Background: Mechanisms by which RBP4 interacts with cells are not completely understood.
Results: 1300002K09Rik (RBPR2) is identified as a Stra6-related protein expressed in liver, intestine, and obese fat that mediates RBP4 binding and retinol transport.
Conclusion: RBPR2 is a novel RBP4 receptor that mediates retinol uptake.
Significance: RBPR2 may be important for whole body retinol homeostasis or cellular actions of RBP4 in certain tissues.
Keywords: Adipokines; Diabetes; Insulin Resistance; Obesity; Receptors; Retinoid-binding Protein; Vitamin A; RBP4, Retinol-binding Protein; Stra6
Abstract
Vitamin A (retinol) is absorbed in the small intestine, stored in liver, and secreted into circulation bound to serum retinol-binding protein (RBP4). Circulating retinol may be taken up by extrahepatic tissues or recycled back to liver multiple times before it is finally metabolized or degraded. Liver exhibits high affinity binding sites for RBP4, but specific receptors have not been identified. The only known high affinity receptor for RBP4, Stra6, is not expressed in the liver. Here we report discovery of RBP4 receptor-2 (RBPR2), a novel retinol transporter expressed primarily in liver and intestine and induced in adipose tissue of obese mice. RBPR2 is structurally related to Stra6 and highly conserved in vertebrates, including humans. Expression of RBPR2 in cultured cells confers high affinity RBP4 binding and retinol transport, and RBPR2 knockdown reduces RBP4 binding/retinol transport. RBPR2 expression is suppressed by retinol and retinoic acid and correlates inversely with liver retinol stores in vivo. We conclude that RBPR2 is a novel retinol transporter that potentially regulates retinol homeostasis in liver and other tissues. In addition, expression of RBPR2 in liver and fat suggests a possible role in mediating established metabolic actions of RBP4 in those tissues.
Introduction
Vitamin A (retinol) is obtained through diet and required for life by mammals and other vertebrates (1, 2). Retinol is catabolized to retinoic acid isomers that act as ligands for multiple nuclear hormone receptors to regulate transcription of hundreds of genes (3–5). Nevertheless, little is known about the molecular basis of the initial step of retinol metabolism, the entry of retinol into cells. Dietary retinol is absorbed in the small intestine, and up to 90% of total body retinol is stored in liver, distributed between hepatocytes and stellate cells specialized for retinyl ester storage (6). In addition, adipocytes can store a significant amount of retinyl esters (7). Retinol is mobilized from liver to extrahepatic tissues by secretion of RBP4 (serum retinol-binding protein) into the circulation (8, 9). RBP4 is the sole specific transport protein for retinol in blood, and nearly all circulating retinol is RBP4-bound (8, 9). Dietary retinol deficiency results in depletion of liver retinyl ester stores and, when severe, manifests as decreased circulating RBP4 and retinol (2). Modeling of intertissue retinol transport kinetics indicates that retinol traffics into and out of hepatocytes multiple times before finally undergoing irreversible catabolism in extrahepatic tissues (10). Consistent with this, binding studies have revealed high affinity sites for RBP4 on hepatocyte membranes (11, 12), and hepatocytes efficiently take up RBP4-bound retinol (13, 14). However, circulating RBP4 is not internalized by the liver and does not appear to mediate secretion (or resecretion) of retinol from liver (15).
Discovery of a high affinity RBP4 receptor, Stra6 (stimulated by retinoic acid-6), provided new insight into the molecular basis of RBP4-dependent retinol transport (16). Stra6 is a multitransmembrane domain protein that mediates binding of RBP4 to cell membranes and transfer of retinol from RBP4 to cells (16, 17). Stra6 is highly conserved among mammals and other vertebrates. Stra6 is expressed in several tissues, including retina, brain, testis, muscle, and placental endothelial cells, but not in liver or intestine (16, 18). This tissue distribution suggests an important role for Stra6 in “directional delivery” of RBP4-bound retinol from liver to extrahepatic tissues or, as in the case of placenta, in directional delivery to the developing fetus. The physiological importance of Stra6 in development has been confirmed by its identification as the causal gene in human micro-ophthalmic syndrome 9 (HMS95; encompassing Mathew-Wood Syndrome and PDAC (19–21)). HMS9 is a congenital complex of micro-ophthalmia, mental retardation, and severe pulmonary, diaphragmatic, gastrointestinal, and in some cases, pancreatic malformations (22, 23); these specific anomalies are consistent with the established role of retinoic acid in organogenesis, embryonic patterning, and limb and nervous system development (24–27). HMS9 results from homozygous missense or truncating mutations of Stra6 that disrupt RBP4 binding and retinol transport or destabilize the protein (21).
Although Stra6 plays an important role in multiple tissues during development, it is not normally expressed in adult intestine or liver (16, 18), the tissues that mediate uptake, storage, and distribution of retinol in the body. Therefore, Stra6 may not play an important role in total body retinol homeostasis. High affinity RPB4 binding and retinol uptake in liver and other cell types not expressing Stra6 imply the existence of a second RBP4 receptor/retinol transporter. Here we report that 1300002K09Rik, a gene with previously unknown function, encodes a novel retinol transporter and RBP4 receptor. We have designated this gene RBPR2 (RBP4 receptor-2). RBPR2 is expressed primarily in liver and small intestine and is induced in adipocytes of obese mice. Our findings identify a potentially important new mechanism for RBP4-dependent retinol transport and other cellular actions of RBP4.
EXPERIMENTAL PROCEDURES
Survey of Mouse Transcriptome for Stra6-related Proteins
The mouse Stra6 open reading frame amino acid sequence was used to perform an unbiased query of the non-redundant nucleotide database collection of the National Institutes of Health National Center for Biotechnology Information, using the phylogeny-based search engine of the Laboratoire d'Informatique, de Robotique et de Microélectronique de Montpellier (Montpellier, France) (28, 29). This method identified two groups of phylogentically related transcripts within mammals: an expected group corresponding to Stra6 and its orthologues and another group corresponding to rat hypothetical protein LOC298077 (RGD1305807; NCBI accession number NM_001025276.1). The UniProt database was searched for related sequences, resulting in identification of multiple non-primate vertebrate sequences related to the expressed mouse transcript 1300002K09Rik. Stra6 exhibits only 17.8% overall homology to 1300002K09 and hence could not be detected by Basic Local Alignment Search Tool (BLAST) queries, even when employing algorithms designed to detect proteins with only partial conservation.
5′- and 3′-RACE Cloning of Human RBPR2
The University of California Santa Cruz Genome Browser was used to identify open reading frames potentially encoding a human RBPR2 orthologue in the human genome. Potential open reading frames were found at two distinct loci on the long and short arms of chromosome 9 (9q and 9p). RACE PCR was used to obtain the 5′- and 3′-end sequences for isolating full-length cDNA of RBPR2 sequences in the human genome. Human liver Marathon-ready cDNA (Clontech) was used as template, and RACE PCR was carried out with the Marathon cDNA amplification kit (Clontech), following instructions from the manufacturer. 5′-RACE and 3′-RACE were carried out using an adaptor primer (AP1) provided by the kit and RBPR2 gene-specific primers (5′-GSP and 3′-GSP; Table 1). The products of 3′- and 5′-RACE were purified and cloned into the pCR4-TOPO vector provided by the TOPO TA cloning kit (Invitrogen), and inserts were sequenced by the core facility at the University of Utah.
TABLE 1.
Primers used for 5′- and 3′-RACE cloning of human RBPR2
| Primer | Chromosome | Human genome location | Sequence |
|---|---|---|---|
| 5′-GSP1a | 9q | chr9:99097837–99097861 | TCCGAGCATGCCATCAGAATTTGGG |
| 5′-GSP2 | 9q | chr9:99097721–99097741 + GC | CTAGCTGAGACATGGGGACCTGC |
| 5′-GSP3 | 9q | chr9:99096170–99096194 | GCCACTCTGCATGATGGTCCTGTCG |
| 5′-GSP4 | 9q | chr9:99093561–99093583 | TGCCCAGCCCTTGCAGCATTTCC |
| 5′-GSP5 | 9q | chr9:99092961–99092988 | ATTTGCCAGCCAGAGTACCTTGCAATGG |
| 5′-GSP6 | 9q | chr9:99083741–99083765 | GACACTGATCAGCTCTTTCACCTGC |
| 5′-GSP7 | 9q | chr9:99078732–99078757 | AACATATGCAGGAAGCGGCCCAGCAG |
| 5′-GSP8 | 9p | chr9:38459926–38459951 | AGCAGAAGCCCAAGATGGAGCTGACC |
| 5′-GSP9 | 9p | chr9:38459842–38459865 | CCTCCAATCAGAAGCTCAAAGGCT |
| 5′-GSP10 | 9p | chr9:38452117–38452139 | GGACCATCGGTTACTGAAAGTGC |
| 3′-GSP1 | 9q | chr9:99097075–99097096 + C | GTGCCTAAATCAATCGGCAGGTC |
| 3′-GSP2 | 9q | chr9:99096129–99096151 | CTCATCAGTTGCCTCCTCGGCAT |
| 3′-GSP3 | 9q | chr9:99093551–99093574 | ACCAGGGCCTGGAAATGCTGCAAG |
| 3′-GSP4 | 9q | chr9:99092688–99092715 | AAGCACATGAAGAGGCTATGGGCAGGAG |
| 3′-GSP5 | 9q | chr9:99078732–99078755 | CTGCTGGGCCGCTTCCTGCATATG |
| 3′-GSP6 | 9p | chr9:38459851–38459877 | GCTTCTGATTGGAGGCATTGAAGTCGG |
| AP1b | CCATCCTAATACGACTCACTATAGGGC |
a GSP, gene-specific primers corresponding to exonic sequences at 9p and 9q loci.
b AP1, adaptor primer 1.
Plasmids
C terminus HA-tagged RBPR2 and Stra6 expression plasmids were generated by amplifying cDNA open reading frames of murine RBPR2 (Genecopoeia; 1300002K09Rik; NCBI accession number NM_028788.1; product Mm11236) or human Stra6 (Genecopoeia; NCBI accession number NM_001142619; product Z6735) into pMEX-HA (Dualsystems Biotech) with 5′-EcoRI and 3′-BamHI overhangs and performing directional ligation into the vector; the 3′ overhangs were constructed to maintain the open reading frame through the HA tag encoded by the plasmid. Expression plasmids pM2 for untagged RBPR2 and Stra6 open reading frames were obtained commercially (Genecopoeia). Thermostable secreted alkaline phosphatase-RBP4 fusion protein (SAP-RBP4) was generated by amplifying RBP4 mRNA (without sequence encoding the secretory signal peptide) with 5′-XhoI and 3′-XbaI overhangs and performing directional ligation into pAPTag5 (GenHunter). Stop codons were included at the 3′ end of all open reading frames. Expression plasmids for transforming growth factor β receptor II (TGFR2), a gift of Dr. Joan Massagué (Memorial Sloan-Kettering Cancer Institute), and receptor-type protein tyrosine phosphatase α (PTPRα), a gift of Dr. Xinmin Zheng (Temple University School of Medicine), were obtained through the Addgene repository.
Cell Culture and Transfections
HEK-293T (HEK-293), 3T3L1, and Hepa1c1c7 (Hepa1) cell lines were obtained from the American Tissue Type Culture Collection (ATCC) and grown in DMEM with 10% heat-inactivated fetal bovine serum supplemented with 1 mm glutamine and primocin (for HEK-293 and Hepa1; InVivoGen) or penicillin-streptomycin (for 3T3L1). 3T3L1 cells were differentiated as described elsewhere (30). H4IIe cells were obtained from ATCC and grown in α-minimum essential medium supplemented with 10% heat-inactivated FBS, glutamine, and penicillin-streptomycin. Lines were re-established from early passage frozen stocks after more than 12 passages. A siRNA pool containing oligonucleotides targeting four separate proprietary sequences in mouse/rat 1300002K09Rik and a pool of four non-targeting control oligonucleotides were obtained commercially (On-Target Plus SmartPool, Dharmacon). Plasmid DNA transfections were performed with Xfect (Invitrogen), and siRNA transfections were performed with Dharmafect (Dharmacon) under reduced serum conditions, following the manufacturers' standard protocols.
Animal Husbandry and Diets
Mice were maintained in a temperature- and humidity-controlled vivarium with 12-h dark/light cycles managed by the University of Utah Office of Comparative Medicine. Teklad 5008 chow was fed (Vitamin A content is 15 IU/g, provided as retinyl acetate). All studies were approved by the University of Utah Institutional Animal Care and Use Committee. Lep(ob/ob) and Lep(?/+) lean littermate controls on the C5Bl6 background (ob/ob and ob/+ control mice) were obtained from Taconic Laboratories and maintained on a normal chow diet. For high fat diet studies, wild type male C57Bl6 mice bred in-house were weaned onto a purified high fat (57 kcal% fat) diet or a matched, purified low fat (11 kcal% fat; Open Source Diets). Mice fed the high fat diet gained on average 8.2 ± 2.3 g more body weight than mice fed the low fat diet over 12 weeks of feeding. Mice were sacrificed at 15–16 weeks of age.
RBPR2 Promoter-Reporter Mice
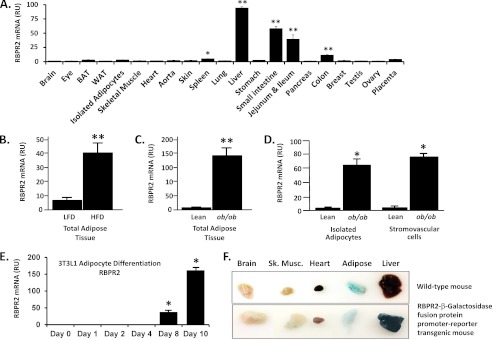
Mouse embryonic stem cells derived from C57Bl6 were engineered by the National Institutes of Health Knock Out Mouse Project with heterozygous knock-in of a conditional gene expression cassette (flox-β-GEO cassette) between exons 5 and 6 of the 1300002K09Rik (RBPR2) allele; the cassette contains β-galactosidase (β-gal) sequence with an upstream 5′ splice acceptor sequence capable of producing a transcript composed of RBPR2 (exons 1–5, amino acids 1–109) fused in-frame to β-gal and a 3′ poly(A) tail after exposure of the cassette to Cre recombinase (31). Embryonic stem cells were injected into C57Alb mouse embryos to obtain chimeric progeny by the University of California Davis Transgenic Mouse Core (via the National Institutes of Health Knockout Mouse Project). Male chimeras were bred with C57Alb female mice to obtain non-albino pups with potential germ line transmission of the modified RBPR2 allele. Founder pups carrying modified RBPR2 were identified by genomic PCR with primers targeting the flox-β-GEO cassette and bred to zona pellucida (Zp3)-Cre transgenic mice (expressing Cre in the oocyte (32)) to generate female digenic RBPR2(floxβGEO/+), Zp3-Cre(Tg/0) mice. Female digenic mice were bred with wild type male C57Bl6 mice to produce pups heterozygous for a Cre/LoxP-recombined RPBR2 promoter-RBPR2/β-gal fusion protein reporter allele (RBPR2 promoter-reporter mice). Cre/LoxP recombination was confirmed by genomic PCR using long range PCR with primers outside the LoxP-recombined region (positive for recombinant allele) and also by short range PCR with primers from within the recombined region (negative for the recombinant allele). Detection of β-gal activity was performed by sacrificing heterozygous promoter-reporter mice and rapidly dissecting tissues and incubating them for 1 h in PBS containing X-gal (Sigma). After 1 h, tissues were transferred to PBS containing 4% paraformaldehyde, 0.1% Triton X-100, and X-gal and stored overnight at 4 ºC. Tissues were arrayed and photographed under white light. Because of reflection problems, the image of the brain for wild type mice was photographed separately and merged with images of other organs using photo editing software (Fig. 4F, top).
FIGURE 4.
Tissue expression pattern of RBPR2. A, RBPR2 mRNA was measured in tissues of wild type FVB mice by qRT-PCR. Five different commercial primer/probe sets were tested with similar results (Solaris assay results are shown). 8–9-week-old male and female mice (n = 3 for each gender) were sacrificed in the early morning fed state. Relative expression of RBPR2 was determined separately for individual tissues and normalized to liver, which was assigned relative units (RU) equal to 100. Bars, average ± S.E. (error bars) of relative expression for six mice except for ovary, testis, and breast (n = 3 of male or female gender). Similar results were obtained using two other commercial primer/probe sets (Applied Biosystems; not shown). *, p < 0.05 versus mean relative expression level for all tissues; **, p < 0.01 versus mean relative expression level for all tissues. B, RBPR2 mRNA was measured by qRT-PCR (Solaris) in whole adipose tissue of C57Bl6 mice fed a high fat diet (HFD) or low fat control diet (LFD) for 12 weeks starting at weaning (n = 8 mice/group, males, age 15–16 weeks). **, p < 0.01 versus LFD. C, RBPR2 mRNA was measured by qRT-PCR (Solaris) in whole adipose tissue of leptin-deficient LepLep(ob/ob) mice or lean Lep(+/?) littermate controls (n = 6 mice/group, males, age 4–5 months) fed normal chow. **, p < 0.01 versus lean control. D, adipocytes and stromovascular cells were isolated by collagenase digestion and buoyancy centrifugation from perigonadal fat pads of leptin-deficient LepLep(ob/ob) mice or lean Lep(+/?) littermate controls (n = 6 mice/group, males, age 4–5 months) fed normal chow. mRNA was measured by qRT-PCR (Solaris). E, RBPR2 mRNA was measured by qRT-PCR (Applied Biosystems) in cultured 3T3L1 preadipocytes just prior to confluence (day 0) and at the indicated time points during differentiation to mature adipocytes containing multiple lipid droplets. Adipocyte differentiation was maximal by day 10 based on lipid accumulation (not shown). Three biological replicates were measured for each condition. F, expression of RBPR2-β-galactosidase fusion protein in RBPR2 promoter-reporter transgenic mice was detected by overnight staining of tissues with X-gal substrate. Blue coloration produced by X-gal cleavage indicates expressed β-galactosidase activity (top, tissues from representative C57Bl6 wild type mouse; bottom, tissues from representative littermate RBPR2 promoter-reporter mouse). A high level of endogenous, nonspecific galactosidase activity was detected in the intestine of both the wild type and transgenic promoter-reporter mice, preventing assessment of reporter expression in that tissue (not shown). A significant amount of endogenous galactosidase activity was also present in adipose tissue (top and bottom panels), and total activity did not differ significantly between wild type (top) and transgenic promoter-reporter mice (bottom).
Quantitative RT-PCR (qRT-PCR)
For primary tissue studies, mRNA was isolated from 126 tissue samples from 6 FVB mice (3 male and 3 female) using TriReagent (MRC) according to the manufacturer's directions. Tissue mRNA was arrayed with a primer-probe set from Solaris (product 74152, targeting the exon 12/13 boundary; Thermo Scientific) on 384-well optical plates, and two-step qRT-PCRs (Qiagen) were performed and analyzed using the Applied Biosystems 7900HT Fast Real-Time PCR system. Two replicate reactions with a single RBPR2 primer-probe set and a parallel β-actin housekeeper reaction for each tissue mRNA sample were included on each plate. Comparison studies were performed for several mice with Applied Biosystems primer-probe sets (product Mm01345317_m1, targeting the exon 1/2 boundary; Mm01345323_m1, targeting the exon 8/9 boundary; Mm01345309_m1, targeting the exon 10/11 boundary; and Mm01345316_g1, targeting the exon 17/18 boundary), producing similar results as the Solaris primer-probe set (not shown). Normalization to β-actin by the ΔCt method did not alter interpretation of the tissue expression pattern; unnormalized data are reported. For analysis of isolated adipocyte and stromovascular adipose tissue fractions, perigonadal adipose tissue was subjected to collagenase digestion and differential buoyancy centrifugation as described elsewhere (33). mRNA was immediately isolated from adipocyte and stromovascular fractions. For cultured cells, mRNA was prepared using TriReagent and reverse transcribed using a Verso kit (Thermo Scientific), and one-step quantitative PCR was performed with standard reagents (Qiagen).
Western Blotting
Cells were washed twice in PBS and once in hypotonic lysis buffer (20 mm Hepes, pH 7.5, 1 mm EDTA) and resuspended in ice-cold hypotonic lysis buffer supplemented with the HALT mixture of protease and phosphatase inhibitors (Pierce). Cells were allowed to swell for 15 min on ice and homogenized (10 strokes with a Dounce type B glass plunger set). Lysates were centrifuged 5 min at 500 × g at 4 ºC to pellet nuclei and other insoluble material. Supernatants were transferred to fresh tubes and centrifuged at 100,000 × g for 1 h to pellet total membranes. The supernatant (cytosol) was removed, and membranes were washed by resuspension in hypotonic lysis buffer and recentrifugation at 16,000 × g for 20 min at 4 ºC. The wash was removed. Membranes were resuspended in 50 μl of PBS, 1% dodecyl maltoside, 20% glycerol (without HALT) and gently vortexed and diluted in 0.450 ml of PBS, 2% zwitterionic detergent ASB14, 10% glycerol, 2 mm β-mercaptoethanol, 1 mg/ml brain polar lipid extract. Membranes were solubilized for 30 min on ice and centrifuged 16,000 × g for 10 min at 4 ºC. 25–50 μg of protein were prepared for SDS-PAGE using Novex sample buffer and gels (Invitrogen) with MES-based running buffer, according to the manufacturer's instructions. Gels were transferred to nitrocellulose (iBlot system, Invitrogen). Western blot buffers contained nonfat milk as the blocking reagent (2% milk for blocking and 1% milk for primary and secondary antibody incubations). Anti-HA antibody was obtained commercially (Sigma). Blots were visualized by HRP-catalyzed chemiluminescence and imaged/quantitated on a Genome system (SynGene). For glycosylation studies, membrane or cytosol fractions were treated for 1 h at 25 ºC with a mix of glycosidases (DeGlycoMx kit, QABio) containing glycopeptide N-glycosidase F sialidase, β-galactosidase, glucosaminidase, and O-glycosidase, according to the manufacturer's instructions; membranes were treated with glycosidases before the ASB14 solubilization step.
Microscopy
HEK-293 cells were seeded on glass coverslips in 6-well plates and transfected with expression plasmids for C terminus-tagged HA-Stra6 or HA-RBPR2. Cells on coverslips were washed in PBS, fixed for 20 min in 4% paraformaldehyde, and permeabilized with 0.1% Triton X-100. Blocking was performed in PBS with 1 mg/ml bovine serum albumin (Sigma). Primary and secondary antibody incubations (1:1000 dilution of anti-HA antibody (Sigma); 1:2000 of Alexa Fluor 660-conjugated anti-rabbit IgG (Invitrogen)) and wheat germ agglutinin-fluorescein incubation (1:5000 dilution (Invitrogen)) were performed in the same buffer. Coverslips were mounted on slides with Prolong Gold antifade reagent containing DAPI nuclear stain (Invitrogen). Imaging was performed on a FV1000 inverted confocal scanning laser fluorescence microscope (Olympus) equipped with appropriate filters. Image analysis was performed with Fluoview software (Olympus, version 2.0). Co-localization studies were performed with ImageJ open source software (National Institutes of Health).
RBP4 Binding Studies
Endotoxin-free recombinant mouse holo-RBP4 was expressed in Escherichia coli and purified using methods described previously (34). Holo-RBP4 and holo-RBP4·transthyretin (TTR) complex purified from human plasma under non-denaturing conditions were obtained commercially (HyTest). N-terminal SAP-RBP4 fusion protein was produced by transfection of HEK-293 cells in 100-mm plates with 5 μg of expression plasmid pAPTAG5. Media were assayed for immunoreactive SAP-RBP4 by anti-RBP4 Western blotting, and heat-resistant alkaline phosphatase activity was assayed by heating at 60 ºC for 20 min to inactivate endogenous galactosidases and measuring room temperature X-gal cleavage activity by optical absorbance of colorimetric product. Binding of recombinant proteins was performed after washing cells two times with serum-free medium and incubating cells for 1–2 h in serum-free medium. Binding was performed for 1 h at 37 ºC at the indicated concentrations in serum-free medium for all experiments. For binding of purified human RBP4 or RBP4·TTR complex to cells, three washes were performed in culture wells in situ, and cells were lysed in radioimmunoprecipitation assay (RIPA) buffer, clarified by centrifugation at 16,000 × g for 10 min at 4 ºC, and analyzed by SDS-PAGE and Western blotting with anti-RBP4 or anti-TTR antibodies (DAKO-Cytomation). A similar method has been used by others to detect RBP4 binding to cells (35). For binding of SAP-RBP4, cells were collected by scraping with a rubber policeman and washed by centrifugation in microtubes (1,000 × g for 5 min to collect cell pellets) and lysed in 1% Triton X-100. Lysates were clarified and assayed for β-galactosidase activity using X-gal substrate (GenHunter) with known quantities of purified β-galactosidase (GenHunter) as a calibrator. Activity was expressed in galactosidase units/mg of lysate protein. SAP-RBP4 binding data were further analyzed using Prism GraphPad software (version 5). Best-fit binding curves were generated (R2 = 0.8531 for Stra6 binding and 0.9488 for RBPR2 binding), enabling interpolation of maximum binding Bmax and estimated binding affinity constants (Kd; data not shown).
[3H]Retinol Uptake Studies
Apo-RBP4 was produced by stripping off retinol with butanol and diisopropyl ether as described (36), repurified over a DEAE spin column (Sartorius), and dialyzed overnight in PBS to remove excess salt. Removal of retinol was confirmed by loss of the 330 nm (retinol) absorbance scan peak (data not shown). Apo-RBP4 (100 μg) was reloaded with retinol in 0.2 ml of PBS by the addition of 100 μm radiolabeled retinol (American Radiolabeled Chemicals; vitamin A alcohol [15-3H(N)Retinol-labeled, adjusted to 1 μCi/nmol specific activity by the addition of cold retinol) and incubating for 1 h at room temperature and then overnight at 4 ºC in light-protected tubes. The mixture was centrifuged 16,000 × g in a refrigerated centrifuge, and supernatant was subjected to gel filtration chromatography (Zeba Spin Columns, Pierce) to remove unbound retinol. A protein assay of RBP4 (BCA method, Sigma) and scintillation counting of [3H]retinol were performed to obtain specific activity (typically 2,000 dpm/pmol of protein). The quality of the [3H]retinol-bound holo-RBP4 was confirmed by reassessing 280 and 330 nm absorbance scan peaks (37–39), which were approximately equal (data not shown), indicating that RBP4 bound retinol in a 1:1 molar ratio based on reported molar extinction coefficients for RBP4 and retinol. The ability of the holo-RBP4 to bind immobilized transthyretin (34) also remained intact (data not shown). A similar method was used to produce [3H]retinol-bound albumin, except that stripping was not necessary because commercially available fatty acid-free BSA (Desert Biologicals) was used. Albumin bound less retinol (280 nm/330 nm peak absorbance ratio of 0.81:1, indicating a molar binding ratio of ∼0.72 based on molar extinction coefficients for BSA and retinol; data not shown), so the specific activity of the retinol bound to BSA was adjusted empirically to match that of holo-RBP4. Cells were washed twice and incubated for 1 h in serum-free medium, at which point [3H]retinol-bound RBP4 was added for 30 min; cells were washed three times in PBS and lysed in PBS, 1% Nonidet P-40. Lysates were homogenized and transferred to scintillation tubes for scintillation counting. Lysates were clarified by centrifugation at 16,000 × g for 10 min, and a protein assay was performed (BCA method, Sigma). In separate experiments, a shorter time course of retinol uptake from holo-RBP4 was similarly tested (2–15 min, at a concentration of 100 nm).
Retinol and Retinyl Ester Measurements
Liver for retinoid measurements was rapidly frozen in liquid N2 and stored at −80 ºC protected from light. Frozen liver was pulverized and homogenized in a liquid N2-cooled mortar/pestle, and ∼30 mg of tissue powder was transferred to microtubes containing acid-washed ceramic disruptor beads and 500 μl of PBS. 500 μl of EtOH containing 0.1% butylated hydroxytoluene was added, and the mixture was bead-homogenized. Homogenates were transferred to glass tubes, and 1 ml of 0.025 m KOH in EtOH and 10 μl of 2 mm retinyl acetate standard in EtOH were added and vortexed. 10 ml hexane was added, and the mixture was vortexed vigorously and then centrifuged at 1,200 rpm in a clinical centrifuge for 3 min at room temperature. The hexane (top phase) was transferred to a fresh glass tube and dried under N2 without heat. The sample was resuspended in 0.6 ml of acetonitrile, and 100- and 10-μl injections over a Zorbax SB-C18 reverse phase column (Agilent) were analyzed on an Agilent 1100 quaternary pump HPLC system by applying multiple linear solvent gradients and diode array UV absorbance detection as described by Napoli and co-workers (40). 325 nm UV absorbance peaks were quantified. Sample duplicates (100- and 10-μl injections) were compared to ensure that retinyl ester peaks did not exceed the detector linear range. The retinyl acetate internal standard (peak elution at 10.2–10.6 min) was used to quantify amounts of retinyl esters (peak elution at 16.5–17 min) and retinol (peak elution at 4.7–5.2 min), as described (40). Two biological replicates (separate aliquots of liver) were analyzed for each mouse and averaged. Replicates with >10% variation were reanalyzed. All extractions and studies were performed in reduced light (semidark room) before solvent extraction of tissues; after extraction, all studies, including HPLC, were performed in a dark room under dim red filtered light (>600 nm).
Statistical Analysis
Student's t test (two-tailed) was performed for group-wise comparisons for sample size (n) > 4 and where data were normally distributed. For sample size (n) ≤ 4 or for non-normally distributed data, the Mann-Whitney U test was performed. Pearson correlation and two-way ANOVA were performed with Prism 5 (GraphPad Software). A p value of <0.05 was considered significant for all tests.
RESULTS
Identification of a Putative Retinol Transporter and RBP4 Receptor, RBPR2, with Short Regions of Homology to Stra6
Because liver and other tissues without Stra6 demonstrate high affinity RBP4 binding and retinol uptake (11–13), we considered the possibility of an unidentified RBP4 receptor and retinol transporter in those tissues. Stra6 is reported to be a unique protein in a “family unto its own” (41). Nevertheless, we reasoned that other retinol transporters may share some structural similarity with Stra6 despite lacking overall homology. We therefore employed an unbiased phylogenetic search strategy to identify transcripts with relatively weak but conserved homology to Stra6. Mouse gene 1300002K09Rik, containing several short amino acid segments with >50% amino acid homology to Stra6 (but <18% overall amino acid homology; Fig. 1), was identified and further studied. We subsequently refer to this gene and its orthologues as RBP4 receptor-2, or RBPR2. Alignment of mouse RBPR2 and Stra6 amino acid sequences reveals unexpected structural similarities. RBPR2 is predicted to have a molecular mass of 70.1 kDa and 9–11 transmembrane domains like Stra6 (73.8 kDa) (16, 17), with similar spacing of the transmembrane domains (Figs. 1 and 2A and Table 2). In addition, both proteins have predicted intracellular C-terminal soluble domains containing 74–75 amino acids (Table 2). Remarkably, five of six amino acids affected by missense mutations in HMS9 (20, 21) are conserved in the alignment of Stra6 with RBPR2, suggesting analogous roles for these residues in the function or structural integrity of the two proteins (Figs. 1 and 2A and Table 2). RBPR2 lacks the canonical Src homology 2 domain present in the Stra6 C terminus (YTLL, amino acids 643–646 (16)) but displays a conserved threonine corresponding to the T644M HMS9 mutation (20) within the Stra6 Src homology 2 domain (Figs. 1 and 2A and Table 2).
FIGURE 1.
Alignment of Stra6, mouse 1300002K09Rik (RBPR2), and human RBPR2 orthologue chains A and B. Novel human RBPR2 chain A and chain B sequences, Hs-RBPR2-A and Hs-RBPR2-B, were cloned by 5′-RACE and 3′-RACE from a liver cDNA library and aligned with human (Hs) Stra6, murine (Mm) Stra6, and murine (Mm) RBPR2 using ClustalW (76). 5 of 6 amino acids with documented missense mutations causing loss of Stra6 function in HMS9 are conserved in the alignment. A proposed RBP4 binding site on Stra6 is also conserved. Homology of individually aligned amino acids is rated as exact (*), conserved (:), or partially conserved (.). Overall homology between murine Stra6 and RBPR2 is 17.8% based on this alignment.
FIGURE 2.
Conserved structural elements in human Stra6, murine 1300002K09Rik (RBPR2), and RBPR2 human orthologues. A, graphical alignment of conserved structural elements. The number of amino acids (aa) in the open reading frame encoding each protein is indicated. Solid black boxes indicate locations of conserved amino acids corresponding to Stra6 HMS9 mutations and proposed RBP4 binding domain. Shaded boxes indicate locations of possible transmembrane domains, based on predictive algorithms. Predicted transmembrane domains 10 and 11 of Stra6 (asterisks) may form part of an extended intracellular hydrophobic domain based on mapping studies by Kawaguchi et al. (17). B, alignment of RBPR2 coding exons (17 exons total) and flanking genes on mouse chromosome 4B1 with corresponding locations of RBPR2 Chain A (5 exons), RBPR2 Chain B (11 exons), and flanking genes at separate loci on human chromosome 9. The schematic illustrates only the relative positions of the exon sequences; distances between the genetic loci are not drawn to scale.
TABLE 2.
Conservation of structure in Stra6 and RBPR2
aa, amino acids; TMD, predicted transmembrane domain; HMS9 site, amino acid residues affected by missense mutations causing Stra6 loss of function in human microophthalmic syndrome 9; RBP4 binding, experimentally determined region of Stra6 involved in RBP4 binding (16, 17); SH2, Src homology 2 domain.
| Length | Mass | TMD1 | HMS9 site 1 | TMD2 | TMD3 | TMD4 | TMD5 | HMS9 site 2 | TMD6 | RBP4 binding | TMD7 | TMD8 | TMD9 | TMD10a | HMS9 Site 3 | TMD11a | SH2 domain | HMS9 site 4 | HMS9 site 5 | C terminus length | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| aa | kDa | aa | |||||||||||||||||||
| Stra6 | 667 | 73.8 | 54–71 | Pro-90 | 101–119 | 148–166 | 172–196 | 205–221 | Pro-293 | 302–318 | 336–338 | 366–382 | 435–451 | 475–491 | 509–528 | Asp-560 | 576–592 | 643–646 | Thr-644 | Arg-655 | 75 |
| 1300002K09Rik (RBPR2) | 621 | 70.9 | 23–43 | Pro-67 | 70–86 | 104–122 | 137–153 | 172–188 | Pro-253 | 256–275 | 294–296 | 323–340 | 391–409 | 425–443 | 486–502 | Asp-522 | 529–547 | Thr-589 | Arg-600 | 74 |
a Predicted TMD10 and TMD11 of Stra6 may be part of an extended intracellular hydrophobic domain based on mapping studies by Kawaguchi et al. (17); according to that model, Stra6 and RBPR2 potentially have nine actual transmembrane domains.
Novel, Unannotated Human Orthologues of RBPR2
RBPR2 is highly conserved throughout vertebrate species, including primates. The mouse RBPR2 gene shares similar exon/intron organization with mouse and human Stra6 (not shown). However, in humans and other primates, open reading frames corresponding to mouse RBPR2 are found to exist as separate sets of exons within separate genes located in regions of human chromosomes 9p and 9q syntenic to the RBPR2 locus at mouse chromosome 4B1 (Fig. 2B). The 9q gene product (which we designate RBPR2 human homolog chain B) is highly homologous to the C-terminal 70% of mouse RBPR2 and has a predicted molecular mass of 48.3 kDa, whereas the 9p gene product (RBPR2 human homolog chain A) is highly homologous to the N-terminal 30% of mouse RBPR2 and has a predicted molecular mass of 18.8 kDa (Figs. 1 and 2).
RBPR2 Membrane Localization
To determine the subcellular localization of RBPR2 we transfected HEK-293 cells with plasmids expressing HA epitope-tagged mouse RBPR2 (HA-RBPR2) or Stra6 (HA-Stra6) for comparison. RBPR2 and Stra6 were detected by immunofluorescence confocal microscopy (Fig. 3A, top panels). A membrane-specific fluorescent label (fluorescein-wheat germ agglutinin; Fig. 3A, middle panels) co-localized with RBPR2 and Stra6 (Fig. 3A, bottom panels) in the plasma membrane and also co-localized with RBPR2 in the Golgi region (Fig. 3A, bottom left panel). Western blot analysis of total membrane and cytosol fractions confirmed membrane localization (Fig. 3B, lanes 1, 2, 5, and 6, labeled M (membrane) and C (cytosol). HA-Stra6 exhibited similar localization by microscopy and Western blotting (Fig. 3B, lanes 3, 4, 7, and 8). Western blotting of both HA-RBPR2 and HA-Stra6 produced immunoreactive doublets for each protein, with major bands running at or near predicted molecular masses of ∼71 kDa (HA-RBPR2) and 75 kDa (HA-Stra6) for the tagged proteins (Fig. 3B, arrows). Both proteins displayed some higher molecular mass laddering (Fig. 3B, lanes 1 and 3, asterisk). HA-RBPR2 additionally displayed prominent lower molecular mass (40–50 kDa) bands that appear to be C terminus proteolytic products (Fig. 3B, lane 5, bracket labeled RBPR2′). Membrane fractions treated in vitro with a mixture of endoglycosidases exhibited reduced higher molecular weight laddering (Fig. 3B, lanes 5 and 7), indicating that each protein is glycosylated to some extent. Interestingly, endoglycosidase treatment of the RBPR2 membrane fraction increased the amount of ∼45-kDa RBPR2 C terminus proteolytic fragment detected by Western blotting (Fig. 3B, lane 5). Hence, glycosylation state could play a role in regulating RBPR2 stability or susceptibility to proteolytic cleavage.
FIGURE 3.
Subcellular localization of RBPR2. A, immunofluorescence confocal microscopy of fixed, permeabilized HEK-293 cells transfected with plasmids expressing C terminus HA-tagged RBPR2 (HA-RBPR2) or Stra6 (HA-Stra6). Top panels, anti-HA immunofluorescence; middle panels, membrane staining (endomembranes and plasma membranes) with wheat germ agglutinin-fluorescein (WGA) in green and DAPI nuclear staining in blue; bottom panels, co-localization of HA-tagged receptors with WGA-stained membranes. Increasing brightness of the pseudocolored images indicates increasing receptor-membrane co-localization. B, Western blotting of cytosol (C) and membrane (M) fractions of HEK-293 cells transfected with HA-RBPR2 (lanes 1, 2, 5, and 6) or HA-Stra6 (lanes 3, 4, 7, and 8). The indicated fractions (lanes 5–8) were treated in vitro with an endoglycosidase mix before SDS-PAGE. Western blots of the same membrane and cytosol fractions for transferrin receptor (TfR) and the IGF1 receptor (IGF1R), two known transmembrane receptors, are shown as markers for the quality of the fractions.
RBPR2 Tissue Expression Pattern
To determine the tissue distribution of RBPR2, we measured RBPR2 mRNA in a survey of normal mouse tissues. RBPR2 mRNA is most highly expressed in the liver, followed by small intestine, including the jejunum and ileum regions involved in dietary retinol absorption (Fig. 4A). The spleen exhibits a smaller but significantly increased amount of RBPR2 expression (Fig. 4A). In other tissues, including adipose tissue and isolated adipocytes, RBPR2 mRNA is near the limit of detection for qRT-PCR (Ct values 35–40; relative expression shown in Fig. 4A). A similar pattern of RBPR2 tissue expression was observed in a survey of tissues obtained from wild type male C57Bl6 mice (data not shown). Interestingly, RBPR2 mRNA is increased in adipose tissue of two different obese mouse models: C57Bl6 mice fed a high fat diet (Fig. 4B) and leptin-deficient ob/ob mice (Fig. 4C). Within obese adipose tissue, RBPR2 mRNA is increased in both isolated adipocytes and stromovascular cells (Fig. 4D). Because the stromovascular fraction is enriched in vascular cells, this observation suggests that RBPR2, like Stra6, might be expressed in the vascular endothelium of certain tissues. In addition, it is possible that RBPR2 is expressed in non-vascular components, such as stromal macrophages, the presence of which is known to increase in the setting of obesity and insulin resistance. Consistent with a potential role in obesity, RBPR2 mRNA is induced in late stages of 3T3L1 preadipocyte to adipocyte differentiation (Fig. 4E). Because we lack antibodies that can quantify in vivo tissue expression of RBPR2 protein, we studied expression of a RBPR2-β-gal fusion protein under the control of the endogenous RPBR2 promoter in transgenic “promoter-reporter” mice. This model confirmed high level expression of RBPR2 in the liver (Fig. 4F). Intestine or adipose expression of RBPR2 could not be confirmed in this model due to significant endogenous galactosidase activity in those tissues (shown for wild type adipose in Fig. 4F). Together, these findings indicate that RBPR2 is most highly expressed in the liver and intestine and strongly induced in obese adipose tissue. Because these tissues mediate uptake, storage, and distribution of retinol, these findings suggest that RBPR2 could play a physiologically relevant role in whole body retinol homeostasis.
RBP4 Binding to RBPR2
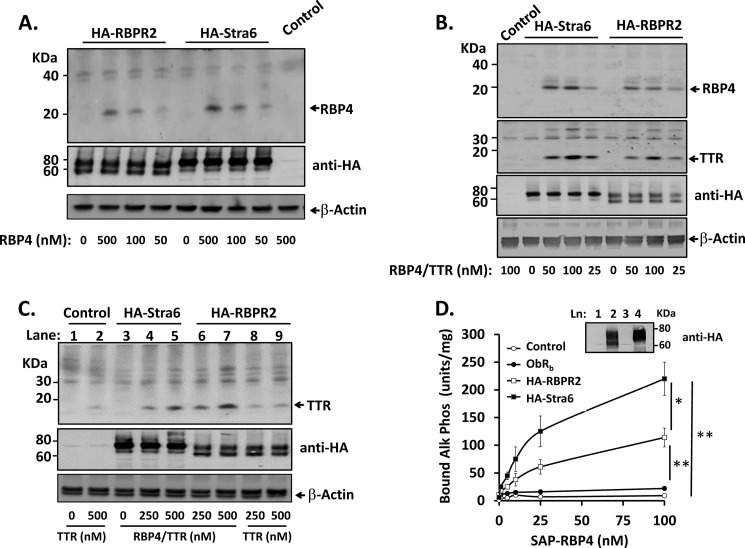
Despite low amino acid homology, RBPR2 shares considerable structural similarity with Stra6 and therefore potentially binds circulating RBP4. We tested whether transfection of RBPR2 in HEK-293 cells confers increased binding of purified RBP4 or RBP4·TTR complex. HEK-293 cells were selected for these studies because they were reported to exhibit low RBP4-dependent retinol transport in prior studies (16); in addition, we do not detect significant expression of Stra6 or RBPR2 in this line, and secreted endogenous RBP4 is not detected in cell-conditioned medium (data not shown). RBP4 circulates as a 1:1 complex with TTR at concentrations of 0.5–2 μm in normal mice and humans (9). In control-transfected cells incubated with purified RBP4 (500 nm), there was no detectable binding of RBP4 (Fig. 5A), whereas in cells transfected with RBPR2 or Stra6, RBP4 binding was detected over a range of concentrations (50–500 nm; Fig. 5A). Incubating cells with purified RBP4·TTR complex produced similar results (Fig. 5B, top), and TTR paralleled RBP4 binding (Fig. 5B, top middle panel). Purified TTR alone exhibited some nonspecific binding activity in control plasmid-transfected cells (Fig. 5C, lane 2); however, there was no significant increase in binding in HA-RBPR2-transfected cells (Fig. 5C, lanes 8 and 9 versus lane 2). In contrast, cells transected with HA-Stra6 or HA-RBPR2 and incubated with RBP4·TTR complex exhibited increased, dose-dependent binding of TTR (Fig. 5C, lanes 4–7). Together, these data indicate that RBP4·TTR complex binding is mediated by RBP4-RBPR2 interactions rather than TTR-RBPR2 interactions.
FIGURE 5.
RBPR2 mediates binding of RBP4 to cells. For A–C, HEK-293 cells transfected with C terminus HA-tagged RBPR2 or Stra6 (HA-RBPR2 and HA-Stra6) or empty plasmid control (pcDNA3.1), as indicated, were incubated in serum-free medium containing different purified proteins (RBP4 alone, RBP4·TTR complex, or TTR alone). Cells were washed to remove unbound proteins, lysed, and analyzed by SDS-PAGE and Western blotting. A, incubations were performed with indicated concentrations of purified human RBP4; Western blots are shown for bound RBP4 (top), transfected receptors (middle), or β-actin as a loading control (bottom). B, incubations were performed with the indicated concentrations of purified human RBP4·TTR complex; Western blots are shown for bound RBP4 (top panel), bound TTR (second panel), transfected receptors (third panel) or β-actin as a loading control (bottom panel). C, incubations were performed with the indicated concentrations of purified human TTR; Western blots are shown for bound TTR (top), transfected receptors (middle), or β-actin as a loading control (bottom). D, HEK-293 cells were transfected with HA-RBPR2, HA-Stra6, or an irrelevant receptor (leptin receptor long form, ObRb) and incubated in serum-free medium with indicated concentrations of recombinant SAP-RBP4 fusion protein. Cells were washed and lysed, and binding of SAP-RBP4 fusion protein was detected by measurement of thermostable alkaline phosphatase activity (units/mg of lysate protein), as described (16). Control-transfected cells (Control) received an empty plasmid (pcDNA3.1). *, p < 0.05; **, p < 0.01 by repeated measures ANOVA for the indicated binding curve comparisons. Inset, Western blot of HA-tagged receptors in membrane preparations of cells transfected in parallel. Error bars, S.E.
To better assess binding affinity, we used recombinant SAP-RBP4. This method can detect RBP4 binding at low nanomolar concentrations (16). Concentration-dependent SAP-RBP4 binding was observed in cells expressing HA-RBPR2 or HA-Stra6 (Fig. 5D). Background SAP-RBP4 binding was very low (5–7% of levels observed for HA-RBPR2) in control plasmid-transfected cells or cells transfected with leptin receptor long form (ObRb), an irrelevant receptor negative control (Fig. 5D). Calculated binding affinities (Kd) of SAP-RBP4 for HA-Stra6 and HA-RBPR2 were 44.4 ± 17.6 and 52.6 ± 14.9 nm, respectively. Bovine Stra6 was previously reported to bind SAP-RBP4 with a Kd of 59 nm in transfected COS1 cells (16). Amounts of total HA-RBPR2 and HA-Stra6 detected in membrane preparations by anti-HA Western blotting were similar (Fig. 5D, inset). However, there was more total RBP4 binding observed for Stra6 than for RBPR2 at each concentration (Fig. 5D), suggesting that a greater percentage of Stra6 sites may be functional in this model. These data indicate that RBPR2 binds RBP4, alone or in complex with TTR, with affinity comparable to that of Stra6.
Binding of RBP4 was non-covalent and peripheral to the membrane (i.e. not associated with endocytosis), because treatment of intact cells with sodium bicarbonate at pH 10 followed by washing in PBS eliminated bound RBP4 (Fig. 6A). As expected, RBP4 binding correlated with the amount of RBPR2 expression plasmid used in transfections (Fig. 6B), and there was no increase of RBP4 binding to cells transfected with HA-tagged membrane proteins other than Stra6 and RBPR2 (Fig. 6, C and D).
FIGURE 6.
Characterization of RBP4 binding to receptor-transfected cells. A, HEK-293 cells transfected with C terminus HA-tagged RBPR2 (HA-RBPR2) or Stra6 (HA-Stra6) were incubated in serum-free medium containing 0.5 μm purified human RBP4. Cells were (i) washed twice with PBS and lysed; (ii) treated with 100 mm sodium bicarbonate, pH 10, for 20 min to extract peripheral membrane proteins and lysed; or (iii) incubated in bicarbonate buffer, washed twice with PBS, and lysed, as indicated. Lysates were analyzed by SDS-PAGE and Western blotting to detect bound RBP4 (arrow). B, HEK-293 cells were transfected with the indicated quantities of plasmid DNA for expression of C terminus HA-tagged RBPR2 (HA-RBPR2); the “0” HA-RBPR2 condition represents cells transfected with an empty vector control plasmid (pcDNA3.1, 5 μg). Binding was performed as described for Fig. 5 and under “Experimental Procedures” and analyzed by SDS-PAGE and Western blotting to detect bound RBP4 (arrow). C and D, HEK-293 cells were transfected with expression plasmids for C terminus HA-tagged Stra6 (HA-Stra6), a positive control for RBP4 binding, or two irrelevant HA-tagged transmembrane proteins: transforming growth factor β receptor II (TGFR2) or receptor-type protein-tyrosine phosphatase α (PTPRα). Each receptor displays an extracellular HA tag based on known topology. C, anti-HA tag Western blot of membrane (M) and cytosol (C) fractions from the transfected cells. Arrows, bands in membrane fraction corresponding to HA-Stra6 (usual mobility 70–80 kDa), HA-TGFR2 (usual mobility 60–80 kDa), and HA-PTPRα (usual mobility 95–115 kDa). D, binding of purified RBP4 to HA-receptor-transfected cells was performed as described above and analyzed by SDS-PAGE and Western blotting to detect bound RBP4 (arrow).
RBPR2-mediated Retinol Uptake from Holo-RBP4
In order to determine whether RBPR2 shares the retinol transport function of Stra6, we studied uptake of [3H]retinol in RBPR2-transfected HEK-293 cells. Cells transfected with HA-Stra6 (Fig. 7A, left) or HA-RBPR2 (Fig. 7A, right) exhibited increased retinol uptake in comparison with cells transfected with an empty control vector (open and closed squares versus open circles); control (basal) values were comparable with those reported by others in similar experiments (16). Increased uptake of retinol by RBPR2 could be detected within 10 min of incubation with holo-RBP4 (Fig. 7B). Co-transfection of the retinol esterifying enzyme, LRAT, increased uptake by both Stra6 and RBPR2 in comparison with cells transfected with either receptor alone (Fig. 7A, closed squares versus open squares). However, LRAT alone did not increase retinol uptake in the absence of receptor expression (Fig. 7A, closed circles). Uptake of albumin-bound [3H]retinol was low (4.4–5.5% of maximal values) in HA-Stra6- or HA-RBPR2-transfected cells (Fig. 7A, closed triangles), indicating that retinol transport favors specific RBP4-receptor interactions, as reported by others (16, 42, 43). Together, these data indicate that RBPR2 functions as an RBP4-dependent retinol transporter like Stra6. In addition, although LRAT alone does not catalyze retinol uptake, it does act cooperatively with RBPR2 to enhance the intracellular accumulation of retinol. These observations suggest that RBPR2 could mediate retinol uptake and storage in liver, where endogenous LRAT is highly expressed.
FIGURE 7.
RBPR2 mediates retinol uptake. A, HEK-293 cells were transfected with C terminus HA-tagged Stra6 (left, open and closed squares) or RBPR2 (right, open and closed squares). Control cells were transfected with an empty plasmid (pcDNA3.1; left and right, open circles). Where indicated, cells were additionally co-transfected with an expression plasmid for LRAT (left and right), the intracellular enzyme that produces the retinyl ester storage form of retinol. Intact cells were incubated for 30 min at 37 ºC in serum-free medium containing purified recombinant murine RBP4 or fatty acid-free bovine serum albumin (a nonspecific ligand control) loaded with [3H]retinol. The x axis shows the concentration of either retinol-bound RBP4 or retinol-bound albumin. Cells were washed and lysed, and retinol uptake was determined by scintillation counting (expressed as fmol/mg of cell lysate protein/min). **, p < 0.01 for indicated comparisons as determined by two-way ANOVA. Inset, Western blot of HA-tagged receptors in membrane preparations of cells transfected in parallel. B, [3H]retinol uptake from holo-RBP4 or bovine serum albumin (100 nm each) was measured in transfected HEK-293 cells at different time points. The mean of four biological replicates is shown for each time point and condition. Inset, anti-HA Western blot of HA-tagged RBPR2 in membrane preparations of cells transfected in parallel. *, p < 0.05 for RBP4-ROH versus control and albumin-ROH, RBPR2 conditions.
Endogenous RBPR2 Knockdown in Cultured Hepatocytes
To study endogenous RBPR2 regulation and function, we made use of hepatoma cell lines H4IIe (rat) and Hepa1 (mouse) that express endogenous RBPR2 at levels comparable with liver; in addition, Stra6 expression in these cell lines (as in liver) is near the limits of detection based on absolute Ct values of qRT-PCR curves (data not shown). Each of these cell types also express endogenous RBP4, although at levels much lower than in liver, such that accumulation of secreted RBP4 protein is not detected in culture medium by Western blotting (data not shown). H4IIe cells express LRAT at levels comparable with liver, whereas Hepa1 cells express much lower levels of LRAT (data not shown); even so, both cell types exhibit rates of retinol uptake similar to those of isolated primary hepatocytes studied ex vivo (14, 44, 45). To determine whether RBPR2 is required for normal retinol transport in hepatocytes, we studied the effect of siRNA-mediated knockdown of RBPR2 in H4IIe. Transfection of RBPR2-specific siRNA but not non-targeting control (NTC) siRNA resulted in >80% knockdown of RBPR2 mRNA (Fig. 8A). RBPR2 knockdown decreased binding of SAP-RBP4 by 48% (Fig. 8B) and decreased [3H]retinol uptake from holo-RBP4 (Fig. 8C) by 73%. These findings indicate that RBPR2 may be an important mediator of RBP4 binding and RBP4-dependent retinol uptake in hepatocytes. In addition, the persistence of retinol uptake after efficient knockdown of RBPR2 suggests that other receptors or mechanisms could mediate some fraction of retinol transport in these cells.
FIGURE 8.
Endogenous RBPR2 mediates [3H]retinol uptake and RBP4 binding in hepatoma cells. H4IIe rat hepatoma cells were treated with a siRNA pool designed to knock down RBPR2 or a non-targeting control siRNA pool (NTC). Effects of siRNAs were compared with treated control cells that did not receive siRNA (Mock). A, RBPR2 knockdown was confirmed by measuring RBPR2 mRNA levels by qRT-PCR (Solaris). **, p < 0.01 for the indicated comparison. B, binding of SAP-RBP4 at the indicated concentrations was measured in control plasmid-transfected cells and cells transfected with RBPR2 siRNA or non-targeting control siRNA, as described in the legend to Fig. 5D and under “Experimental Procedures.” **, p < 0.01 for the indicated comparison by two-way ANOVA. C, [3H]retinol uptake was measured in mock-transfected cells and cells transfected with RBPR2 siRNA or NTC siRNA, as described in the legend to Fig. 7A and under “Experimental Procedures.” *, p < 0.05 for the indicated comparison. For A, three biological replicates were measured for each condition; data are representative of two repeated experiments. For B and C, six biological replicates were measured in data combined from two repeated experiments. Error bars, S.E.
Regulation of RBPR2 Expression and Relation to Liver Retinol Stores
To determine whether RBPR2 expression is associated with retinol homeostasis in the liver in vivo, we compared RBPR2 mRNA with retinol and retinyl ester content in livers of normal adult mice (6–7 months of age) fed chow containing 15 IU/g retinol. RBPR2 mRNA correlated inversely with liver total retinol content (retinol plus retinyl esters) across the range of observed values (Fig. 9A). Because RBPR2 mediates cellular uptake of retinol from holo-RBPR2, this finding could indicate that RBPR2 expression is negatively coupled to intracellular retinol/retinoic acid. Several important regulators of liver retinol homeostasis are controlled by retinoic acid (RA) at the transcriptional level, including LRAT, CRBP1, RALDH1, RARβ, and Cyp26a (46–49). In addition, the proximal promoter of RBPR2 contains a conserved DR2 retinoic acid response element (at −685 base pairs from start) that may confer regulation by retinoic acid (50). To determine whether retinoids regulate RBPR2 expression, we treated mouse Hepa1 cultured hepatocytes overnight with equal amounts of holo-RBP4 or free retinol; both treatments caused dose-dependent reductions in RBPR2 mRNA (Fig. 9B). The treatments did not appear to induce toxicity because they did not significantly alter cell adherence, cellular protein content, or expression of individual housekeeping genes (not shown). To determine whether metabolism of retinol to retinoic acid mediates these effects, we treated Hepa1 cells with all-trans-retinoic acid (ATRA) under conditions of normal (10%) or reduced (0.2%) fetal bovine serum. ATRA strongly suppressed RBPR2 mRNA under both serum conditions (Fig. 9C). In addition, RBPR2 expression was decreased 77% in medium with reduced fetal bovine serum, indicating that serum factors other than retinol, potentially growth factors, additionally regulate RBPR2 expression (Fig. 9C). To determine if suppression of RBPR2 by retinoids could alter retinol uptake in Hepa1, we measured uptake of [3H]retinol from holo-RBP4 in cells pretreated with retinol or retinoic acid; both treatments suppressed [3H]retinol uptake (Fig. 9D). Together, these findings suggest the possible existence of a short loop negative feedback mechanism by which increased availability of exogenous retinol might down-regulate RBPR2 mRNA in a retinoic acid-dependent manner.
FIGURE 9.
Liver RBPR2 expression in vivo and regulation of RBPR2 mRNA by retinol and retinoic acid. A, RBPR2 mRNA was measured by qRT-PCR targeting the exon 10/11 junction (Solaris) in the liver of wild type C57Bl6 mice (n = 7, males, age 6–7 months). Retinol and retinyl ester were measured in parallel by HPLC in aliquots of the same liver samples, and averages were correlated; R is the Pearson correlation co-efficient. A two-tailed p value was calculated. Correlation with retinol and retinyl ester was also obtained (R = 0.77, p < 0.05; data not shown) for qRT-PCR targeting the exon 1/2 junction (Applied Biosystems). B, RBPR2 mRNA was measured in Hepa1 mouse hepatoma cells treated overnight in reduced serum (0.2% FBS) medium containing the indicated concentrations of human holo-RBP4 or equimolar free retinol added directly to the medium. *, p < 0.05; **, p < 0.01 versus vehicle-treated control. C, RBPR2 mRNA was measured in Hepa1 cells treated overnight with the indicated concentrations of ATRA in medium containing normal or reduced serum (10 or 0.2% FBS, respectively). **, p < 0.01 for the indicated comparisons; three biological replicates were measured. For B and C, RBPR2 was normalized to housekeeping gene GAPDH; results without normalization were comparable (not shown). D, [3H]retinol uptake from mouse holo-RBP4 (100 nm) was determined in cells pretreated overnight with 1 μm retinol or ATRA, as described in the legend to Fig. 7A and under “Experimental Procedures.” *, p < 0.05; **, p < 0.01 versus vehicle-treated control. Six biological replicates were measured in data combined from two separate experiments. Error bars, S.E.
DISCUSSION
Liver has been recognized as the primary site of retinol storage in the body since the discovery of vitamin A in the early 20th century (1, 51, 52). However, the molecular basis of retinol transport in liver has remained elusive. We now report discovery of RBPR2, a retinol transporter and high affinity RBP4 receptor expressed highly in liver. RBPR2 is also expressed in the intestine, suggesting a potential role in dietary retinol absorption. In addition, RBPR2 is induced during adipocyte differentiation, and RBPR2 mRNA is increased in adipose tissue in obesity. RBPR2 shares structural homology with Stra6 and exhibits comparable RBP4-binding and retinol uptake kinetics. Therefore, RBPR2 appears to be a previously unrecognized member of the Stra6 family.
RBPR2 and Stra6 differ in several ways. RBPR2 and Stra6 exhibit fundamentally different patterns of tissue expression. Stra6 is not expressed in liver or intestine (16, 18), tissues where RBPR2 is most highly expressed. RBPR2 lacks the Stra6 C terminus Src homology 2 domain recently found to mediate RBP4-dependent Jak-STAT signaling in certain cell types (53, 54). It remains to be determined whether RBP4 binding to RBPR2 can activate signaling in the absence of an intact Src homology 2 domain. RA suppresses RBPR2 and up-regulates Stra6 (55, 56). Because RA production is in part controlled by retinol availability, opposite regulation of the two proteins by RA could be a mechanism for directing RBP4 or retinol flux as needed either to liver (expressing RBPR2 when circulating RBP4, retinol, and/or intracellular RA levels are low) or to extrahepatic tissues (expressing Stra6 when RBP4, retinol, and/or RA are high). Consistent with that possibility, compartmental modeling of retinol delivery in humans has shown that utilization/metabolism of retinol is greatest when liver retinol stores and circulating holo-RBP4 concentrations are high (57). Future studies are needed to determine whether inverse regulation of RBPR2 and Stra6 by RA plays a role in coordinating in vivo retinol homeostasis.
RBPR2 is highly conserved in vertebrates, including mammals. In humans and other primates, RBPR2 appears to have evolved as two separate genes; exons encoding a longer “RBPR2 chain B” are located on 9q and contain open reading frames corresponding to the C terminus 70% of the mouse protein, whereas exons encoding a shorter “RBPR2 Chain A” are located on 9p and contain open reading frames corresponding to the N terminus 30% of the mouse protein (Fig. 2B). Each of these regions of human chromsome 9 is syntenic with mouse chromosome 4B1. Several of the nearest genes located centromeric to RBPR2 on mouse 4B1 are also present in tandem with the RBPR2 Chain A gene on human 9p (Aldh1b, Igfbpl1, Shb, and Mcart1), indicating that the RBPR2 locus was itself the breakpoint for a chromosomal inversion during primate evolution (Fig. 2B). Interestingly, mouse RBPR2 expressed in HEK-293 cells exhibits a distinct C-terminal ∼45-kDa proteolytic product on Western blotting that is increased by endoglycosidase treatment (Fig. 3B, lane 5), suggesting that under some conditions RBPR2 may undergo proteolysis to produce a fragment similar in size to the predicted human RBPR2 chain B protein (48.3 kDa). Additional studies are needed to determine the extent to which human RBPR2 chains A and B may be co-regulated, whether the chains interact to assemble a functional retinol transporter and RBP4 receptor, and whether the mouse RBPR2 protein undergoes regulated post-translational processing by proteolysis.
These studies have limitations. We have attempted to develop several different polyclonal antibodies to RBPR2, but have not produced an antibody that reliably detects RBPR2 protein in endogenous tissues. Transfected HA-RBPR2 is highly insoluble (like Stra6) and requires use of zwitterionic detergents and phospholipids for solubilization from membranes of transfected cells (see “Experimental Procedures”). Antibody design and solubilization methods for primary tissues will require optimization before we can accurately determine endogenous RBP4 protein expression by Western blotting. Nevertheless, we are able to confirm high level expression of a RBPR2-alkaline phosphatase fusion protein in the liver of RBPR2 promoter-reporter mice, confirming qRT-PCR findings that RBPR2 expression is highest in the liver. Consistent with this, RBPR2 mRNA is detected in H4IIe and Hepa1 hepatoma cells, lines known to express markers of differentiated hepatocytes (58, 59). In addition, the diffuse pattern of β-gal expression in intact liver of RBPR2 promoter-reporter mice suggests that hepatocytes, which make up 80% of liver by mass (60), are the principal cell type in which RBPR2 is expressed. However, it remains possible that other liver cell types, including stellate cells, ductal cells, endothelial cells, and Kupffer cells, express RBPR2. Stellate cells play a particularly important role in liver retinol homeostasis, because they store the majority of retinyl esters in neutral lipid droplets (61, 62). The process by which retinol is transferred between stellate cells and hepatocytes is not well defined, and future studies are needed to determine if RBPR2 plays a role.
The inverse relationship between liver retinol stores and RBPR2 expression (Fig. 9A) and the inhibitory effects of holo-RBP4, retinol, or retinoic acid on RBPR2 expression (Fig. 9, B and C) suggest that RBPR2 may respond in a physiologically relevant manner to circulating retinol, dietary retinol, or liver retinol stores. To test this possibility, further studies assessing in vivo regulation of liver RBPR2 during manipulation of dietary retinol may be informative. Because in vivo retinol homeostasis involves coordination of multiple tissues and cell types (1), much information may be gained in the future through the use of in vivo models for studying tissue-specific expression or genetic deletion of RBPR2. Toward that end, we are engineering mice with a Cre/LoxP conditional allele of RBPR2.
We did not study all possible functions of RBPR2 that may exist by analogy to Stra6. For instance, it recently has been proposed that Stra6 functions as a bidirectional transporter to mediate release of retinol under certain conditions (63, 64); hence, RBPR2 might similarly mediate bidirectional shuttling of retinol between different cell types in liver or in other tissues. Consistent with this possibility, it appears that LRAT expression is required to drive Stra6-mediated retinol transport directionally toward uptake rather than efflux in certain tissues in vivo (65). Future studies are needed to determine whether RBPR2 binds apo-RBP4 or mediates retinol “reverse transport” and whether RBPR2 similarly requires LRAT for uptake of retinol in vivo.
In addition to its important role in retinol transport, RBP4 has been implicated in the pathogenesis of diseases associated with insulin resistance (34). Circulating RBP4 is increased in obesity, metabolic syndrome, and type 2 diabetes, and excess RBP4 produced by adipocytes may play a causal role in the pathogenesis of type 2 diabetes (66–74). Injection of mice with purified holo-RBP4 causes insulin resistance and glucose intolerance, in part through induction of liver phosphoenolpyruvate carboxykinase, a rate-controlling enzyme of gluconeogenesis (34). Treatment of cultured hepatocytes, 3T3L1 adipocytes, and isolated primary adipocytes with purified RBP4 also causes insulin resistance via regulation of several intracellular signaling pathways (34, 53, 75). Our findings raise the possibility that RBPR2, in the capacity of RBP4 receptor, mediates effects of RBP4 on insulin action and glucose homeostasis. In addition, the strong induction of RBPR2 observed in obese adipose tissue suggests a potential role for RBPR2 in adaptations of adipose tissue that enable production of excess RBP4 by adipocytes in insulin-resistant states. Future studies assessing the metabolic phenotype of mice genetically engineered for altered expression of RBPR2 are planned.
Acknowledgments
We thank Dr. E. Dale Abel, Dr. Dean Li, Dr. Curt Hagedorn, and Dr. Don McClain (University of Utah) for technical assistance and advice. We thank Dr. Jared Rutter (University of Utah) for sharing rat liver mRNA and for useful insight throughout the project.
This work was supported, in whole or in part, by National Institutes of Health Grant R03 DK080195 (to T. E. G.). This work was also supported by a Biomedical Laboratory Research and Development Merit Review Award from the Department of Veterans Affairs (01BX000937) (to T. E. G.) and a Physician Scientist Early Career Award from the Howard Hughes Medical Institute (to T. E. G.). Cloning of human RBPR2 orthologues (and no animal-related work) was supported by a Clinical Scientist Development Award from the Doris Duke Charitable Foundation (to T. E. G.).
- HMS9
- human micro-ophthalmic syndrome 9
- ANOVA
- analysis of variance
- ATRA
- all-trans-retinoic acid
- β-gal
- β-galactosidase
- LRAT
- lecithin retinol acyltransferase (phosphatidylcholine-retinol O-acyltransferase)
- RACE
- rapid amplification of cDNA ends
- SAP
- secreted alkaline phosphatase
- TTR
- transthyretin, also known as prealbumin
- X-gal
- 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside
- qRT-PCR
- quantitative RT-PCR
- RA
- retinoic acid.
REFERENCES
- 1. Norum K. R., Blomhoff R. (1992) McCollum Award Lecture, 1992. Vitamin A absorption, transport, cellular uptake, and storage. Am. J. Clin. Nutr. 56, 735–744 [DOI] [PubMed] [Google Scholar]
- 2. Tanumihardjo S. A. (2011) Vitamin A. Biomarkers of nutrition for development. Am. J. Clin. Nutr. 94, 658S–665S [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chambon P. (1996) A decade of molecular biology of retinoic acid receptors. FASEB J. 10, 940–954 [PubMed] [Google Scholar]
- 4. McGrane M. M. (2007) Vitamin A regulation of gene expression. Molecular mechanism of a prototype gene. J. Nutr. Biochem. 18, 497–508 [DOI] [PubMed] [Google Scholar]
- 5. Mendoza-Parra M. A., Walia M., Sankar M., Gronemeyer H. (2011) Dissecting the retinoid-induced differentiation of F9 embryonal stem cells by integrative genomics. Mol. Syst. Biol. 7, 538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shirakami Y., Lee S. A., Clugston R. D., Blaner W. S. (2012) Hepatic metabolism of retinoids and disease associations. Biochim. Biophys. Acta 1821, 124–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tsutsumi C., Okuno M., Tannous L., Piantedosi R., Allan M., Goodman D. S., Blaner W. S. (1992) Retinoids and retinoid-binding protein expression in rat adipocytes. J. Biol. Chem. 267, 1805–1810 [PubMed] [Google Scholar]
- 8. Blaner W. S. (1989) Retinol-binding protein. The serum transport protein for vitamin A. Endocr. Rev. 10, 308–316 [DOI] [PubMed] [Google Scholar]
- 9. Zanotti G., Berni R. (2004) Plasma retinol-binding protein. Structure and interactions with retinol, retinoids, and transthyretin. Vitam. Horm. 69, 271–295 [DOI] [PubMed] [Google Scholar]
- 10. Lewis K. C., Green M. H., Green J. B., Zech L. A. (1990) Retinol metabolism in rats with low vitamin A status. A compartmental model. J. Lipid Res. 31, 1535–1548 [PubMed] [Google Scholar]
- 11. Gjøen T., Bjerkelund T., Blomhoff H. K., Norum K. R., Berg T., Blomhoff R. (1987) Liver takes up retinol-binding protein from plasma. J. Biol. Chem. 262, 10926–10930 [PubMed] [Google Scholar]
- 12. Smeland S., Bjerknes T., Malaba L., Eskild W., Norum K. R., Blomhoff R. (1995) Tissue distribution of the receptor for plasma retinol-binding protein. Biochem. J. 305, 419–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Blomhoff R., Norum K. R., Berg T. (1985) Hepatic uptake of [3H]retinol bound to the serum retinol-binding protein involves both parenchymal and perisinusoidal stellate cells. J. Biol. Chem. 260, 13571–13575 [PubMed] [Google Scholar]
- 14. Yamamoto Y., Yoshizawa T., Kamio S., Aoki O., Kawamata Y., Masushige S., Kato S. (1997) Interactions of transthyretin (TTR) and retinol-binding protein (RBP) in the uptake of retinol by primary rat hepatocytes. Exp. Cell Res. 234, 373–378 [DOI] [PubMed] [Google Scholar]
- 15. Quadro L., Blaner W. S., Hamberger L., Novikoff P. M., Vogel S., Piantedosi R., Gottesman M. E., Colantuoni V. (2004) The role of extrahepatic retinol binding protein in the mobilization of retinoid stores. J. Lipid Res. 45, 1975–1982 [DOI] [PubMed] [Google Scholar]
- 16. Kawaguchi R., Yu J., Honda J., Hu J., Whitelegge J., Ping P., Wiita P., Bok D., Sun H. (2007) A membrane receptor for retinol binding protein mediates cellular uptake of vitamin A. Science 315, 820–825 [DOI] [PubMed] [Google Scholar]
- 17. Kawaguchi R., Yu J., Wiita P., Ter-Stepanian M., Sun H. (2008) Mapping the membrane topology and extracellular ligand binding domains of the retinol binding protein receptor. Biochemistry 47, 5387–5395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bouillet P., Sapin V., Chazaud C., Messaddeq N., Décimo D., Dollé P., Chambon P. (1997) Developmental expression pattern of Stra6, a retinoic acid-responsive gene encoding a new type of membrane protein. Mech. Dev. 63, 173–186 [DOI] [PubMed] [Google Scholar]
- 19. Golzio C., Martinovic-Bouriel J., Thomas S., Mougou-Zrelli S., Grattagliano-Bessieres B., Bonniere M., Delahaye S., Munnich A., Encha-Razavi F., Lyonnet S., Vekemans M., Attie-Bitach T., Etchevers H. C. (2007) Matthew-Wood syndrome is caused by truncating mutations in the retinol-binding protein receptor gene STRA6. Am. J. Hum. Genet. 80, 1179–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pasutto F., Sticht H., Hammersen G., Gillessen-Kaesbach G., Fitzpatrick D. R., Nürnberg G., Brasch F., Schirmer-Zimmermann H., Tolmie J. L., Chitayat D., Houge G., Fernández-Martínez L., Keating S., Mortier G., Hennekam R. C., von der Wense A., Slavotinek A., Meinecke P., Bitoun P., Becker C., Nürnberg P., Reis A., Rauch A. (2007) Mutations in STRA6 cause a broad spectrum of malformations including anophthalmia, congenital heart defects, diaphragmatic hernia, alveolar capillary dysplasia, lung hypoplasia, and mental retardation. Am. J. Hum. Genet. 80, 550–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chassaing N., Golzio C., Odent S., Lequeux L., Vigouroux A., Martinovic-Bouriel J., Tiziano F. D., Masini L., Piro F., Maragliano G., Delezoide A. L., Attié-Bitach T., Manouvrier-Hanu S., Etchevers H. C., Calvas P. (2009) Phenotypic spectrum of STRA6 mutations. From Matthew-Wood syndrome to non-lethal anophthalmia. Hum. Mutat. 30, E673–681 [DOI] [PubMed] [Google Scholar]
- 22. Chitayat D., Sroka H., Keating S., Colby R. S., Ryan G., Toi A., Blaser S., Viero S., Devisme L., Boute-Bénéjean O., Manouvrier-Hanu S., Mortier G., Loeys B., Rauch A., Bitoun P. (2007) The PDAC syndrome (pulmonary hypoplasia/agenesis, diaphragmatic hernia/eventration, anophthalmia/microphthalmia, and cardiac defect) (Spear syndrome, Matthew-Wood syndrome). Report of eight cases including a living child and further evidence for autosomal recessive inheritance. Am. J. Med. Genet. A 143A, 1268–1281 [DOI] [PubMed] [Google Scholar]
- 23. Slavotinek A. M. (2011) Eye development genes and known syndromes. Mol. Genet. Metab. 104, 448–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schnitzer J. J. (2004) Control and regulation of pulmonary hypoplasia associated with congenital diaphragmatic hernia. Semin. Pediatr. Surg. 13, 37–43 [DOI] [PubMed] [Google Scholar]
- 25. Rhinn M., Dollé P. (2012) Retinoic acid signalling during development. Development 139, 843–858 [DOI] [PubMed] [Google Scholar]
- 26. Paschaki M., Lin S. C., Wong R. L., Finnell R. H., Dollé P., Niederreither K. (2012) Retinoic acid-dependent signaling pathways and lineage events in the developing mouse spinal cord. PLoS One 7, e32447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Robinson J. F., Verhoef A., Pennings J. L., Pronk T. E., Piersma A. H. (2012) A comparison of gene expression responses in rat whole embryo culture and in vivo. Time-dependent retinoic acid-induced teratogenic response. Toxicol. Sci. 126, 242–254 [DOI] [PubMed] [Google Scholar]
- 28. Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J. F., Guindon S., Lefort V., Lescot M., Claverie J. M., Gascuel O. (2008) Phylogeny.fr. Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36, W465–W469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dereeper A., Audic S., Claverie J. M., Blanc G. (2010) BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol. Biol. 10, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Venable C. L., Frevert E. U., Kim Y. B., Fischer B. M., Kamatkar S., Neel B. G., Kahn B. B. (2000) Overexpression of protein-tyrosine phosphatase-1B in adipocytes inhibits insulin-stimulated phosphoinositide 3-kinase activity without altering glucose transport or Akt/protein kinase B activation. J. Biol. Chem. 275, 18318–18326 [DOI] [PubMed] [Google Scholar]
- 31. Testa G., Schaft J., van der Hoeven F., Glaser S., Anastassiadis K., Zhang Y., Hermann T., Stremmel W., Stewart A. F. (2004) A reliable lacZ expression reporter cassette for multipurpose, knockout-first alleles. Genesis 38, 151–158 [DOI] [PubMed] [Google Scholar]
- 32. de Vries W. N., Binns L. T., Fancher K. S., Dean J., Moore R., Kemler R., Knowles B. B. (2000) Expression of Cre recombinase in mouse oocytes. A means to study maternal effect genes. Genesis 26, 110–112 [PubMed] [Google Scholar]
- 33. Shepherd P. R., Gnudi L., Tozzo E., Yang H., Leach F., Kahn B. B. (1993) Adipose cell hyperplasia and enhanced glucose disposal in transgenic mice overexpressing GLUT4 selectively in adipose tissue. J. Biol. Chem. 268, 22243–22246 [PubMed] [Google Scholar]
- 34. Yang Q., Graham T. E., Mody N., Preitner F., Peroni O. D., Zabolotny J. M., Kotani K., Quadro L., Kahn B. B. (2005) Serum retinol-binding protein contributes to insulin resistance in obesity and type 2 diabetes. Nature 436, 356–362 [DOI] [PubMed] [Google Scholar]
- 35. Kawaguchi R., Yu J., Ter-Stepanian M., Zhong M., Cheng G., Yuan Q., Jin M., Travis G. H., Ong D., Sun H. (2011) Receptor-mediated cellular uptake mechanism that couples to intracellular storage. ACS Chem. Biol. 6, 1041–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cham B. E., Knowles B. R. (1976) A solvent system for delipidation of plasma or serum without protein precipitation. J. Lipid Res. 17, 176–181 [PubMed] [Google Scholar]
- 37. Goodman D. S., Raz A. (1972) Extraction and recombination studies of the interaction of retinol with human plasma retinol-binding protein. J. Lipid Res. 13, 338–347 [PubMed] [Google Scholar]
- 38. Ganguly J. (1989) Biochemistry of Vitamin A, pp. 63–85, CRC Press, Inc., Boca Raton, FL [Google Scholar]
- 39. Abelson J. N., Simon M. I., Sies H. (1990) Retinoids. Part A. Molecular and metabolic aspects. Methods Enzymol. 189, 3–536 [PubMed] [Google Scholar]
- 40. Kane M. A., Folias A. E., Napoli J. L. (2008) HPLC/UV quantitation of retinal, retinol, and retinyl esters in serum and tissues. Anal. Biochem. 378, 71–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sun H., Kawaguchi R. (2011) The membrane receptor for plasma retinol-binding protein, a new type of cell-surface receptor. Int. Rev. Cell Mol. Biol. 288, 1–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen C. C., Heller J. (1977) Uptake of retinol and retinoic acid from serum retinol-binding protein by retinal pigment epithelial cells. J. Biol. Chem. 252, 5216–5221 [PubMed] [Google Scholar]
- 43. Sivaprasadarao A., Findlay J. B. (1988) The mechanism of uptake of retinol by plasma-membrane vesicles. Biochem. J. 255, 571–579 [PMC free article] [PubMed] [Google Scholar]
- 44. Vogel S., Piantedosi R., Frank J., Lalazar A., Rockey D. C., Friedman S. L., Blaner W. S. (2000) An immortalized rat liver stellate cell line (HSC-T6). A new cell model for the study of retinoid metabolism in vitro. J. Lipid Res. 41, 882–893 [PubMed] [Google Scholar]
- 45. Randolph R. K., Ross A. C. (1991) Regulation of retinol uptake and esterification in MCF-7 and HepG2 cells by exogenous fatty acids. J. Lipid Res. 32, 809–820 [PubMed] [Google Scholar]
- 46. Matsuura T., Ross A. C. (1993) Regulation of hepatic lecithin. Retinol acyltransferase activity by retinoic acid. Arch. Biochem. Biophys. 301, 221–227 [DOI] [PubMed] [Google Scholar]
- 47. Faraonio R., Galdieri M., Colantuoni V. (1993) Cellular retinoic-acid-binding-protein and retinol-binding-protein mRNA expression in the cells of the rat seminiferous tubules and their regulation by retinoids. Eur. J. Biochem. 211, 835–842 [DOI] [PubMed] [Google Scholar]
- 48. Niederreither K., McCaffery P., Dräger U. C., Chambon P., Dollé P. (1997) Restricted expression and retinoic acid-induced downregulation of the retinaldehyde dehydrogenase type 2 (RALDH-2) gene during mouse development. Mech. Dev. 62, 67–78 [DOI] [PubMed] [Google Scholar]
- 49. Wang Y., Zolfaghari R., Ross A. C. (2002) Cloning of rat cytochrome P450RAI (CYP26) cDNA and regulation of its gene expression by all-trans-retinoic acid in vivo. Arch. Biochem. Biophys. 401, 235–243 [DOI] [PubMed] [Google Scholar]
- 50. Leid M., Kastner P., Chambon P. (1992) Multiplicity generates diversity in the retinoic acid signalling pathways. Trends Biochem. Sci. 17, 427–433 [DOI] [PubMed] [Google Scholar]
- 51. Capper N. S. (1930) Carotene and vitamin A. The transformation of carotene into vitamin A as shown by a study of the absorption spectra of rat-liver oils. Biochem. J. 24, 980–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ahmad B., Drummond J. C. (1930) The relative vitamin A value of the body and liver oils of certain fish. Biochem. J. 24, 27–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Berry D. C., Jin H., Majumdar A., Noy N. (2011) Signaling by vitamin A and retinol-binding protein regulates gene expression to inhibit insulin responses. Proc. Natl. Acad. Sci. U.S.A. 108, 4340–4345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Berry D. C., Noy N. (2012) Signaling by vitamin A and retinol-binding protein in regulation of insulin responses and lipid homeostasis. Biochim. Biophys. Acta 1821, 168–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Szeto W., Jiang W., Tice D. A., Rubinfeld B., Hollingshead P. G., Fong S. E., Dugger D. L., Pham T., Yansura D. G., Wong T. A., Grimaldi J. C., Corpuz R. T., Singh J. S., Frantz G. D., Devaux B., Crowley C. W., Schwall R. H., Eberhard D. A., Rastelli L., Polakis P., Pennica D. (2001) Overexpression of the retinoic acid-responsive gene Stra6 in human cancers and its synergistic induction by Wnt-1 and retinoic acid. Cancer Res. 61, 4197–4205 [PubMed] [Google Scholar]
- 56. Tice D. A., Szeto W., Soloviev I., Rubinfeld B., Fong S. E., Dugger D. L., Winer J., Williams P. M., Wieand D., Smith V., Schwall R. H., Pennica D., Polakis P. (2002) Synergistic induction of tumor antigens by Wnt-1 signaling and retinoic acid revealed by gene expression profiling. J. Biol. Chem. 277, 14329–14335 [DOI] [PubMed] [Google Scholar]
- 57. Cifelli C. J., Green J. B., Wang Z., Yin S., Russell R. M., Tang G., Green M. H. (2008) Kinetic analysis shows that vitamin A disposal rate in humans is positively correlated with vitamin A stores. J. Nutr. 138, 971–977 [DOI] [PubMed] [Google Scholar]
- 58. Pitot H. C., Peraino C., Morse P. A., Jr., Potter V. R. (1964) Hepatomas in tissue culture compared with adapting liver in vivo. Natl. Cancer Inst. Monogr. 13, 229–245 [PubMed] [Google Scholar]
- 59. Bernhard H. P., Darlington G. J., Ruddle F. H. (1973) Expression of liver phenotypes in cultured mouse hepatoma cells. Synthesis and secretion of serum albumin. Dev. Biol. 35, 83–96 [DOI] [PubMed] [Google Scholar]
- 60. St. George S., Friedman M., Byers S. O. (1954) Mass separation of reticuloendothelial and parenchymal cells of rat's liver. Science 120, 463–465 [DOI] [PubMed] [Google Scholar]
- 61. Senoo H., Kojima N., Sato M. (2007) Vitamin A-storing cells (stellate cells). Vitam. Horm. 75, 131–159 [DOI] [PubMed] [Google Scholar]
- 62. Blaner W. S., O'Byrne S. M., Wongsiriroj N., Kluwe J., D'Ambrosio D. M., Jiang H., Schwabe R. F., Hillman E. M., Piantedosi R., Libien J. (2009) Hepatic stellate cell lipid droplets: a specialized lipid droplet for retinoid storage. Biochim. Biophys. Acta 1791, 467–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Isken A., Golczak M., Oberhauser V., Hunzelmann S., Driever W., Imanishi Y., Palczewski K., von Lintig J. (2008) RBP4 disrupts vitamin A uptake homeostasis in a STRA6-deficient animal model for Matthew-Wood syndrome. Cell Metab. 7, 258–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kawaguchi R., Zhong M., Kassai M., Ter-Stepanian M., Sun H. (2012) STRA6-catalyzed vitamin A influx, efflux, and exchange. J. Membr. Biol. 245, 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Amengual J., Golczak M., Palczewski K., von Lintig J. (2012) Lecithin:retinol acyltransferase is critical for cellular uptake of vitamin A from serum retinol-binding protein. J. Biol. Chem. 287, 24216–24227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Graham T. E., Smith U., Kahn B. B. (2006) Retinol-binding protein 4 and insulin resistance (authors' reply). N. Engl. J. Med. 355, 1394–1395 [PubMed] [Google Scholar]
- 67. Munkhtulga L., Nakayama K., Utsumi N., Yanagisawa Y., Gotoh T., Omi T., Kumada M., Erdenebulgan B., Zolzaya K., Lkhagvasuren T., Iwamoto S. (2007) Identification of a regulatory SNP in the retinol binding protein 4 gene associated with type 2 diabetes in Mongolia. Hum. Genet. 120, 879–888 [DOI] [PubMed] [Google Scholar]
- 68. Klöting N., Graham T. E., Berndt J., Kralisch S., Kovacs P., Wason C. J., Fasshauer M., Schön M. R., Stumvoll M., Blüher M., Kahn B. B. (2007) Serum retinol-binding protein is more highly expressed in visceral than in subcutaneous adipose tissue and is a marker of intra-abdominal fat mass. Cell Metab. 6, 79–87 [DOI] [PubMed] [Google Scholar]
- 69. Kovacs P., Geyer M., Berndt J., Klöting N., Graham T. E., Böttcher Y., Enigk B., Tönjes A., Schleinitz D., Schön M. R., Kahn B. B., Blüher M., Stumvoll M. (2007) Effects of genetic variation in the human retinol binding protein-4 gene (RBP4) on insulin resistance and fat depot-specific mRNA expression. Diabetes 56, 3095–3100 [DOI] [PubMed] [Google Scholar]
- 70. Graham T. E., Kahn B. B. (2007) Tissue-specific alterations of glucose transport and molecular mechanisms of intertissue communication in obesity and type 2 diabetes. Horm. Metab. Res. 39, 717–721 [DOI] [PubMed] [Google Scholar]
- 71. Hahn S., Backhaus M., Broecker-Preuss M., Tan S., Dietz T., Kimmig R., Schmidt M., Mann K., Janssen O. E. (2007) Retinol-binding protein 4 levels are elevated in polycystic ovary syndrome women with obesity and impaired glucose metabolism. Eur. J. Endocrinol. 157, 201–207 [DOI] [PubMed] [Google Scholar]
- 72. Qi Q., Yu Z., Ye X., Zhao F., Huang P., Hu F. B., Franco O. H., Wang J., Li H., Liu Y., Lin X. (2007) Elevated retinol-binding protein 4 levels are associated with metabolic syndrome in Chinese people. J. Clin. Endocrinol. Metab. 92, 4827–4834 [DOI] [PubMed] [Google Scholar]
- 73. van Hoek M., Dehghan A., Zillikens M. C., Hofman A., Witteman J. C., Sijbrands E. J. (2008) An RBP4 promoter polymorphism increases risk of type 2 diabetes. Diabetologia 51, 1423–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wu Y., Li H., Loos R. J., Qi Q., Hu F. B., Liu Y., Lin X. (2009) RBP4 variants are significantly associated with plasma RBP4 levels and hypertriglyceridemia risk in Chinese Hans. J. Lipid Res. 50, 1479–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ost A., Danielsson A., Lidén M., Eriksson U., Nystrom F. H., Strålfors P. (2007) Retinol-binding protein-4 attenuates insulin-induced phosphorylation of IRS1 and ERK1/2 in primary human adipocytes. FASEB J. 21, 3696–3704 [DOI] [PubMed] [Google Scholar]
- 76. Thompson J. D., Higgins D. G., Gibson T. J. (1994) CLUSTAL W. Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]