Abstract
The ubiquitin-proteasome system has numerous crucial roles in physiology and pathophysiology. Fundamental to the specificity of this system are ubiquitin-protein ligases (E3s). Of these, the majority are RING finger and RING finger-related E3s. Many RING finger E3s have roles in processes that are central to the maintenance of genomic integrity and cellular homeostasis, such as the anaphase promoting complex/cyclosome (APC/C), the SKP1–cullin 1–F-box protein (SCF) E3s, MDM2, BRCA1, Fanconi anaemia proteins, CBL proteins, von Hippel–Lindau tumour suppressor (VHL) and SIAH proteins. As a result, many RING finger E3s are implicated in either the suppression or the progression of cancer. This Review summarizes current knowledge in this area.
Consequent to the determination that many RING finger-containing proteins are ubiquitin-protein ligases (E3)1, and therefore the primary determinants of substrate specificity in ubiquitylation (also known as ubiquitination; Box 1, Box 2 and Box 3), it has become evident that many RING finger E3s are implicated in malignancy. Oncogenic transformation is characterized by dysregulated cell growth signals leading to limitless autonomous proliferation, insensitivity to anti-growth or pro-apoptotic signals, dysregulation of the cell cycle and genomic instability. Solid tumors also acquire the ability to induce angiogenesis, which allows for expansion of the primary tumour and facilitates metastasis2. RING finger E3s are implicated in all of these steps. Some are bona fide oncogenes whereas others are products of tumor suppressor genes (TSGs). However, a single E3 can also have opposing functions in malignancy owing to multiple substrates or multiple roles of a single substrate (Table 1 and below).
Box 1 Ubiquitylation by RING finger E3s.
Ubiquitylation is a multienzyme process (see part a of the figure) in which ubiquitin (Ub) is first activated in an ATP-dependent reaction that leads to a high-energy thiolester linkage between the carboxy-terminal glycine residue of ubiquitin and the active site cysteine residue on the E1 (ubiquitin-activating enzyme) protein. Ubiquitin is transferred to the active site cysteine of one of approximately 40 E2s (ubiquitin-conjugating enzymes or ubiquitin carrier proteins), where a second high-energy thiolester linkage is formed. RING finger (RF) E3s interact with the substrate and a ubiquitin-charged E2 and mediate the direct transfer of ubiquitin from E2 to the substrate, most frequently either forming an isopeptide bond between the C terminus of ubiquitin with an internal lysine or forming a peptide bond with the amino terminus of the substrate. Less frequently observed are linkages with internal serine, threonine or cysteine residues of the substrates. Ubiquitin can also be added to the growing end of a nascent ubiquitin chain on a substrate (see right side of part a of the figure), usually through linkages between internal lysines of the most distal of the substrate-bound ubiquitins and the C terminus of the newly added ubiquitin. Linkages through the N terminus of a bound ubiquitin chain can also occur. After multiple rounds, polyubiquitin (also known as multiubiquitin) chains are formed. RING finger E3s can mediate the addition of a single ubiquitin to the substrate (monoubiquitylation; see part b of the figure), the addition of single ubiquitin molecules to multiple different sites on the substrate (multi-monoubiquitylation) or the addition of polyubiquitin chains, potentially on any of the lysines of ubiquitin. Lys48- linked chains, which are best known for their role in targeting proteins for degradation by the 26S proteasome, are often depicted (based on structure) as having a kinked appearance. By contrast, Lys63-linked chains, which are implicated in a number of nonproteasomal functions, are often depicted as having a linear arrangement. Ubiquitylation is central to diverse processes that regulate protein fate and function, most prominently proteasomal degradation, but also processes such as endocytosis, activation of signal transduction cascades and DNA repair. The outcome of ubiquitylation is specified by several factors, including the type of ubiquitylation of the substrate, cellular location and context, interaction with specific ubiquitin receptors, and by the opposing activity of specific deubiquitylating enzymes. Figure reproduced with permission © Macmillan Publishers (2011).
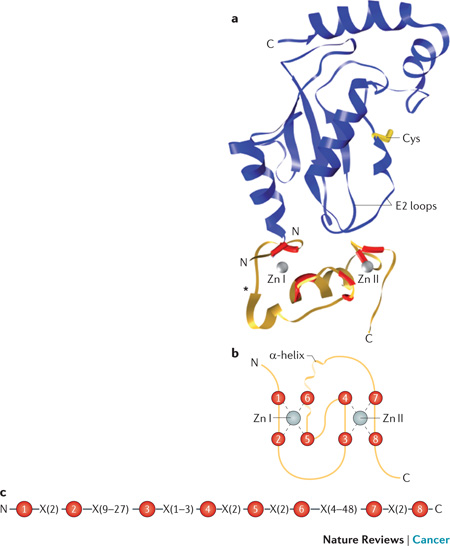
Box 2 RING finger E3 Structure.
The vast majority of known E3s contain a RING finger, which is a small (~40–60 amino acid) domain that coordinates two zinc (Zn) ions. Active RING finger domains interact with a subset of the ~40 E2s expressed in humans and mediate the transfer of ubiquitin from the active site of the E2 to acceptor residues on target proteins or the growing end of ubiquitin chains. A ribbon diagram, based on a model of the RING finger of CNOT4 bound to the E2 UBCH5B (blue; catalytic cysteine of E2 in yellow) is shown in the figure above (N and C termini indicated; PDB ID: 1UR6)155 with a schematic of the RING finger directly below. The RING finger includes eight residues (red in model and schematic) that coordinates the two Zn ions (silver) in a cross-braced pattern. Residues 1, 2, 5 and 6 interact with one Zn and residues 3, 4, 7 and 8 interact with the second. Most coordinating amino acids are cysteines, there are frequently 1–2 histidines, generally not found in the first or last pair of coordinating residues, and in rare cases (e.g., RBX1) an acidic residue can function as a coordinating residue. The Zn I region interacts with the N-terminus and other regions (E2 loops) of the E2. The α-helix, which generally includes the final coordinating residue for Zn I (i.e., residue 6) and the Zn II region interact with the E2 loops. RING fingers have conserved spacing between most of the coordinating residues but there is considerable variability in the loop between the second and third coordinating residues (*) and in the length of the region containing the α-helix between the sixth and seventh residues (see linear representation below schematic: coordinating residues numbered in red, X indicates intervening amino acids followed by spacing in numbers). The leukocyte-association protein- or plant homeodomain (LAP/PHD) is a variation on the RING finger. The U-box resembles the RING finger but its structure is determined by interactions between U-box amino acids rather than coordination of Zn ions. Many of both of these are established E3s. Figure reproduced with permission © Macmillan Publishers (2011).
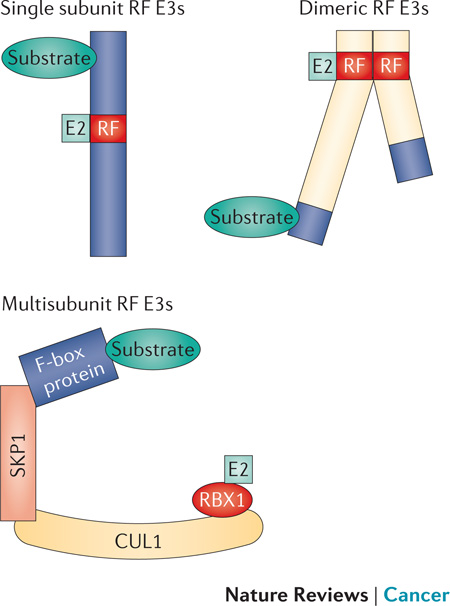
Box 3 RING finger E3 types.
RING finger E3s can be single subunit E3s in which the RING finger (red) is surrounded by protein-interacting motifs (dark blue). A number of RING finger E3s exist as homo-or heterodimers where the RING finger and/or surrounding regions serve as the site of dimerization and enhances or directs E3 activity (for example, BRCA1-BARD1 and MDM2-MDMX). For a number of heterodimeric E3s only one RING finger functionally interacts with E2 (e.g., BRCA1 and MDM2). The most complex RING finger E3s are multisubunit E3s, which include the cullin RING ligase (CRL) superfamily (including the SCF and CRL2 E3 families), the anaphase promoting complex/cyclosome (APC/C) and the Fanconi anaemia (FANC) E3 complex. In CRLs a cullin protein serves as a scaffold to assemble multiple proteins including a small RING finger protein RBX1, adaptor proteins (such as SKP1 in the SCF, elongin B-elongin C in CRL2), and substrate targeting proteins (such as F-box proteins for the SCF complex, von Hippel Lindau tumour suppressor protein (VHL) and SOCS box proteins for the CRL2 family). RING finger E3s recognize substrates through diverse protein-protein interactions. This regulated recognition is modulated by other post-translational modifications that include phosphorylation, glycosylation and sumoylation, and the activity of CRLs is generally activated by neddylation. There are not general consensus sites for ubiquitylation, although the binding sites for E3s on target proteins can be highly specific and often dictate sites of ubiquitylation. Proteins may be targeted by multiple E3s and conversely E3s can have multiple substrates, including themselves. Moreover, different E3s might recognize the same or different sites on proteins and a single E3 can have multiple ways to recognize target proteins. Figure reproduced with permission © Macmillan Publishers (2011).
Table 1.
RING finger E3s as oncogenes and tumour suppressor genes
| E3 | Function | Role in Cancer* | Refs |
|---|---|---|---|
| Cell Cycle | |||
| APC/C ligases | Multisubunit E3s that regulate the cell cycle. | Tumor suppressor complexes. Evidence of mutations in several subunits that disrupt function in colon cancer cells, resulting in increased accumulation of cyclins and progression of the cell cycle. Some experimental data suggest that compromised function of APC/C by loss of substrate recognition subunits, CDC20 or CDH1, can result in genomic instability consistent with a role for APC/C as a tumour suppressor | 7–9 |
| β–TrCP (FBW1A) | F-box protein, functions as a substrate recognition component of the SCF E3 complex, which is involved in degradation of phosphorylated cell cycle and signaling molecules. β–TrCP targets the SCF complex to multiple proteins that have both pro- and anti-proliferative effects such as BCL2L11 (also known as BimEL), CDC25A, β-catenin, IκB, PDCD4, SMAD3, SMAD4 and WEE1 | A potential oncogene. Transgenic mice overexpressing β–TrCP have an increased incidence of epithelial tumors. Consistent with its function as an oncogene, β–TrCP is overexpressed and associated with poor prognosis in many human epithelial cancers. However, β–TrCP mutations described in GI malignancies are associated with stabilization and the accumulation of nuclear β-catenin, consistent with a role as a TSG. This suggests that the oncogenic or tumour suppressive effects of β–TrCP are context-dependent | 5,13 |
| COP1 (RFWD2) | RING finger component of a multisubunit E3 that targets multiple transcription factors including members of the JUN and ETS family for proteasomal degradation. Also reported to target p53 for proteasomal degradation (see Supplementary information supplementary table 1) | A TSG. Rfwd2 hypomorphic mice develop thymic lymphomas, teratomas and uterine tumors. Rare deletions of RFWD2 have been described in lymphoblastic lymphoma, melanoma and prostate cancer. There is an inverse correlation between low COP1 expression and high expression of JUN and ETS family members in prostate cancer. Translocations of ETS family genes that delete COP1 binding sites stabilize ETS proteins | 158,159 |
| EMI1 (FBXO5) | Inhibits activity of APC/CCDH1 and allows progression from G1 to S phase. In some cells it also functions to inactivate APC/CCDC20 | A potential oncogene. EMI1 is overexpressed in breast, colon, ovarian, uterine and lung cancer. Predicted to allow cell cycle progression when APC/CCDH1 is inhibited | 160–162 |
| FBXW7 | F-box and WD repeat containing protein that functions as a recognition component of an SCF E3 complex. Substrates include cyclin E, MYC, NOTCH and JUN | A haploinsufficient TSG. Loss-of-function mutations identified in cholangiocarcinoma, T-ALL, breast, bladder, ovarian, liver, lung, bone and endometrial cancers. There is evidence that mutant alleles dimerize with and inhibit wild-type alleles | 10,163 |
| SKP2 | F-box protein that functions as a recognition component of the SCF E3 complex that targets p27 and other cell cycle proteins | An oncogene. SKP2 is amplified in human epithelial cancers including small cell and non-small-cell lung cancer, glioblastoma, squamous cell esophageal cancer, cervical cancer and thyroid cancer and overexpressed in many human tumors. Cooperates with activated RAS in transformation assays. Transgenic mice expressing SKP2 and activated NRAS develop lymphomas with increased frequency and decreased latency compared to activated NRAS alone. High expression of SKP2 correlates with high-grade lymphoma | 3,164–166 |
| Genomic integrity | |||
| BARD1 | RING finger protein without E3 activity. Heterodimerizes with BRCA1 and enhances BRCA1 E3 activity. Functions in HR DNA repair pathway | A TSG. Patients with familial breast cancer have been described with homozygous deletion or inactivating mutations | 56,167–169 |
| BRCA1 | RING finger E3. Functions in transcription and HR DNA repair pathway. | A TSG. Deleted or inactivated in patients with familial breast and ovarian cancer. | 56,170 |
| CUL7 | Cullin protein. Forms an SCF-like multisubunit E3 complex. Heterodimerizes with PARC (also known as CUL9) and inactivates p53 | A potential oncogene. Prevents MYC-induced apoptosis and cooperates with MYC in transformation assays. Appears to function by inactivating p53 | 171–173 |
| FANC core complex | Multisubunit E3 that monoubiquitylates FANCD2 and FANCI and regulates DNA repair. FANCL contains a RING finger-like PHD domain with E3 activity | A tumour suppressor. Mutations in individual FANC E3 subunits lead to Fanconi anaemia, which is associated with an increased risk of cancer | 73 |
| MDM2 | RING finger E3. Inactivates p53 by ubiquitylation and proteasomal degradation. Other substrates identified (see Supplementary information) | An oncogene. Transforming protein when overexpressed, it is amplified in human cancers.. | 15,174 |
| MDMX (MDM4) | RING finger protein without E3 activity. Dimerizes with MDM2 and enhances p53 ubiquitylation. | An oncogene. Enhances degradation of p53 by MDM2. Transforming protein when overexpressed. Amplified in human cancers. | 31,175–177 |
| PARC (CUL9) | Cullin-like protein that forms a multisubunit E3 complex with the RING finger protein RBX1. | A potential oncogene. Sequesters p53 in the cytoplasm and prevents p53 activation. | 172,178 |
| PIRH2 (RCHY) | RING finger E3 that ubiquitylates and targets p53 for proteasomal degradation | A potential oncogene. Overexpressed in lung cancer resulting in p53 degradation. For additional information see Supplementary Information (supplementary table 1). | 179,180 |
| Signal Transduction | |||
| CBLs | RING finger E3s that target activated kinases for ubiquitylation and degradation. | Oncogenes. Dominant-negative forms of Cbl act as oncogenes in mice and NIH-3T3 cells. Mutations that create dominant-negative forms of CBL have been found in human myeloid neoplasms. Mutations in CBL binding sites on kinases or in negative regulators of CBL have been described in various cancers. | 113 |
| FBXW5 | F-box and WD repeat protein. Substrate binding component of CRL4 E3 (CRL4FBW5) | A potential oncogene. Targets the tumour suppressor protein TSC2 for proteasomal degradation | 181 |
| Hakai (CBLL1) | Single subunit RING finger E3 that targets E-cadherin for degradation | A potential oncogene or prometastatic gene. Promotes cell migration, proliferation and anchorage-independent growth | 88,182 |
| IAPs | RING finger E3s that autoubiquitylate and ubiquitylate caspases and TRAFs. They regulate NF-κB signaling and also negatively regulate caspase activation | Oncogenes and/or TSGs that inhibit apoptosis and promote cell proliferation. Overexpressed in many malignancies. Translocations creating MALT1-cIAP2 fusion protein are seen in 25% of MALT lymphomas. These fusions delete the RING finger of cIAP2 and overexpression activates the NF-κB pathway. Homozygous deletions of the chromosome region containing BIRC2 and BIRC3 (which encode cIAP1 and cIAP2, respectively) described in multiple myeloma are associated with increased NF-κB activity | 95,183–187 |
| TRAFs | A family of RING finger E3s that positively and negatively regulate NF-κB activation | Oncogenes and/or TSGs. Missense mutations identified in TRAF2 and TRAF5 in 2–5% of B-cell lymphomas. Overexpression experiments of one such mutation in TRAF2 demonstrated increased NF-κB activity, although the mechanism is not defined. By contrast, inactivating mutations or homozygous deletions of TRAF2 and TRAF3 have been described in multiple myeloma associated with increased NF-κB activity and are consistent with a TSG function for these TRAFs | 183,186,188 |
| TRC8 (RNF139) | RING finger E3 with sterol sensing domain involved in protein biosynthesis | A TSG. Disruption of RNF139 by translocations is found in patients with familial clear cell renal cancer, patients with thyroid cancer, and a patient with dysgerminoma. Overexpression of TRC8 suppresses tumor cell growth | 189–191 |
| Hypoxia | |||
| VHL | Recognition component of the cullin based CRL2VHL E3 complex. Targets HIF transcription factors for degradation under conditions of normoxia | A TSG lost in von-Hippel Lindau syndrome, which is associated with CNS tumors, haemangioblastomas, pheochromocytomas and clear cell kidney cancer. VHL is inactivated in 40–80% of sporadic clear cell kidney cancer | 131–133 |
| SIAHs | RING finger E3s | Potential oncogenes. Ubiquitylates and targets proline hydroxylases for proteasomal degradation, which results in stabilization of HIFα transcription factors | 152,153 |
| Metastasis | |||
| gp78(AMFR) | RING finger E3 implicated in endoplasmic reticulum-associated degradation (ERAD). | A potential oncogene or prometastatic gene. Promotes sarcoma metastasis by degrading metastasis suppressor KAI1 (also known as CD82). | 192 |
β-TrCP, β-transducin repeat containing protein; AMFR, human tumor autocrine motility factor receptor; APC/C, anaphase-promoting complex/cyclosome; BARD1, BRCA1 associated RING domain 1; BIRC, baculoviral IAP repeat containing; CDC, cell division cycle; CNS, central nervous system; COP1, constitutive photomorphogenesis protein 1 homologue; CRL, cullin RING ligase; CUL, cullin; EMI1, early mitotic inhibitor 1; FANC, Fanconi anaemia; FBXW, F-box/WD-repeat containing protein; GI, gastrointestinal; HIF, hypoxia-inducible factor; HR, homologous recombination; KAI1, kangai 1; MALT, mucosa-associated lymphoid tissue lymphoma translocation; NF-κB, nuclear factor-κB; PDCD4, programmed cell death protein 4; PHD, plant homeodomain; RCHY, ring finger and CHY zinc finger domain containing 1; RFWD2, RING finger and WD repeat domain protein 2; RNF139, RING finger protein 139; SCF, SKP1-cullin 1-F-box protein; SIAH, seven in absentia homologue; SKP2, S-phase kinase-associated protein 2; T-ALL, T-cell acute lymphoblastic leukaemia; TRAF, TNF receptor-associated factor; TSC2, tuberin; TSG, tumour suppressor gene; VHL, von Hippel Lindau tumour suppressor.
Genes are designated as oncogenes or TSGs when there is functional evidence for their role in cancer and genetic mutations consistent with that role have been found in human cancers. For oncogenes this generally consists of activating mutations and/or gene amplification. For TSGs, this generally consists of inactivating mutations and/or deletions. Genes are designated as potential oncogenes or TSGs when functional studies are consistent with that role but no mutations have been reported in human tumors to date. In some instances a protein may act as an oncoprotein or a tumor suppressor depending on the cellular context.
We focus below on specific RING finger E3s that are of particular therapeutic interest related to their roles in maintaining genomic stability, signalling and in responses to hypoxia with a more comprehensive listing of RING finger E3s associated with cancer provided in Table 1.
RING fingers and the genome
A hallmark of cancer is altered genomic stability. This can be manifest in ways that include a failure to repair mutations or other DNA damage, as well as gross chromosome abnormalities including aneuploidy.
Cell Cycle
The cell cycle must proceed in an orderly fashion to ensure genomic integrity and to prevent dysregulated proliferation. Fundamental to this is precisely-timed degradation of key regulators including cyclins, cyclin-dependent kinase inhibitors, securin, and transcription factors such as MYC and JUN. Two multi-subunit RING finger ubiquitin ligase families are critical to cell cycle progression and regulation3–6. The anaphase promoting complex/cyclosome (APC/C) consists of at least 13 subunits. Although the APC/C is substantially more complex, it shares properties with the cullin RING ligase (CRL) (Box 3) superfamily including having at its core a cullin-like subunit, a small RING finger protein and interchangeable substrate recognition subunits. Two substrate recognition elements have been identified for the APC/C: cell division cycle 20 (CDC20) and CDH1 (also known as FZR1 or CDC homologue 1). APC/CCDC20 and APC/CCDH1 are activated sequentially with the former active from prometaphase to telophase and the latter active primarily in G1 phase of the cell cycle. From late G1 until prometaphase the APC/C is inactive. The regulation of the APC/C is complex and includes temporal ubiquitin-mediated degradation of CDC20, CDH1 and an E2 UBCH10 (also known as UBE2C, UBCX and UBCC), which is one of several employed by the APC/C. Also contributing to regulation of the APC/C is the phosphorylation-dependent degradation of the APC/C pseudosubstrate, early mitotic inhibitor 1 (EMI1; also known as FBX05) (discussed below) 6.
Hemizygous frame shift and point mutations in several subunits of APC/C have been found in colon cancer cell lines and tumors7. Overexpression studies of one mutant subunit suggest that APC/C mutations can act in a dominant negative manner to inhibit function and cause inappropriate progression of the cell cycle7. Also, altered APC/C function can lead to genomic instability owing to decreased degradation of securin or cyclin B1 and cyclin B2 by APC/CCDC20 and consequently to aberrant chromosome segregation8. In addition, CDH1-deficient mice (Fzr1−/−) display genomic instability and Fzr1+/− mice develop epithelial tumors supporting a role for FZR1 as a haploinsufficient TSG9.
A second ligase family strongly implicated in cell cycle regulation is the S-phase kinase-associated protein 1 (SKP1)-cullin 1 (CUL1)-F-box protein (SCF) family of the CRL superfamily, in which each of up to 69 F-box proteins (in humans) can potentially serve as substrate recognition elements (Box 3), although to date less than 20% of these have defined substrates5. The activity of SCF E3s towards substrates during the cell cycle can be regulated by the level of the F-box protein, as is the case for SKP2. SCFSKP2 targets the G1/S cyclin-dependent kinase inhibitor p27 for proteasomal degradation. Therefore increased SKP2 levels would be predicted to result in a failure to appropriately control the G1/S checkpoint and to be oncogenic3. Indeed, amplification of SKP2 is found in multiple carcinomas5. For some F-box proteins, including both F-box/WD repeat-containing protein 7 (FBW7) and β-transducin repeat containing (β-TrCP; also known as FBW1A), regulation of activity occurs by substrate phosphorylation. SCFFBW7 targets substrates that promote cell cycle progression, and therefore proliferation, including cyclin E, MYC, JUN and NOTCH. Consistent with this, FBW7 is a TSG and loss of FBW7 function is seen in a number of carcinomas3,10. SCFβ-TrCP has a substantial number of phosphorylation-dependent substrates and plays a more complex role in the cell cycle and in tumorigenesis. SCFβ-TrCP targets the APC/C inhibitor EMI1 during late G2, thereby relieving one level of APC/C inhibition. Among its other substrates are proteins that either oppose (for example IκB and WEE1) or promote (for example CDC25 and β -catenin (see also transmembrane signalling section below)) cell cycle progression and proliferation. In addition, if Fanconi anaemia group M protein (FANCM) is not degraded prior to mitosis by SCFβ-TrCP this can lead to inappropriate activity of the FANC ubiquitin ligase (see below) and result in chromosome abnormalities. SCFβ-TrCP also targets the transcriptional repressor RE1 silencing transcription factor (REST; also known as NRSF), which can potentially function as either a tumor suppressor or oncogenic protein4,11,12. Additionally, SCF β-TrCP targets multiple pro-apoptotic proteins for degradation (such as BCL2L11 (also known as BimEL), NF-κB inhibitor α IκBα and IκBβ and programmed cell death protein 4 (PDCD4))5,13. Given the diverse roles of its targets, SCFβ-TrCP can function as an oncogene primarily as a consequence of its prosurvival roles and potentially also as a tumor suppressor in a context-dependent manner. The E3 activity of many SCF E3s, including SCFβ-TrCP, towards substrates is regulated by serine and threonine phosphorylation of the substrate. For this reason, whether a particular SCF E3 functions as an oncogene or tumor suppressor may be determined by what substrates are phosphorylated in a particular cell.
MDM2: keeping the ‘guardian of the genome’ in check
The cellular response to genomic damage is largely mediated by the tumor suppressor p53, which blocks cell proliferation by cell cycle arrest or can induce apoptosis, particularly in transformed cells. p53 is best characterized as a transcription factor, although there is increasing evidence for cytoplasmic roles14. Its importance is underscored by the finding of mutations that inactivate p53 in up to 50% of human cancers and that cellular alterations that suppress p53 activity are found in many other cancers15.
The RING finger E3 and oncogene product mouse double minute 2 (MDM2; also know as HDM2 in humans) is a major regulator of p53 that is under the direct transcriptional control of p5316, 17. MDM2 binds directly to p53 and targets itself and p53 for ubiquitylation and proteasomal degradation (Figure 1)18–22. Amplification or overexpression of MDM2 and mutations in p53 represent alternative means of escaping growth control in cancer16. The relationship between MDM2 and p53 was established by early embryonic lethality in Mdm2−/− mice and their rescue by crossing to Trp53−/− mice 23,24. The exquisitely sensitive nature of the p53-MDM2 relationship is underscored by the decrease in tumor formation in mice with reduced MDM2 and dramatic apoptosis in multiple tissues seen with an inducible p53 in mice lacking Mdm2 (REFS 25, 26).
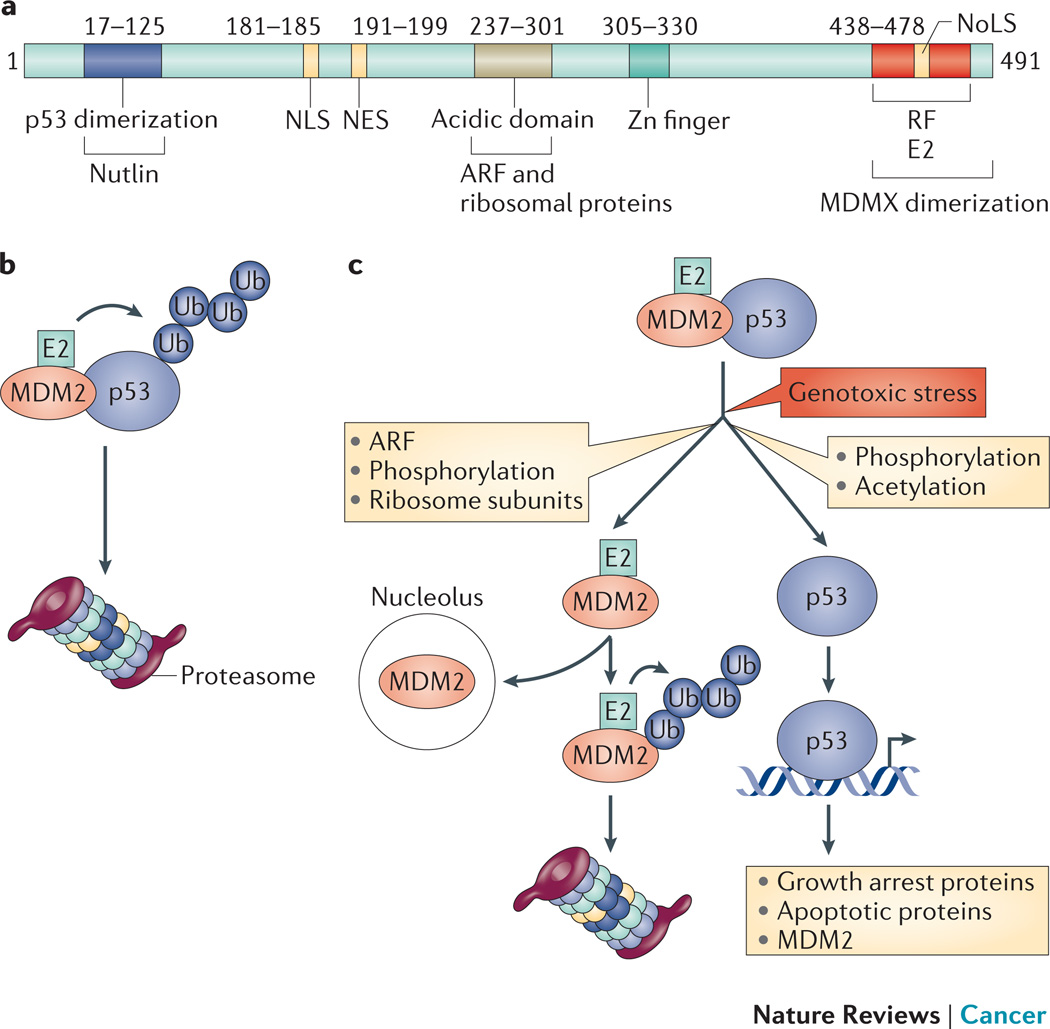
Figure 1. MDM2.
a | A linear representation of MDM2 showing defined domains (boundaries are approximate). MDMX (also known as MDM4) has a similar domain structure but lacks the nuclear localization signal (NLS), the nucleolar localization signal (NoLS), and the nuclear export signal (NES) found in MDM2. Interacting proteins are listed below the domains of MDM2 with which they interact, site of Nutlin binding is also shown. b | MDM2, predominantly as a dimer with MDMX (not shown), binds in vivo to and ubiquitylates p53, which exists largely as a tetramer. This results in the proteasomal degradation of p53. c | In response to genotoxic and other stresses, kinases and acetylases promote the modification of p53 as well as MDM2 such that their binding is reduced and sites of ubiquitylation are unavailable. Additionally, expression of ARF is increased and small ribosome subunits become available. These inhibit the activity of MDM2 towards p53 and result in targeting MDM2 to the nucleolus and potentially increase autoubiquitylation and degradation of MDM2 in the context of the homodimer. This degradation may be further facilitated by stress-induced phosphorylation of MDM2 leading to decreased binding of herpes virus-associated ubiquitin-specific protease (HAUSP; also known as USP7; not shown). As a consequence of its stabilization and dissociation from MDM2 p53 activity increases, resulting in the induction of growth arrest proteins (such as p21) or pro-apoptotic proteins (such as the BH3 only proteins p53-upregulated modulator of apoptosis (PUMA) and NOXA) as well as increased MDM2 expression. Figure reproduced with permission © Macmillan Publishers (2011).
There are other critical players in this relationship. The significance of MDMX (also known as MDM4; HDMX in humans) was established through genetic studies analogous to those for MDM2 (REFS 15,17). MDMX shares the domain structure of MDM2 and can bind p53, however it lacks E3 activity and does not include the subcellular localization signals found in MDM2 (Figure 1). MDM2 can either form homodimers or heterodimerize with MDMX through their RING fingers27–29 and both types of dimers are active as E3s; MDMX has little tendency to form homodimers15,17,30. MDMX plays critical roles in MDM2 stability, presumably by decreasing MDM2 homodimers and thereby limiting MDM2 homodimer ubiquitylation in trans. Importantly, the relative levels of these proteins influences p53 activity in ways that remain to be fully established but are suggested to include enhanced ubiquitylation of p53 when MDMX is expressed31,32. Additionally, the capacity of MDM2 to export p53 to the cytoplasm for degradation is expected to be negatively affected by competition with MDMX33. The p53-MDM2-MDMX relationship is further complicated by all three being substrates for the deubiquitylating enzyme (DUB; also known as deubiquitinating enzyme) herpes virus-associated ubiquitin-specific protease (HAUSP; also known as USP7). Genotoxic stress-induced phosphorylation of MDM2 and MDMX decreases their binding to HAUSP leading to rapid MDM2 and MDMX degradation, thereby contributing to p53 stabilization15.
The levels or availability of other proteins that increase with genotoxic stress bind to MDM2 and decrease its activity towards p53. The tumor suppressor ARF, which is the product of the CDKN2A locus, binds to the central acidic domain of MDM2 and inhibits its activity towards p5334,35; ARF is inactive in a high percentage of tumors that retain wild-type p53 (REF. 36). Several ribosomal proteins, L5 (also known as RPL5), L11 (also known as RPL11) and L23 (also known as RPL23)37, bind MDM2 in a region similar to that described for ARF and function cooperatively37. Free levels of these proteins increase with inhibition of ribosomal biogenesis during genomic stress (REFS 38, 39 and references therein).
Both ARF and these ribosomal proteins can localize to the nucleolus, where MDM2 is similarly re-localized by ARF. This has led to the hypothesis that increases in ARF sequesters MDM2 in the nucleolus, which thereby prevents its interaction with p53. A cryptic nucleolar localization signal located in the MDM2 RING finger is revealed by ARF binding40, which suggests that ARF has the potential to induce significant conformational changes in MDM2. However, the story is more complex as L11 does not appear to affect E3 activity. Moreover both L5 and an ARF peptide inhibit p53 ubiquitylation by MDM2. Furthermore, ARF inhibition of p53 degradation does not require nucleolar localization36,37. A unifying hypothesis is that ARF and these ribosomal proteins bind MDM2 in a region distinct from the RING finger, which leads to a conformational change in the MDM2 RING finger that both inhibits E3 activity and reveals the MDM2 nucleolar localization site.
In addition to the direct effects of interacting proteins, the E3 activity of MDM2 towards p53 is affected by other protein modifiers. Phosphorylation of p53, MDM2 and MDMX, which are often induced in response to DNA damage or other genomic stress, can affect interactions and ubiquitylation15. Acetylation of carboxy-terminal lysines on p53 competes for MDM2-mediated ubiquitylation and thereby activates p53; recently, acetylation of other lysine residues of p53 has been shown to activate p53 by blocking MDM2 binding14. A provocative observation is that MDM2 can mediate p53 neddylation on lysines overlapping those modified with ubiquitin and that this inhibits p53-mediated transcription16. In addition to MDM2 and MDMX, >10 other E3s (most of which are RING fingers) have been implicated in ubiquitylating p53, although for a number of these this role has not been confirmed (Table 1 and Supplemental Table 1). One consideration is that some of these function either cooperatively with MDM2-MDMX or act to further ubiquitylate p53 either in the nucleus or perhaps after export to the cytoplasm17.
Up to 50% of malignancies retain wild type p53 and in most of these there is increased MDM2 activity towards p53 as a consequence of either amplification of MDM2, increased MDM2 expression owing to polymorphisms, alterations in ARF activity or other factors16,36. Thus, inhibiting the physical and functional interactions between these proteins is of great therapeutic interest. The small molecule RITA binds to p53 and blocks interactions with MDM2. This results in p53-dependent apoptosis in cell lines and inhibition of growth in xenografts41. The function of RITA has been extended to human papillomavirus (HPV) E6-dependent degradation of p53 by the Homologous to E6-AP Carboxy Terminus (HECT) domain E3 E6-Associated Protein (E6-AP; also known as UBE3A), consistent with RITA inducing a conformational alteration in p53 (REF. 42). Nutlins are small molecules with IC50s of ~100 nM that bind MDM2 and competitively inhibit its binding to p53 (Figure 1) and have been shown to have activity in preclinical models either alone43 or, in combination with other treatments in xenografts of prostate cancer and neuroblastoma44,45. With Nutlins validating inhibition of proteinprotein interactions as a way of activating p53, and the crystal structure of the MDM2-p53 interface well defined, there is the potential for structure-based refinement of this approach to targeting p53-MDM2 binding46. Another approach to reactivate p53 is through inhibiting the E3 activity of MDM2, ideally through small molecule ‘ARF mimetics’. There are several small molecules that inhibit MDM2 activity either in general or specifically towards p53 (REFS 47–49). Although these establish proof-of-principle for the inhibition of E3 activity, neither the molecular basis for their action nor their in vivo efficacy is known. With knowledge of the MDM2-MDMX RING finger dimer structure29 there is the potential to design inhibitors that disrupt the active dimeric E3. However, as the animal studies described above demonstrate, there is a fine line between achieving a therapeutic level of p53 activation and causing undesirable widespread apoptosis25,26.
DNA repair-associated RING fingers and cancer
Together with DNA damage-activated kinases and other protein modifying enzymes, RING finger and RING finger-like E3s play critical roles in sensing and repairing DNA damage, regulating cell cycle progression and minimizing the propagation of damaged DNA and chromosome abnormalities (Supplemental Table 2). RING finger E3s perform essential roles in the five best-characterized forms of DNA repair (Supplemental Table 3). Particularly illustrative of the importance of RING finger E3s in repair processes are BRCA1 and the FANC E3 complex.
BRCA1 is a tumor suppressor that is frequently mutated in familial breast and ovarian cancer50. The role of BRCA1 in DNA repair was established as a consequence of its co-localization with RAD51 in nuclear foci during S-phase and co-localization with RAD51, and proliferating cell nuclear antigen (PCNA) upon DNA damage51,52. This led to the discovery that cells deficient in BRCA1 have a defect in the repair of double stranded breaks by homologous recombination (HR)53,54. The central role of BRCA1 in DNA repair is underscored by its recruitment to DNA as a dimer with BRCA1-associated RING domain 1 (BARD1; discussed below) in several different protein complexes that perform functions in sensing DNA damage, controlling both G2/M and DNA replication checkpoints and in recruiting DNA repair enzymes55. A key BRCA1-BARD1 complex that accumulates at DNA damage foci and is involved in the G2/M checkpoint is comprised of abraxas (also known as FAM175A), RAP80, MERIT40 (also known as BABAM1), BRE (also known as BRCC45) and the DUB BRCC36. A notable feature of all five of these complex members is that they each include ubiquitin-binding domains and bind ubiquitylated proteins55. Such associations are a recurrent theme in DNA damage complexes, as is the presence of DUBs (see Fanconi anemia below). Thus, an important concept is that protein ubiquitylation at sites of DNA damage, primarily by RING finger and related E3s, provides a means of recruiting other specific proteins with ubiquitin-binding domains and this accumulation can be regulated in part by DUBs.
BRCA1 includes an N-terminal RING finger, a nuclear localization sequence, a coiled-coil domain and two C-terminal BRCA1 C-terminal (BRCT) domains55. The BRCA1 RING finger has intrinsic E3 activity1. However, in cells it exists as a stable heterodimer with the related RING finger protein BARD1, which greatly increases the activity of the BRCA1 RING finger in vitro, but which by itself lacks E3 activity56,57. BRCA1 and BARD1 dimerize through their RFs and in cells loss of expression of either protein results in the degradation of the other58–60. How the BARD1 RING finger enhances the activity of BRCA1 in vitro and stabilizes it in cells is unknown. Interestingly, it was recently shown that BRCA1 sumoylation by protein inhibitor of activated STAT (PIAS) proteins is critical to the cellular and in vitro activity of the BRCA1-BARD1 complex61.
The nature of BRCA1, including its large size and involvement as a common player in multiple forms of DNA damage repair can account, in part, for its prominent association with cancer. However, the plethora of germline cancer-associated missense and nonsense mutations throughout the coding region do not provide a ready explanation for its differential association with familial breast and ovarian cancer. Similarly, while the BRCA1-BARD1 dimer associates with a large number of proteins, the most critical substrates for ubiquitylation may remain to be determined.
Among established BRCA1 substrates is CtBP interacting protein (CTIP; also known as RBBP8), the binding partner of the transcriptional repressor carboxy-terminal binding protein (CtBP)62. CTIP is considered a TSG based on it being mutated in a number of cancers and its association with other tumor suppressors including BRCA1 63. CTIP binds the BRCT domains of BRCA1 in a phosphorylation-dependent manner. In response to ionizing radiation ubiquitylated CTIP is found in an insoluble chromatincontaining fraction of cell lysates, suggesting a role for BRCA1-mediated ubiquitylation of CTIP in checkpoint arrest in response to DNA damage62. Based on negative data from cycloheximide chase experiments it was concluded that BRCA1-mediated CTIP ubiquitylation does not result in proteasomal degradation. This would be consistent with the capacity of BRCA1 to form non-canonical Lys6 polyubiquitin chains.
BRCA1 binds both progesterone receptor (PR) and oestrogen receptor-α (ERα also known as ESR1) through interactions primarily involving its N terminal regions64–66. BRCA1 decreases the transcriptional activity of both PR and ERα but the role that ubiquitylation and proteasomal degradation plays remains to be fully determined. Convincing evidence exists that PR is a target for polyubiquitylation and degradation in breast cancer cells67, however other studies suggest the role of BRCA1 is to decrease PR activity without degradation68.
BRCA1 has been shown to target ERα for monoubiquitylation66. The specific Lys in ERα that is ubiquitylated by BRCA1 in vitro overlaps with lysines in a region of ERα that is acetylated by the histone acetyltransferase p300. Activated BRCA1, but not RING mutant BRCA1, induces decreased ERα acetylation and there is evidence that acetylation is directly correlated with ERα transcriptional activity. Thus, an emerging model is that BRCA1-mediated monoubiquitylation competes with acetylation to inhibit ERα transcriptional activity69. However a role for BRCA1 in the proteasome-mediated degradation of ERα cannot be excluded.
An apparent paradox is that, most commonly, BRCA1 mutant breast cancers do not express ERα or PR, yet both of these receptors interact with BRCA1. Currently a satisfying explanation is lacking. However, several lines of evidence suggest that despite the ERα− PR− phenotype, ovary-derived hormones and ERα play critical roles in the genesis of BRCA1-mutant tumors. For example, the incidence of mammary carcinomas in mice with conditional Brca1 deletion targeted to the mammary gland is increased by exogenous oestrogen. Consistent with the suppression of ERα-mediated transcription by BRCA1, the loss of BRCA1 results in increased sensitivity of cells to oestrogen-induced proliferation70,71. In adult patients with germline BRCA1 mutations, oophorectomy results in a 56% decrease in the incidence of breast cancer72. Thus, it may be that early in tumorigenesis BRCA1 loss or mutation alters the regulation of these receptors and thereafter the manifestation of BRCA1 loss is primarily on genomic stability.
In addition to the well-established involvement of BRCA1 in breast and ovarian cancer, there is also an association of an E3 containing a RING finger-like domain (PHD domain E3; Box 2) with Fanconi anemia. This recessive genetic disease of childhood is caused by defective DNA repair proteins. Children who survive its early multi-system effects are susceptible to a number of malignancies73. This rare genetic disease occurs as a consequence of mutations in any of at least 13 different FANC genes, the loss of which results in failure to repair DNA crosslinks during DNA synthesis. Failure to repair crosslinks has catastrophic consequences for the surrounding DNA. Crosslink repair requires the sequential action of several different DNA damage response pathways including translesion synthesis (TLS; which is intrinsically mutagenic in itself), nucleotide excision repair (NER) and HR. Accordingly, DNA damage response foci show reactivity for other proteins involved in DNA repair, including RAD51, PCNA and BRCA1 (REF. 73). Beyond the 13 FANC genes, biallelic mutations in either of two other genes have recently been identified as being associated with Fanconi anaemia74,75.
The 13 FANC proteins can be subdivided accordingly: eight components of the core FANC E3 ubiquitin ligase, two substrates for transient monoubiquitylation and three other effector proteins (including BRCA2, which is also known as FANCD1). These effector proteins are recruited to sites of DNA damage together with the ubiquitylation substrates. FANCM plays a central role as it is recruited to sites of DNA damage and serves to recruit the other FANC proteins. Crucial to the activity of the FANC E3 ligase is FANCL, which has a RING finger-like PHD domain that is responsible for its activity. FANCL functions together with the E2 UBE2T to add a single ubiquitin to specific lysine residues on FANCD2 and FANCI. This transient monoubiquitylation leads to their binding to chromatin, with FANCD2 ubiquitylation being essential for DNA repair while FANCI ubiquitylation possibly enhancing repair73. An interesting feature of this E2-E3 pair is that a relatively limited region of FANCL, which includes its PHD domain, is sufficient for a high affinity interaction with UBE2T76. Such an interaction is unusual for E2s, which have to dissociate from the RING finger to reload with ubiquitin77. However, this finding might make intuitive sense as the FANCL substrates only undergo monoubiquitylation.
Monoubiquitylated FANCD2 is required to ‘unhook’ the crosslinked nucleotide from one of the parental strands as well as for nucleotide insertion on the opposite strand78. Deubiquitylation of FANCD2 and FANCI is mediated by the ubiquitin specific peptidase 1 (USP1) and USP1-associated factor 1 (UAF1; also known as WDR48) complex73. This reversibility is essential for crosslink repair. Recently a critical link between FANC-mediated ubiquitylation and repair was made with the discovery that the endonuclease FANCD2/FANCI-associated nuclease 1 (FAN1) binds specifically to ubiquitylated FANCD2 (REFS 79–81). The FANC repair pathway must be inactivated during mitosis to prevent the formation of radial chromosomes and other abnormalities. This is accomplished by ubiquitylation and degradation of FANCM. FANCM is phosphorylated by kinases that include polo-like kinase 1 (PLK1), which leads to ubiquitylation by SCF β-TrCP and proteasomal degradation82.
The defects in DNA repair in tumors with BRCA1 or BRCA2 loss has led to a synthetic lethal approach to targeting these tumors. The inhibition of poly(ADP-ribose) polymerase (PARP), which is involved in the repair of single-stranded DNA breaks, leads to the formation of double-stranded DNA breaks, which are usually repaired by HR. Cells deficient in BRCA1 or BRCA2 (and thereby HR) are thus selectively sensitive to PARP inhibition83,84. Clinical trials using PARP inhibitors in patients with BRCA1- or BRCA2-deficient breast and ovarian cancers have shown promising single agent activity 85–87.
RING fingers and transmembrane signalling
Regulated transmembrane signalling is essential for normal homeostasis and development. Many RING Finger E3s are implicated in the regulation of signal transduction. For example, the E3 CBL (discussed below) mediates ubiquitylation and downregulation of many activated receptor tyrosine kinases (RTKs) such as the epidermal growth factor receptor (EGFR), Hakai (also known as CBLL1) downregulates E-cadherin, neuregulin receptor degradation protein 1 (NRDP1) downregulates ERBB3, and the RING finger-related U-box protein carboxy-terminus of HSP70 interacting protein (CHIP) mediates-chaperone dependent ubiquitylation and downregulation of ERBB2 (REFS 88–91). Similarly, the SCF β-TrCP RING finger E3 ubiquitylates and negatively regulates activated prolactin receptor, it also regulates the endocytosis of the growth hormone receptor and negatively regulates WNT signalling by targeting β -catenin for proteasomal degradation92–94. Multiple RING finger E3s both positively and negatively regulate nuclear factor-κB (NF-κB) signaling and some are implicated in the development of lymphoid malignancies95 (Table 1).
CBL
Dysregulated signalling in cancer cells is frequently caused by activating mutations and/or amplification of RTKs96. This results in autonomous activation of growth and survival signalling pathways. Activated RTKs are negatively regulated by their internalization and intracellular trafficking to lysosomes97. This is driven by RTK ubiquitylation, which serves as a docking site for ubiquitin binding proteins that mediate the trafficking of RTKs from the early endosome to the multivesicular body, leading ultimately to lysosomal degradation97. The CBL proteins mediate ubiquitylation and lysosomal degradation of many RTKs and thereby down regulate their function98–103 (Figure 2).
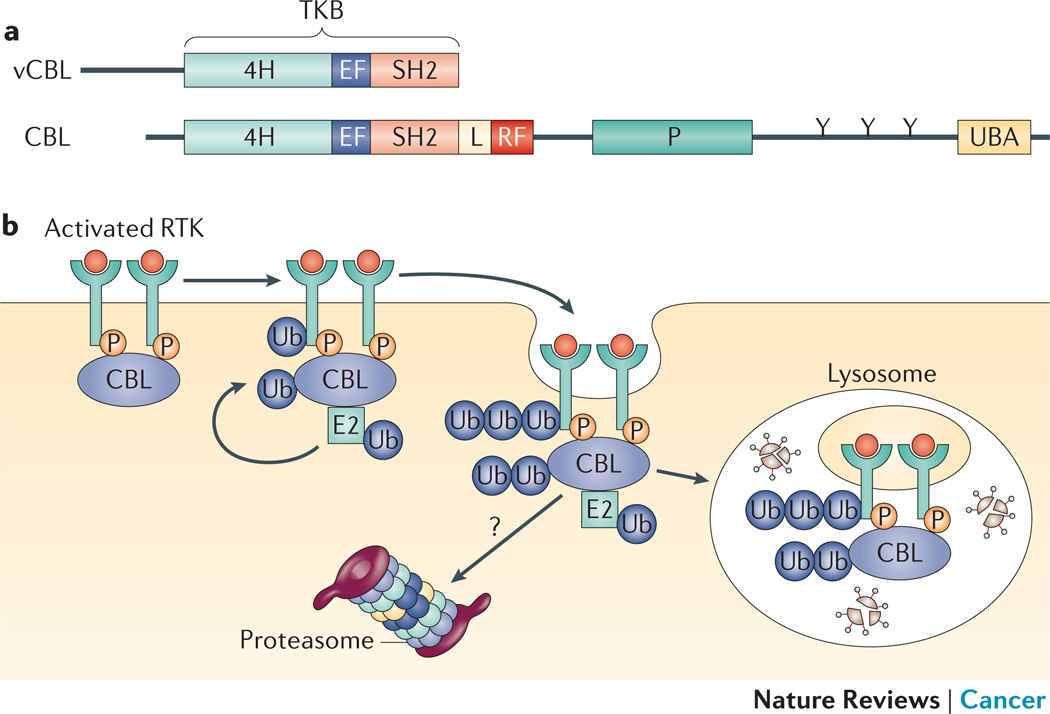
Figure 2. CBL.
a | Schematic structure of the vCBL and CBL proteins. The tyrosine kinase-binding (TKB) domain is comprised of a 4-helix bundle (4H), an EF hand (EF), and a variant SH2 domain (SH2). The linker (L), RING finger (RF), proline-rich (P), and the ubiquitin-associated (UBA) domains are indicated. The tyrosines (Y) at sites of phosphorylation in the C-terminal of CBL are also indicated. b | CBL proteins mediate ubiquitylation and downregulation of receptor tyrosine kinases (RTKs). Whether CBL proteins associated with activated complexes are degraded in lysosomes or proteasomes is unresolved. Figure reproduced with permission © Macmillan Publishers (2011).
The CBL proteins (in mammals, CBL, CBLB and CBLC) are a family of single subunit RING finger E3s that contain multiple protein interaction motifs surrounding the RING finger104. All CBL proteins have a highly conserved N-terminal tyrosine kinase binding (TKB) domain composed of a 4-helix bundle, a calcium-binding EF hand and an atypical SH2 domain104. The TKB domain mediates interactions between CBL proteins and phosphorylated tyrosine residues on CBL substrates. The RING finger and TKB domain are separated by an α-helical linker region, which is critical for the regulation of CBL E3 function and is implicated in the transforming mutations discussed below. Based on the crystal structure, the linker region contacts the TKB, the RING finger and the E2 (REF.105). Phosphorylation of a conserved tyrosine residue in the linker region activates E3 activity by causing a conformational change in CBL that modulates affinity for the E2 (REFS 102,106,107). C-terminal to the RING finger, CBL proteins have proline-rich domains, which mediate interactions with SH3-containing proteins, and tyrosines that become phosphorylated and mediate interactions with SH2 proteins (such as the p85 subunit of PI3K).
The CBL proteins catalyze the formation of monoubiquitin or Lys63-linked polyubiquitin chains on EGFR – modes of ubiquitylation that are associated with endocytic trafficking and not with proteasomal degradation108,109. Also, the CBL proteins function as adaptor proteins through a diverse array of interactions110. This adaptor function contributes to the E3 dependent activities of CBL proteins by targeting specific substrates for ubiquitylation and degradation. However, there are also non-E3 dependent activities served by the adaptor function of the CBL proteins such as the recruitment of proteins involved in internalization of the target RTK, localization of the CBL proteins to specific compartments within the cell and activation of signalling pathways110.
vCbl was identified as the product of the transforming gene of the Cas NS-1 murine retrovirus, which causes leukaemia and lymphomas in mice111,. The molecular basis for the oncogenic nature of vCBL is due to a dominant-negative function of this truncated CBL protein, which lacks an intact RING finger112. Mutations that abrogate the E3 activity of CBL are found in ~5% of human myeloid neoplasms113. This loss of activity is largely due to homozygous missense mutations, frameshifts or deletions in and around the linker and RING finger domains. These associations underscore the importance of the RING finger in the essential function of CBL proteins. As other CBL isoforms are expressed and active in these cells these mutants are also likely to function in a dominant-negative manner. Further evidence supporting a tumor suppressor role for CBL proteins derives from studies where mice null for both Cbl and Cblb in haematopoietic stem cells develop early-onset myeloid neoplasms114. However, several studies suggest that oncogenic CBL proteins may also exhibit a gain-of-function activity resulting in aberrant activation of the JAK-STAT and PI3K-AKT pathways in particular115–117. Whether the apparent gain-of-function is due predominantly to a dominant-negative effect on other CBL proteins or due to the adaptor function of CBL is not clear.
The RTK fms-like tyrosine kinase 3 (FLT3) has been implicated in the pathogenesis of myeloid leukaemias and is frequently the target of activating mutations118. Interestingly, cells expressing mutant CBL proteins show increased activity of FLT3, consistent with FLT3 being the relevant target of the CBL proteins, which thus contributes to the oncogenic activity of mutant CBL proteins115,116,119. This is supported by the finding that loss of FLT3 suppresses the development of myeloid neoplasms in mice harboring a RING finger mutation in CBL120. However, many RTKs interact with and are ubiquitylated by CBL, thus there are other substrates that may contribute to the oncogenic effects of CBL in myeloid neoplasms110.
The role of CBL in the development of other malignancies requires further study. In non-small-cell lung cancer mutations in CBL that do not disrupt E3 activity have been found, but the importance of these is yet to be determined121. CBL function could also be disrupted by mutations in the substrates of the CBL proteins or by aberrant activity or overexpression of negative regulators of CBL122,123. Since oncogenic mutations in CBL proteins predominantly lead to loss of function, novel strategies will be needed to develop therapies for tumors with CBL mutations. One approach could be to target FLT3 or downstream signaling pathways such as PI3K and AKT that are activated in myeloid neoplasms containing CBL mutations115–117,119,120,124.
A causative oncogenic role for loss of function mutations of CBLB has not been demonstrated. However, the function of CBLB in negatively regulating the immune responses provides a potential therapeutic opportunity for CBLB inhibition. In vitro studies of T-lymphocytes isolated from Cblb-deficient mice have demonstrated that CBLB negatively regulates the CD28 co-stimulation pathway. Thus CBLB positively controls the induction of anergy125–127. Interestingly, CD8+ cytotoxic T lymphocytes in Cblb−/− mice mediate enhanced regression of tumors128,129. Similarly, knockin mice expressing a RING finger mutant form of CBLB show enhanced antitumor immunity, demonstrating the essential role of the RING finger E3 activity in regulating antitumor immunity130. Together, these data suggest that inhibition of CBLB, alone or in combination with adoptive immunotherapy may enhance immunotherapy of tumours.
Responses to hypoxia
VHL
VHL was first discovered as the TSG that is inactivated in the familial kidney cancer syndrome von Hippel-Lindau disease131. This syndrome is characterized by the development of benign and malignant tumors of multiple organs, most notably clear cell kidney cancers. Also, approximately 57% of sporadic clear cell cancers of the kidney contain inactivating mutations of VHL and 98% have loss of heterozygosity (LOH) at the VHL locus 132. Additional studies suggest that VHL is epigenetically silenced in some clear cell kidney cancers 133. Xenograft studies using a renal carcinoma cell line that expresses a VHL mutant have demonstrated that reconstitution of the wild type VHL protein into the cell line suppresses tumorigenicity, consistent with the proposed function of VHL as a TSG 134.
VHL forms a complex with elongin B and elongin C, cullin 2 and the small RING finger protein RBX1 (REFS 135–137). This architectural similarity to the multisubunit SCF RING finger E3s (Figure 3) led to the demonstration that the VHL complex is an E3 (REFS 138,139). VHL is a substrate targeting subunit of the CRL2 or cullin 2–elongin B–elongin C (CBC) family of RING finger E3s140,141. The identification of the critical CRL2VHL targets, the three hypoxia inducible factor-α (HIFα) proteins, stemmed from the observations that the mRNA for hypoxia-associated genes such as vascular endothelial growth factor A (VEGFA), solute carrier family 2 member 1 (SLC2A1; which encodes GLUT1) and platelet-derived growth factor-β (PDGFB) were expressed in at similar levels in VHL-deficient cells grown in either normoxic or hypoxic conditions and that the levels of the mRNA for these proteins did not change with hypoxia140,142,143. By contrast, in cells expressing WT VHL the mRNAs for these genes were upregulated by hypoxia. Reintroduction of VHL into VHL-deficient cells reduced expression of these genes under normoxic conditions and restored their hypoxia-induced upregulation140,142,143. Many of these genes were previously shown to be regulated by the HIFα family of transcription factors (HIF1α-3α), and subsequent work demonstrated that VHL interacted with HIFα proteins and induced rapid oxygen-dependent proteasomal degradation of HIFα proteins140,141. In normal cells, HIFα proteins are stable under hypoxic conditions. This allows for their dimerization with the constitutively expressed HIF1β (also known as ARNT) and their translocation to the nucleus. The complex binds to hypoxia responsive elements associated with target genes and enhances their transcription. In the absence of VHL, these genes are constitutively expressed and can lead to tumor development.
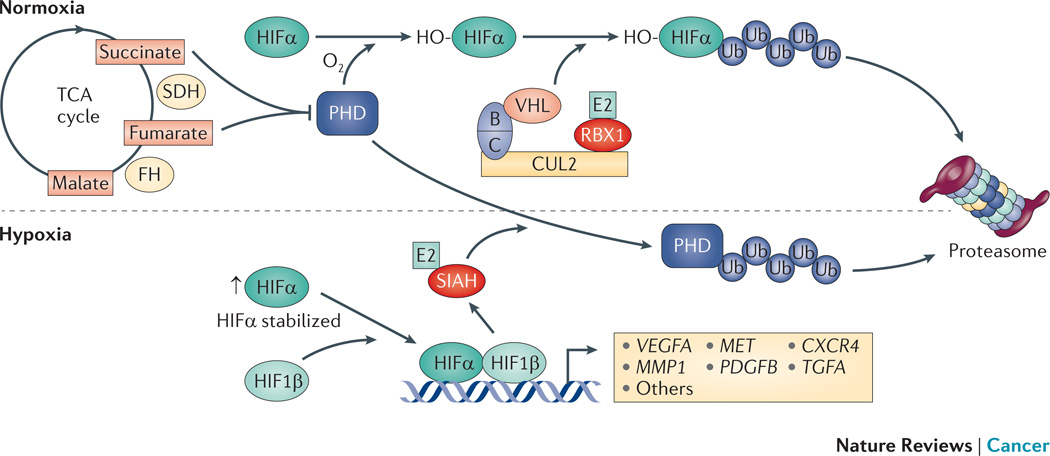
Figure 3. Hypoxia.
Hypoxia-inducible factor-α (HIFα) proteins are subunits of the HIF transcription factors, which regulate the expression of many genes associated with the response to hypoxia and proliferation. Under normoxic conditions the prolyl hydroxylase (PHD) proteins hydoxylate HIFα proteins, which creates a binding site recognized by the cullin RING ligase 2 (CRL2) – von Hippel Lindau tumor suppressor protein (VHL) E3 complex (CRL2VHL). This leads to proteasomal degradation of HIFα proteins. The PHD proteins are ubiquitylated and degraded by the seven in absentia homologue (SIAH) RING finger E3s that are themselves transcriptionally upregulated by HIF transcription factors in response to hypoxia152. The stability of HIFα proteins is also regulated by fumarate hydratase (FH) and succinate dehyrogenase (SDH), which are tumor suppressors associated with kidney cancer156. The loss of FH and SDH results in the accumulation of fumarate and succinate, respectively, which competitively inhibit PHD activity and prevent hydroxylation and VHL-mediated degradation of HIFα proteins leading to inappropriate upregulation of hypoxia inducible genes157. Figure reproduced with permission © Macmillan Publishers (2011).
Under normoxic conditions, HIFα proteins are hydroxylated on a specific proline residue creating a binding site for the hydroxyproline binding pocket of the VHL protein leading to CRL2VHL-mediated ubiquitylation and proteasomal degradation144–146. Three oxygen-dependent prolyl hydroxylases (PHD1–3; also known as EGLN1–3) have been identified in mammalian cells that catalyze proline hydroxylation of HIFα proteins and thus enhance VHL binding under normoxic conditions147 (Figure 3).
Although CRL2VHL can target all three HIFα proteins for degradation, the degradation of HIF2α appears to be the most critical target for CRL2VHL (REFS 148–151). Whether there are other substrates of the CRL2VHL RING finger E3 that contribute to its role as a TSG has not been demonstrated.
SIAHs
A second class of RING finger E3s, the seven in absentia homologue (SIAH) proteins1, regulates HIFα protein stability by functioning upstream of VHL (Figure 3). SIAH1 and SIAH2 bind PHD1 and PHD3 and target them for proteasomal degradation 152. SIAH2 expression is transcriptionally upregulated under hypoxic conditions. Accordingly, Siah2−/− cells have an impaired response to hypoxia owing to the stabilization of PHD proteins, ongoing hydroxylation of HIFα proteins and constitutive recognition and degradation of HIFα proteins by CRL2VHL (REF 152). Consistent with a role for SIAH2 in mediating the response to hypoxia, Siah2-null mice have impaired increase in red blood cells in response to chronic hypoxia152. SIAH proteins have other roles that are potentially both oncogenic and tumour suppressive153.
Conclusions and perspectives
Since the discovery that RING finger proteins are, in general, ubiquitin-protein ligases over 12 years ago it became apparent that the ubiquitin conjugating system plays a far greater role in both malignancy and tumour suppression than previously appreciated. We now understand that many processes that influence the course of cancer development and progression are regulated by RING finger proteins. The trajectory of publications in this area suggests we are just seeing the tip of the iceberg. Yet even among some of the best-studied, cancer relevant, RING finger E3s and substrates, there are many questions yet to be answered. We understand relatively little about how MDM2-MDMX is regulated in vivo and we know even less about the functional interactions between this E3 and the numerous other RING finger E3s that have been identified for p53. While we are beginning to ‘drill down’ on the ligases and ubiquitin chain linkages involved in DNA repair, there is still much to be learned about how ubiquitylation by RING finger E3s leads to recruitment of the DNA repair complexes and the importance of the dynamics of ubiquitylation-deubiquitylation and what the overall role of proteasomal degradation is in these processes. In cell signalling we understand that ubiquitylation can induce protein trafficking to the lysosome, yet in the context of RTK signaling complexes it is unclear to what extent proteasomal degradation and lysosomal degradation intersect in the degradation of complexes that include transmembrane proteins and associated signaling complexes and the extent to which types of ubiquitin linkages generated by RING finger E3s specify fate.
The therapeutic success of the proteasome inhibitor bortezomib demonstrates the potential efficacy of targeting the ubiquitin-proteasome system 154. A question of great importance is the extent to which all of the information we are now garnering about RING finger function can be applied towards the development of targeted therapeutics of greater specificity than proteasome inhibitors. Pre-clinical data using nutlins provides encouragement that targeting RING finger E3-substrate interactions might be of benefit in the treatment of cancer, and one might envisage similar approaches to limit cell proliferation by interfering, for example, with SKP2-p27 interactions. Related to this, as a number of RING finger-E3 interactions are phosphorylation dependent, an alternative means of disrupting interactions is the inhibition of specific kinases. However, when it comes to inhibiting RING finger E3 activity itself, the jury is out. Proof-of-principle has been established for MDM2 but 12 years on the number of reports of inhibitors specific for RING finger E3s is limited. Part of this challenge probably derives from the fact that RING finger domains function to activate ubiquitin transfer from the E2 directly to the substrate, but they are not bona fide catalysts with active sites. Thus, approaches would probably have to be focused on disrupting the RING structure or the RING-E2 interface, which appears to be relatively highly conserved among E2-E3 pairs. In many instances where the loss of activity of RING finger or RING finger-like E3s is associated with the development of cancer (e.g. VHL, BRCA1, FANCL and CBL) novel approaches may be necessary. The promising results with PARP inhibition in BRCA1- and BRCA2-deficient tumors suggests that such a synthetic lethal approach may be applicable in some instances where tumors arise because of loss of E3 activity. Additionally, ubiquitylation is a reversible process involving, in many cases, specific deubiquitylating enzymes. Therefore in situations where compromised ubiquitylation plays a role in cancer, inhibition of specific deubiquitylating enzymes may represent a viable therapeutic modality. With the accumulation of structural and functional data and elucidation of the pathways controlled by RING finger E3s, there is great potential for seeing this knowledge used in the development of new, targeted therapeutics.
ON LINE MATERIALS
At a Glance
RING finger ubiquitin-protein ligases (E3s) are the most abundant class of E3 that mediate protein ubiquitylation (also known as ubiquitination). They regulate critical cellular functions such as the cell cycle, DNA repair, cell signalling and responses to hypoxia. Genetic alterations including activating and inactivating mutations, gene amplifications, translocations and deletions have been described for many RING finger E3s. RING finger E3s are validated oncogenes (such as MDM2) or tumor suppressor genes (such as BRCA1 and von Hippel-Lindau tumor suppressor (VHL)) because of their role in regulating critical cell functions.
The cell cycle is regulated by the S-phase kinase-associated protein 1 (SKP1)- cullin 1 (CUL1)-F-box protein (SCF) and anaphase-promoting complex/cyclosome (APC/C) multisubunit RING finger E3s. These complexes are targeted to specific substrates via interchangeable substrate recognition subunits, including F-box proteins for SCF and cell division cycle 20 (CDC20) and CDH1 (also known as FZR) for APC/C. These multisubunit E3s have a large number of substrates with oncogenic and tumour suppressive effects. Genetic alterations to components of these E3 complexes resulting in loss of function (such as FBW7, CDH1 and CDC20) or gain of function ((such as S-phase kinase-associated protein 2 (SKP2) and β-transducin repeat containing protein (β-TrCP; also known as FBW1A)) are implicated in the development of cancer.
RING finger E3s play central roles in DNA damage responses and DNA repair. For example, MDM2 targets p53 for degradation. MDM2 is amplified, overexpressed or activated in other ways in cancers and is a means of inactivating the tumor suppressor p53. The BRCA1 and the Fanconi anaemia (FANC) E3s have essential roles in the repair of DNA damage. Both E3s function as tumor suppressors.
RING finger E3s have important roles in both positively and negatively regulating signal transduction. A prominent example of negative regulation is the CBL family of RING finger E3s that target activated receptor tyrosine kinases (RTKs) for degradation. Mutations that inactivate CBL E3 function have been described in myeloid neoplasms and result in hyperactivation of RTKs and intracellular signaling pathways.
The response to hypoxia is regulated by the multisubunit CRL2VHL RING finger E3 and the single subunit RING finger E3 seven in absentia homologue (SIAH). The VHL complex targets the hypoxia-inducible factor-α (HIFα) transcription factors for proteasomal degradation, which prevents expression of angiogenic and growth promoting genes under normoxic conditions. Inactivating mutations of VHL are found in familial and sporadic clear cell cancer of the kidney, resulting in stabilization of the HIFα transcription factor subunits and consequently abnormally high expression of angiogenic and growth genes. By contrast, the SIAH RING finger E3s stabilize HIFα under hypoxic conditions.
Targeting RING finger E3s for the treatment of cancer is being actively explored. For example, small molecule inhibitors have been developed that interfere with the MDM2-p53 interaction or inhibit MDM2 E3 activity, thus stabilizing p53. These approaches have demonstrated antitumor activity in preclinical studies but the clinical efficacy of interfering with MDM2 function remains to be determined. Targeting the loss of activity of RING finger E3s that are tumor suppressors will require novel approaches such as the synthetic lethality induced by poly(ADPribose) polymerase (PARP) inhibition in cells that are deficient in BRCA1 or BRCA2 (also known as FANCD1).
Supplementary Material
ACKNOWLEDGEMENTS
The authors apologize to the many scientists whose contributions to this extensive and exponentially growing field of research could not be directly cited because of space limitations. They are indebted to their colleagues whose outstanding reviews on individual RING finger E3s and RING finger E3 families summarize and cite much of this important research. The authors thank Ranabir Das (Structural Biophysics Laboratory, National Cancer Institute, USA) for invaluable assistance in generating the RING finger-E2 ribbon diagram. The authors’ research programmes are supported by the National Institutes of Health, National Cancer Institute, Center for Cancer Research, USA.
GLOSSARY
- Amplification
Increased copy number of a gene within the genome; this is a common mechanism to increase the activity of oncogenes.
- Anergy
When lymphocytes fail to mount an immune response to an antigen.
- Aneuploidy
Abnormal number of chromosomes resulting in more or less than the normal diploid number of chromosomes. Cancer cells are frequently aneuploid.
- Cyclins
Proteins that control progression of the cell cycle by activating cyclin-dependent kinases.
- Haploinsufficient tumor suppressor gene
A gene for which loss of one allele is sufficient to promote cancer development.
- Oncogenes
Genes with protein products that can promote cancer development. Oncogenes frequently undergo amplification or activating mutations and act in a genetically dominant fashion.
- Securin
A protein that forms a complex with separase and thereby inhibits separase activity and prevents chromosome separation at anaphase. Securin is dephosphorylated and degraded by APC/CCDC20 at the onset of anaphase.
- Tumor suppressor genes
(TSGs). Genes whose protein products, when lost or mutated, are permissive for the development of cancer. Tumor suppressor genes frequently undergo deletion or inactivating mutations of both alleles and act in a genetically recessive fashion.
Biographies
SL completed an M.D and Ph.D. in Biomedical Sciences from Weill Cornell Medical College, New York, NY, USA. He is currently a Senior Investigator in the Laboratory of Cellular and Molecular Biology, Center for Cancer Research, National Cancer Institute, Bethesda, MD, USA. His lab investigates the structure and function of CBL proteins.
AMW obtained his B.S. in Biochemistry from Stony Brook University, New York and his M.D. from Albert Einstein College of Medicine, New York. Following training in Internal Medicine at Washington University he came to the National Institute of Health (USA) where he is now a Laboratory Chief in the National Cancer Institute. His group focuses on physiological and pathological roles of the ubiquitin-proteasome system and on understanding structure-function relationships for ubiquitin-carrier enzymes (E2s) and ubiquitin ligases (E3s).
Footnotes
AUTHORS’ DISCLAIMER
This publically available version of the manuscript represents the authors’ edits to the accepted manuscript and has significant variations from the published version.
REFERENCES
- 1. Lorick KL, et al. RING fingers mediate ubiquitin-conjugating enzyme (E2)- dependent ubiquitination. Proc. Natl. Acad. Sci. U S A. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. This study determined that RING fingers are E2-binding ubiquitin ligase domains and that RING finger-containing proteins including SIAH and BRCA1 have ubiquitin ligase activity.
- 2.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat. Rev. Cancer. 2006;6:369–381. doi: 10.1038/nrc1881. [DOI] [PubMed] [Google Scholar]
- 4.Skaar JR, Pagano M. Control of cell growth by the SCF and APC/C ubiquitin ligases. Curr. Opin. Cell Biol. 2009;21:816–824. doi: 10.1016/j.ceb.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat. Rev. Cancer. 2008;8:438–449. doi: 10.1038/nrc2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat. Rev. Mol. Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. References 3–6 review the SCF and APC/C family of E3 ubiquitin ligases and their role in cell cycle control and cancer and cite key primary literature that is not directly cited herein except in Supplemental Tables owing to space limitations.
- 7.Wang Q, et al. Alterations of anaphase-promoting complex genes in human colon cancer cells. Oncogene. 2003;22:1486–1490. doi: 10.1038/sj.onc.1206224. [DOI] [PubMed] [Google Scholar]
- 8.Wasch R, Engelbert D. Anaphase-promoting complex-dependent proteolysis of cell cycle regulators and genomic instability of cancer cells. Oncogene. 2005;24:1–10. doi: 10.1038/sj.onc.1208017. [DOI] [PubMed] [Google Scholar]
- 9.Garcia-Higuera I, et al. Genomic stability and tumour suppression by the APC/C cofactor Cdh1. Nat. Cell Biol. 2008;10:802–811. doi: 10.1038/ncb1742. [DOI] [PubMed] [Google Scholar]
- 10.Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat. Rev. Cancer. 2008;8:83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- 11.Guardavaccaro D, et al. Control of chromosome stability by the beta-TrCP-RESTMad2 axis. Nature. 2008;452:365–369. doi: 10.1038/nature06641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Westbrook TF, et al. SCFbeta-TRCP controls oncogenic transformation and neural differentiation through REST degradation. Nature. 2008;452:370–374. doi: 10.1038/nature06780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wiggins CM, et al. BIM(EL), an intrinsically disordered protein, is degraded by 20S proteasomes in the absence of poly-ubiquitylation. J. Cell Sci. 2011;124:969–977. doi: 10.1242/jcs.058438. [DOI] [PubMed] [Google Scholar]
- 14.Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 15.Wade M, Wang YV, Wahl GM. The p53 orchestra: Mdm2 and Mdmx set the tone. Trends Cell Biol. 2010;20:299–309. doi: 10.1016/j.tcb.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marine JC, Lozano G. Mdm2-mediated ubiquitylation: p53 and beyond. Cell Death Differ. 2010;17:93–102. doi: 10.1038/cdd.2009.68. [DOI] [PubMed] [Google Scholar]
- 17. Lee JT, Gu W. The multiple levels of regulation by p53 ubiquitination. Cell Death Differ. 2010;17:86–92. doi: 10.1038/cdd.2009.77. References 14–17 review much of the relevant primary literature on p53, MDM2 and MDMX and their regulation they also include references not cited herein.
- 18.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 19.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 20.Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 21.Honda R, Yasuda H. Activity of MDM2, a ubiquitin ligase, toward p53 or itself is dependent on the RING finger domain of the ligase. Oncogene. 2000;19:1473–1476. doi: 10.1038/sj.onc.1203464. [DOI] [PubMed] [Google Scholar]
- 22. Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J. Biol. Chem. 2000;275:8945–8951. doi: 10.1074/jbc.275.12.8945. References 18–22 collectively establish that MDM2 is responsible for the degradation of p53 and is a RING finger-dependent E3 ubiquitin ligase for p53 and itself.
- 23.Jones SN, Roe AE, Donehower LA, Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 1995;378:206–208. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- 24.Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 25.Mendrysa SM, et al. Tumor suppression and normal aging in mice with constitutively high p53 activity. Genes Dev. 2006;20:16–21. doi: 10.1101/gad.1378506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ringshausen I, O'Shea CC, Finch AJ, Swigart LB, Evan GI. Mdm2 is critically and continuously required to suppress lethal p53 activity in vivo. Cancer Cell. 2006;10:501–514. doi: 10.1016/j.ccr.2006.10.010. References 25 and 26 demonstrate the importance of the p53-MDM2 relationship in cancer and apoptosis using mouse models. They provide evidence supporting the concept that inducing moderate changes in p53 activity may be optimal in cancer therapies.
- 27.Uldrijan S, Pannekoek WJ, Vousden KH. An essential function of the extreme C-terminus of MDM2 can be provided by MDMX. EMBO J. 2007;26:102–112. doi: 10.1038/sj.emboj.7601469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poyurovsky MV, et al. The Mdm2 RING domain C-terminus is required for supramolecular assembly and ubiquitin ligase activity. EMBO J. 2007;26:90–101. doi: 10.1038/sj.emboj.7601465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Linke K, et al. Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans. Cell Death Differ. 2008;15:841–848. doi: 10.1038/sj.cdd.4402309. This paper elucidates the structure of the MDM2-MDMX RING finger dimer and provides important insights into function. The structure bears substantial similarity to the cIAP2 (also known as BIRC3) dimer, which was also solved by the Day laboratory.
- 30.Tanimura S, et al. MDM2 interacts with MDMX through their RING finger domains. FEBS Lett. 1999;447:5–9. doi: 10.1016/s0014-5793(99)00254-9. [DOI] [PubMed] [Google Scholar]
- 31.Okamoto K, Taya Y, Nakagama H. Mdmx enhances p53 ubiquitination by altering the substrate preference of the Mdm2 ubiquitin ligase. FEBS Lett. 2009;583:2710–2714. doi: 10.1016/j.febslet.2009.07.021. [DOI] [PubMed] [Google Scholar]
- 32.Linares LK, Hengstermann A, Ciechanover A, Muller S, Scheffner M. HdmX stimulates Hdm2-mediated ubiquitination and degradation of p53. Proc. Natl. Acad. Sci. U S A. 2003;100:12009–12014. doi: 10.1073/pnas.2030930100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stad R, et al. Mdmx stabilizes p53 and Mdm2 via two distinct mechanisms. EMBO Rep. 2001;2:1029–1034. doi: 10.1093/embo-reports/kve227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pomerantz J, et al. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell. 1998;92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- 35.Weber JD, Taylor LJ, Roussel MF, Sherr CJ, Bar-Sagi D. Nucleolar Arf sequesters Mdm2 and activates p53. Nat. Cell Biol. 1999;1:20–26. doi: 10.1038/8991. [DOI] [PubMed] [Google Scholar]
- 36.Saporita AJ, Maggi LBJ, Apicelli AJ, Weber JD. Therapeutic targets in the ARF tumor suppressor pathway. Curr. Med. Chem. 2007;14:1815–1827. doi: 10.2174/092986707781058869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y, Lu H. Signaling to p53: ribosomal proteins find their way. Cancer Cell. 2009;16:369–377. doi: 10.1016/j.ccr.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horn HF, Vousden KH. Cooperation between the ribosomal proteins L5 and L11 in the p53 pathway. Oncogene. 2008;27:5774–5784. doi: 10.1038/onc.2008.189. [DOI] [PubMed] [Google Scholar]
- 39.Dai MS, Lu H. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J. Biol. Chem. 2004;279:44475–44482. doi: 10.1074/jbc.M403722200. [DOI] [PubMed] [Google Scholar]
- 40.Lohrum MA, Ashcroft M, Kubbutat MH, Vousden KH. Identification of a cryptic nucleolar-localization signal in MDM2. Nat. Cell Biol. 2000;2:179–181. doi: 10.1038/35004057. [DOI] [PubMed] [Google Scholar]
- 41. Issaeva N, et al. Small molecule RITA binds to p53, blocks p53-HDM-2 interaction and activates p53 function in tumors. Nat. Med. 2004;10:1321–1328. doi: 10.1038/nm1146. This paper demonstrates that binding a small molecule to p53 can inhibit its degradation and activate a p53 response in tumors.
- 42.Zhao CY, Szekely L, Bao W, Selivanova G. Rescue of p53 function by small-molecule RITA in cervical carcinoma by blocking E6-mediated degradation. Cancer Res. 2010;70:3372–3381. doi: 10.1158/0008-5472.CAN-09-2787. [DOI] [PubMed] [Google Scholar]
- 43. Vassilev LT, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. This paper demonstrates that a small molecule that blocks the p53 binding site on MDM2 can activate a p53 response in tumors.
- 44.Patterson DM, et al. Effect of MDM2 and vascular endothelial growth factor inhibition on tumor angiogenesis and metastasis in neuroblastoma. Angiogenesis. 2011 doi: 10.1007/s10456-011-9210-8. [DOI] [PubMed] [Google Scholar]
- 45.Tovar C, et al. MDM2 antagonists boost antitumor effect of androgen withdrawal: implications for therapy of prostate cancer. Mol. Cancer. 2011;10:49. doi: 10.1186/1476-4598-10-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bernal F, Tyler AF, Korsmeyer SJ, Walensky LD, Verdine GL. Reactivation of the p53 tumor suppressor pathway by a stapled p53 peptide. J. Am. Chem. Soc. 2007;129:2456–2457. doi: 10.1021/ja0693587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lai Z, et al. Differentiation of Hdm2-mediated p53 ubiquitination and Hdm2 autoubiquitination activity by small molecular weight inhibitors. Proc. Natl. Acad. Sci. U S A. 2002;99:14734–14739. doi: 10.1073/pnas.212428599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yang Y, et al. Small molecule inhibitors of HDM2 ubiquitin ligase activity stabilize and activate p53 in cells. Cancer Cell. 2005;7:547–559. doi: 10.1016/j.ccr.2005.04.029. This paper provides proof-of-principle that blocking the ubiquitin ligase activity of MDM2 can activate a p53 response in cells and kill tumor cells.
- 49.Kitagaki J, Agama KK, Pommier Y, Yang Y, Weissman AM. Targeting tumor cells expressing p53 with a water-soluble inhibitor of Hdm2. Mol. Cancer Ther. 2008;7:2445–2454. doi: 10.1158/1535-7163.MCT-08-0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Welcsh PL, King MC. BRCA1 and BRCA2 and the genetics of breast and ovarian cancer. Hum. Mol. Genet. 2001;10:705–713. doi: 10.1093/hmg/10.7.705. [DOI] [PubMed] [Google Scholar]
- 51.Scully R, et al. Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell. 1997;90:425–435. doi: 10.1016/s0092-8674(00)80503-6. [DOI] [PubMed] [Google Scholar]
- 52.Scully R, et al. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell. 1997;88:265–275. doi: 10.1016/s0092-8674(00)81847-4. [DOI] [PubMed] [Google Scholar]
- 53.Moynahan ME, Chiu JW, Koller BH, Jasin M. Brca1 controls homologydirected DNA repair. Mol. Cell. 1999;4:511–518. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- 54.Snouwaert JN, et al. BRCA1 deficient embryonic stem cells display a decreased homologous recombination frequency and an increased frequency of nonhomologous recombination that is corrected by expression of a brca1 transgene. Oncogene. 1999;18:7900–7907. doi: 10.1038/sj.onc.1203334. [DOI] [PubMed] [Google Scholar]
- 55. Huen MS, Sy SM, Chen J. BRCA1 and its toolbox for the maintenance of genome integrity. Nat. Rev. Mol. Cell Biol. 2010;11:138–148. doi: 10.1038/nrm2831. This review summarizes the role of BRCA1 in DNA repair, including many important primary references not cited herein.
- 56. Hashizume R, et al. The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J. Biol. Chem. 2001;276:14537–14540. doi: 10.1074/jbc.C000881200. This paper establishes the importance of the BRCA1-BARD1 dimer in ubiquitin ligase function and demonstrates that a disease-associated mutation in the BRCA1 RING finger results in loss of activity.
- 57.Xia Y, Pao GM, Chen HW, Verma IM, Hunter T. Enhancement of BRCA1 E3 ubiquitin ligase activity through direct interaction with the BARD1 protein. J. Biol. Chem. 2003;278:5255–5263. doi: 10.1074/jbc.M204591200. [DOI] [PubMed] [Google Scholar]
- 58.Joukov V, Chen J, Fox EA, Green JB, Livingston DM. Functional communication between endogenous BRCA1 and its partner, BARD1, during Xenopus laevis development. Proc. Natl. Acad. Sci. U S A. 2001;98:12078–12083. doi: 10.1073/pnas.211427098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meza JE, Brzovic PS, King MC, Klevit RE. Mapping the functional domains of BRCA1. Interaction of the ring finger domains of BRCA1 and BARD1. J. Biol. Chem. 1999;274:5659–5665. doi: 10.1074/jbc.274.9.5659. [DOI] [PubMed] [Google Scholar]
- 60. Brzovic PS, Rajagopal P, Hoyt DW, King MC, Klevit RE. Structure of a BRCA1-BARD1 heterodimeric RING-RING complex. Nat. Struct. Biol. 2001;8:833–837. doi: 10.1038/nsb1001-833. This paper elucidates the structure of the BRCA1-BARD1 heterodimer by nuclear magnetic resonance (NMR). This is the first structure of a RING finger dimer.
- 61.Morris JR, et al. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature. 2009;462:886–890. doi: 10.1038/nature08593. [DOI] [PubMed] [Google Scholar]
- 62.Yu X, Fu S, Lai M, Baer R, Chen J. BRCA1 ubiquitinates its phosphorylation-dependent binding partner CtIP. Genes Dev. 2006;20:1721–1726. doi: 10.1101/gad.1431006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barber LJ, Boulton SJ. BRCA1 ubiquitylation of CtIP: Just the tIP of the iceberg? DNA Repair (Amst) 2006;5:1499–1504. doi: 10.1016/j.dnarep.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 64.Ma Y, et al. The breast cancer susceptibility gene BRCA1 regulates progesterone receptor signaling in mammary epithelial cells. Mol. Endocrinol. 2006;20:14–34. doi: 10.1210/me.2004-0488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fan S, et al. Role of direct interaction in BRCA1 inhibition of estrogen receptor activity. Oncogene. 2001;20:77–87. doi: 10.1038/sj.onc.1204073. [DOI] [PubMed] [Google Scholar]
- 66.Eakin CM, Maccoss MJ, Finney GL, Klevit RE. Estrogen receptor alpha is a putative substrate for the BRCA1 ubiquitin ligase. Proc. Natl. Acad. Sci. U S A. 2007;104:5794–5799. doi: 10.1073/pnas.0610887104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Poole AJ, et al. Prevention of Brca1-mediated mammary tumorigenesis in mice by a progesterone antagonist. Science. 2006;314:1467–1470. doi: 10.1126/science.1130471. [DOI] [PubMed] [Google Scholar]
- 68.Katiyar P, Ma Y, Riegel A, Fan S, Rosen EM. Mechanism of BRCA1-mediated inhibition of progesterone receptor transcriptional activity. Mol. Endocrinol. 2009;23:1135–1146. doi: 10.1210/me.2008-0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ma Y, et al. BRCA1 regulates acetylation and ubiquitination of estrogen receptoralpha. Mol. Endocrinol. 2010;24:76–90. doi: 10.1210/me.2009-0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li W, Xiao C, Vonderhaar BK, Deng CX. A role of estrogen/ERalpha signaling in BRCA1-associated tissue-specific tumor formation. Oncogene. 2007;26:7204–7212. doi: 10.1038/sj.onc.1210527. [DOI] [PubMed] [Google Scholar]
- 71.Jones LP, et al. Activation of estrogen signaling pathways collaborates with loss of Brca1 to promote development of ERalpha-negative and ERalpha-positive mammary preneoplasia and cancer. Oncogene. 2008;27:794–802. doi: 10.1038/sj.onc.1210674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Eisen A, et al. Breast cancer risk following bilateral oophorectomy in BRCA1 and BRCA2 mutation carriers: an international case-control study. J. Clin. Oncol. 2005;23:7491–7496. doi: 10.1200/JCO.2004.00.7138. [DOI] [PubMed] [Google Scholar]
- 73. Moldovan GL, D'Andrea AD. How the fanconi anemia pathway guards the genome. Annu. Rev. Genet. 2009;43:223–249. doi: 10.1146/annurev-genet-102108-134222. This review article summarizes a large body of work on the FANC ubiquitin ligase and its function including many important primary references not cited herein.
- 74.Vaz F, et al. Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat. Genet. 2010;42:406–409. doi: 10.1038/ng.570. [DOI] [PubMed] [Google Scholar]
- 75.Stoepker C, et al. SLX4, a coordinator of structure-specific endonucleases, is mutated in a new Fanconi anemia subtype. Nat. Genet. 2011;43:138–141. doi: 10.1038/ng.751. [DOI] [PubMed] [Google Scholar]
- 76.Machida YJ, et al. UBE2T is the E2 in the Fanconi anemia pathway and undergoes negative autoregulation. Mol. Cell. 2006;23:589–596. doi: 10.1016/j.molcel.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 77.Eletr ZM, Huang DT, Duda DM, Schulman BA, Kuhlman B. E2 conjugating enzymes must disengage from their E1 enzymes before E3-dependent ubiquitin and ubiquitin-like transfer. Nat. Struct. Mol. Biol. 2005;12:933–934. doi: 10.1038/nsmb984. [DOI] [PubMed] [Google Scholar]
- 78.Knipscheer P, et al. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science. 2009;326:1698–1701. doi: 10.1126/science.1182372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.MacKay C, et al. Identification of KIAA1018/FAN1, a DNA repair nuclease recruited to DNA damage by monoubiquitinated FANCD2. Cell. 2010;142:65–76. doi: 10.1016/j.cell.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Smogorzewska A, et al. A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Mol. Cell. 2010;39:36–47. doi: 10.1016/j.molcel.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kratz K, et al. Deficiency of FANCD2-associated nuclease KIAA1018/FAN1 sensitizes cells to interstrand crosslinking agents. Cell. 2010;142:77–88. doi: 10.1016/j.cell.2010.06.022. [DOI] [PubMed] [Google Scholar]
- 82.Kee Y, Kim JM, D'Andrea AD. Regulated degradation of FANCM in the Fanconi anemia pathway during mitosis. Genes Dev. 2009;23:555–560. doi: 10.1101/gad.1761309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Farmer H, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 84. Bryant HE, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. References 83 and 84 demonstrate the synthetic lethality of PARP inhibition in cells deficiency in BRCA1 or BRCA2 and are the basis of clinical trials using PARP inhibitors in patients with BRCA1 and BRCA2 mutations.
- 85.Fong PC, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl. J. Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 86.Audeh MW, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet. 2010;376:245–251. doi: 10.1016/S0140-6736(10)60893-8. [DOI] [PubMed] [Google Scholar]
- 87.Tutt A, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet. 2010;376:235–244. doi: 10.1016/S0140-6736(10)60892-6. [DOI] [PubMed] [Google Scholar]
- 88.Fujita Y, et al. Hakai, a c-Cbl-like protein, ubiquitinates and induces endocytosis of the E-cadherin complex. Nat. Cell Biol. 2002;4:222–231. doi: 10.1038/ncb758. [DOI] [PubMed] [Google Scholar]
- 89.Qiu XB, Goldberg AL. Nrdp1/FLRF is a ubiquitin ligase promoting ubiquitination and degradation of the epidermal growth factor receptor family member, ErbB3. Proc. Natl. Acad. Sci. U S A. 2002;99:14843–14848. doi: 10.1073/pnas.232580999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xu W, et al. Chaperone-dependent E3 ubiquitin ligase CHIP mediates a degradative pathway for c-ErbB2/Neu. Proc. Natl. Acad. Sci. U S A. 2002;99:12847–12852. doi: 10.1073/pnas.202365899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhou P, et al. ErbB2 degradation mediated by the co-chaperone protein CHIP. J. Biol. Chem. 2003;278:13829–13837. doi: 10.1074/jbc.M209640200. [DOI] [PubMed] [Google Scholar]
- 92.Li Y, Kumar KG, Tang W, Spiegelman VS, Fuchs SY. Negative regulation of prolactin receptor stability and signaling mediated by SCF(beta-TrCP) E3 ubiquitin ligase. Mol. Cell. Biol. 2004;24:4038–4048. doi: 10.1128/MCB.24.9.4038-4048.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.van Kerkhof P, Putters J, Strous GJ. The ubiquitin ligase SCF(betaTrCP) regulates the degradation of the growth hormone receptor. J. Biol. Chem. 2007;282:20475–20483. doi: 10.1074/jbc.M702610200. [DOI] [PubMed] [Google Scholar]
- 94.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 95.Staudt LM. Oncogenic activation of NF-kappaB. Cold Spring Harb Perspect Biol. 2010;2:a000109. doi: 10.1101/cshperspect.a000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 97.Acconcia F, Sigismund S, Polo S. Ubiquitin in trafficking: the network at work. Exp Cell Res. 2009;315:1610–1618. doi: 10.1016/j.yexcr.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 98.Ettenberg SA, et al. cbl-b inhibits EGF-receptor-induced apoptosis by enhancing ubiquitination and degradation of activated receptors. Mol Cell Biol Res Commun. 1999;2:111–118. doi: 10.1006/mcbr.1999.0157. [DOI] [PubMed] [Google Scholar]
- 99.Levkowitz G, et al. c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev. 1998;12:3663–3674. doi: 10.1101/gad.12.23.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Miyake S, Mullane-Robinson KP, Lill NL, Douillard P, Band H. Cbl-mediated negative regulation of platelet-derived growth factor receptor-dependent cell proliferation. A critical role for Cbl tyrosine kinase-binding domain. J. Biol. Chem. 1999;274:16619–16628. doi: 10.1074/jbc.274.23.16619. [DOI] [PubMed] [Google Scholar]
- 101.Joazeiro CA, et al. The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science. 1999;286:309–312. doi: 10.1126/science.286.5438.309. [DOI] [PubMed] [Google Scholar]
- 102.Levkowitz G, et al. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol. Cell. 1999;4:1029–1040. doi: 10.1016/s1097-2765(00)80231-2. [DOI] [PubMed] [Google Scholar]
- 103. Yokouchi M, et al. Ligand-induced ubiquitination of the epidermal growth factor receptor involves the interaction of the c-Cbl RING finger and UbcH7. J. Biol. Chem. 1999;274:31707–31712. doi: 10.1074/jbc.274.44.31707. References 100–103 demonstrate that CBL proteins are RING finger E3 ubiquitin ligases that ubiquitylate and target activated RTKs for degradation.
- 104.Nau MM, Lipkowitz S. In: Cbl proteins. Tsygankov A, editor. New York: Nova Science Publishers Inc; 2008. pp. 3–25. [Google Scholar]
- 105. Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin- protein ligases. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. This paper describes the first crystal structure of a RING finger – E2 complex. The sites of RING finger-E2 interaction have held up through subsequent structure and structure-function analyses.
- 106.Kassenbrock CK, Anderson SM. Regulation of ubiquitin protein ligase activity in c-Cbl by phosphorylation-induced conformational change and constitutive activation by tyrosine to glutamate point mutations. J. Biol. Chem. 2004;279:28017–28027. doi: 10.1074/jbc.M404114200. [DOI] [PubMed] [Google Scholar]
- 107.Ryan PE, Sivadasan-Nair N, Nau MM, Nicholas S, Lipkowitz S. The N-terminus of Cbl-c regulates ubiquitin ligase activity by modulating affinity for the ubiquitin conjugating enzyme. J. Biol. Chem. 2010 doi: 10.1074/jbc.M109.091157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Huang F, Kirkpatrick D, Jiang X, Gygi S, Sorkin A. Differential regulation of EGF receptor internalization and degradation by multiubiquitination within the kinase domain. Mol. Cell. 2006;21:737–748. doi: 10.1016/j.molcel.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 109.Mosesson Y, et al. Endocytosis of receptor tyrosine kinases is driven by monoubiquitylation, not polyubiquitylation. J. Biol. Chem. 2003;278:21323–21326. doi: 10.1074/jbc.C300096200. [DOI] [PubMed] [Google Scholar]
- 110.Schmidt MH, Dikic I. The Cbl interactome and its functions. Nat. Rev. Mol. Cell Biol. 2005;6:907–918. doi: 10.1038/nrm1762. [DOI] [PubMed] [Google Scholar]
- 111.Langdon WY, Hartley JW, Klinken SP, Ruscetti SK, Morse HC., III v-cbl, an oncogene from a dual-recombinant murine retrovirus that induces early Blineage lymphomas. Proc. Natl. Acad. Sci. U S A. 1989;86:1168–1172. doi: 10.1073/pnas.86.4.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bonita DP, Miyake S, Lupher ML, Jr, Langdon WY, Band H. Phosphotyrosine binding domain-dependent upregulation of the platelet-derived growth factor receptor alpha signaling cascade by transforming mutants of Cbl: implications for Cbl's function and oncogenicity. Mol. Cell. Biol. 1997;17:4597–4610. doi: 10.1128/mcb.17.8.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kales SC, Ryan PE, Nau MM, Lipkowitz S. Cbl and human myeloid neoplasms: the Cbl oncogene comes of age. Cancer Res. 2010;70:4789–4794. doi: 10.1158/0008-5472.CAN-10-0610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Naramura M, Nandwani N, Gu H, Band V, Band H. Rapidly fatal myeloproliferative disorders in mice with deletion of Casitas B-cell lymphoma (Cbl) and Cbl-b in hematopoietic stem cells. Proc. Natl. Acad. Sci. U S A. 2010;107:16274–16279. doi: 10.1073/pnas.1007575107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Reindl C, et al. CBL exon 8/9 mutants activate the FLT3 pathway and cluster in core binding factor/11q deletion acute myeloid leukemia/myelodysplastic syndrome subtypes. Clin Cancer Res. 2009;15:2238–2247. doi: 10.1158/1078-0432.CCR-08-1325. [DOI] [PubMed] [Google Scholar]
- 116.Sanada M, et al. Gain-of-function of mutated C-CBL tumour suppressor in myeloid neoplasms. Nature. 2009;460:904–908. doi: 10.1038/nature08240. [DOI] [PubMed] [Google Scholar]
- 117.Sargin B, et al. Flt3-dependent transformation by inactivating c-Cbl mutations in AML. Blood. 2007 doi: 10.1182/blood-2007-01-066076. [DOI] [PubMed] [Google Scholar]
- 118.Gilliland DG, Griffin JD. Role of FLT3 in leukemia. Curr. Opin. Hematol. 2002;9:274–281. doi: 10.1097/00062752-200207000-00003. [DOI] [PubMed] [Google Scholar]
- 119.Grand FH, et al. Frequent CBL mutations associated with 11q acquired uniparental disomy in myeloproliferative neoplasms. Blood. 2009;113:6182–6192. doi: 10.1182/blood-2008-12-194548. [DOI] [PubMed] [Google Scholar]
- 120.Rathinam C, Thien CB, Flavell RA, Langdon WY. Myeloid leukemia development in c-Cbl RING finger mutant mice is dependent on FLT3 signaling. Cancer Cell. 2010;18:341–352. doi: 10.1016/j.ccr.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 121.Tan YH, et al. CBL Is Frequently Altered in Lung Cancers: Its Relationship to Mutations in MET and EGFR Tyrosine Kinases. PLoS ONE. 2010;5:e8972. doi: 10.1371/journal.pone.0008972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Peschard P, Park M. Escape from Cbl-mediated downregulation: a recurrent theme for oncogenic deregulation of receptor tyrosine kinases. Cancer Cell. 2003;3:519–523. doi: 10.1016/s1535-6108(03)00136-3. [DOI] [PubMed] [Google Scholar]
- 123.Ryan PE, Davies GC, Nau MM, Lipkowitz S. Regulating the regulator: negative regulation of Cbl ubiquitin ligases. Trends Biochem. Sci. 2006;31:79–88. doi: 10.1016/j.tibs.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 124.Thien CB, et al. c-Cbl promotes T cell receptor-induced thymocyte apoptosis by activating the phosphatidylinositol 3-kinase/Akt pathway. J. Biol. Chem. 2010;285:10969–10981. doi: 10.1074/jbc.M109.094920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bachmaier K, et al. Negative regulation of lymphocyte activation and autoimmunity by the molecular adaptor Cbl-b. Nature. 2000;403:211–216. doi: 10.1038/35003228. [DOI] [PubMed] [Google Scholar]
- 126.Chiang YJ, et al. Cbl-b regulates the CD28 dependence of T-cell activation. Nature. 2000;403:216–220. doi: 10.1038/35003235. [DOI] [PubMed] [Google Scholar]
- 127.Jeon MS, et al. Essential role of the E3 ubiquitin ligase Cbl-b in T cell anergy induction. Immunity. 2004;21:167–177. doi: 10.1016/j.immuni.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 128.Chiang JY, Jang IK, Hodes R, Gu H. Ablation of Cbl-b provides protection against transplanted and spontaneous tumors. J. Clin. Invest. 2007;117:1029–1036. doi: 10.1172/JCI29472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Loeser S, et al. Spontaneous tumor rejection by cbl-b-deficient CD8+ T cells. J. Exp. Med. 2007;204:879–891. doi: 10.1084/jem.20061699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Paolino M, et al. Essential role of E3 ubiquitin ligase activity in Cbl-b-regulated T cell functions. J. Immunol. 2011;186:2138–2147. doi: 10.4049/jimmunol.1003390. [DOI] [PubMed] [Google Scholar]
- 131.Latif F, et al. Identification of the von Hippel-Lindau disease tumor suppressor gene. Science. 1993;260:1317–1320. doi: 10.1126/science.8493574. [DOI] [PubMed] [Google Scholar]
- 132.Gnarra JR, et al. Mutations of the VHL tumour suppressor gene in renal carcinoma. Nat. Genet. 1994;7:85–90. doi: 10.1038/ng0594-85. [DOI] [PubMed] [Google Scholar]
- 133.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc. Natl. Acad. Sci. U S A. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Iliopoulos O, Kibel A, Gray S, Kaelin WG., Jr Tumour suppression by the human von Hippel-Lindau gene product. Nat. Med. 1995;1:822–826. doi: 10.1038/nm0895-822. [DOI] [PubMed] [Google Scholar]
- 135. Kamura T, et al. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science. 1999;284:657–661. doi: 10.1126/science.284.5414.657. This paper demonstrates the importance of the small RING finger protein RBX1 (also known as ROC1, Hrt1 in yeast) in the activity of the CRL2VHL E3 ubiquitin ligase. Contemporaneous papers from the Deshaies, Elledge, Harper, Pan and Xiong labs established the importance of this protein for the SCF family of E3s.
- 136.Kibel A, Iliopoulos O, DeCaprio JA, Kaelin WG., Jr Binding of the von Hippel-Lindau tumor suppressor protein to Elongin B and C. Science. 1995;269:1444–1446. doi: 10.1126/science.7660130. [DOI] [PubMed] [Google Scholar]
- 137.Pause A, et al. The von Hippel-Lindau tumor-suppressor gene product forms a stable complex with human CUL-2, a member of the Cdc53 family of proteins. Proc. Natl. Acad. Sci. U S A. 1997;94:2156–2161. doi: 10.1073/pnas.94.6.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Iwai K, et al. Identification of the von Hippel-lindau tumor-suppressor protein as part of an active E3 ubiquitin ligase complex. Proc. Natl. Acad. Sci. U S A. 1999;96:12436–12441. doi: 10.1073/pnas.96.22.12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Lisztwan J, Imbert G, Wirbelauer C, Gstaiger M, Krek W. The von Hippel-Lindau tumor suppressor protein is a component of an E3 ubiquitin-protein ligase activity. Genes Dev. 1999;13:1822–1833. doi: 10.1101/gad.13.14.1822. References 138 and 139 demonstrate that the VHL complex is an E3 ubiquitin ligase and that disease-associated mutations of the VHL protein abrogate ubiquitin ligase activity.
- 140.Maxwell PH, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 141.Ohh M, et al. Ubiquitination of hypoxia-inducible factor requires direct binding to the beta-domain of the von Hippel-Lindau protein. Nat. Cell Biol. 2000;2:423–427. doi: 10.1038/35017054. [DOI] [PubMed] [Google Scholar]
- 142.Gnarra JR, et al. Post-transcriptional regulation of vascular endothelial growth factor mRNA by the product of the VHL tumor suppressor gene. Proc. Natl. Acad. Sci. U S A. 1996;93:10589–10594. doi: 10.1073/pnas.93.20.10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Iliopoulos O, Levy AP, Jiang C, Kaelin WG, Jr, Goldberg MA. Negative regulation of hypoxia-inducible genes by the von Hippel-Lindau protein. Proc. Natl. Acad. Sci. U S A. 1996;93:10595–10599. doi: 10.1073/pnas.93.20.10595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ivan M, et al. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 145.Jaakkola P, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 146.Hon WC, et al. Structural basis for the recognition of hydroxyproline in HIF-1 alpha by pVHL. Nature. 2002;417:975–978. doi: 10.1038/nature00767. [DOI] [PubMed] [Google Scholar]
- 147.Epstein AC, et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 148.Kondo K, Kim WY, Lechpammer M, Kaelin WG., Jr Inhibition of HIF2alpha is sufficient to suppress pVHL-defective tumor growth. PLoS Biol. 2003;1:E83. doi: 10.1371/journal.pbio.0000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kondo K, Klco J, Nakamura E, Lechpammer M, Kaelin WG., Jr Inhibition of HIF is necessary for tumor suppression by the von Hippel-Lindau protein. Cancer Cell. 2002;1:237–246. doi: 10.1016/s1535-6108(02)00043-0. [DOI] [PubMed] [Google Scholar]
- 150.Maranchie JK, et al. The contribution of VHL substrate binding and HIF1-alpha to the phenotype of VHL loss in renal cell carcinoma. Cancer Cell. 2002;1:247–255. doi: 10.1016/s1535-6108(02)00044-2. [DOI] [PubMed] [Google Scholar]
- 151.Zimmer M, Doucette D, Siddiqui N, Iliopoulos O. Inhibition of hypoxiainducible factor is sufficient for growth suppression of VHL−/− tumors. Mol. Cancer Res. 2004;2:89–95. [PubMed] [Google Scholar]
- 152.Nakayama K, et al. Siah2 regulates stability of prolyl-hydroxylases, controls HIF1alpha abundance, and modulates physiological responses to hypoxia. Cell. 2004;117:941–952. doi: 10.1016/j.cell.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 153.House CM, Moller A, Bowtell DD. Siah proteins: novel drug targets in the Ras and hypoxia pathways. Cancer Res. 2009;69:8835–8838. doi: 10.1158/0008-5472.CAN-09-1676. [DOI] [PubMed] [Google Scholar]
- 154.Crawford LJ, Walker B, Irvine AE. Proteasome inhibitors in cancer therapy. J. Cell Commun. Signal. 2011 doi: 10.1007/s12079-011-0121-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Dominguez C, et al. Structural model of the UbcH5B/CNOT4 complex revealed by combining NMR mutagenesis, and docking approaches. Structure. 2004;12:633–644. doi: 10.1016/j.str.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 156.Linehan WM, Srinivasan R, Schmidt LS. The genetic basis of kidney cancer: a metabolic disease. Nat Rev Urol. 2010;7:277–285. doi: 10.1038/nrurol.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Isaacs JS, et al. HIF overexpression correlates with biallelic loss of fumarate hydratase in renal cancer: novel role of fumarate in regulation of HIF stability. Cancer Cell. 2005;8:143–153. doi: 10.1016/j.ccr.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 158.Vitari AC, et al. COP1 is a tumour suppressor that causes degradation of ETS transcription factors. Nature. 2011;474:403–406. doi: 10.1038/nature10005. [DOI] [PubMed] [Google Scholar]
- 159.Migliorini D, et al. Cop1 constitutively regulates c-Jun protein stability and functions as a tumor suppressor in mice. J. Clin. Invest. 2011;121:1329–1343. doi: 10.1172/JCI45784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hsu JY, Reimann JD, Sorensen CS, Lukas J, Jackson PK. E2F-dependent accumulation of hEmi1 regulates S phase entry by inhibiting APC(Cdh1) Nat. Cell Biol. 2002;4:358–366. doi: 10.1038/ncb785. [DOI] [PubMed] [Google Scholar]
- 161.Lehman NL, Verschuren EW, Hsu JY, Cherry AM, Jackson PK. Overexpression of the anaphase promoting complex/cyclosome inhibitor Emi1 leads to tetraploidy and genomic instability of p53-deficient cells. Cell Cycle. 2006;5:1569–1573. doi: 10.4161/cc.5.14.2925. [DOI] [PubMed] [Google Scholar]
- 162.Reimann JD, et al. Emi1 is a mitotic regulator that interacts with Cdc20 and inhibits the anaphase promoting complex. Cell. 2001;105:645–655. doi: 10.1016/s0092-8674(01)00361-0. [DOI] [PubMed] [Google Scholar]
- 163.O'Neil J, et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. J. Exp. Med. 2007;204:1813–1824. doi: 10.1084/jem.20070876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Bloom J, Pagano M. Deregulated degradation of the cdk inhibitor p27 and malignant transformation. Semin. Cancer Biol. 2003;13:41–47. doi: 10.1016/s1044-579x(02)00098-6. [DOI] [PubMed] [Google Scholar]
- 165.Gstaiger M, et al. Skp2 is oncogenic and overexpressed in human cancers. Proc. Natl. Acad. Sci. U S A. 2001;98:5043–5048. doi: 10.1073/pnas.081474898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Latres E, et al. Role of the F-box protein Skp2 in lymphomagenesis. Proc. Natl. Acad. Sci. U S A. 2001;98:2515–2520. doi: 10.1073/pnas.041475098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.De Brakeleer S, et al. Cancer predisposing missense and protein truncating BARD1 mutations in non-BRCA1 or BRCA2 breast cancer families. Hum Mutat. 2010;31:E1175–E1185. doi: 10.1002/humu.21200. [DOI] [PubMed] [Google Scholar]
- 168.Ratajska M, et al. Cancer predisposing BARD1 mutations in breast-ovarian cancer families. Breast Cancer Res. Treat. 2011 doi: 10.1007/s10549-011-1403-8. [DOI] [PubMed] [Google Scholar]
- 169.Sabatier R, et al. BARD1 homozygous deletion, a possible alternative to BRCA1 mutation in basal breast cancer. Genes Chromosomes Cancer. 2010;49:1143–1151. doi: 10.1002/gcc.20822. [DOI] [PubMed] [Google Scholar]
- 170.Ruffner H, Joazeiro CA, Hemmati D, Hunter T, Verma IM. Cancer-predisposing mutations within the RING domain of BRCA1: loss of ubiquitin protein ligase activity and protection from radiation hypersensitivity. Proc. Natl. Acad. Sci. U S A. 2001;98:5134–5139. doi: 10.1073/pnas.081068398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Dias DC, Dolios G, Wang R, Pan ZQ. CUL7: A DOC domain-containing cullin selectively binds Skp1.Fbx29 to form an SCF-like complex. Proc. Natl. Acad. Sci. U S A. 2002;99:16601–16606. doi: 10.1073/pnas.252646399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Skaar JR, et al. PARC and CUL7 form atypical cullin RING ligase complexes. Cancer Res. 2007;67:2006–2014. doi: 10.1158/0008-5472.CAN-06-3241. [DOI] [PubMed] [Google Scholar]
- 173.Kim SS, et al. CUL7 is a novel antiapoptotic oncogene. Cancer Res. 2007;67:9616–9622. doi: 10.1158/0008-5472.CAN-07-0644. [DOI] [PubMed] [Google Scholar]
- 174.Oliner JD, Kinzler KW, Meltzer PS, George DL, Vogelstein B. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature. 1992;358:80–83. doi: 10.1038/358080a0. [DOI] [PubMed] [Google Scholar]
- 175.Bo MD, et al. MDM4 (MDMX) is overexpressed in chronic lymphocytic leukaemia (CLL) and marks a subset of p53wild-type CLL with a poor cytotoxic response to Nutlin-3. Br. J. Haematol. 2010;150:237–239. doi: 10.1111/j.1365-2141.2010.08185.x. [DOI] [PubMed] [Google Scholar]
- 176.Danovi D, et al. Amplification of Mdmx (or Mdm4) directly contributes to tumor formation by inhibiting p53 tumor suppressor activity. Mol. Cell. Biol. 2004;24:5835–5843. doi: 10.1128/MCB.24.13.5835-5843.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Riemenschneider MJ, et al. Amplification and overexpression of the MDM4 (MDMX) gene from 1q32 in a subset of malignant gliomas without TP53 mutation or MDM2 amplification. Cancer Res. 1999;59:6091–6096. [PubMed] [Google Scholar]
- 178.Nikolaev AY, Li M, Puskas N, Qin J, Gu W. Parc: a cytoplasmic anchor for p53. Cell. 2003;112:29–40. doi: 10.1016/s0092-8674(02)01255-2. [DOI] [PubMed] [Google Scholar]
- 179.Duan W, et al. Expression of Pirh2, a newly identified ubiquitin protein ligase, in lung cancer. J. Natl. Cancer Inst. 2004;96:1718–1721. doi: 10.1093/jnci/djh292. [DOI] [PubMed] [Google Scholar]
- 180.Leng RP, et al. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell. 2003;112:779–791. doi: 10.1016/s0092-8674(03)00193-4. [DOI] [PubMed] [Google Scholar]
- 181.Hu J, et al. WD40 protein FBW5 promotes ubiquitination of tumor suppressor TSC2 by DDB1-CUL4-ROC1 ligase. Genes Dev. 2008;22:866–871. doi: 10.1101/gad.1624008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Figueroa A, et al. Novel roles of hakai in cell proliferation and oncogenesis. Mol. Biol. Cell. 2009;20:3533–3542. doi: 10.1091/mbc.E08-08-0845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Annunziata CM, et al. Frequent engagement of the classical and alternative NF-kappaB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell. 2007;12:115–130. doi: 10.1016/j.ccr.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Danson S, Dean E, Dive C, Ranson M. IAPs as a target for anticancer therapy. Curr Cancer Drug Targets. 2007;7:785–794. doi: 10.2174/156800907783220471. [DOI] [PubMed] [Google Scholar]
- 185.Huang H, et al. The inhibitor of apoptosis, cIAP2, functions as a ubiquitin-protein ligase and promotes in vitro monoubiquitination of caspases 3 and 7. J. Biol. Chem. 2000;275:26661–26664. doi: 10.1074/jbc.C000199200. [DOI] [PubMed] [Google Scholar]
- 186.Keats JJ, et al. Promiscuous mutations activate the noncanonical NF-kappaB pathway in multiple myeloma. Cancer Cell. 2007;12:131–144. doi: 10.1016/j.ccr.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Yang Y, Fang S, Jensen JP, Weissman AM, Ashwell JD. Ubiquitin protein ligase activity of IAPs and their degradation in proteasomes in response to apoptotic stimuli. Science. 2000;288:874–877. doi: 10.1126/science.288.5467.874. [DOI] [PubMed] [Google Scholar]
- 188.Compagno M, et al. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature. 2009;459:717–721. doi: 10.1038/nature07968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Gemmill RM, et al. The hereditary renal cell carcinoma 3;8 translocation fuses FHIT to a patched-related gene, TRC8. Proc. Natl. Acad. Sci. U S A. 1998;95:9572–9577. doi: 10.1073/pnas.95.16.9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Gimelli S, et al. The tumor suppressor gene TRC8/RNF139 is disrupted by a constitutional balanced translocation t(8;22)(q24.13;q11.21) in a young girl with dysgerminoma. Mol. Cancer. 2009;8:52. doi: 10.1186/1476-4598-8-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Lee JP, et al. The TRC8 ubiquitin ligase is sterol regulated and interacts with lipid and protein biosynthetic pathways. Mol. Cancer Res. 2010;8:93–106. doi: 10.1158/1541-7786.MCR-08-0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Tsai YC, et al. The ubiquitin ligase gp78 promotes sarcoma metastasis by targeting KAI1 for degradation. Nat. Med. 2007;13:1504–1509. doi: 10.1038/nm1686. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.