Background: Protein biosynthesis and secretion are essential biological processes for therapeutic protein production.
Results: Generating functional anti-IL17A and anti-IL22 peptide-antibody bispecific therapeutic proteins requires pyroglutamate addition and O-linked glycan removal.
Conclusion: Post-translational modifications play critical roles in determining structure and function of therapeutic proteins.
Significance: The peptide-antibody genetic fusion is promising for targeting multiple antigens in a single antibody-like molecule.
Keywords: Biosynthesis, Glycosylation, Peptides, Post-translational Modification, Protein Engineering, Protein Folding, O-Linked Glycan, Antibody, Bispecific, Pyroglutamate
Abstract
Protein biosynthesis and extracellular secretion are essential biological processes for therapeutic protein production in mammalian cells, which offer the capacity for correct folding and proper post-translational modifications. In this study, we have generated bispecific therapeutic fusion proteins in mammalian cells by combining a peptide and an antibody into a single open reading frame. A neutralizing peptide directed against interleukin-17A (IL17A) was genetically fused to the N termini of an anti-IL22 antibody, through either the light chain, the heavy chain, or both chains. Although the resulting fusion proteins bound and inhibited IL22 with the same affinity and potency as the unmodified anti-IL22 antibody, the peptide modality in the fusion scaffold was not active in the cell-based assay due to the N-terminal degradation. When a glutamine residue was introduced at the N terminus, which can be cyclized to form pyroglutamate in mammalian cells, the IL17A neutralization activity of the fusion protein was restored. Interestingly, the mass spectroscopic analysis of the purified fusion protein revealed an unexpected O-linked glycosylation modification at threonine 5 of the anti-IL17A peptide. The subsequent removal of this post-translational modification by site-directed mutagenesis drastically enhanced the IL17A binding affinity and neutralization potency for the resulting fusion protein. These results provide direct experimental evidence that post-translational modifications during protein biosynthesis along secretory pathways play critical roles in determining the structure and function of therapeutic proteins produced by mammalian cells. The newly engineered peptide-antibody genetic fusion is promising for therapeutically targeting multiple antigens in a single antibody-like molecule.
Introduction
Ever since human tissue plasminogen activator was first approved by the Food and Drug Administration in 1986, cultivated mammalian cells have become important host cells for the production of recombinant proteins for clinical applications, due to their relevant post-translational modifications and stringent protein quality control mechanisms for correct protein folding and assembly (1, 2). Protein synthesis and extracellular secretion of therapeutic proteins in mammalian cells are complicated biological processes. Physiological and pharmacological properties of therapeutic proteins produced therein can vary significantly due to the impacts on cellular biosynthetic machineries by cell culture conditions and cell lineages. A better understanding of these fundamental biological processes and their relationships with the primary sequences of the proteins can potentially improve the quality, efficacy, and safety of the next generation of therapeutic products through protein engineering and process engineering.
Recombinant therapeutic proteins generally include therapeutic antibodies and Fc-like fusions, therapeutic enzymes, and small protein therapeutics (3–5), whereas therapeutic peptides comprise another important class of biotherapeutic drugs. Chemically synthesized peptide therapeutics have been successfully marketed for many years (6, 7). They include highly potent, naturally occurring peptide hormones, hormone analogues, and fragments of larger proteins. Recent examples of these therapeutic peptides include a 36-amino acid enfuvirtide for HIV treatment (7), a hirudin-based thrombin inhibitor bivalirudin (8), and exenatide (9), a 39-amino acid synthetic peptide of exendin-4 found in the saliva of Gila monsters. However, very few peptide-based drugs are derived from recombinant display technologies (10), although they are attracting increasing attention in recent years. The only known examples on the market are ecallantide, a recombinant protein inhibitor of plasma kallikrein for the treatment of acute attacks of hereditary angioedema (11), romiplostim, a recombinant thrombopoietin peptide mimetic Fc fusion for chronic idiopathic thrombocytopenic purpura (12), and peginesatide, a recombinant erythropoientin-receptor peptide agonist for chronic kidney disease (13, 14).
One of the major obstacles for peptide therapeutics development is short serum half-life in vivo due to a rapid clearance from circulation and little resistance to serum and tissue proteases. There are a number of recent technology developments addressing these issues. Chemical attachment of a polyethylene glycol moiety is commonly used for half-life extension (15), but intrinsic heterogeneity, renal toxicity (16), and induction of anti-PEG antibodies (17) are potential drawbacks. Peptibodies (12) or mimetibodies (18) consist of a peptide fusion with an immunoglobulin Fc domain for peptide half-life extension. Albumin-peptide fusion proteins (19) take advantage of the long half-life of human albumin. Most recently, a randomly engineered de novo polypeptide XTEN (20) is reported to extend half-life of the exenatide. The CovX body, consisting of synthetic peptides chemically conjugated to a humanized catalytic aldolase antibody (21), has also been found, effectively extending the half-life of bispecific peptide heterodimers (22). Although these advanced modalities attempt to solve the issue of poor pharmacokinetics of peptides, they do not utilize existing therapeutic antibodies, and some have manufacturing complexity, requiring an additional in vitro production step.
We have developed an approach to combine one or more peptides with an antibody using genetic engineering to make use of the well established mammalian production system for recombinant proteins. This bispecific peptide-antibody fusion protein should extend the peptide half-life while targeting two unique and potentially synergistic therapeutic targets. CVX-5484 is a 15-amino acid peptide optimized from a lead peptide isolated by phage display,3 which can potently neutralize human homodimeric cytokine IL17A (24). Because IL17A and another CD4+ T helper 17 cell lineage (25, 26) cytokine, IL22, both exhibit potent proinflammatory properties in animal models of autoimmunity (27, 28) and may have complementary and synergistic effects in the etiology of rheumatoid arthritis and psoriasis, we have decided to investigate the genetic fusion of this neutralizing IL17A peptide to a previously developed anti-IL22 human antibody (IL22-104) (28). The data from the study reveal that the peptide modality in the resulting fusion proteins is much more susceptible to loss of function than antibody modality when expressed in human embryonic kidney 293 cells (HEK293). For generating a bispecific fusion molecule fully functional with both in vitro binding and cell-based activities, N-terminal pyroglutamate addition and removal of an unexpected O-linked glycan are required.
EXPERIMENTAL PROCEDURES
Expression Constructs
A PCR fragment encoding a murine IgG1 leader sequence, anti-IL17A CVX-5484, six-glycine linker, and antibody IL22-104 mature light chain was cloned into a murine cytomegalovirus promoter containing vector pSMED2 (pWZ1237, G6-LC). A similar PCR fragment encoding a murine IgG1 leader sequence, CVX-5484, GSE21 linker (GSGEGEGSEGGGEGEGSEGSG), and antibody IL22-104 mature light chain, was cloned into pSMED2 vector (pWZ1236, GSE21-LC). G6-HC (pWZ1238) encodes a murine IgG1 leader sequence, CVX-5484, a G6 linker, and antibody IL22-104 mature heavy chain in pSMED2. GSE21-HC (pWZ1239) encodes a murine IgG1 leader sequence, CVX-5484, GSE21 linker, and antibody IL22-104 mature heavy chain in pSMED2. Q-G6-HC (pWZ1246) and Q-GSE21-HC (pWZ1247) inserted a glutamine codon 5′ to the CVX-5484 coding fragment in pWZ1238 and pWZ1239, respectively. Q-GSE12-HC (pWZ1248) replaced the G6 linker in Q-G6-HC with GSE 12 (GSGEGEGSEGSG). Q-GSE21-LC (pWZ1256) inserted a glutamine codon 5′ to CVX-5484 coding fragment in GSE21-LC. Q-G-HC (pWZ1257) replaced the G6 linker in Q-G6-HC with glycine. Q-HC (pWZ1258) removed glycine from Q-G-HC. Q-G6-LC (pWZ1260) inserted a glutamine codon 5′ to the CVX-5484 coding fragment in G6-LC. Q-G-LC (pWZ1263) replaced the G6 linker in Q-G6-LC with glycine. Q-GSE21-LC-T5A (pWZ1269) and Q-GSE21-HC-T5A (pWZ1274) contained a TCC → GCC change in Q-GSE21-LC and Q-GSE21-HC, respectively. All constructs were confirmed by DNA sequencing.
Cell Culture and Transfections
HEK293F cells (American Type Culture Collection (ATCC), Manassas, VA) were cultured in freestyle 293 medium (Invitrogen). Cells were grown and maintained in a humidified incubator with 7% CO2 at 37 °C. Conditioned medium for the peptide-antibody constructs were produced from a large scale transient HEK293 transfection process. Essentially, for the culture volume of 10 liters, 5 mg of heavy chain and light chain plasmid DNAs were mixed with 20 mg of polyethyleneimine (25 kDa, linear, neutralized to pH 7.0 by HCl, 1 mg/ml, Polysciences (Warrington, PA)) in 500 ml of Opti-MEM medium (Invitrogen). Then the mixtures were mixed with 10 liters of HEK293F cells preseeded in a wave bioreactor at a cell density of 1.25 × 106 cells/ml. 0.5% tryptone N1 (Organotechnie, Quebec, Canada) was added 24 h post-transfection. The wave bioreactors were incubated at 37 °C with a rocking rate of 20 rpm for 120 h before harvest. The conditioned media were filtered at 0.22 μm prior to purification.
Protein Purification
The conditioned medium was batch-bound to 4 ml of rmpProtein A resin (GE Healthcare) equilibrated with 137 mm NaCl, 2.7 mm KCl, 8.1 mm Na2HPO4, 2.7 mm KH2PO4, pH 7.2 (PBS), at 4 °C overnight. The column was washed with 10 column volumes of PBS and then with 10 column volumes of PBS + 0.02% Tween 20. The column was then washed with an additional 30 column volumes of PBS before a step elution with 100 mm citric acid, 150 mm NaCl, pH 3. Peak fractions were neutralized to pH 7.0 with 2 m Tris, pH 9.0, pooled, buffer-exchanged into PBS, and then concentrated to 1.5 mg/ml using a 50,000 molecular weight cut-off centrifugal device.
Biacore
Kinetic analysis was performed using a Biacore 2000 or 3000 (GE Healthcare). All experiments were performed at 25 °C, and all reagents were coupled using N-hydroxysuccinimide and N-ethyl-N′-(3-dimethylaminopropyl) carbodiimide to activate the surface, and the surface was subsequently blocked following immobilization with ethanolamine. For IL22 kinetics experiments, the antibodies/fusions were directly immobilized. Antibody/fusion immobilization levels ranged from 75 to 1,700 RU. Kinetic measurements were performed at 30 μl/min. Various concentrations of IL22, ranging from 15.6 to 1 μm, or buffer were prepared in PBS-NET buffer (8.1 mm Na2HPO4, 1.47 mm KH2PO4, pH 7.2, 287 mm NaCl, 2.7 mm KCl, 3.2 mm EDTA, 0.005% Tween 20). All surfaces were regenerated using one 30-s pulse of 10 mm glycine HCl at pH 1.7, followed by PBS-NET. Two independent experiments were acquired, with injections acquired in duplicate, on two different surfaces of immobilized antibody or fusion.
For measurement of peptide kinetics to IL17A, IL17A was directly immobilized. Kinetic measurements were performed at 50 μl/min. Various concentrations of each peptide, ranging from 7.8 to 500 nm, or buffer were prepared in PBS-NET buffer. All surfaces were regenerated at 50 μl/min using three 30-s pulses of a solution of 33 mm NaPO4, 333 mm NaCl, 6.7% DMSO, 6.7% formamide, 6.7% ethanol, 6.7% acetonitrile, and 6.7% 1-butanol, followed by PBS-NET. Three independent experiments were acquired, with injections acquired in duplicate, each over three surfaces of immobilized IL17A.
For IL17A binding kinetics to the genetic fusions, anti-human IgG antibody (GE Healthcare) was coupled to a research grade CM5 sensor chip by amine coupling as instructed by the manufacturer at surface densities between 4,000 and 7,500 RU. Genetic fusions were captured to the anti-human IgG surface at surface densities between 60 and 560 RU. Kinetic measurements were performed at 50 μl/min. Various concentrations of the human IL17A, ranging from 3.9 nm to 4 μm, or buffer were prepared in PBS-NET. All surfaces were regenerated at 50 μl/min using three 30-s pulses of 3 m MgCl2, followed by 2 mm glycine HCl at pH 1.7, followed by PBS-NET. Two independent experiments were acquired, with injections acquired in duplicate or in triplicate, each over three surfaces of captured genetic fusion.
For all experiments, data were double referenced using Scrubber version 2.0c software (BioLogic Software) (29). The transformed data were fit to a 1:1 binding model in BiaEvaluation version 4.1.1 (GE Healthcare).
BJ Cell Assay
Human foreskin fibroblast cells and BJ cells (American Type Culture Collection) were maintained in DMEM, 10% FCS, 2 mm glutamine, 1 mm sodium pyruvate, 0.1 mm MEM nonessential amino acids, 100 units/ml penicillin, 100 μg/ml streptomycin. BJ cells were seeded at 5 × 103 cells/well into 96-well flat bottom microtiter plates in which IL17A had been prediluted in culture medium at a final concentration between 1 and 5 ng/ml. In treatments where IL17A inhibitors were used, genetic fusions were added to the wells and serially diluted 3× in a 150-μl total volume. Cells were incubated at 37 °C for 16–24 h, and supernatants were collected and analyzed for growth-related oncogene-α (GRO-α) production by ELISA.
HT29 Assay
GRO-α assays were performed to assess the antibody's ability to block the IL22-induced GRO-α secretion from HT29 cells. HT29 cells where seeded in a 96-well flat bottom tissue culture plate (catalog no. 3595, Corning Inc.) in DMEM medium (DMEM, 10% FBS, 100 units/ml penicillin and streptomycin, 2 mm glutamine) at 5 × 104/well. 10 ng/ml IL22 was mixed with serially diluted antibody in DMEM medium and incubated for 30 min at 37 °C. 24 h after seeding, medium was removed from HT29 cells, and premixed IL22 and antibody were added to the cells in a 96-well plate. After 48 h of incubation at 37 °C with 5% CO2, medium was collected, and secreted GRO-α was tested using a human GRO-α immunoassay kit (catalog no. DGR00, R&D Systems), according to the manufacturer's directions.
ELISA
The ELISA was run according to the manufacturer's protocol (catalog no. DY275E, R&D Systems). Plates were coated with anti-human GRO-α (catalog no. MAB275, R&D Systems) at 4 μg/ml. A standard curve was run from a 4 ng/ml recombinant human GRO-α standard (catalog no. 275-GR, R&D Systems) diluted serially 2×. Samples were added in supernatant from assay plate, diluted 2× into PBS plus 1% BSA, pH 7.2. Bound GRO-α was detected with anti-human GRO-α biotin (catalog no. BAF275, R&D Systems) at 40 ng/ml, followed by streptavidin-HRP (catalog no. DY998, R&D Systems), followed by a 1:1 mixture of H2O2 and tetramethylbenzidine substrate (catalog no. DY999, R&D Systems), followed by 2 n H2SO4. An end point measurement was taken using the SpectraMax instrument at 450–550 nm. GRO-α levels were calculated from ELISA absorbance units by fitting the binding curve for the GRO-α internal standard to a four-parameter fit using the XLfit Excel Add-In version 4.3.2 build 11 and MathIQ version 2.2.2 build 152. IC50 values were determined by fitting sample data to a four-parameter dose-response curve. Data shown are for a single representative experiment. Reported values are averages ± S.D. from two independent experiments, one of which was run in duplicate.
Deglycosylation of N-Linked Glycans and Disulfide Bond Reduction
Peptide-antibody fusion protein samples were deglycosylated by peptide:N-glycosidase F (New England BioLabs, Ipswich, MA) and further reduced by TCEP (Thermo Fisher). The samples were acidified by diluting 1:1 with 0.1% formic acid (Sigma-Aldrich) followed by liquid chromatography mass spectrometry analysis.
β-Elimination and Michael Addition of O-Linked Glycan
Peptide-antibody fusion protein samples were reduced with DTT (20 mm) and alkylated with iodoacetamide (50 mm) and separated on a SDS-polyacrylamide gel. The band corresponding to the fusion protein containing the anti-IL17A peptide was excised and incubated with 70% ethylamine at 50 °C for 18 h. The band was then digested with trypsin and the resulting peptides were analyzed by liquid chromatography mass spectrometry/mass spectrometry analysis.
Liquid Chromatography Mass Spectrometry
Liquid chromatography mass spectrometry (LC-MS) analysis was performed using a Waters Xevo Q-TOF G2 mass spectrometer (Waters, Milford, MA) coupled to an Agilent (Santa Clara, CA) 1100 capillary HPLC. The deglycosylated and reduced samples were separated over an Agilent Poroshell 300SB-C8 (0.5 × 75-mm) column maintained at 80 °C with a flow rate of 20 μl/min. Mobile phase A was water with 2% acetonitrile and 0.1% formic acid, and mobile phase B was acetonitrile with 2% water and 0.1% formic acid. Proteins are eluted from the column using a gradient: 2–20% B in 0.5 min, 20–40% B in 6 min, and 40–100% B in 4 min. The mass spectrometer was run in positive MS only mode, scanning from 800 to 2,000 m/z, and data were acquired with MassLynx (Waters) version 4.1 software. The TOF-MS signals corresponding to the light and heavy chains were summarized and deconvoluted using the MaxEnt1 (Waters) program.
For LC-MS/MS analysis, recovered peptides were loaded onto a Pico-frit column (New Objective, Woburn, MA) packed with reversed-phase Magic (Michrom, Auburn, CA) C18 material (5 μm, 200 Å, 75 μm × 10 cm) coupled to an LTQ-Orbitrap XL mass spectrometer (ThermoElectron, Waltham, MA). Peptides were separated at a flow rate of 0.2 μl/min using a 90-min linear gradient ranging from 2 to 40% B (mobile phase A: 0.1% formic acid, 2% acetonitrile; mobile phase B: 90% acetonitrile, 0.1% formic acid). The instrumental method consisted of a full MS scan (scan range 375–2,000 m/z, with 30,000 full width at half-maximum resolution at m/z 400, target value 2 × 106, maximum ion injection time of 500 ms) followed by data-dependent collision-induced dissociation scans of the four most intense precursor ions. Peptide precursor ions were selected with an isolation window of 2.5 Da and a target value of 1 × 105. The normalized collision energy was set to 35% for collision-induced dissociation and 40% for higher energy collisional dissociation, respectively. The mass spectra were searched against a public antibody database, with the sequences of our constructs added, using the Proteome Discoverer version 1.2 search algorithm (ThermoElectron). The mass accuracy was set to 5 ppm for precursor ions and to 0.5 Da tolerances for fragment ions. The search parameters took into account two missed cleavages for trypsin, static modification of carboxamidomethylation at cysteine (+57.0215 Da), and dynamic modification for methionine oxidation (Met +15.9949 Da), and the addition of ethylamine was also considered on serine and threonine (+27.0473 Da).
RESULTS
Generating Peptide-Antibody Genetic Fusions between Anti-IL17A Peptide and Anti-IL22 Antibody
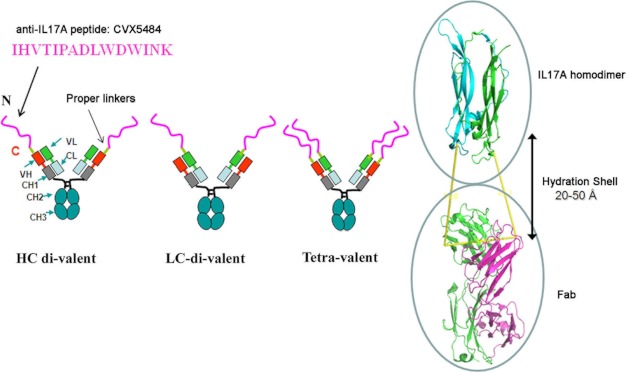
As shown in Fig. 1, the 15-amino acid peptide CVX5484 was fused genetically to IL22-104 antibody. Because the peptide requires a free N terminus for its anti-IL17A neutralization activity, it was only fused to N-terminal ends of the antibody polypeptides.
FIGURE 1.
Diagram of peptide-antibody genetic fusions between anti-IL17A peptide and anti-IL22 antibody.
As described under “Experimental Procedures,” the DNA sequence encoding the peptide was inserted into the 5′ DNA sequence encoding either the heavy chain (HC-divalent), the light chain (LC-divalent), or both chains (tetravalent) of the IL22-104 antibody downstream of the signal sequence. According to structural modeling (Fig. 1), the hydration shell (30) between the IL17A structure (31) and a Fab molecule of an antibody is about 20–50 Å. We therefore added three different linker lengths (6 glycines (G6) and glycine-serine-glutamate repeats (GSE12 and GSE21)) between the peptide and antibody in order to minimize any loss of binding for either the peptide to IL17A or the antibody to IL22. The resulting DNA constructs were expressed in HEK293 cells, and the proteins were purified from the conditioned media, as described under “Experimental Procedures.”
The Peptide-Antibody Genetic Fusion Neutralized IL22 Activity Equivalent to Parental Anti-IL22 Antibody While Lacking Anti-IL17A Neutralizing Activity
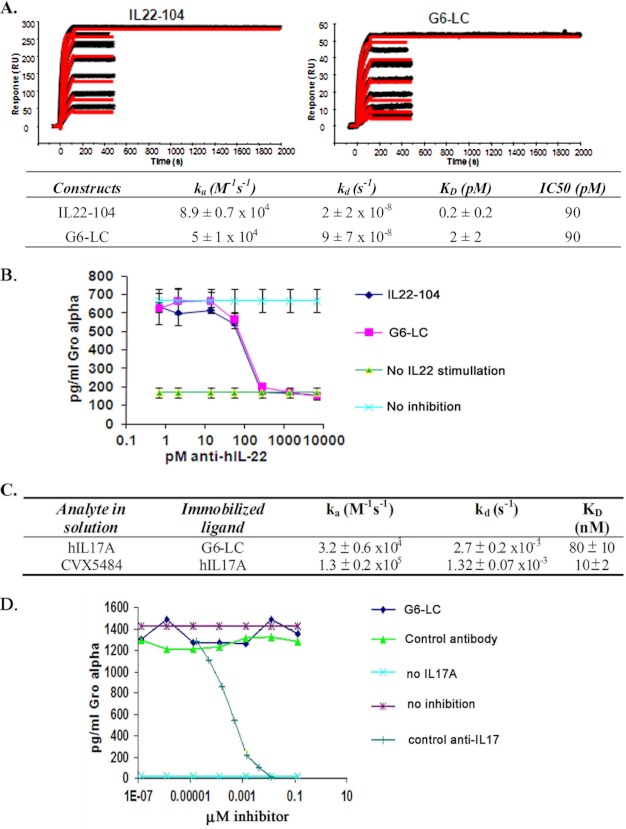
We first determined if the peptide-antibody fusion retained any anti-IL22 binding activity. A kinetic analysis with parental IL22-104 antibody and peptide-antibody fusions was performed using a Biacore 2000 by directly immobilizing the antibody and fusions on the sensor chips, as described under “Experimental Procedures.” The peptide-antibody fusion proteins (G6-LC and others) retained full binding to IL22 (2 ± 2 pm) compared with the parental IL22 antibody, IL22-104 (0.2 ± 0.2 pm) (Fig. 2A).
FIGURE 2.
The peptide-antibody genetic fusion neutralizes the IL22 activity equivalent to parental anti-IL22 antibody while lacking anti-IL17A neutralizing activity. A, kinetic analysis with human IL22 using a Biacore 2000. B, IL22-stimulated release of GRO-α production in HT29 cells. C, table summary of kinetic analysis with human IL17A using a Biacore 2000. D, IL17A-stimulated release of GRO-α in BJ human foreskin fibroblast cells. Error bars, S.D.
To characterize the cell-based neutralizing activity of the peptide-antibody fusions, IL22-stimulated release of GRO-α production (32–34) in HT29 cells was measured as described under “Experimental Procedures.” The peptide-antibody fusion G6-LC showed no loss of neutralization activity for IL22-stimulated HT29 cells with an IC50 of 90 pm (Fig. 2B). These data together demonstrate that the presence of an anti-IL17A peptide at the N terminus of the IL22-104 antibody does not affect its binding and neutralizing activities.
Next we assessed the IL17A binding ability of the peptide-antibody fusions. Kinetic Biacore analysis with an anti-human Fc-coated sensor chip immobilizing the peptide-antibody G6-LC was performed as described under “Experimental Procedures” (Fig. 2C). Various concentrations of human IL17A (125–1,000 nm) were injected over the sensor chip surface. The peptide-antibody has a KD of 80 nm. When measuring the CVX5484 peptide kinetics to IL-17A, IL-17A was directly immobilized on a CM5 sensor chip via amine coupling, followed by injection of various concentrations of the peptide. The CVX5484 peptide has a KD of 10 nm (Table 1). These data indicate that, although the affinity of the peptide-antibody G6-LC for IL17A is reduced, it retains the binding activity with IL17A in vitro.
TABLE 1.
Binding affinity and cell-based potency values of different peptides and peptide-antibody genetic fusions for IL17A
| Modalities | ka | kd | KD | IC50 |
|---|---|---|---|---|
| m−1 s−1 | s−1 | nm | nm | |
| IHVTIPADLWDWINK-NH2 (CVX5484) | 1.3 ± 0.2 × 105 | 1.32 ± 0.07 × 10−3 | 10 ± 2 | No activity |
| PyrEIHVTIPADLWDWINK-NH2 | 1.7 ± 0.2 × 105 | 1.0 ± 0.2 × 10−3 | 6.0 ± 0.6 | 1.4 |
| PyrEAIHVTIPADLWDWINK-NH2 | 1.3 ± 0.3 × 105 | 1.0 ± 0.2 × 10−3 | 8 ± 2 | 1.7 |
| PyrEGIHVTIPADLWDWINK-NH2 | 1.5 ± 0.4 × 105 | 1.1 ± 0.2 × 10−3 | 7.2 ± 0.8 | 10 |
| PyrEGGIHVTIPADLWDWINK-NH2 | 1.6 ± 0.3 × 105 | 1.1 ± 0.3 × 10−3 | 7 ± 2 | 2.4 |
| Q-G6-HC | 2.5 ± 0.9 × 104 | 1.4 ± 0.2 × 10−3 | 60 ± 30 | 7.0 |
| Q-GSE12-HC | 3.0 ± 1.0 × 104 | 1.9 ± 0.5 × 10− | 80 ± 70 | 12 |
| Q-GSE21-HC | 1.84 ± 0.02 × 104 | 1.7 ± 0.3 × 10−3 | 100 ± 20 | >133 |
| Q-GSE21-LC | 5.0 ± 3.0 × 103 | 1.9 ± 0.3 × 10−3 | 500 ± 300 | 18.4 |
| Q-GSE21-LC-T5A | 2.2 ± 0.5 × 105 | 2.5 ± 0.5 × 10−4 | 1.2 ± 0.2 | 0.25 |
To examine the IL17A cell-based neutralizing activity of the peptide-antibody, IL17A-stimulated release of GRO-α (35) in human foreskin fibroblast BJ cells was measured as described under “Experimental Procedures.” As shown in Fig. 2D, the control anti-IL17 antibody blocked IL17A-stimulated GRO-α release, but no inhibition activity was observed for the peptide-antibody fusion G6-LC. The anti-IL17A chemically synthesized peptide CVX-5484 without N-acetylation cap, which mimics naturally occurring peptide, also showed no neutralizing activity in the BJ cell-based assays, but this peptide did show 10 nm KD binding (Table 1). These data indicate that the peptide modality in the peptide-antibody scaffold was more sensitive to loss of function when expressed in mammalian cells as part of a genetic fusion than the corresponding antibody modality.
N Terminus of the Anti-IL17A and Anti-IL22 Peptide-Antibody Fusion Was Degraded
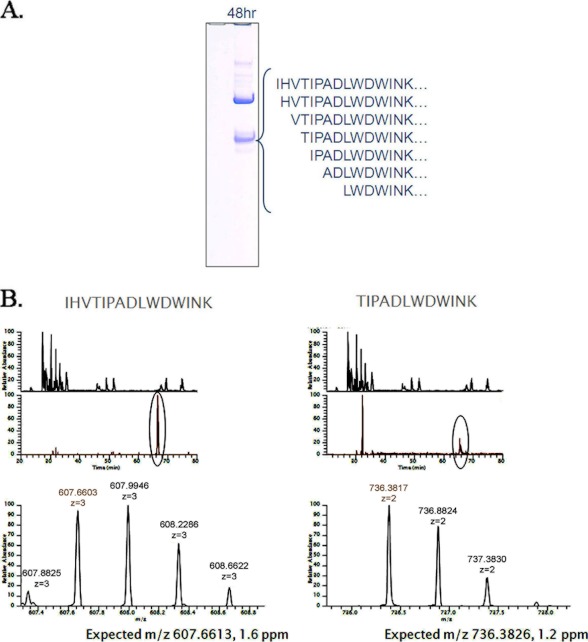
We suspected that this lack of IL17A cell-based neutralizing activity for the peptide-antibodies was probably due to N-terminal degradation of the anti-IL17A peptide in the fusion, because the N terminus of the peptide is important for its activity.3 To better understand this susceptibility to loss of function, we examined the integrity of the N terminus in the peptide-antibody fusions. As shown in Fig. 3, degradation at the N terminus of the peptide G6-LC fusion protein was indeed observed following incubation in mouse serum. Fig. 3A showed a range of smaller than expected polypeptides in SDS-PAGE, and LC-MS/MS data indicated that they were degraded at the N terminus (Fig. 3B).
FIGURE 3.
Characterization of the peptide-antibody fusion G6-LC in serum. A, SDS-PAGE of G6-LC peptide-antibody fusion protein following incubation in mouse serum for 48 h. The IL17A peptide was cleaved nonspecifically at numerous positions. B, mass spectroscopic extracted ion chromatograms of the intact anti-IL17A peptide CVX5484 of the peptide-antibody fusion G6-LC and one example of the detected truncations, as described under “Experimental Procedures.”
Engineering an N-terminal Pyroglutamate Helped Restore IL17A Neutralization Activity
To further delineate the function of the N terminus, several anti-IL17A peptides that varied in size and N terminus (Table 1, peptide modality rows 2–5) were chemically synthesized with no N-acetylation (Table 1). In these peptides, non-natural amino acid pyroglutamate was added to the N terminus. This pyroglutamate modification could presumably prevent N-terminal degradation of polypeptide chains. As shown in Table 1, the addition of pyroglutamate either alone or with additional amino acids did not affect binding to IL17A. All of the peptides with the N-terminal pyroglutamate regained their ability to neutralize IL17A in the BJ cell-based assay with IC50 values ranging from 1 to 10 nm (Table 1), indicating that engineering a pyroglutamate can indeed rescue the anti-IL17A neutralizing activity of the CVX-5484 peptides that lack N-acetylation protection. Because pyroglutamate can be formed naturally from the cyclization of an N-terminal glutamine residue in a polypeptide when expressed in mammalian cells (36, 37), new peptide-antibody fusions with an engineered glutamine N-terminal to the CVX-5484 peptide were generated. LC-MS data confirmed the cyclization rate of the N-terminal glutamine of the peptide-antibodies in HEK293 cells was nearly 100% (data not shown). Two of these peptide-antibody fusions, Q-G6-HC and Q-G12-HC, inhibited IL17A in the cell-based assay with a potency of 7 and 12 nm IC50, respectively (Table 1). Thus, the presence of pyroglutamate at the N terminus restored potency for IL17A cell-based activity, although this modification was not essential for the binding.
Mass Spectroscopic Analysis Revealed an Unexpected O-Linked Glycosylation Modification at Threonine 5 of the Anti-IL17A Peptide
Although the presence of a pyroglutamate at the N terminus restored the cell-based activity, this scaffold was still less potent than the pyroglutamate-CVX-5484 peptide alone (Table 1). To further optimize the scaffold, we shortened the linker and constructed a tetravalent scaffold by fusing the CVX-5484 peptide to both the heavy and light chains. However, no significant improvement in binding was observed (Table 2).
TABLE 2.
Summary of binding kinetics for different peptide-antibody fusion proteins to human IL17A
| Peptide-Antibodies | ka | kd | KD |
|---|---|---|---|
| m−1 s−1 | s−1 | nm | |
| Q-G-HC | 2.0 ± 0.5 × 104 | 1.0 ± 0.2 × 10−3 | 51 ± 6 |
| Q-G6-HC | 1.8 ± 0.5 × 104 | 9.2 ± 0.9 × 10−4 | 50 ± 10 |
| Q-GSE12-HC | 2.1 ± 0.8 × 104 | 1.5 ± 0.3 × 10−3 | 70 ± 40 |
| Q-G-LC | 3.1 ± 0.6 × 104 | 7 ± 2 × 10−4 | 23 ± 6 |
| Q-G6-LC | 3 ± 1 × 104 | 1.1 ± 0.2 × 10−3 | 32 ± 9 |
| Q-GSE12-LC | 2.8 ± 0.9 × 104 | 1.4 ± 0.1 × 10−3 | 50 ± 10 |
| Q-G-Tetravalent | 2.0 ± 0.8 × 104 | 1.0 ± 0.1 × 10−3 | 50 ± 20 |
| Q-G6-Tetravalent | 1.3 ± 0.5 × 104 | 1.0 ± 0.1 × 10−3 | 80 ± 20 |
| Q-GSE12-Tetravalent | 2 ± 1 × 104 | 1.5 ± 0.3 × 10−3 | 80 ± 70 |
| Q-GSE21-Tetravalent | 4 ± 2 × 104 | 1.9 ± 0.8 × 10−3 | 50 ± 20 |
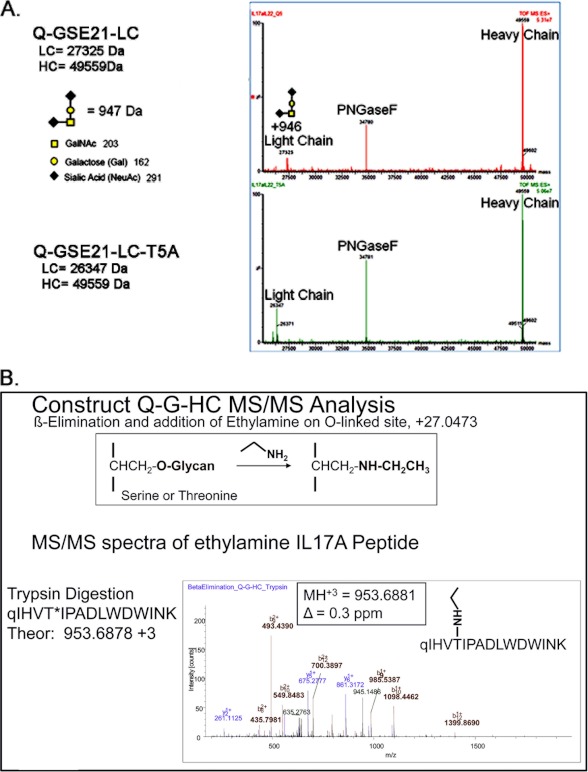
Unexpectedly, the LC-MS mass spectroscopic data (Fig. 4A) of the peptide:N-glycosidase F deglycosylated and reduced sample had a mass increase of 946 Da on the light chain of the Q-GSE21-LC fusion, the same chain that contained the anti-IL17A peptide CVX5484. We rationalized that the anti-IL17A peptide was post-translationally modified. Such a large mass of modification typically indicates a glycan modification. There is no N-linked glycosylation site present in the peptide (NX(S/T) motif), and the treatment of peptide:N-glycosidase F, which was used to remove N-linked glycan from IgG heavy chain, did not eliminate this modification. We therefore hypothesize that this modification is probably an O-linked glycan, based on the molecular mass increase (946 Da) matching that of the core 1 structure with two additional sialic acids. There is no consensus sequence for O-linked modification (38), but any serine or threonine residue is a potential site, and O-linked glycans are frequently clustered in short regions of peptide chains that contain repeating units of serine, threonine, and proline. For the CVX-5484 peptide, a proline residue was found at the +2-position to threonine 5 (Fig. 1), making it a potential site.
FIGURE 4.
Mass spectroscopic analysis revealed an unexpected O-linked glycosylation modification at threonine 5 of the anti-IL17A peptide. A, TOF-MS deconvoluted spectra of Q-GSE21-LC and Q-GSE21-LC-T5A, as described under “Experimental Procedures.” B, MS/MS fragmentation of the anti-IL17A trypsin generated peptide, after β-elimination and Michael addition with ethylamine, identified the site of O-linked glycosylation to be at position threonine 5, as described under “Experimental Procedures.”
To test this hypothesis and to identify the site of modification, we performed β-elimination and Michael addition using ethylamine on the Q-G-HC construct. Treatment with ethylamine can remove the O-linked glycan and also add an ethylamine in its place (39). A molecular ion with m/z of 953.6881 (z = 3) was detected, corresponding to the anti-IL17A peptide with an addition of ethylamine. Further MS/MS analysis confirmed the identity and location of the modification on Thr-5 of the anti-IL17A peptide (Fig. 4B). Both protein level and peptide mapping results suggest the presence of an O-linked glycan modification, and the occupancy rate of the modification was greater than 90% on threonine 5.
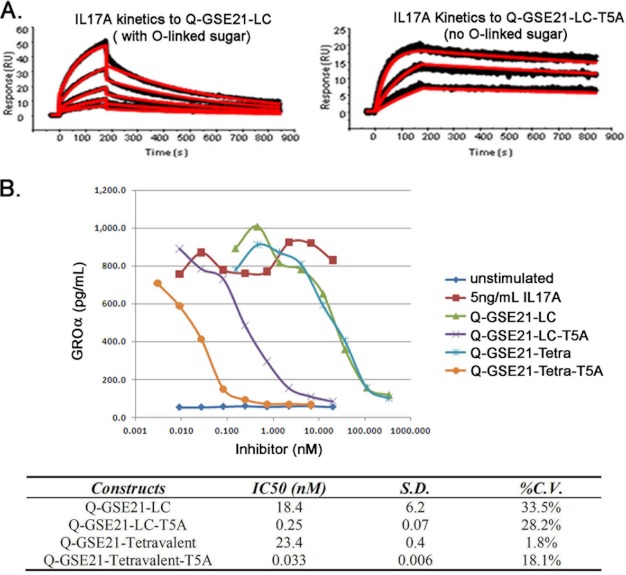
The Removal of O-Linked Glycan by Site-directed Mutagenesis Drastically Enhanced the IL17A Binding Affinity and Neutralization Potency for the Resulting Fusion Protein
We therefore hypothesized that the presence of a glycan at the threonine 5 position blocked the interaction between the CVX-5484 peptide and IL17A, thus lowering the ability of the peptide-antibody fusion to neutralize IL17A.
To confirm this hypothesis, we mutated threonine 5 to an alanine in the peptide-antibody Q-GSE21-LC. The resulting mutant peptide-antibody, Q-GSE21-LC-T5A, was expressed, purified, and confirmed to lack the O-linked modification (Fig. 4A). The peptide-antibody fusion, Q-GSE21-LC-T5A, showed a 400-fold improved KD compared with Q-GSE21-LC (1.2 versus 500 nm, respectively) (Table 1 and Fig. 5A). The Q-GSE21-LC-T5A lacking O-linked modification had a binding affinity that was better than or at least similar to that of the pyroglutamate-CVX-5484 peptide (1.2 nm versus 6 nm, respectively) (Table 1). In addition, a significant improvement in potency was observed in the IL17A cell-based assay (Table 1 and Fig. 5B). The peptide-antibody fusion, Q-GSE21-LC-T5A, showed a 70-fold improved IC50 compared with Q-GSE21-LC (0.25 versus 18.4 nm, respectively). The peptide-antibody tetravalent fusion, Q-GSE21-tetravalent-T5A, also showed a similar IC50 improvement over Q-GSE21-tetravalent (0.033 versus 23.4 nm, respectively; Fig. 5B). Both T5A peptide-antibody fusions have an IC50 value that is significantly improved over that of the CVX-5484 peptide (0.033 and 0.25 nm versus 1.4 nm) (Fig. 5B and Table 1). Thus, the peptide-antibody fusions lacking the O-linked post-translational modification in the CVX-5484 peptide region are now fully functional against two different human targets, IL17A and IL22.
FIGURE 5.
The removal of an unexpected O-linked glycosylation modification is crucial to retaining the full anti-IL17A activity of the peptide-antibody scaffold. A, kinetic analysis with human IL17A. B, IL17A inhibition curves for the peptide-antibody fusion with either the original sequence (Q-GSE21-LC) or the threonine mutation to alanine (Q-GSE21-LC-T5A). Data shown are from a single representative experiment.
DISCUSSION
This study has produced for the first time in mammalian cells a single fusion molecule utilizing both therapeutic peptides and therapeutic antibodies. Because both peptides and antibodies are versatile pharmacophores, the potential diversity of the combination of these two modalities makes this bispecific scaffold attractive. In the literature, an anti-fungal peptide fusion with a chicken-derived single-chain variable fragment made by bacteria was previously shown to be functional for plant fungal protection (40). The peptide-antibody format in this study is with full antibody structures and produced by mammalian cells, therefore presenting a clear advantage. The data in this study have demonstrated that post-translational modifications play critical roles in generating a fully active biotherapeutic molecule. Engineering an N-terminal pyroglutamate is essential in preventing peptide degradation in the fusion protein, and removing an unexpected O-linked glycan drastically enhances binding affinity and neutralization potency. These findings on protein biosynthesis may offer new insights and strategies for protein engineering and process engineering.
For the past 2 decades, molecular selection technologies have enabled identification of many novel therapeutic peptide candidates. These technologies utilize large, diverse collections of nucleic acid molecules (DNA or RNA), which include phage display (41), peptides on plasmid (42), ribosome display (43, 44), mRNA display (45, 46), and DNA display (47, 48). The peptide-antibody format allows the generation of new lead bispecific molecules by partnering various therapeutic antibodies with novel peptide hits identified through the recombinant screening technologies. Also, different peptide fusions with either heavy chain or light chain permit a multispecific targeting option. A unique feature of the peptide-antibody format is that the peptide and antibody modalities can be optimized separately for potency and affinity. Although a number of multispecific formats have been reported (i.e. tandem single-chain variable fragment (49), diabodies (50), tandem diabodies (51), two-in-one antibody (52), and dual variable domain antibodies (53)), none of these formats have made use of a peptide modality. A recently reported bispecific CovX body takes advantage of chemically conjugated peptides (22), yet its antibody modality is for half-life extension, not for therapeutic targeting. In addition, a peptide-antibody fusion is more antibody-like due to its similar protein size. A genetic fusion format makes the manufacturing process simple and low in cost. As a promising solution to diseases driven by multiple mechanisms, this peptide-antibody scaffold may provide a novel platform for multispecific protein therapeutics.
One of the major findings in this study is that the data have provided direct evidence for the impact of two distinct post-translational modifications on the properties, quality, and efficacy of therapeutic proteins. Both pyroglutamate and O-linked glycans are a form of post-translational modification commonly found in the majority of protein-based biotherapeutics (54). Pyroglutamate is frequently detected in the heavy chain and/or light chain sequences beginning with glutamine in antibodies (36, 37). Although it remains unclear whether or not glutamine to pyroglutamate conversion in vivo is nonenzymatic, pyroglutamate formation has been proposed as a stabilization mechanism, because it is found resistant to amino peptidases (23). Our data here clearly support this hypothesis. Considering that the pyroglutamate conversion rate in mammalian cells is nearly 100% and that it does not affect binding and efficacy, the N-terminal glutamine introduction can serve well as a protein engineering strategy to prevent N-terminal degradation.
O-Linked glycan modification is a less common form of glycosylation modification than N-linked oligosaccharides (38, 54). There is no consensus sequence, and therefore it is not readily predicted. The result of identifying an O-linked glycan in a short 15-amino acid peptide is indeed surprising. Moreover, glycan modification normally has positive impacts on protein properties (i.e. better folding and increased stability solubility and half-life). In this study, an O-linked glycan had a detrimental effect by inhibiting ligand binding and significantly reducing the efficacy. Removing this O-linked modification increased the affinity over 400-fold (Fig. 5B). The adverse impact posed by O-linked sugars can be explained by the fact that threonine 5 in the anti-IL17A peptide probably resides in the interacting pocket with IL17A. The mode of action of this neutralizing peptide against human IL-17A remains unknown and must wait for the crystal structures of peptide-IL17A complex. One possible action mode might be through the interaction between aromatic amino acids. A sizable O-linked glycan can surely block the interaction between the peptide and the ligand. It is noteworthy that the O-linked occupancy rate in threonine 5 is more than 90%, indicating that the modification is very efficient. Because the O-linked site is not predictable, special attention needs to be paid to threonine and serine residues during the protein engineering process. Despite the progress in understanding the structure and function of post-translational modifications in the context of biotherapeutic proteins, the importance of any given modification can only be truly assessed by direct experimentation.
In conclusion, we have generated a novel bispecific genetic fusion between a peptide and an antibody that maintains high affinity binding against two distinct and potentially synergistic targets, IL17A and IL22. The study has revealed two post-translational modifications during protein biosynthesis in mammalian cells that greatly impact the activity of the anti-IL17A peptide in the fusion scaffold. As the first multispecific modality utilizing both therapeutic peptides and antibodies in a single molecule, the peptide-antibody genetic fusion could allow the generation of new multispecific molecules by partnering various therapeutic antibodies with novel peptide hits identified through the recombinant screening technologies.
Acknowledgment
We thank Dr. Lynette Fouser for helpful discussions.
Footnotes
P. Jin, B. Oates, L. Wood, R. Murphy, G. Dello Iacono, J. Del Rosario, J. Jimenez, G. Chen, A. Bhat, C. Bradshaw, G. Woodnut, R. Lappe, and J. Desharnais, manuscript in preparation.
REFERENCES
- 1. Wurm F. M. (2004) Production of recombinant protein therapeutics in cultivated mammalian cells. Nat. Biotechnol. 22, 1393–1398 [DOI] [PubMed] [Google Scholar]
- 2. Omasa T., Onitsuka M., Kim W. D. (2010) Cell engineering and cultivation of Chinese hamster ovary (CHO) cells. Curr. Pharm. Biotechnol. 11, 233–240 [DOI] [PubMed] [Google Scholar]
- 3. Carter P. J. (2006) Potent antibody therapeutics by design. Nat. Rev. Immunol. 6, 343–357 [DOI] [PubMed] [Google Scholar]
- 4. Leader B., Baca Q. J., Golan D. E. (2008) Protein therapeutics. A summary and pharmacological classification. Nat. Rev. Drug Discov. 7, 21–39 [DOI] [PubMed] [Google Scholar]
- 5. Leavy O. (2010) Therapeutic antibodies. Past, present and future. Nat. Rev. Immunol. 10, 297. [DOI] [PubMed] [Google Scholar]
- 6. Kelley W. S. (1996) Therapeutic peptides. The devil is in the details. Biotechnology 14, 28–31 [DOI] [PubMed] [Google Scholar]
- 7. Bray B. L. (2003) Large-scale manufacture of peptide therapeutics by chemical synthesis. Nat. Rev. Drug Discov. 2, 587–593 [DOI] [PubMed] [Google Scholar]
- 8. Stone G. W., McLaurin B. T., Cox D. A., Bertrand M. E., Lincoff A. M., Moses J. W., White H. D., Pocock S. J., Ware J. H., Feit F., Colombo A., Aylward P. E., Cequier A. R., Darius H., Desmet W., Ebrahimi R., Hamon M., Rasmussen L. H., Rupprecht H. J., Hoekstra J., Mehran R., Ohman E. M., and ACUITY Investigators (2006) Bivalirudin for patients with acute coronary syndromes. N. Engl. J. Med. 355, 2203–2216 [DOI] [PubMed] [Google Scholar]
- 9. Drucker D. J., Buse J. B., Taylor K., Kendall D. M., Trautmann M., Zhuang D., Porter L., and DURATION-1 Study Group (2008) Exenatide once weekly versus twice daily for the treatment of type 2 diabetes. A randomised, open-label, non-inferiority study. Lancet 372, 1240–1250 [DOI] [PubMed] [Google Scholar]
- 10. McGregor D. P. (2008) Discovering and improving novel peptide therapeutics. Curr. Opin. Pharmacol. 8, 616–619 [DOI] [PubMed] [Google Scholar]
- 11. Zuraw B., Yasothan U., Kirkpatrick P. (2010) Ecallantide. Nat. Rev. Drug Discov. 9, 189–190 [DOI] [PubMed] [Google Scholar]
- 12. Cohn C. S., Bussel J. B. (2009) Romiplostim. A second-generation thrombopoietin agonist. Drugs Today 45, 175–188 [DOI] [PubMed] [Google Scholar]
- 13. Macdougall I. C., Rossert J., Casadevall N., Stead R. B., Duliege A. M., Froissart M., Eckardt K. U. (2009) A peptide-based erythropoietin-receptor agonist for pure red-cell aplasia. N. Engl. J. Med. 361, 1848–1855 [DOI] [PubMed] [Google Scholar]
- 14. Wrighton N. C., Farrell F. X., Chang R., Kashyap A. K., Barbone F. P., Mulcahy L. S., Johnson D. L., Barrett R. W., Jolliffe L. K., Dower W. J. (1996) Small peptides as potent mimetics of the protein hormone erythropoietin. Science 273, 458–464 [DOI] [PubMed] [Google Scholar]
- 15. Fishburn C. S. (2008) The pharmacology of PEGylation. Balancing PD with PK to generate novel therapeutics. J. Pharm. Sci. 97, 4167–4183 [DOI] [PubMed] [Google Scholar]
- 16. Bendele A., Seely J., Richey C., Sennello G., Shopp G. (1998) Short communication. Renal tubular vacuolation in animals treated with polyethylene-glycol-conjugated proteins. Toxicol. Sci. 42, 152–157 [DOI] [PubMed] [Google Scholar]
- 17. Sroda K., Rydlewski J., Langner M., Kozubek A., Grzybek M., Sikorski A. F. (2005) Repeated injections of PEG-PE liposomes generate anti-PEG antibodies. Cell Mol. Biol. Lett. 10, 37–47 [PubMed] [Google Scholar]
- 18. Huang C. (2009) Receptor-Fc fusion therapeutics, traps, and MIMETIBODY technology. Curr. Opin. Biotechnol. 20, 692–699 [DOI] [PubMed] [Google Scholar]
- 19. Subramanian G. M., Fiscella M., Lamousé-Smith A., Zeuzem S., McHutchison J. G. (2007) Albinterferon α-2b. A genetic fusion protein for the treatment of chronic hepatitis C. Nat. Biotechnol. 25, 1411–1419 [DOI] [PubMed] [Google Scholar]
- 20. Schellenberger V., Wang C. W., Geething N. C., Spink B. J., Campbell A., To W., Scholle M. D., Yin Y., Yao Y., Bogin O., Cleland J. L., Silverman J., Stemmer W. P. (2009) A recombinant polypeptide extends the in vivo half-life of peptides and proteins in a tunable manner. Nat. Biotechnol. 27, 1186–1190 [DOI] [PubMed] [Google Scholar]
- 21. Rader C., Turner J. M., Heine A., Shabat D., Sinha S. C., Wilson I. A., Lerner R. A., Barbas C. F. (2003) A humanized aldolase antibody for selective chemotherapy and adaptor immunotherapy. J. Mol. Biol. 332, 889–899 [DOI] [PubMed] [Google Scholar]
- 22. Doppalapudi V. R., Huang J., Liu D., Jin P., Liu B., Li L., Desharnais J., Hagen C., Levin N. J., Shields M. J., Parish M., Murphy R. E., Del Rosario J., Oates B. D., Lai J. Y., Matin M. J., Ainekulu Z., Bhat A., Bradshaw C. W., Woodnutt G., Lerner R. A., Lappe R. W. (2010) Chemical generation of bispecific antibodies. Proc. Natl. Acad. Sci. U.S.A. 107, 22611–22616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rink R., Arkema-Meter A., Baudoin I., Post E., Kuipers A., Nelemans S. A., Akanbi M. H., Moll G. N. (2010) To protect peptide pharmaceuticals against peptidases. J. Pharmacol. Toxicol. Methods 61, 210–218 [DOI] [PubMed] [Google Scholar]
- 24. Fouser L. A., Wright J. F., Dunussi-Joannopoulos K., Collins M. (2008) Th17 cytokines and their emerging roles in inflammation and autoimmunity. Immunol. Rev. 226, 87–102 [DOI] [PubMed] [Google Scholar]
- 25. Miossec P., Korn T., Kuchroo V. K. (2009) Interleukin-17 and type 17 helper T cells. N. Engl. J. Med. 361, 888–898 [DOI] [PubMed] [Google Scholar]
- 26. Korn T., Bettelli E., Oukka M., Kuchroo V. K. (2009) IL-17 and Th17 cells. Annu. Rev. Immunol. 27, 485–517 [DOI] [PubMed] [Google Scholar]
- 27. Liang S. C., Tan X. Y., Luxenberg D. P., Karim R., Dunussi-Joannopoulos K., Collins M., Fouser L. A. (2006) Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J. Exp. Med. 203, 2271–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ma H. L., Liang S., Li J., Napierata L., Brown T., Benoit S., Senices M., Gill D., Dunussi-Joannopoulos K., Collins M., Nickerson-Nutter C., Fouser L. A., Young D. A. (2008) IL-22 is required for Th17 cell-mediated pathology in a mouse model of psoriasis-like skin inflammation. J. Clin. Invest. 118, 597–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Myszka D. G. (1999) Improving biosensor analysis. J. Mol. Recognit. 12, 279–284 [DOI] [PubMed] [Google Scholar]
- 30. Ebbinghaus S., Kim S. J., Heyden M., Yu X., Heugen U., Gruebele M., Leitner D. M., Havenith M. (2007) An extended dynamical hydration shell around proteins. Proc. Natl. Acad. Sci. U.S.A. 104, 20749–20752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gerhardt S., Abbott W. M., Hargreaves D., Pauptit R. A., Davies R. A., Needham M. R., Langham C., Barker W., Aziz A., Snow M. J., Dawson S., Welsh F., Wilkinson T., Vaugan T., Beste G., Bishop S., Popovic B., Rees G., Sleeman M., Tuske S. J., Coales S. J., Hamuro Y., Russell C. (2009) Structure of IL-17A in complex with a potent, fully human neutralizing antibody. J. Mol. Biol. 394, 905–921 [DOI] [PubMed] [Google Scholar]
- 32. Liang S. C., Nickerson-Nutter C., Pittman D. D., Carrier Y., Goodwin D. G., Shields K. M., Lambert A. J., Schelling S. H., Medley Q. G., Ma H. L., Collins M., Dunussi-Joannopoulos K., Fouser L. A. (2010) IL-22 induces an acute-phase response. J. Immunol. 185, 5531–5538 [DOI] [PubMed] [Google Scholar]
- 33. Sonnenberg G. F., Fouser L. A., Artis D. (2010) Functional biology of the IL-22-IL-22R pathway in regulating immunity and inflammation at barrier surfaces. Adv. Immunol. 107, 1–29 [DOI] [PubMed] [Google Scholar]
- 34. Brand S., Beigel F., Olszak T., Zitzmann K., Eichhorst S. T., Otte J. M., Diepolder H., Marquardt A., Jagla W., Popp A., Leclair S., Herrmann K., Seiderer J., Ochsenkühn T., Göke B., Auernhammer C. J., Dambacher J. (2006) IL-22 is increased in active Crohn's disease and promotes proinflammatory gene expression and intestinal epithelial cell migration. Am. J. Physiol. Gastrointest. Liver Physiol. 290, G827–G838 [DOI] [PubMed] [Google Scholar]
- 35. Liang S. C., Long A. J., Bennett F., Whitters M. J., Karim R., Collins M., Goldman S. J., Dunussi-Joannopoulos K., Williams C. M., Wright J. F., Fouser L. A. (2007) An IL-17F/A heterodimer protein is produced by mouse Th17 cells and induces airway neutrophil recruitment. J. Immunol. 179, 7791–7799 [DOI] [PubMed] [Google Scholar]
- 36. Dick L. W., Jr., Kim C., Qiu D., Cheng K. C. (2007) Determination of the origin of the N-terminal pyro-glutamate variation in monoclonal antibodies using model peptides. Biotechnol. Bioeng. 97, 544–553 [DOI] [PubMed] [Google Scholar]
- 37. Liu Y. D., Goetze A. M., Bass R. B., Flynn G. C. (2011) N-terminal glutamate to pyroglutamate conversion in vivo for human IgG2 antibodies. J. Biol. Chem. 286, 11211–11217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Van den Steen P., Rudd P. M., Dwek R. A., Opdenakker G. (1998) Concepts and principles of O-linked glycosylation. Crit. Rev. Biochem. Mol. Biol. 33, 151–208 [DOI] [PubMed] [Google Scholar]
- 39. Hanisch F. G., Jovanovic M., Peter-Katalinic J. (2001) Glycoprotein identification and localization of O-glycosylation sites by mass spectrometric analysis of deglycosylated/alkylaminylated peptide fragments. Anal. Biochem. 290, 47–59 [DOI] [PubMed] [Google Scholar]
- 40. Peschen D., Li H. P., Fischer R., Kreuzaler F., Liao Y. C. (2004) Fusion proteins comprising a Fusarium-specific antibody linked to antifungal peptides protect plants against a fungal pathogen. Nat. Biotechnol. 22, 732–738 [DOI] [PubMed] [Google Scholar]
- 41. Smith G. P. (1985) Filamentous fusion phage. Novel expression vectors that display cloned antigens on the virion surface. Science 228, 1315–1317 [DOI] [PubMed] [Google Scholar]
- 42. Cull M. G., Miller J. F., Schatz P. J. (1992) Screening for receptor ligands using large libraries of peptides linked to the C terminus of the lac repressor. Proc. Natl. Acad. Sci. U.S.A. 89, 1865–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mattheakis L. C., Bhatt R. R., Dower W. J. (1994) An in vitro polysome display system for identifying ligands from very large peptide libraries. Proc. Natl. Acad. Sci. U.S.A. 91, 9022–9026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zahnd C., Amstutz P., Pluckthun A. (2007) Ribosome display. Selecting and evolving proteins in vitro that specifically bind to a target. Nat. Methods 4, 269–279 [DOI] [PubMed] [Google Scholar]
- 45. Roberts R. W., Szostak J. W. (1997) RNA-peptide fusions for the in vitro selection of peptides and proteins. Proc. Natl. Acad. Sci. U.S.A. 94, 12297–12302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Huang B. C., Liu R. (2007) Comparison of mRNA-display-based selections using synthetic peptide and natural protein libraries. Biochemistry 46, 10102–10112 [DOI] [PubMed] [Google Scholar]
- 47. Odegrip R., Coomber D., Eldridge B., Hederer R., Kuhlman P. A., Ullman C., FitzGerald K., McGregor D. (2004) CIS display. In vitro selection of peptides from libraries of protein-DNA complexes. Proc. Natl. Acad. Sci. U.S.A. 101, 2806–2810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yonezawa M., Doi N., Higashinakagawa T., Yanagawa H. (2004) DNA display of biologically active proteins for in vitro protein selection. J. Biochem. 135, 285–288 [DOI] [PubMed] [Google Scholar]
- 49. Hagemeyer C. E., von Zur Muhlen C., von Elverfeldt D., Peter K. (2009) Single-chain antibodies as diagnostic tools and therapeutic agents. Thromb. Haemost. 101, 1012–1019 [PubMed] [Google Scholar]
- 50. Hudson P. J., Kortt A. A. (1999) High avidity scFv multimers. Diabodies and triabodies. J. Immunol. Methods 231, 177–189 [DOI] [PubMed] [Google Scholar]
- 51. Kipriyanov S. M. (2009) Generation of bispecific and tandem diabodies. Methods Mol. Biol. 562, 177–193 [DOI] [PubMed] [Google Scholar]
- 52. Bostrom J., Yu S. F., Kan D., Appleton B. A., Lee C. V., Billeci K., Man W., Peale F., Ross S., Wiesmann C., Fuh G. (2009) Variants of the antibody herceptin that interact with HER2 and VEGF at the antigen binding site. Science 323, 1610–1614 [DOI] [PubMed] [Google Scholar]
- 53. Wu C., Ying H., Grinnell C., Bryant S., Miller R., Clabbers A., Bose S., McCarthy D., Zhu R. R., Santora L., Davis-Taber R., Kunes Y., Fung E., Schwartz A., Sakorafas P., Gu J., Tarcsa E., Murtaza A., Ghayur T. (2007) Simultaneous targeting of multiple disease mediators by a dual-variable-domain immunoglobulin. Nat. Biotechnol. 25, 1290–1297 [DOI] [PubMed] [Google Scholar]
- 54. Walsh G., Jefferis R. (2006) Post-translational modifications in the context of therapeutic proteins. Nat. Biotechnol. 24, 1241–1252 [DOI] [PubMed] [Google Scholar]