Background: Bnip3 is both a pro-apoptotic BH3-only protein and a mitochondrial autophagy receptor.
Results: Serine phosphorylation of the Bnip3 LC3-interacting region (LIR) increased binding to Atg8 members and consequently mitophagy, in a manner positively regulated by Bcl-xL.
Conclusion: The Bnip3 LIR activity state determines either pro-survival mitophagy or mitochondrial apoptosis.
Significance: Bnip3-induced mitophagy is serine kinase-regulated and thus a targetable pathway.
Keywords: Apoptosis, Autophagy, Bcl-2 Family, Mitochondria, Mitochondrial Apoptosis, Bcl-xL, Bnip3, LC3, LIR Domain, Mitophagy
Abstract
BH3-only proteins integrate apoptosis and autophagy pathways, yet regulation and functional consequences of pathway cross-talk are not fully resolved. The BH3-only protein Bnip3 is an autophagy receptor that signals autophagic degradation of mitochondria (mitophagy) via interaction of its LC3-interacting region (LIR) with Atg8 proteins. Here we report that phosphorylation of serine residues 17 and 24 flanking the Bnip3 LIR promotes binding to specific Atg8 members LC3B and GATE-16. Using quantitative multispectral image-based flow cytometry, we demonstrate that enhancing Bnip3-Atg8 interactions via phosphorylation-mimicked LIR mutations increased mitochondrial sequestration, lysosomal delivery, and degradation. Importantly, mitochondria were targeted by mitophagy prior to cytochrome c release, resulting in reduced cellular cytochrome c release capacity. Intriguingly, pro-survival Bcl-xL positively regulated Bnip3 binding to LC3B, sequestration, and mitochondrial autophagy, further supporting an anti-apoptotic role for Bnip3-induced mitophagy. The ensemble of these results demonstrates that the phosphorylation state of the Bnip3 LIR signals either the induction of apoptosis or pro-survival mitophagy.
Introduction
BH3-only proteins are activated by diverse intracellular stresses to induce Bax/Bak-mediated release of mitochondrial apoptotic factors (1). Concomitantly, BH3-only proteins disrupt anti-apoptotic Bcl-2 family protein suppression of the autophagy activator Beclin-1 (2, 3) and thereby stimulate the lysosomal degradative pathway of autophagy. Directly linking autophagy to apoptosis are furthermore members of the autophagy-induction machinery, including Atg4D (4), Atg5 (5), and Atg12 (6), which can function as BH3-only proteins to promote cytochrome c release and thus are described as having “dual-functional” activities. In addition, a selective mode of macroautophagy (i.e. mitochondrial autophagy, or mitophagy) may attenuate (7), amplify (8), or operate independently from mitochondrial apoptosis during differentiation (9).
BH3-only proteins with multiple functionalities include Bnip3 and Bnip3L/Nix, homologous members of the BCL2/adenovirus E1B 19-kDa interacting protein (BNIP) family, which activate mitochondria-mediated cell death as well as autophagy and mitophagy (reviewed in Ref. 10). Bnip3 and Nix localize to mitochondria (11), and upon activation, BH3 and transmembrane domains (12, 13) are required to induce caspase-dependent (14) and -independent (15) cell death. In vivo and ex vivo studies in mice demonstrate that Bnip3 and Nix are causative for cell death in cardiovascular disease (14, 16, 17), whereas Nix signals mitophagy during the maturation of red blood cells in the absence of cell death (9, 18).
It is well understood that BH3-only protein-induced autophagy and apoptosis signaling events converge at mitochondria. However, mechanistic insight discriminating the function of Bnip3-induced apoptosis versus autophagy and mitophagy is lacking. This is relevant because clinical cancer studies report conflicting roles for Bnip3. For example, Bnip3 expression is a good prognostic indicator in pancreatic cancer (19), is either a poor or good prognosis indicator in breast cancer (20), and indicates poor prognosis in cervical cancer (21).
Mitophagy ranges from mitochondrial pruning (16) to full clearance (9, 18), suggesting mechanisms regulating activity. Although BH3 domains of Bnip3 and Nix are sufficient to induce the general autophagy response (22), induction of mitophagy requires the LC3-interacting region (LIR)4 of Nix (23) and Bnip3 (24).
Intriguingly, phosphorylation regulates the activity of the selective autophagy receptor p62 (25) and mitophagy via the yeast functional Bnip homologue Atg32 (26). Moreover, serine phosphorylation of the LIR of optineurin is required for selective autophagy of bacteria (27). Bnip3 LIR, an N-terminal WVEL tetrapeptide motif identical to Nix, lies within a predicted serine-rich PEST domain associated with protein degradation (28). Thus, the aim of this work was to investigate whether the phosphorylation status of serines within Bnip3 LIR regulates Bnip3-mediated induction of mitophagy and to determine the function of mitophagy in the context of apoptosis signaling.
We report that phosphorylation of serine residues flanking the Bnip3 LIR is required for selective Bnip3-Atg8 homologue interactions and mitochondrial autophagy. Moreover, Bcl-xL enhances Bnip3-induced mitophagy, in a manner requiring Bnip3 LIR and BH3 activities. Functionally, we show that Bnip3-induced mitophagy proactively lowers the cell's cytochrome c release capacity, demonstrating a pro-survival function.
EXPERIMENTAL PROCEDURES
Chemicals and Antibodies
λ-Protein phosphatase was purchased from New England Biolabs. K252c was purchased from Sigma. Okadaic acid was purchased from Calbiochem. CCCP was purchased from Calbiochem. Rhodamine 123 was purchased from Invitrogen. TNFα was a gift of BASF. G418 was purchased from Carl Roth. Cell culture reagents were obtained from Invitrogen, Sigma, Lonza, and Pan Biotech. JetPRIME was obtained from Polyplus. Electron microscopy grade paraformaldehyde was obtained from EMS. The GasPakTM EZ system for hypoxia was purchased from BD Biosciences. Complete EDTA-free protease inhibitor mixture and PhosSTOP phosphatase inhibitor mixture were purchased from Roche Applied Science. Antibodies used were against active caspase-3 (Cell Signaling catalog no. 9664), β-actin (GeneTex catalog no. GTX26276, Cell Signaling catalog no. 4970), Bnip3 (Santa Cruz Biotechnology, Inc., catalog no. sc-56167), GFP (Roche Applied Science catalog no. 11814460001 for immunoprecipitations; Cell Signaling catalog no. 2555 for Western blotting), LC3B (Cell Signaling catalog no. 2775), RFP (Chromotek catalog no. 5F8), Beclin-1 (Cell Signaling catalog no. 3738), RhoGDI (BD Biosciences catalog no. 610255 and Santa Cruz Biotechnology, Inc., catalog no. H0510), Tim23 (Santa Cruz Biotechnology, Inc., catalog no. sc-1329), COX IV (Cell Signaling catalog no. 4850S), Tom20 (Santa Cruz Biotechnology, Inc., catalog no. sc-11415), VDAC (Cell Signaling catalog no. 4661), Bcl-xL (Santa Cruz Biotechnology, Inc., catalog no. sc-8392), and cytochrome c (Santa Cruz Biotechnology, Inc., catalog no. sc-13156). Horseradish peroxidase-conjugated secondary antibodies (Cell Signaling) were used for chemiluminescence detection for Western blotting. Highly cross-absorbed Alexa Fluor 488, 546, and 647 secondary antibodies (Invitrogen) were used for immunofluorescence staining.
Plasmids Used in Study
Bnip3 (NM_004052), LC3B (NM_022818), and GABARAP-L2/GATE-16 (NM_007285) were obtained from DKFZ Core Facilities, PCR-amplified, and inserted into either pmCherry-C1 (RFP) or pEGFP-C1 (GFP). Mutations were inserted by site-directed mutagenesis. DNA correctness was verified by sequencing. Additional protein expression vectors were used as described previously: GFP-Rab7 WT (29); GFP-GABARAP and GFP-GABARAP-L1 (23); and GFP-cytochrome c (30). Wild type Bcl-xL was obtained from Morishima et al. (31). RFP-Bcl-xL was generated using pmCherry-C1, and mutants were generated by site-directed mutagenesis. shRNA pLKO lentiviral vectors against Beclin-1 (GACAGTTTGGCACAATCAATA) and control (AATTGCCAGCTGGTTCCATCA) were generated. Throughout this study, mCherry is referred to as RFP (red fluorescent protein).
Cell Culture
Human MCF-7 (Cell Lines Services, Heidelberg, Germany), Panc-1 (obtained from the Department of General Surgery, University of Heidelberg, Germany), and HeLa Kyoto cancer cell lines (32) were maintained in full medium, consisting of DMEM, 10% FBS, l-glutamine, non-essential amino acids, penicillin, streptomycin, amphotericin B. Clonal MCF-7 cell lines stably expressing fusion proteins or shRNA were generated via selection with 1 mg/ml G418. The HL-1 cardiac myocyte cell line was maintained in Claycomb medium supplemented with 10% FBS, 0.1 mm norepinephrine, 2 mm l-glutamine, penicillin/streptomycin/amphotericin B. Transient transfections were performed using JetPRIME (Polyplus) transfection reagents. To generate stable HL-1 cells, pWIPI lentiviral vectors containing GFP-Rab7 and GFP-LC3B were generated in 293T cells. Cells were infected using lentivirus-containing 293T cell supernatant. For shRNA experiments, pLKO lentiviral vectors containing shRNA were generated in 293T cells, and cells were infected using lentivirus-containing 293T cell supernatant.
Hypoxia Induction
Hypoxia/reoxygenation experiments were performed in glucose-containing HBSS (Invitrogen catalog no. 14025) supplemented with NaHCO3 (25 mm), PSF, and 10 mm Hepes. Cells were incubated under hypoxia conditions using the (GasPakTM EZ, BD Biosciences) at 37 °C for 42 h (Panc-1, MCF7, and HeLa) or 18 h (HL-1). Normoxic conditions were reintroduced through incubation at 37 °C in 5% CO2 for 6 h (cancer cell lines) or 2 h (HL-1 cardiac cells) and immediately paraformaldehyde-fixed for imaging or harvested for Western blot analysis.
Fluorescence Imaging
Cells were plated in 8-well microscopy μ-slides (Ibidi), treated as indicated, and subjected to either live cell imaging or fixed with 4% paraformaldehyde in PBS. Wide field fluorescence microscopy was performed with a DeltaVision RT microscope system (Applied Precision) using a ×60 oil immersion objective to generate Z-stacks (0.3-μm step sizes). Images of representative cells are total intensity projections of Z-stacks prepared using ImageJ (National Institutes of Health).
Autophagosome formation and mitochondrial fragmentation result in a high degree of intensity variation within images of single cells. Because of the difficulty in representing colocalizations using RGB colors, images were inverted to generate negative gray scale (i.e. white background, with black signifying the highest intensity). In the merged imaged, the convergence of organelles yields a higher intensity pseudocolor (dark blue). RGB plot profiles indicate protein and organelle colocalization.
Colocalization analysis was performed using the ImageJ colocalization color map plugin (33). Colocalization was performed on single, segmented cells. The index of colocalization (Icorr value) is represented on the y axis.
Quantification of Mitochondrial Sequestration, Degradation, and Cytochrome c Content by Multispectral Imaging-based Flow Cytometry (MIFC)
MIFC was performed using the ImageStream X flow cytometer (Amnis, Seattle, WA). MIFC combines flow cytometry with high content microscopic analysis. Large cell populations are acquired, and fluorescent markers are analyzed at the single-cell level, from which population responses are quantified.
In experiments to measure cytochrome c and mitochondrial mass, cells were transfected with mito-RFP or the indicated RFP-fused Bnip3 WT and mutants and analyzed at 48 h of expression, as indicated. Cells were trypsinized and fixed with 4% paraformaldehyde in PBS. Immunofluorescence staining was performed using α-Tom20 and α-cytochrome c and Alexa Fluor 488 or 647 secondary antibodies, respectively. Alexa Fluor 488, RFP, and Alexa 647 were excited using the corresponding 488, 561, and 657-nm lasers. To quantify cellular responses, channel compensation and image analysis were performed using IDEAS 4.0. Briefly, single/focused/RFP-positive (RFP+) cells were selected, yielding ∼1000 cells/condition for analysis.
The subpopulation of cells with high mitophagy (i.e. reduced mass of Tom20+ mitochondria) was calculated from total cellular intensity and the bright detail intensity of Tom20 staining. The bright detail intensity feature provided by the IDEAS 4.0 software computes the intensity of localized bright spots within the cell. This feature was used as a second parameter of mitochondrial mass. The subpopulation of cells with low cellular cytochrome c content was determined on the basis of a low value of intensity and the bright detail intensity of cytochrome c immunostaining.
In experiments to measure mitochondrial sequestration by autophagosomes and autolysosomes, cells were co-transfected with RFP-Bnip3 WT and mutants and either GFP-LC3B or GFP-Rab7, respectively. In single/focused/RFP-positive/GFP-positive cells, colocalization of Bnip3 WT and mutants with LC3B and Rab7 was quantified using the IDEAS colocalization analysis function. The subpopulation with high colocalization is reported.
Co-immunoprecipitation
Cells were scraped, centrifuged at 550 × g for 5 min, and lysed in 200 μl of CHAPS lysis buffer (2% CHAPS, 1% Triton X-100, 137 mm NaCl, 2 mm EDTA, 10% glycerol, and 20 mm Tris-HCl, pH 8.0) containing protease and phosphatase inhibitors. 190 μl of cell lysate was incubated with 12.5 μl of α-GFP (1:40; Roche Applied Science)-coupled Dynabeads Protein G (Invitrogen) for 2 h at 4 °C. Then beads were washed with 0.02% Tween 20 plus PBS, and immunoprecipitated proteins were eluted from Dynabeads by adding 2× LDS sample and reducing buffer to the beads and heating at 70 °C for 10 min. Samples were analyzed by Western blotting.
Western Blotting
Protein samples were electrophoresed using BisTris NuPAGE gels (Invitrogen) and transferred to nitrocellulose using the iBlot dry blotting system (Invitrogen). Subsequently, membranes were blocked and incubated with primary antibodies. Horseradish peroxidase-conjugated secondary antibodies were used, and the membranes were developed using SuperSignal West Pico chemiluminescent substrate (Pierce) and a chemiluminescence imager (Intas). Blots shown are representative of at least three independent experiments. Densitometric band quantifications were performed using ImageJ software. ClustalW multiple-sequence alignment was performed using Jalview (34).
Survival Curve Generation
Bnip3 and Fundc1 survival data are presented as Kaplan Meyer plots and were generated using the R2: Microarray Analysis and Visualization Platform (available on the Academic Medical Center (Amsterdam, The Netherlands) Web site).
Statistical Analyses
The probability of statistically significant increases or decreases between conditions was determined using Student's t test. One-tailed t tests were performed, paired for matched data sets and unpaired for unmatched data sets. Data with a p value less than or equal to 0.05 were accepted as statistically significant. Values are expressed as mean ± S.E. of at least three independent experiments unless otherwise stated.
RESULTS
Hypoxia/Reoxygenation Induction of Bnip3 Correlates with Mitophagy
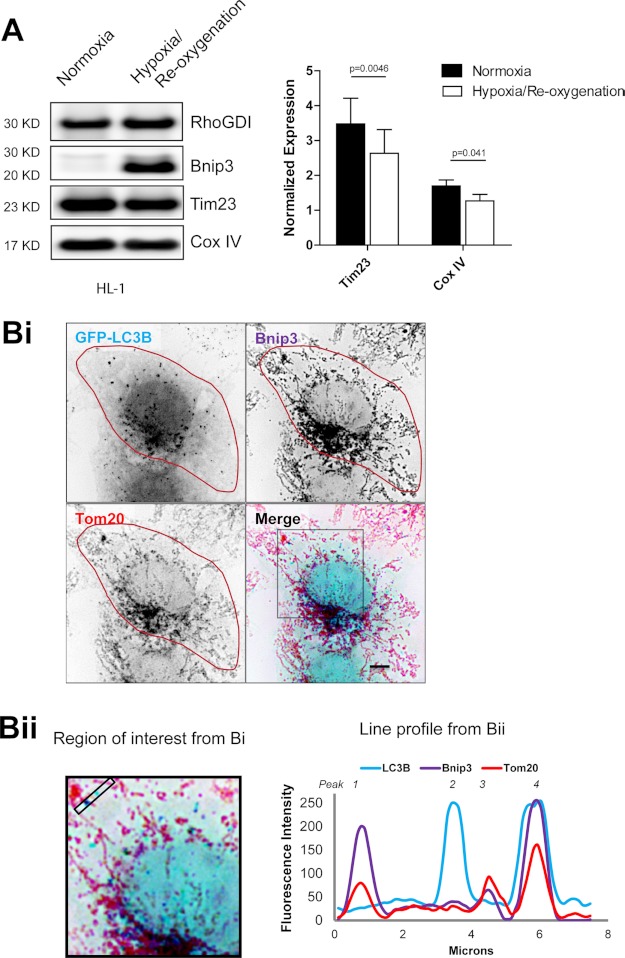
Bnip3 is strongly expressed in response to hypoxia (35) and is activated by oxidative conditions during cardiac hypoxia/reoxygenation (14). In HL-1 cardiac cells, which exhibit mitophagy in response to ectopic Bnip3 expression (14), 18 h of hypoxia followed by 2 h of reoxygenation resulted in decreased mitochondrial content, concomitant with increased Bnip3 levels (Fig. 1A). Whereas hypoxia promotes mitochondrial fragmentation (36), in HL-1 cells following reoxygenation, Bnip3-coated mitochondria existed in both fragmented and networked states (Fig. 1Bi). Notably, a subset of fragmented mitochondria in both the cell periphery and the perinuclear region were sequestered by GFP-LC3B-positive autophagosomes, whereas Bnip3-coated, networked mitochondria did not colocalize with GFP-LC3B (Fig. 1Bii).
FIGURE 1.
Bnip3 up-regulation correlates with mitochondrial autophagy in cardiac HL-1 cells. A, hypoxia/reoxygenation increased Bnip3 expression and decreased mitochondrial content. HL-1 cells were submitted to 18 h of hypoxia, followed by 2 h of reoxygenation Western blot detection of mitochondrial markers Tim23, COXIV, Bnip3, and RhoGDI as a loading control. Shown is densitometric quantification of Tim23 and COXIV, normalized to RhoGDI. B, immunodetection of endogenous Bnip3 targeted to mitochondria, where it colocalized with GFP-LC3B following hypoxia/reoxygenation. Bi, HL-1 cells expressing GFP-LC3B were submitted to 18 h of hypoxia, followed by 2 h of reoxygenation. GFP-LC3B-labeled autophagosomes (pseudocolored light blue) and antibody labeled Tom20 (red) and Bnip3 (purple) were imaged by high resolution microscopy. Bnip3-targeted mitochondria that colocalized with autophagosomes appear dark blue. Bii, the region of interest from Bi is magnified, and the ImageJ plot profile function was used to illustrate colocalizations within the bounded region between autophagosomes (blue), Bnip3 (purple), and Tom20 (red). Error bars, S.E.
Similarly, in pancreatic, breast, and cervical cancer cell lines, 42 h of hypoxia under nutrient deprivation conditions, followed by 6 h of reoxygenation, resulted in increased Bnip3 expression and decreased mitochondrial content, to varying extents between cancer cell lines (supplemental Fig. S1A). Together, these findings suggest that increased expression of Bnip3 correlates with different degrees of mitophagy activation.
Bnip3 LIR Activity Is Serine Phosphorylation-regulated
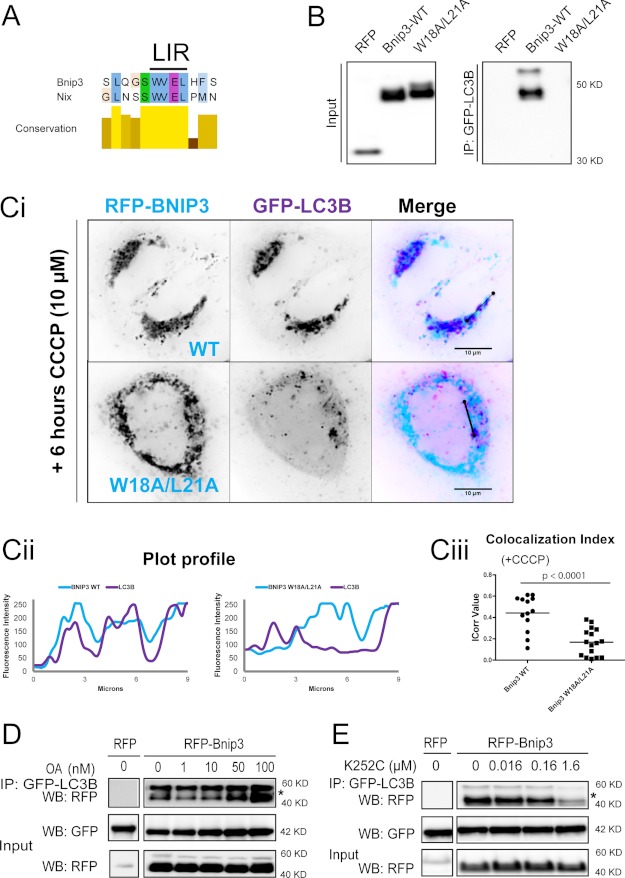
Bnip3 contains a WVEL LIR motif identical to Nix (23) (Fig. 2A) and was immunoprecipitated by LC3B (Fig. 2B). Selective autophagy requires the interaction of the receptor's LIR core with Atg8 members at a hydrophobic binding site. Mutation of either acidic residues proximal to the LIR or of the hydrophobic core WXXL LIR residues to neutral alanines is sufficient to abolish this interaction (37). We therefore generated Bnip3 W18A/L21A, containing inactivating mutations of the putative LIR. Bnip3-W18A/L21A did not bind to LC3B (Fig. 2B), confirming a receptor function for Bnip3.
FIGURE 2.
The Bnip3 LIR determines the interaction with LC3B, in a manner dependent on serine/threonine phosphorylation. A, sequence alignment demonstrating that Bnip3 contains a core tetrapeptide LIR, identical to Nix. B and C, Bnip3 WVEL is a functioning LIR and is responsible for interactions with LC3B. B, RFP-tagged Bnip3 WT and LIR mutant W18A/L21A were expressed in MCF-7 cells stably expressing GFP-LC3B. At 48 h of expression, immunoprecipitations were performed with α-GFP. Shown is Western blot detection of RFP. C, CCCP-enhanced mitophagy is dependent on Bnip3 LIR. Ci, RFP-tagged Bnip3 WT and Bnip3 W18A/L21A co-expressed with GFP-LC3B in HeLa cells for 24 h, treated with CCCP (10 μm; 6 h), and imaged by high resolution microscopy. Bnip3 is pseudocolored light blue, and LC3B is pseudocolored purple. Dark blue indicates colocalization due to sequestration of mitochondria. Cii, plot profiles of Bnip3 (light blue) and LC3B (purple) intensities demonstrating localizations of Bnip3-targeted mitochondria and autophagosomes. Ciii, colocalization of Bnip3 WT and Bnip3 W18A/L21A with GFP-LC3B was quantified using the ImageJ colocalization color map plugin. Dot plots indicate the data set distribution and mean correlation index (Icorr) ± S.E. of at least 10 cells (Z-stacks) per condition. D, the serine/threonine phosphatase inhibitor okadaic acid (OA) increases binding of Bnip3 to LC3B. RFP-Bnip3 WT was expressed in MCF-7 cells stably expressing GFP-LC3B. At 42 h of expression, cells were treated with increasing concentrations of okadaic acid (1, 10, 50, and 100 nm) for 6 h, and immunoprecipitations (IP) were then performed with α-GFP. Western blot (WB) detection of RFP and GFP. *, both bands correspond to RFP-Bnip3. E, binding of Bnip3 to LC3B is dependent on serine/threonine kinase activities. RFP-Bnip3 WT was expressed in MCF-7 cells stably expressing GFP-LC3B. At 42 h of expression, cells were treated with concentrations of K252c (staurosporine aglycone; 16, 160, and 1600 nm) for 6 h, and immunoprecipitations were then performed with α-GFP. Shown is Western blot detection of RFP and GFP. *, both bands correspond to RFP-Bnip3.
The mitochondrial uncoupler CCCP, which collapses the mitochondrial membrane potential, enhances Nix-driven mitophagy in a LIR-dependent manner (23). Similarly, we observed prominent autophagic sequestration of wild-type (WT) Bnip3-targeted mitochondria in response to CCCP (10 μm, 6 h). However, sequestration of mitochondria targeted by Bnip3-W18A/L21A was significantly less responsive to CCCP (Fig. 2C), further evidence that Bnip3 functions as a LIR-dependent mitochondrial autophagy receptor, as recently reported (24).
Because Bnip3 apoptotic activity is correlated with high levels of serine/threonine phosphorylation in vivo (38), we sought to determine the effect of increased Bnip3 phosphorylation state on its binding to LC3B, initially using inhibitors. Inhibition of serine/threonine phosphatases with okadaic acid increased the strength of the association between LC3B and Bnip3 in a dose-dependent manner (Fig. 2D). Note that increased binding did not correlate with mobility shifts of RFP-Bnip3, indicating that only a fraction of the phosphorylated residue is undergoing rapid dephosphorylation. Conversely, the serine/threonine kinase inhibitor K252c (39) reduced LC3B binding (Fig. 2E). Bnip3 is highly phosphorylated and therefore runs by SDS-PAGE as multiple bands (38). Here two main bands were observed, both responsive to phosphatase and kinase inhibitors. Both bands are attributed to Bnip3 because cross-linking was performed to minimize elution of the IgG heavy chain (also see supplemental Fig. S2).
Together, these results pointed toward a role for phosphorylation in regulating the affinity of Bnip3 for LC3B. Subsequently, we undertook molecular studies in order to exclude nonspecific effects of the chemical inhibitors on general autophagy (40, 41) and investigated the identity of the phosphorylated residues.
Serine Residue 17 Preceding the Bnip3 LIR Is Essential for LC3B Binding
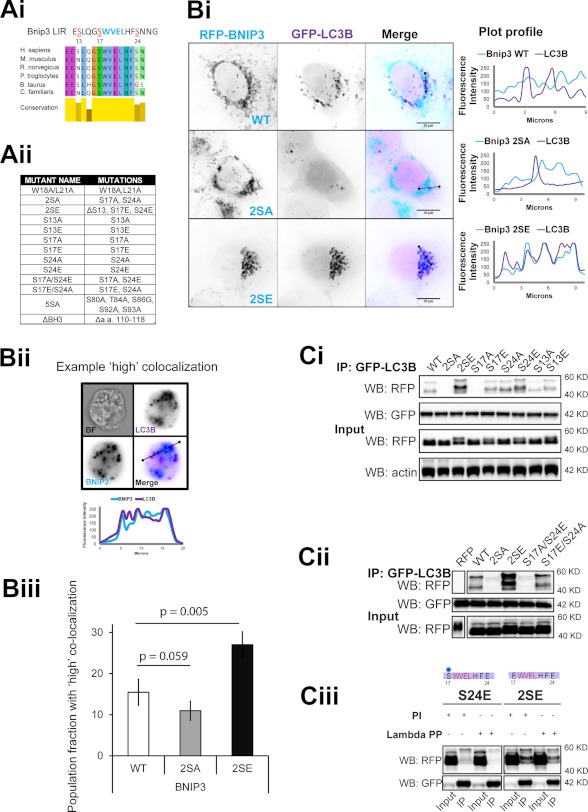
Docking of ubiquitin-like proteins SUMO (42) and LC3 (27) is locally promoted by serine phosphorylation of their corresponding receptors. Notably, both Bnip3 and Nix LIR motifs are flanked by conserved and partially conserved serine residues (Fig. 3Ai). To localize residues responsible for phosphorylation regulation of Bnip3-LC3B interaction, we generated single and combinatorial phosphorylation mimicking serine-to-glutamate and non-phosphorylatable serine-to-alanine mutations flanking the Bnip3 LIR. Mutations and designations are listed in Fig. 3Aii.
FIGURE 3.
Serine phosphorylation of the Bnip3 LIR motif determines its affinity for LC3B. A, putative serine phosphorylation sites of interest and generated mutants. Ai, alignment of the region containing the Bnip3 LIR across mammalian species. Aii, table describing Bnip3 mutants used in this study. B, phosphorylation state of the LIR determines autophagosomal sequestration of mitochondria. Bi, representative high resolution images of GFP-LC3B colocalization with RFP-Bnip3 WT and the indicated multisite LIR mutants at mitochondria in HeLa cells at 24 h. Plot profiles illustrate colocalization between Bnip3-targeted mitochondria and autophagosomes. Bii, MIFC analysis of Bnip3 WT, 2SA, and 2SE induction of mitophagy. The indicated Bnip3 constructs were expressed with GFP-LC3B in cardiac HL-1 cells for 24 h and imaged using the ImageStream X flow cytometer. Representative images and plot profiles are shown. Biii, using MIFC, quantification of the subpopulation of Bnip3 WT-targeted mitochondria with high GFP-LC3B colocalization was achieved using the colocalization function and gating on the fraction of cells with high colocalization. The identical analysis was applied to Bnip3 mutant populations. Population fractions are indicated as well as the percentage change of the two mutants compared with the wild type. C, localization of serine residues, which control binding affinity with LC3B. Ci, the indicated RFP-Bnip3 WT and multisite and single site mutant constructs were expressed in MCF-7 cells stably expressing GFP-LC3B. At 48 h of expression, immunoprecipitations (IP) were performed with α-GFP. Shown is Western blot detection of RFP, GFP, and actin. Cii, double mutant S17A/S24E and S17E/S24A Bnip3 constructs were expressed in MCF-7 stably expressing GFP-LC3B. At 48 h of expression, immunoprecipitations were performed with α-GFP. Shown are Western blot detection of RFP and GFP. Ciii, in vitro analysis of serine 17 phosphorylation during LC3B binding to Bnip3. Bnip3 S24E and 2SE mutants were expressed for 48 h in MCF-7 cells stably expressing GFP-LC3B. Cell lysates were incubated with λ-phosphatase (800 units) prior to immunoprecipitation with α-GFP. Shown is Western blot detection of RFP and GFP. PI, phosphatase inhibitor.
WT and LIR mutant Bnip3 localized entirely at Tom20-immunolabeled mitochondria in HeLa cells (supplemental Fig. S3Ai). At early time points (18–24 h), Bnip3 WT and the 2SA mutant localized at networked mitochondria, distributed throughout the cell. However, mitochondria targeted by 2SE mutants were fragmented and clustered (supplemental Fig. S3Aii). Bnip3 WT colocalized with GFP-LC3B-labeled autophagosomes, but no colocalization was detected in cells expressing the Bnip3 2SA mutant. Autophagic sequestration of Bnip3 2SE-targeted mitochondria was remarkably greater than with Bnip3 WT (Fig. 3Bi). Using MIFC, we directly quantified the colocalization of Bnip3 WT and mutants with LC3B in populations of HL-1 cardiac cells (Fig. 3, Bii and Biii). At 24 h of expression, the fraction of cells exhibiting prominent autophagic sequestration of Bnip3-targeted mitochondria (gated as high colocalization of Bnip3 and LC3) was reduced by 30% in response to Bnip3 2SA and enhanced by ∼75% in response to Bnip3 2SE, relative to Bnip3 WT. Similar results were obtained in HeLa cells at 24 h (supplemental Fig. S3B).
Binding of WT and mutant Bnip3 to LC3B was determined by co-immunoprecipitation. In HL-1 cells, Bnip3 WT and Bnip3 2SE were similarly immunoprecipitated by LC3B, whereas much less interaction with Bnip3 2SA was detected (supplemental Fig. S3Ci). Remarkably, in both MCF-7 (Fig. 3, Ci and Cii) and HeLa cell lines (supplemental Fig. S3Cii), Bnip3 2SE was more strongly associated with LC3B than Bnip3 WT. Conversely, LC3B did not immunoprecipitate the Bnip3 2SA mutant, further indicating that Bnip3 LIR activity is phosphorylation-regulated, with phosphorylation of serine 17 and/or 24 required for binding.
Single and double E/A combinatorial LIR mutants were used to investigate the individual contributions of serines 13, 17, and 24 (Fig. 3Ci). Bnip3 S13A exhibited a minor reduction in binding, whereas Bnip3 S13E had no impact. Bnip3 S17A most strongly inhibited binding to LC3B, whereas Bnip3 S17E slightly enhanced LC3B binding. In contrast, Bnip3 S24A binding was similar to Bnip3 WT, whereas Bnip3 S24E most strongly enhanced binding to LC3B. Putative regulation by multisite phosphorylation was then investigated, using the most influencing single mutations, at serines 17 and 24. Bnip3 S17A/S24E did not bind LC3B, and Bnip3 S17E/S24A binding was comparable with Bnip3 S17E and WT (Fig. 3Cii), indicating that serine 17 phosphorylation is required for binding to LC3B. However, the most effective interaction was obtained with mimicked phosphorylation of both serines 17 and 24.
Importantly, the Bnip3 binding to LC3B was phosphatase-sensitive in vitro. Treatment of cell lysates with λ-phosphatase prior to immunoprecipitation of GFP-LC3B did not alter binding to the dual phosphorylation mimic Bnip3 2SE, which is independent of endogenous phosphorylation of either serine 17 or 24 (Fig. 3Ciii). However, binding was lost to Bnip3 S24E, which presumably is endogenously phosphorylated at serine 17, compared with the control lysate treated with phosphatase inhibitors. These results suggest that Bnip3 binding to LC3B is dependent on endogenous phosphorylation at serine 17.
Bnip3 is reported to be highly phosphorylated at regions downstream of the LIR (supplemental Fig. S4A), and Atg32 and p62 autophagy activities are positively regulated by serine phosphorylation downstream of LIR (25, 26). Potential roles for additional phosphorylation sites were investigated using Bnip3–5SA, which is mutated at phosphorylated sites downstream from the LIR (Fig. 3Aii). The non-phosphorylatable Bnip3–5SA mutant bound to LC3B similar to Bnip3 WT (supplemental Fig. S3Bi). Together, these results indicate that the strength of interaction between LC3B and Bnip3 is dependent on specific phosphorylation of residues flanking the Bnip3 LIR.
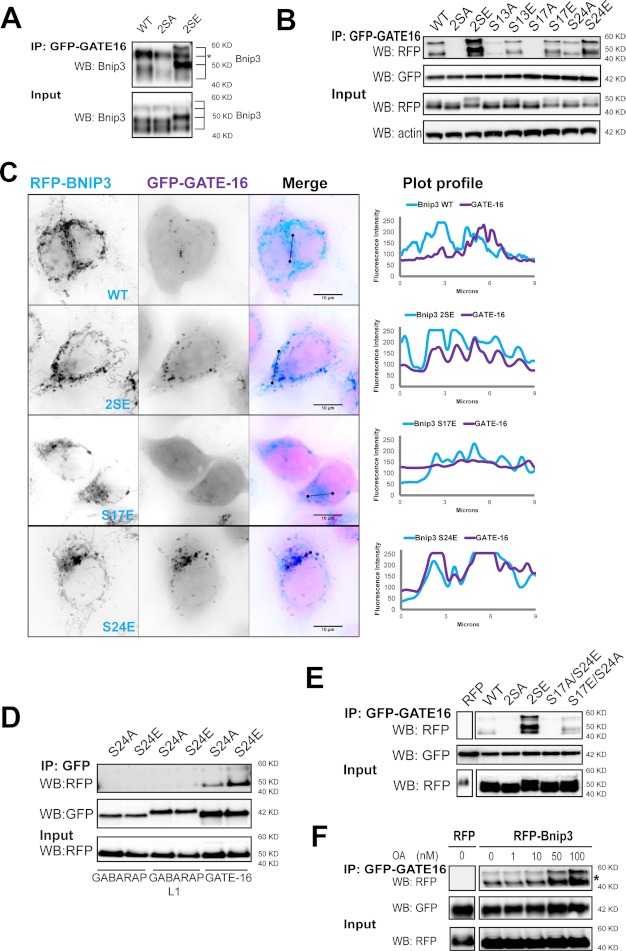
Serine Residues 17 and 24 Flanking the Bnip3 LIR Are Essential for GATE-16 Binding
Whereas LC3B weakly binds Nix, other Atg8 family members exhibit higher binding affinities (23). Compared with interactions with LC3B, Bnip3 WT weakly bound GABARAP subfamily member GATE-16 (Fig. 4A), and only few RFP-Bnip3 WT-targeted mitochondria were sequestered by GATE-16-labeled autophagosomes (Fig. 4C). We therefore determined a role of LIR phosphorylation on Bnip3 binding to GATE-16. Bnip3 2SA and S17A mutations abolished binding with GATE-16. Bnip3 S17E and, surprisingly, S24A mutants bound GATE-16 similar to Bnip3 WT. The 2SE and S24E mutants similarly enhanced binding to GATE-16 (Fig. 4B), and consistent with these findings, both mutants colocalized with GATE-16 at higher levels than Bnip3 WT and S17E (Fig. 4C).
FIGURE 4.
Serine phosphorylation of the Bnip3 LIR motif determines its affinity for GATE-16. A, LIR serine residues control binding affinity for GATE-16. The indicated RFP-Bnip3 WT and multisite mutant constructs were expressed in MCF-7 stably expressing GFP-GATE-16. At 48 h of expression, immunoprecipitations (IP) were performed with α-GFP. Shown is Western blot (WB) detection of Bnip3. *, this band of RFP-Bnip3 overlaps with IgG heavy chain. B, phosphorylation at serine 17 is required for, whereas phosphorylation at serine 24 promotes, Bnip3 binding to GATE-16. The indicated RFP-Bnip3 WT and multisite and single site mutant constructs were expressed in MCF-7 stably expressing GFP-GATE-16. At 48 h of expression, immunoprecipitations were performed with α-GFP. Shown is Western blot detection of RFP, GFP, and actin. C, LIR phosphorylation enhances GATE-16 sequestration of Bnip3-targeted mitochondria. Shown is high resolution imaging of GFP-GATE-16 colocalization with WT and multisite LIR mutant Bnip3 at mitochondria in HeLa cells at 24 h. Plot profiles identify colocalizations between Bnip3-targeted mitochondria and autophagosomes. D, LIR phosphorylation does not enhance binding of GABARAP and GABARAP-L1. Strongly binding RFP-Bnip3 S24E and non-binding RFP-Bnip3 S24A were co-expressed with GFP-fused GABARAP, GABARAP-L1, or GABARAP-L2/GATE-16 in HeLa cells. At 24 h of expression, immunoprecipitations were performed with α-GFP antibody. Shown is Western blot detection of RFP and GFP. E, dual phosphorylation at serine 17 and serine 24 enhances GATE-16 binding. Double mutant S17A/S24E and S17E/S24A Bnip3 constructs were expressed in MCF-7 cells stably expressing GFP-GATE-16. At 48 h of expression, immunoprecipitations were performed with α-GFP. Shown is Western blot detection of RFP and GFP. F, okadaic acid treatment increases binding of Bnip3 to GATE-16. RFP-Bnip3 WT was expressed in MCF-7 cells stably expressing GATE-16. At 42 h of expression, cells were treated with increasing concentrations of okadaic acid (1, 10, 50, and 100 nm) for 6 h, and immunoprecipitations were then performed with α-GFP. Shown is Western blot detection of RFP and GFP.
Notably, the S24E mutation increased Bnip3 binding to GATE-16 but not to GABARAP or GABARAP-L1 (Fig. 4D). Furthermore, similar to LC3B binding, regulation of GATE-16 binding was localized to the Bnip3 LIR region, because Bnip3–5A, rendered non-phosphorylatable downstream of the LIR, did not display altered binding to GATE-16 (supplemental Fig. S4Bii). As with LC3B, Bnip3 S17A/S24E did not bind GATE-16, and Bnip3 S17E/S24A binding to GATE-16 was comparable with Bnip3 WT (Fig. 4E). Similar to the interaction between Bnip3 and LC3B, treatment with okadaic acid increased the association between GATE-16 and Bnip3 in a dose-dependent manner (Fig. 4F).
Taken together, this mutational analysis indicates that phosphorylation of Bnip3 at serine 17 is prerequisite for LC3B and GATE-16 binding, whereas phosphorylation at serine 24 positively regulates the affinity for LC3B and GATE-16.
Both Autophagy Induction and LIR Phosphorylation Are Required for Bnip3 Binding to LC3B
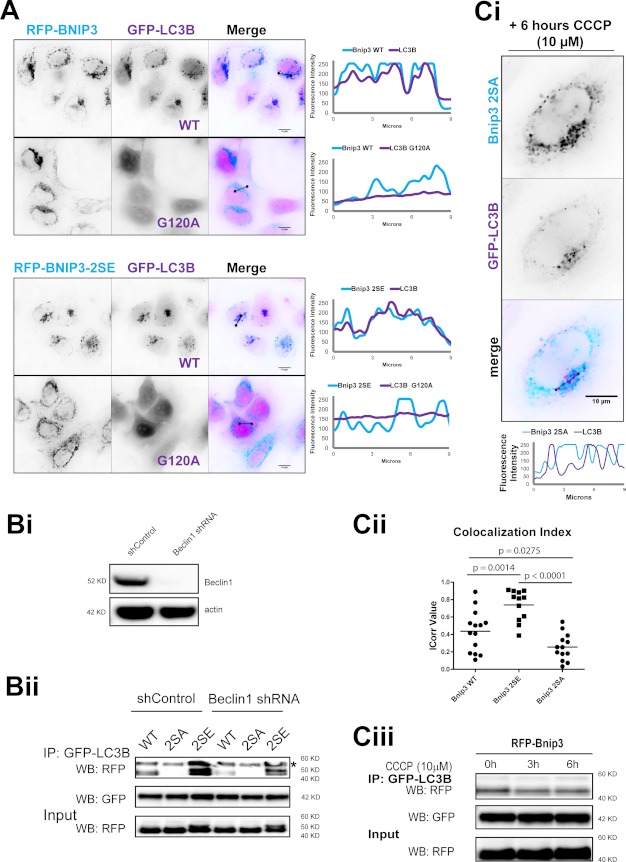
We next sought to determine the relationship between autophagy induction and mitophagy regulation. The LC3B (G120A) mutant, which is not converted to the membrane-inserting form LC3B-II (43), did not localize to either Bnip3 WT or 2SE mutant (Fig. 5A). Likewise, decreasing autophagic capacity via shRNA knockdown of Beclin-1 (Fig. 5Bi) reduced the binding affinity of both Bnip3 WT and the phosphorylation mimic 2SE for LC3B (Fig. 5Bii). Thus, mitophagy engagement via the LC3B interaction with Bnip3 requires autophagy induction.
FIGURE 5.
Autophagy induction is required for Bnip3 binding to LC3B. A, Bnip3 binding to LC3B depends on LC3B lipidation. GFP-LC3B and GFP-LC3B (G120A) were co-expressed with either RFP-Bnip3 WT or Bnip3 2SE. At 18 h of expression, high resolution imaging was performed. Plot profiles illustrate colocalizations between Bnip3-targeted mitochondria and autophagosomes. B, Beclin-1 knockdown inhibits the interaction between Bnip3 and LC3B. Bi, stable control and Beclin-1 shRNA knockdown were performed in MCF7 cells stably expressing GFP-LC3B. Bii, Bnip3 WT, Bnip3 2A, and Bnip3 2E were expressed in MCF-7/GFP-LC3B with stable Beclin-1 knockdown or control knockdown. At 48 h of expression, immunoprecipitations (IP) were performed with α-GFP. Shown is Western blot (WB) detection of RFP and GFP. *, this band of RFP-Bnip3 overlaps with IgG heavy chain. C, CCCP drives mitochondrial sequestration in a LIR-dependent manner. Ci, Bnip3 2SA was co-expressed with GFP-LC3B in HeLa cells for 24 h, treated with CCCP (10 μm, 6 h), and imaged by high resolution microscopy. Cii, colocalization of Bnip3 WT, Bnip3 2A, and Bnip3 2E with GFP-LC3B in HeLa cells treated with CCCP (10 μm, 6 h) was quantified using the ImageJ colocalization color map plugin. Dot plots indicate the data set distribution and mean correlation index (Icorr) ± S.E. of at least 10 cells (Z-stacks) per condition. Ciii, RFP-tagged Bnip3 WT was expressed in MCF-7 cells stably expressing GFP-LC3B for 48 h. Cells were treated with CCCP as indicated, and immunoprecipitations were performed with α-GFP. Shown is Western blot detection of RFP and GFP.
The energy sensor AMP-activated protein kinase directly induces autophagy (36) and is potently activated by CCCP-mediated mitochondrial uncoupling (45). In cells expressing Bnip3 2SA, autophagosomes formed following CCCP treatment (Fig. 5Ci), yet mitochondrial sequestration was not detected (Fig. 5, Ci and Cii). In contrast to the prominent sequestration observed for Bnip3 WT and 2SE (Fig. 3Bi), formed autophagosomes were juxtaposed with Bnip3 2SA-targeted mitochondria but not sequestering mitochondria.
Notably, relative to Bnip3 WT, sequestration was reduced by the 2SA mutant and enhanced by the 2SE mutant, (Fig. 5Cii). Furthermore, Bnip3 WT binding to LC3B was not affected by CCCP (Fig. 5Ciii). These results indicate that whereas mitophagy requires LIR phosphorylation for the coupling of Bnip3-targeted mitochondria to autophagosomal membranes, the Atg8-LIR interactions act as a rate-limiting step during mitophagy, in parallel to autophagy.
Bnip3 LIR Phosphorylation Regulates Maturation and Autophagic Degradation of Mitochondria
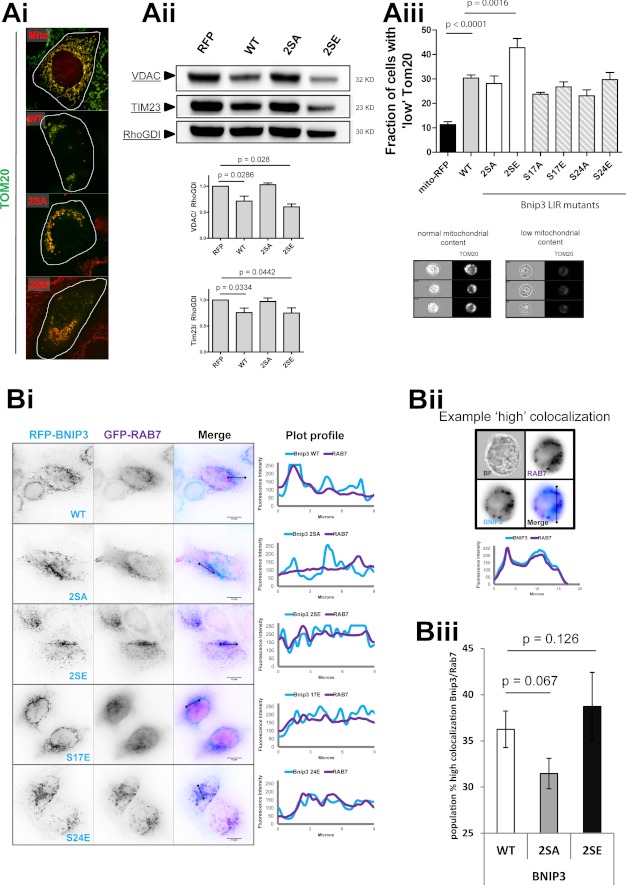
In addition to sequestration, the impact of phosphorylation-mimicking and non-phosphorylatable LIR mutants on mitochondrial degradation was measured using multiple approaches. In HeLa cells at 48 h of expression, Bnip3 WT and 2SE mutant reduced the cellular mitochondrial content, detected by immunofluorescence of mitochondrial outer membrane protein Tom20 (46) (Fig. 6Ai). Western blot analysis of mitochondrial markers VDAC and Tim23 revealed a significant reduction in mitochondrial content in response to Bnip3 WT and 2SE, but not Bnip3 2SA (Fig. 6Aii). The loss of mitochondria was further quantified by MIFC, based on the fraction of the cellular population exhibiting a standardized loss of Tom20, relative to control cells (Fig. 6Aiii). This direct and quantitative measurement of mitochondrial degradation is more sensitive than population-averaged Western blotting and demonstrated that, relative to Bnip3 WT, Bnip3 2SE significantly increased mitochondrial loss, whereas Bnip3 2SA induced less mitochondrial loss. Single phosphorylation-mimicking mutants induced similar levels of mitophagy as Bnip3 WT, whereas non-phosphorylatable single mutants had reduced mitophagy. These results indicate that dual phosphorylation underlies not only sequestration but also degradation (i.e. mitophagy flux).
FIGURE 6.
Bnip3 LIR phosphorylation regulates maturation and autophagic degradation of mitochondria. A, mitophagy is enhanced by Bnip3 LIR phosphorylation. RFP-Bnip3 WT and the indicated multisite and single site mutant constructs were expressed in HeLa cells for 48 h. Ai, high resolution imaging of RFP-Bnip3-targeted mitochondria immunolabeled for Tom20 (green). Aii, Western blot analysis of mitochondrial protein levels. HeLa cells expressing RFP or the indicated RFP-Bnip3 construct were harvested 48 h after transfection and analyzed by Western blot. The levels of Tim23 and VDAC were quantified by densitometric analysis and normalized to RhoGDI. Aiii, use of MIFC to quantify subpopulations of cells with high mitophagy (low mitochondrial mass) through detection of mitochondrial Tom20 content in cell populations. Example phenotypes of high and low Tom20-labeled cells are shown below the graph. B, LIR phosphorylation enhances Bnip3 targeting to lysosomes. Bi, RFP-Bnip3 WT and indicated multisite and single site mutant constructs were co-expressed with the endolysosomal marker GFP-Rab7 in HeLa cells for 24 h. Shown are plot profiles of Bnip3 (light blue) and Rab7 (purple) demonstrating localizations of Bnip3-targeted mitochondria and autolysosomes. Bii, MIFC analysis of Bnip3 WT, 2SA, and 2SE colocalization with the endo-/lysosomal marker Rab7. The indicated Bnip3 constructs were expressed with GFP-LC3B in cardiac HL-1 cells for 24 h and imaged using the ImageStream X flow cytometer. Representative images and plot profiles are shown. Biii, using MIFC, the subpopulation of Bnip3 WT-targeted mitochondria with high GFP-Rab7 colocalization was quantified, gating on the fraction of cells with high colocalization. Population fractions are indicated as well as the percentage change of the two Bnip3 mutants compared with Bnip3 WT. Error bars, S.E.
As an additional index of mitophagy flux, the delivery of Bnip3-targeted mitochondria to lysosomes was measured. In MCF-7 cells at 24 h, Bnip3 WT-targeted mitochondria did not yet localize to GFP-Rab7-labeled endolysosomes and only partly in response to the S17E mutation. Remarkably, even at 24 h, colocalization of Rab7 was high with 2SE and S24E mutants (Fig. 6Bi). Colocalization of Bnip3 and Rab7 was quantified using MIFC (Fig. 6, Bii and Biii). At 24 h of expression, the fraction of HL-1 cardiac cells with Rab7 sequestration of Bnip3-targeted mitochondria was increased by the 2SE mutant and decreased by the 2SA mutant (Fig. 6Biii). Thus, mitochondrial sequestration promoted by mimicking LIR phosphorylation results in enhanced mitochondrial delivery to and degradation by lysosomes.
Bcl-xL Enhances Bnip3-induced Mitophagy, Depending on Bnip3 BH3 Domain and LIR Activity
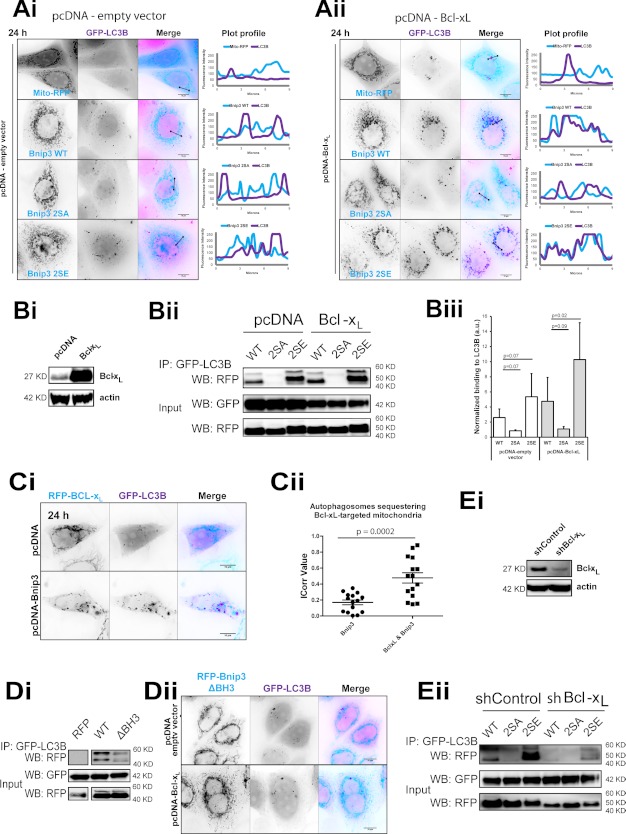
Bcl-2 inhibits Beclin-1 induction of autophagy (2), yet Bcl-xL has been shown to promote autophagosome production during mitochondrial apoptosis (47). Moreover, mitophagy can be active under conditions of high expression levels of Bcl-xL (9). Remarkably, increased Bcl-xL expression promoted the sequestration of Bnip3 WT- and 2SE-targeted mitochondria (Fig. 7A) and significantly enhanced LIR-dependent Bnip3 binding to LC3B of both Bnip3 WT and 2SE in Western blot analysis (Fig. 7B). Strikingly, Bcl-xL significantly localized within autophagosomes when co-expressed with WT Bnip3 (Fig. 7, Ci and Cii).
FIGURE 7.
Bcl-xL overexpression enhances autophagic sequestration of Bnip3-targeted mitochondria. A, Bcl-xL enhances Bnip3-induced mitochondrial sequestration. RFP-Bnip3 WT and the indicated multisite mutants were co-expressed with GFP-LC3B for 24 h and imaged by high resolution microscopy. As a control, MCF-7 cells were stably transfected with pcDNA3.1 (Ai), and MCF-7 cells were generated to stably overexpress Bcl-xL (Aii), shown in Bi. B, Bcl-xL enhances LIR-dependent Bnip3 binding to LC3B. RFP, RFP-Bnip3 WT, and 2SE were expressed in MCF-7 cells stably expressing GFP-LC3B, together with either Bcl-xL or vector control pcDNA3.1. Bi, Western blot detection of Bcl-xL expression in control pcDNA- and pcDNA-Bcl-xL-expressing MCF-7 cells. Bii, at 48 h of expression immunoprecipitations (IP) were performed with α-GFP. Shown is Western blot (WB) detection of RFP and GFP. Biii, immunoprecipitated Bnip3 was quantified by densitometric analysis and normalized to input. C, Bnip3 promotes Bcl-xL localization to autophagosomes. Ci, GFP-LC3B and RFP-Bcl-xL were co-expressed with either pcDNA3.1 or pcDNA-Bnip3 in HeLa cells for 24 h and imaged by high resolution microscopy. Plot profiles illustrate colocalizations between Bcl-xL-targeted mitochondria and autophagosomes. Cii, autophagosome sequestration of Bcl-xL-targeted mitochondria in the absence and presence of Bnip3 was quantified using the ImageJ colocalization color map plugin. Dot plots indicate the data set distribution and mean correlation index (Icorr) ± S.E. of at least 10 cells (Z-stacks) per condition. D, the Bnip3 BH3 domain is required for LC3B binding. RFP-fused WT Bnip3 and Bnip3 deleted for its BH3 domain (Bnip3ΔBH3) were expressed in MCF-7 cells stably expressing GFP-LC3B. Di, at 48 h of expression, immunoprecipitations were performed with α-GFP. Shown is Western blot detection of RFP and GFP. Dii, RFP-Bnip3ΔBH3 and GFP-LC3B were expressed in HeLa cells, in the presence and absence of pcDNA3.1-Bcl-xL. High resolution imaging was performed at 24 h. Plot profiles illustrate colocalizations between Bnip3ΔBH3-targeted mitochondria and autophagosomes. E, shRNA depletion of Bcl-xL reduced Bnip3 binding to LC3B. Ei, Western blot detection of Bcl-xL expression in control shRNA and Bcl-xL knock-down shRNA HeLa cells. Eii, at 24 h of expression, immunoprecipitations were performed with α-GFP. Shown is Western blot detection of RFP and GFP. Error bars, S.E.
We subsequently sought to elucidate the relationship between the anti-apoptotic functions of Bcl-xL and Bnip3-induced mitophagy. WT Bcl-xL and the Bax-binding mutant Bcl-xL-F131V (Mut1) (48) similarly promoted sequestration of Bnip3-targeted mitochondria (supplemental Fig. S5). In contrast, Bcl-xL-G138E/R139L/I140N (Mut2), which does not bind BH3-only proteins (49), uncoupled Bnip3-targeted mitochondria from autophagic sequestration. Bnip3 2SA did not induce autophagic sequestration of mitochondria even in the presence of Bcl-xL (Fig. 7, A and B). Although the BH3 domain may (13) or may not (50) be required for interactions with Bcl-xL, Bnip3 without a functional BH3 domain (Bnip3ΔBH3, described in Fig. 3Aii) had reduced binding to LC3B (Fig. 7Di). Furthermore, Bcl-xL overexpression did not enhance mitophagy in Bnip3ΔBH3-expressing cells (Fig. 7Dii). In addition, Bcl-xL was depleted in HeLa cells by shRNA (Fig. 7Ei). Consistent with a role for Bcl-xL in promoting mitophagy, Bcl-xL knockdown reduced Bnip3 2SE binding to GFP-LC3B (Fig. 7Eii).
These results indicate that Atg8-mediated mitophagy is dependent on BH3-binding anti-apoptotic activity of Bcl-xL as well as the Bnip3 BH3 domain and LIR phosphorylation.
Bnip3-induced Mitophagy Counteracts Apoptosis via Reduction of Cellular Cytochrome c Release Capacity
The relation between Bnip3-induced mitophagy and apoptosis could involve either pro-death or pro-survival functions. (i) Mitophagy may destabilize mitochondria and contribute to release of cytochrome c. (ii) Pro-apoptotic mitochondria may be recognized due to the presence of active Bnip3 as a receptor, and mitophagy may thereby serve as a reactive anti-apoptotic mechanism to scavenge dysfunctional mitochondria. (iii) Mitophagy may degrade mitochondria prior to their conversion to pro-apoptotic organelles to limit cytochrome c release (i.e. acting as a counteracting pathway to apoptosis).
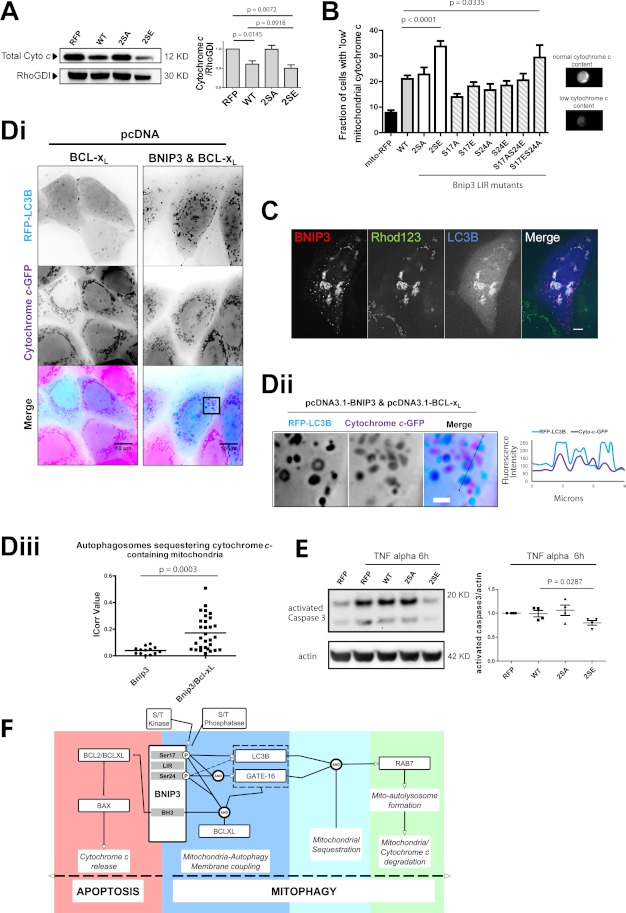
Cytochrome c was monitored as an index of mitochondrial apoptosis. Western blot analysis of total cellular lysates revealed that cytochrome c content was equivalent between control and Bnip3 2SA-transfected cells. Cytochrome c content was significantly decreased by Bnip3 WT and, to a slightly greater extent, Bnip3 2SE (Fig. 8A). MIFC analysis of mitochondrially localized cytochrome c provided further evidence of LIR regulation of cytochrome c content. The quantitative assessment of cell populations showed that mitochondrial cytochrome c was significantly reduced in response to Bnip3 2SE. In response to Bnip3 2SA, cytochrome c levels were comparable with WT. Notably, all single mutations maintained cytochrome c slightly above Bnip3 WT (Fig. 8B).
FIGURE 8.
Mimicked phosphorylation of Bnip3 LIR promotes mitochondrial cytochrome c degradation. A, quantification of cellular cytochrome c levels by Western blot. HeLa cells expressing RFP or the indicated Bnip3 construct were harvested 48 h after transfection and analyzed via Western blot. The levels of cytochrome c were quantified by densitometric analysis and normalized to RhoGDI. Cellular cytochrome c content was found decreased in RFP-Bnip3 WT and RFP-Bnip3 2SE-expressing cells, compared with cells expressing RFP. B, cellular cytochrome c content regulation by LIR phosphorylation. RFP-Bnip3 WT and indicated multisite and single site mutant constructs were expressed in HeLa cells for 48 h. Subpopulations of cells with low mitochondrial cytochrome c content were quantified by MIFC detection of mitochondrial cytochrome c. Example phenotypes of high and low cytochrome c-positive cells are shown below the graph. C, MCF-7 cells expressing RFP-Bnip3 2SE and BFP-LC3B for 24 h were stained with rhodamine 123 (Rhod123; 50 nm) to assess mitochondrial polarization state. D, Bnip3-induced mitophagy of cytochrome c-containing mitochondria is enhanced by Bcl-xL. Di, RFP-LC3B and Bcl-xL alone or in combination with Bnip3 were expressed in MCF-7 cells stably expressing cytochrome c-GFP. At 24 h, high resolution imaging was performed. Dii, enlarged region of Bnip3/Bcl-xL cell in i. The plot profile illustrates colocalizations between Bnip3-targeted mitochondria and autophagosomes in the enlarged region. iii, autophagosome sequestration of cytochrome c-containing mitochondria, in the presence of Bnip3 and Bnip3/Bcl-xL expression, was quantified using the ImageJ colocalization color map plugin. Dot plots indicate the data set distribution and mean correlation index (Icorr) ± S.E. of at least 10 cells (Z-stacks) per condition. E, effect of Bnip3-induced mitophagy on caspase 3 activation. Following 24 h of expression of RFP, RFP-Bnip3, RFP-Bnip3 2SA, or RFP-Bnip3 2SE, HeLa cells were treated with TNFα for 6 h and analyzed by Western blot. The levels of active caspase-3 were quantified by densitometric analysis and normalized to actin. F, schematic for Bnip3 regulation and interactions that form the cell fate decision of apoptosis versus mitophagy. Error bars, S.E.
Correlating these results with mitochondrial autophagy (Fig. 7) suggests that LIR-driven mitophagy reduces cellular contents of cytochrome c. Notably, whereas Bnip3 WT and 2SA similarly reduced mitochondrial levels of cytochrome c (Fig. 8B), total cytochrome c was the same between control and 2SA mutant (Fig. 8A). Furthermore, enhancing mitophagy with Bnip3 2SE further depleted cytochrome c from the cell, supporting a pro-survival function for Bnip3-induced mitophagy.
Next, we determined whether Bnip3-targeted mitochondria were sequestered prior to or following mitochondrial membrane permeabilization. Staining with rhodamine 123, which specifically labels actively respiring mitochondria, revealed polarized Bnip3 2SE-targeted mitochondria within LC3B-labeled mitophagosomes (Fig. 8C). In addition, MCF-7 cells were generated to stably express cytochrome c-GFP (30), and mitochondrial outer membrane permeabilization was blocked via overexpression of Bcl-xL. In response to Bcl-xL expression alone, cytochrome c-containing mitochondria did not colocalize with autophagosomes (Fig. 8Di). Remarkably, co-expression of Bnip3 and Bcl-xL resulted in significant sequestration of cytochrome c-containing mitochondria (Fig. 8, Dii and Diii). Thus, rather than a response to Bnip3-targeted mitochondria undergoing outer membrane permeabilization, Bnip3 LIR-driven mitophagy is a distinct pathway to reduce cellular cytochrome c release capacity.
Finally, HeLa cells were transfected with Bnip3 WT, 2SE, or 2SA for 24 h and treated for 6 h with TNFα to enhance apoptosis signaling. The reduction of cellular mitochondrial cytochrome c content, through expression of Bnip3 2SE, resulted in reduced activation of executioner caspase-3 (Fig. 8E), supporting our hypothesis that Bnip3-induced mitophagy limits mitochondrial amplification of apoptosis.
DISCUSSION
We here report, in cancer and non-cancer cells, that Bnip3 functions as a bona fide mitochondrial autophagy receptor via its LIR and that LIR phosphorylation positively regulates mitophagy flux. Our data suggest that LIR serine 17 phosphorylation of monomeric Bnip3 is required for Atg8 member binding and that LIR serine 24 phosphorylation increases LC3B binding and engages GATE-16. These interactions may serve as a seeding process for mitophagy, because CCCP enhanced mitochondrial sequestration in a LIR-dependent manner but did not alter LC3B binding to Bnip3. Thus, the process of Bnip3-induced mitophagy can be mechanistically classified according to the initial coupling of mitochondrial and autophagosomal membranes via Bnip3-Atg8 interactions, followed by sequestration and ultimately lysosomal entry and degradation of mitochondria (Fig. 8F).
LIR regulation by phosphorylation may be a fundamental regulatory mechanism for LIR-containing autophagy receptors. TBK1 (TANK-binding kinase 1)-mediated phosphorylation of the LIR of optineurin, a bacterial autophagy receptor, regulates autophagy of intracellular bacteria (27), and yeast Atg32 mitophagy requires phosphorylation distal to the LIR (26). Atg8/LC3 homologues are responsible for preautophagosomal structure nucleation and elongation, and Atg8/GABARAP homologues function in autophagosomal membrane closure (51), suggesting that different LIR phosphorylation states may coordinate individual steps of mitochondrial sequestration. Although we identified the Bnip3 LIR as a target for regulating mitophagy, the kinase and phosphatase responsible remain to be determined. Importantly, phosphorylation can also negatively alter LIR activity. During hypoxia, the mitochondrial outer membrane protein Fundc1 functions as an autophagy receptor in the absence of tyrosine phosphorylation within its LIR (52). Kinase and phosphatase thus may be considered as novel targets for positive and negative manipulation of specific autophagy activities.
Importantly, many autophagy receptors contain additional, post-transcriptionally regulated and pathway-integrated signaling domains (e.g. BH3 domains of Nix and Bnip3). As such, within a dynamic population of mitochondria, subcellular heterogeneities may generate dueling pathway behaviors, which decide the overall cellular response. Indeed, MIFC quantification of cell-to-cell variability revealed shifts in the population fraction of mitophagic cells, indicating that mitophagy constitutes a cell fate decision process. Moreover, the autophagy receptor Nix can promote Parkin-mediated recruitment of an additional autophagy receptor, p62, to mitochondria in response to CCCP (53), indicating that multiparametric and quantitative single-cell methodologies will be necessary to elucidate roles, function of cross-talk, and hierarchies between distinct modes of autophagy.
Dual functionality for apoptosis and autophagy-classified proteins is an emerging paradigm in programmed cell death pathways, with several proteins involved in autophagosome initiation also functioning to promote apoptosis (4–6). Here we demonstrate that Bnip3, a pro-death BH3-only protein, can likewise exhibit dual functionality, with pro-survival activity via LIR-dependent autophagy interactions. Our results directly evidence that by specifically enhancing LIR-mediated mitophagy, a cell can negatively regulate its cytochrome c release capacity. Previously, mitophagy was hypothesized to constitute a defense mechanism via removal of mitochondria following their conversion to pro-apoptotic organelles. We here show that Bnip3 proactively induces mitophagic removal of mitochondria prior to dysfunction (i.e. depolarization and release of cytochrome c), thereby lowering the cell's intrinsic apoptotic potential. The cell fate decision elucidated here (Fig. 8F) is a function of the LIR activity state and may be relevant to explain suggested contradicting cell survival versus cell death roles of Bnip3 in vivo ((19–21) (supplemental Fig. S6). Moreover, the presence of multiple, distinct modes of mitophagy indicate varied roles, also suggested by the opposing prognostic indicator values for Fundc1 and Bnip3 (supplemental Fig. S6).
It is remarkable that Bcl-xL, which binds Beclin-1 (2, 54), positively regulated Bnip3-induced mitophagy. Bcl-2 negatively regulates autophagy when targeted to the endoplasmic reticulum (55, 56). However, Bcl-xL mainly localizes to mitochondria (57) and was detected at mitochondria sequestered by autophagosomes only in the presence of Bnip3 with a functional LIR. As such, the Bcl-2 rheostat model (2) can be integrated with our findings; whereas the Bnip3 LIR is required to couple the membranes of autophagosome and mitochondria, canonical BH3-driven autophagy may drive sequestration. Our findings then support the emerging model of cross-talk whereby pro-survival Bcl-2 member action on autophagy is determined based on their subcellular distribution and activity (47). Moreover, the pro-survival activities of Bcl-xL are not only compatible but are fully integrated with pro-survival mitophagy.
In conclusion, phosphorylation regulation of LIR-Atg8 interactions may be a fundamental component of cell fate decisions involving mitochondria (23, 44, 52). Given the expanding number of mitophagy receptors, the elucidation of mode-specific, enhancing mechanisms, including phosphorylation and ubiquitination events, provides new targets for improved manipulation of cell death pathways.
Supplementary Material
Acknowledgments
We acknowledge the kind gifts of GFP-Rab7 from Dr. B. van Deurs (Copenhagen University), Bcl-xL from Dr. N. Morishima (RIKEN), GFP-LC3B from Dr. T. Yoshimori (Osaka University), and cytochrome c-GFP from Dr. D. Green (St. Jude Children's Research Hospital). We thank Dr. W. Claycomb for the HL-1 cells (Louisiana State University Health Sciences Center, New Orleans, LA). We thank Dr. A. Chiramel and X. Huang for technical assistance.
This work was supported through SBCancer within the Helmholtz Alliance on Systems Biology funded by the Initiative and Networking Fund of the Helmholtz Association and through the MedSys Project Multi-Scale Models (FKZ 0315416C), FORSYS-ViroQuant (0313923), and e:Bio (0316191) funded by the Federal Ministry of Education and Research (BMBF), Germany.

This article contains supplemental Figs. S1–S6.
- LIR
- LC3-interacting region
- VDAC
- voltage-dependent anion channel
- CCCP
- carbonyl cyanide p-chlorophenylhydrazone
- MIFC
- multispectral imaging-based flow cytometry
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Willis S. N., Adams J. M. (2005) Life in the balance. How BH3-only proteins induce apoptosis. Curr. Opin. Cell Biol. 17, 617–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pattingre S., Tassa A., Qu X., Garuti R., Liang X. H., Mizushima N., Packer M., Schneider M. D., Levine B. (2005) Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 122, 927–939 [DOI] [PubMed] [Google Scholar]
- 3. Maiuri M. C., Le Toumelin G., Criollo A., Rain J. C., Gautier F., Juin P., Tasdemir E., Pierron G., Troulinaki K., Tavernarakis N., Hickman J. A., Geneste O., Kroemer G. (2007) Functional and physical interaction between Bcl-XL and a BH3-like domain in Beclin-1. EMBO J. 26, 2527–2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Betin V. M., Lane J. D. (2009) Caspase cleavage of Atg4D stimulates GABARAP-L1 processing and triggers mitochondrial targeting and apoptosis. J. Cell Sci. 122, 2554–2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yousefi S., Perozzo R., Schmid I., Ziemiecki A., Schaffner T., Scapozza L., Brunner T., Simon H. U. (2006) Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat. Cell Biol. 8, 1124–1132 [DOI] [PubMed] [Google Scholar]
- 6. Rubinstein A. D., Eisenstein M., Ber Y., Bialik S., Kimchi A. (2011) The autophagy protein Atg12 associates with antiapoptotic Bcl-2 family members to promote mitochondrial apoptosis. Mol. Cell 44, 698–709 [DOI] [PubMed] [Google Scholar]
- 7. Hamacher-Brady A., Brady N. R., Gottlieb R. A., Gustafsson A. B. (2006) Autophagy as a protective response to Bnip3-mediated apoptotic signaling in the heart. Autophagy 2, 307–309 [DOI] [PubMed] [Google Scholar]
- 8. Yee K. S., Wilkinson S., James J., Ryan K. M., Vousden K. H. (2009) PUMA- and Bax-induced autophagy contributes to apoptosis. Cell Death Differ. 16, 1135–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schweers R. L., Zhang J., Randall M. S., Loyd M. R., Li W., Dorsey F. C., Kundu M., Opferman J. T., Cleveland J. L., Miller J. L., Ney P. A. (2007) NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc. Natl. Acad. Sci. U.S.A. 104, 19500–19505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang J., Ney P. A. (2009) Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ. 16, 939–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yasuda M., Han J. W., Dionne C. A., Boyd J. M., Chinnadurai G. (1999) BNIP3α. A human homolog of mitochondrial proapoptotic protein BNIP3. Cancer Res. 59, 533–537 [PubMed] [Google Scholar]
- 12. Chen G., Ray R., Dubik D., Shi L., Cizeau J., Bleackley R. C., Saxena S., Gietz R. D., Greenberg A. H. (1997) The E1B 19K/Bcl-2-binding protein Nip3 is a dimeric mitochondrial protein that activates apoptosis. J. Exp. Med. 186, 1975–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yasuda M., Theodorakis P., Subramanian T., Chinnadurai G. (1998) Adenovirus E1B-19K/BCL-2 interacting protein BNIP3 contains a BH3 domain and a mitochondrial targeting sequence. J. Biol. Chem. 273, 12415–12421 [DOI] [PubMed] [Google Scholar]
- 14. Hamacher-Brady A., Brady N. R., Logue S. E., Sayen M. R., Jinno M., Kirshenbaum L. A., Gottlieb R. A., Gustafsson A. B. (2007) Response to myocardial ischemia/reperfusion injury involves Bnip3 and autophagy. Cell Death Differ. 14, 146–157 [DOI] [PubMed] [Google Scholar]
- 15. Chen G., Cizeau J., Vande Velde C., Park J. H., Bozek G., Bolton J., Shi L., Dubik D., Greenberg A. (1999) Nix and Nip3 form a subfamily of pro-apoptotic mitochondrial proteins. J. Biol. Chem. 274, 7–10 [DOI] [PubMed] [Google Scholar]
- 16. Dorn G. W., 2nd. (2010) Mitochondrial pruning by Nix and BNip3: an essential function for cardiac-expressed death factors. J. Cardiovasc. Transl. Res. 3, 374–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen Y., Lewis W., Diwan A., Cheng E. H., Matkovich S. J., Dorn G. W., 2nd. (2010) Dual autonomous mitochondrial cell death pathways are activated by Nix/BNip3L and induce cardiomyopathy. Proc. Natl. Acad. Sci. U.S.A. 107, 9035–9042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sandoval H., Thiagarajan P., Dasgupta S. K., Schumacher A., Prchal J. T., Chen M., Wang J. (2008) Essential role for Nix in autophagic maturation of erythroid cells. Nature 454, 232–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Erkan M., Kleeff J., Esposito I., Giese T., Ketterer K., Büchler M. W., Giese N. A., Friess H. (2005) Loss of BNIP3 expression is a late event in pancreatic cancer contributing to chemoresistance and worsened prognosis. Oncogene 24, 4421–4432 [DOI] [PubMed] [Google Scholar]
- 20. Tan E. Y., Campo L., Han C., Turley H., Pezzella F., Gatter K. C., Harris A. L., Fox S. B. (2007) BNIP3 as a progression marker in primary human breast cancer. Opposing functions in in situ versus invasive cancer. Clin. Cancer Res. 13, 467–474 [DOI] [PubMed] [Google Scholar]
- 21. Leo C., Horn L. C., Höckel M. (2006) Hypoxia and expression of the proapoptotic regulator BNIP3 in cervical cancer. Int. J. Gynecol. Cancer 16, 1314–1320 [DOI] [PubMed] [Google Scholar]
- 22. Bellot G., Garcia-Medina R., Gounon P., Chiche J., Roux D., Pouysségur J., Mazure N. M. (2009) Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol. Cell Biol. 29, 2570–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Novak I., Kirkin V., McEwan D. G., Zhang J., Wild P., Rozenknop A., Rogov V., Löhr F., Popovic D., Occhipinti A., Reichert A. S., Terzic J., Dötsch V., Ney P. A., Dikic I. (2010) Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 11, 45–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hanna R. A., Quinsay M. N., Orogo A. M., Giang K., Rikka S., Gustafsson Å. B. (2012) Microtubule-associated protein 1 light chain 3 (LC3) interacts with Bnip3 protein to selectively remove endoplasmic reticulum and mitochondria via autophagy. J. Biol. Chem. 287, 19094–19104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Matsumoto G., Wada K., Okuno M., Kurosawa M., Nukina N. (2011) Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Mol. Cell 44, 279–289 [DOI] [PubMed] [Google Scholar]
- 26. Aoki Y., Kanki T., Hirota Y., Kurihara Y., Saigusa T., Uchiumi T., Kang D. (2011) Phosphorylation of serine 114 on Atg32 mediates mitophagy. Mol. Biol. Cell 22, 3206–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wild P., Farhan H., McEwan D. G., Wagner S., Rogov V. V., Brady N. R., Richter B., Korac J., Waidmann O., Choudhary C., Dötsch V., Bumann D., Dikic I. (2011) Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 333, 228–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rogers S., Wells R., Rechsteiner M. (1986) Amino acid sequences common to rapidly degraded proteins. The PEST hypothesis. Science 234, 364–368 [DOI] [PubMed] [Google Scholar]
- 29. Bucci C., Thomsen P., Nicoziani P., McCarthy J., van Deurs B. (2000) Rab7. A key to lysosome biogenesis. Mol. Biol. Cell 11, 467–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Goldstein J. C., Waterhouse N. J., Juin P., Evan G. I., Green D. R. (2000) The coordinate release of cytochrome c during apoptosis is rapid, complete, and kinetically invariant. Nat. Cell Biol. 2, 156–162 [DOI] [PubMed] [Google Scholar]
- 31. Morishima N., Nakanishi K., Tsuchiya K., Shibata T., Seiwa E. (2004) Translocation of Bim to the endoplasmic reticulum (ER) mediates ER stress signaling for activation of caspase-12 during ER stress-induced apoptosis. J. Biol. Chem. 279, 50375–50381 [DOI] [PubMed] [Google Scholar]
- 32. Neumann B., Held M., Liebel U., Erfle H., Rogers P., Pepperkok R., Ellenberg J. (2006) High-throughput RNAi screening by time-lapse imaging of live human cells. Nat. Methods 3, 385–390 [DOI] [PubMed] [Google Scholar]
- 33. Jaskolski F., Mulle C., Manzoni O. J. (2005) An automated method to quantify and visualize colocalized fluorescent signals. J. Neurosci. Methods 146, 42–49 [DOI] [PubMed] [Google Scholar]
- 34. Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., Barton G. J. (2009) Jalview version 2. A multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Guo K., Searfoss G., Krolikowski D., Pagnoni M., Franks C., Clark K., Yu K. T., Jaye M., Ivashchenko Y. (2001) Hypoxia induces the expression of the pro-apoptotic gene BNIP3. Cell Death Differ. 8, 367–376 [DOI] [PubMed] [Google Scholar]
- 36. Kim J., Kundu M., Viollet B., Guan K. L. (2011) AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 13, 132–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Johansen T., Lamark T. (2011) Selective autophagy mediated by autophagic adapter proteins. Autophagy 7, 279–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Graham R. M., Thompson J. W., Wei J., Bishopric N. H., Webster K. A. (2007) Regulation of Bnip3 death pathways by calcium, phosphorylation, and hypoxia-reoxygenation. Antioxid. Redox Signal. 9, 1309–1315 [DOI] [PubMed] [Google Scholar]
- 39. Nakanishi S., Matsuda Y., Iwahashi K., Kase H. (1986) K-252b, c and d, potent inhibitors of protein kinase C from microbial origin. J. Antibiot. 39, 1066–1071 [DOI] [PubMed] [Google Scholar]
- 40. Yoon S. Y., Choi J. E., Kweon H. S., Choe H., Kim S. W., Hwang O., Lee H., Lee J. Y., Kim D. H. (2008) Okadaic acid increases autophagosomes in rat neurons. Implications for Alzheimer's disease. J. Neurosci. Res. 86, 3230–3239 [DOI] [PubMed] [Google Scholar]
- 41. Holen I., Gordon P. B., Seglen P. O. (1993) Inhibition of hepatocytic autophagy by okadaic acid and other protein phosphatase inhibitors. Eur. J. Biochem. 215, 113–122 [DOI] [PubMed] [Google Scholar]
- 42. Stehmeier P., Muller S. (2009) Phospho-regulated SUMO interaction modules connect the SUMO system to CK2 signaling. Mol. Cell 33, 400–409 [DOI] [PubMed] [Google Scholar]
- 43. Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. (2000) LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19, 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mathew R., Karp C. M., Beaudoin B., Vuong N., Chen G., Chen H. Y., Bray K., Reddy A., Bhanot G., Gelinas C., Dipaola R. S., Karantza-Wadsworth V., White E. (2009) Autophagy suppresses tumorigenesis through elimination of p62. Cell 137, 1062–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kwon K. Y., Viollet B., Yoo O. J. (2011) CCCP induces autophagy in an AMPK-independent manner. Biochem. Biophys. Res. Commun. 416, 343–348 [DOI] [PubMed] [Google Scholar]
- 46. Narendra D., Tanaka A., Suen D. F., Youle R. J. (2008) Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183, 795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Luo S., Rubinsztein D. C. (2010) Apoptosis blocks Beclin 1-dependent autophagosome synthesis. An effect rescued by Bcl-xL. Cell Death Differ. 17, 268–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cheng E. H., Levine B., Boise L. H., Thompson C. B., Hardwick J. M. (1996) Bax-independent inhibition of apoptosis by Bcl-XL. Nature 379, 554–556 [DOI] [PubMed] [Google Scholar]
- 49. Cheng E. H., Wei M. C., Weiler S., Flavell R. A., Mak T. W., Lindsten T., Korsmeyer S. J. (2001) BCL-2, BCL-XL sequester BH3 domain-only molecules preventing BAX- and BAK-mediated mitochondrial apoptosis. Mol. Cell 8, 705–711 [DOI] [PubMed] [Google Scholar]
- 50. Ray R., Chen G., Vande Velde C., Cizeau J., Park J. H., Reed J. C., Gietz R. D., Greenberg A. H. (2000) BNIP3 heterodimerizes with Bcl-2/Bcl-XL and induces cell death independent of a Bcl-2 homology 3 (BH3) domain at both mitochondrial and nonmitochondrial sites. J. Biol. Chem. 275, 1439–1448 [DOI] [PubMed] [Google Scholar]
- 51. Weidberg H., Shvets E., Shpilka T., Shimron F., Shinder V., Elazar Z. (2010) LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 29, 1792–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liu L., Feng D., Chen G., Chen M., Zheng Q., Song P., Ma Q., Zhu C., Wang R., Qi W., Huang L., Xue P., Li B., Wang X., Jin H., Wang J., Yang F., Liu P., Zhu Y., Sui S., Chen Q. (2012) Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat. Cell Biol. 14, 177–185 [DOI] [PubMed] [Google Scholar]
- 53. Ding W. X., Ni H. M., Li M., Liao Y., Chen X., Stolz D. B., Dorn G. W., 2nd, Yin X. M. (2010) Nix is critical to two distinct phases of mitophagy, reactive oxygen species-mediated autophagy induction and Parkin-ubiquitin-p62-mediated mitochondrial priming. J. Biol. Chem. 285, 27879–27890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Oberstein A., Jeffrey P. D., Shi Y. (2007) Crystal structure of the Bcl-XL-Beclin 1 peptide complex. Beclin 1 is a novel BH3-only protein. J. Biol. Chem. 282, 13123–13132 [DOI] [PubMed] [Google Scholar]
- 55. Høyer-Hansen M., Bastholm L., Szyniarowski P., Campanella M., Szabadkai G., Farkas T., Bianchi K., Fehrenbacher N., Elling F., Rizzuto R., Mathiasen I. S., Jäättelä M. (2007) Control of macroautophagy by calcium, calmodulin-dependent kinase kinase-β, and Bcl-2. Mol. Cell 25, 193–205 [DOI] [PubMed] [Google Scholar]
- 56. Brady N. R., Hamacher-Brady A., Yuan H., Gottlieb R. A. (2007) The autophagic response to nutrient deprivation in the hl-1 cardiac myocyte is modulated by Bcl-2 and sarco/endoplasmic reticulum calcium stores. FEBS J. 274, 3184–3197 [DOI] [PubMed] [Google Scholar]
- 57. Kaufmann T., Schlipf S., Sanz J., Neubert K., Stein R., Borner C. (2003) Characterization of the signal that directs Bcl-xL, but not Bcl-2, to the mitochondrial outer membrane. J. Cell Biol. 160, 53–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.