Background: How transmembrane (TM) proteins interact with ordered membrane domains (rafts) remains unknown.
Results: The raft affinity of a multi-TM segment protein, PFO, is increased by matching between TM segment lengths and bilayer width.
Conclusion: Hydrophobic mismatch strongly influences the raft affinity of PFO, in contrast to results with single TM segment proteins.
Significance: This study defines one mechanism controlling raft-TM protein association.
Keywords: Fluorescence, Fluorescence Resonance Energy Transfer (FRET), Membrane Bilayer, Membrane Biophysics, Membrane Proteins, Cytolysin
Abstract
The hypothesis that mismatch between transmembrane (TM) length and bilayer width controls TM protein affinity for ordered lipid domains (rafts) was tested using perfringolysin O (PFO), a pore-forming cholesterol-dependent cytolysin. PFO forms a multimeric barrel with many TM segments. The properties of PFO mutants with lengthened or shortened TM segments were compared with that of PFO with wild type TM sequences. Both mutant and wild type length PFO exhibited cholesterol-dependent membrane insertion. Maximal PFO-induced pore formation occurred in vesicles with wider bilayers for lengthened TM segments and in thinner bilayers for shortened TM segments. In diC18:0 phosphatidylcholine (PC)/diC14:1 PC/cholesterol vesicles, which form ordered domains with a relatively thick bilayer and disordered domains with a relatively thin bilayer, affinity for ordered domains was greatest with lengthened TM segments and least with shortened TM segments as judged by FRET. Similar results were observed by microscopy in giant vesicles containing sphingomyelin in place of diC18:0 PC. In contrast, in diC16:0 PC/diC14:0 PC/diC20:1 PC/cholesterol vesicles, which should form ordered domains with a relatively thin bilayer and disordered domains with a relatively thick bilayer, relative affinity for ordered domains was greatest with shortened TM segments and least with lengthened TM segments. The inability of multi-TM segment proteins (unlike single TM segment proteins) to adapt to mismatch by tilting may explain the sensitivity of raft affinity to mismatch. The difference in width sensitivity for single and multi-TM helix proteins may link raft affinity to multimeric state and thus control the assembly of multimeric TM complexes in rafts.
Introduction
The heterogeneity of biological membranes plays an important role in cellular functions (1, 2). It has been proposed that cellular membranes contain liquid-ordered (Lo)2 lipid domains enriched in sphingolipids and cholesterol (membrane (or lipid) rafts) that co-exist with disordered (Ld) domains rich in unsaturated glycerophospholipids (1, 3, 4). Raft domains are believed to control numerous protein-protein and lipid-protein interactions (5, 6). This may be important because partitioning between different lipid domains may control membrane protein function and the formation of functional complexes (7–9). Studies have shown that the raft affinity of peripheral proteins is dependent upon their attachment to saturated lipid anchors, which pack well in ordered domains (10). For example, nonreceptor tyrosine kinases can be reversibly palmitoylated and lose their raft association after depalmitoylation (11). However, the physical basis through which transmembrane (TM) proteins interact with lipid rafts remains unknown. Unlike a saturated lipid anchoring a peripheral membrane protein, the lipid-facing surface of TM sequences would disrupt lipid-lipid van der Waals interactions in tightly packed ordered domains without replacing them with equivalent lipid-protein interactions (12–14). This should oppose facile incorporation of TM proteins into ordered domains.
A possible explanation for TM protein affinity for rafts could involve hydrophobic mismatch. Hydrophobic mismatch can be defined as the energetically unfavorable situation encountered when the lengths of hydrophobic TM protein segments exceed (positive mismatch) or are less than (negative mismatch) the thickness/width of the hydrophobic core of the bilayer. A substantial body of data suggests that hydrophobic match influences the functioning of membrane channels, pumps, and transporters (15–17). In reconstitution studies on cytochrome c oxidase (18), melibiose permease (19), and different ATPases (16, 17, 20, 21), enzymatic activity was found to be lower when enzymes were reconstituted in bilayers with mismatching hydrophobic thickness. Mismatch also influences how integral membrane proteins are inserted into, secreted through, and folded within the membrane (22, 23). Mismatch could also influence interactions with lipid membrane domains. Lo domains are thicker than Ld domains, due to the loss of gauche rotamers (kinks) in acyl chains (24). Long hydrophobic TM segments would extend beyond the hydrophobic part of the lipid bilayer in Ld domains, resulting in unfavorable exposure of hydrophobic residues to the aqueous phase or unfavorable distortion of lipids to locally increase bilayer width. Such energetically unfavorable behaviors would not occur in thicker Lo domains, resulting in increased affinity of long hydrophobic sequences for Lo domains.
However, previous studies of single TM helices have not detected a significant effect of hydrophobic mismatch upon raft affinity (25, 26). The lack of an effect of mismatch may reflect the ability of long TM helices to tilt in order to avoid positive mismatch (27). However, tilting should be energetically more costly for membrane proteins with multiple, rigid TM segments (see “Discussion”). Therefore, to test the effect of mismatch upon raft affinity, we investigated interactions between the TM β-barrel protein perfringolysin O (PFO) and model membrane vesicles with co-existing ordered and disordered lipid domains. PFO is a member of the cholesterol-dependent cytolysin family, a family that requires cholesterol for membrane insertion, for oligomerization, and for pore formation (28–30). PFO (56 kDa) is a protein with four domains. It exists as a monomer in solution but in membranes forms an oligomer with 35–40 subunits, as judged by molecular weight on gels. Domain 4 binds to cholesterol, whereas sequences in domain 3 form two TM β-hairpins after membrane insertion. In this report, the raft affinity of PFO with wild type, lengthened, and shortened TM segment lengths was measured by both fluorescence resonance energy transfer (FRET) and confocal microscopy of giant unilamellar vesicles (GUVs). The results indicate that the affinity of PFO for ordered lipid domains is increased by matching between TM segment lengths and bilayer width in the ordered domains. Thus, hydrophobic mismatch and multimeric state can control TM protein association with lipid rafts, and based on this, we propose a mechanism by which the assembly of multi-TM protein complexes can be controlled by rafts.
EXPERIMENTAL PROCEDURES
Reagents
Unlabeled phospholipids, cholesterol (ovine wool), ganglioside M1 (GM1), sphingomyelin (egg), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-pyrenesulfonyl (pyrene-DOPE), 1,2-dipalimitoyl-sn-glycero-3-phosphoethanolamine-N-pyrenesulfonyl (pyrene-DPPE), 1,2-diphytanoyl-sn-glycero-3-phosphoethanolamine-N-(7-nitro-2-1,3-benzoxadiazol-4-yl) (NBD-DPhPE), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (Rho-DOPE), and 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (Rho-DPPE) were purchased from Avanti Polar Lipids (Alabaster, AL). Lipids were stored in ethanol or chloroform at −20 °C. Concentrations were determined by dry weight or by absorbance, using an ϵ of 35,000 cm−1 m−1 at 350 nm for pyrene-DOPE in methanol, 95,000 cm−1 m−1 at 560 nm for rhodamine lipids in methanol, or 21,000 cm−1 m−1 at 463 nm for NBD-DPhPE in methanol (31). The labeling reagents N-(4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-yl)methyl)iodoacetamide (BODIPY-FL) and 6-acryloyl-2-dimethylaminonaphthalene (acrylodan) were purchased from Invitrogen. Acetyl-K2W2L8AL8W2K2-amide (LW peptide) was purchased from Anaspec, Inc. (San Jose, CA) and used without further purification. Custom synthesized 10-doxylnonadecane (10-DN) was purchased from Invitrogen. Cholera toxin B subunit was purchased from EMD Chemicals (Gibbstown, NJ). TALON bead resin was purchased from Clontech (Mountain View, CA). All other chemicals were reagent grade.
Generation of PFO Mutants
A functional cysteineless derivative of wild type PFO (PFO C459A) and a prepore mutant of PFO (PFO C459A/Y181A), both in the pRSET B vector (32, 33), were kind gifts of A. Heuck (University of Massachusetts, Amherst, MA). PFO with lengthened or shortened TM sequences was generated from the former protein using site-directed ligase-independent mutagenesis (SLIM) (34, 35) with the two pairs of primers to lengthen or shorten each transmembrane helix (TMH) and using the plasmid C459A PFO-pRSET B as template. Long-tailed primers Slim-Ft (5′-GCTGCTCTTGAAGCTGCTAACTCACTTGGAGTAGACTTTA-3′) and Slim-Rt (5′-AGCAGCTTCAAGAGCAGCGACTTTAGCATTAACATTA-3′) and short corresponding primers Slim-Fs (5′-AACTCACTTGGAGTAGACTTTA-3′) and Slim-Rs (5′-GACTTTAGCATTAACATTA-3′) were designed to specifically insert two Ala between residues Val202 and Leu203 and between residues Glu204 and Asn205 in TMH1, and long-tailed primers Slim-Ft (5′-GCTGCTAAGAACGCTGCTACTGATATAAAAAATAGTCAACA-3′) and Slim-Rt (5′-AGCAGCGTTCTTAGCAGCTATAAGAGCTTTGAAAGCAGCT-3′) and short corresponding primers Slim-Fs (5′-ACTGATATAAAAAATAGTCAACA-3′) and Slim-Rs (5′-TATAAGAGCTTTGAAAGCAGCTTGTACA-3′) were designed to insert two Ala between residues Ile298 and Lys299 and between residues Asn300 and Thr301 in TMH2 for the lengthened mutant. Likewise, long-tailed primers Slim-Ft (5′-ATGTTAATGCTCTTGAACTTGGAGTAGACTTTAATGCAG-3′) and Slim-Rt (5′-GTCTACTCCAAGTTCAAGAGCATTAACATTAAGGGCA-3′) and short corresponding primers Slim-Fs (5′-CTTGGAGTAGACTTTAATGC-3′) and Slim-Rs (5′-AGCATTAACATTAAGGGCACTTG-3′) were used to delete Lys201-Val202 and Asn205-Ser206 in TMH1; long-tailed primers Slim-Ft (5′-CTTTCAAAGCTAAGAACATAAAAAATAGTCAACAATATA-3′) and Slim-Rt (5′-GACTATTTTTATGTTCTTAGCTTTGAAAGCAGCTTGTACA-3′) and short corresponding primers Slim-Fs (5′-ATAAAAAATAGTCAACAATA-3′) and Slim-Rs (5′-AGCTTTGAAAGCAGCTTGTACA-3′) were used to delete Leu297-Ile298 and Thr301-Asp302 in TMH2 for the shortened mutant. PCR conditions used to amplify the plasmid were as follows: 30 s at 98 °C; 25 cycles of 15 s at 98 °C, 20 s at 55 °C, and 4 min at 72 °C, with a final 10 min extension step at 72 °C. The template plasmid was removed by restriction with DpnI. The DpnI-treated reaction products were then mixed to set up SLIM hybridization by three cycles of 5 min at 65 °C and 40 min at 65 °C. After transformation of Escherichia coli DH5α with PCR products obtained by following the SLIM PCR protocol, plasmids bearing the insertions or deletions were picked and confirmed by sequencing. Variants of PFO with an Ala to Cys substitution at residue 215 were generated using the QuikChange site-directed mutagenesis kit (Stratagene). PCR products were transformed into E. coli DH5α, and positive colonies were selected and confirmed by sequencing. (For simplicity, the term PFO will be used to encompass all of the PFO variants studied in this report, unless referring to a specific variant of PFO, in which case the variant will be specified.)
Purification of PFO
PFO was expressed in E. coli BL21(DE3)pLysS and purified in a manner similar to that described previously (31).
Labeling of PFO
Labeling of PFO variants containing Cys residues with BODIPY-FL and acrylodan was carried out in a manner similar to that described previously (31). Briefly, stock solutions with 0.5–1 mg/ml PFO were thawed and dialyzed against 4 liters of PBS overnight to remove excess DTT. BODIPY-FL (dissolved in DMSO) or acrylodan (dissolved in N,N-dimethylformamide) were added to a concentration providing a ∼15-fold molar excess of reagent over PFO. After labeling at room temperature for 2 h (for BODIPY labeling) or 4 h (for acrylodan labeling), the reaction mixtures were centrifuged to remove precipitated dye and then chromatographed in PBS (10 mm sodium phosphate, 1 mm potassium phosphate, 137 mm sodium chloride, 13 mm potassium chloride) buffer (pH 7.4) on a 1 × 20-cm length column containing 10 ml of Sephadex-G50, collecting 1-ml fractions. Fractions containing labeled protein were dialyzed against 4 liters of PBS, pH 7.4, overnight at 4 °C to remove any excess fluorescent dye. The extent of covalent reaction with the fluorescent dye was estimated using an ϵ of 76,000 m−1 cm−1 at 502 nm for BODIPY and 16,400 m−1 cm−1 at 372 nm for acrylodan.
Preparation of Lipid Vesicles
Multilamellar vesicles (MLVs) were prepared with the desired mixture of lipids. Lipids in solvent were mixed and then dried with N2. They were then redissolved in CHCl3 and redried under N2 and then high vacuum for at least 1 h. The redried lipid mixtures were then dispersed in PBS, pH 5.1, at 70 °C to give the desired final concentration and agitated at 55 °C for 15 min using a VWR multitube vortexer (Westchester, PA) placed within a convection oven (GCA Corp., Precision Scientific, Chicago, IL). Samples were then cooled to room temperature. Large unilamellar vesicles (LUVs) were prepared from MLV by subjecting the MLV to seven cycles of freezing in a mixture of dry ice and acetone and thawing at room temperature.
Fluorescence Intensity Measurements
Fluorescence emission intensity was measured (unless otherwise noted) at room temperature on a SPEX Fluorolog 3 spectrofluorimeter. For fixed wavelength measurements, excitation and emission wavelength sets used (in nm) were (280, 340) for tryptophan, (488, 515) for BODIPY-FL-labeled streptavidin, (490, 510) for BODIPY-FL-labeled PFO, and (334, 384) for pyrene-DPPE. Unless otherwise noted, fluorescence intensity in single background samples lacking fluorophore was subtracted. For acrylodan-labeled PFO emission spectra, samples and backgrounds were excited at 350 nm, and emission was acquired from 420 to 560 nm.
Vesicle Binding by Centrifugation
The ability of PFO to associate with vesicles was assessed by ultracentrifugation. MLVs (500 μm lipid) composed of different lipid species with 40 or 45 mol % cholesterol were prepared in 1 ml of PBS, pH 5.1, and incubated with 25 nm BODIPY-labeled PFO for 1 h at room temperature. Samples were then spun for 45 min in a Beckman L8-85 ultracentrifuge at 84,000 × g at 4 °C. After spinning, supernatants containing the unbound PFO were removed, and pellets containing the MLV and bound PFO were resuspended in 1 ml of PBS, pH 5.1. Then BODIPY fluorescence was measured for both the supernatant and the pellet.
Vesicle Binding and Insertion Experiments
PFO-membrane interaction was monitored by the changes of fluorescence intensity and λmax of PFO labeled with acrylodan on Cys215. Residue 215 becomes buried in the bilayer when PFO inserts into the bilayer and forms TM strands (36). To do this, 25 nm acrylodan-labeled PFO was added to 1 ml of (500 μm lipid) MLVs composed of DOPC (containing different percentages of cholesterol) in PBS, pH 5.1. After a 1-h incubation at room temperature, acrylodan emission spectra were measured as described above.
FRET Measurement of the Proximity of BODIPY-labeled Cys215 to the Membrane
To measure the location of BODIPY-labeled Cys 215 relative to the bilayer surface, 100 μl of MLVs containing 5 mm DOPC or 6:4 (mol/mol) DOPC/cholesterol in PBS, pH 5.1, were incubated with 50 nm BODIPY-labeled PFO carrying the A215C substitution. “F sample” vesicles contained 1 mol % Rho-DOPE as FRET acceptor. “Fo sample” vesicles lacked an energy transfer acceptor. After 1 h of incubation at room temperature, samples were diluted with PBS, pH 5.1, to 1 ml, and then BODIPY fluorescence emission intensity was measured for each sample and backgrounds lacking protein. We used BODIPY rather than acrylodan in these experiments because the efficiency of the energy transfer depends not only on the distance of the donor and acceptor but also on the extent of the overlap between the emission of the donor and the absorption spectra of the acceptor. The quantum yield and emission spectrum of BODIPY fluorescence is insensitive to environment (31, 37), unlike acrylodan, so that interpretation of FRET is simpler using BODIPY. Fluorescence intensities were corrected for fluorescence intensities in background samples lacking protein.
Quenching Experiments to Measure Membrane Penetration by Acrylodan-labeled Cysteine 215
To measure the penetration of acrylodan-labeled Cys215 into the bilayer, quenching of acrylodan fluorescence by 10-DN was measured. Samples were prepared by incorporating 50 nm acrylodan-labeled PFO with the A215C substitution into 1 ml of MLVs containing 500 μm 65:35 (mol/mol) DOPC/cholesterol in PBS, pH 5.1, as described above except that 5 mol % 10-DN (replacing an equivalent amount of DOPC) was mixed with the lipids prior to vesicle formation for F samples. Fo samples contained no 10-DN. Acrylodan fluorescence in the presence of 10-DN was then compared with that in its absence. Quenching by 10-DN was calculated using the fluorescence intensity at the intensity peak of the emission spectra. Fluorescence intensities were corrected for fluorescence intensities in background samples lacking protein.
Assay for Pore Formation
PFO-induced pore formation was measured by assaying the reaction of vesicle-trapped biocytin with externally added BODIPY-labeled streptavidin via the increase in the BODIPY fluorescence emission intensity upon binding of biocytin to BODIPY-labeled streptavidin located in the external solution, as described previously (32). To confirm that biocytin was trapped in the vesicles in cases in which PFO did not appear to form pores, BODIPY-labeled streptavidin fluorescence was measured before and after the addition of 50 μl of 1:4 Triton X-100/water to dissolve the vesicles and release trapped biocytin. The -fold level of BODIPY intensity increase was always between 3 and 3.5.
Detection of Domain Formation by FRET
FRET measurements were made in 1-ml MLV samples dispersed in PBS pH 5.1 prepared as described previously (38). Samples contained 500 μm lipid with 0.05 mol % pyrene-DPPE as a donor and 2 mol % Rho-DOPE as an acceptor in F samples. The Fo samples contained only donor. Backgrounds for Fo samples (containing only lipid and lacking donor) and for F samples (containing lipid plus acceptor) were also prepared. Samples were prepared at 70 °C and then incubated at room temperature for 1 h, after which they were cooled to 15 °C, and the fluorescence measurements were initiated. Cuvette temperature, measured as described previously (38), was increased at a rate of ∼0.5 °C/min, and readings were taken every 5 °C. In addition, background fluorescence was measured and then subtracted from the FRET sample values. The ratio of fluorescence intensity in the presence of acceptor to its absence (F/Fo) was calculated.
FRET Assay of Raft Affinity
FRET experiments were carried out to assess PFO affinity for raft domains similarly as described by Nelson et al. (31), except that 1 mol % pyrene-DOPE or 2 mol % NBD-DPhPE were used as FRET acceptor. Briefly, 100 μl of MLVs prepared in PBS, pH 5.1, at a total lipid concentration of 5 mm were incubated with 3 μg PFO. Two types of samples were prepared. F sample vesicles also contained FRET acceptor (either 1 mol % pyrene-DOPE or 2 mol % NBD-DPhPE of total lipid). Fo sample vesicles lacked acceptor. To measure cholera toxin B chain (CT-B) raft affinity, the same procedure was followed as for PFO, but vesicles contained an additional 2 mol % ganglioside GM1 and 5 μg of CT-B chain. For samples with LW peptide, peptide was added from an ethanolic stock solution to lipids prior to preparation of the MLV at a concentration of 0.25 mol % relative to lipid. Lipid composition and concentration was the same as in PFO-containing samples. After preparation, samples were incubated at room temperature for at least 1 h (for PFO or CT-B) or at least 15 min (for LW peptide) and then diluted with PBS, pH 5.1, to 1 ml. Tryptophan fluorescence emission intensity was then measured at room temperature for each sample and for background samples lacking proteins, as described above. Uncorrected F/Fo is the ratio of measured fluorescence in the presence of acceptor to that in its absence (equal to Fmeasured/Fomeasured). Corrected F/Fo = (Fmeasured − xFunbound)/(Fomeasured − xFunbound), where Funbound is the Trp fluorescence the amount of PFO in the samples would have if it were fully unbound to vesicles, and x represents the fraction of unbound protein. For all of these experiments, the fraction of PFO bound to membranes was measured, and was generally >95% for WT and long PFO, and close to 50% for short PFO.
Raft Affinity Derived from FRET
FRET was used to assay relative localization of PFO in Lo or Ld domains. If there is partitioning of a FRET donor (i.e. PFO or other Trp-containing protein) and a FRET acceptor into the same type of domain, there will be an increase in local acceptor concentration around the donor relative to that in a homogeneous membrane. If a donor and acceptor are segregated into different types of domains, then there will be a decrease in local acceptor concentration around the donor. To approximate local FRET acceptor concentration around donor, the Perrin equation for quenching in two dimensions was used (39). This equation states that F/Fo = exp −πRc2C, where F/Fo is the ratio of fluorescence of a sample containing FRET acceptor to the fluorescence of a sample lacking FRET acceptor, C is the concentration of acceptors in the membrane, and Rc is the effective radius of quenching (∼1.1Ro). Solving for C gives C = ln(F/Fo)/−πRc2. Because Rc should not be significantly dependent upon lipid composition, the ratio of the local concentration of acceptor around the donor in a membrane containing both Ld and Lo domains (CLoLd) relative to that in a homogeneous membrane lacking domains (Ch) will be given by the equation, CLoLd/CLh = ln(F/Fo)LoLd/ln(F/Fo)Lh. The ratio CLoLd/CLh will be high if the FRET acceptor and the donor partition into the same type of domains and low if they are segregated into different types of domains.
Laser Scanning Microscopy
GUVs composed of 1:1 (mol/mol) egg SM/DMoPC with 37 mol % cholesterol were prepared using the electroformation method (40). The inclusion in the GUVs of the fluorescently labeled lipid Rho-DPPE (0.05 mol %), a marker for disordered domains despite having saturated acyl chains (41), allowed the optical visualization of bilayer domains. To prepare GUVs, the lipids were dissolved in chloroform at 10 mg/ml, and a small volume (∼1.2 μl) was spread on indium tin oxide-covered coverslips. The solvent was then completely evaporated, and two coverslips were positioned in a home-built flow chamber at a distance of ∼2 mm from each other. After 200–300 μl of 320 mm trehalose was added to the chamber, and a voltage of 1.2 V at 10 Hz was applied for 2 h at 60 °C. Following electroformation, the content of the chamber was gently exchanged with 200–300 μl of pH 5.1 PBS (which is equiosmolar with the trehalose) containing 10–20 μl of 2–3 μm BODIPY-labeled PFO. The protein was incubated with the vesicles for 1 h at room temperature and then observed under confocal laser scanning microscopy. For confocal laser scanning microscopy, the excitation light of an argon laser at 488 nm and a helium-neon laser at 543 nm was reflected by a dichroic mirror (HFT 488/543/633) and focused onto the sample by a Zeiss C-Apochromat ×40, numerical aperture 1.2 water immersion objective. Fluorescence signal was then recollected by the same objective and separated in two different paths using a secondary dichroic beam splitter (NFT 545). After passing through a 525-nm filter with 50-nm bandwidth (equivalent to a 525/50 bandpass filter, green channel, to detect BODIPY-labeled PFO) or a 580/60 bandpass filter (red channel, to detect Rho-DPPE), the fluorescence signal was measured by two photomultipliers. The confocal geometry was ensured by a 70-μm pinhole in front of the green channel photomultiplier and an 80-μm pinhole for the red channel photomultiplier. All of the measurements were performed at room temperature.
The partition of the fluorescent protein constructs between ordered and disordered domains was calculated from confocal sections of GUVs using a self-written Matlab algorithm (MathWorks, Natick, MA) routine. For each sample/protein construct, we imaged ∼20 vesicles and observed the lateral distribution of Rho-DPPE (marker for Ld phase, red channel) and the fluorescent protein (green channel). For each GUV image, we also calculated and subtracted the background. Afterward, we extracted five cross-sections of the green channel fluorescence signal through the membrane in the Lo phase and in the Ld phase, respectively. The Ld phase could be easily identified through the strong partition of the Rho-DPPE observed in the red channel. To quantitatively analyze the Lo/Ld cross-section pairs for each GUV, we used two methods. The first method consisted of simply integrating numerically each cross-section to estimate the total fluorescence intensity (i.e. the area under the cross-section curve). For each pair of Lo-Ld cross-sections, we calculated the partition coefficient K(Lo/Ld) as the ratio between the total fluorescence intensity in the Lo phase and the total fluorescence intensity in the Ld phase. Finally, we averaged K(Lo/Ld) over all of the cross-sections recorded for the sample. However, we noticed that in some cases, the brightest pixels had a saturated intensity. To correct for this, a second method was used. This consisted of fitting each cross-section using a Gaussian intensity versus pixel position model in which it was assumed that only the non-saturated pixels fall on the Gaussian curve, whereas the saturated pixels underestimate true intensity. For each pair of Lo/Ld cross-sections, we calculated the partition coefficient K(Lo/Ld) as the ratio between the area under the Gaussian curve fitting the Lo phase cross-section and the area under the Gaussian curve fitting the Ld phase cross-section. Finally, we averaged K(Lo/Ld) over all of the cross-sections recorded for the sample. Both methods require the condition that, for each analyzed cross-section, the fluorescence signal originates exclusively from one GUV membrane (i.e. background signal is weak enough to be accurately subtracted, and one cross-section cannot include multiple membranes/vesicles or other bright objects).
RESULTS
PFO Mutants with Lengthened or Shortened TM Segments
To investigate the effect of hydrophobic match/mismatch upon membrane protein affinity for ordered membrane domains, PFO molecules were prepared in which the membrane-inserting segments were either two residues longer (“long PFO”) or two residues shorter (“short PFO”) than normal PFO (“WT PFO”). The ability of the long and short PFO variants to interact with and insert into vesicles was compared with that of WT PFO. As a control, the behavior of a prepore PFO mutant, which binds to membranes but in which the TM strand-forming sequences do not move into the membrane, was also characterized. PFO with either acrylodan- or BODIPY-labeled residue Cys215 was used to probe PFO membrane interaction. Residue 215 is a sensitive probe of PFO conformation because it moves from about 62 Å above the membrane surface to a position located within the hydrophobic core in the bilayer during the conversion of PFO from the prepore state to the TM barrel-containing pore-forming state (36). In the TM β-barrel, residue 215 faces the hydrophobic core of the membrane bilayer (42, 43). Cysteine substitution and fluorophore labeling at residue 215 affect neither PFO topology nor hemolytic activity (33). We first examined whether the PFO variants would interact with membranes. As judged by centrifugation, binding to 6:4 (mol/mol) DOPC/cholesterol was complete (95–100%) for WT, long, and prepore PFO, with the long PFO showing slightly less binding than WT or prepore PFO. However, binding of short PFO to membranes was incomplete (∼65% bound). Experiments in which FRET from a BODIPY-labeled Cys215 to rhodamine on 1 mol % Rho-DOPE incorporated into the vesicles was measured confirmed these results. FRET was strong (∼65–70%) for the long and WT protein and weaker for the short PFO (∼40%). Interestingly, FRET was also weak (∼30%) for the prepore PFO. This is expected because, in the prepore state, residue 215 should not have moved into the membrane (36).
Long, WT, and Short PFO Show Cholesterol-dependent Deep Insertion and Pore Formation
Wild type PFO undergoes cholesterol-dependent deep insertion into membranes. To see if the PFO variants also underwent cholesterol-dependent deep membrane insertion, the fluorescence of PFO acrylodan-labeled on residue 215 was characterized. Acrylodan fluorescence can be used to assay insertion because, in an aqueous milieu, it has highly red-shifted emission λmax, whereas in a hydrophobic environment, such as the hydrophobic core of the lipid bilayer, it has a highly blue-shifted emission λmax. Fig. 1A shows that in 6:4 (mol/mol) DOPC/cholesterol vesicles, prepore PFO, in which residue 215 is located far from the membrane surface, had highly red-shifted fluorescence, whereas short, WT, and long PFO had highly blue-shifted fluorescence, indicative of deep membrane insertion. (The shoulder seen at longer wavelengths for short PFO is probably due to protein unbound to the vesicles.) To confirm insertion, we measured the quenching of acrylodan fluorescence by 10-DN embedded in the core of the membrane bilayer (Fig. 1B). 10-DN has a nitroxide group that is able to strongly quench fluorescent groups buried within the core of the lipid bilayer (44). The fluorescence of acrylodan-labeled residue 215 was strongly quenched for short, WT, and long PFO, indicating that they underwent similar levels of membrane insertion, but only very weakly quenched for prepore PFO, indicating a lack of membrane insertion, as expected.
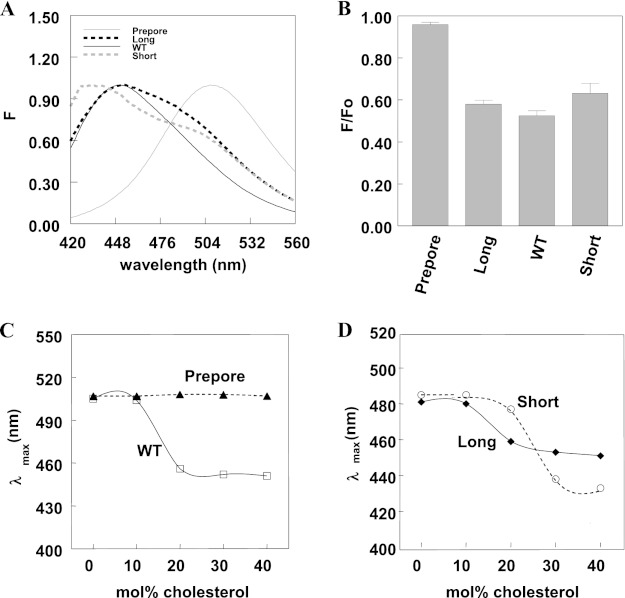
FIGURE 1.
Long, WT, and short PFO show cholesterol-dependent deep insertion into vesicles. A, emission spectra of acrylodan-labeled PFO in vesicles containing cholesterol. Samples contained 25 nm prepore; long, WT, or short PFO with acrylodan-labeled cysteine 215; and 6:4 (mol/mol) DOPC/cholesterol vesicles (500 μm total lipid) in PBS, pH 5.1. Solid black line, WT PFO; solid gray line, prepore PFO; dashed black line, long PFO; dashed gray line, short PFO. B, quenching of acrylodan fluorescence by 10-DN. Samples contained 50 nm acrylodan-labeled PFO and 65:35 (mol/mol) DOPC/cholesterol vesicles (500 μm total lipid) in PBS, pH 5.1. F/Fo is the ratio of acrylodan fluorescence at λmax in the presence of vesicles containing 5 mol % 10-DN (F) (replacing 5 mol % DOPC) to that in the presence of vesicles lacking 10-DN (Fo). Average (mean) values and S.D. values (error bars) were obtained from four samples. The F/Fo values in these experiments were uncorrected for incomplete binding to vesicles. C and D, samples contained 25 nm WT (open square), prepore (filled triangle), long (filled diamond), or short (open circle) PFO with acrylodan-labeled cysteine 215 and vesicles composed of DOPC and increasing amounts of cholesterol (500 μm total lipid) in PBS, pH 5.1. All measurements were made at room temperature in this and the following figures unless otherwise noted.
Next, the dependence of membrane insertion upon cholesterol concentration in DOPC vesicles was examined. Prepore PFO showed no membrane insertion at any cholesterol concentration, as expected. WT PFO underwent membrane insertion that was roughly half-maximal at 16 mol % cholesterol (Fig. 1C). (This is actually a lower limit to the half-maximal value because the acrylodan residue is generally much more fluorescent in the membrane-inserted state.) Long and short PFO also exhibited a cholesterol dependence for insertion (Fig. 1D). Long PFO inserted at a cholesterol concentration (∼17 mol %) similar to that of WT PFO, whereas short PFO required a higher cholesterol concentration for insertion (∼29 mol %).
The cholesterol dependence of PFO-induced pore formation was also characterized. Pore formation was assayed by measuring the reaction of biocytin encapsulated inside LUVs with BODIPY-streptavidin added externally to the vesicles. Pore formation can be detected by the increase in BODIPY fluorescence that occurs when biocytin binds to BODIPY-streptavidin (32, 45, 46). Experiments showed that pore formation can be induced by WT, short, and long PFO but not prepore PFO, which does not form pores (Fig. 2). The extent of pore formation was dependent on cholesterol concentration, with the increase in BODIPY emission intensity being half-maximal at 16, 17, and 29 mol % cholesterol for WT, long, and short PFO, respectively. These values are identical to the cholesterol concentrations needed for insertion of acrylodan-labeled residue 215. Note that short PFO induces less pore formation per μg than WT or long PFO. This is partly due to the fact that only about half of short PFO inserts into membranes.
FIGURE 2.
Formation of pores by PFO in LUV is cholesterol-dependent. The y axis shows the extent of increase of external BODIPY-streptavidin fluorescence after the addition of PFO relative to that before the PFO addition. Samples contained LUV composed of DOPC/cholesterol (100 μm total lipid) containing entrapped biocytin and 10 nm externally added BODIPY-tagged streptavidin. BODIPY fluorescence was measured 40 min after the addition of 5 μg/ml WT PFO (open squares), 7 μg/ml long PFO (filled diamonds), 20 μg/ml short PFO (open circles), or 5 μg/ml prepore PFO (filled triangles). Prepore PFO pore formation was only assayed in DOPC and 6:4 (mol/mol) DOPC/cholesterol vesicles. Average (mean) values and S.D. values (error bars) were obtained from four samples. Error bars in this and the following figures are not shown where they were small relative to symbol size.
Long, WT, and Short PFO Exhibit Mismatch-dependent Pore Formation
To define how increasing or decreasing the number of residues in the membrane-inserting segments affects hydrophobic mismatch, the effect of varying bilayer width upon pore formation was measured. An effect of mismatch upon pore formation is expected because prior studies have shown that TM proteins have an optimal bilayer thickness for carrying out function, and the optimal thickness is near the value giving hydrophobic match (see the Introduction). To vary hydrophobic mismatch, PFO-induced pore formation was measured in LUVs composed of various phosphatidylcholines with monounsaturated fatty acyl chains ranging from 14 to 24 carbon atoms, plus cholesterol (40 mol %). Fig. 3 shows the normalized dependence of the extent of pore formation upon bilayer width for short, long, and WT PFO. In all three cases, there was a dependence of pore formation upon acyl chain length, with maximal pore formation at different acyl chain lengths. The acyl chain length at which pore formation was maximal was 16 carbon atoms for short PFO, 19 carbon atoms for WT PFO, and 21 carbon atoms for long PFO. This indicates that PFO pore formation is sensitive to mismatch and that decreasing or increasing the number of residues in the membrane-inserting segments of PFO decreases or increases PFO TM length, respectively. These changes in PFO TM length are close to that predicted for insertion or deletion of two amino acid residues in TM β-strands (see “Discussion”). It should be noted that the decrease in pore formation at wide or thin bilayer widths was not due to a loss of membrane insertion, because the fluorescence emission of acrylodan-labeled residue 215 was highly blue-shifted at all bilayer widths for WT, long, and short PFO. As expected, prepore PFO exhibited highly red-shifted emission at all bilayer widths, indicative of a lack of insertion.
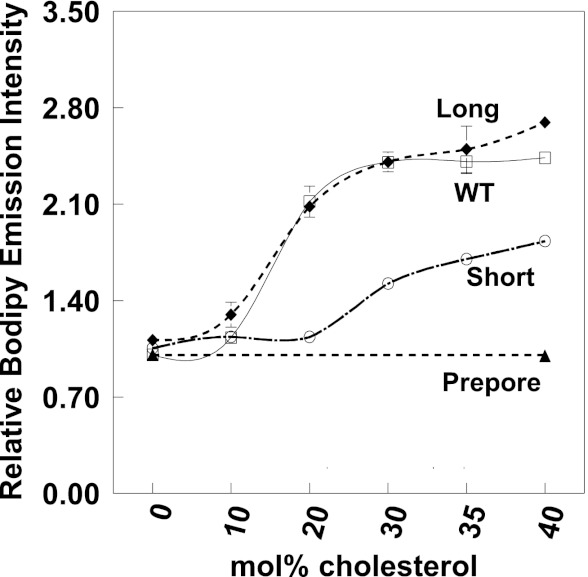
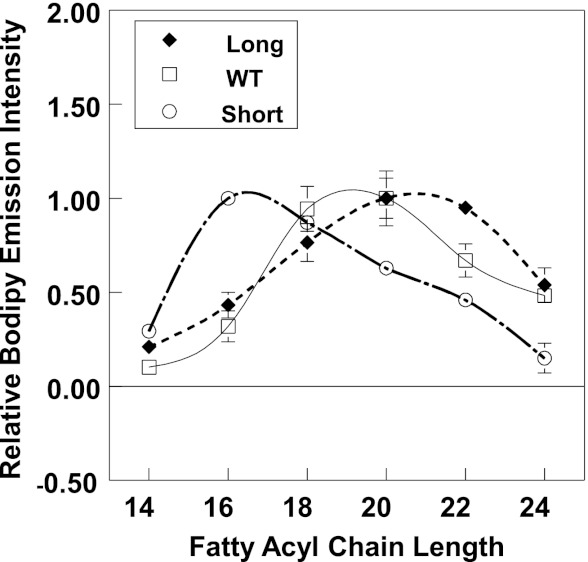
FIGURE 3.
Hydrophobic mismatch between PFO TM domain and lipid controls PFO pore formation activity. Samples contained LUV composed of 6:4 (mol/mol) PC/cholesterol (100 μm total lipid) with entrapped biocytin. Vesicles were dispersed in PBS, pH 5.1, plus 10 nm BODIPY-tagged streptavidin added externally. BODIPY fluorescence was measured 40 min after the addition of 5 μg/ml WT PFO (open squares), 7 μg/ml long PFO (filled diamonds), or 20 μg/ml short PFO (open circles). Normalized values are shown with maximal release (BODIPY fluorescence) assigned as 1. The x axis shows the acyl chain length (n) of the di-Cn:1 PCs used. Average (mean) values and S.D. values (error bars) were obtained from four samples.
FRET Assay of Protein Partition between Ordered and Disordered Domains
To study the effect of TM length on PFO affinity for ordered lipid domains (rafts), a FRET assay that we developed previously was used (31). The FRET donor was protein Trp, and the acceptor was either pyrene-DOPE or NBD-DPhPE, probe lipids that favorably partition into Ld domains (31). The basis of the assay is that in membranes with co-existing Lo and Ld domains, FRET should be weak when PFO associates with Lo domains and strong when it associates with acceptor-rich Ld domains. Experiments with two different acceptors were carried out to test the possibility that specific interactions between the acceptor group and donor molecule might influence FRET.
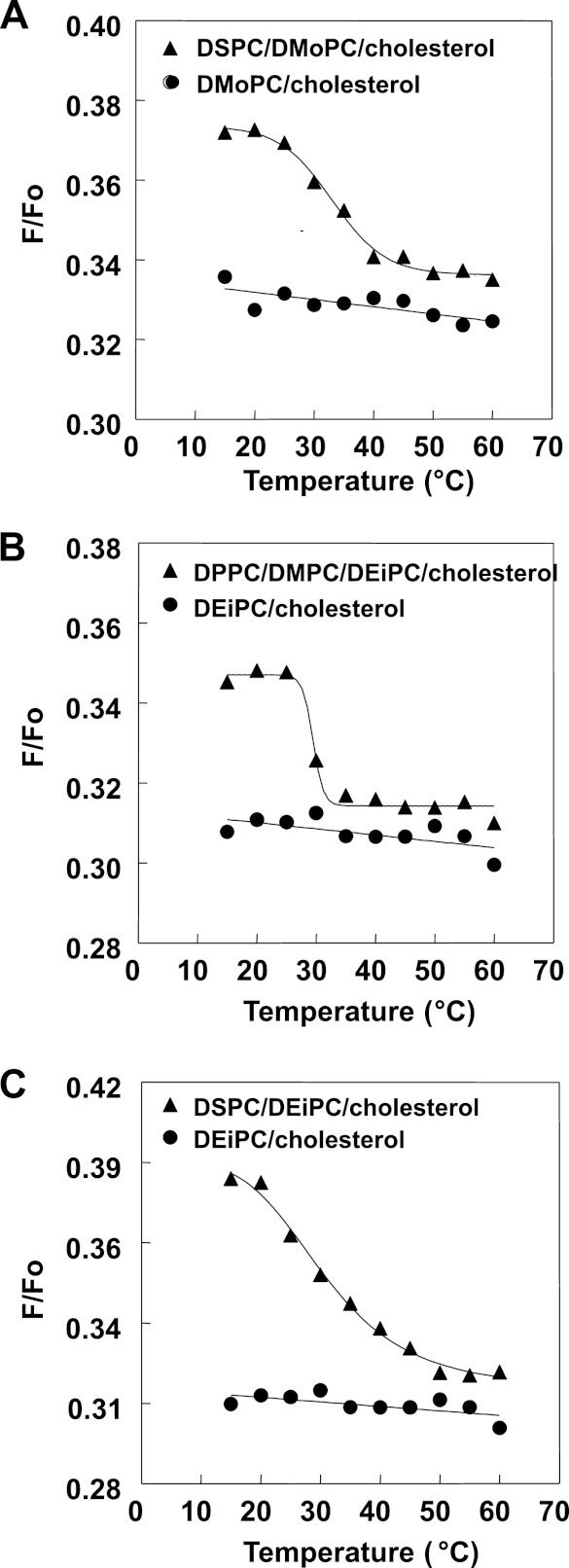
Vesicles with different lipid mixtures having a tendency to form co-existing Lo and Ld domains were prepared. The first contained 1:1 (mol/mol) DSPC/DMoPC (diC18:0 PC/diC14:1 PC) with 45 mol % cholesterol. In these vesicles, the DSPC-rich Lo domains will form a much wider bilayer than the DMoPC-rich Ld domains. The second type of vesicles were composed of 1:1:2 (mol/mol/mol) DPPC/DMPC/DEiPC (diC16:0 PC/diC14:0 PC/diC20:1 PC) with 45 mol % cholesterol. In these vesicles, the DPPC/DMPC-rich Lo domains will be thinner than the DEiPC-rich Ld domains. FRET from pyrene-DPPE (which associates moderately with Lo domains (38)) to Rho-DOPE confirmed that these mixtures formed co-existing ordered and disordered domains (Fig. 4). As shown previously (38), vesicles with co-existing Lo and Ld domains show weaker pyrene to rhodamine FRET (higher F/Fo) than homogeneous vesicles lacking domains. Samples with domains can also be detected by the characteristic disappearance of domains upon melting at high temperature, such that FRET increases (F/Fo decreases) to levels very close to that in homogeneous vesicles. The ternary mixtures we studied all show this behavior, confirming domain formation at 23 °C.
FIGURE 4.
Detection of domain formation by FRET. Samples were composed of MLVs containing 500 μm lipid composed of the following: 55:45 (mol/mol) DMoPC/cholesterol or 27.5:27.5:45 (mol/mol/mol) DSPC/DMoPC/cholesterol (A); 55:45 (mol/mol) DEiPC/cholesterol or 13.75:13.75:27.5:45 (mol/mol/mol/mol) DPPC/DMPC/DEiPC/cholesterol (B); and 55:45 (mol/mol) DEiPC/cholesterol or 27.5:27.5:45 (mol/mol/mol) DSPC/DEiPC/cholesterol (C). Samples were dispersed in PBS, pH 5.1. F samples contained both FRET donor (0.05 mol % pyrene-DPPE) and FRET acceptor (2 mol % Rho-DOPE). Fo samples only contained unlabeled lipids plus FRET donor (0.05 mol % pyrene-DPPE). The ratio of donor fluorescence in the presence of acceptor to that in its absence (F/Fo) is graphed. Average (mean) values were obtained from duplicate samples.
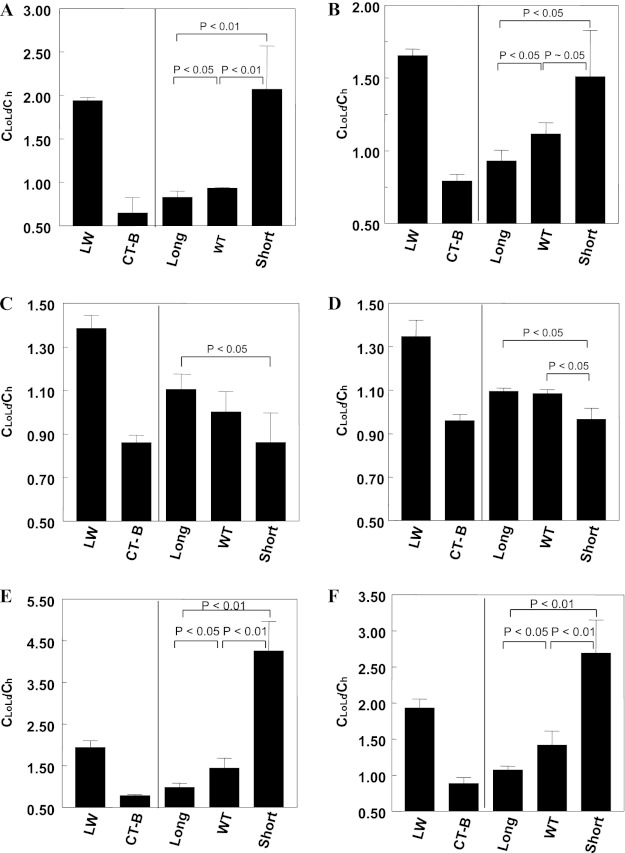
FRET from PFO Trp to acceptor labeled lipids was measured for short, WT, and long PFO as well as two standard proteins: cholera toxin B (CT-B), a marker protein associating with ordered domains (47), and LW peptide, a TM helix that associates with disordered domains (48). To be able to compare FRET values for proteins with different inherent FRET efficiencies (due to different Trp locations relative to the membrane bilayer), FRET values in the domain-containing lipid mixtures were normalized to FRET in homogeneous vesicles lacking domains. For the latter case, vesicles containing 45 mol % cholesterol and DMoPC were used for comparison with domain-forming mixtures containing DMoPC; vesicles containing 45 mol % cholesterol and DEiPC were used for comparison with domain-forming mixtures containing DEiPC. FRET was then used to calculate the effective local acceptor concentration around PFO molecules in domain-containing membranes (CLoLd) relative to that in the homogeneous membranes (Ch) (see “Experimental Procedures” for details). When a protein partitions into Ld domains, the local acceptor concentration is higher in domain-containing membranes than in homogeneous membranes (CLoLd/Ch > 1), and lower than in homogeneous membranes (CLoLd/Ch < 1) when PFO is in the Lo domains. It is important to note that these values give a relative, not absolute, value for the concentration of protein in ordered and disordered domains because FRET is a function of several unknown variables, such as the exact partition coefficient of FRET acceptor between Lo and Ld domains and the fraction of the bilayer in the Lo and Ld states (31, 38) (see “Discussion”).
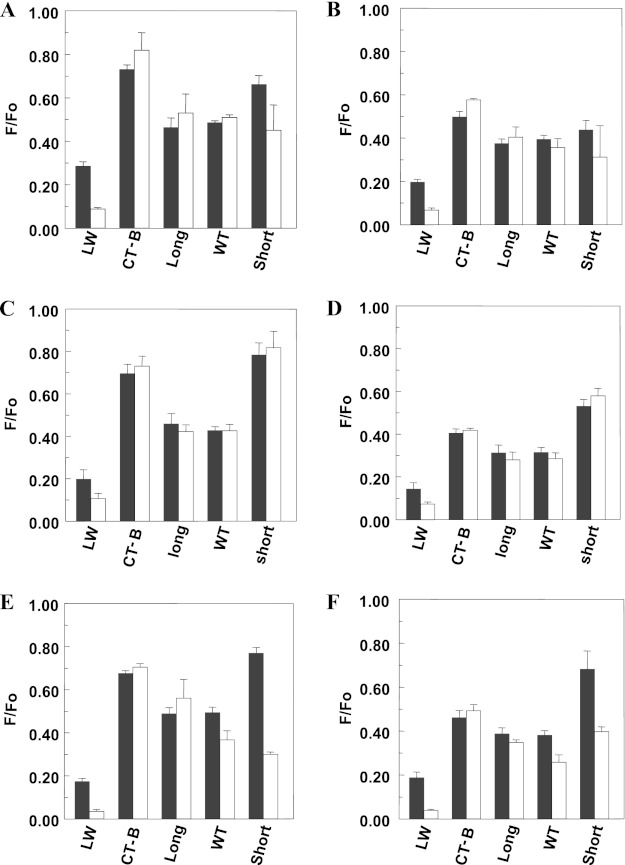
The expected FRET pattern was confirmed using LW peptide and CT-B (Fig. 5 shows the raw fluorescence data, and Fig. 6 shows CLoLd/Ch values). For LW peptide, CLoLd/Ch was >1, reflecting preferential location of LW peptide in disordered domains. In contrast, for CT-B, CLoLd/Ch was <1, consistent with its affinity for ordered domains. This pattern was observed both in bilayers having thick Ld domains and thin Lo domains and in bilayers containing thick Lo domains and thin Ld domains, reflecting an association of CT-B and LW peptide with Ld and Lo domains that was not strongly affected by mismatch. The pattern was also similar using the two different acceptors.
FIGURE 5.
Raw F/Fo values for FRET detection of PFO raft affinity in vesicles containing co-existing Lo/Ld domains. A and B, the F/Fo for LW peptide, CT-B, and PFO in MLVs (500 μm total lipid) composed of 27.5:27.5:45 (mol/mol/mol) DSPC/DEiPC/cholesterol (white bar) or 55:45 (mol/mol) DMoPC/cholesterol (gray bar). C and D, F/Fo for LW peptide, CT-B, and PFO in MLVs (500 μm total lipid) composed of 13.75:13.75:27.5:45 (mol/mol/mol/mol) DPPC/DMPC/DEiPC/cholesterol (white bar) or 55:45 (mol/mol) DEiPC/cholesterol (gray bar). E and F, the F/Fo for LW peptide, CT-B, and PFO in MLVs (500 μm total lipid) composed of 27.5:27.5:45 (mol/mol/mol) DSPC/DEiPC/cholesterol (white bar) or 55:45 (mol/mol) DEiPC/cholesterol (gray bar). Samples were prepared in PBS, pH 5.1. F/Fo is the ratio of fluorescence in the presence of FRET acceptor to that in its absence. A, C, and E, 2 mol % NBD-DPhPE as FRET acceptor. B, D, and F, 1 mol % pyrene-DOPE as FRET acceptor. Average (mean) values and S.D. values (error bars) were obtained from four separate FRET experiments, each having triplicate samples. The F/Fo values shown have been corrected for incomplete binding of PFO to membranes.
FIGURE 6.
FRET-detected raft affinity of PFO with different TM domain length in vesicles containing co-existing Lo and Ld domains. A and B, raft affinity of LW peptide, CT-B, and PFO in membranes composed of 1:1 (mol/mol) DSPC/DMoPC with 45 mol % cholesterol. C and D, raft affinity of LW peptide, CT-B, and PFO in membranes composed of 1:1:2 (mol/mol/mol) DPPC/DMPC/DEiPC with 45 mol % cholesterol. E and F, raft affinity of LW peptide, CT-B, and PFO in membranes composed of 1:1 (mol/mol) DSPC/DEiPC with 45 mol % cholesterol. A, C, and E, 2 mol % NBD-DPhPE as FRET acceptor. B, D, and F, 1 mol % pyrene-DOPE as FRET acceptor. The CLoLd/Ch ratio represents the average local concentration of acceptor around the donor (protein) in vesicles containing Lo and Ld domains (CLoLd) relative to that in a homogeneous bilayer lacking domains (Ch). CLoLd/Ch is high for a protein in Ld domains and low for a protein in Lo domains. Average (mean) values and S.D. values (error bars) were obtained from four separate FRET experiments, each having triplicate samples. The F/Fo values shown have been corrected for incomplete binding of PFO to membranes.
FRET Assay of PFO Affinity for Ordered Domains Is Dependent upon Mismatch
Next, we applied the FRET assay to PFO-containing vesicles. Centrifugation and fluorescence measurements similar to those described above measured the amount of PFO binding and confirmed PFO insertion into the vesicles, which was found to be similar to that in 6:4 (mol/mol) DOPC/cholesterol (data not shown). Because short PFO binding was incomplete, it was important to correct FRET values to eliminate the influence of PFO molecules not bound to the vesicles (see “Experimental Procedures”).
In DSPC/DMoPC/cholesterol vesicles, PFO association with Lo domains increased as TM segment length increased. The CLoLd/Ch ratio for WT and long PFO was between that of LW and CT-B but more similar to CT-B, indicating a significant association with Lo domains. The association of WT PFO with Lo domains was slightly weaker than that of long PFO. The CLoLd/Ch ratio for short PFO was close to that of LW peptide, indicating relatively strong association with Ld domains. Results using pyrene-DOPE or NBD-DPhPE as acceptors were similar.
The pattern of PFO association with Lo domains was basically reversed in vesicles composed of DPPC/DMPC/DEiPC/cholesterol (i.e. Lo affinity generally increased as TM segment length decreased (although the difference between long PFO and WT PFO was not statistically significant)). Most strikingly, short PFO associated with Lo domains to a similar extent as did CT-B in these vesicles. Nevertheless, long and WT PFO retained a considerable affinity for Lo domains in vesicles composed of this lipid mixture. Combined, these results show that TM segment length has a strong influence upon PFO association with Lo domains, such that there is a tendency for association with the domains that have the least hydrophobic mismatch with PFO TM segments.
We also measured PFO location using FRET in vesicles composed of 1:1 (mol/mol) DSPC/DEiPC (diC18:0 PC/diC20:1 PC) with 45 mol % cholesterol. This type of vesicle should closely resemble natural membranes in that DSPC-rich Lo domains should form a bilayer that is only slightly wider than that formed in the DEiPC-rich Ld domains (see “Discussion”). In these vesicles, PFO affinity for Lo domains increased as TM segment length increased, similar to the pattern in DSPC/DMoPC/cholesterol vesicles.
Microscopy Assay of Partition Behavior of PFO Derivatives with Different Length of TM Segments between Ordered and Disordered Domains
Next, we measured the PFO domain location via microscopy. PFO labeled with BODIPY-FL at residue 215 was added to GUVs composed of 1:1 egg SM/DMoPC with 37 mol % cholesterol. This mixture should have thick Lo domains and thin Ld domains. (These experiments were carried out at 37% cholesterol because domains large enough to detect by microscopy were not present at 45 mol % cholesterol. Although at 37 mol %, cholesterol PFO may not fully bind to membranes, which is problematic for FRET, this is not a problem for microscopy.)
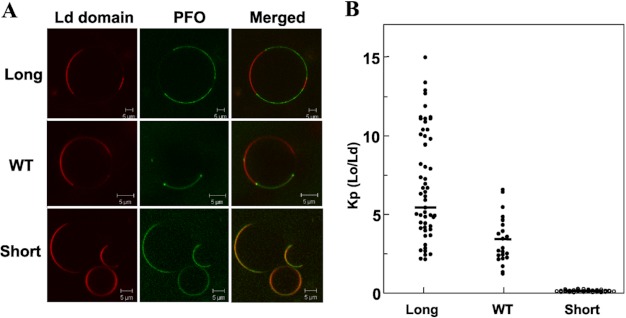
There was a clear influence of TM segment length on domain affinity (Fig. 7A). Long PFO segregated into different domains than the Ld marker Rho-DPPE, showing that it partitioned favorably into the thick egg SM Lo domains. WT PFO also partitioned favorably into Lo domains but also frequently concentrated at the boundary between the Lo and Ld domains, consistent with our previous observations, and suggestive of highest affinity for a bilayer of width intermediate between that of the Lo and Ld domains in this lipid mixture (31). In contrast, short PFO was primarily localized in Ld domains. Quantitatively, even when ignoring PFO located at domain boundaries, PFO affinity for Lo domains, as given by the partition coefficient KP, decreased in the order long PFO > WT PFO ≫ short PFO (Fig. 7B), the same order observed in domain forming DMoPC-containing samples by FRET. It should be noted that omitting correction for possible signal saturation in the brightest image pixels did not alter the pattern of differences between Kp values for the different forms of PFO (data not shown).
FIGURE 7.
Fluorescence imaging of PFO with different TM domain length binding to GUVs. A, confocal image of the equatorial plane of a typical GUV composed of 1:1 (mol/mol) egg SM/DMoPC with 37 mol % cholesterol after PFO binding. The red channel shows the fluorescence signal originating from the Ld marker Rho-DPPE. The dark portion of the bilayer corresponds to Lo domains. The green channel shows the signal from BODIPY-labeled PFO. B, protein partition coefficient calculated from image intensity analysis for fluorescent PFO (see “Experimental Procedures”) (long PFO, n = 83; WT PFO, n = 33; short PFO, n = 55). K(Lo/Ld) equals the ratio of fluorescence intensity in Lo domains divided by that in Ld domains, not counting protein at domain boundaries. All microscopy experiments were carried out at room temperature. Horizontal bars show mean values. The differences between the long, WT, and short PFO were all significant to the level of p < 0.01.
DISCUSSION
Designing Experiments to Test the Effect of Hydrophobic Mismatch upon the Affinity of Multi-TM Segment Proteins for Ordered Domains
There are two potential strategies for varying mismatch: altering bilayer width and altering TM segment length. Altering bilayer width by changing lipid acyl chain length is relatively simple, but changing acyl chain length in domain-containing samples not only changes bilayer width but also alters the lipid compositions of each type of domain, the fraction of the membrane in the form of ordered domains, and domain size. As a result, interpreting the results of such experiments solely in terms of mismatch can be misleading. For that reason, we concentrated upon the strategy of varying TM segment length to define the effects of hydrophobic mismatch. This approach has significant challenges because for a multi-TM segment, length changes must be made in every TM segment, and the resulting mutant protein must fold into the appropriate structure. For such studies, TM β-barrels are a good choice because they are relatively rigid structures. This rigidity may explain why it has been observed that the lipid binding constant of β-barrel porin OmpF is more restricted to matching of membrane thickness than that of α-helical K+ channel KcsA and Ca2+-ATPase (15, 17, 49–51). PFO forms a β-barrel with many TM segments (4 per monomer, with about 35–40 monomers per PFO oligomer (30)) and is known to have a significant affinity for ordered domains due to its binding to cholesterol (30, 31). Furthermore, it was possible to prepare mutant PFO molecules that preserved the most essential features of PFO-membrane interaction. Thus, it was an excellent choice for these studies.
Two different methods were used to detect PFO location, FRET and microscopy. Each has strengths and weaknesses that should be mentioned. Microscopy requires conditions in which domains are large. This greatly restricts the range of lipid compositions that can be used. FRET does not have this problem, because it can detect segregation even when very small domains are present (38). On the other hand, several factors prevent calculation of precise partition coefficients (Kp) giving protein domain location from FRET. This is because FRET values depend on the partition coefficient of the acceptor between Lo and Ld domains, the amount of ordered domains, bilayer width, and differences in donor fluorescence intensity and lifetime in different domains, as well as the value of the partition of the protein into different domains. We minimized these issues by using standard protein markers to identify FRET values characteristic of location in the Lo or Ld domains and by emphasizing the differences between the Lo affinity of different forms of PFO under conditions in which lipid composition is kept constant. Fortunately, there was good agreement between FRET and microscopy experiments. This suggests that the effect of mismatch is similar in microscopic and submicroscopic domains.
Effect of Mismatch upon PFO Ordered Domain Affinity
The results of this study show that hydrophobic mismatch strongly influences the affinity of PFO for ordered membrane domains. In a bilayer containing thick Lo domains and thin Ld domains, PFO with lengthened TM segments has the highest affinity for Lo domains, and PFO with shortened TM segments has the lowest. The relationship of PFO TM length and Lo affinities reverses when Lo domains are thin and Ld domains are thick. From the results, it is possible to roughly quantify the dependence of domain affinity upon TM length. Changing strand length by 2 residues is equivalent to a change of ∼6–7 Å for a strand that is not tilted relative to the bilayer normal. Strands tilted at an angle of 30º would correspond to a change in TM length of 5.6 Å, whereas a tilt of 45º would correspond to a change in TM length of 4.6 Å. These numbers can be compared with the change in effective TM lengths derived from the bilayer width giving maximal pore formation, which should be near the bilayer width giving hydrophobic match. The difference in maximal pore formation width for the wild type and short TM PFO was about 3 carbon atoms, which is equivalent to a change in optimal bilayer width of 5.4 Å (52, 53). This is close to the above-noted 4.6–6.5 Å estimate based on the change in sequence length. Thus, a ∼5-Å change in TM length, which is ∼2 residues in a TM β-strand or ∼3.5–4 residues in a TM α-helix, can induce up to a 10-fold change in domain affinity based on PFO behavior in egg SM/DMoPC/cholesterol.
The difference between the effective TM length of long PFO and that of WT PFO was significantly smaller than that between the short PFO and WT PFO, despite the fact that the mutation involved a length change of 2 residues in both cases. This is most likely to reflect an inability of the inserted Ala residues in long PFO to maximally extend the β-strands. The difference in bilayer width for maximal pore formation for long and TM PFO (about 2 carbon atoms) gives an estimate for the increase in TM length of ∼3.6 Å. This is equivalent to increasing a hydrophobic segment by ∼1–1.5 amino acid residues for β-strand or 2–2.5 residues for a TM helix. Because long and WT PFO exhibited a difference in Lo affinity of roughly 2-fold in egg SM/DMoPC/cholesterol, this implies that even this small change in TM sequence length can have a significant effect upon raft affinity.
Another question raised by this study is how the effect of TM length on raft affinity depends on the difference between Lo and Ld domain bilayer width. The observation that long and WT PFO had a much higher Lo affinity than short PFO in DSPC/DEiPC/cholesterol vesicles implies that even a small difference in domain width can have a large influence on raft affinity. Previous studies in Ld state bilayers indicate that a lipid with saturated acyl chains forms bilayers with a width close to that of a lipid with dimonounsaturated acyl chains 2 carbons longer (53, 54). This means that DSPC and DEiPC in the Ld state should have similar bilayer widths. As a result, the DSPC/DEiPC/cholesterol mixture should have DSPC-rich Lo domains wider than DEiPC-rich Ld domains only due to tighter packing (because of straighter acyl chains) in the Lo state. This difference in width would only be a few Å and similar to what might be encountered in a biological membrane.
Relationship of Mismatch and Domain Affinity for Multi-TM Segment Versus Single TM Segment Proteins
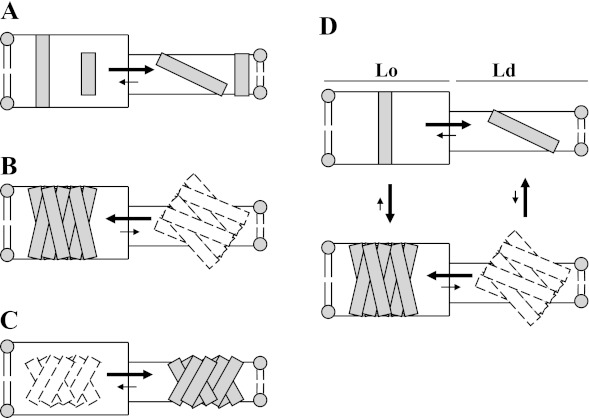
The results of this study have interesting implications for the regulation of TM protein behavior and function in membranes. Fig. 8 schematically summarizes the interactions of single and multi-TM segment proteins with domains of different width and order. In the case of single TM segments in a membrane with co-existing thick Lo domains and thin Ld domains, a short TM segment will tend to partition into the thinner Ld domains to minimize mismatch and to minimize unfavorable packing with lipid. A single long TM segment would also partition into Ld domains to avoid unfavorable packing because it could easily avoid the consequences of mismatch by tilting to avoid exposing hydrophobic surface to aqueous solution in thin domains (Fig. 8A). Thus, there is no situation in which hydrophobic mismatch tends to drive single TM segment proteins into Lo domains. In contrast, for a multi-TM segment protein with long TM sequences, tilting would be ineffective for burying long hydrophobic sequences within the lipid bilayer (Fig. 8B), forcing it into Lo domains, whereas a protein with short TM segments would locate in Ld domains (Fig. 8C). As a result, partitioning of a multi-TM segment protein will be very strongly influenced by mismatch, as observed in this report.
FIGURE 8.
Schematic illustration of how mismatch and TM protein dimensions affect affinity for domains of different widths and of the linkage between ordered domain affinity and TM protein multimerization. TM segments are shown as individual rectangles. Dashed rectangles indicate TM species unlikely to form to a significant extent. A, partitioning behavior of single short or long TM segments between thick Lo and thin Ld domains. B, partitioning behavior of protein or complex with multiple long TM segments between thick Lo domains and thin Ld domains. C, partitioning behavior of protein or complex with multiple short TM segments between thick Lo domains and thin Ld domains. D, linkage between multimerization and ordered domain affinity for proteins or complexes with long TM segments in membranes with thick Lo domains and thin Ld domains. For simplicity, the figure does not illustrate the likely local distortions in bilayer width adjacent to mismatched protein segments.
This difference between single TM segment and multi-TM segment proteins has important functional implications for how co-existing membrane domains with different widths could influence the formation of multi-TM segment complexes from proteins with single TM segments. As shown in Fig. 8D, based on the ideas above, for proteins with single long TM segments, the association with ordered domains will favor formation of complexes with multiple TM segments. Simultaneously, complex formation favors association with Lo domains. This could explain why TM protein complexes forming in signal transduction processes appear have an affinity for rafts. (In addition, when the membrane bilayer has a borderline tendency to form co-existing ordered and disordered domains, the formation of complexes containing long TM segments would be a driving force for lipid domain formation.)
The model above assumes that the TM segments of a multi-TM segment protein/complex have a fixed relationship to each other (and thus fixed hydrophobic width). For cases in which there is a more flexible arrangement of TM segments, multiple conformations/states with different hydrophobic widths are possible, and ordered domains could control which conformation/state forms.
Acknowledgment
We thank Dr. Salvatore Chiantia for assistance in the analysis of microscopy data, including development of the algorithm for correcting for signal saturation.
This work was supported by National Science Foundation Grant MCB 1019986.
- Lo
- liquid-ordered
- Ld
- liquid-disordered
- TM
- transmembrane
- PFO
- perfringolysin O
- GUV
- giant unilamellar vesicle
- LUV
- large unilamellar vesicle
- pyrene-DOPE
- 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-pyrenesulfonyl
- pyrene-DPPE
- 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-pyrenesulfonyl
- NBD-DPhPE
- 1,2-diphytanoyl-sn-glycero-3-phosphoethanolamine-N-(7-nitro-2-1,3-benzoxadiazol-4-yl)
- Rho-DOPE
- 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl)
- Rho-DPPE
- 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl)
- BODIPY-FL
- N-(4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-yl)methyl)iodoacetamide
- acrylodan
- 6-acryloyl-2-dimethylaminonaphthalene
- GM1
- ganglioside M1
- LW peptide
- acetyl-K2W2L8AL8W2K2-amide
- 10-DN
- 10-doxylnonadecane
- SLIM
- site-directed ligase-independent mutagenesis
- TMH
- transmembrane helix
- MLV
- multilamellar vesicle
- DOPC
- 1,2-dioleoyl-sn-glycero-3-phosphocholine
- PC
- phosphatidylcholine
- CT-B
- cholera toxin B chain
- SM
- sphingomyelin
- DMPC
- 1,2-dimyristoyl-sn-glycero-3-phosphocholine
- DMoPC
- 1,2-dimyristoleoyl-sn-glycero-3-phosphocholine
- DEiPC
- 1,2-dieicosenoyl-sn-glycero-3-phosphocholine
- DPPC
- 1,2-dipalmitoyl-sn-glycero-3-phosphocholine.
REFERENCES
- 1. Simons K., Ikonen E. (1997) Functional rafts in cell membranes. Nature 387, 569–572 [DOI] [PubMed] [Google Scholar]
- 2. Lingwood D., Simons K. (2010) Lipid rafts as a membrane-organizing principle. Science 327, 46–50 [DOI] [PubMed] [Google Scholar]
- 3. Simons K., Toomre D. (2000) Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 1, 31–39 [DOI] [PubMed] [Google Scholar]
- 4. Brown D. A., London E. (1998) Structure and origin of ordered lipid domains in biological membranes. J. Membr. Biol. 164, 103–114 [DOI] [PubMed] [Google Scholar]
- 5. Phillips R., Ursell T., Wiggins P., Sens P. (2009) Emerging roles for lipids in shaping membrane-protein function. Nature 459, 379–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sachs J. N., Engelman D. M. (2006) Introduction to the membrane protein reviews. The interplay of structure, dynamics, and environment in membrane protein function. Annu. Rev. Biochem. 75, 707–712 [DOI] [PubMed] [Google Scholar]
- 7. Sprong H., van der Sluijs P., van Meer G. (2001) How proteins move lipids and lipids move proteins. Nat. Rev. Mol. Cell Biol. 2, 504–513 [DOI] [PubMed] [Google Scholar]
- 8. Hanzal-Bayer M. F., Hancock J. F. (2007) Lipid rafts and membrane traffic. FEBS Lett. 581, 2098–2104 [DOI] [PubMed] [Google Scholar]
- 9. van Meer G., Voelker D. R., Feigenson G. W. (2008) Membrane lipids. Where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9, 112–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schroeder R., London E., Brown D. (1994) Interactions between saturated acyl chains confer detergent resistance on lipids and glycosylphosphatidylinositol (GPI)-anchored proteins. GPI-anchored proteins in liposomes and cells show similar behavior. Proc. Natl. Acad. Sci. U.S.A. 91, 12130–12134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zacharias D. A., Violin J. D., Newton A. C., Tsien R. Y. (2002) Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 296, 913–916 [DOI] [PubMed] [Google Scholar]
- 12. London E., Feigenson G. W. (1981) Fluorescence quenching in model membranes. 2. Determination of local lipid environment of the calcium adenosinetriphosphatase from sarcoplasmic reticulum. Biochemistry 20, 1939–1948 [DOI] [PubMed] [Google Scholar]
- 13. Florine K. I., Feigenson G. W. (1987) Protein redistribution in model membranes. Clearing of M13 coat protein from calcium-induced gel-phase regions in phosphatidylserine/phosphatidylcholine multilamellar vesicles. Biochemistry 26, 2978–2983 [DOI] [PubMed] [Google Scholar]
- 14. de Planque M. R., Killian J. A. (2003) Protein-lipid interactions studied with designed transmembrane peptides. Role of hydrophobic matching and interfacial anchoring. Mol. Membr. Biol. 20, 271–284 [DOI] [PubMed] [Google Scholar]
- 15. East J. M., Lee A. G. (1982) Lipid selectivity of the calcium and magnesium ion dependent adenosinetriphosphatase, studied with fluorescence quenching by a brominated phospholipid. Biochemistry 21, 4144–4151 [DOI] [PubMed] [Google Scholar]
- 16. Johannsson A., Smith G. A., Metcalfe J. C. (1981) The effect of bilayer thickness on the activity of (Na+ + K+)-ATPase. Biochim. Biophys. Acta 641, 416–421 [DOI] [PubMed] [Google Scholar]
- 17. Caffrey M., Feigenson G. W. (1981) Fatty-acyl-chain characteristics of phosphatidylcholines affect Ca2+-dependent ATPase enzymic activity but not the affinity of the protein for these different lipid species. Biochem. Soc. Trans. 9, 155–156 [DOI] [PubMed] [Google Scholar]
- 18. Montecucco C., Smith G. A., Dabbeni-sala F., Johannsson A., Galante Y. M., Bisson R. (1982) Bilayer thickness and enzymatic activity in the mitochondrial cytochrome c oxidase and ATPase complex. FEBS Lett. 144, 145–148 [DOI] [PubMed] [Google Scholar]
- 19. Dumas F., Tocanne J. F., Leblanc G., Lebrun M. C. (2000) Consequences of hydrophobic mismatch between lipids and melibiose permease on melibiose transport. Biochemistry 39, 4846–4854 [DOI] [PubMed] [Google Scholar]
- 20. Johannsson A., Keightley C. A., Smith G. A., Richards C. D., Hesketh T. R., Metcalfe J. C. (1981) The effect of bilayer thickness and n-alkanes on the activity of the (Ca2+ + Mg2+)-dependent ATPase of sarcoplasmic reticulum. J. Biol. Chem. 256, 1643–1650 [PubMed] [Google Scholar]
- 21. Caffrey M., Feigenson G. W. (1981) Fluorescence quenching in model membranes. 3. Relationship between calcium adenosinetriphosphatase enzyme activity and the affinity of the protein for phosphatidylcholines with different acyl chain characteristics. Biochemistry 20, 1949–1961 [DOI] [PubMed] [Google Scholar]
- 22. Ridder A. N., van de Hoef W., Stam J., Kuhn A., de Kruijff B., Killian J. A. (2002) Importance of hydrophobic matching for spontaneous insertion of a single-spanning membrane protein. Biochemistry 41, 4946–4952 [DOI] [PubMed] [Google Scholar]
- 23. Hong H., Tamm L. K. (2004) Elastic coupling of integral membrane protein stability to lipid bilayer forces. Proc. Natl. Acad. Sci. U.S.A. 101, 4065–4070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Marsh D., Páli T. (2004) The protein-lipid interface. Perspectives from magnetic resonance and crystal structures. Biochim. Biophys. Acta 1666, 118–141 [DOI] [PubMed] [Google Scholar]
- 25. van Duyl B. Y., Rijkers D. T., de Kruijff B., Killian J. A. (2002) Influence of hydrophobic mismatch and palmitoylation on the association of transmembrane α-helical peptides with detergent-resistant membranes. FEBS Lett. 523, 79–84 [DOI] [PubMed] [Google Scholar]
- 26. Vidal A., McIntosh T. J. (2005) Transbilayer peptide sorting between raft and nonraft bilayers. Comparisons of detergent extraction and confocal microscopy. Biophys. J. 89, 1102–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stopar D., Spruijt R. B., Hemminga M. A. (2009) Membrane protein frustration. Protein incorporation into hydrophobic mismatched binary lipid mixtures. Biophys. J. 96, 1408–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tweten R. K., Parker M. W., Johnson A. E. (2001) The cholesterol-dependent cytolysins. Curr. Top. Microbiol. Immunol. 257, 15–33 [DOI] [PubMed] [Google Scholar]
- 29. Giddings K. S., Johnson A. E., Tweten R. K. (2003) Redefining cholesterol's role in the mechanism of the cholesterol-dependent cytolysins. Proc. Natl. Acad. Sci. U.S.A. 100, 11315–11320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hotze E. M., Tweten R. K. (2012) Membrane assembly of the cholesterol-dependent cytolysin pore complex. Biochim. Biophys. Acta 1818, 1028–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nelson L. D., Chiantia S., London E. (2010) Perfringolysin O association with ordered lipid domains. Implications for transmembrane protein raft affinity. Biophys. J. 99, 3255–3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nelson L. D., Johnson A. E., London E. (2008) How interaction of perfringolysin O with membranes is controlled by sterol structure, lipid structure, and physiological low pH. Insights into the origin of perfringolysin O-lipid raft interaction. J. Biol. Chem. 283, 4632–4642 [DOI] [PubMed] [Google Scholar]
- 33. Shepard L. A., Heuck A. P., Hamman B. D., Rossjohn J., Parker M. W., Ryan K. R., Johnson A. E., Tweten R. K. (1998) Identification of a membrane-spanning domain of the thiol-activated pore-forming toxin Clostridium perfringens perfringolysin O. An α-helical to β-sheet transition identified by fluorescence spectroscopy. Biochemistry 37, 14563–14574 [DOI] [PubMed] [Google Scholar]
- 34. Chiu J., March P. E., Lee R., Tillett D. (2004) Site-directed, ligase-independent mutagenesis (SLIM). A single-tube methodology approaching 100% efficiency in 4 h. Nucleic Acids Res. 32, e174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chiu J., Tillett D., Dawes I. W., March P. E. (2008) Site-directed, ligase-independent mutagenesis (SLIM) for highly efficient mutagenesis of plasmids greater than 8kb. J. Microbiol. Methods 73, 195–198 [DOI] [PubMed] [Google Scholar]
- 36. Ramachandran R., Tweten R. K., Johnson A. E. (2005) The domains of a cholesterol-dependent cytolysin undergo a major FRET-detected rearrangement during pore formation. Proc. Natl. Acad. Sci. U.S.A. 102, 7139–7144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Johnson I. D., Kang H. C., Haugland R. P. (1991) Fluorescent membrane probes incorporating dipyrrometheneboron difluoride fluorophores. Anal. Biochem. 198, 228–237 [DOI] [PubMed] [Google Scholar]
- 38. Pathak P., London E. (2011) Measurement of lipid nanodomain (raft) formation and size in sphingomyelin/POPC/cholesterol vesicles shows TX-100 and transmembrane helices increase domain size by coalescing preexisting nanodomains but do not induce domain formation. Biophys. J. 101, 2417–2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chattopadhyay A., London E. (1987) Parallax method for direct measurement of membrane penetration depth utilizing fluorescence quenching by spin-labeled phospholipids. Biochemistry 26, 39–45 [DOI] [PubMed] [Google Scholar]
- 40. Chiantia S., Kahya N., Schwille P. (2007) Raft domain reorganization driven by short- and long-chain ceramide. A combined AFM and FCS study. Langmuir 23, 7659–7665 [DOI] [PubMed] [Google Scholar]
- 41. Chiantia S., Schwille P., Klymchenko A. S., London E. (2011) Asymmetric GUVs prepared by MbetaCD-mediated lipid exchange. An FCS study. Biophys. J. 100, L1–L3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shatursky O., Heuck A. P., Shepard L. A., Rossjohn J., Parker M. W., Johnson A. E., Tweten R. K. (1999) The mechanism of membrane insertion for a cholesterol-dependent cytolysin. A novel paradigm for pore-forming toxins. Cell 99, 293–299 [DOI] [PubMed] [Google Scholar]
- 43. Heuck A. P., Savva C. G., Holzenburg A., Johnson A. E. (2007) Conformational changes that effect oligomerization and initiate pore formation are triggered throughout perfringolysin O upon binding to cholesterol. J. Biol. Chem. 282, 22629–22637 [DOI] [PubMed] [Google Scholar]
- 44. Wang J., Rosconi M. P., London E. (2006) Topography of the hydrophilic helices of membrane-inserted diphtheria toxin T domain. TH1-TH3 as a hydrophilic tether. Biochemistry 45, 8124–8134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nicol F., Nir S., Szoka F. C., Jr. (1999) Orientation of the pore-forming peptide GALA in POPC vesicles determined by a BODIPY-avidin/biotin binding assay. Biophys. J. 76, 2121–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nir S., Nicol F., Szoka F. C., Jr. (1999) Surface aggregation and membrane penetration by peptides. Relation to pore formation and fusion. Mol. Membr. Biol. 16, 95–101 [DOI] [PubMed] [Google Scholar]
- 47. Lencer W. I., Saslowsky D. (2005) Raft trafficking of AB5 subunit bacterial toxins. Biochim. Biophys. Acta 1746, 314–321 [DOI] [PubMed] [Google Scholar]
- 48. Fastenberg M. E., Shogomori H., Xu X., Brown D. A., London E. (2003) Exclusion of a transmembrane-type peptide from ordered-lipid domains (rafts) detected by fluorescence quenching. Extension of quenching analysis to account for the effects of domain size and domain boundaries. Biochemistry 42, 12376–12390 [DOI] [PubMed] [Google Scholar]
- 49. Lee A. G. (2003) Lipid-protein interactions in biological membranes. A structural perspective. Biochim. Biophys. Acta 1612, 1–40 [DOI] [PubMed] [Google Scholar]
- 50. O'Keeffe A. H., East J. M., Lee A. G. (2000) Selectivity in lipid binding to the bacterial outer membrane protein OmpF. Biophys. J. 79, 2066–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Williamson I. M., Alvis S. J., East J. M., Lee A. G. (2002) Interactions of phospholipids with the potassium channel KcsA. Biophys. J. 83, 2026–2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ren J., Lew S., Wang J., London E. (1999) Control of the transmembrane orientation and interhelical interactions within membranes by hydrophobic helix length. Biochemistry 38, 5905–5912 [DOI] [PubMed] [Google Scholar]
- 53. Lewis B. A., Engelman D. M. (1983) Lipid bilayer thickness varies linearly with acyl chain length in fluid phosphatidylcholine vesicles. J. Mol. Biol. 166, 211–217 [DOI] [PubMed] [Google Scholar]
- 54. Kucerka N., Nagle J. F., Sachs J. N., Feller S. E., Pencer J., Jackson A., Katsaras J. (2008) Lipid bilayer structure determined by the simultaneous analysis of neutron and x-ray scattering data. Biophys. J. 95, 2356–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]