---|
| Overlapping deletion | ||||||||
| Position | chr5:137,750,721-138,687,057 | chr5:136,159,881-138,277,839 | chr5:136,886,534-138,717,853 | chr5:138,011,434-139,592,820 | chr5:137,538,788-142,574,719 | chr5:138,234,725-145,172,065 | chr5:131,857,587-139,915,446 | chr5:132,413,216-138,489,956 |
| Subband | 5q31.2 | 5q31.2 | 5q31.2 | 5q31.2q31.3 | 5q31.2q31.3 | 5q31.2q32 | 5q31.1q31.3 | 5q31.1q31.2 |
| Size, Mb | 0.9 | 2.1 | 1.8 | 1.6 | 5.0 | 7.0 | 8.1 | 6.1 |
| Number of protein coding RefSeq genes | 9 | 21 | 23 | 16 | 95 | 89 | 84 | 52 |
| Additional de novo imbalances | none | duplication | none | none | none | deletion | none | deletion |
| Position | chr6:29,032,508-29,333,774 | chr5:135,257,630-136,982,620 | chr5:53,474,049-54,593,863 | |||||
| Subband | 6p22.1 | 5q31.1q31.2 | 5qll.2 | |||||
| Size, Mb | 0.3 | 1.7 | 1.1 | |||||
| Number of protein coding RefSeq genes | 5 | 8 | 10 | |||||
| Age at examination | 7 years 3 months | 1 year | 1 year 11 months | < 1 year | 1 year 6 months | n.r. | newborn | 10 years |
| Sex | male | female | female | male | male | female | male | male |
| Developmental delay | + | + | + | +* | + | + | n.a. | + |
| Speech | delayed | delayed | delayed | n.a. | n.a. | n.r. | n.a. | delayed |
| First words at age | 18 months | n.r. | 14 months | n.r. | ||||
| Walking age | 21 months | 16 months | n.a. | n.a. | n.r. | n.a. | 3 years 6 months | |
| Length/height | proportionate short stature | |||||||
| At birth | P 3-10 | P 25-50 | n.r. | +0.5 SD | n.r. | n.r. | P 10 | |
| At examination | P 3 | <P 3 | n.r. | −1.6 SD | n.r. | n.a. | ≪P 3 | |
| Weight | ||||||||
| At birth | P 3 | n.r. | P 3-10 | n.r. | −0.3 SD | n.r. | P 10-25 | <P 3 |
| At examination | P 25-50 | n.r. | <P 3 | n.r. | −1.7 SD | n.r. | n.a. | <P 3 |
| OFC | ||||||||
| At birth | P 3-10 | n.r. | P 3-10 | n.r. | + 1.3 SD | n.r. | P 10-25 | P 50 |
| At examination | <P 3 | n.r. | >P 25 | n.r. | −3.0 SD | n.r. | n.a. | <P 3 |
| Cranial MRI/CT findings | bilateral hypoplasia of cerebellar tonsil | n.r. | − | n.r. | severely delayed reduced cerebral volume myelination | n.r. | − | hydrocephalus, otherwise normal |
| Palate anomalies | (+) (high) | n.r. | − | n.r. | (+) (high) | n.r. | + (cleft) | + (cleft) |
| Muscular hypotonia | (+) | n.r. | n.r. | + | + | + | + | + |
| Feeding difficulties | (+) | n.r. | n.r. | + | + | n.r. | n.r. | + |
| Congenital heart defect | − | n.r. | − | n.r. | PDA, small VSD | PDA | multiple VSDs, ASD, PDA | VSD |
| Skeletal anomalies | − | n.r. | n.r. | n.r. | − | n.r. | + widely open anterior fontanelle, sacral anomaly, joint hypermobility | + hip dysplasia, neuromuscular scoliosis, sternal asymmetry, delayed bone age |
| Hand anomalies | − | n.r. | − | n.r. | + postaxial Polydactyly | n.r. | + preaxial Polydactyly, long fingers/thumbs, single flexion crease of 5th finger, ulnar deviation of hands | + distally broadened, long fingers |
| Foot anomalies | + bilateral mild syndactyly of 2nd/3rd toes | n.r. | − | n.r. | − | n.r. | + club feet, long toes, with hypoplastic nails, bulbous tips of great toes, duplicated left hallux | + long toes, small great toenails, bilateral metatarsus adductus |
| Facial anomalies | deep-set eyes, thin upper lip, mildly anteverted nares, prominent canine teeth (also in the father), mild microretrognathi | upslanting palpebral fissures, small mandible/micrognathia | upslanting palpebral fissures, hypertelorism, anteverted nares, down-turned corners of the mouth, microretrognathia | n.r. | narrow forehead, bilateral ptosis, epicanthic folds, low-set and abnormal auricles, depressed nasal bridge, anteverted nares, long philtrum, edematous cheeks, tented upper vermilion | coarse facial features | broad forehead, hypertelorism, low-set ears, reduced upper helix, severe micrognathia, short neck | downslanting palpebral fissures, epicanthal folds, mild hypertelorism, posteriorly rotated ears, prominent helices, prominent nasal bridge, long philtrum, peaked upper lip, micrognathia |
| Ophthalmological findings | hyperopia, astigmatism | n.r. | − | n.r. | − | n.r. | − | strabismus, corneal clouding, myopia, astigmatism |
| Others | 3 café-au-lait spots, autoaggressive episodes, formerly: nocturnal awakenings | n.r. | n.r. | hyporeflexia, history of apnea | peripheral neuropathy | trachea or laryngeal anomalies | microphallus, hydronephrosis | growth-hormone deficiency, mild high-frequency hearing loss |
ASD = Atrial septal defect; EEG = electroencephalography; P = percentile; PDA = persistent ductus arteriosus; VSD = ventricular septal defect; * = personal communication; + = present; (+) = present transiently or mildly; − = absent; n.r. = not reported; n.a. = not analyzed; bold characters = common clinical findings; positions according to GRCh36.
Two patients were identified in DECIPHER. Male patient 248784 has an overlapping 1.6-Mb deletion in 5q31.2, with reported clinical signs including muscular hypotonia and feeding difficulties, as well as DD (pers. commun.). The 5q31.2 deletion in female patient 249097 spans 2.1 Mb, with reported clinical signs including DD/speech delay, short stature, and micrognathia. This patient also carries a second, likely benign 300-kb de novo duplication in 6p22.1. Three factors argue in support of a negligible or comparably small phenotypical influence of this second CNV: (1) duplications are widely believed to influence gene dosage less frequently than deletions [Veitia and Birchler, 2010]; (2) evidence of a de novo occurrence of a CNV alone is insufficient for its causality, especially for small CNVs such as the 300-kb duplication in question [Vermeesch et al., 2011]; (3) the duplication contains only 6 RefSeq genes, 4 of which are olfactory receptor genes which are known to be very copy-number variable [Waszak et al., 2010], and 2 of which are only provisional/predicted genes.
Taken together with the clinical similarities to patients with ‘pure’ deletions, the phenotypic influence of the 5q31.2 deletion in DECIPHER patient 249097 is likely predominant enough to include this patient, although a minor influence of the second CNV on the patient's phenotype cannot be excluded entirely.
The phenotype associated with larger overlapping deletions that extend distally into 5q31.3 is more severe and appears distinguishable from the clinical picture reported here and by Mosca et al. [2007] for deletions concerning only 5q31.2. Patient 1 reported by Shimojima et al. [2011] carries a 5-Mb deletion in 5q31.2q31.3 and has severe ID/DD, encephalopathy, severe hypotonia, pronounced postnatal microcephaly, congenital heart defects, and distinctive facial features (table 1). Patient 2 in the same report displayed a similar, severe phenotype, but had a more distal deletion that does not overlap the deletion in our patient. Thus, Shimojima and colleagues attributed the severe phenotype common to these 2 patients to a more distal region in 5q31.3, which does not overlap the deletion found in our patient. DECIPHER contains an entry for a second patient with an overlapping microdeletion that extends distally: patient 249028 is reported to display ID/DD, hypotonia, patent ductus arteriosus, trachea/laryngeal anomalies, and coarse facial features. Here, a genotype-phenotype correlation is complicated by a second, non-overlapping de novo deletion in 5q31.1q31.2 containing 11 RefSeq genes.
Rosenfeld et al. [2011] report a patient (patient 1) with a larger deletion that extends proximally into 5q31.1 (fig. 2, table 1). Besides DD, he also presented with hydrocephalus, cleft palate, bilateral hip dysplasia, pronounced postnatal short stature, microcephaly, and dysmorphic features. In addition to a 6.1-Mb deletion in 5q31.1q31.2, he also carried a 1.1-Mb deletion of 5q11.2 that contains 14 genes and could likely influence his phenotype. Patient 3 in the same report was diagnosed with an 8.1-Mb deletion in 5q31.1q31.3 that extends further both proximally and distally compared to our patient [Rosenfeld et al., 2011]. This newborn male displayed multiple congenital anomalies including a large cleft palate, severe micrognathia, preaxial polydactyly, club feet, a duplicated hallux, multiple septal heart defects, significant hypotonia, and several dysmorphic features.
Although these tentative genotype-phenotype correlations are based on a small number of patients, the clinical pictures of 5q31 microdeletions may fall into 3 distinguishable categories. The phenotype of small medial microdeletions concerning 5q31.2 only, such as the deletion reported here and by Mosca et al. [2007], seems to be relatively mild, with mild DD, no malformations, mild or borderline short stature, and rather nonspecific dysmorphic signs. In contrast, in patients with larger deletions also affecting proximal subband 5q31.1 [Rosenfeld et al., 2011], postnatal short stature appears to be more pronounced, and congenital heart defects, cleft palate, and skeletal abnormalities of hands and feet may be present. Microdeletions that extend distally into subband 5q31.3 appear to be associated with a combination of severe ID/DD, typical cranial MRI findings, and characteristic facial features. Although the 8 patients with overlapping microdeletions in 5q31 do not have a common recognizable phenotype, all patients in which the psychomotor development could be assessed displayed ID/DD, which often especially concerns language development.
The SRO of the 6 ‘pure’ microdeletions is 240 kb and contains only 2 genes: CTNNA1 (partial) and LRRTM2 (fig. 2). CTNNA1 is an unlikely candidate gene for ID due to its function as a tumor suppressor gene [Shibata et al., 2007] and its low probability for haploinsufficiency (score 22%) [Huang et al., 2010]. However, LRRTM2 (leucine-rich repeat transmembrane neuronal protein 2) is an excellent candidate gene for DD/ID, as it is a key regulator of excitatory synapse development and function and has a predicted very high probability for haploinsufficiency (score 7.4%) [Huang et al., 2010]. Lrrtm2 is expressed in the neural tube during mouse embryogenesis in mid-gestation [Haines and Rigby, 2007]. It induces presynaptic differentiation and LRRTM2-Neurexin 1 interaction plays a critical role in regulating excitatory synapse development [de Wit et al., 2009]. Neurexin 1 plays an important role in autism and schizophrenia [Rujescu et al., 2009] as well as in ID/DD [Zweier et al., 2009]. We therefore propose LRRTM2 as a promising candidate gene for ID/DD in patients with 5q31.2 microdeletions.
We did not identify any additional patients with overlapping deletions amongst the 1,891 patients with ID/DD in the German MRNET (Mental Retardation Network Database, http://www.german-mrnet.de). Therefore, we performed mutational screening of LRRTM2 in 330 patients with ID/DD. One known SNP (rs11744283) was found in 20 patients and 1 novel intronic alteration in 6 patients; we determined that the novel sequence alteration is likely benign. Thus, we did not identify a causative mutation in the coding region of LRRTM2, excluding mutations as a frequent cause of ID/DD in patients without microdeletions. Although no additional pathogenic mutations of LRRTM2 were detected, we consider LRRTM2 haploinsufficiency as the most probable cause for ID/DD in our patient and the patients with small overlapping deletions. Mutation screening in a larger cohort may be necessary to detect rare LRRTM2 mutations.
In conclusion, we present the smallest microdeletion in 5q31 reported to date, and present evidence for a 240-kb critical region. We propose that LRRTM2 is a candidate gene for mild to moderate ID/DD in 5q31.2 microdeletions. The phenotype of small microdeletions concerning 5q31.2 only, such as the deletion in our patient, seems to be relatively mild, with mild ID/DD, no malformations, and rather nonspecific dysmorphic signs. The analysis presented herein contributes considerably to the genotype-phenotype correlation for microdeletions in 5q31.
Acknowledgements
We thank the patient and his family for their kind cooperation. A.M.Z., U.M., G.A.R., E.W., and H.E. are members of the ‘German Mental Retardation Network’ (MRNET), which is funded by the German Federal Ministry of Education and Research (BMBF) as a part of the National Genome Research Network (NGFNplus, www.ngfn.de/en/ngfnplus.html, project reference numbers 01GS08164, 01GS08160).
References
- Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu T, et al. A copy number variation morbidity map of developmental delay. Nat Genet. 2011;43:838–846. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit J, Sylwestrak E, O'sullivan ML, Otto S, Tiglio K, et al. LRRTM2 interacts with Neurexin1 and regulates excitatory synapse formation. Neuron. 2009;64:799–806. doi: 10.1016/j.neuron.2009.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels H, Wohlleber E, Zink A, Hoyer J, Ludwig KU, et al. A novel microdeletion syndrome involving 5q14.3–q15: clinical and molecular cytogenetic characterization of three patients. Eur J Hum Genet. 2009;17:1592–1599. doi: 10.1038/ejhg.2009.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines BP, Rigby PWJ. Developmentally regulated expression of the LRRTM gene family during mid-gestation mouse embryogenesis. Gene Expr Patterns. 2007;7:23–29. doi: 10.1016/j.modgep.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Huang N, Lee I, Marcotte EM, Hurles ME. Characterising and predicting haploinsufficiency in the human genome. PLoS Genet. 2010;6:e1001154. doi: 10.1371/journal.pgen.1001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86:749–764. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosca AL, Callier P, Leheup B, Marle N, Jalloul M, et al. Fortuitous FISH diagnosis of an interstitial microdeletion (5)(q31.1q31.2) in a girl suspected to present a cri-du-chat syndrome. Am J Med Genet A. 2007;134A:1342–1347. doi: 10.1002/ajmg.a.31742. [DOI] [PubMed] [Google Scholar]
- Rosenfeld JA, Drautz JM, Clericuzio CL, Cushing T, Raskin S, et al. Deletions and duplications of developmental pathway genes in 5q31 contribute to abnormal phenotypes. Am J Med Genet A. 2011;155A:1906–1916. doi: 10.1002/ajmg.a.34100. [DOI] [PubMed] [Google Scholar]
- Rujescu D, Ingason A, Cichon S, Pietiläinen OP, Barnes MR, et al. Disruption of the neurexin 1 gene is associated with schizophrenia. Hum Mol Genet. 2009;18:988–996. doi: 10.1093/hmg/ddn351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata H, Takano H, Ito M, Shioya H, Hirota M, et al. Alpha-catenin is essential in intestinal adenoma formation. Proc Natl Acad Sci USA. 2007;104:18199–18204. doi: 10.1073/pnas.0705730104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimojima K, Isidor B, Le Caignec C, Kondo A, Sakata S, et al. A new microdeletion syndrome of 5q31.3 characterized by severe developmental delays, distinctive facial features, and delayed myelination. Am J Med Genet A. 2011;155A:732–736. doi: 10.1002/ajmg.a.33891. [DOI] [PubMed] [Google Scholar]
- Veitia RA, Birchler JA. Dominance and gene dosage balance in health and disease: why levels matter! J Pathol. 2010;220:174–185. doi: 10.1002/path.2623. [DOI] [PubMed] [Google Scholar]
- Vermeesch JR, Balikova I, Schrander-Stumpel C, Fryns J-P, Devriendt K. The causality of de novo copy number variants is overestimated. Eur J Hum Genet. 2011;19:1112–1113. doi: 10.1038/ejhg.2011.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waszak SM, Hasin Y, Zichner T, Olender T, Keydar I, et al. Systematic inference of copy-number genotypes from personal genome sequencing data reveals extensive olfactory receptor gene content diversity. PLoS Comput Biol. 2010;6:e1000988. doi: 10.1371/journal.pcbi.1000988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweier C, de Jong EK, Zweier M, Orrico A, Ousager LB, et al. CNTNAP2 and NRXN1 are mutated in autosomal-recessive Pitt-Hopkins-like mental retardation and determine the level of a common synaptic protein in Drosophila. Am J Hum Genet. 2009;85:655–666. doi: 10.1016/j.ajhg.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]