Abstract
It is a great privilege to contribute to the Reflections essays. In my particular case, this essay has allowed me to weave some of my major scientific contributions into a tapestry held together by what I have learned from three colleagues (Robert Letsinger, Gobind Khorana, and George Rathmann) who molded my career at every important junction. To these individuals, I remain eternally grateful, as they always led by example and showed many of us how to break new ground in both science and biotechnology. Relative to my scientific career, I have focused primarily on two related areas. The first is methodologies we developed for chemically synthesizing DNA and RNA. Synthetic DNA and RNA continue to be an essential research tool for biologists, biochemists, and molecular biologists. The second is developing new approaches for solving important biological problems using synthetic DNA, RNA, and their analogs.
Keywords: Genomics, Proteomics, DNA, RNA, DNA Synthesis, RNA Synthesis
Introduction
The excitement of learning about biology, chemistry, physics, astronomy, and so on, has always been part of my life. I remember in the third grade when my parents gave me a chemistry set with real chemicals that could be used for all kinds of fun stuff (not the watered down, bland kits that are available now and approved by every government agency you care to name). I did all of the experiments in the manual and then decided that I knew everything there was to know about chemistry. Little did I realize that my future would involve new discoveries in this very same field. In the sixth grade, my best friend and I put on a chemistry magic show for our class in which we had liquids changing colors, chemicals exploding, and so on. Every month, I eagerly awaited the arrival of the latest Scientific American while reading all of the science fiction books in the Des Moines Public Library. There was never any doubt that my life's work would somehow be involved with one of the natural or biological sciences, and of course, this is what happened.
Throughout the formative years of my scientific education, I was blessed with a truly outstanding high school science teacher, Paul Sloan. He not only provided a solid scientific education for me and my classmates, but he helped us discover the fruits of a scientific education. He formed a science club for us, took us on numerous field trips to laboratories and medical facilities throughout the Des Moines area, and encouraged us to participate in various scientific activities such as the Westinghouse Science Talent Search (now sponsored by Intel), where Ted Leffler and I received recognition at the state level. Several of my classmates (of a total of twenty-eight), including me, Ted (M.D./Ph.D., The Johns Hopkins University), Robert Kinsey (Ph.D. in nuclear physics, University of California, Berkeley), Stan Perrin (aeronautical and space engineering, Iowa State University), and many others, are forever in debt to Paul for leading us to the starting gates of our scientific careers.
Among those who have helped mold my professional career, three stand out: Robert Letsinger, Gobind Khorana, and George Rathmann (Fig. 1). Each contributed significantly to who I am today and how I arrived at this juncture of my life (Fig. 2).
FIGURE 1.
From left: George Rathmann, Robert Letsinger, and H. Gobind Khorana.
FIGURE 2.
The author, Marv Caruthers. This photograph was taken in Washington, D.C., at the time I received the National Medal of Science (2006).
During my senior year of study for a B.S. degree at Iowa State University, I decided to continue my education and applied to the chemistry/biochemistry Ph.D. program at Northwestern University. During my interview, I met Robert Letsinger, and he told me how they were attempting to synthesize macromolecules on an insoluble support. This was an exciting new concept that had never been explored (1962), and I decided not only to choose Northwestern University but also to work with Bob. Once I visited with him, I had no interest in shopping around either at Northwestern or elsewhere for an alternative research director. Upon my arrival, I immediately joined Bob's laboratory and learned that Milt Kornet had synthesized a dipeptide on a polystyrene support. However, about one month later, our celebrations of this momentous event quickly turned to disappointment, as Robert Merrifield published his tetrapeptide synthesis, also on polystyrene, in the Journal of the American Chemical Society (1). (Neither knew that the other had embarked on the development of this concept.) The major difference between the two approaches was the direction of peptide synthesis and, hence, the choice of protecting groups. Bob and Milt quickly wrote up their synthesis, and it was published the same year, also in the Journal of the American Chemical Society (2). However, only Robert Merrifield was recognized with the Nobel Prize, which was a great disappointment, as we believed that Bob Letsinger should have shared this honor.
Meanwhile, Bob accepted a new postdoctoral associate, V. Mahadevan (Devan), whose project was to attempt the synthesis of oligonucleotides on the same type of support. Later, after our course work and other academic requirements such as oral examinations had been completed, Don Jerina and I, as graduate students, joined Devan on this project. Using different approaches, each of us had considerable success, as we were able to synthesize oligomers up to five in length. The research constituted a major breakthrough in this field, as it laid the foundation for a new oligonucleotide synthesis strategy that later became known as the phosphotriester method (3–5 nucleotides in length). This procedure remained the method of choice for synthesizing both RNA and DNA until many years later, when my laboratory introduced 2′-deoxynucleoside 3′-phosphoramidites as synthons for DNA synthesis. Although I learned an enormous amount of chemistry from Bob, he excelled in many other ways that I have tried to emulate over the years. For example, he showed me how to be a good teacher and how to motivate students in a positive and productive manner. I have also found myself following his research style. Thus, with very few exceptions, I assign each graduate or postdoctoral student an individual research project for which he or she has complete responsibility. As also the case with Bob, I usually have a meeting about once a week with each student, which may last for a few minutes or an hour if there are problems. However, under no circumstance do I dictate a research pathway, another approach that I learned from Bob. Instead, we discuss alternatives, and the students carry forward with paths of their choosing. My favorite comment is, “Once you leave here, I won't be around to help, so you might as well learn now how to do good research,” again, a lesson I learned from Bob. Over the years, as I observed how others direct their programs, I consider myself one of the most fortunate of graduate students. Somehow, I found a mentor who was patient, who allowed students to explore their potential, and who focused his research on extremely challenging problems.
My education in how to do quality research continued when I chose Gobind Khorana as my postdoctoral mentor. Gobind taught me how to focus on problems that everyone else had yet to discover were truly important. I joined Gobind's laboratory in February 1968, three years into the yeast alanine tRNA gene synthesis project and eight months prior to the announcement of the Nobel Prize that he shared with Marshall Nirenberg for solving the genetic code. At that time, most biologists, biochemists, and chemists had a hard time understanding why Gobind wanted to synthesize a gene. Gobind's response (6) was, “We would like to know, for example, what the initiation and the termination signals for RNA polymerase are, what kind of sequences are recognized by repressors, by host modification and host restrictive enzymes, and by enzymes involved in genetic recombination and so on. For these studies ultimately what is required is the ability to synthesize long chains of DNA with specific, non-repeating sequences. With this should come the ability to ‘manipulate’ DNA for different types of studies.” In other words, he was using the synthesis of this gene as a template for developing methodologies useful for chemically preparing DNA so that biologists could investigate a large number of important problems.
As the historical record demonstrates, approximately forty years later, Gobind's foresight was provocative, accurate, and very conservative relative to what synthetic DNA now allows us to do. For example, much of biology, biochemistry, and molecular biology depends upon synthetic DNA as an indispensable component of basic research. It is used for massive DNA sequencing of genes and organismic DNA; polymerase chain reactions (PCRs); functional studies on genes, proteins, and chromosomes; DNA diagnostics; synthesizing DNA chips containing 105–106 unique sequences/chip to profile gene expression; generating modified genes or oligomers useful for monitoring chromosome epigenetic activities; and controlling gene expression using antisense DNA, interfering RNA, and microRNA antimers. Among the many additional applications for synthetic DNA, the cloning of heterologous DNAs in prokaryotic and eukaryotic cells is also noteworthy.
In December 1972, the total synthesis of the alanine tRNA gene was published as a complete issue (thirteen manuscripts) in the Journal of Molecular Biology (7). This work was a major milestone in biology, as it demonstrated what could be done by one of the great leaders in science, one who had the courage to work at the very edge of the possible and to point the way for others to follow in his footsteps.
Armed with a wealth of knowledge on how to conduct fundamental important research and the laboratory expertise to make sure we carried through with the right experiments, I accepted an academic appointment as an assistant professor in the Department of Chemistry and Biochemistry at the University of Colorado Boulder. Our initial research focused on an important unresolved problem in molecular biology: how proteins recognize nucleic acid sequences. Just prior to leaving Gobind's laboratory, I was fortunate to attend Walter Gilbert's lecture at Harvard University, where he presented the sequence of lac operator, which he, Nancy Maizels, and Allan Maxam had just obtained using Fred Sanger's RNA sequencing method. The results intrigued me because there was a large amount of genetic data on the lac operon, including many Oc mutants, and the lac operator formed a very high affinity binding complex with the lac repressor. Thus, we reasoned that we could change individual base pairs within the lac operator by chemical synthesis and then determine, by a combination of kinetics and binding constants, how these changes altered the recognition of the lac operator by the lac repressor. However, our focus was not merely to create a series of transitions and transversions. Instead, we carried out a series of experiments in which we altered one functional group on a base pair per each operator that we synthesized and then studied the kinetics and binding constant. We called this approach “functional group mutagenesis.” I was very fortunate that two excellent graduate students, Dave Goeddel and Dan Yansura, chose to work on this project.
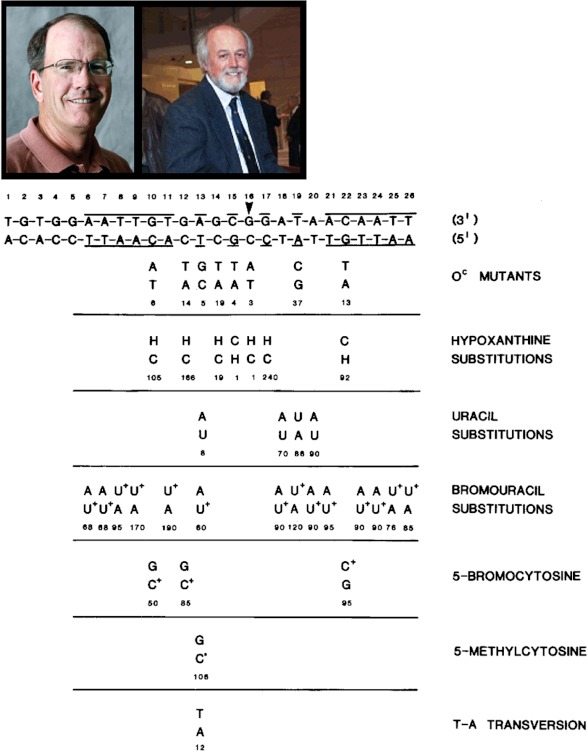
A summary of our results for the lac operator (8) are presented in Fig. 3. One example illustrates the power of the approach. Consider the base pair defined as position 13 in the synthetic operator. The natural constitutive mutant at this site is the transition of A-T to G-C. When we measure the binding constant for this mutant, it is 5% of the unmodified complex. If we substitute deoxyuridine for thymidine in the A-T base pair, the binding constant is 8% of the wild type, the same binding constant as observed for the A-T to G-C transition. If we now substitute 5-methyldeoxycytidine for deoxycytidine in the constitutive mutant G-C base pair, the binding constant is the same as for an A-T base pair (106% of the wild-type operator/repressor binding constant). Thus, the lac repressor recognizes only a methyl group at position 5 on a pyrimidine. Otherwise, either an A-T or G-C base pair will suffice. Using several other substitutions, we identified additional contacts and also concluded that the lac repressor recognized only one face of the DNA. (It does not wrap around the DNA.) Using alkylation protection experiments, Walter Gilbert demonstrated that the lac repressor bound to only one face of the operator. His results and ours were presented as back-to-back lectures at the 1977 Gordon Research Conference on Nucleic Acids. It was rewarding to discover that the face of the lac operator as covered by the repressor from Gilbert's laboratory was also the same region of the operator as defined by our functional group mutagenesis studies.
FIGURE 3.
Summary of single-site, functional group mutagenesis alterations in the lac operator sequence. The lac operator sequence is shown at the top. The bold lines delineate the 2-fold symmetric positions. The dyad axis is shown by the arrowhead. Various sequence changes are listed below the lac operator base pairs. A number located directly below each modification is the corresponding affinity of the repressor for the modified operator expressed as a percentage of the affinity of the repressor for the unmodified operator with the same length and sequence. H, hypoxanthine; U, uracil; U+, 5-bromouracil; C+, 5-bromocytosine; C0, 5-methylcytosine. Recent photographs are shown of Dave Goeddel (left) and Dan Yansura (right).
Using this approach, we also mapped protein-DNA recognition sites in an Escherichia coli RNA polymerase promoter (9) as well as the operators recognized by the Cro and CI repressors. Cloning and site-specific mutagenesis of the Cro repressor was also used to derive contact sites within the Cro operator (10). A particularly interesting exercise was changing Cro repressor amino acids to generate a new contact site with a mutated Cro operator and therefore regenerate the wild-type phenotype (11). The combination of these various studies produced for the first time a map of one mechanism whereby proteins can recognize DNA. Since this early work, we now know of many additional pathways that are used for specific recognition.
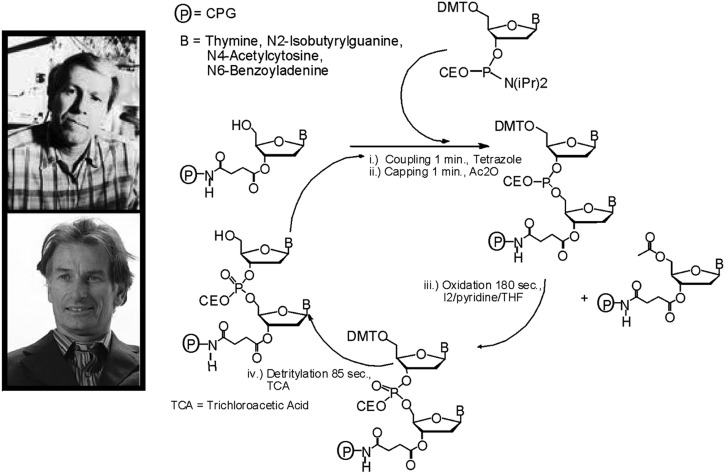
In 1976, while we were deeply involved in the functional group mutagenesis work, a new graduate student, Mark Matteucci, approached me seeking a research project focused not on biochemistry but on organic chemistry. I proposed to Mark that we try to use silica supports for DNA synthesis. Initially, I wanted to attach the growing deoxyoligonucleotide to sintered glass funnels. We would then simply pass reagents sequentially through the funnel to grow the DNA. The problem was in evaluating results because each funnel would have to be broken, the glass crushed, and then some type of analysis carried out. This was unacceptable, so we searched for alternatives. At that time, high performance liquid chromatography (HPLC)-grade controlled pore glass had just become available, not as a synthesis support but as a matrix for separating and purifying organic reaction products. We turned to this material, however, as a polymer support for oligonucleotide synthesis and discovered that, when coupled with activated phosphites, DNA segments 10–15 nucleotides in length could be rapidly synthesized in very high yield. Then, in 1978, Serge Beaucage joined my laboratory as a postdoctoral student. Based upon Mark's success, we knew that the next step was to tame the appropriately protected and activated deoxynucleotide phosphites so that they could be used routinely in machines and, most importantly, by non-chemists. Our initial research focused on earlier manuscripts that had shown how amino phosphines could be activated by an insertion reaction with CO2 to generate a mixed anhydride (in phosphorus chemistry and not the nucleic acid field). We tried very hard without success to repeat this work. However, these failures eventually led to our development of appropriately protected 2′-deoxynucleoside 3′-phosphoramidites as synthons for DNA synthesis. We discovered that these synthons could be easily prepared, stored without decomposition, and then readily activated with tetrazole to generate an internucleotide linkage in very high yield (98%). These results were unprecedented in the nucleic acid field and far exceeded anything previously accomplished. By incorporating Mark's work on controlled pore glass, we developed the synthesis approach (12, 13) outlined in Fig. 4. This approach is often called the phosphoramidite method of DNA synthesis. Once this procedure was published, others immediately started using our chemistry. The first step is to remove the 5′-dimethoxytrityl protecting group from a 2′-deoxynucleoside covalently attached to controlled pore glass. Next, an appropriately protected 2′-deoxynucleoside 3′-phosphoramidite and tetrazole as an activator are introduced to the support, and a phosphite triester internucleotide linkage forms within 30 s. Acetic anhydride in pyridine is then used to remove phosphite adducts from the bases and to acylate any unreacted 2′-deoxynucleoside. Oxidation follows in order to convert the phosphite internucleotide linkage to phosphate. The cycle is then repeated many times to produce deoxyoligonucleotides containing at least 20–30 mononucleotides. This procedure continues, thirty years later, to be the method of choice for the chemical synthesis of DNA and RNA.
FIGURE 4.
DNA phosphoramidite synthesis cycle. The photographs are of Serge Beaucage (upper) and Mark Matteucci (lower). CPG, controlled pore glass; CEO, beta-cyanoethyl; DMTO, dimethoxytrityl.
During the time we were developing our polynucleotide synthesis chemistry (1978–1980), several other important technological breakthroughs were being discovered. These included DNA sequencing, restriction modification of DNA, cloning heterologous DNA elements into plasmids and phages, and rapid protein sequencing. (Later, PCR and site-specific mutagenesis, which are technologies that are completely dependent on synthetic DNA, were developed.) Together, these new, very powerful technologies, including synthetic DNA and RNA, reinvigorated biological research and gave birth to the biotechnology industry. I also found myself in the middle of this revolution both in research and as a cofounder of two biotechnology companies, Amgen and Applied Biosystems.
In February 1980, late one afternoon, I received a phone call from Winston Salser. He told me how he, along with three venture capitalists (Sam Wohlstadter, Pitch Johnson, and Bill Bowes), were contemplating the formation of a biotechnology company and asked if I wanted to join these discussions. At the time, I was receiving, almost daily, phone calls about potential business opportunities that would utilize our new DNA synthesis chemistry. Winston's proposal was especially intriguing to me, as he also had approached several other well known scientists, including Norm Davidson, Lee Hood, Arnie Berk, John Carbon, Bob Schimke, Arno Motulsky, and Dave Gibson. During our initial meetings at the University of California, Los Angeles, and Stanford University, we decided to move forward, and Winston located a small unoccupied laboratory in Thousand Oaks, California. At our first meeting in Thousand Oaks, Lee and I proposed that this new company, which was to be called Applied Molecular Genetics, should have an instrument division that would sell protein sequencers and DNA synthesizers based upon Lee's and my work, respectively. However, the other scientists were concerned that Applied Molecular Genetics would become primarily an instrumentation company rather than one focused on genetic engineering and molecular biology. As a result, the proposal was dropped, but later that same morning, Lee and I, in discussion with the venture capitalists who were starting Applied Molecular Genetics, decided to move forward with the formation of a new instrument-focused company, which became known as Applied Biosystems.
We located Applied Biosystems in Foster City, California, and hired Sam Eletr as our first chief executive officer (spring 1981). Sam proved to be an excellent entrepreneurial choice. We started with only three million dollars but were able to convince a large number of pharmaceutical and biotechnology companies and academic laboratories to advance large deposits (half of the proposed sale price) on future protein sequencers and DNA synthesizers (that had yet to be designed, let alone manufactured) simply so they would be in the queue for the first machines. With this capital, Sam hired two or three scientists each from Lee's and my laboratories to design and build our first instruments. From my laboratory, Bill Efcavitch and Curt Becker were among the first employees (later, Lincoln McBride as well). Bill and Curt started with a series of valves, tubing, small HPLC-grade silica columns, a tank of liquid nitrogen, and a large piece of plywood. Within a few months, they were synthesizing DNA on this platform. Sam asked John Bridger, an engineer hired from Hewlett-Packard, to design a DNA synthesizer, which became known as our 380A machine. By December 1982, Bill installed the first commercial synthesizer in my laboratory (Fig. 5), and Applied Biosystems began shipping the instruments in 1983.
FIGURE 5.

Bill Efcavitch delivers the first commercial Applied Biosystems 380A DNA synthesizer to the Caruthers laboratory in December 1982.
Meanwhile, Winston Salser and Bill Bowes recruited George Rathmann as the first chief executive officer of Applied Molecular Genetics, and they raised 18.9 million dollars, which, at that time, was the largest initial private placement ever put together to start a new venture capital-funded company. Later, George changed the company's name to Amgen. Almost immediately, George and I decided to place a small group of scientists in Boulder so that we could most effectively utilize my time in directing a DNA synthesis group at Amgen, as I had no interest in resigning my position at the University of Colorado and moving to Thousand Oaks. Over the next few years, this proved to be a very important and positive decision for Amgen. The group in Boulder (ten scientists) organized very quickly, and we began manually synthesizing DNA for the molecular biologists we hired in Thousand Oaks. Manual synthesis was important, as the first Applied Biosystems DNA synthesizers were not available to us for two more years, and we did not want to wait. During this time, the group in Boulder synthesized and expressed several genes in addition to preparing a large number of probes. Several accomplishments are noteworthy, but I will describe only two. From the very beginning, we were focused on several potential therapeutic proteins, including erythropoietin (the protein that stimulates the production of erythrocytes). Using crude, partially purified erythropoietin isolated from urine (from Gene Goldwasser's laboratory at The University of Chicago), we obtained preliminary peptide sequences from an instrument in Lee's laboratory at the California Institute of Technology. On the basis of these complex amino acid results (many peptides (most, unfortunately) with a large number of amino acids having 4- and 6-fold redundant codons), Haki and Zippi Stabinsky began manually synthesizing DNA at our facility in Boulder. These probes were shipped to Fu-Kuen Lin in Thousand Oaks, where he used them in his work focused on isolating the erythropoietin gene. After two years and the manual synthesis of about 700 probes, Fu-Kuen isolated this gene. We then cloned and expressed it in Chinese hamster ovary (CHO) cells, purified the protein, carried out successful clinical studies, and began marketing this therapeutic as Epogen in 1989. One of the most rewarding and exciting phases of my scientific career occurred as a result of this project. Our first clinical study was with a cohort of patients who had severe kidney disease and were not capable of producing their own erythropoietin. They were therefore anemic, completely dependent on blood transfusions, and bedridden because their hematocrit could be maintained only at about one-half of the normal. They were very weak and sick. However, within one month of receiving Epogen, all were walking, some were hiking, and others had returned to their jobs. This created a problem for us, relative to the clinical trial, as we had to track them down and obtain statements on the benefits of the drug. As a result of this phase 1 study, we knew we had a wonderful drug that would significantly improve the well-being and health of a large number of patients. This discovery was possible because we had learned how to synthesize DNA rapidly and with a purity that allowed us to prepare the very large number of probes for locating this gene. One paradox of this story involves George Rathmann. During the final years of his life, he suffered from severe kidney disease and was dependent upon dialysis and erythropoietin to remain alive.
A second major milestone at Amgen during these early days was our work with granulocyte colony-stimulating factor. This project also began with Haki and Zippi Stabinsky in Boulder. Working with Larry Souza in Thousand Oaks, they manually prepared ∼500 probes that were used to isolate the gene for this protein. After cloning and expression, this time in E. coli, we successfully isolated the protein, conducted clinical trials, and marketed the drug as Neupogen. This was our second blockbuster drug; Epogen was the first. Neupogen was unique, as it stimulated the production of leukocytes. Thus, the initial major application of the drug was as an adjunct to chemotherapy. It protected against the loss of white blood cells and therefore dramatically reduced infections associated with chemotherapy. Patients were able to complete their chemotherapy and have a far better outcome for control of their cancer. Again, I was very proud of our contribution to the development of this drug.
George Rathmann was an absolutely spectacular choice to lead Amgen through the first ten years of our existence. He was everything you expected from the leader of a new biotechnology company: visionary; accomplished scientist; a communicator with the scientists in the laboratory, his management team, and investors; and a kind gentle giant. I have often said that if I wanted to manage a business activity, I would want to emulate George in every way possible. Along with Bob Letsinger and Gobind Khorana, I would add George Rathmann as the third individual who positively shaped my scientific career.
For many years, the synthesis approach as outlined in Fig. 4 proved to be satisfactory for all biological applications in which segments 20 or so nucleotides in length were adequate. However, over the past ten years, many new applications required oligomers up to 300 nucleotides in length. This development led me to collaborate with Emily LeProust and others at Agilent to design a synthesis protocol that could lead to DNA of this size (14). We now have a procedure in which 240,000 unique DNA segments are prepared on a glass chip using a modified inkjet printer to deposit the activated phosphoramidites on the glass surface. Agilent currently has several DNA chip synthesizers operating 24/7 producing oligomers 250–300 nucleotides in length. Each day, these machines produce the equivalent of the human genome (3 billion base pairs or 6 billion chemical couplings). Although results of this type are unprecedented, there are several steps in the chemistry where improvements can be made. The synthesis of oligomers containing 600 or so nucleotides should be possible in the not too distant future.
Once we had developed a methodology for synthesizing DNA, our focus turned to RNA. The problem for many years with RNA has been identifying appropriate protection of reactive sites on the nucleosides, especially an orthogonal set that includes protection of the 2′-hydroxyl during synthesis. After various false starts, Steve Scaringe (Fig. 6) developed the 5′-O-silyl-2′-O-orthoester RNA synthesis chemistry in my laboratory (15). We had considerable success with this approach, as we could routinely synthesize RNA with up to 50 mononucleotides in each segment. The chemistry was so promising that Steve started a very successful biotechnology company called Dharmacon, which was sold to Fisher Scientific for approximately seventy million dollars in 2004. A serious problem was that the approach required, as one critical step, the use of fluoride, which was highly corrosive to the RNA synthesizers. Eventually, this limitation led Doug Dellinger (Fig. 6) and me to develop an alternative approach with thionocarbamate protection at the 2′-hydroxyl (16). This method appears to be very robust. We can routinely synthesize RNA containing 150 nucleotides on glass slides containing up to 100,000 segments per array. This was an extremely challenging research problem that was successfully solved by several excellent colleagues, primarily Steve and Doug.
FIGURE 6.
From left: Peter Seeberger, Steve Scaringe, and Doug Dellinger.
Among my graduate and postdoctoral students, many were willing to accept the challenge of developing new exciting projects. Over the years, they have gone on to productive careers in teaching, research, and business. One is now a member of the United States National Academy of Science (Dave Goeddel), and another has his own institute in Potsdam, Germany (Peter Seeberger) (Fig. 6). My students and I always have maintained a very warm, respectful, and admiring relationship with one another. Several years ago, they organized a three-day symposium in my honor. I was pleasantly surprised that at least 90% of my former colleagues were there and that they came from all over the world.
I have also been fortunate to have parents who always encouraged me in any endeavors, even though, without a scientific background, they had no idea what I was doing. My wife of thirty-four years, Jennie (deceased January 8, 2006), was there for me and with me in everything we wanted to do. Without her love and encouragement over the years, I am sure that I would never have attained what success I have had. And now Fiona, to whom I have been married the past few years, has helped me through the dark times and provided the path to get me going again, as I now have a new research group doing unexplored and exciting science.
REFERENCES
- 1. Merrifield R. B. (1963) Solid phase peptide synthesis. I. The synthesis of a tetrapeptide. J. Am. Chem. Soc. 85, 2149–2154 [Google Scholar]
- 2. Letsinger R. L., Kornet M. J. (1963) Popcorn polymer as a support in multistep syntheses. J. Am. Chem. Soc. 85, 3045–3046 [Google Scholar]
- 3. Letsinger R. L., Mahadevan V. (1965) Oligonucleotide synthesis on a polymer support. J. Am. Chem. Soc. 87, 3526–3527 [DOI] [PubMed] [Google Scholar]
- 4. Letsinger R. L., Caruthers M. H., Jerina D. M. (1967) Reactions of nucleosides on polymer supports. Synthesis of thymidylylthymidylylthymidine. Biochemistry 6, 1379–1388 [DOI] [PubMed] [Google Scholar]
- 5. Letsinger R. L., Caruthers M. H., Miller P. S., Ogilvie K. K. (1967) Oligonucleotide syntheses utilizing β-benzoylpropionyl, a blocking group with a trigger for selective cleavage. J. Am. Chem. Soc. 89, 7146–7147 [DOI] [PubMed] [Google Scholar]
- 6. Khorana H. G. (1968) Synthesis in the study of nucleic acids. The Fourth Jubilee Lecture. Biochem. J. 109, 709–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Khorana H. G., Agarwal K. L., Büchi H., Caruthers M. H., Gupta N. K., Kleppe K., Kumar A., Otsuka E., RajBhandary U. L., Van de Sande J. H., Sgaramella V., Terao T., Weber H., Yamada T. (1972) Studies on polynucleotides. 103. Total synthesis of the structural gene for an alanine transfer ribonucleic acid from yeast. J. Mol. Biol. 72, 209–217 [DOI] [PubMed] [Google Scholar]
- 8. Goeddel D. V., Yansura D. G., Caruthers M. H. (1978) How lac repressor recognizes lac operator. Proc. Natl. Acad. Sci. U.S.A. 75, 3578–3582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dubendorff J. W., deHaseth P. L., Rosendahl M. S., Caruthers M. H. (1987) DNA functional groups required for formation of open complexes between Escherichia coli RNA polymerase and the λ PR promoter. Identification via base analog substitutions. J. Biol. Chem. 262, 892–898 [PubMed] [Google Scholar]
- 10. Hubbard A. J., Bracco L. P., Eisenbeis S. J., Gayle R. B., Beaton G., Caruthers M. H. (1990) Role of the Cro repressor carboxy terminal domain and flexible dimer linkage in operator and nonspecific DNA binding. Biochemistry 29, 9241–9249 [DOI] [PubMed] [Google Scholar]
- 11. Caruthers M. H., Gottlieb P., Bracco L. P., Cummins L. (1987) in Structure and Expression (Sarma R., Sarma M., eds) pp. 157–166, Adenine Press, Guilderland, NY [Google Scholar]
- 12. Matteucci M. D., Caruthers M. H. (1981) Synthesis of deoxyoligonucleotides on a polymer support. J. Am. Chem. Soc. 103, 3185–3191 [PubMed] [Google Scholar]
- 13. Beaucage S. L., Caruthers M. H. (1981) Deoxynucleoside phosphoramidites–a new class of key intermediates for deoxypolynucleotide synthesis. Tetrahedron Lett. 22, 1859–1862 [Google Scholar]
- 14. LeProust E. M., Peck B. J., Spirin K., McCuen H. B., Moore B., Namsaraev E., Caruthers M. H. (2010) Synthesis of high-quality libraries of long (150mer) oligonucleotides by a novel depurination controlled process. Nucleic Acids Res. 38, 2522–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Scaringe S. A., Wincott F. E., Caruthers M. H. (1998) Novel RNA synthesis method using 5′-O-silyl-2′-O-orthoester protecting groups. J. Am. Chem. Soc. 120, 11820–11821 [Google Scholar]
- 16. Dellinger D. J., Timár Z., Myerson J., Sierzchala A. B., Turner J., Ferreira F., Kupihár Z., Dellinger G., Hill K. W., Powell J. A., Sampson J. R., Caruthers M. H. (2011) Streamlined process for the chemical synthesis of RNA using 2′-O-thionocarbamate-protected nucleoside phosphoramidites in the solid phase. J. Am. Chem. Soc. 133, 11540–11556 [DOI] [PubMed] [Google Scholar]