Background: The γ complex loads the ring-shaped β sliding clamp onto DNA.
Results: The rate of β clamp closing is faster than the rate of β release on DNA, and the first turnover of ATP hydrolysis is faster than β closing.
Conclusion: Clamp closing occurs before clamp release but after a burst of ATP hydrolysis.
Significance: These results demonstrate that clamp release around DNA is a two-step process.
Keywords: ATPases, DNA Enzymes, DNA Replication, Enzyme Kinetics, Pre-steady-state Kinetics, Protein-Protein Interactions, Clamp Loader, Sliding Clamp
Abstract
Escherichia coli γ complex clamp loader functions to load the β sliding clamp onto sites of DNA replication and repair. The clamp loader uses the energy of ATP binding and hydrolysis to drive conformational changes allowing for β binding and opening, DNA binding, and then release of the β·DNA complex. Although much work has been done studying the sliding clamp and clamp loader mechanism, kinetic analysis of the events following β·γ complex·DNA formation is not complete. Using fluorescent clamp closing and release assays, we show that β closing is faster than β release, indicating that γ complex closes β before releasing it around DNA. Using a fluorescent ATP hydrolysis assay, we show that there is a burst of ATP hydrolysis before β closing and that β release may be the rate-limiting step in the overall clamp loading reaction. The combined use of these fluorescent assays provides a unique perspective into the E. coli clamp loader by providing a measure of the relative timing of different events in the clamp loading reaction, helping to elucidate the complicated clamp loading mechanism.
Introduction
Sliding clamps and clamp loaders are present in all kingdoms of life performing vital functions in both DNA replication and repair (reviewed in Ref. 1). Sliding clamps are ring-shaped proteins that encircle DNA to secure proteins to and coordinate enzyme activity at a specific region of DNA (2, 3). The main function of the sliding clamp is to secure DNA polymerase to the replication fork, increasing the processivity of nucleotide polymerization from tens of nucleotides per polymerase binding event to thousands of nucleotides per polymerase binding event (reviewed in Refs. 4 and 5). The sliding clamp also plays a role in coordinating proteins involved in Okazaki fragment maturation and translesion synthesis, as well as many other functions in DNA metabolism.
The sliding clamp in Escherichia coli, β, is a homodimer composed of three globular domains in each monomer. The monomers are arranged head to tail creating a conserved toroidal structure with an interior hole large enough to fit duplex DNA (2, 3). The two faces of β are asymmetric, and proteins, including the clamp loader and DNA polymerase, bind to a hydrophobic pocket on one face of β via a conserved sequence motif (6–8). Multiple lines of evidence support a closed conformation for the β clamp in solution. In numerous crystal structures, the β clamp is in a closed conformation (2, 3, 9–11). Molecular dynamics simulations demonstrate that β is most stable in its closed conformation (12). β has a high half-life on closed circular DNA of ∼72 min, indicating that transient β opening allowing dissociation from DNA occurs infrequently (13).
Due to the stable closed ring structure of the β clamp, a clamp loader is required to open β and place it around DNA in the appropriate conformation as well as direct it to the appropriate location for DNA replication or repair. The clamp loader, γ complex, is a member of the AAA+ family of ATPase enzymes, which are characterized by the use of ATP binding and hydrolysis to drive reactions that typically relocate or rearrange macromolecules (reviewed in Refs. 14–16). The γ complex is a heptamer, composed of three copies of the γ subunit and one copy each of δ, δ′, χ, and ψ, with the three γ subunits binding and hydrolyzing ATP (17, 18). The clamp loader core (γ3δδ′) forms a highly conserved cap-like structure with subunits arranged in a circular fashion and a gap between the δ and δ′ subunits (18). The γ complex loads β on primed template junctions with a 3′ recessed end. The duplex region of DNA sits inside the cap, stabilized by positively charged residues, and the single-stranded overhang bends out of the cap through the gap between δ and δ′ (19).
The β clamp is estimated to be loaded every 2–3 s during lagging strand synthesis, one clamp for every Okazaki fragment. This reaction must happen in a timely and ordered fashion so that lagging strand synthesis does not become uncoupled from leading strand synthesis, causing replication delays and increasing the probability of replication fork collapse. Therefore, to maximize efficiency, an ordered mechanism for γ complex loading β likely exists that is driven by conformational changes associated with ATP binding and hydrolysis and interactions with β and DNA. In general, ATP binding by γ complex drives β binding and opening and DNA binding (20–22). DNA binding triggers ATP hydrolysis and results in release of β·DNA (22–24). Pre-steady-state kinetic experiments have given a more detailed view of the clamp loading cycle. β binding occurs before β opening, indicating that γ complex does not simply trap open clamps, but instead opens clamps (11). γ complex binding to β occurs at a rate limited by diffusion, whereas binding to DNA is slower and limited by a conformational change in γ complex (25). A burst of ATP hydrolysis occurs before γ complex releases β, and DNA release occurs before β release (24, 26, 27).
The high stability of clamps in the closed conformation suggests the need for clamp loaders to open and stabilize the open clamps long enough to allow loading around DNA instead of simply capturing open clamps (11). The high stability of the β clamp in a closed ring conformation, with a Kd in the pm range and a half-life on DNA of over an hour (13, 28), also implies that clamp loaders are not needed to close the clamps around DNA. Instead, open clamps could be released first and “snap shut” after release as implied by computational studies, which indicate that replication factor C simply stabilizes the open conformation of proliferating cell nuclear antigen (29). This scenario could potentially result in an open clamp slipping away from DNA before it snaps shut, resulting in an unsuccessful attempt at loading the clamp, which would reduce the overall efficiency of clamp loading. In contrast, molecular dynamics simulations propose that closed β molecules are under “spring tension,” which facilitates opening by the clamp loader (12). If this were the case, then this would also suggest that the β clamp could not simply snap shut after being released and that γ complex would need to exert energy to close the clamp around DNA.
If required, energy to promote clamp closing could potentially come from hydrolysis of some or all of the bound molecules of ATP, which raises the question of the relative timing of ATP hydrolysis and β closing. A recent structural study with the T4 bacteriophage clamp loader suggested that hydrolysis of one ATP molecule causes a conformational change in the clamp loader·clamp complex that closes the clamp (30), and studies with the E. coli replisome suggest that hydrolysis of one ATP molecule is sufficient to form initiation complexes but hydrolysis of 3 molecules accelerates the process (31, 32). Studies have shown that ATP hydrolysis is required for clamp release (24, 26, 33), but the question here is whether ATP hydrolysis is required for clamp closing or whether binding of the clamp loader·clamp complex to DNA is sufficient to promote clamp closing. To address the timing of β closing and how it correlates with the other steps in the clamp loading cycle, most notably ATP hydrolysis and β release, a fluorescent β closing assay was used to measure clamp closing rates for comparison with rates of other events.
EXPERIMENTAL PROCEDURES
Buffers, Proteins, and DNA
Final buffer concentrations in the reactions were 20 mm Tris·HCl, pH 7.5, 50 mm sodium chloride, 8 mm magnesium chloride, 0.5 mm EDTA, 4% glycerol, and 2 mm DTT. Storage buffer for β was 20 mm Tris·HCl, pH 7.5, 10% glycerol, 0.5 mm EDTA, and 2 mm DTT. Storage buffer for γ complex was the same as for β, except that 50 mm sodium chloride was added. γ complex subunits (γ, δ, δ′, χ, and ψ) were purified and reconstituted as described previously (27, 34–37). Wild type β was purified as described previously (38). Purification and labeling of the β clamp mutants for the β closing and release assays are discussed in more detail below.
Synthetic oligonucleotides (Integrated DNA Technology) were purified using 10–12% denaturing polyacrylamide gel electrophoresis. Two different p/t-DNA2 substrates were used. The first substrate was made by annealing two 60-mers to create a symmetrical structure with a 30-nucleotide duplex region and two 30-nucleotide 5′-single-stranded overhangs (see Figs. 1–4). The second p/t-DNA substrate consisted of a 30-nucleotide primer annealed to a 60-nucleotide template to create a 30-nucleotide duplex region and a 30-nucleotide 5′-single-stranded overhang (see Fig. 5).
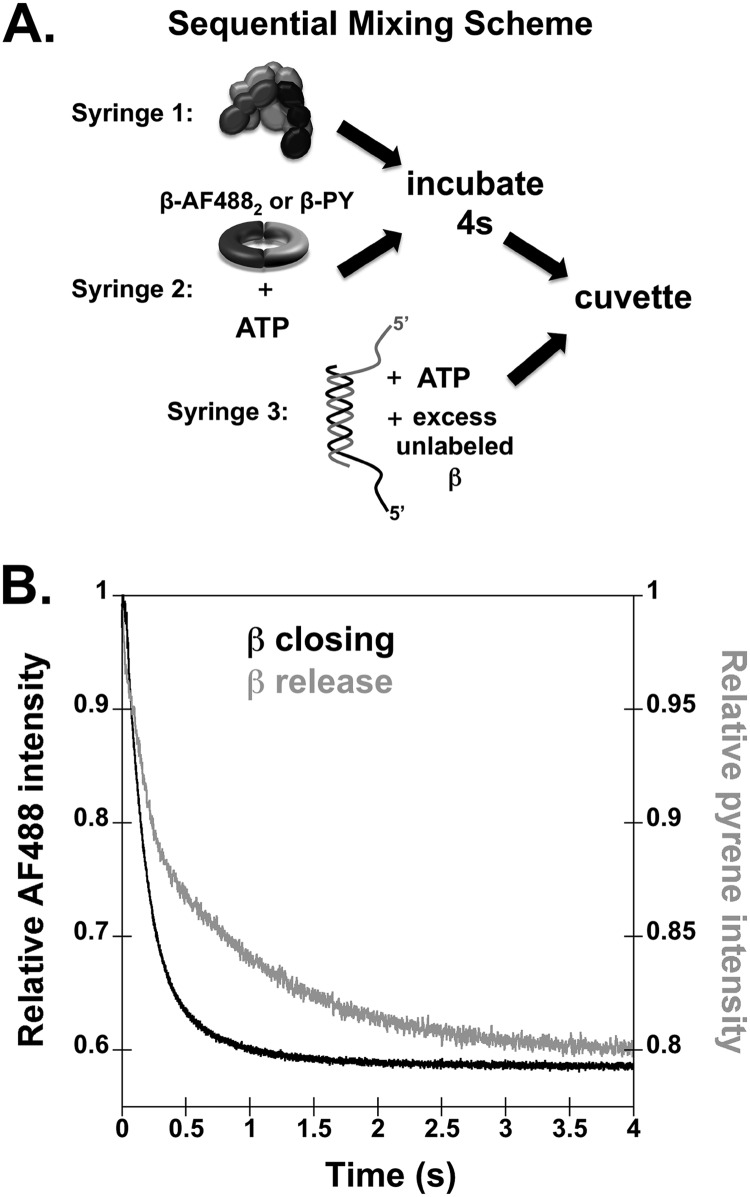
FIGURE 1.
Time courses for β sliding clamp closing and release onto DNA. A, a diagram of the sequential mix stopped-flow reaction scheme is shown. B, β closing and release reactions were performed with final concentrations of 20 nm γ complex, 20 nm labeled β, 40 nm p/t-DNA, 0.5 mm ATP, and 200 nm unlabeled β trap. Symmetrical p/t-DNA substrates were made by annealing two 60-mers to create a 30-nucleotide duplex and two 30-nucleotide 5′ template overhangs. Representative reaction time courses are shown for clamp closing measured in reactions with β-AF4882 (black trace) and for clamp release measured with β-PY (gray trace).
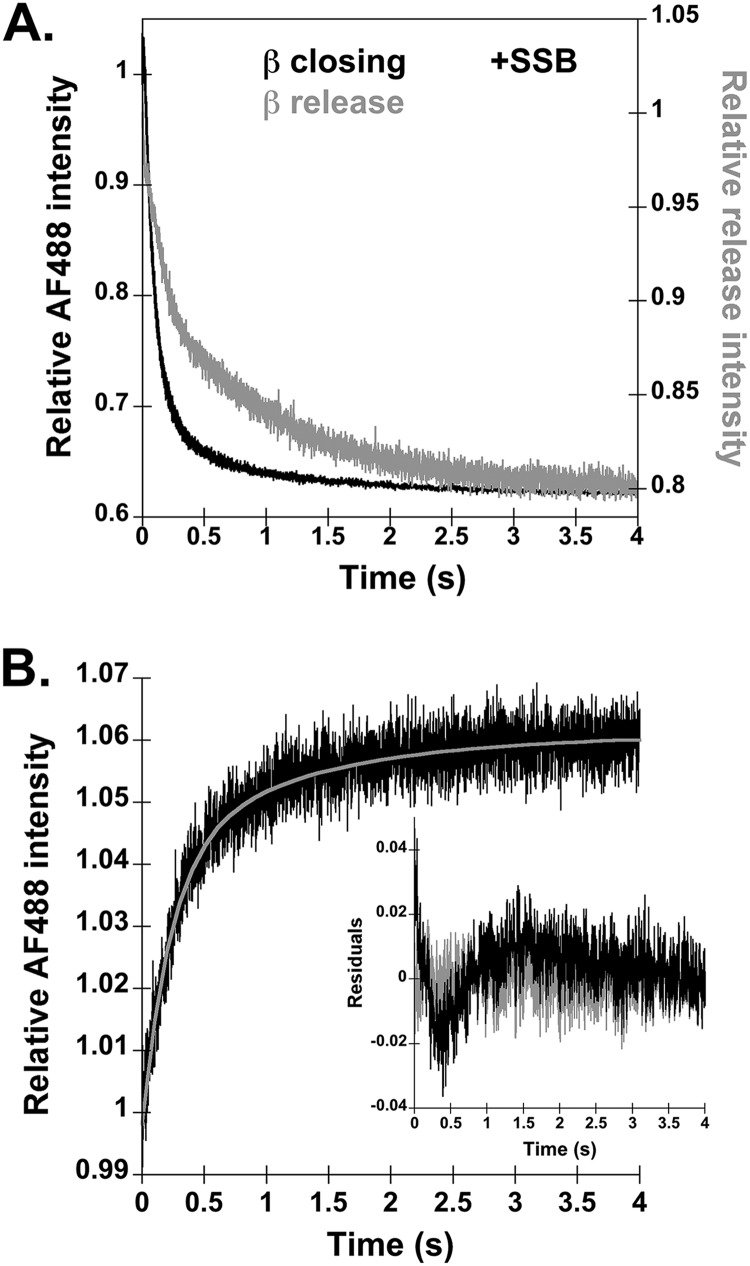
FIGURE 2.
Time courses for β release measured in reactions with SSB and measured by a FRET assay. A, β closing and release reactions from Fig. 1 were repeated except that 400 nm SSB was added to block the single-stranded DNA ends. Representative time courses are shown for β closing measured in reactions with β-AF4882 (black trace) and for β release measured with β-PY (gray trace). B, β release was measured with an alternative FRET-based assay, where γ complex is labeled with the fluorophore and β is labeled with a nonfluorescent quencher. Release reactions were performed with final concentrations of 20 nm γ complex-AF488, 20 nm β-QSY9, 40 nm p/t-DNA, 0.5 mm ATP, and 200 nm unlabeled β trap. The inset graph shows the residuals of both single (black) and double (gray) exponential fits of the FRET-based β release experiment.
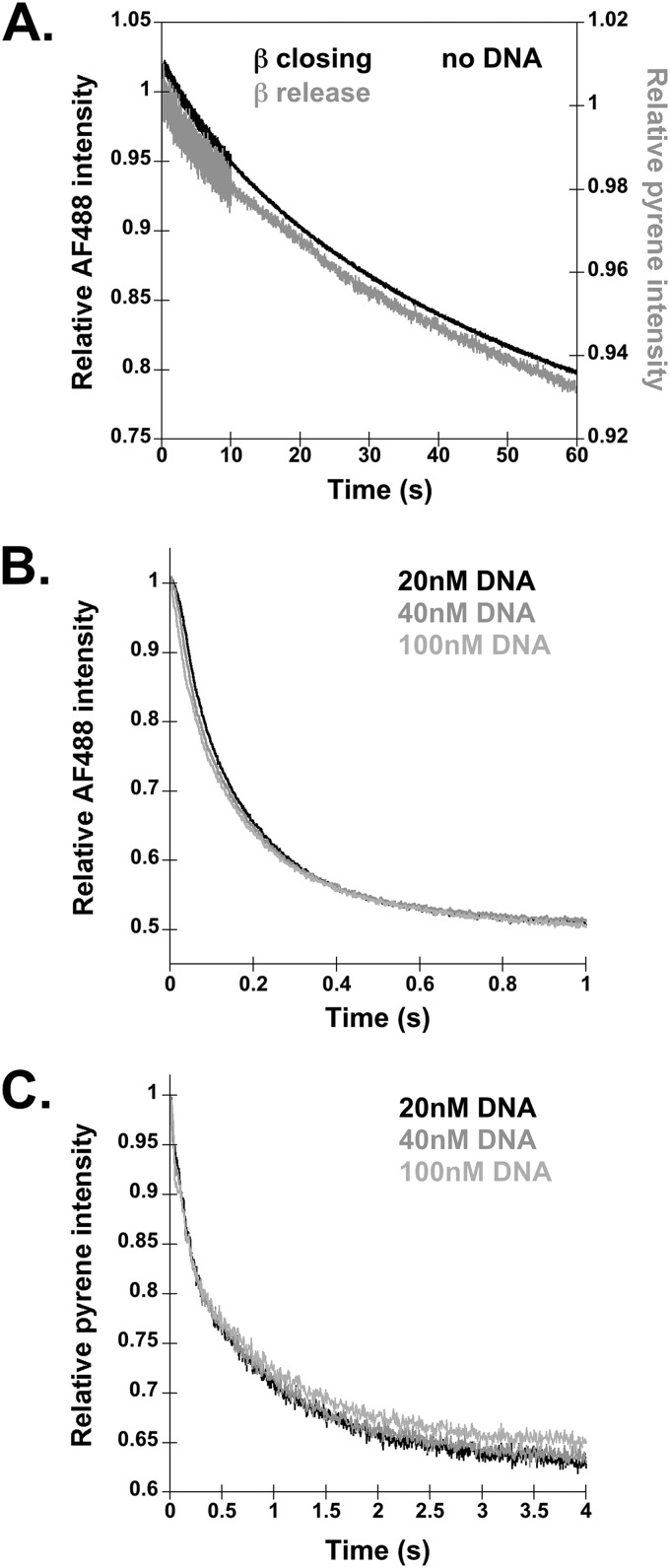
FIGURE 3.
Time courses for β closing and release in the absence of DNA and as a function of DNA concentration. A, β closing and release assays were performed omitting DNA, using the same sequential mixing scheme outlined in Fig. 1A. Final reactions contained 20 nm γ complex, 20 nm labeled β, 0.5 mm ATP, and 200 nm unlabeled β trap. Representative reaction time courses for clamp closing and clamp release are shown in black and gray, respectively. B and C, β closing (B) and β release (C) were measured in clamp loading reactions with varying concentrations of DNA. Representative traces for β closing (B) and release (C) with 20 nm (black trace), 40 nm (gray trace), and 100 nm DNA (light gray trace) are shown.
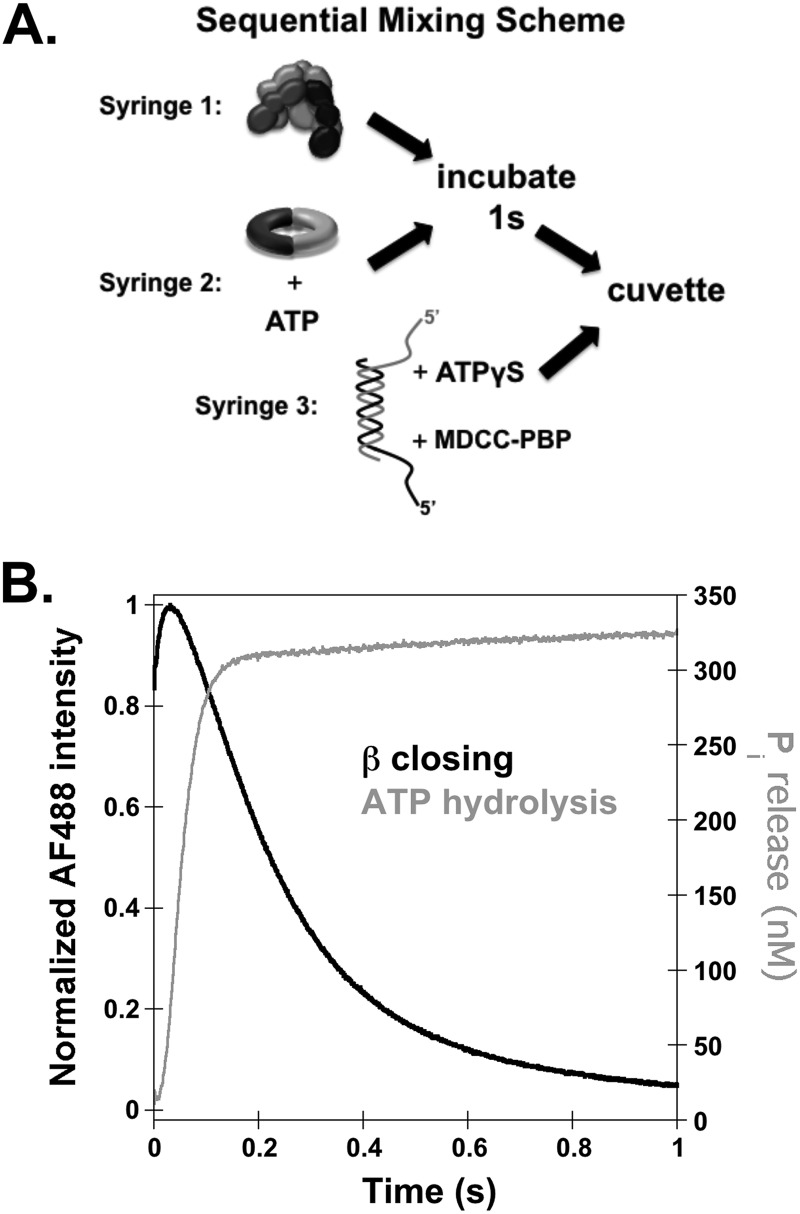
FIGURE 4.
Temporal correlation of β closing and ATP hydrolysis in single-turnover reactions. A, a diagram of the sequential mix stopped-flow reaction scheme for measuring ATP hydrolysis is shown. Final reactions contained 200 nm γ complex, 200 nm β, 400 nm p/t-DNA, 0.2 mm ATP, 2 μm MDCC-PBP, and 2 mm ATPγS. B, a representative trace of ATP hydrolysis as measured by MDCC fluorescence and converted to concentration of inorganic phosphate released (gray trace) was overlaid with a trace of β closing (black trace) at a final concentration of 200 nm γ complex, 200 nm labeled β, 400 nm p/t-DNA, 0.5 mm ATP, and 2 μm unlabeled β trap.
FIGURE 5.
Temporal order of β closing and ATP hydrolysis in multiple-turnover reactions. Multiple-turnover β closing (black trace) and ATP hydrolysis (gray trace) representative traces are shown. All reactions were performed using the stopped flow sequential mixing scheme illustrated in Figs. 1A and 4A but omitting the unlabeled β and ATPγS chases. Final concentrations are 200 nm γ complex, 200 nm labeled β, 400 nm p/t-DNA, and 0.5 mm ATP for the β closing reactions and are the same for ATPase assays except for the inclusion of 2 μm PBP-MDCC and 200 nm unlabeled β in place of the labeled β.
Pre-steady-state Stopped Flow
Pre-steady-state reactions were performed using an Applied Photophysics SX20 MV stopped-flow apparatus at 20 °C with 3.72-nm band pass. Assays were performed in sequential mix mode in which a solution of γ complex was mixed with a solution of β and ATP and incubated for 1–4 s before adding a solution of DNA and ATP.
Clamp Closing and Release Assays
The β closing assay uses clamps labeled with 2 Alexa Fluor 488 (AF488) molecules covalently attached to Cys-103 and Cys-305 on either side of the monomer interfaces (11). The β release assay uses clamps covalently labeled with pyrene (PY) at Cys-299 on the surface of the clamp to which γ complex binds (25). The FRET-based β release assay uses γ complex labeled with AF488 on the δ′ subunit and a β clamp labeled with the nonfluorescent quencher QSY9 (27). Pre-steady-state clamp closing and release reactions were limited to a single turnover by including an unlabeled β trap with the DNA and ATP solution. The final concentrations in these reactions were: 20 nm γ complex, 20 nm β, 40 nm DNA (when present), 0.5 mm ATP, and 200 nm unlabeled β unless otherwise noted. β-PY was excited at 345 nm, and emission was measured with a 365-nm cut-on filter. β-AF4882 and γ complex-AF488 were excited at 490 nm, and emission was measured with a 515-nm cut-on filter. Observed rates were calculated for β closing reactions using a single exponential decay equation (Equation 1), and observed rates for β release reactions were calculated using a double exponential decay equation (Equation 2). Observed rates for FRET β release assays were calculated using a double exponential increase equation (Equation 3).
ATP Hydrolysis Assay
The ATP hydrolysis assay uses phosphate-binding protein labeled with MDCC (PBP-MDCC, Invitrogen) to quantify the inorganic phosphate product of ATP hydrolysis (39, 40). PBP-MDCC was included in the DNA and ATP solution along with an excess of ATPγS as a trap. Final concentrations were: 200 nm γ complex, 200 nm β, 400 nm DNA, 0.2 mm ATP, 2 μm ATPγS, and 2 μm PBP-MDCC. MDCC was excited at 425 nm, and emission was measured with a 455-nm cut-on filter. Steady-state rates of ATP hydrolysis were calculated by first converting the fluorescent signal into the concentration of inorganic phosphate released. The steady-state phase was fit to a line by linear regression.
RESULTS
β Clamps Are Closed before They Are Released
To determine the relative timing of β closing and release, fluorescence-based assays were used to report on these aspects of the clamp loading cycle. β closing was measured using a β clamp labeled on each monomer at the interface with AF488. When the clamp, β-AF4882, is open, the AF488 fluorophores are far enough apart that there is no interaction and they fluoresce, but when the clamp closes, the fluorophores return to a position in which they are close enough to self-quench, decreasing the signal (11). β release is measured using a clamp that is covalently labeled with PY on the surface to which the clamp loader binds. When β-PY is bound by γ complex, PY is about two times more fluorescent than when the clamp is released and unbound (25). Reactions were initiated by adding a solution of γ complex, labeled β, and ATP to a solution of DNA, ATP, and a 10-fold excess of unlabeled β (Fig. 1A). The unlabeled β limited reactions to a single turnover. Experiments were done three separate times, and a representative time course for β closing is shown in Fig. 1B (black trace). In the first ∼40 ms of each β closing time course, there is a small increase in fluorescence signal (see Fig. 3B for an expanded time scale). It is unclear what causes this increase, but it is unlikely to arise from the interaction with DNA because it is also present in experiments in which DNA is omitted (data not shown). It is more pronounced when protein concentrations are higher (see Fig. 4B). Because a unique fit to the rate of the small rise was difficult to obtain, the time course was fit to a single exponential decay to calculate an observed rate of release. The average rate calculated from three separate closing reactions was 6.2 ± 1.3 s−1.
Time courses for β release gave biphasic decreases in PY fluorescence (Fig. 1B, gray trace). Fits of the data to a double exponential decay gave an initial rate of 6.3 ± 0.3 s−1 and a second rate of 0.85 ± 0.04 s−1. The rapid decrease in PY fluorescence occurs at the same rate as closing measured in β-AF4882 assays and is interpreted as being associated with a conformational change in the clamp loader·clamp complex that allows the clamp to close. The second, slower rate of 0.85 s−1 is interpreted as reflecting the rate of β release by the clamp loader. This interpretation of the biphasic decrease in PY fluorescence is consistent with previous equilibrium binding studies that show that the fluorescence of PY is sensitive to the γ complex·β conformation (11, 25). In the absence of ATP, γ complex can bind but not open β (11, 21, 22). In this closed conformation of γ complex·β, PY fluorescence is weaker (i.e. lower quantum yield) than PY fluorescence in an open clamp loader·clamp complex (11, 25). Similarly, PY fluorescence is lower in complexes of arginine finger mutants of γ complex and β, where the γ complex mutants are defective in β opening even in the presence of ATP. The amplitudes of the two phases in the β-PY release assay are consistent with the relative quantum yields of PY in open and closed clamp loader·clamp complexes.
The biphasic release rates are not likely to represent two populations of γ complex because amplitudes of the two rates are consistent between multiple preparations of γ complex. If there were two populations of γ complex, one that reacted more rapidly than the second, then two phases would also be expected in clamp closing (AF488) assays and in other assays, but this was not observed.
An alternative interpretation of the biphasic nature of the β-PY release traces is that the first rate reflects β-PY release, and the second rate reflects loss of the interaction between β-PY and DNA as the clamp slides off the short linear p/t-DNA substrates. To test this possibility, two experiments were performed. In the first experiment, β closing and release were measured on p/t-DNA bound by SSB to block the ends and prevent β from sliding off the DNA. In vitro DNA replication assays demonstrated that an ssDNA overhang of 25 nucleotides is of sufficient length to bind SSB and stabilize β on DNA to form a preinitiation complex to which the core polymerase can bind (41). In assays with SSB-bound p/t-DNA, the decrease in PY fluorescence remained biphasic (Fig. 2A). Moreover, the calculated closing rates measured in the β-AF4882 assay and closing and release rates measured in the β-PY assay were about the same as measured in the absence of SSB (Table 1). If the slower rate measured in the β-PY assay had corresponded to β-PY sliding off DNA, this rate should have decreased in assays with SSB. This experiment also shows that protein-protein interactions with SSB via the χ subunit (42, 43) of the γ complex have little influence on the rates of β closing and release.
TABLE 1.
Rate constants calculated for β closing and release reactions
Standard deviations are given for rate constants calculated from three independent experiments.
| Substrates | kobs, closing | kobs, release,1 | kobs, release,2 |
|---|---|---|---|
| s−1 | s−1 | s−1 | |
| p/t-DNA | 6.2 ± 1.3 | 6.3 ± 0.3 | 0.85 ± 0.04 |
| p/t-DNA + SSB | 8.7 ± 0.6 | 5.2 ± 1.3 | 0.5 ± 0.3 |
| No DNA | 0.03 ± 0.01 | 0.04 ± 0.02 | NAa |
| p/t-DNA + ATPγSb | 0.078 ± 0.002 | 0.055 ± 0.007 | NAa |
a NA, not applicable.
b ATPγS was substituted for ATP.
A second experiment to show that the slow phase in β-PY time courses is likely due to β release and not due to interactions with DNA used an alternative FRET-based assay to measure β release. In this assay, the δ′ subunit of γ complex is labeled with a fluorescent AF488 donor, and the β clamp is labeled with a nonfluorescent QSY9 quencher (27). When β-QSY9 is bound to γ complex-AF488, the quencher is in close proximity to the AF488 fluorophore, so the fluorescent signal is quenched, but when the clamp is released, the QSY9 is no longer close enough to quench the AF488 signal. This reaction was performed as illustrated in Fig. 1A by incubating γ complex-AF488 with β-QSY9 for 4 s before the addition of DNA and excess unlabeled β. Time courses for this reaction were also biphasic, with an initial calculated rate of 4.1 ± 0.2 s−1 and a second calculated rate of 0.82 ± 0.06 s−1 (Fig. 2B). Again, the rapid increase is interpreted as clamp closing, which moves AF488 farther from the QSY9 quencher, and the slower increase is interpreted as dissociation of β. Because β-QSY9 is not fluorescent, interactions between β and DNA cannot be responsible the slower phase of the reaction. Taken together, these results show that the rapid decrease in fluorescence in β-PY assays reflects β closing and that the slow decrease reflects β release.
β Closing before Release Requires DNA-dependent ATP Hydrolysis
Clamps can dissociate from the γ complex by two mechanisms, a productive clamp loading reaction in which β is released on DNA and a passive dissociation reaction reflecting the equilibrium interaction between β and the γ complex. In the absence of DNA, passive dissociation is the only mechanism by which β can dissociate from the γ complex. To determine whether clamp closing is also faster than clamp release when the clamp passively dissociates from the clamp loader, closing and release assays were performed in the absence of DNA (Fig. 3A). When DNA is omitted, both the closing and the release rates are much slower than for a productive clamp loading reaction on DNA. The closing rate is about 200 times slower in assays without DNA (Table 1). Interestingly, rates calculated for β closing and release in the absence of DNA are the same within experimental error. In addition, the clamp (β-PY) release trace was no longer biphasic, but could be fit by a single rate. These data show that clamp closing is not faster than release in the passive dissociation reaction. β closing and release may be simultaneous, or more likely, the clamp may rapidly close on its own after dissociation from the γ complex.
These data show that DNA binding promotes β closing prior to β release. Given that DNA binding is a bimolecular reaction, the rates of clamp closing and release may increase with increasing DNA concentrations. To determine how clamp closing and release rates are influenced by DNA concentration, both reactions were measured at DNA concentrations of 20, 40, and 100 nm (Fig. 3, B and C). Changing the DNA concentration had no effect on rates of clamp closing or release, showing that these rates must be limited by the rate of some intramolecular reaction such as a DNA-induced conformational change in the clamp loader·clamp complex or ATP hydrolysis.
DNA binding triggers ATP hydrolysis by the γ complex to promote release of β on DNA in a productive clamp loading reaction (24, 26). To determine whether DNA binding alone is required to activate clamp closing prior to release or whether ATP hydrolysis is required, β-AF4882 closing and β-PY release were measured in assays in which the nonhydrolyzable analog, ATPγS, was substituted for ATP. Time courses for reactions with ATPγS were similar to time courses for reactions lacking DNA. Calculated rates for clamp closing were about the same in reactions with ATPγS as in reactions lacking DNA (Table 1), and the rate of clamp closing was the same as the rate of clamp release. Most likely, clamp closing and release occur via a passive dissociation mechanism rather than an active clamp loading mechanism in reactions with ATPγS (44).
Together, these data show that DNA-dependent ATP hydrolysis is required for β to close prior to release by the γ complex and that the rate of clamp closing is limited by an intramolecular reaction in the ternary clamp loader·ATP·clamp·DNA complex. When DNA is omitted from the reactions or ATPγS is present, β closing and release occur with the same observed rates, about 200-fold slower than when DNA is present.
A Burst of ATP Hydrolysis Occurs before the β Clamp Closes
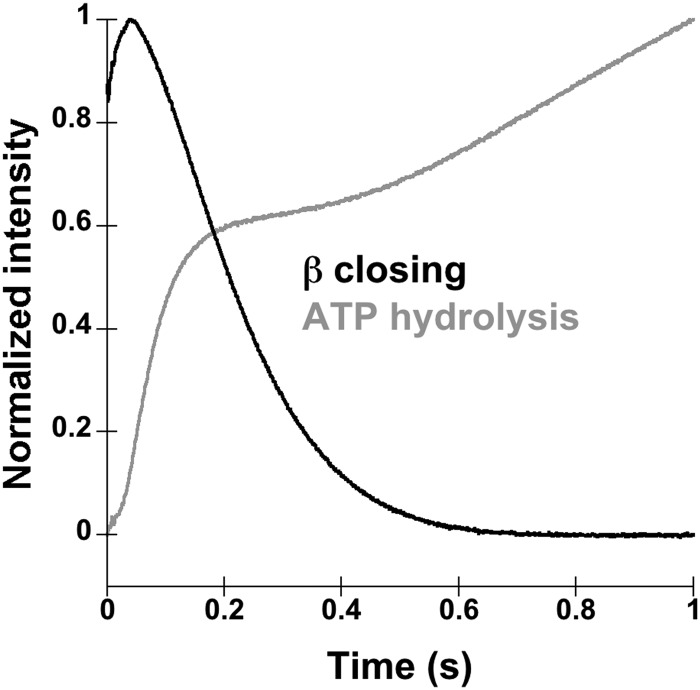
A fluorescent ATPase assay was used to measure the timing of ATP hydrolysis relative to β closing. ATP hydrolysis is quantified using phosphate-binding protein (PBP) labeled with MDCC, which increases in fluorescence when the inorganic phosphate product of ATP hydrolysis binds to MDCC-PBP (39, 45). ATPase assays were done using a sequential mixing scheme in which a solution of γ complex and a solution of β and ATP were incubated for 1 s before adding a solution of p/t-DNA, PBP-MDCC, and excess ATPγS (Fig. 4A). ATP hydrolysis reactions were limited to a single turnover by the addition of nonhydrolyzable ATPγS. Final concentrations were 200 nm γ complex, 200 nm β, 400 nm p/t-DNA, 200 μm ATP, 2 mm ATPγS, and 2 μm PBP. Clamp closing assays were done using identical concentrations except that 200 nm β-AF4882 and 2 μm unlabeled β were used instead of PBP-MDCC and ATPγS. Fig. 4B shows the time course for ATP hydrolysis by γ complex (gray trace) overlaid on a time course for β closing (black trace). A burst of ATP hydrolysis is complete in just over 0.1 s, whereas the decrease in β-AF488 fluorescence is only about 20% complete in the same time, showing that ATP is hydrolyzed before the clamp closes.
The γ complex contains three active ATP sites, one in each γ subunit, and can hydrolyze up to 3 molecules of ATP. Based on the amplitude of the burst of ATP hydrolysis, 1.7 molecules of Pi per γ complex were released in the rapid burst. Previous work has shown that the γ complex binds 3 molecules of ATP (46) and that all ATP sites hydrolyze ATP within this burst phase (26, 27, 40). It is not clear why our burst amplitudes correspond to fewer than 3 molecules of ATP. Possibilities include that some of the sites exchange ATP for ATPγS faster than ATP is hydrolyzed, a fraction of our clamp loaders or ATP sites are not active for hydrolysis, or the last inorganic phosphate molecule is released too slowly to be seen in our assay. Regardless of the number of ATP molecules hydrolyzed, the important point here is that ATP is hydrolyzed prior to clamp closing by at least some of the ATP sites. Any molecules of ATP hydrolyzed after clamp closing would have given rise to a phase of ATP hydrolysis limited by the slower 6 s−1 closing rate, or possibly would been exchanged with ATPγS prior to hydrolysis.
The Temporal Correlation of Multiple Turnovers of β Closing and ATP Hydrolysis
Experiments in Figs. 1–4 were limited to a single turnover by inclusion of a trap, unlabeled β or ATPγS. In multiple-turnover reactions on the short linear p/t-DNA substrates, clamps slide off DNA after being loaded and are reloaded by the clamp loader, setting up a steady-state cycle (41). To determine how reaction kinetics change when the clamp loaders catalyze multiple rounds of clamp loading, pre-steady-state kinetic assays were repeated in the absence of these trapping agents (Fig. 5). These clamp closing and ATP hydrolysis reactions contained the same concentrations of substrates as in Fig. 4B. Multiple-turnover β closing time courses (black trace) have the same basic shape as the single-turnover experiments, but reach a limiting fluorescence intensity at earlier times as the reaction reaches steady state. The multiple-turnover ATPase time courses (gray trace) contain three phases, a sigmoid-shaped burst of hydrolysis present in the single-turnover experiments, a lag, and a linear increase in fluorescence as the reaction reaches steady state. Both β closing and ATP hydrolysis time courses transition into the steady-state phase over the same time period between about 0.4 and 0.6 s. The burst amplitude in the multiple-turnover ATPase reactions is the same as in the single-turnover reactions, showing that the amplitude of the burst phase in single-turnover ATPase reactions is not reduced by rapid exchange of ATP for ATPγS. Steady-state rates of ATP hydrolysis determined previously yielded kcat values of ∼2 s−1 (22, 24, 47), which agrees with the value of 1.8 ± 0.2 s−1 calculated from the ATP hydrolysis reactions performed here. When this kcat value is corrected for the number of active ATP sites based on the burst amplitudes, a kcat of about 1 s−1 per active ATP site is obtained. This kcat value is about the same as the rate of release of the β clamp, suggesting that clamp release may be the rate-limiting step in the clamp loading reaction cycle.
DISCUSSION
The E. coli clamp loader, γ complex, performs the vital function of loading the β sliding clamp onto DNA for various DNA replication and repair functions. To load clamps, the γ complex must modulate its affinity for multiple substrates, first by having a high affinity for β and DNA to bring these macromolecules together and then by decreasing its affinity for β and DNA to release them. The γ complex uses ATP binding and hydrolysis to regulate the different affinities by driving conformational changes that make interactions with β and DNA more or less favorable. Clamp loading must be quick and efficient to complete these steps in the timetable required for DNA replication, so there is an ordered mechanism to prevent unproductive clamp loading events.
This work uses fluorescent kinetic assays reporting on individual interactions and reaction steps in the clamp loading cycle to measure the temporal correlation of events catalyzed by the E. coli γ complex. The main focus was on the relative timing of clamp closing and release. Given the stability of the ring-shaped β dimer in solution and the requirement for the clamp loader to stabilize the clamp in an open conformation to be loaded onto DNA, it was quite possible that the clamp simply shut rapidly upon release by the clamp loader. However, in single-turnover fluorescent β closing and release assays, β closing is faster than β release, showing that the β clamp closes while still bound to the γ complex (Fig. 6). This order of events may ensure that β clamps are closed around DNA and reduce the possibility that DNA slips out of an open clamp before closure. A two-step clamp closing/release reaction is unique to the situation in which clamps are loaded on DNA. In the passive clamp dissociation reaction in the absence of DNA, clamp closing and release rates are the same, albeit much slower than in the clamp loading reaction. Similarly, in assays with nonhydrolyzable ATPγS, clamp closing and release rates are slower but the same. In these reactions, either clamp closing and release occur at the same time or the clamp may rapidly snap shut on release.
FIGURE 6.
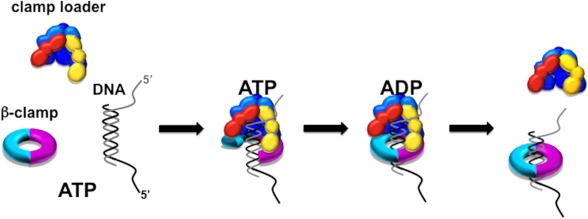
Model for the temporal order of events in loading the clamp on DNA. On the left, γ complex, with ATP, forms a ternary complex composed of β in an open conformation and DNA. DNA binding triggers hydrolysis of ATP followed by closing of β around DNA. On the right, once β is closed, γ complex releases the β·DNA complex, resulting in a loaded clamp and freeing γ complex to load another clamp.
There are several possible mechanisms that would facilitate β closing prior to release from the γ complex; two possibilities are described below. The first is that an ATP hydrolysis-induced conformational change in the γ complex forces the clamp closed. This type of mechanism is supported by molecular dynamics simulations on closed β clamps showing that β is held closed under spring tension. This spring tension would facilitate clamp opening by the δ subunit of the clamp loader, but would require energy to close the clamp (12, 48). It is possible that it takes more effort for the γ complex to close the β clamp than to open it. On the other hand, although the δ subunit can transiently open clamps to unload clamps from DNA (12), the δ subunit alone does not stabilize the clamp in an open conformation in solution sufficiently to produce a measurable population of open clamps (11). Formation of open clamps in solution requires intact clamp loaders (11). Therefore, a second possible mechanism for β closure is that ATP hydrolysis-induced conformational changes in the γ complex remove clamp loader-clamp interactions that stabilize an open conformation of the clamp, without destabilizing the complex enough to promote clamp dissociation. This type of mechanism is supported by the high stability of the closed β conformation and the requirement for binding to the clamp loader to maintain a relatively large fraction of clamps in an open conformation (11). This model is supported by molecular dynamics simulations with the eukaryotic clamp loader, replication factor C, and proliferating cell nuclear antigen clamp that show that clamp loader-clamp interactions simply stabilize the open conformation of the clamp (29). The corollary to this is that removing these interactions would destabilize the open conformation and allow clamp closure.
DNA binding triggers a burst of ATP hydrolysis by the clamp loader (24, 26). Here, we show that clamp closing is slower than ATP hydrolysis, supporting the idea that ATP hydrolysis-induced conformational changes in the clamp loader are required for clamp closure regardless of whether the reactions are measured in single- or multiple-turnover situations (Figs. 4 and 5). It will be interesting to determine the relative timing of DNA release, clamp closure, and hydrolysis of ATP molecules at individual sites. Previous work suggested that DNA may be released prior to clamp release (27). Do the same conformational changes that allow clamp closure also promote release of DNA, or do clamp closure and DNA release occur sequentially, possibly regulated by ATP hydrolysis at individual clamp loader sites? In a recent crystal structure of the bacteriophage T4 clamp loader bound to a closed clamp, ADP was bound at one of the sites, whereas ATP was bound to the others, suggesting that hydrolysis of ATP at one site promotes clamp closure (30). This opens the possibility that sequential clamp closure and DNA release could be regulated by sequential ATP hydrolysis.
Clamp release following DNA-dependent ATP hydrolysis (active clamp loading) is on the order of 10–20 times faster than in the absence of DNA-dependent ATP hydrolysis (passive dissociation). Structural studies yield an explanation for this result. If γ complex bound to closed β resembles the structures for both the closed bacteriophage T4 and the Saccharomyces cerevisiae clamp loader·clamp complexes, then γ complex would have a lower affinity for closed β simply because there are fewer contacts between γ complex subunits and the clamp than are present in the open clamp loader·clamp complex (30, 49). Passive dissociation of the open clamp loader·clamp complex may be slower because it would have to break more contacts.
The clamp loading reaction cycle is complex and composed of multiple steps driven by ATP binding and hydrolysis at multiple sites and interactions with two other ligands, the β clamp and DNA. These interactions with the clamp, DNA, and ATP likely promote conformational changes in the clamp loader that facilitate the next step in the reaction cycle to generate an ordered clamp loading mechanism that ensures that β is loaded quickly, in the correct position, and with as little wasted effort as possible. This type of mechanism could potentially give the clamp loading reaction the efficiency required to keep pace with the moving replication fork. This work, through the use of unique fluorescent assays, helps to fill in the gaps of the known γ complex clamp loading mechanism and give a better understanding of how this remarkable enzyme, as well as the highly conserved clamp loaders from other organisms, functions in the cell.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM082849 (to L. B. B.) and the Training Grant in Cancer Biology Grant 5 T32 CA009126-32 (to J. N. H.).
- p/t-DNA
- primed template DNA
- AF488
- Alexa Fluor 488
- MDCC
- 7-diethylamino-3-((((2-maleimidyl)ethyl)amino)carbonyl)coumarin
- PBP
- phosphate-binding protein
- SSB
- single-stranded DNA-binding protein
- PY
- pyrene
- ATPγS
- adenosine 5′-O-(thiotriphosphate).
REFERENCES
- 1. Indiani C., O'Donnell M. (2006) The replication clamp-loading machine at work in the three domains of life. Nat. Rev. Mol. Cell Biol. 7, 751–761 [DOI] [PubMed] [Google Scholar]
- 2. Kong X. P., Onrust R., O'Donnell M., Kuriyan J. (1992) Three-dimensional structure of the β subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell 69, 425–437 [DOI] [PubMed] [Google Scholar]
- 3. Georgescu R. E., Kim S.-S., Yurieva O., Kuriyan J., Kong X.-P., O'Donnell M. (2008) Structure of a sliding clamp on DNA. Cell 132, 43–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Johnson A., O'Donnell M. (2005) Cellular DNA replicases: components and dynamics at the replication fork. Annu. Rev. Biochem. 74, 283–315 [DOI] [PubMed] [Google Scholar]
- 5. McHenry C. S. (2011) DNA replicases from a bacterial perspective. Annu. Rev. Biochem. 80, 403–436 [DOI] [PubMed] [Google Scholar]
- 6. Naktinis V., Turner J., O'Donnell M. (1996) A molecular switch in a replication machine defined by an internal competition for protein rings. Cell 84, 137–145 [DOI] [PubMed] [Google Scholar]
- 7. Dalrymple B. P., Kongsuwan K., Wijffels G., Dixon N. E., Jennings P. A. (2001) A universal protein-protein interaction motif in the eubacterial DNA replication and repair systems. Proc. Natl. Acad. Sci. U.S.A. 98, 11627–11632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. López de Saro F., Georgescu R. E., Leu F., O'Donnell M. (2004) Protein trafficking on sliding clamps. Philos. Trans. R Soc. Lond. B Biol. Sci. 359, 25–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Georgescu R. E., Yurieva O., Kim S.-S., Kuriyan J., Kong X.-P., O'Donnell M. (2008) Structure of a small-molecule inhibitor of a DNA polymerase sliding clamp. Proc. Natl. Acad. Sci. U.S.A. 105, 11116–11121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Oakley A. J., Prosselkov P., Wijffels G., Beck J. L., Wilce M. C. J., Dixon N. E. (2003) Flexibility revealed by the 1.85 Ä crystal structure of the β sliding-clamp subunit of Escherichia coli DNA polymerase III. Acta Crystallogr. D Biol. Crystallogr. 59, 1192–1199 [DOI] [PubMed] [Google Scholar]
- 11. Paschall C. O., Thompson J. A., Marzahn M. R., Chiraniya A., Hayner J. N., O'Donnell M., Robbins A. H., McKenna R., Bloom L. B. (2011) The Escherichia coli clamp loader can actively pry open the β-sliding clamp. J. Biol. Chem. 286, 42704–42714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jeruzalmi D., Yurieva O., Zhao Y., Young M., Stewart J., Hingorani M., O'Donnell M., Kuriyan J. (2001) Mechanism of processivity clamp opening by the δ subunit wrench of the clamp loader complex of E. coli DNA polymerase III. Cell 106, 417–428 [PubMed] [Google Scholar]
- 13. Yao N., Turner J., Kelman Z., Stukenberg P. T., Dean F., Shechter D., Pan Z. Q., Hurwitz J., O'Donnell M. (1996) Clamp loading, unloading and intrinsic stability of the PCNA, β, and gp45 sliding clamps of human, E. coli, and T4 replicases. Genes Cells 1, 101–113 [DOI] [PubMed] [Google Scholar]
- 14. Ogura T., Wilkinson A. J. (2001) AAA+ superfamily ATPases: common structure–diverse function. Genes Cells 6, 575–597 [DOI] [PubMed] [Google Scholar]
- 15. Davey M. J., Jeruzalmi D., Kuriyan J., O'Donnell M. (2002) Motors and switches: AAA+ machines within the replisome. Nat. Rev. Mol. Cell Biol. 3, 826–835 [DOI] [PubMed] [Google Scholar]
- 16. Erzberger J. P., Berger J. M. (2006) Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct. 35, 93–114 [DOI] [PubMed] [Google Scholar]
- 17. Pritchard A. E., Dallmann H. G., Glover B. P., McHenry C. S. (2000) A novel assembly mechanism for the DNA polymerase III holoenzyme DnaX complex: association of δδ′ with DnaX4 forms DnaX3δδ′. EMBO J. 19, 6536–6545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jeruzalmi D., O'Donnell M., Kuriyan J. (2001) Crystal structure of the processivity clamp loader gamma (γ) complex of E. coli DNA polymerase III. Cell 106, 429–441 [DOI] [PubMed] [Google Scholar]
- 19. Simonetta K. R., Kazmirski S. L., Goedken E. R., Cantor A. J., Kelch B. A., McNally R., Seyedin S. N., Makino D. L., O'Donnell M., Kuriyan J. (2009) The mechanism of ATP-dependent primer-template recognition by a clamp loader complex. Cell 137, 659–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Naktinis V., Onrust R., Fang L., O'Donnell M. (1995) Assembly of a chromosomal replication machine: two DNA polymerases, a clamp loader, and sliding clamps in one holoenzyme particle. II. Intermediate complex between the clamp loader and its clamp. J. Biol. Chem. 270, 13358–13365 [PubMed] [Google Scholar]
- 21. Hingorani M. M., O'Donnell M. (1998) ATP binding to the Escherichia coli clamp loader powers opening of the ring-shaped clamp of DNA polymerase III holoenzyme. J. Biol. Chem. 273, 24550–24563 [DOI] [PubMed] [Google Scholar]
- 22. Turner J., Hingorani M. M., Kelman Z., O'Donnell M. (1999) The internal workings of a DNA polymerase clamp-loading machine. EMBO J. 18, 771–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bertram J. G., Bloom L. B., Turner J., O'Donnell M., Beechem J. M., Goodman M. F. (1998) Pre-steady state analysis of the assembly of wild type and mutant circular clamps of Escherichia coli DNA polymerase III onto DNA. J. Biol. Chem. 273, 24564–24574 [DOI] [PubMed] [Google Scholar]
- 24. Hingorani M. M., Bloom L. B., Goodman M. F., O'Donnell M. (1999) Division of labor–sequential ATP hydrolysis drives assembly of a DNA polymerase sliding clamp around DNA. EMBO J. 18, 5131–5144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Thompson J. A., Paschall C. O., O'Donnell M., Bloom L. B. (2009) A slow ATP-induced conformational change limits the rate of DNA binding but not the rate of β clamp binding by the Escherichia coli γ complex clamp loader. J. Biol. Chem. 284, 32147–32157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bertram J. G., Bloom L. B., Hingorani M. M., Beechem J. M., O'Donnell M., Goodman M. F. (2000) Molecular mechanism and energetics of clamp assembly in Escherichia coli: the role of ATP hydrolysis when γ complex loads β on DNA. J. Biol. Chem. 275, 28413–28420 [DOI] [PubMed] [Google Scholar]
- 27. Anderson S. G., Thompson J. A., Paschall C. O., O'Donnell M., Bloom L. B. (2009) Temporal correlation of DNA binding, ATP hydrolysis, and clamp release in the clamp loading reaction catalyzed by the Escherichia coli γ complex. Biochemistry 48, 8516–8527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Leu F. P., O'Donnell M. (2001) Interplay of clamp loader subunits in opening the β sliding clamp of Escherichia coli DNA polymerase III holoenzyme. J. Biol. Chem. 276, 47185–47194 [DOI] [PubMed] [Google Scholar]
- 29. Tainer J. A., McCammon J. A., Ivanov I. (2010) Recognition of the ring-opened state of proliferating cell nuclear antigen by replication factor C promotes eukaryotic clamp-loading. J. Am. Chem. Soc. 132, 7372–7378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kelch B. A., Makino D. L., O'Donnell M., Kuriyan J. (2011) How a DNA polymerase clamp loader opens a sliding clamp. Science 334, 1675–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wieczorek A., Downey C. D., Dallmann H. G., McHenry C. S. (2010) Only one ATP-binding DnaX subunit is required for initiation complex formation by the Escherichia coli DNA polymerase III holoenzyme. J. Biol. Chem. 285, 29049–29053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Downey C. D., Crooke E., McHenry C. S. (2011) Polymerase chaperoning and multiple-ATPase sites enable the E. coli DNA polymerase III holoenzyme to rapidly form initiation complexes. J. Mol. Biol. 412, 340–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gomes X. V., Burgers P. M. (2001) ATP utilization by yeast replication factor C. I. ATP-mediated interaction with DNA and with proliferating cell nuclear antigen. J. Biol. Chem. 276, 34768–34775 [DOI] [PubMed] [Google Scholar]
- 34. Onrust R., Finkelstein J., Naktinis V., Turner J., Fang L., O'Donnell M. (1995) Assembly of a chromosomal replication machine: two DNA polymerases, a clamp loader, and sliding clamps in one holoenzyme particle. I. Organization of the clamp loader. J. Biol. Chem. 270, 13348–13357 [DOI] [PubMed] [Google Scholar]
- 35. Dong Z., Onrust R., Skangalis M., O'Donnell M. (1993) DNA polymerase III accessory proteins. I. holA and holB encoding δ and δ′. J. Biol. Chem. 268, 11758–11765 [PubMed] [Google Scholar]
- 36. Olson M. W., Dallmann H. G., McHenry C. S. (1995) DnaX complex of Escherichia coli DNA polymerase III holoenzyme: the X Ψ complex functions by increasing the affinity of τ and γ for δ·δ′ to a physiologically relevant range. J. Biol. Chem. 270, 29570–29577 [PubMed] [Google Scholar]
- 37. Maki S., Kornberg A. (1988) DNA polymerase III holoenzyme of Escherichia coli. I. Purification and distinctive functions of subunits τ and γ, the dnaZX gene products. J. Biol. Chem. 263, 6547–6554 [PubMed] [Google Scholar]
- 38. Johanson K. O., Haynes T. E., McHenry C. S. (1986) Chemical characterization and purification of the β subunit of the DNA polymerase III holoenzyme from an overproducing strain. J. Biol. Chem. 261, 11460–11465 [PubMed] [Google Scholar]
- 39. Brune M., Hunter J. L., Corrie J. E., Webb M. R. (1994) Direct, real-time measurement of rapid inorganic phosphate release using a novel fluorescent probe and its application to actomyosin subfragment 1 ATPase. Biochemistry 33, 8262–8271 [DOI] [PubMed] [Google Scholar]
- 40. Williams C. R., Snyder A. K., Kuzmic P., O'Donnell M., Bloom L. B. (2004) Mechanism of loading the Escherichia coli DNA polymerase III sliding clamp. I. Two distinct activities for individual ATP sites in the γ complex. J. Biol. Chem. 279, 4376–4385 [DOI] [PubMed] [Google Scholar]
- 41. Bloom L. B., Turner J., Kelman Z., Beechem J. M., O'Donnell M., Goodman M. F. (1996) Dynamics of loading the β sliding clamp of DNA polymerase III onto DNA. J. Biol. Chem. 271, 30699–30708 [DOI] [PubMed] [Google Scholar]
- 42. Glover B. P., McHenry C. S. (1998) The X Ψ subunits of DNA polymerase III holoenzyme bind to single-stranded DNA-binding protein (SSB) and facilitate replication of an SSB-coated template. J. Biol. Chem. 273, 23476–23484 [DOI] [PubMed] [Google Scholar]
- 43. Kelman Z., Yuzhakov A., Andjelkovic J., O'Donnell M. (1998) Devoted to the lagging strand-the subunit of DNA polymerase III holoenzyme contacts SSB to promote processive elongation and sliding clamp assembly. EMBO J. 17, 2436–2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Downey C. D., McHenry C. S. (2010) Chaperoning of a replicative polymerase onto a newly assembled DNA-bound sliding clamp by the clamp loader. Mol. Cell 37, 481–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Brune M., Hunter J. L., Howell S. A., Martin S. R., Hazlett T. L., Corrie J. E., Webb M. R. (1998) Mechanism of inorganic phosphate interaction with phosphate binding protein from Escherichia coli. Biochemistry 37, 10370–10380 [DOI] [PubMed] [Google Scholar]
- 46. Johnson A., O'Donnell M. (2003) Ordered ATP hydrolysis in the γ complex clamp loader AAA+ machine. J. Biol. Chem. 278, 14406–14413 [DOI] [PubMed] [Google Scholar]
- 47. Chen S., Coman M. M., Sakato M., O'Donnell M., Hingorani M. M. (2008) Conserved residues in the δ subunit help the E. coli clamp loader, γ complex, target primer-template DNA for clamp assembly. Nucleic Acids Res. 36, 3274–3286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fang J., Engen J. R., Beuning P. J. (2011) Escherichia coli processivity clamp β from DNA polymerase III is dynamic in solution. Biochemistry 50, 5958–5968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bowman G. D., O'Donnell M., Kuriyan J. (2004) Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature 429, 724–730 [DOI] [PubMed] [Google Scholar]