Background: RecBCD helicase is involved in repair of double-stranded DNA breaks.
Results: The 5′ to 3′ ssDNA translocation rate of RecBCD is faster than the 3′ to 5′ rate in the absence of a CHI site, and the rates are coupled asymmetrically.
Conclusion: RecBC controls 3′ to 5′ and 5′ to 3′ translocation, but RecD controls only 5′ to 3′ translocation.
Significance: Asymmetric regulation may explain how RecBCD is regulated after CHI recognition.
Keywords: ATPases, DNA Helicase, DNA Recombination, DNA Repair, Kinetics, DNA, RecBCD, Fluorescence, Translocation, Unwinding
Abstract
Repair of double-stranded DNA breaks in Escherichia coli is initiated by the RecBCD helicase that possesses two superfamily-1 motors, RecB (3′ to 5′ translocase) and RecD (5′ to 3′ translocase), that operate on the complementary DNA strands to unwind duplex DNA. However, it is not known whether the RecB and RecD motors act independently or are functionally coupled. Here we show by directly monitoring ATP-driven single-stranded DNA translocation of RecBCD that the 5′ to 3′ rate is always faster than the 3′ to 5′ rate on DNA without a crossover hotspot instigator site and that the translocation rates are coupled asymmetrically. That is, RecB regulates both 3′ to 5′ and 5′ to 3′ translocation, whereas RecD only regulates 5′ to 3′ translocation. We show that the recently identified RecBC secondary translocase activity functions within RecBCD and that this contributes to the coupling. This coupling has implications for how RecBCD activity is regulated after it recognizes a crossover hotspot instigator sequence during DNA unwinding.
Introduction
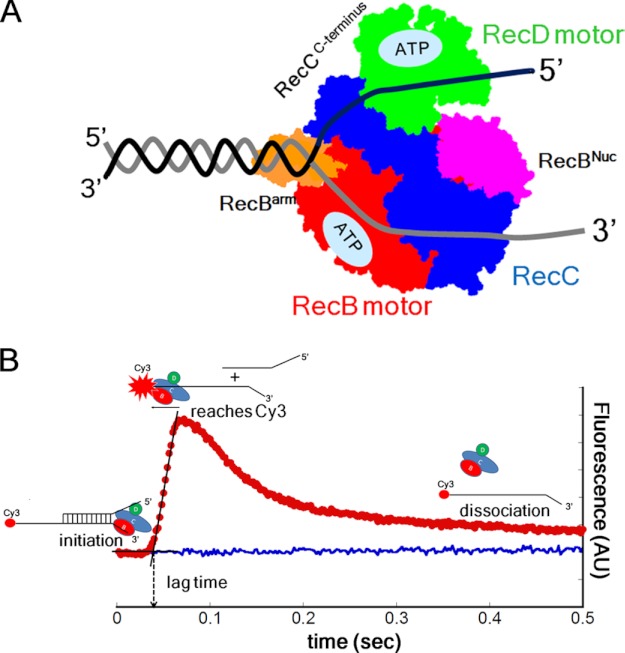
The Escherichia coli RecBCD enzyme possesses DNA helicase and nuclease activities and is involved in repair of double-stranded (ds) DNA breaks (1–6). The heterotrimeric bipolar helicase (see Fig. 1A) possesses two translocating motors, RecB, a 3′ to 5′ SF1 helicase and nuclease, and RecD, a 5′ to 3′ SF1 helicase (7–10). RecC is structurally similar to RecB but with no ATPase or helicase activities (11) and functions as a processivity and regulatory factor. After initiating DNA unwinding, the two motors within the RecBCD holoenzyme move in the same net direction by operating along complementary strands of the unwound DNA duplex (7, 10).
FIGURE 1.
Fluorescence assay to monitor RecBCD translocation in the 3′ to 5′ and 5′ to 3′ directions. A, schematic of RecBCD bound to a DNA end. RecB motor (red), RecC (blue), and RecD motor (green), RecB nuclease (Nuc) domain (purple), and RecB arm region (orange). B, stopped-flow time course (red) monitoring RecBCD translocation along ssDNA in the 3′ to 5′ direction using DNA III. RecBCD binds at the loading site, upon addition of ATP and heparin (trap for free RecBCD) unwinds the 24-bp duplex, and then translocates along the (dT)L extension until it reaches the Cy3 fluorophore (red circle), resulting in Cy3 fluorescence enhancement. Dissociation of RecBCD returns the Cy3 fluorescence to the free DNA level (blue). AU, arbitrary units.
RecBCD uses its helicase and nuclease activities to degrade foreign DNA, but upon encountering a crossover hotspot instigator (“CHI”)2 sequence (5′-GCTGGTGG-3′) within the E. coli genome, RecBCD promotes a recombination event by loading RecA onto single-stranded (ss) DNA (1, 12). One current model proposes that before CHI recognition RecD is the faster motor (10, 13), and the nuclease preferentially degrades the 3′-terminated ssDNA but cleaves the 5′-terminated strand infrequently (13, 14). However, after CHI is recognized by the RecC subunit, DNA unwinding becomes slower, and RecB is proposed to act as the lead motor (13, 15), whereas the nuclease preferentially degrades the 5′-terminated ssDNA. RecBCD then loads RecA protein onto the 3′ ssDNA to initiate recombinational DNA repair (12, 14, 16).
It has been suggested that intersubunit communication exists between RecB and RecD (17). However, although it has been shown that the RecD and RecB motors do not act concertedly within RecBCD (10, 13), it is not known whether the translocation activities of the two motors are independent or are functionally coupled. This is because the translocation rates of the individual RecB and RecD motors have not been measured within a functioning RecBCD holoenzyme but have only been inferred from DNA unwinding studies of RecBCD and variants possessing mutations within the RecB and RecD motors by assuming motor independence (10, 16, 18).
Using a fluorescence assay that allows direct monitoring of translocation along each of the single strands, it has recently been shown that RecBC alone (without RecD) possesses two distinct translocase activities that are controlled by the single ATPase motor within RecB (19, 20). The primary translocase enables RecBC to move along ssDNA in the expected 3′ to 5′ direction, consistent with the directionality of RecB on ssDNA, whereas a secondary translocase facilitates translocation along the other DNA strand in the 5′ to 3′ direction, although this secondary translocase is not sensitive to the polarity of the ssDNA backbone (19). As such, RecBC can move along two non-complementary strands of ssDNA at the same rates in a concerted mechanism in which both translocases are tightly coupled to ATP hydrolysis within the RecB motor (20). Using an assay that enables direct measurement of the rates of ssDNA translocation of the RecBCD holoenzyme along each DNA strand in both the 3′ to 5′ and 5′ to 3′ directions, we show that the translocation rates along each DNA strand driven by the RecB and RecD motors are functionally coupled due to the action of the secondary RecBC translocase that operates within RecBCD.
EXPERIMENTAL PROCEDURES
Buffers and Reagents
Buffers were prepared with reagent grade chemicals and double distilled water that was further deionized using a Milli-Q purification system (Millipore Corp., Bedford, MA) and filtered through 0.2-μm filters. Buffer M is 20 mm MOPS-KOH (pH 7.0 at 25 °C), 1 or 10 mm MgCl2 (as indicated), 1 mm 2-mercaptoethanol, 5% (v/v) glycerol (spectrum grade). Buffer T is 25 mm Tris-HCl (pH 7.5 at 20 °C), 1 mm 2-mercaptoethanol, 1 or 10 mm Mg2+ (as indicated). A subscript (e.g. Buffer M250) indicates the concentration of NaCl (250 mm) in the buffer.
Heparin stock solutions were prepared by dissolving heparin sodium salt (Sigma) in buffer M30 and dialyzing extensively against buffer M using 3500 molecular weight-cutoff dialysis tubing. Heparin stock solutions were stored at 4 °C until use, and its concentration was determined by titration with Azure A as described (21). ATP stock solutions were prepared and stored at −20 °C as described (22).
Proteins
Wild type E. coli RecBCD was expressed and purified as a heterotrimer and stored in −80 °C as described (23, 24). The mutants RecBD1080ACD, RecBK29QCD, RecBY803HCD, RecBCDK177Q, and RecBCDY567H were expressed in E. coli strain V2831 (a gift from Dr. Gerald R. Smith) in which the genes encoding wild type RecBCD have been deleted from the E. coli chromosome (17, 25–27). RecBCD and mutant enzymes were purified as described (24, 28) except that the Mono Q column (Amersham Biosciences) was used as the final step to prevent possible RecBC contamination (29). RecB and RecC were purified separately as described previously (24, 28). RecBC was reconstituted by mixing RecB and RecC at equimolar concentrations on ice. RecBC was shown to fully form heterodimers by sedimentation velocity experiments in buffer M. The enzymes were dialyzed against buffer M30 at 4 °C before use, and enzyme concentrations were determined spectrophotometrically using an extinction coefficient of ϵ280 = 3.9 × 105 m−1 cm−1 for the RecBC heterodimer and ϵ280 = 4.5 × 105 m−1 cm−1 for the RecBCD heterotrimer (24, 30).
Oligodeoxynucleotides
Oligodeoxynucleotides, either unlabeled or labeled covalently with Cy3 or fluorescein, were synthesized and purified, and their concentrations were determined as described (22). DNA stocks were stored in buffer M30 in the absence of Mg2+ at −20 °C until use. The sequences of the DNA substrates used are given in supplemental Table S1.
Stopped-flow Fluorescence Experiments
Stopped-flow kinetics experiments were performed in buffer M250 plus 10 mm MgCl2 at 25 °C unless otherwise indicated using an SX.18MV stopped-flow apparatus (Applied Photophysics Ltd., Leatherhead, UK). RecBCD (or RecBC) was preincubated with DNA on ice for 20 min before loading into the stopped-flow syringe. Reactions were initiated by rapid 1:1 mixing of preformed enzyme-DNA complex in one syringe versus ATP and heparin in the other syringe. Final concentrations in the cuvette after mixing were 37.5 nm RecBCD, 50 nm DNA, and 7.5 mg/ml heparin in buffer M250 plus 10 mm MgCl2 and 5 mm ATP unless indicated otherwise. Cy3 fluorescence was excited at 515 nm, and its emission was monitored using a >570 nm-cutoff filter (Oriel Corp., Stratford, CT). Fluorescein fluorescence was excited at 492 nm, and its emission was monitored using a >520 nm-cutoff filter (Oriel Corp.).
Stopped-flow DNA unwinding experiments were performed in buffer M250 plus 10 mm MgCl2 at 25 °C. DNA substrates with various duplex lengths possessing a high affinity binding site (a fork with a 3′-(dT)6 and a 5′-(dT)10 tail) were described previously (22). Final concentrations after mixing were 150 nm WT and mutant RecBCD, 50 nm DNA, and 7.5 mg/ml heparin in buffer M250 plus 10 mm MgCl2 and 5 mm ATP. Cy3 fluorescence was excited at 515 nm, its emission was monitored using a 570 nm interference filter (Oriel Corp.), and Cy5 fluorescence was monitored simultaneously using a >665 nm-cutoff filter (Oriel Corp.).
Analysis of DNA Translocation Time Courses
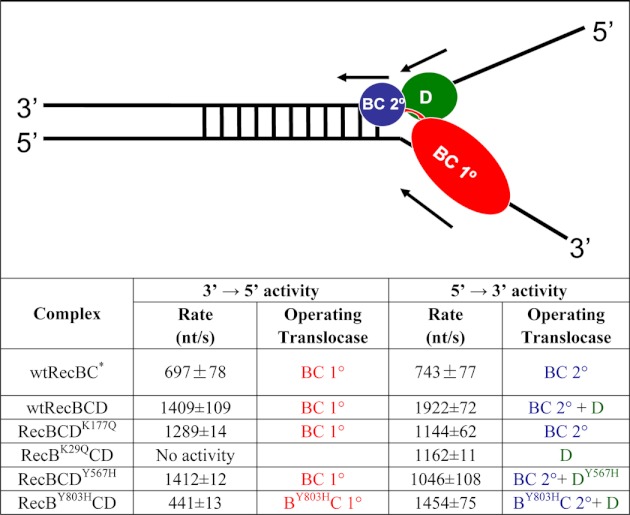
Rates of ssDNA translocation were obtained by performing translocation experiments as a function of ssDNA extension length and determining the inverse of the slope of a plot of lag time versus ssDNA extension length as described (19, 31). We measure the “lag time” or duration of the lag phase as the time at the intersection of the two linear fits of the time course (see Fig. 1B). By performing this experiment with DNA substrates differing in the length of the oligodeoxythymidylate, (dT)L, extension, the translocation rate can be calculated from the inverse of the slope of a plot of lag time versus L. Table 1 lists the average rates ±S.D. based on three or four measurements where S.D. is calculated as √(Σ(x − x̄)2)/(n − 1) where x̄ is the average rate. Any rate reported without uncertainties is based on a single set of measurements. The plot of the macroscopic translocation rate (Vtrans) versus [ATP] was fit to Equation 1 using KaleidaGraph (Synergy Software, Reading, PA) to obtain Km and Vmax.
TABLE 1.
Asymmetric contributions of the three translocase activities to RecBCD translocation along ssDNA
All rates were determined in buffer M250 plus 10 mm Mg2+, 5 mm ATP, and 7.5 mg/ml heparin. DNA I and DNA II were used to determine 3′ to 5′ and 5′ to 3′ rates, respectively. Three translocases, RecBC primary (BC 1°) (in red), RecBC secondary (BC 2°) (in blue), and RecD (D) (in green), are indicated in the schematic.
* Time courses are shown in supplemental Fig. S3.
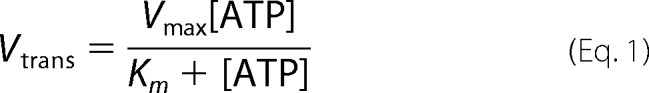 |
Analysis of DNA Unwinding Time Courses
Global non-linear least square analysis of DNA unwinding time courses was performed using Conlin (kindly provided by Dr. Jeremy Williams and modified by Dr. Chris Fischer) and the International Mathematics and Statistics Library C Numerical Libraries (Visual Numeric Inc., Houston, TX) as described previously (22, 24, 28, 32). The entire time courses (up to 2 s) for the production of ssDNA, fss(t), were fit to Scheme 1,
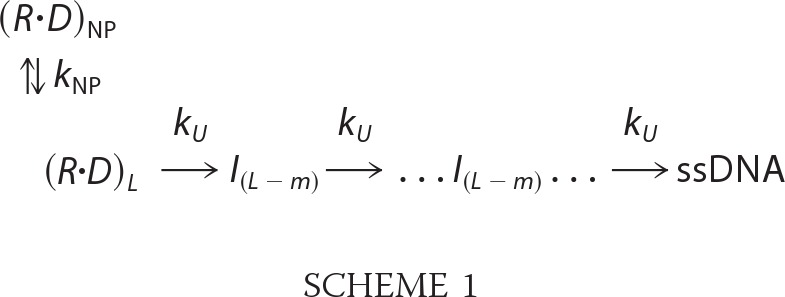 |
using Equation 2 using numerical methods as described previously (22, 24, 33),
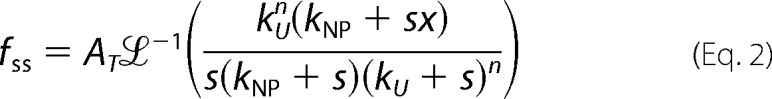 |
where ℒ−1 is the inverse Laplace transform operator with s as the Laplace variable, AT is the unwinding amplitude for a given DNA substrate, n is the number of DNA unwinding steps, kU is the unwinding step rate constant, and kNP is the rate constant for isomerization from a non-productive to a productive helicase-DNA complex. The average kinetic step size m is defined as the average number of unwound base pairs unwound between two consecutive rate-limiting steps with rate constant kU. The kinetic step size for unwinding, m, is obtained as the inverse of the slope of a plot of duplex length versus n.
RESULTS
DNA Substrates and Fluorescence Assay to Monitor ssDNA Translocation of RecBCD
The DNA substrates used (DNA sequences are given in supplemental Table S1) consist of a 24-base pair (bp) duplex possessing a high affinity loading site for RecBCD (a fork with a 3′-(dT)6 and a 5′-(dT)10 tail) or RecBC (a fork with a 3′-(dT)6 and a 5′-(dT)6 tail) (18, 19, 30, 34). At the other end of the duplex, either one or both of the DNA strands possesses a (dT)L extension where L is the nucleotide length. Each DNA is labeled covalently with a Cy3 fluorophore on the terminal end of one (dT)L extension. We refer to the two DNA strands as the 3′-terminated strand or the 5′-terminated strand where the 3′ or 5′ designation refers to the DNA end containing the enzyme loading site. When RecBCD reaches the Cy3 fluorophore, the Cy3 fluorescence increases (19, 22); hence, ssDNA translocation rates can be monitored independently either in the 5′ to 3′ direction or in the 3′ to 5′ direction depending on which (dT)L extension is labeled as described below.
All translocation experiments were performed in 250 mm NaCl (buffer M250 at 25 °C) with DNA (50 nm) in excess over RecBCD (37.5 nm). Under these conditions, RecBCD binds exclusively at the high affinity loading site and upon addition of ATP initiates unwinding of the 24-bp duplex (19) followed by ssDNA translocation along the (dT)L extension (supplemental Fig. S1 and supplemental Discussion). An excess of heparin (7.5 mg/ml after mixing) added with the ATP serves to trap any free RecBCD, thus ensuring a single round of translocation (i.e. no rebinding of dissociated enzyme). Fig. 1B shows a stopped-flow fluorescence time course from an experiment with DNA substrate (L = 51 nucleotides) showing the expected “lag phase” reflecting the average time for RecBCD to translocate (3′ to 5′) to the Cy3 label followed by a rapid increase in Cy3 fluorescence when the population of RecBCD reaches the Cy3 fluorophore. The final slow decrease in Cy3 fluorescence reflects RecBCD dissociation from the DNA.
We measure the lag time or duration of the lag phase as the time at the intersection of the two linear fits of the time course (Fig. 1B). By performing this experiment with a set of DNA substrates differing in ssDNA (i.e. (dT)L) extension length but with a constant short duplex length (24 bp), the average rate of translocation along the (dT)L extension can be calculated from the inverse of the slope of a plot of lag time versus L (19, 31). The amplitude of the fluorescence peak is dependent upon the fraction of DNA labeled with Cy3 as well as the processivity of the translocase. The Cy3 labeling of the DNA can vary from 75 to 98%. These differences have no effect on the lag times or the calculated rates of translocation (supplemental Fig. S2); however, they do preclude accurate estimates of translocation processivity.
5′ to 3′ ssDNA Translocation of RecBCD Is Faster than 3′ to 5′ Translocation
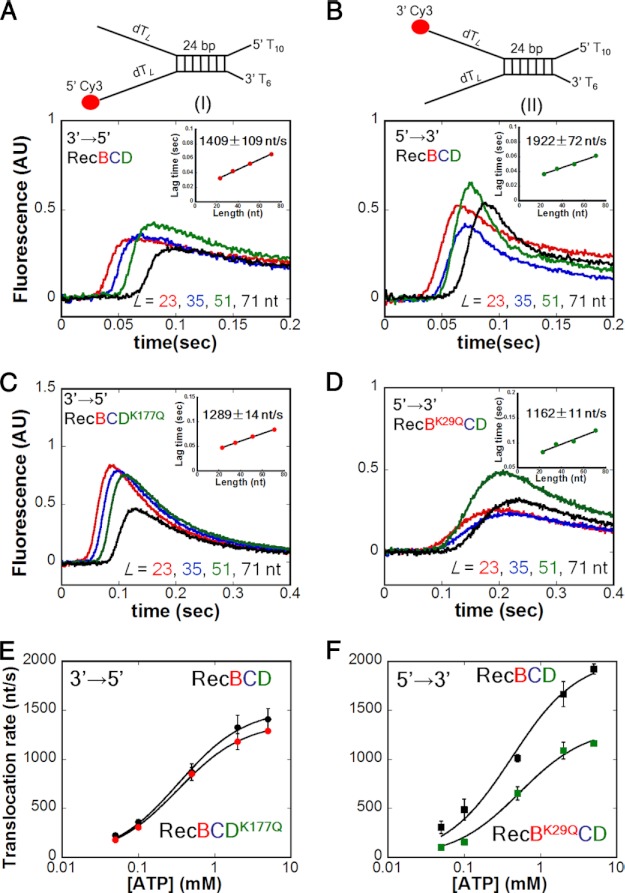
We first used the double (dT)L extension substrates I and II (Fig. 2) with L = 23, 35, 51, and 71 nucleotides to examine RecBCD translocation along ssDNA at saturating [ATP] (5 mm) and 10 mm Mg2+. The resulting RecBCD time courses independently monitoring ssDNA translocation of RecBCD in the 3′ to 5′ and 5′ to 3′ directions are shown in Fig. 2, A and B, respectively. For both substrates, the lag time increases linearly with increasing ssDNA length L (Fig. 2, A and B, insets) from which we calculate the average rate of ssDNA translocation (Table 1). Under these conditions, the 5′ to 3′ rate (1922 ± 72 nt/s) is faster than the 3′ to 5′ rate (1409 ± 109 nt/s). Experiments comparing RecBC with RecBCD under these same conditions (buffer M250) with the same DNA substrate (III) indicate that the 3′ to 5′ translocation rate of RecBCD (1627 ± 103 nt/s) is faster than that of RecBC (909 ± 51 nt/s) (19), and both are faster than the 3′ to 5′ translocation rate of the RecB motor alone (∼800 nt/s) (19) (supplemental Table S2 and supplemental Fig. S4A). Hence, the 3′ to 5′ translocation rate of RecBC is enhanced upon forming a complex with RecD. The 5′ to 3′ rate is also faster than the 3′ to 5′ rate with the single (dT)L extension substrates (III and IV) under the same conditions, and the same rates are observed for RecBD1080ACD (a nuclease-deficient mutant) (supplemental Table S3). Experiments performed with the double (dT)L extension substrates (I and II) under other conditions (ATP (5 mm) in excess over [Mg2+] (1 mm) or in buffer T (supplemental Table S4)) also show the 5′ to 3′ rate to be faster than the 3′ to 5′ rate.
FIGURE 2.
The rate of RecBCD translocation along ssDNA in the 5′ to 3′ direction is faster than in the 3′ to 5′ direction, and the rates are coupled asymmetrically. Stopped-flow experiments were performed by mixing a preformed complex of RecBCD (37.5 nm) and DNA (50 nm) with 5 mm ATP, 10 mm MgCl2, and 7.5 mg/ml heparin (concentrations after mixing) in buffer M250 at 25 °C. The lengths of the twin ssDNA extensions, L, are indicated. All insets show the linear dependence of lag time on ssDNA extension length L. A, monitoring 3′ to 5′ ssDNA translocation of RecBCD using DNA I. Inset, rate = 1409 ± 109 nt/s. B, monitoring 5′ to 3′ ssDNA translocation of RecBCD using DNA II. Inset, rate = 1922 ± 72 nt/s. C, monitoring 3′ to 5′ ssDNA translocation of RecBCDK177Q using DNA I. Inset, rate = 1289 ± 14 nt/s. D, monitoring 5′ to 3′ ssDNA translocation of RecBK29QCD using DNA II. Inset, rate = 1162 ± 11 nt/s. E, [ATP] dependence of the 3′ to 5′ translocation rates of RecBCDK177Q (red) and WT RecBCD (black). For WT RecBCD, Km = 337 ± 30 μm and Vmax = 1515 ± 33 nt/s; for RecBCDK177Q, Km = 327 ± 14 μm and Vmax = 1377 ± 14 nt/s. F, [ATP] dependence of the 5′ to 3′ translocation rates of RecBK29QCD (green) and WT RecBCD (black). For WT RecBCD, Km = 420 ± 89 μm and Vmax = 2037 ± 109 nt/s; for RecBK29QCD, Km = 534 ± 84 μm and Vmax = 1325 ± 56 nt/s. AU, arbitrary units.
The 3′ to 5′ and 5′ to 3′ Translocation Rates Are Coupled Asymmetrically
We next examined whether the 3′ to 5′ and 5′ to 3′ ssDNA translocase activities within RecBCD are independent or show coupling. We examined the behavior of two well studied mutants (25, 26), RecBK29QCD and RecBCDK177Q, in which the conserved Lys within the Walker A sequence (helicase motif I) is mutated to Gln, thus eliminating the ATPase activity of the mutated motor. Hence, only the RecD ATPase is active in RecBK29QCD, and only the RecB ATPase is active in RecBCDK177Q. As expected, no 3′ to 5′ translocase activity was detected for RecBK29QCD, and initiation of DNA unwinding and translocation by RecBK29QCD requires the 5′-(dT) tail of the enzyme loading site to be at least 5′-(dT)10 (supplemental Fig. S5), consistent with the need for a 5′-ssDNA tail to reach the RecD subunit (18, 30, 34).
The 3′ to 5′ translocase activity of RecBCDK177Q (Fig. 2C) and the 5′ to 3′ translocase activity of RecBK29QCD (Fig. 2D) were examined at saturating ATP (5 mm). Loss of the RecD ATPase activity has only a minor effect (if any) on the 3′ to 5′ translocase rate (1289 ± 14 nt/s for RecBCDK177Q versus 1409 ± 109 nt/s for WT RecBCD in Table 1). In contrast, loss of the RecB ATPase activity causes a nearly 40% reduction in the 5′ to 3′ translocase rate (1162 ± 11 nt/s for RecBK29QCD versus 1922 ± 72 nt/s for WT RecBCD in Table 1). This asymmetry indicates that the RecB motor influences not only the 3′ to 5′ translocation rate as expected because RecB is a 3′ to 5′ translocase (19) but also the 5′ to 3′ translocation rate. However, the RecD motor only affects the 5′ to 3′ translocation rate. These conclusions are supported by experiments performed on the single extension DNA substrates III and IV (supplemental Figs. S5 and S6 and supplemental Table S3).
We next examined translocation as a function of ATP concentration at constant [Mg2+] (10 mm). Fig. 2E shows little effect of knocking out the RecD ATPase (RecBCDK177Q) on the 3′ to 5′ translocation rate. In contrast, deactivating the RecB ATPase (RecBK29QCD) reduces the 5′ to 3′ translocation rate at all [ATP] (Fig. 2F). Therefore, the asymmetric coupling of the two translocation rates driven by the RecB and RecD motors is maintained at all [ATP]. Furthermore, deactivating the ATPase of one motor does not affect the Km for ATP of the other motor. Therefore, the reduction in the 5′ to 3′ translocase rate upon inactivating the RecB ATPase (RecBK29QCD) does not appear to result from an effect on the ATPase activity of the RecD motor.
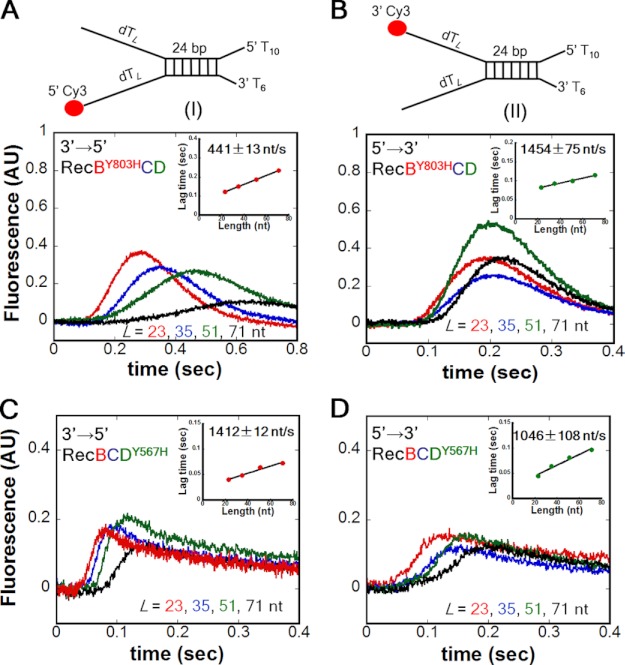
To further investigate the coupling of the two translocation rates, we examined two additional mutants, RecBY803HCD and RecBCDY567H. These mutations are in equivalent positions within helicase motif VI of the RecB and RecD motors (17, 35). Previous studies showed that the DNA unwinding rate of RecBY803HCD is slowed significantly (17), and this mutation also slows the rates of both the primary and secondary translocases within RecBY803HC (20). As anticipated, the translocation rate of RecBY803HCD in the 3′ to 5′ direction (Fig. 3A) is reduced to 441 ± 13 from 1409 ± 109 nt/s for WT RecBCD. In addition, the translocation rate of RecBY803HCD in the 5′ to 3′ direction (Fig. 3B) is also reduced to 1454 ± 75 from 1922 ± 72 nt/s for WT RecBCD (Table 1). Therefore, this single mutation within RecB shows reductions in both the 3′ to 5′ and 5′ to 3′ translocation rates, providing further evidence that the RecB motor regulates both translocation rates.
FIGURE 3.
Slowing the RecB motor (RecBY803H) slows both the 3′ to 5′ and 5′ to 3′ translocation rates, whereas slowing the RecD motor (RecDY567H) slows only the 5′ to 3′ translocation rate. A, monitoring 3′ to 5′ ssDNA translocation of RecBY803HCD using DNA I. Inset, rate = 441 ± 13 nt/s. B, monitoring 5′ to 3′ ssDNA translocation of RecBY803HCD using DNA II. Inset, rate = 1454 ± 75 nt/s. C, monitoring 3′ to 5′ ssDNA translocation of RecBCDY567H using DNA I. Inset, rate = 1412 ± 12 nt/s. D, monitoring 5′ to 3′ ssDNA translocation of RecBCDY567H using DNA II. Inset, rate = 1046 ± 108 nt/s. AU, arbitrary units.
We next examined the effect of the equivalent mutation (Y567H) within RecD. As anticipated, the 5′ to 3′ translocation rate of RecBCDY567H (Fig. 3C) is reduced to 1046 ± 12 from 1922 ± 72 nt/s for WT RecBCD. However, the 3′ to 5′ translocation rate of RecBCDY567H (Fig. 3D) is unchanged (1412 ± 12 nt/s) relative to WT RecBCD (1409 ± 109 nt/s). Hence, reducing the rate of RecD translocation only reduces the 5′ to 3′ translocation rate (Table 1). These effects were also observed using the single extension DNA substrates (supplemental Table S3). These experiments show asymmetric coupling within RecBCD such that the 3′ to 5′ translocation rate depends solely on the RecB motor, whereas the 5′ to 3′ translocation activity is controlled by both the RecB and RecD motors.
The Secondary Translocase Activity of RecBC Is Functional within RecBCDK177Q and RecBCD
Although the 5′ to 3′ translocation activity of RecBCD depends on both the RecB and RecD motors, this coupling does not appear to be due to communication between their ATP binding sites. Recently, Wu et al. (19) have shown that RecBC has both a primary (3′ to 5′) translocase activity and a secondary translocase activity that can operate in the 5′ to 3′ direction along the other DNA strand, although it is not sensitive to the ssDNA polarity. Hence, a possible explanation for the asymmetric coupling of the translocase activities driven by the RecB and RecD motors is that the secondary translocase activity of RecBC is functional within RecBCD so that the 5′ to 3′ translocation rate is controlled by both RecD and the secondary RecBC translocase.
To determine whether the secondary RecBC translocase is functional within RecBCD, we examined whether any 5′ to 3′ translocation activity is observed for RecBCDK177Q. Because RecDK177Q has no ATPase activity, any 5′ to 3′ translocation activity observed for RecBCDK177Q could result only from a functional secondary RecBC translocase activity. Using the single extension DNA substrate IV, the results in Fig. 4A show that RecBCDK177Q can undergo 5′ to 3′ translocation (1393 ± 10 nt/s), indicating that the secondary RecBC translocase can function when the RecD translocase is inactivated. The rate of 5′ to 3′ translocation using the double extension DNA substrate II is 1144 ± 62 nt/s (Table 1 and supplemental Fig. S7), similar to the 3′ to 5′ rate (1289 ± 14 nt/s) (Table 1 and Fig. 2C). For comparison, RecBC shows the same rates in both directions (Table 1 and supplemental Fig. S3).
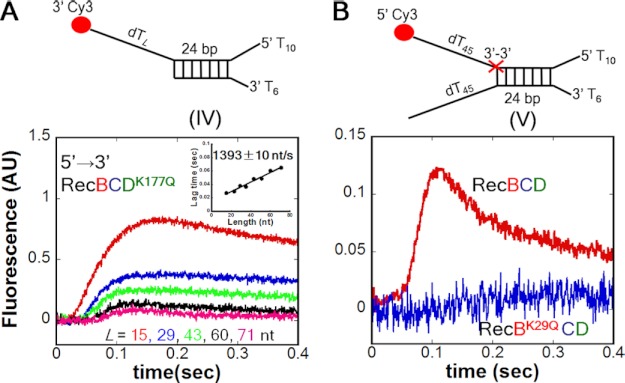
FIGURE 4.
The secondary translocase activity of RecBC is functional within RecBCDK177Q and WT RecBCD. A, time courses monitoring the secondary RecBC (5′ to 3′) ssDNA translocation of RecBCDK177Q using DNA substrate IV. Inset, rate = 1393 ± 10 nt/s. B, time course monitoring the secondary RecBC translocase within WT RecBCD (red trace) on DNA substrate V. The Cy3-labeled ssDNA extension (dT)45 contains a reverse polarity (3′ to 3′) linkage (red X). No translocation is observed for RecBK29QCD (blue trace) on the same DNA. AU, arbitrary units.
To examine whether the secondary RecBC translocase is functional within WT RecBCD, we used DNA substrate V (Fig. 4B). This substrate contains a 3′-3′-phosphodiester linkage in the top strand (red X) after the 24-bp duplex that reverses the DNA backbone polarity and thus should block the 5′ to 3′ translocation activity of the RecD motor at that point. However, such a reverse polarity linkage does not block the RecBC secondary translocase activity; hence, any Cy3 signal observed using this DNA substrate could only result if the secondary RecBC translocase activity functions within RecBCD. Fig. 4B (red curve) shows 5′ to 3′ translocase activity of RecBCD on this substrate. In contrast, RecBK29QCD shows no 5′ to 3′ translocase activity on this same substrate (Fig. 4B, blue curve), indicating that the RecD motor cannot bypass the reverse polarity linkage. Hence, the 5′ to 3′ translocation activity observed for WT RecBCD on this DNA requires an active RecB ATPase, consistent with translocation being due to the secondary translocase activity within RecBC.
The RecB and RecD Slow Translocation Mutants Also Reduce DNA Unwinding Rates
RecBCD is a dual motor helicase, and it has been shown that inactivating either motor by mutating the ATPase site (RecBK29Q or RecDK177Q) decreases the observed rate of duplex DNA unwinding (16, 18). Our current results as well as a previous study (20) show that a mutation within RecD (Y567H) that slows its 5′ to 3′ ssDNA translocation rate does not affect the 3′ to 5′ translocation rate of RecB, whereas a mutation within RecB (Y803H) that slows its primary 3′ to 5′ translocation rate also slows the secondary 5′ to 3′ ssDNA translocation rate of RecBC. We therefore examined how these mutations affect the DNA unwinding rates of RecBCD.
We examined the DNA unwinding kinetics using a stopped-flow fluorescence assay under single round DNA unwinding conditions (22, 24, 28, 36) under the same solution conditions used in the ssDNA translocation studies. The time courses were analyzed to obtain average macroscopic rates of DNA unwinding as well as a kinetic unwinding step size using a uniform n-step sequential kinetic model (Scheme 1 and Equation 2 under “Experimental Procedures”) as described (22, 24, 28, 36). DNA unwinding was performed using a series of DNA substrates with varying duplex lengths with each possessing a high affinity RecBCD binding site (a fork with a 3′-(dT)6 and a 5′-(dT)10 tail) on one end. The time courses are shown in supplemental Fig. S8, and the best fit kinetic parameters are summarized in Table 2.
TABLE 2.
DNA unwinding rates and kinetic parameters for WT and mutant RecBCD
All parameters except RecBC were determined in buffer M250 plus 10 mm Mg2+, 5 mm ATP, and 7.5 mg/ml heparin. Reaction time courses were fit to Scheme 1 using non-linear least square methods as described previously (22). DNA unwinding rates were calculated by kU timing step size m. Standard deviations are calculated based on three or four observations.
| Enzyme | DNA unwinding rate | Step size m | kU | kNP |
|---|---|---|---|---|
| bp/s | bp | s−1 | s−1 | |
| RecBCa | 320 ± 7 | 3.5 ± 0.1 | 92 ± 12 | 1.9 ± 0.6 |
| RecBCD | 539 ± 27 | 3.1 ± 0.7 | 175 ± 40 | 4.0 ± 1.7 |
| RecBK29QCD | 161 ± 36 | 3.2 ± 0.8 | 51 ± 12 | 2.2 ± 0.6 |
| RecBY803HCD | 212 ± 36 | 5.3 ± 1.7 | 43 ± 8 | 2.3 ± 1.4 |
| RecBCDK177Q | 391 ± 34 | 4.3 ± 0.5 | 90 ± 4 | 4.7 ± 1.6 |
| RecBCDY567H | 452 ± 31 | 4.5 ± 1.3 | 106 ± 26 | 2.2 ± 1.0 |
a The DNA unwinding rate of RecBC was determined in buffer M30 plus 10 mm Mg2+ and 5 mm ATP (22).
We first examined WT RecBCD as well as the RecBK29QCD and RecBCDK177Q mutants that knock out the respective ATPase activities of the individual motors. In agreement with previous reports at high Mg2+ concentration (16, 18), we observe that RecBCDK177Q unwinds faster (391 ± 34 bp/s) than does RecBK29QCD (161 ± 36 bp/s), and both are slower than WT RecBCD (539 ± 27 bp/s). Although mutants in RecD (RecBCDK177Q and RecBCDY567H) only slow down 5′ to 3′ ssDNA translocation without affecting 3′ to 5′ translocase activity, DNA unwinding rates are decreased. The RecBY803H and RecDY567H mutations that slow the ssDNA translocation rates also slow the DNA unwinding rate. RecBY803HCD unwinds with a rate (212 ± 36 bp/s) that is intermediate between those of WT RecBCD and RecBK29QCD but closer to that of RecBK29QCD, whereas RecBCDY567H unwinds with a rate (452 ± 31 bp/s) that is also intermediate between those of WT RecBCD and RecBCDK177Q but closer to that of WT RecBCD (Table 2). Hence, both of the slow motor mutants also slow DNA unwinding, although slowing the RecB motor has a greater impact than slowing the RecD motor. Therefore, both motors facilitate DNA unwinding, and the DNA unwinding rate of the complex is not solely determined by either motor.
DNA unwinding kinetic step sizes ranging from 3.1 ± 0.7 to 5.3 ± 1.7 bp were found for all of the RecBCD mutants (Table 2), and these are in the same range as observed previously for RecBCD and RecBC (22, 37, 38). This suggests that the mutant RecBCD enzymes use the same kinetic mechanism for DNA unwinding regardless of the speed of the individual motors and that the decreased unwinding rate is due to the decreased ssDNA translocation rate.
DISCUSSION
The Bipolar Translocation Rates within RecBCD Are Coupled Asymmetrically Due to the Presence of Three Translocase Activities
The methods reported here enabled us to monitor directly the rates of ssDNA translocation by RecBCD in both the 3′ to 5′ and 5′ to 3′ directions. We infer the following conclusions from the results of our studies, summarized in Table 1. First, under all conditions examined, including a range of ATP and Mg2+ concentrations (supplemental Table S4), the 5′ to 3′ translocase rate is always faster than the 3′ to 5′ translocase rate on ssDNA without a CHI sequence. This agrees with previous DNA unwinding studies showing that an ssDNA loop forms ahead of RecBCD in the 3′-terminated strand along which the primary RecB translocase operates (10, 13). Second, the secondary translocase activity previously identified within RecBC (19) also functions within RecBCD. Hence, RecBCD possesses three translocase activities: the primary RecBC (3′ to 5′) translocase, the RecD (5′ to 3′) translocase, and the recently identified secondary RecBC translocase that normally operates 5′ to 3′ and is driven by the RecB motor but is insensitive to ssDNA polarity (19). Third, the 3′ to 5′ and 5′ to 3′ translocase activities within the RecBCD holoenzyme are coupled asymmetrically. That is, the 3′ to 5′ translocation rate is regulated only by the RecB motor, whereas the 5′ to 3′ translocation rate is regulated by both the RecD and RecB motors due to the presence of the secondary RecBC translocase activity. If the RecD translocase activity is eliminated as in RecBCDK177Q, then the 3′ to 5′ and 5′ to 3′ translocation rates become equal because both rates are tightly controlled by the single RecB ATPase motor as shown for RecBC (19, 20). Separately, neither the RecD nor the secondary RecBC translocation rate (5′ to 3′) appears to be faster than the primary RecBC translocation rate (3′ to 5′) (Table 1). Therefore, before CHI, the 5′ to 3′ translocation rate of RecBCD may be faster simply due to the fact that two translocase activities (RecD and secondary RecBC) operate in the 5′ to 3′ direction, whereas only one translocase activity (primary RecBC) operates in the 3′ to 5′ direction. How the two translocase activities (RecD and secondary RecBC) coordinate to move at a rate (1922 ± 72 nt/s) that is faster than the rates of the individual translocases (1144 ± 62 and 1162 ± 11 nt/s) remains to be determined.
As discussed (19), the likely candidates are either the “arm” of RecB that interacts with the duplex region of DNA in the crystal structure (34) and/or the “dead nuclease domain” within RecC that interacts with the 5′-ssDNA tail that eventually threads into the RecD motor (Fig. 1A). The fact that the secondary RecBC ssDNA translocase activity is insensitive to the ssDNA backbone polarity (19) differentiates it from the canonical primary translocase activities of RecB and RecD. This suggests that the secondary translocase activity may normally operate during DNA unwinding as a dsDNA translocase ahead of the fork through the RecB arm.
Relationship between ssDNA Translocase Activity and Helicase Activity
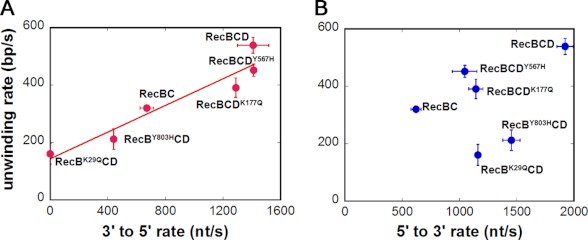
Based on the experiments reported here, we can compare the DNA unwinding rates of RecBCD, RecBC, and the various mutants with both the 3′ to 5′ and 5′ to 3′ ssDNA translocation rates, and these are shown in Fig. 5. Fig. 5A suggests that the DNA unwinding rate correlates with the 3′ to 5′ ssDNA translocation rate, which represents both the primary and secondary RecBC ssDNA translocation rates because these are equal. Fig. 5B shows no correlation between the 5′ to 3′ ssDNA translocation rate, which represents contributions from both the secondary RecBC and RecD translocase activities, and the DNA unwinding rate. This suggests that the DNA unwinding rate under these conditions is influenced by the primary and secondary RecBC translocase rates and not by the RecD translocase rate. The fact that the RecBK29QCD rate falls on the line in Fig. 5A even though only RecD can contribute to translocation may be fortuitous.
FIGURE 5.
Comparison between ssDNA translocase activities and helicase activity within RecBCD. A, 3′ to 5′ translocase activity is correlated with the helicase activity. The linear fit is introduced to show the correlation. B, 5′ to 3′ translocase activity is not correlated with the helicase activity. Error bars represent S.D.
Previous ensemble studies (18) have attempted to infer information about the relative rates of the RecB and RecD motors by comparing DNA unwinding rates of WT RecBCD with the same mutants that we have studied here containing individual motor knockouts, RecBK29QCD and RecBCDK177Q. Those studies (16, 18) showed and our own experiments (Table 2) confirm that RecBK29QCD unwinds DNA more slowly (161 ± 36 bp/s) than RecBCDK177Q (391 ± 34 bp/s) (when [Mg2+] > [ATP]) from which it was inferred that the RecB motor is faster than the RecD motor (18). However, when [ATP] > [Mg2+], it was observed that RecBCDK177Q unwinds DNA slightly more slowly than RecBK29QCD, although both are much slower than WT RecBCD (16). Hence, it was also inferred from those studies that the relative rates of the two motors could be reversed by changing the ratio of ATP to Mg2+ in the buffer (16). In the experiments reported here that monitor ssDNA translocation rates directly rather than DNA unwinding rates, we observe that the 5′ to 3′ rate is always faster than the 3′ to 5′ rate in WT RecBCD regardless of the ATP to Mg2+ ratio (supplemental Table S4). The conclusions concerning the relative rates of the two motors drawn from the DNA unwinding studies were based on the assumption that the translocation rates of RecB and RecD are independent. However, we show here that the translocation rates are not independent but are coupled. The main reason that the DNA unwinding rate is reduced more when the RecB ATPase is inactivated is likely that RecB motor inactivation knocks out both the primary and secondary translocase activities, whereas inactivating the RecD ATPase only affects the 5′ to 3′ translocation rate. Thus, the fact that the 5′ to 3′ translocation rate is regulated by both RecB and RecD makes it impossible to infer rates for the two motors from DNA unwinding rates alone.
The observation that DNA unwinding activity is maintained even in mutants that knock out the ATPase activities of either of the individual motors (i.e. RecBK29QCD and RecBCDK177Q) has also been interpreted as an indication that either motor (RecB or RecD) can function as the helicase. However, DNA unwinding by RecBCD requires more than just the ability to translocate along ssDNA, and it is possible (likely) that DNA unwinding (bp separation) and translocation are separate processes (20). It is possible that base pair separation occurs in multiple (4–6) base pair steps via the ATP-independent melting reaction that has been observed for initiation and that uses only the free energy from RecBCD (37) or RecBC (38) binding to the DNA end. If this mechanism applies, the ATPase activities of the motors would be used only for directional translocation along the ssDNA that is formed by the protein-induced DNA melting reaction. In such a mechanism, DNA melting is facilitated by a part of RecBC that is independent of its ATPase activities and thus remains functional regardless of whether one motor is knocked out. As such, the only role of the ATPase activity is to effect ssDNA translocation to move the enzyme to the new ss/dsDNA junction at which point the enzyme is reset so that the binding free energy can be used again to melt out another 4–6-bp region of the DNA. Hence, it is not likely that the helicase activity switches between motors when one motor is knocked out but rather that a region of RecBC distinct from its ATPase provides the DNA melting activity and that the RecB and RecD ATPase motors only provide translocase activity, and this can be accomplished (albeit at different rates) by either only one or both motors.
How Might CHI Recognition Affect the Three Translocase Activities within RecBCD?
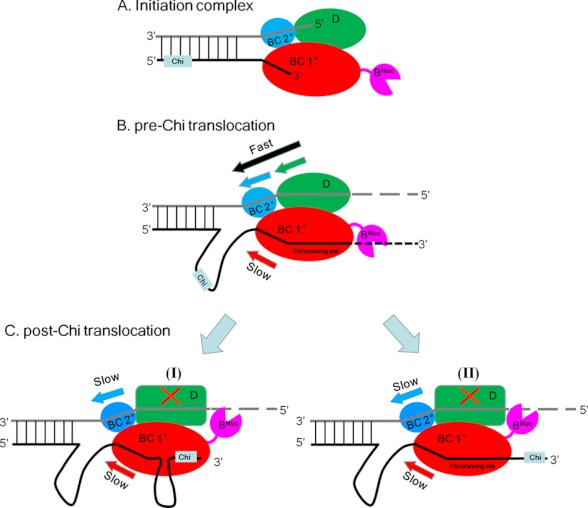
The finding that RecBCD possesses three translocase activities on non-CHI-containing DNA substrates raises the question of how these might be affected after RecBCD recognizes a CHI site. One current view is that, before CHI, RecD is the faster motor and thus operates as the helicase, whereas after CHI, recognition by RecB becomes the faster motor and thus serves as the helicase (13). However, the results reported here suggest that the faster 5′ to 3′ translocation rate of RecBCD before CHI is due to the activity of two translocases (RecD plus secondary RecBC) working 5′ to 3′, whereas only one translocase (primary RecBC) works 3′ to 5′ (Fig. 6).
FIGURE 6.
Schematic of RecBCD-DNA complexes pre- and post-CHI. A, RecBCD-DNA initiation complex. RecD (green) and the secondary RecBC translocation site (blue) operate on the 5′-ssDNA tail, and the primary RecBC translocase site (red) operates on the 3′-ssDNA tail. B, pre-CHI DNA unwinding by RecBCD. The 5′ to 3′ rate is faster due to the action of two translocases (RecD (green) and the secondary RecBC (blue)) versus only one translocase (primary RecBC) acting in the 3′ to 5′ direction, resulting in a loop in the 3′-terminated strand ahead of the enzyme. C, post-CHI DNA unwinding by RecBCD. Upon deactivation of RecD, the 3′ to 5′ and 5′ to 3′ translocation rates are equal due to the concerted action of the RecBC secondary (5′ to 3′) (blue) and primary (3′ to 5′) (red) translocases, resulting in maintenance of the loop formed in the 3′-terminated strand ahead of the enzyme. Two alternative pathways (I versus II) result depending on whether CHI remains bound to RecC (I) wherein a second loop would form at the interface between RecB and RecC as proposed (13, 41). Alternatively, if the 3′-tail does not remain bound to RecC (II), then the 3′-ssDNA will pass through RecC, and a second loop would not form.
Post-CHI RecBCD unwinds DNA at half its pre-CHI rate (13), similar to the rate of RecBCDK177Q. As suggested previously (14, 39), we also propose that post-CHI the RecD motor is inactivated and thus no longer contributes its translocase activity to RecBCD, resulting in a decrease in unwinding rate. However, our results (19, 20) suggest that the primary and secondary RecBC translocase activities will both remain operational because they are powered by the RecB ATPase and will continue to move at the same rates post-CHI; hence, both unwound DNA strands will be fed through RecBCD at the same rate. As a result, the ssDNA loop in the 3′-tailed strand that was formed ahead of the fork due to the faster pre-CHI 5′ to 3′ translocase activity (Fig. 6B) would remain in place (Fig. 6C). This ssDNA loop could serve as the site for loading RecA protein. As shown in Fig. 6C, two alternative pathways could ensue depending on whether the CHI site remains bound to RecC. If the CHI site remains bound to RecC as has been proposed (13), a second ssDNA loop would then be formed at the interface between RecB and RecC (Fig. 6C, I), and this could serve as the loading site for RecA protein (13). Alternatively, if the 3′ tail does not remain bound to RecC, then the 3′-ssDNA tail would pass through RecC, and a second loop would not form (Fig. 6C, II).
The 3′ to 5′ Translocation Rate of RecB Is Enhanced by Both RecC and RecD Binding
The experiments reported here and in previous studies (19) allow us to compare the primary 3′ to 5′ ssDNA translocation rates of RecB, RecBC, RecBCDK177Q, and RecBCD (supplemental Table S2). Formation of a RecBC complex increases the 3′ to 5′ translocase rate of RecB slightly from ∼800 to ∼1000 nt/s. However, a more substantial increase in the 3′ to 5′ rate to ∼1600 nt/s occurs upon interaction of RecBC with the RecD motor. In fact, even RecDK177Q with an inactive ATPase enhances the 3′ to 5′ translocation rate of RecBC to ∼1400 nt/s. Based on the crystal structures of RecBCD (34, 40), there does not appear to be any direct contact between RecD and RecB, although not all of the residues are observable within RecD. Therefore, the observation that RecD binding increases the rate of RecB translocation appears to result from a long range communication through the RecC subunit. This allosteric effect may occur via the same pathway but in reverse when RecC recognizes a CHI sequence, resulting in inhibition or deactivation of the translocase activity of RecD.
Supplementary Material
Acknowledgments
We thank R. Galletto, T. Ha, P. de Haseth, R. Pappu, A. Lucius, and members of the Lohman laboratory for discussions; T. Ho for the DNA substrates; and J. Brettmann and J. Rammohan for preliminary studies.
This work was supported, in whole or in part, by National Institutes of Health Grant GM045948 (to T. M. L.).

This article contains supplemental Figs. S1–S8, Tables S1–S4, and Discussion.
- CHI
- crossover hotspot instigator
- ss
- single-stranded
- nt
- nucleotides.
REFERENCES
- 1. Anderson D. G., Kowalczykowski S. C. (1997) The translocating RecBCD enzyme stimulates recombination by directing RecA protein onto ssDNA in a χ-regulated manner. Cell 90, 77–86 [DOI] [PubMed] [Google Scholar]
- 2. Kowalczykowski S. C., Dixon D. A., Eggleston A. K., Lauder S. D., Rehrauer W. M. (1994) Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev. 58, 401–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dillingham M. S., Kowalczykowski S. C. (2008) RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol. Mol. Biol. Rev. 72, 642–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Amundsen S. K., Neiman A. M., Thibodeaux S. M., Smith G. R. (1990) Genetic dissection of the biochemical activities of RecBCD enzyme. Genetics 126, 25–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Smith G. R. (1989) Homologous recombination in prokaryotes: enzymes and controlling sites. Genome 31, 520–527 [DOI] [PubMed] [Google Scholar]
- 6. Smith G. R. (2012) How RecBCD enzyme and Chi promote DNA break repair and recombination: a molecular biologist's view. Microbiol. Mol. Biol. Rev. 76, 217–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dillingham M. S., Spies M., Kowalczykowski S. C. (2003) RecBCD enzyme is a bipolar DNA helicase. Nature 423, 893–897 [DOI] [PubMed] [Google Scholar]
- 8. Finch P. W., Storey A., Brown K., Hickson I. D., Emmerson P. T. (1986) Complete nucleotide sequence of recD, the structural gene for the α subunit of exonuclease V of Escherichia coli. Nucleic Acids Res. 14, 8583–8594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Finch P. W., Storey A., Chapman K. E., Brown K., Hickson I. D., Emmerson P. T. (1986) Complete nucleotide sequence of the Escherichia coli recB gene. Nucleic Acids Res. 14, 8573–8582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Taylor A. F., Smith G. R. (2003) RecBCD enzyme is a DNA helicase with fast and slow motors of opposite polarity. Nature 423, 889–893 [DOI] [PubMed] [Google Scholar]
- 11. Rigden D. J. (2005) An inactivated nuclease-like domain in RecC with novel function: implications for evolution. BMC Struct. Biol. 5, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Arnold D. A., Kowalczykowski S. C. (2000) Facilitated loading of RecA protein is essential to recombination by RecBCD enzyme. J. Biol. Chem. 275, 12261–12265 [DOI] [PubMed] [Google Scholar]
- 13. Spies M., Amitani I., Baskin R. J., Kowalczykowski S. C. (2007) RecBCD enzyme switches lead motor subunits in response to χ recognition. Cell 131, 694–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Spies M., Bianco P. R., Dillingham M. S., Handa N., Baskin R. J., Kowalczykowski S. C. (2003) A molecular throttle: the recombination hotspot χ controls DNA translocation by the RecBCD helicase. Cell 114, 647–654 [DOI] [PubMed] [Google Scholar]
- 15. Handa N., Bianco P. R., Baskin R. J., Kowalczykowski S. C. (2005) Direct visualization of RecBCD movement reveals cotranslocation of the RecD motor after χ recognition. Mol. Cell 17, 745–750 [DOI] [PubMed] [Google Scholar]
- 16. Spies M., Dillingham M. S., Kowalczykowski S. C. (2005) Translocation by the RecB motor is an absolute requirement for χ-recognition and RecA protein loading by RecBCD enzyme. J. Biol. Chem. 280, 37078–37087 [DOI] [PubMed] [Google Scholar]
- 17. Amundsen S. K., Taylor A. F., Reddy M., Smith G. R. (2007) Intersubunit signaling in RecBCD enzyme, a complex protein machine regulated by Chi hot spots. Genes Dev. 21, 3296–3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dillingham M. S., Webb M. R., Kowalczykowski S. C. (2005) Bipolar DNA translocation contributes to highly processive DNA unwinding by RecBCD enzyme. J. Biol. Chem. 280, 37069–37077 [DOI] [PubMed] [Google Scholar]
- 19. Wu C. G., Bradford C., Lohman T. M. (2010) Escherichia coli RecBC helicase has two translocase activities controlled by a single ATPase motor. Nat. Struct. Mol. Biol. 17, 1210–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wu C. G., Xie F., Lohman T. M. (2012) The primary and secondary translocase activities within E. coli RecBC helicase are tightly coupled to ATP hydrolysis by the RecB motor. J. Mol. Biol. 423, 303–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mascotti D. P., Lohman T. M. (1995) Thermodynamics of charged oligopeptide-heparin interactions. Biochemistry 34, 2908–2915 [DOI] [PubMed] [Google Scholar]
- 22. Wu C. G., Lohman T. M. (2008) Influence of DNA end structure on the mechanism of initiation of DNA unwinding by the Escherichia coli RecBCD and RecBC helicases. J. Mol. Biol. 382, 312–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Taylor A. F., Smith G. R. (1995) Monomeric RecBCD enzyme binds and unwinds DNA. J. Biol. Chem. 270, 24451–24458 [DOI] [PubMed] [Google Scholar]
- 24. Lucius A. L., Vindigni A., Gregorian R., Ali J. A., Taylor A. F., Smith G. R., Lohman T. M. (2002) DNA unwinding step-size of E. coli RecBCD helicase determined from single turnover chemical quenched-flow kinetic studies. J. Mol. Biol. 324, 409–428 [DOI] [PubMed] [Google Scholar]
- 25. Hsieh S., Julin D. A. (1992) Alteration by site-directed mutagenesis of the conserved lysine residue in the consensus ATP-binding sequence of the RecB protein of Escherichia coli. Nucleic Acids Res. 20, 5647–5653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Korangy F., Julin D. A. (1992) Alteration by site-directed mutagenesis of the conserved lysine residue in the ATP-binding consensus sequence of the RecD subunit of the Escherichia coli RecBCD enzyme. J. Biol. Chem. 267, 1727–1732 [PubMed] [Google Scholar]
- 27. Yu M., Souaya J., Julin D. A. (1998) Identification of the nuclease active site in the multifunctional RecBCD enzyme by creation of a chimeric enzyme. J. Mol. Biol. 283, 797–808 [DOI] [PubMed] [Google Scholar]
- 28. Lucius A. L., Wong C. J., Lohman T. M. (2004) Fluorescence stopped-flow studies of single turnover kinetics of E. coli RecBCD helicase-catalyzed DNA unwinding. J. Mol. Biol. 339, 731–750 [DOI] [PubMed] [Google Scholar]
- 29. Bianco P. R., Kowalczykowski S. C. (1997) The recombination hotspot Chi is recognized by the translocating RecBCD enzyme as the single strand of DNA containing the sequence 5′-GCTGGTGG-3′. Proc. Natl. Acad. Sci. U.S.A. 94, 6706–6711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wong C. J., Lucius A. L., Lohman T. M. (2005) Energetics of DNA end binding by E. coli RecBC and RecBCD helicases indicate loop formation in the 3′-single-stranded DNA tail. J. Mol. Biol. 352, 765–782 [DOI] [PubMed] [Google Scholar]
- 31. Tomko E. J., Fischer C. J., Lohman T. M. (2010) Ensemble methods for monitoring enzyme translocation along single stranded nucleic acids. Methods 51, 269–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Williams D. J., Hall K. B. (2000) Monte Carlo applications to thermal and chemical denaturation experiments of nucleic acids and proteins. Methods Enzymol. 321, 330–352 [DOI] [PubMed] [Google Scholar]
- 33. Lucius A. L., Maluf N. K., Fischer C. J., Lohman T. M. (2003) General methods for analysis of sequential “n-step” kinetic mechanisms: application to single turnover kinetics of helicase-catalyzed DNA unwinding. Biophys. J. 85, 2224–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Singleton M. R., Dillingham M. S., Gaudier M., Kowalczykowski S. C., Wigley D. B. (2004) Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature 432, 187–193 [DOI] [PubMed] [Google Scholar]
- 35. Caruthers J. M., McKay D. B. (2002) Helicase structure and mechanism. Curr. Opin. Struct. Biol. 12, 123–133 [DOI] [PubMed] [Google Scholar]
- 36. Lucius A. L., Lohman T. M. (2004) Effects of temperature and ATP on the kinetic mechanism and kinetic step-size for E. coli RecBCD helicase-catalyzed DNA unwinding. J. Mol. Biol. 339, 751–771 [DOI] [PubMed] [Google Scholar]
- 37. Farah J. A., Smith G. R. (1997) The RecBCD enzyme initiation complex for DNA unwinding: enzyme positioning and DNA opening. J. Mol. Biol. 272, 699–715 [DOI] [PubMed] [Google Scholar]
- 38. Wong C. J., Lohman T. M. (2008) Kinetic control of Mg2+-dependent melting of duplex DNA ends by Escherichia coli RecBC. J. Mol. Biol. 378, 761–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Anderson D. G., Churchill J. J., Kowalczykowski S. C. (1997) Chi-activated RecBCD enzyme possesses 5′ → 3′ nucleolytic activity, but RecBC enzyme does not: evidence suggesting that the alteration induced by Chi is not simply ejection of the RecD subunit. Genes Cells 2, 117–128 [DOI] [PubMed] [Google Scholar]
- 40. Saikrishnan K., Griffiths S. P., Cook N., Court R., Wigley D. B. (2008) DNA binding to RecD: role of the 1B domain in SF1B helicase activity. EMBO J. 27, 2222–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wong C. J., Rice R. L., Baker N. A., Ju T., Lohman T. M. (2006) Probing 3′-ssDNA loop formation in E. coli RecBCD/RecBC-DNA complexes using non-natural DNA: a model for “Chi” recognition complexes. J. Mol. Biol. 362, 26–43 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.