Background: Tumor suppressor p53 plays a pivotal role in the regulation of DNA damage response.
Results: RUNX1 enhances p53 activity in response to DNA damage through elevation of p53 acetylation.
Conclusion: RUNX1 acts as a co-activator for p53 during DNA damage response.
Significance: This study provides novel insight into understanding the molecular mechanisms behind DNA damage-mediated activation of p53.
Keywords: Apoptosis, DNA Damage, p300, p53, Tumor Suppressor Gene, Acetylation, Adriamycin, RUNX1
Abstract
Representative tumor suppressor p53 plays a critical role in the regulation of proper DNA damage response. In this study, we have found for the first time that Runt-related transcription factor 1 (RUNX1) contributes to p53-dependent DNA damage response. Upon adriamycin (ADR) exposure, p53 as well as RUNX1 were strongly induced in p53-proficient HCT116 and U2OS cells, which were closely associated with significant transactivation of p53 target genes, such as p21WAF1, BAX, NOXA, and PUMA. RUNX1 was exclusively expressed in the cell nucleus and formed a complex with p53 in response to ADR. Chromatin immunoprecipitation assay demonstrated that p53 together with RUNX1 are efficiently recruited onto p53 target gene promoters following ADR exposure, indicating that RUNX1 is involved in p53-mediated transcriptional regulation. Indeed, forced expression of RUNX1 stimulated the transcriptional activity of p53 in response to ADR. Consistent with these observations, knockdown of RUNX1 attenuated ADR-mediated induction of p53 target genes and suppressed ADR-dependent apoptosis. Furthermore, RUNX1 was associated with p300 histone acetyltransferase, and ADR-dependent acetylation of p53 at Lys-373/382 was markedly inhibited in RUNX1 knockdown cells. In addition, knockdown of RUNX1 resulted in a significant decrease in the amount of p53-p300 complex following ADR exposure. Taken together, our present results strongly suggest that RUNX1 is required for the stimulation of p53 in response to DNA damage and also provide novel insight into understanding the molecular mechanisms behind p53-dependent DNA damage response.
Introduction
The appropriate DNA damage response, which monitors and ensures genomic integrity, has been considered to be a critical barrier to tumorigenesis (1). The representative tumor suppressor p53, which plays an integral role in the regulation of DNA damage response, acts as a nuclear transcription factor. p53 is organized into several well defined functional domains, including NH2-terminal transactivation domain, proline-rich domain, highly conserved central DNA-binding domain, COOH-terminal oligomerization domain, and three nuclear localization sequence motifs. Proper conformation of the DNA-binding domain of p53 is required for its sequence-specific transcriptional activation (2). Importantly, it has been shown that p53 is highly mutated in various human primary tumors, and over 90% of p53 mutations are detected within the genomic region encoding its central sequence-specific DNA-binding domain (3). Mutant p53, which exhibits the prolonged half-life, lacks the sequence-specific transactivation function and displays a dominant-negative behavior toward wild-type p53 (4). Mutant p53 in turn acquires oncogenic potential, and certain cancerous cells carrying p53 mutations exhibit the chemo-resistant phenotypes (5–7). Indeed, the early genetic studies demonstrated that p53-deficient mice develop spontaneous tumors (8). Based on these findings, it is likely that the sequence-specific transcriptional activity of p53 is tightly linked to its tumor-suppressive function (2, 9).
Under normal physiological conditions, p53 is kept at extremely low levels. Upon DNA damage, p53 is quickly induced and activated in the cell nucleus through the sequential post-translational modifications, including phosphorylation and acetylation (4, 10, 11). It has been well documented that DNA damage-induced accumulation of p53 is largely regulated by its rate of degradation. MDM2, which acts as an E3 ubiquitin protein ligase for p53, binds to the NH2-terminal region of p53 and facilitates its proteolytic degradation by the proteasome (12–14). DNA damage-induced NH2-terminal phosphorylation of p53 leads to a dissociation of MDM2 from p53 (15), and the COOH-terminal acetylation suppresses MDM2-mediated ubiquitination of p53 (16). These chemical modifications repress the ubiquitin-dependent proteasomal degradation of p53, and thereby p53 becomes stable. Because MDM2 is one of p53 target gene products, MDM2 and p53 form a negative feedback loop in which p53 transactivates MDM2, which in turn down-regulates p53. Thus, the intracellular amount of MDM2 available appears to be critical in determining the expression level of p53.
In response to the severe DNA damage, p53 induces the irreversible apoptosis to eliminate cells with damaged DNA through transactivating its downstream target gene products involved in the regulation of mitochondrial apoptotic pathways, such as BAX, NOXA, and PUMA (17). Induction of apoptosis in developing tumors might clearly be an efficient inhibitor of tumor development. When cells receive repairable DNA damage, p53 instead promotes the reversible cell cycle arrest by stimulating the expression of p21WAF1, GADD45, and 14-3-3σ to save time to repair damaged DNA, and then cells with repaired DNA re-enter into the normal cell cycle (17). Therefore, p53 stands at the crossroad between cell survival and cell death in response to DNA damage.
Because the transcriptional activity of p53 is strongly related to its DNA damage-induced biological outcomes such as cell cycle arrest or apoptosis, numerous studies have concentrated on the elucidation of the regulatory mechanisms that contribute to the activation of p53 in response to DNA damage. Accumulating evidence suggests that DNA damage-mediated chemical modification as well as interaction with cellular co-activator proteins are highly involved in the modulation of p53. For example, p53 is extensively phosphorylated at NH2-terminal Ser-15, Ser-20, and/or Ser-46 following DNA damage (4, 10, 11). Among them, phosphorylation of p53 at Ser-46, which might be mediated by HIPK2 and PKC (18, 19), has been shown to contribute to the transactivation of a specific subset of pro-apoptotic genes (20). In addition to DNA damage-dependent phosphorylation of p53, the p300/CBP3 family of acetyltransferase-mediated acetylation of p53 at Lys-373/Lys-382 led to enhance its stability and activity (21, 22). Recently, Kim et al. (23) found that Wilms tumor suppressor WTX has the ability to increase p300/CBP-dependent acetylation level of p53. Ivanov et al. (24) reported that DNA damage-mediated methylation of p53 at Lys-372 by Set7/9 is important for transcriptional activation and stabilization of p53. For co-activator proteins of p53, it has been described that ASPP1/ASPP2 interact with the DNA-binding domain of p53 to allow induction of its target pro-apoptotic genes (25). Yang et al. (26) demonstrated that 14-3-3σ forms a complex with p53 in response to DNA damage and enhances the transcriptional activity of p53.
RUNX1 belongs to a small family of transcription factors, including RUNX1, RUNX2, and RUNX3, and is composed of NH2-terminal DNA-binding runt homology domain followed by the transcriptional activation domain and COOH-terminal negative regulatory domain (27). RUNX1 has been initially identified at a breakpoint of human chromosome 21 in the t(8; 21) translocation, which is commonly observed in human leukemia (28, 29). Considering that RUNX1 is frequently deregulated in human leukemia and is a target for loss of heterozygosity, it is likely that RUNX1 acts as a classical tumor suppressor (30, 31). Subsequent genetic studies revealed that RUNX1-deficient mice display no definitive hematopoiesis, suggesting that RUNX1 plays a critical role in the regulation of normal blood development (32, 33). Consistent with these observations, RUNX1 stimulates the transcription of a number of myeloid and lymphoid-related genes (34, 35). In addition to hematopoiesis-specific genes, it has been shown that RUNX1 is also implicated in the regulation of cell cycle-related genes, such as p21WAF1 (36).
Several lines of evidence indicate that the post-translational modifications have a significant impact on the transcriptional activity of RUNX1. Guo and Friedman (37) described that cyclin-dependent protein kinase-mediated phosphorylation of RUNX1 at Ser-48, Ser-303, and Ser-424 strengthens the transcriptional ability of RUNX1. According to their results, RUNX1 phosphorylation resulted in a reduction of its interaction with the transcriptional repressor histone deacetylase. Yamaguchi et al. (38) found that p300-mediated acetylation enhances the transcriptional activity of RUNX1. Zhao et al. (39) reported that RUNX1 interacts with arginine methyltransferase PRMT1. Based on their observations, PRMT1-dependent methylation of RUNX1 promoted the dissociation of the co-repressor SIN3A complex from RUNX1, thereby enhancing RUNX1 transcriptional activity.
Although numerous studies with respect to RUNX1 have focused largely on its functional significance in hematopoietic system, it has been described that RUNX1 induces senescence-like growth arrest in primary murine fibroblasts, and this response is lost in cells lacking functional p53 (40, 41). Intriguingly, Li et al. (42) reported that HIPK2, which has the ability to promote p53-dependent apoptosis in response to DNA damage, is a part of the RUNX1 transcription complex. These observations strongly suggest the presence of a functional link between RUNX1 and p53. In this study, we have found for the first time that RUNX1 acts as a co-activator for p53 in response to DNA damage.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
Human colon carcinoma HCT116, human lung carcinoma H1299, and human osteosarcoma U2OS cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS) (Invitrogen) and penicillin/streptomycin at 37 °C in 5% CO2. For transfection, cells were transfected with the indicated combinations of the expression plasmids using Lipofectamine 2000 transfection reagent according to the manufacturer's instructions (Invitrogen).
Colony Formation Assay
H1299 cells were seeded at a density of 1 × 105 cells/6-well tissue culture plate and transfected with the indicated combinations of the expression plasmids. The total amount of plasmid DNA per transfection was kept constant (2 μg) with pcDNA3. Forty eight hours after transfection, cells were maintained in fresh medium containing G418 (at a final concentration of 800 μg/ml). After 2 weeks of the incubation, drug-resistant colonies were fixed in methanol and stained with Giemsa solution.
MTT Assay
HCT116 cells were seeded at a final density of 3,000 cells/96-well plate and allowed to attach overnight. Cells were then treated with the indicated concentrations of adriamycin (ADR). Twenty four hours after ADR exposure, 10 μl of a modified 3-(4,5-dimethylthiazol-2-yl) 2,5-diphenyltetrazolium bromide (MTT) solution (Dojindo, Kumamoto, Japan) was added to the culture, and reaction mixtures were incubated at 37 °C for 2 h. The absorbance readings for each well were carried out at 570 nm using the microplate reader (Model 450, Bio-Rad).
FACS Analysis
HCT116 cells were treated with the indicated concentrations of ADR. Twenty four hours after ADR treatment, floating and attached cells were collected, washed in ice-cold PBS, and fixed in 70% ethanol at −20 °C. Following incubation in PBS containing 25 μg/ml propidium iodide and 200 μg/ml RNase A for 1 h at room temperature in the dark, stained nuclei were analyzed on a FACScan machine (BD Biosciences).
RT-PCR
For RT-PCR, total RNA was prepared by using an RNeasy mini kit according to the manufacturer's instructions (Qiagen, Valencia, CA) and reverse-transcribed into cDNA with random primers using SuperScript II reverse transcriptase (Invitrogen). The resultant cDNA was subjected to PCR-based amplification. The primer sets used in this study were as follows: RUNX1, 5′-CCGAGAACCTCGAAGACATC-3′ (sense) and 5′-GATGGTTGGATCTGCCTTGT-3′ (antisense); p53, 5′-CTGCCCTCAACAAGATGTTTTG-3′ (sense) and 5′-CTATCTGAGCAGCGCTCATGG-3′ (antisense); p21WAF1, 5′-ATGAAATTCACCCCCTTTCC-3′ (sense) and 5′-CCCTAGGCTGTGCTCACTTC-3′ (antisense); BAX, 5′-TTTGCTTCAGGGTTTCATCC-3′ (sense) and 5′-CAGTTGAAGTTGCCGTCAGA-3′ (antisense); NOXA, 5′-CTGGAAGTCGAGTGTGCTACT-3′ (sense) and 5′-TCAGGTTCCTGAGCAGAAGAG-3′ (antisense); PUMA, 5′-GCCCAGACTGTGAATCCTGT-3′ (sense) and 5′-TCCTCCCTCTTCCGAGATTT-3′ (antisense); and GAPDH, 5′-ACCTGACCTGCCGTCTAGAA-3′ (sense) and 5′-TCCACCACCCTGTTGCTGTA-3′ (antisense). The PCR products were electrophoresed on 1.5% agarose gels, and their amounts were evaluated by staining with ethidium bromide.
Immunoblotting
Cells were lysed in Triton X-100 lysis buffer containing 25 mm Tris-HCl (pH 7.5), 137 mm NaCl, 2.7 mm KCl, 1% Triton X-100, and protease inhibitor mixture (Sigma). The lysates were sonicated briefly and clarified by centrifugation at 4 °C for 10 min. Protein concentrations of the lysates were determined by Bradford reagent (Bio-Rad). Equal amounts of the lysates were separated by 10% standard SDS-PAGE and transferred onto polyvinylidene difluoride membranes (Millipore, Billerica, MA). The membranes were blocked with 5% dry milk in Tris-buffered saline (TBS) plus 0.1% Tween 20 and incubated with anti-p53 (DO-1, Santa Cruz Biotechnology, Santa Cruz, CA), anti-phospho-p53 at Ser-15 (Cell Signaling Technologies, Beverly, MA), anti-acetyl-p53 at Lys-373/382 (Millipore), anti-p21WAF1 (H-164, Santa Cruz Biotechnology), anti-BAX (Cell Signaling Technologies), anti-NOXA (Abcam, Cambridge, UK), anti-PUMA (Abcam), anti-poly(ADP-ribose)polymerase (Cell Signaling Technologies), anti-γH2AX (2F3, BioLegend, San Diego), anti-RUNX1 (Epitomics, Burlingame, CA), and anti-p300 (N-15, Santa Cruz Biotechnology) or with anti-actin antibody (20–33, Sigma), followed by the incubation with HRP-conjugated anti-mouse IgG or with anti-rabbit IgG (Cell Signaling Technology). The membranes were extensively washed with TBS plus 0.1% Tween 20, and the proteins were then visualized by enhanced chemiluminescence (ECL, Amersham Biosciences).
Cell Fractionation
Cells were fractionated into nuclear and cytoplasmic fractions as described previously (43). In brief, cells were washed with PBS and lysed in lysis buffer containing 10 mm Tris-HCl (pH 7.5), 1 mm EDTA, 0.5% Nonidet P-40, and protease inhibitor mixture at 4 °C for 30 min. Cell lysates were centrifuged at 15,000 rpm for 15 min at 4 °C to separate soluble (cytoplasmic) from insoluble (nuclear) fractions. The pellets were washed extensively with lysis buffer and then dissolved in SDS sample buffer. The cytoplasmic and nuclear fractions were analyzed by immunoblotting with anti-lamin B (Millipore) or with anti-tubulin-α antibody (NeoMarkers, Fremont, CA).
Co-immunoprecipitation
Equal amounts of cell lysates were pre-absorbed with protein G-Sepharose beads (Amersham Biosciences) at 4 °C for 1 h, and the precleared lysates were incubated with the indicated antibodies at 4 °C for 2 h, followed by incubation with protein G-Sepharose beads for an additional 1 h at 4 °C. The immune complexes were then washed extensively with lysis buffer, eluted by boiling in SDS sample buffer for 5 min, and subjected to immunoblot analysis.
Immunofluorescence
Cells were plated on glass coverslips and fixed in freshly prepared 3.7% formaldehyde in PBS at room temperature for 15 min. After washing with PBS, the cells were permeabilized with 0.1% Triton X-100 in PBS at room temperature for 5 min and then blocked with 3% bovine serum albumin (BSA) in PBS at room temperature for 1 h. After washing with PBS, the cells were simultaneously incubated with anti-p53 and anti-RUNX1 antibodies at room temperature for 1 h, followed by the incubation with rhodamine-conjugated anti-mouse IgG and FITC-conjugated anti-rabbit IgG (Invitrogen) at room temperature for 1 h. The coverslips were washed with PBS and mounted in VectaShield containing DAPI (Vector Laboratories, Peterborough, UK). Fluorescent images were captured using a confocal microscope (Leica, Milton Keynes, UK).
siRNA-mediated Knockdown
Control siRNA or siRNA against RUNX1 (Dharmacon, Chicago) was introduced into HCT116 cells by the use of Lipofectamine RNAiMAX transfection reagent according to the manufacturer's instructions (Invitrogen). Forty eight hours after transfection, total RNA and cell lysates were prepared and processed for RT-PCR and immunoblotting, respectively.
Chromatin Immunoprecipitation (ChIP) Assay
ChIP assay was performed using a kit from Millipore following the manufacturer's procedure. In brief, cross-linking was achieved by incubating cells on a 10-cm plate with 10 ml of 1% formaldehyde in fresh medium at 37 °C for 15 min. After cross-linking, cells were washed with PBS and harvested by centrifugation at 4,000 rpm for 5 min at 4 °C. Cell pellets were suspended in 200 μl of lysis buffer and incubated at 4 °C for 10 min. The cell lysates were diluted with immunoprecipitation buffer and then sonicated to share DNA to an average size of 500 bp. The chromatin solutions were precleared with protein A-agarose beads and incubated with normal rabbit serum (NRS) or with polyclonal anti-RUNX1 antibody at 4 °C overnight. Protein A-agarose beads were added, and the reaction mixtures were incubated for another 2 h at 4 °C. After the incubation, the beads were washed with the appropriate buffers, and the immune complexes were eluted from the beads with elution buffer containing 1% SDS and 0.1 m NaHCO3. The DNA-protein complexes were then treated with proteinase K at 50 °C for 1 h, followed by reverse cross-linking at 65 °C for 4 h. DNA was extracted with phenol/chloroform, precipitated with ethanol, dissolved in 25 μl of Tris/EDTA buffer, and analyzed by PCR.
Trypan Blue Exclusion Assay
Twenty four hours after siRNA transfection, HCT116 cells were exposed to ADR (at a final concentration of 1 μm) or left untreated. Twenty four hours after ADR treatment, both floating and adherent cells were suspended in 0.4% trypan blue in PBS, and the number of live and dead cells was measured using hemocytometer. The cells that excluded the blue dye and displayed a well defined cellular outline were scored as live.
RESULTS
RUNX1 Suppresses Cell Growth in Collaboration with p53
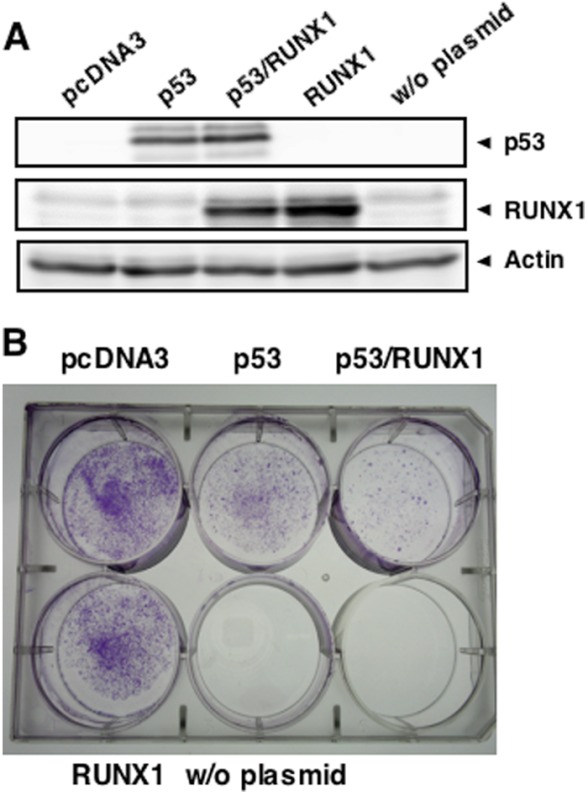
To ask whether there could exist a functional interaction between RUNX1 and p53, we performed colony formation assay. p53-deficient human lung carcinoma H1299 cells were transfected with the expression plasmid for p53, p53 plus RUNX1, or RUNX1 and maintained in the presence of G418 for 2 weeks. Drug-resistant colonies were then stained with Giemsa solution. Transient expression levels of the exogenous p53 and RUNX1 were examined by immunoblotting (Fig. 1A). Consistent with our previous observations (44), forced expression of p53 in H1299 cells significantly reduced the number of drug-resistant colonies (Fig. 1B). Of note, co-expression of p53 with RUNX1 resulted in more of a decrease in the number of viable colonies as compared with that caused by p53 alone, whereas ectopic expression of RUNX1 alone had a marginal effect on colony formation. These results imply that RUNX1 might collaborate with p53 to suppress cell growth.
FIGURE 1.
Collaboration of RUNX1 with p53 to suppress cell growth. A, exogenous expression of p53 and RUNX1. p53-deficient H1299 cells were transiently transfected with the indicated combinations of the expression plasmids. Forty eight hours after transfection, cell lysates were prepared and analyzed for the expression levels of p53 and RUNX1 by immunoblotting. The expression level of actin was examined as a loading control. B, colony formation assay. H1299 cells were transfected with the indicated combinations of the expression plasmids. Forty eight hours after transfection, cells were cultured in fresh medium containing G418 (at a final concentration of 800 μg/ml). Two weeks after the selection, drug-resistant colonies were fixed in methanol and stained with Giemsa solution. w/o, without.
RUNX1 Is Induced in Response to ADR
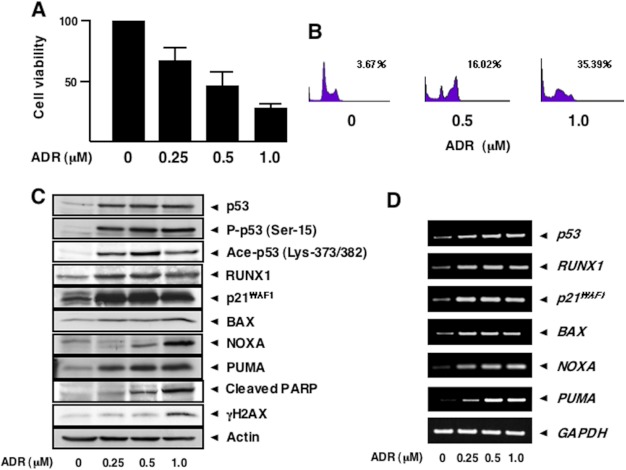
Because it has been well documented that p53 plays a critical role in the regulation of DNA damage response (4, 10, 11), we sought to examine the expression patterns of p53 as well as RUNX1 in response to DNA damage. To this end, p53-proficient human colon carcinoma HCT116 cells were exposed to the indicated concentrations of the anti-cancer drug adriamycin. Twenty four hours after ADR treatment, cell lysates and total RNA were prepared and analyzed by immunoblotting and RT-PCR, respectively. MTT cell survival assay revealed that cell viability is reduced in a dose-dependent manner (Fig. 2A), and FACS analysis showed that the number of cells with sub-G1 DNA content is increased in the presence of ADR (Fig. 2B). As seen in Fig. 2C, a dose-dependent accumulation of γH2AX and also a proteolytic cleavage of poly(ADP-ribose)polymerase were detected, indicating that DNA damage-mediated apoptosis takes place under our experimental conditions. In a good agreement with our recent observations (45), ADR-dependent accumulation of p53 and phosphorylation of p53 at Ser-15 were detectable in association with a remarkable induction of p53 target gene products such as p21WAF1, BAX, NOXA, and PUMA (Fig. 2C), suggesting that HCT116 cells undergo apoptosis in a p53-dependent manner. ADR-mediated phosphorylation of p53 at Ser-20 and Ser-46 was undetectable under our experimental conditions (data not shown). In addition to ADR-dependent phosphorylation of p53 at Ser-15, ADR treatment caused acetylation of p53 at Lys-373/382. Intriguingly, RUNX1 was clearly up-regulated following ADR exposure. Time course experiments demonstrated that ADR-mediated induction of RUNX1 was regulated in a time-dependent fashion (data not shown). RT-PCR analysis indicated that ADR-dependent induction of RUNX1 is regulated at an mRNA level (Fig. 2D). The similar response was also apparent in p53-proficient human osteosarcoma U2OS cells, as a consequence of treating with ADR (data not shown). Thus, it is likely that ADR-mediated induction of RUNX1 is not restricted to HCT116 cells.
FIGURE 2.
RUNX1 is induced in response to ADR. A, MTT cell survival assay. p53-proficient HCT116 cells were exposed to ADR at the indicated concentrations. Twenty four hours after ADR treatment, cells were subjected to MTT cell survival assay. B, FACS analysis. HCT116 cells were treated with ADR at the indicated concentrations. Twenty four hours after ADR exposure, floating and attached cells were collected, fixed in ethanol, stained with propidium iodide, and the number of cells with sub-G1 DNA content was measured by FACS. C and D, ADR-mediated induction of RUNX1. HCT116 cells were exposed to ADR as in A. Twenty four hours after ADR treatment, cell lysates and total RNA were prepared and analyzed by immunoblotting (C) and RT-PCR (D), respectively. The expression levels of actin and GAPDH were examined as a loading and an internal control, respectively. PARP, poly(ADP-ribose) polymerase.
The clear correlation between the expression levels of RUNX1 and p53 in response to ADR prompted us to test whether RUNX1 could be a direct transcriptional target gene of p53. However, forced expression of p53 or siRNA-mediated knockdown of p53 in HCT116 cells had a negligible effect on the expression level of endogenous RUNX1 (data not shown), suggesting that RUNX1 is not a direct transcriptional target gene of p53.
ADR-dependent Nuclear Accumulation of RUNX1
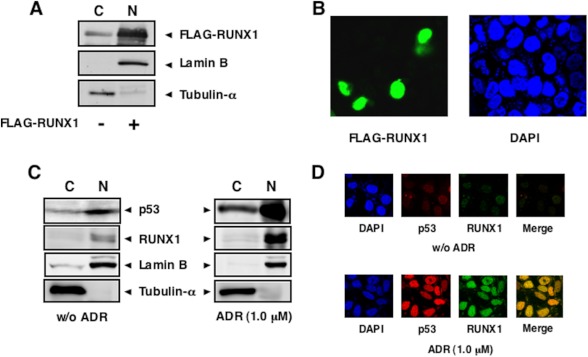
To determine the subcellular distribution of RUNX1, HCT116 cells were transiently transfected with the expression plasmid for FLAG-RUNX1 for 48 h, after which cells were fractionated into cytoplasmic and nuclear fractions, and the fractions obtained were analyzed by immunoblotting with anti-FLAG antibody. The purity of the cytoplasmic and nuclear fractions was verified by immunoblotting with anti-tubulin-α and anti-lamin B antibodies, respectively. As shown in Fig. 3A, FLAG-RUNX1 was largely detectable in nuclear fractions. As expected, confocal microscopy of immunostained HCT116 cells expressing FLAG-RUNX1 revealed that FLAG-RUNX1 is exclusively expressed in the cell nucleus (Fig. 3B).
FIGURE 3.
Nuclear accumulation of RUNX1 upon ADR treatment. A and B, nuclear accumulation of the exogenous RUNX1. HCT116 cells were transiently transfected with FLAG-RUNX1 expression plasmid. Forty eight hours after transfection, cells were fractionated into cytoplasmic (C) and nuclear (N) fractions. These fractions were analyzed by immunoblotting with anti-FLAG antibody. The amounts of tubulin-α and lamin B were also examined as a cytoplasmic nuclear and a nuclear marker, respectively (A). For immunostaining, HCT116 cells were transiently transfected with the expression plasmid encoding FLAG-RUNX1. Forty eight hours after transfection, cells were fixed and incubated with anti-FLAG antibody (green). Cell nuclei were stained with DAPI (blue) (B). C and D, ADR-mediated nuclear accumulation of the endogenous RUNX1. HCT116 cells were treated with 1 μm ADR or left untreated. Twenty four hours after ADR treatment, cells were fractionated as in A, and each fraction was analyzed by immunoblotting with the indicated antibodies (C). For immunostaining, HCT116 cells were treated with 1 μm ADR or left untreated. Twenty four hours after ADR exposure, cells were simultaneously incubated with anti-p53 (red) and anti-RUNX1 (green) antibodies. The merged image (yellow) shows the nuclear co-localization of RUNX1 and p53. Cell nuclei were stained with DAPI (blue) (D). w/o, without.
We then sought to investigate the subcellular localization of the endogenous RUNX1 in response to ADR. HCT116 cells were treated with ADR (1 μm) or left untreated. Twenty four hours after ADR treatment, cells were fractionated into cytoplasmic and nuclear fractions and analyzed by immunoblotting with anti-RUNX1 antibody. As shown in Fig. 3C, like p53, ADR treatment resulted in a strong nuclear accumulation of the endogenous RUNX1. In support of these observations, indirect immunofluorescence staining experiments demonstrated that RUNX1 and p53 are induced to accumulate and co-localize in the cell nucleus following ADR exposure (Fig. 3D).
Complex Formation between RUNX1 and p53
To ask whether RUNX1 could be associated with p53 in cells, HCT116 cells were simultaneously transfected with the expression plasmids for RUNX1 and p53. Forty eight hours after transfection, cell lysates were prepared and immunoprecipitated with normal mouse serum (NMS) or with monoclonal anti-p53 antibody. The immune complexes were analyzed by immunoblotting with polyclonal anti-RUNX1 antibody. As shown in Fig. 4A, the anti-p53 immunoprecipitates contained RUNX1. The reciprocal experiments using NRS or anti-RUNX1 antibody revealed that p53 is co-immunoprecipitated with RUNX1.
FIGURE 4.
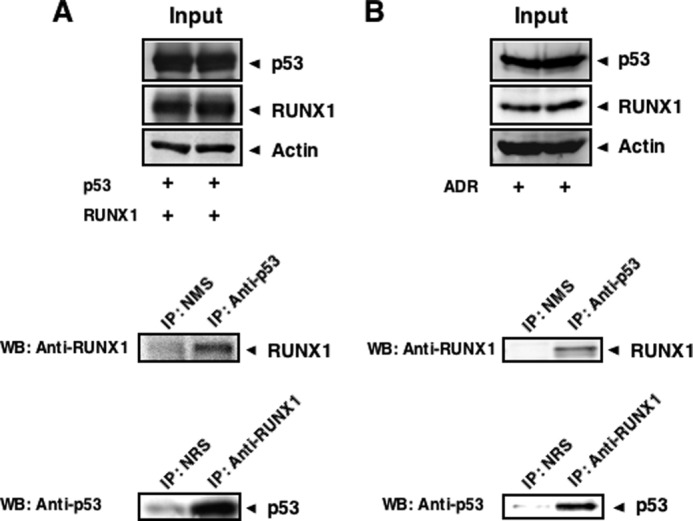
Complex formation between RUNX1 and p53. A, exogenous interaction of RUNX1 with p53. HCT116 cells were transiently transfected with the indicated combinations of the expression plasmids. Forty eight hours after transfection, cell lysates were prepared and immunoprecipitated (IP) with NMS or with anti-p53 antibody. The immunoprecipitates were analyzed by immunoblotting with anti-RUNX1 antibody. The reciprocal experiments using NRS and anti-RUNX1 antibodies are also shown. WB, Western blot. B, endogenous interaction of RUNX1 with p53. HCT116 cells were treated with 0.5 μm ADR. Twenty four hours after ADR treatment, cell lysates were prepared and immunoprecipitated with NMS or with anti-p53 antibody, followed by immunoblotting with anti-RUNX1 antibody. The reciprocal experiments are also shown.
To assess the endogenous interaction between RUNX1 and p53, cell lysates were prepared from ADR-treated HCT116 cells and subjected to the immunoprecipitation experiments using NMS or anti-p53 antibody. As shown in Fig. 4B, the endogenous RUNX1 was detectable in the anti-p53 immunoprecipitates. The reciprocal experiments demonstrated that the anti-RUNX1 immunoprecipitates contain the endogenous p53. We failed to detect the p53-RUNX1 complex in untreated HCT116 (data not shown), which might be due to the quite low base-line expression of the endogenous p53 and RUNX1. Considering that RUNX1 and p53 are exclusively expressed and co-localize in cell nucleus following ADR exposure (Fig. 3), these results strongly suggest that RUNX1 has the ability to interact with p53 in cell nucleus in an ADR-dependent manner.
ADR-mediated Recruitment of RUNX1 and p53 onto p53 Target Promoters
Our present finding that RUNX1 interacts with p53 in ADR-treated cells led us to examine whether RUNX1 together with p53 could be recruited onto p53 target promoters in the presence of ADR. To this end, we performed ChIP assays. HCT116 cells were treated with ADR or left untreated. Twenty four hours after ADR treatment, cells were cross-linked with formaldehyde, and chromatin DNA was immunoprecipitated with the indicated antibodies followed by PCR-based amplification with p21WAF1 or BAX promoter-specific primers flanking their p53-responsive elements. Because p21WAF1 promoter contains two independent p53-responsive elements (distal and proximal sites) (9), we checked both sites of the p21WAF1 promoter. As shown in Fig. 5, A and B, p53 as well as RUNX1 were detected on p21WAF1 and BAX promoters in the absence of ADR. It was worth noting that ADR treatment leads to a remarkable increase in the amounts of p53 and RUNX1 associated with those p53 target promoters. Control experiments demonstrated that NMS and NRS are not able to immunoprecipitate genomic DNA fragments containing the p53-responsive elements, and the amounts of input DNA are similar for all samples. Similar results were also obtained in ADR-treated U2OS cells (data not shown). Thus, it is likely that RUNX1 has a crucial role in the regulation of the transcriptional activity of p53 in response DNA damage.
FIGURE 5.
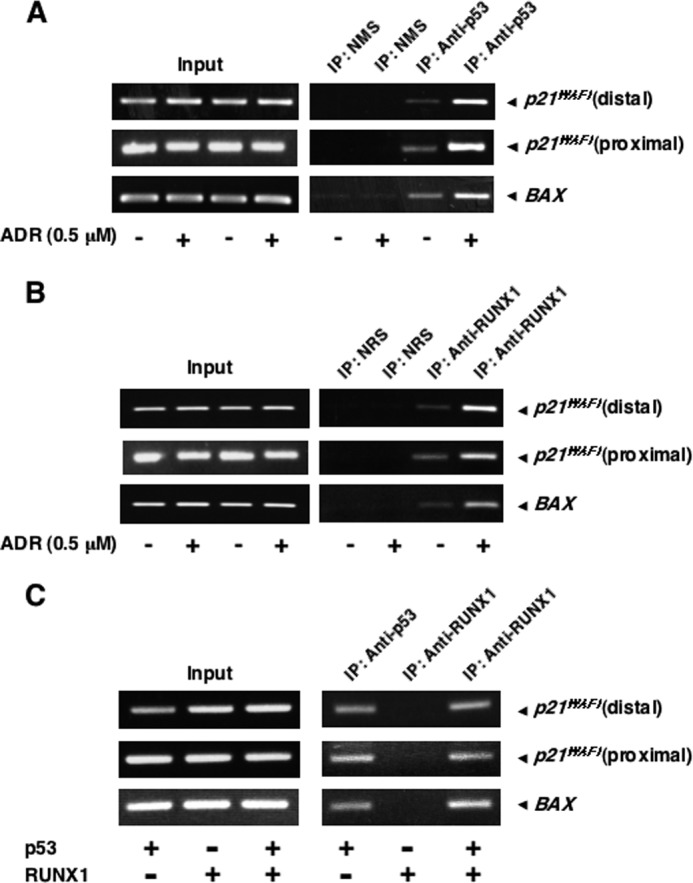
ADR-mediated recruitment of p53 and RUNX1 onto p53 target promoters. A--C, ChIP assay. HCT116 cells were treated with 0.5 μm ADR or were left untreated. Twenty four hours after ADR treatment, cells were fixed in formaldehyde and lysed in SDS-lysis buffer, and chromatin DNA-protein complexes were immunoprecipitated (IP) with anti-p53 (A) or with anti-RUNX1 (B) antibody. The immunoprecipitated DNA was purified and subjected to RCR analysis. Alternatively, p53-deficient H1299 cells were transiently transfected with the indicated combinations of the expression plasmids. Forty eight hours after transfection, cells were fixed in formaldehyde and lysed in SDS-lysis buffer, and chromatin DNA-protein complexes were immunoprecipitated with anti-p53 or with anti-RUNX1 antibody. The immunoprecipitated DNA was purified and analyzed by PCR (C).
Next, we examined whether the recruitment of RUNX1 onto the p53 target promoters could be dependent on p53. For this purpose, p53-deficient H1299 cells were transiently transfected with the indicated combinations of the expression plasmids. Forty eight hours after transfection, cells were cross-linked, and cell lysates were processed for ChIP assay. As shown in Fig. 5C, RUNX1 was associated with p53 target promoters in the presence of p53 but not in the absence of p53, suggesting that the recruitment of RUNX1 onto the p53 target promoters is regulated in a p53-dependent manner.
RUNX1 Elevates the Transcriptional Activity and Acetylation Level of p53 Following ADR Treatment
To gain insight into the functional significance of RUNX1 during p53-dependent DNA damage response, we addressed whether RUNX1 could affect the transcriptional activity of p53 in the presence of ADR. HCT116 cells were transiently transfected with the empty plasmid or with the expression plasmid for RUNX1 followed by ADR treatment. Twenty four hours after ADR treatment, cell lysates and total RNA were prepared and processed for immunoblotting and RT-PCR, respectively. Although RUNX1 had an undetectable effect on phosphorylation level of p53 at Ser-15 in response to ADR, ADR-mediated acetylation level of p53 at Lys-373/382 was increased in cells expressing exogenous RUNX1 (Fig. 6). Consistent with these observations, ADR-mediated transcriptional induction of p53 target genes, including p21WAF1, BAX, NOXA, and PUMA, was markedly enhanced in the presence of exogenous RUNX1, suggesting that RUNX1 has the ability to stimulate the transcriptional activity of p53 through the up-regulation of ADR-mediated acetylation level of p53.
FIGURE 6.
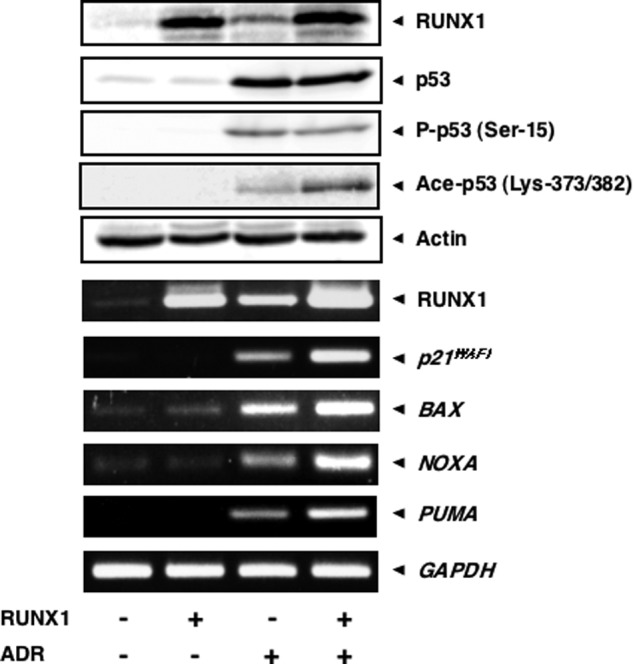
RUNX1 enhances the transcriptional activity of p53 in response to ADR and ADR-mediated acetylation of p53. HCT116 cells were transiently transfected with the empty plasmid or with the expression plasmid for RUNX1 for 24 h, after which cells were treated with ADR (at a final concentration of 0.5 μm) or left untreated. Twenty four hours after the treatment with ADR, cell lysates and total RNA were prepared and subjected to immunoblotting (upper panels) and RT-PCR (lower panels), respectively
Knockdown of RUNX1 Down-regulates ADR-induced Transcriptional Activity and the Acetylation Level of p53
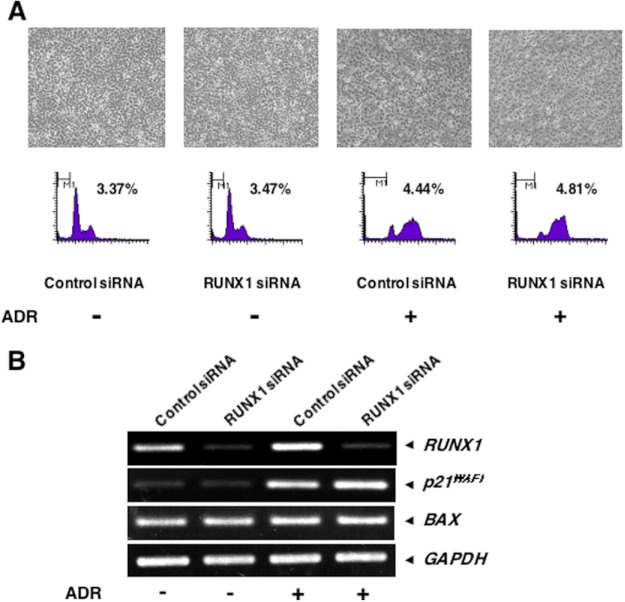
To further confirm the contribution of RUNX1 to p53-mediated transcriptional activation following ADR exposure, we undertook siRNA-mediated knockdown of the endogenous RUNX1. HCT116 cells were transiently transfected with control siRNA or with siRNA targeting RUNX1 for 24 h and incubated in the presence or absence of ADR. Twenty four hours after ADR treatment, cell lysates and total RNA were prepared and subjected to immunoblotting and RT-PCR, respectively. As shown in Fig. 7, knockdown of RUNX1 had a negligible effect on ADR-dependent accumulation of p53, whereas ADR-mediated acetylation of p53 at Lys-373/382 was strongly suppressed in RUNX1-depleted cells. Silencing of RUNX1 also resulted in a reduction of ADR-mediated phosphorylation of p53 at Ser-15, but to a lesser degree. In accordance with these observations, ADR-dependent transcriptional activation of p53 target genes, such as p21WAF1, BAX, NOXA, and PUMA, was significantly attenuated in RUNX1 knockdown cells.
FIGURE 7.
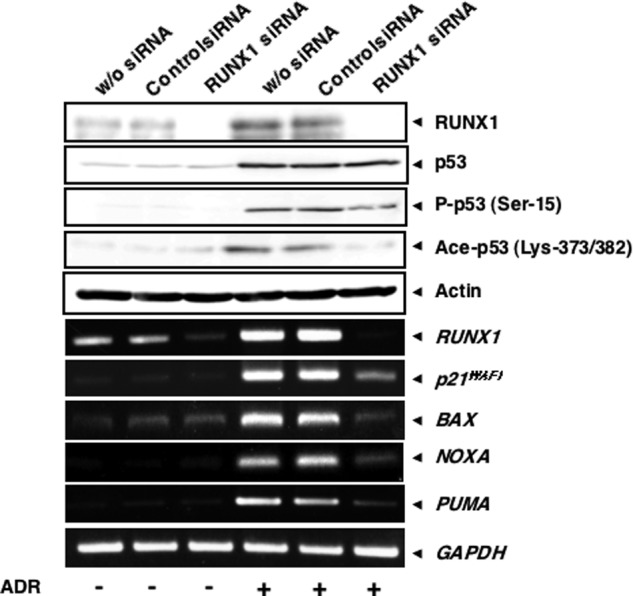
Knocking down RUNX1 suppresses ADR-mediated acetylation and transcriptional activity of p53. HCT116 cells were transiently transfected with control siRNA or with siRNA against RUNX1. Twenty four hours after transfection, cells were incubated in the presence or absence of ADR (at a final concentration of 0.5 μm). Twenty four hours after ADR exposure, cell lysates and total RNA were extracted and processed for immunoblotting (upper panels) and RT-PCR (lower panels), respectively. w/o, without.
In view of the above observations showing that depletion of RUNX1 efficiently inhibits ADR-mediated induction of p53 target genes, we wondered whether knockdown of RUNX1 could suppress ADR-mediated apoptosis. To address this question, we checked the cells by phase-contrast microscopy following RUNX1 siRNA transfection and ADR treatment, and we then performed trypan blue exclusion assay. When HCT116 cells were transiently transfected with control siRNA followed by ADR exposure, a significant decrease in the number of adherent cells and also a remarkable increase in number of trypan blue-positive cells were observed (Fig. 8, A and B). As expected, we found a substantial number of adherent cells following RUNX1 siRNA transfection and ADR treatment. Subsequent trypan blue exclusion assay demonstrated that silencing of RUNX1 causes a detectable decrease in the number of trypan blue-positive cells in response to ADR exposure relative to ADR-treated control cells. These results correlated with the down-regulation of ADR-mediated induction of p53 target genes in RUNX1 knockdown cells, and thus strongly suggest that RUNX1 is required for DNA damage-mediated stimulation of transcriptional as well as pro-apoptotic activity of p53.
FIGURE 8.
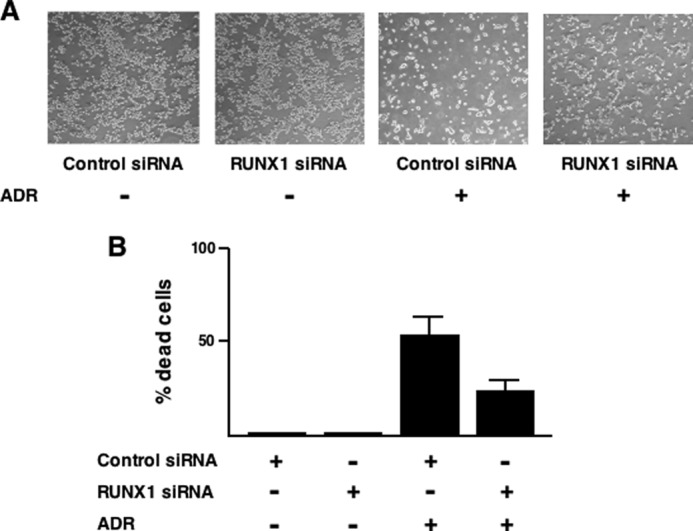
Knocking down RUNX1 suppresses ADR-mediated apoptosis in HCT116 cells. A, phase-contrast micrograph. HCT116 cells were transiently transfected with control siRNA or with siRNA targeting RUNX1 for 24 h, after which cells were exposed to ADR (at a final concentration of 1 μm) or left untreated. Twenty four hours after ADR treatment, cells were examined by phase-contrast microscope. B, trypan blue exclusion assay. RUNX1 knocked down HCT116 cells were exposed to 1 μm ADR or left untreated. Twenty four hours after ADR treatment, floating and attached cells were collected and stained with 0.4% trypan blue. After trypan blue staining, the number of trypan blue-positive cells was measured.
To ask whether the contribution of RUNX1 to ADR-mediated apoptosis could be dependent on p53, we sought to examine a possible effect of RUNX1 on p53-deficient H1299 cells in response to ADR. H1299 cells were transiently transfected with control siRNA or with siRNA targeting RUNX1 for 24 h, and incubated in the presence or absence of ADR. Twenty four hours after ADR treatment, attached and floating cells were collected and analyzed by FACS. As shown in Fig. 9A, a number of cells with sub-G1 DNA content remained unchanged regardless of ADR treatment, and depletion of RUNX1 had an undetectable effect on H1299 cells in the presence or absence of ADR. Consistent with these observations, RT-PCR performed under the same experimental conditions demonstrating that RUNX1 is induced in H1299 cells exposed to ADR, RUNX1 knockdown has a negligible effect on the expression levels of p21WAF1 and BAX in the presence or absence of ADR (Fig. 9B). Collectively, these results indicate that RUNX1 regulates DNA damage-mediated apoptotic response in a p53-dependent manner.
FIGURE 9.
RUNX1 has an undetectable effect on H1299 cells in response to ADR. A, phase-contrast micrograph and FACS analysis. H1299 cells were transiently transfected with control siRNA or with siRNA targeting RUNX1 for 24 h, after which cells were exposed to ADR (at a final concentration of 1 μm) or left untreated. Twenty four hours after ADR treatment, cells were examined by phase-contrast microscope (upper panels) and subjected to FACS analysis (lower panels). B, RT-PCR. H1299 cells were treated as in A. Twenty four hours after ADR treatment, total RNA was extracted and processed for RT-PCR.
RUNX1 Forms a Complex with p300
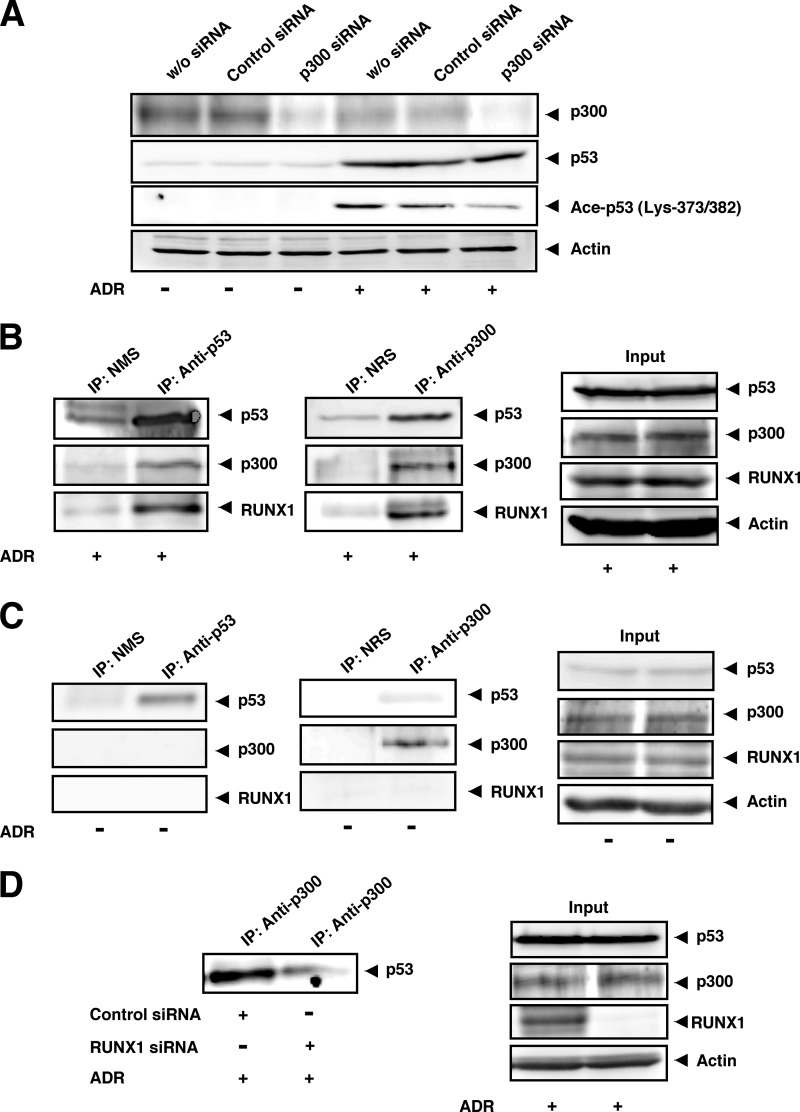
It has been well known that histone acetyltransferase p300 has then ability to acetylate p53 in response to DNA damage (21, 22). Intriguingly, Kitabayashi et al. (46) reported that p300 is associated with RUNX1 in myeloid cells. These observations prompted us to investigate whether p300 could be involved in ADR-mediated p53 acetylation, and RUNX1 could be associated with p300 under our experimental conditions. To this end, HCT116 cells were transiently transfected with control siRNA or with siRNA against p300. Twenty four hours after transfection, cells were treated with ADR for 24 h or left untreated. Cell lysates were then prepared and analyzed by immunoblotting. As shown Fig. 10A, p300 knockdown decreased ADR-mediated p53 acetylation at Lys-373/382, whereas ADR-dependent accumulation of p53 was unaffected in the presence of siRNA targeting p300, suggesting that p300 is at least in part required for ADR-mediated p53 acetylation at Lys-373/382 under our experimental conditions.
FIGURE 10.
RUNX1 as well as p53 interacts with p300 histone acetyltransferase. A, p300 knockdown. HCT116 cells were transiently transfected with control siRNA or with siRNA against p300. Twenty four hours after transfection, cells were treated with ADR (at a final concentration of 0.5 μm) or left untreated. Twenty four hours after ADR exposure, cell lysates were prepared and analyzed by immunoblotting with the indicated antibodies. w/o, without. B and C, interaction between RUNX1 and p300. HCT116 cells were exposed to 0.5 μm ADR (B) or left untreated (C). After 48 h, cell lysates were prepared from ADR-treated and -untreated cells, and immunoprecipitated (IP) with the indicated antibodies followed by immunoblotting with the indicated antibodies. 1/20 of inputs are also shown (right panels). D, complex formation of p53 with p300 is dependent on RUNX1. HCT116 cells were transiently transfected with control siRNA or with siRNA against RUNX1. Twenty four hours after transfection, cells were treated with 0.5 μm ADR. Twenty four hours after ADR treatment, cell lysates were prepared and immunoprecipitated with anti-p300 antibody. The immunoprecipitates were analyzed by immunoblotting with anti-p53 antibody. 1/20 of inputs are also shown (right panels).
For immunoprecipitation, cell lysates prepared from HCT116 cells exposed to ADR for 24 h were immunoprecipitated with NMS or with monoclonal anti-p53 antibody followed by immunoblotting with anti-p53, anti-p300, or with anti-RUNX1 antibody. As clearly shown in Fig. 10B, p300 and RUNX1 were co-precipitated with p53. Similarly, the anti-p300 immunoprecipitates contained p53 and RUNX1. Because p300 was exclusively expressed in the cell nucleus (data not shown), it is likely that RUNX1 interacts with p300 in the cell nucleus. In contrast, we failed to detect the RUNX1-p300 complex in the absence of ADR (Fig. 10C). Furthermore, the additional immunoprecipitation experiments revealed that RUNX1 knockdown causes a significant decrease in the amount of p53-p300 complex in the presence of ADR (Fig. 10D). These results indicate that RUNX1 is required for a complex formation between p53 and p300 in cells exposed to ADR and raise the possibility that RUNX1 might facilitate p300-mediated acetylation of p53 in response to DNA damage, thereby activating p53.
DISCUSSION
In this study, we found for the first time that there exists a physical and functional interaction between RUNX1 and tumor suppressor p53 during DNA damage response. According these results, RUNX1 formed a ternary complex with p53 and histone acetyltransferase p300 and enhanced p300-mediated acetylation of p53 at Lys-373/382 in the presence of ADR. This acetylation status of p53 was positively correlated with its transcriptional and pro-apoptotic activities in response to ADR. Thus, it is likely that RUNX1 has the ability to regulate p53 function by stimulating its p300-mediated acetylation following DNA damage.
Previously, it has been shown that p300 interacts with the NH2-terminal region of p53 and acetylates its COOH-terminal cluster of Lys residues, including Lys-373 and Lys-382 (21, 22, 47). These acetylations dramatically enhanced the sequence-specific transactivation ability of p53, which might be due to the acetylation-induced conformational change of p53. In addition to the transcriptional activation, DNA damage-mediated acetylation has been shown to be essential for the pro-apoptotic activity of p53 (48). In support of these observations, SIRT1-mediated deacetylation of p53 at Lys-382 significantly attenuated DNA damage-induced apoptosis (49). Under our experimental conditions, p300 knockdown led to a remarkable down-regulation of ADR-mediated p53 acetylation at Lys-373/382. Moreover, our co-immunoprecipitation experiments demonstrated that the anti-p300 immunoprecipitates prepared from ADR-treated HCT116 cells contains not only p53 but also RUNX1, and both p300 and RUNX1 are detectable in the anti-p53 immunoprecipitates prepared from HCT116 cells exposed to ADR. Because RUNX1-p53 and RUNX1-p300 complexes were undetectable in the absence of ADR, it is likely that these interactions are dependent on ADR. Thus, these observations strongly indicate that RUNX1 enhances p53 acetylation at Lys-373/382 catalyzed by p300 following ADR treatment.
Recently, Wen et al. (50) reported that orphan nuclear receptor PNR/NR2E3 forms a complex with p53 and p300 and stimulates p53 function by elevating its acetylation level. According to their results, PNR/NR2E3 promoted the intermolecular interaction between p53 and p300 and then increased p300-mediated acetylation of p53 at Lys-373/382. Similarly, Kim et al. (23) found that Wilms tumor suppressor WTX enhances the interaction between p53 and CBP, thereby increasing CBP-mediated acetylation of p53 at Lys-373/382. Under their experimental conditions, siRNA-mediated knockdown of WTX markedly suppressed etoposide-dependent p53 acetylation at Lys-373/382. Both of these observations suggest that PNR/NR2E3 as well as WTX play an important role in the regulation of p300/CBP-mediated p53 acetylation through the complex formation. According to our present results, silencing of RUNX1 efficiently abrogated ADR-induced p53 acetylation at Lys-373/382 mediated by p300, and forced expression of RUNX1 led to the increase in acetylation levels of p53 at Lys-373/382 in the presence of ADR. Although the exact molecular mechanisms behind the contribution of RUNX1 to p300-mediated p53 acetylation following DNA damage are presently unknown, it is possible that, like PNR/NR2E3 and WTX, RUNX1 acts as a molecular bridge or a scaffolding molecule for p53-p300 binding, thereby enabling p300-mediated acetylation of p53 in response to DNA damage. Indeed, RUNX1 knockdown significantly reduced the amount of p53-p300 complex in cells exposed to ADR. Further studies should be required to adequately address this issue.
Another important finding of this study was that the acetylation status of p53 at Lys-373/382 is correlated with ADR-mediated p53 activation but not with p53 accumulation in response to ADR. Knocking down RUNX1 caused a significant suppression of ADR-mediated p53 acetylation at Lys-373/382 in association with a remarkable reduction of ADR-induced transcriptional activity of p53, whereas ADR-dependent accumulation of p53 was still observed in RUNX1-depleted cells. Similarly, ADR-mediated acetylation of p53 at Lys-373/382 as well as activation of p53-dependent transcriptional program were further enhanced by forced expression of RUNX1, whereas ectopic expression of RUNX1 had an undetectable effect on ADR-mediated accumulation of p53. Previously, Yuan et al. (16) reported that p300 is required for DNA damage-dependent accumulation of p53. Intriguingly, Kawai et al. (51) found that p300 contributes to the accumulation of p53 in an acetylase-independent manner. According to their results, the p300 mutant deficient in acetylase activity failed to acetylate p53, whereas the p53 protein level was increased in the presence of p300 mutant. Zhao et al. (52) described that the acetylated p53 at Lys-373/382 is recruited onto the p21WAF1 promoter and enhanced the expression of p21WAF1, indicating that p53 acetylation at Lys-373/382 has an important role in the regulation of its transcriptional activity but not of its accumulation. Together with their observations, our results suggest that DNA damage-mediated accumulation of p53 might be a distinct event from the DNA damage-induced p53 acetylation at Lys-373/382 catalyzed by p300 and activation of p53.
It has been shown that pro-apoptotic Bim (Bcl-2-interacting mediator of cell death) is a key mediator of TGFβ-dependent apoptotic response (53). Subsequently, Wildey and Howe (54) described that RUNX1 is induced in response to TGFβ and cooperates with FoxO3a to stimulate the transcription of Bim in hepatic cells, indicating that RUNX1 is directly involved in the initiation of TGFβ-mediated apoptosis. Previous studies demonstrated that another RUNX family member termed RUNX3 also collaborates with FoxO3a to transcriptionally activate Bim in gastric epithelial cells exposed to TGFβ (55). These observations imply that RUNX1 as well as RUNX3 modulate TGFβ-mediated apoptosis through the direct induction of Bim in a cell type-dependent manner. Although the molecular mechanisms underlying the pro-apoptotic effects of TGFβ appear to be cell type-dependent, it is likely that RUNX family members such as RUNX1 and RUNX3 are the key components during this cellular process. Recently, we have found that RUNX3 participates in p53-dependent DNA damage response (56). Based on our results, RUNX3 was induced to access the cell nucleus following DNA damage and formed a complex with p53 to enhance its transcriptional as well as pro-apoptotic activity. Together with these findings, it is conceivable that, like RUNX3, RUNX1 plays a pivotal role in the regulation of apoptosis in response to a wide variety of cellular pro-apoptotic stimuli such as DNA damage and TGFβ.
Acknowledgment
We thank Dr. Hiroki Nagase for the fruitful discussions and suggestions.
This work was supported in part by a grant-in-aid from the Ministry of Education, Culture, Sports, Science and Technology, Japan, and the Hamaguchi Foundation.
- CBP
- cAMP-response element-binding protein-binding protein
- ADR
- adriamycin
- NMS
- normal mouse serum
- NRS
- normal rabbit serum
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide.
REFERENCES
- 1. Zhou B. B., Elledge S. J. (2000) The DNA damage response. Putting checkpoints in perspective. Nature 408, 433–439 [DOI] [PubMed] [Google Scholar]
- 2. Pietenpol J. A., Tokino T., Thiagalingam S., el-Deiry W. S., Kinzler K. W., Vogelstein B. (1994) Sequence-specific transcriptional activation is essential for growth suppression by p53. Proc. Natl. Acad. Sci. U.S.A. 91, 1998–2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Harris C. C. (1993) p53. At the crossroads of molecular carcinogenesis and risk assessment. Science 262, 1980–1981 [DOI] [PubMed] [Google Scholar]
- 4. Vousden K. H., Lu X. (2002) Live or let die. The cell's response to p53. Nat. Rev. Cancer 2, 594–604 [DOI] [PubMed] [Google Scholar]
- 5. Vogelstein B., Kinzler K. W. (1992) p53 function and dysfunction. Cell 70, 523–526 [DOI] [PubMed] [Google Scholar]
- 6. Velculescu V. E., El-Deiry W. S. (1996) Biological and clinical importance of the p53 tumor suppressor gene. Clin. Chem. 42, 858–868 [PubMed] [Google Scholar]
- 7. Vogelstein B., Lane D., Levine A. J. (2000) Surfing the p53 network. Nature 408, 307–310 [DOI] [PubMed] [Google Scholar]
- 8. Donehower L. A., Harvey M., Slagle B. L., McArthur M. J., Montgomery C. A., Jr., Butel J. S., Bradley A. (1992) Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 356, 215–221 [DOI] [PubMed] [Google Scholar]
- 9. el-Deiry W. S., Kern S. E., Pietenpol J. A. (1992) Definition of a consensus binding site for p53. Nat. Genet. 1, 45–49 [DOI] [PubMed] [Google Scholar]
- 10. Sionov R. V., Haupt Y. (1999) The cellular response to p53. The decision between life and death. Oncogene 18, 6145–6157 [DOI] [PubMed] [Google Scholar]
- 11. Prives C., Hall P. A. (1999) The p53 pathway. J. Pathol. 187, 112–126 [DOI] [PubMed] [Google Scholar]
- 12. Haupt Y., Maya R., Kazaz A., Oren M. (1997) Mdm2 promotes the rapid degradation of p53. Nature 387, 296–299 [DOI] [PubMed] [Google Scholar]
- 13. Kubbutat M. H., Jones S. N., Vousden K. H. (1997) Regulation of p53 stability by Mdm2. Nature 387, 299–303 [DOI] [PubMed] [Google Scholar]
- 14. Honda R., Tanaka H., Yasuda H. (1997) Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 420, 25–27 [DOI] [PubMed] [Google Scholar]
- 15. Piette J., Neel H., Maréchal V. (1997) Mdm2. Keeping p53 under control. Oncogene 15, 1001–1010 [DOI] [PubMed] [Google Scholar]
- 16. Yuan Z. M., Huang Y., Ishiko T., Nakada S., Utsugisawa T., Shioya H., Utsugisawa Y., Yokoyama K., Weichselbaum R., Shi Y., Kufe D. (1999) Role for p300 in stabilization of p53 in the response to DNA damage. J. Biol. Chem. 274, 1883–1886 [DOI] [PubMed] [Google Scholar]
- 17. Oren M. (2003) Decision making by p53. Life, death, and cancer. Cell Death Differ. 10, 431–442 [DOI] [PubMed] [Google Scholar]
- 18. D'Orazi G., Cecchinelli B., Bruno T., Manni I., Higashimoto Y., Saito S., Gostissa M., Coen S., Marchetti A., Del Sal G., Piaggio G., Fanciulli M., Appella E., Soddu S. (2002) Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nat. Cell Biol. 4, 11–19 [DOI] [PubMed] [Google Scholar]
- 19. Yoshida K., Liu H., Miki Y. (2006) Protein kinase Cδ regulates Ser46 phosphorylation of p53 tumor suppressor in the apoptotic response to DNA damage. J. Biol. Chem. 281, 5734–5740 [DOI] [PubMed] [Google Scholar]
- 20. Oda K., Arakawa H., Tanaka T., Matsuda K., Tanikawa C., Mori T., Nishimori H., Tamai K., Tokino T., Nakamura Y., Taya Y. (2000) p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell 102, 849–862 [DOI] [PubMed] [Google Scholar]
- 21. Shikama N., Lee C. W., France S., Delavaine L., Lyon J., Krstic-Demonacos M., La Thangue N. B. (1999) A novel cofactor for p300 that regulates the p53 response. Mol. Cell 4, 365–376 [DOI] [PubMed] [Google Scholar]
- 22. Lacroix M., Toillon R. A., Leclercq G. (2006) p53 and breast cancer, an update. Endocr. Relat. Cancer 13, 293–325 [DOI] [PubMed] [Google Scholar]
- 23. Kim W. J., Rivera M. N., Coffman E. J., Haber D. A. (2012) The WTX tumor suppressor enhances p53 acetylation by CBP/p300. Mol. Cell 45, 587–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ivanov G. S., Ivanova T., Kurash J., Ivanov A., Chuikov S., Gizatullin F., Herrera-Medina E. M., Rauscher F., 3rd, Reinberg D., Barlev N. A. (2007) Methylation-acetylation interplay activates p53 in response to DNA damage. Mol. Cell. Biol. 27, 6756–6769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Samuels-Lev Y., O'Connor D. J., Bergamaschi D., Trigiante G., Hsieh J. K., Zhong S., Campargue I., Naumovski L., Crook T., Lu X. (2001) ASPP proteins specifically stimulate the apoptotic function of p53. Mol. Cell 8, 781–794 [DOI] [PubMed] [Google Scholar]
- 26. Yang H. Y., Wen Y. Y., Chen C. H., Lozano G., Lee M. H. (2003) 14-3-3 sigma positively regulates p53 and suppresses tumor growth. Mol. Cell. Biol. 23, 7096–7107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ito Y. (2008) RUNX genes in development and cancer. Regulation of viral gene expression and the discovery of RUNX family genes. Adv. Cancer Res. 99, 33–76 [DOI] [PubMed] [Google Scholar]
- 28. Miyoshi H., Shimizu K., Kozu T., Maseki N., Kaneko Y., Ohki M. (1991) t(8;21) breakpoints on chromosome 21 in acute myeloid leukemia are clustered within a limited region of a single gene, AML1. Proc. Natl. Acad. Sci. U.S.A. 88, 10431–10434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Golub T. R., Barker G. F., Bohlander S. K., Hiebert S. W., Ward D. C., Bray-Ward P., Morgan E., Raimondi S. C., Rowley J. D., Gilliland D. G. (1995) Fusion of the TEL gene on 12p13 to the AML1 gene on 21q22 in acute lymphoblastic leukemia. Proc. Natl. Acad. Sci. U.S.A. 92, 4917–4921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Silva F. P., Morolli B., Storlazzi C. T., Anelli L., Wessels H., Bezrookove V., Kluin-Nelemans H. C., Giphart-Gassler M. (2003) Identification of RUNX1/AML1 as a classical tumor suppressor gene. Oncogene 22, 538–547 [DOI] [PubMed] [Google Scholar]
- 31. Roumier C., Fenaux P., Lafage M., Imbert M., Eclache V., Preudhomme C. (2003) New mechanisms of AML1 gene alteration in hematological malignancies. Leukemia 17, 9–16 [DOI] [PubMed] [Google Scholar]
- 32. Okuda T., van Deursen J., Hiebert S. W., Grosveld G., Downing J. R. (1996) AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 84, 321–330 [DOI] [PubMed] [Google Scholar]
- 33. Wang Q., Stacy T., Binder M., Marin-Padilla M., Sharpe A. H., Speck N. A. (1996) Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc. Natl. Acad. Sci. U.S.A. 93, 3444–3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Takahashi A., Satake M., Yamaguchi-Iwai Y., Bae S. C., Lu J., Maruyama M., Zhang Y. W., Oka H., Arai N., Arai K., et al. (1995) Positive and negative regulation of granulocyte-macrophage colony-stimulating factor promoter activity by AML1-related transcription factor, PEBP2. Blood 86, 607–616 [PubMed] [Google Scholar]
- 35. Taniuchi I., Osato M., Egawa T., Sunshine M. J., Bae S. C., Komori T., Ito Y., Littman D. R. (2002) Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell 111, 621–633 [DOI] [PubMed] [Google Scholar]
- 36. Lutterbach B., Westendorf J. J., Linggi B., Isaac S., Seto E., Hiebert S. W. (2000) A mechanism of repression by acute myeloid leukemia-1, the target of multiple chromosomal translocations in acute leukemia. J. Biol. Chem. 275, 651–656 [DOI] [PubMed] [Google Scholar]
- 37. Guo H., Friedman A. D. (2011) Phosphorylation of RUNX1 by cyclin-dependent kinase reduces direct interaction with HDAC1 and HDAC3. J. Biol. Chem. 286, 208–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yamaguchi Y., Kurokawa M., Imai Y., Izutsu K., Asai T., Ichikawa M., Yamamoto G., Nitta E., Yamagata T., Sasaki K., Mitani K., Ogawa S., Chiba S., Hirai H. (2004) AML1 is functionally regulated through p300-mediated acetylation on specific lysine residues. J. Biol. Chem. 279, 15630–15638 [DOI] [PubMed] [Google Scholar]
- 39. Zhao X., Jankovic V., Gural A., Huang G., Pardanani A., Menendez S., Zhang J., Dunne R., Xiao A., Erdjument-Bromage H., Allis C. D., Tempst P., Nimer S. D. (2008) Methylation of RUNX1 by PRMT1 abrogates SIN3A binding and potentiates its transcriptional activity. Genes Dev. 22, 640–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Linggi B., Müller-Tidow C., van de Locht L., Hu M., Nip J., Serve H., Berdel W. E., van der Reijden B., Quelle D. E., Rowley J. D., Cleveland J., Jansen J. H., Pandolfi P. P., Hiebert S. W. (2002) The t(8;21) fusion protein, AML1 ETO, specifically represses the transcription of the p14(ARF) tumor suppressor in acute myeloid leukemia. Nat. Med. 8, 743–750 [DOI] [PubMed] [Google Scholar]
- 41. Wotton S. F., Blyth K., Kilbey A., Jenkins A., Terry A., Bernardin-Fried F., Friedman A. D., Baxter E. W., Neil J. C., Cameron E. R. (2004) RUNX1 transformation of primary embryonic fibroblasts is revealed in the absence of p53. Oncogene 23, 5476–5486 [DOI] [PubMed] [Google Scholar]
- 42. Li X. L., Arai Y., Harada H., Shima Y., Yoshida H., Rokudai S., Aikawa Y., Kimura A., Kitabayashi I. (2007) Mutations of the HIPK2 gene in acute myeloid leukemia and myelodysplastic syndrome impair AML1- and p53-mediated transcription. Oncogene 26, 7231–7239 [DOI] [PubMed] [Google Scholar]
- 43. Hanamoto T., Ozaki T., Furuya K., Hosoda M., Hayashi S., Nakanishi M., Yamamoto H., Kikuchi H., Todo S., Nakagawara A. (2005) Identification of protein kinase A catalytic subunit beta as a novel binding partner of p73 and regulation of p73 function. J. Biol. Chem. 280, 16665–16675 [DOI] [PubMed] [Google Scholar]
- 44. Li Y., Ozaki T., Kikuchi H., Yamamoto H., Ohira M., Nakagawara A. (2008) A novel HECT-type E3 ubiquitin protein ligase NEDL1 enhances the p53-mediated apoptotic cell death in its catalytic activity-independent manner. Oncogene 27, 3700–3709 [DOI] [PubMed] [Google Scholar]
- 45. Yoshihara Y., Wu D., Kubo N., Sang M., Nakagawara A., Ozaki T. (2012) Inhibitory role of E2F-1 in the regulation of tumor suppressor p53 during DNA damage response. Biochem. Biophys. Res. Commun. 421, 57–63 [DOI] [PubMed] [Google Scholar]
- 46. Kitabayashi I., Yokoyama A., Shimizu K., Ohki M. (1998) Interaction and functional cooperation of the leukemia-associated factors AML1 and p300 in myeloid cell differentiation. EMBO J. 17, 2994–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sakaguchi K., Herrera J. E., Saito S., Miki T., Bustin M., Vassilev A., Anderson C. W., Appella E. (1998) DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 12, 2831–2841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tang Y., Zhao W., Chen Y., Zhao Y., Gu W. (2008) Acetylation is indispensable for p53 activation. Cell 133, 612–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Michishita E., Park J. Y., Burneskis J. M., Barrett J. C., Horikawa I. (2005) Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol. Biol. Cell 16, 4623–4635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wen Z., Pyeon D., Wang Y., Lambert P., Xu W., Ahlquist P. (2012) Orphan nuclear receptor PNR/NR2E3 stimulates p53 functions by enhancing p53 acetylation. Mol. Cell. Biol. 32, 26–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kawai H., Nie L., Wiederschain D., Yuan Z. M. (2001) Dual role of p300 in the regulation of p53 stability. J. Biol. Chem. 276, 45928–45932 [DOI] [PubMed] [Google Scholar]
- 52. Zhao Y., Lu S., Wu L., Chai G., Wang H., Chen Y., Sun J., Yu Y., Zhou W., Zheng Q., Wu M., Otterson G. A., Zhu W. G. (2006) Acetylation of p53 at lysine 373/382 by the histone deacetylase inhibitor depsipeptide induces expression of p21(Waf1/Cip1). Mol. Cell. Biol. 26, 2782–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wildey G. M., Patil S., Howe P. H. (2003) Smad3 potentiates transforming growth factor β (TGFβ)-induced apoptosis and expression of the BH3-only protein Bim in WEHI 231 B lymphocytes. J. Biol. Chem. 278, 18069–18077 [DOI] [PubMed] [Google Scholar]
- 54. Wildey G. M., Howe P. H. (2009) Runx1 is a co-activator with FOXO3 to mediate transforming growth factor β (TGFβ)-induced Bim transcription in hepatic cells. J. Biol. Chem. 284, 20227–20239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yano T., Ito K., Fukamachi H., Chi X. Z., Wee H. J., Inoue K., Ida H., Bouillet P., Strasser A., Bae S. C., Ito Y. (2006) The RUNX3 tumor suppressor up-regulates Bim in gastric epithelial cells undergoing transforming growth factor β-induced apoptosis. Mol. Cell. Biol. 26, 4474–4488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yamada C., Ozaki T., Ando K., Suenaga Y., Inoue K., Ito Y., Okoshi R., Kageyama H., Kimura H., Miyazaki M., Nakagawara A. (2010) RUNX3 modulates DNA damage-mediated phosphorylation of tumor suppressor p53 at Ser-15 and acts as a co-activator for p53. J. Biol. Chem. 285, 16693–16703 [DOI] [PMC free article] [PubMed] [Google Scholar]