Background: Although Rad14p has well known functions in NER, its role in transcription is not clearly known.
Results: We demonstrate that Rad14p associates with active gene in absence of lesion and promotes transcription.
Conclusion: Our results support a new role of Rad14p in transcriptional regulation in addition to its well known function in NER.
Significance: This study implicates Rad14p as a new regulator of gene expression.
Keywords: Chromatin Immunoprecipitation (ChiP), DNA Repair, DNA Transcription, RNA Polymerase II, Transcription, GAL, Rad14
Abstract
Rad14p is a DNA damage recognition factor in nucleotide excision repair. Intriguingly, we show here that Rad14p associates with the promoter of a galactose-inducible GAL1 gene after transcriptional induction in the absence of DNA lesion. Such an association of Rad14p facilitates the recruitment of TBP, TFIIH, and RNA polymerase II to the GAL1 promoter. Furthermore, the association of RNA polymerase II with the GAL1 promoter is significantly decreased in the absence of Rad14p, when the coding sequence was deleted. These results support the role of Rad14p in transcriptional initiation. Consistently, the level of GAL1 mRNA is significantly decreased in the absence of Rad14p. Similar results are also obtained at other galactose-inducible GAL genes such as GAL7 and GAL10. Likewise, Rad14p promotes transcription of other non-GAL genes such as CUP1, CTT1, and STL1 after transcriptional induction. However, the effect of Rad14p on the steady-state levels of transcription of GAL genes or constitutively active genes such as ADH1, PGK1, PYK1, and RPS5 is not observed. Thus, Rad14p promotes initial transcription but does not appear to regulate the steady-state level. Collectively, our results unveil a new role of Rad14p in stimulating transcription in addition to its well-known function in nucleotide excision repair.
Introduction
Genomic DNA is a target of incessant attack from an immense number of sources comprising radiation, chemicals, and mutagens as external agents along with various internal biological byproducts. Thus, damaged DNA has to be repaired swiftly and efficiently for the cell to survive and function effectively. Therefore, various repair mechanisms are employed by the cell to maintain its genomic stability. Nucleotide excision repair (NER)2 is one such repair mechanism where many proteins work synchronously in a multistep process to coordinate the overall repair pathway (1–8). The importance of this repair mechanism is evidenced by the severe human diseases that result from in-born genetic mutations of NER proteins including xeroderma pigmentosum (XP) and Cockayne syndrome (5, 6, 9–14).
NER is conserved among eukaryotes. In yeast (Saccharomyces cerevisiae), the DNA damage recognition step is mediated by a combination of factors, namely Rad14p, Rad4p, Rad23p, and RPA (replication protein A) (1, 15). Then Rad3p and Rad25p DNA helicases (components of TFIIH) unwind the DNA duplex around the lesion (1). Incision of the damaged strand on the 5′ site of the lesion is brought about by the Rad1p-Rad10p complex, whereas Rad2p incises the 3′ end of the lesion, thus releasing ∼30-nucleotide fragment of damaged DNA strand (1). Neither Rad1p-Rad10p nor Rad2p nuclease shows a specificity for binding to the UV-damaged DNA (16, 17). Rad14p plays a crucial role in targeting Rad1p-Rad10p nuclease to the DNA (2, 18), whereas TFIIH promotes the targeting of Rad2p nuclease (19). The targeting of Rad1p-Rad10p to the lesion by Rad14p is mediated via physical interaction of Rad14p with Rad1p but not Rad10p (2, 20). Rad14p has a zinc finger domain and recognizes DNA lesion (21, 22). Thus, Rad14p plays a crucial role in NER, and its absence dramatically impairs NER (1, 18, 20–23). Rad14p is highly conserved from yeast to humans. Its human homolog is XPA (XP group A) (24–26). Like in yeast, human XPA recognizes DNA lesion and plays a central role in NER (21, 24–28). Mutations in XPA manifest neurological disorders and XP (a high sun-sensitivity predisposing to skin cancer at an early age). In addition, a polymorphic site in the 5′-untranslated region of XPA has been identified. This polymorphism consists of an A to G substitution in the fourth nucleotide before the ATG start codon and has been implicated to be associated with different types of cancers including lung, skin, breast, gastric, head, and neck cancers (25, 29–50). Thus, XPA and its yeast homologue play critical roles in maintaining genome integrity and, hence, normal cellular functions.
Although Rad14p (or its human homologue) has well known functions in NER, its role in transcription has not been clearly elucidated. A previous study (51) has demonstrated the interaction of Rad14p with RPA and transcription factors such as RNA polymerase II and TFIIH. Furthermore, a recent study (52) has implicated the role of RPA in transcriptional regulation. These studies suggest that Rad14p might be involved in transcriptional regulation through its interaction with RPA, TFIIH, and RNA polymerase II. However, it is not clearly known whether Rad14p regulates transcription. Here, we have carried out a series of experiments including formaldehyde-based in vivo cross-linking and chromatin immunoprecipitation (ChIP), transcriptional, and mutational analyses in S. cerevisiae to determine Rad14p's function in transcription. Intriguingly, our results reveal that Rad14p associates with the active gene in the absence of lesion and promotes initial transcription. However, the steady-state or constitutive transcription is not altered in the absence of Rad14p. These results unveil a new role of Rad14p in transcriptional regulation in addition to its well-known function in NER.
EXPERIMENTAL PROCEDURES
Plasmids
The plasmids pRS403 and pRS406 (53) were used in the PCR-based disruption of RAD14 and coding sequence of GAL1. The plasmid pFA6a-13Myc-KanMX6 (54) was used for genomic tagging of the proteins of interest by Myc epitope.
Strains
The yeast (S. cerevisiae) strain FM391 (Research Genetics) was obtained from the Shilatifard laboratory (Ali Shilatifard; Stowers Institute for Medical Research). The RAD14 deletion mutant (YMR201C; Open Biosystems) and wild type (BY4741; Open Biosystems) strains were obtained from the Davie laboratory (Judith K. Davie, Southern Illinois University School of Medicine). The wild type strain W303a was obtained from the Green laboratory (Michael R. Green, University of Massachusetts Medical School). The yeast strain PCY25 (Δrad14::HIS3) was generated by deleting RAD14 from the ZDY2 (W303a expressing Myc-tagged Rad26p) strain using the pRS403 plasmid following PCR-based gene disruption protocol (54). Multiple Myc epitope tags were added to the C terminus of Rad14p at its chromosomal locus in W303a using the pFA6a-13Myc-KanMX6 plasmid to generate the ZDY3 strain. The coding sequence of GAL1 was deleted in the ZDY2 and PCY25 strains to generate the PCY31 and PCY32 strains, respectively, using the pRS406 plasmid. Multiple Myc epitope tags were added at the C terminus in the chromosomal locus of RAD3 in the FM391 strain to generate PCY2 using the pFA6a-13Myc-KanMX6 plasmid. The PCY35 strain was generated by deleting the RAD14 gene in the PCY2 strain following PCR-based gene disruption protocol (54). All these strains with genotypes are listed below in Table 1.
TABLE 1.
List of strains used in this study
| Strain name | Genotype | Source |
|---|---|---|
| BY4741 | MATa his3Δ1 TRP1 leu2Δ0 ura3Δ0 LYS2 met15Δ0 | Open Biosystems |
| YMR201C | MATa his3Δ1 TRP1 leu2Δ0 ura3Δ0 LYS2 met15Δ0 rad14Δ | Open Biosystems |
| FM391 | MATa his1Δ leu2Δ0 met15Δ0 ura3Δ0 | Research Genetics |
| PCY2 | MATa his1Δ leu2Δ0 met15Δ0 ura3Δ0 Rad3-myc(KanMX) | Malik et al. (59) |
| PCY35 | MATa his1Δ leu2Δ0 met15Δ0 ura3Δ0 Rad3-myc(KanMX) rad14Δ::URA3(pRS406) | This study |
| W303a | MATa his3-11, 15 trp1-1 leu2-3,112 ura3-1 ade2-1 can1-100 | The Green lab, UMass. |
| ZDY2 | MATa his3-11, 15 trp1-1 leu2-3,112 ura3-1 ade2-1 can1-100 Rad26-myc(KanMX) | Malik et al. (59) |
| PCY25 | MATa his3-11, 15 trp1-1 leu2-3,112 ura3-1 ade2-1 can1-100 Rad26-myc(KanMX) rad14Δ::HIS3(pRS403) | This study |
| ZDY3 | MATa his3-11, 15 trp1-1 leu2-3,112 ura3-1 ade2-1 can1-100 Rad14-myc(KanMX) | This study |
| PCY31 | MATa his3-11, 15 trp1-1 leu2-3,112 ura3-1 ade2-1 can1-100 Rad26-myc(KanMX) GAL1-ORFΔ::URA3(pRS406) | This study |
| PCY32 | MATa his3-11, 15 trp1-1 leu2-3,112 ura3-1 ade2-1 can1-100 Rad26-myc(KanMX) rad14Δ::HIS3(pRS403) GAL1-ORFΔ::URA3(pRS406) | This study |
Growth Media
For transcriptional induction of GAL1, GAL7, and GAL10, yeast strains were initially grown in YPR (yeast extract, peptone plus 2% raffinose) up to an A600 of 0.9 (without dilution) and then switched to YPG (yeast extract, peptone plus 2% galactose) for 90 min before formaldehyde-based in vivo cross-linking. For continuous induction of GAL genes, yeast cells were inoculated in YPG and grown at 30 °C up to an A600 of 1.0 before cross-linking for the ChIP assay or harvesting for mRNA analysis. For studies at ADH1, PGK1, PYK1, and RPS5, yeast cells were grown in YPD (yeast extract, peptone plus 2% dextrose) up to an A600 of 1.0. The CUP1 gene was induced by 1 mm CuSO4 for 15 min in synthetic complete medium (yeast nitrogen base and complete amino acid mixture plus 2% dextrose) at 30 °C. For studies at the CTT1 and STL1 genes, yeast cells were initially grown in synthetic complete medium up to an A600 of 0.9 and then were induced by 0.45 m NaCl for 7 min before cross-linking or harvesting for mRNA analysis.
ChIP Assay
The ChIP assay was performed as described previously (55–58). Briefly, yeast cells were treated with 1% formaldehyde, collected, and resuspended in lysis buffer. Subsequently, yeast cells were lysed by vortexing with the volume equivalent of glass beads at a maximum speed in a Tomy (MT-360) Vortex mixer at 4 °C for 30 min. The whole cell extract was collected by punching a hole at the bottom of the Eppendorf tube followed by centrifugation and sonicated by a Misonix sonicator for 10 s. The sonication was repeated 4 times with at least a 2 min pause in ice. After sonication, cell lysate (400 μl of lysate from 50 ml of yeast culture) was precleared by centrifugation, and then 100 μl lysate was used for each immunoprecipitation. Immunoprecipitated protein-DNA complexes were treated with proteinase K, the cross-links were reversed, and DNA was purified. Immunoprecipitated DNA was dissolved in 20 μl of TE 8.0 (10 mm Tris-HCl, pH 8.0, and 1 mm EDTA), and 1 μl of immunoprecipitated DNA was analyzed by PCR. PCR reactions contained [α-32P]dATP (2.5 μCi for 25 μl reaction), and the PCR products were detected by autoradiography after separation on a 6% polyacrylamide gel. As a control, “input” DNA was isolated from 5 μl of lysate without going through the immunoprecipitation step and dissolved in 100 μl of TE 8.0. To compare PCR signal arising from the immunoprecipitated DNA with the input DNA, 1 μl of input DNA was used in the PCR analysis. The PCR analysis was carried out within linear range using 23 PCR cycles as done previously (55, 56). The associations of Rad3p and Rad14p with gene were analyzed by modified ChIP assay as described in our recent publication (59). Briefly, 800 μl of lysate was prepared from 100 ml of yeast culture. 400 μl of lysate was used for each immunoprecipitation (using 10 μl of anti-HA (2 μg) or anti-myc (2 μg) antibody and 100 μl of protein A/G plus-agarose suspension (25 μl agarose beads) from Santa Cruz Biotechnology, Inc.), and immunoprecipitated DNA sample was dissolved in 10 μl of TE 8.0 for PCR analysis. In parallel, the PCR for input DNA was performed using 1 μl of DNA that was prepared by dissolving purified DNA from 5 μl of lysate in 100 μl of TE 8.0.
The primer pairs used for PCR analysis were as follows: GAL1(UAS), 5′-CGCTTAACTGCTCATTGCTATATTG-3′ and 5′-TTGTTCGGAGCAGTGCGGCGC-3′; GAL1 (Promoter), 5′-ATAGGATGATAATGCGATTAGTTTTTTAGCCTT-3′ and 5′-GAAAATGTTGAAAGTATTAGTTAAAGTGGTTATGCA-3′; GAL1 (ORF), 5′-CAGTGGATTGTCTTCTTCGGCCGC-3′ and 5′-GGCAGCCTGATCCATACCGCCATT-3′; GAL7 (Promoter), 5′-CTATGTTCAGTTAGTTTGGCTAGC-3′ and 5′-TTGATGCTCTGCATAATAATGCCC-3′; GAL7 (ORF), 5′-AAAGTGCAATCTGTGAGAGGCAATT-3′ and 5′-TTTTCTCTTGCTTCTCTGGAGAGAT-3′; GAL10 (Promoter), 5′-GCTAAGATAATGGGGCTCTTTACAT-3′ and 5′-TTTCACTTTGTAACTGAGCTGTCAT-3′; GAL10 (ORF), 5′-TTAATGCGAATCATAGTAGTATCGG-3′ and 5′-TTACCAATAGATCACCTGGAAATTC-3′; ADH1 (Promoter), 5′-GGTATACGGCCTTCCTTCCAGTTAC-3′ and 5′-GAACGAGAACAATGACGAGGAAACAAAAG-3′; ADH1 (ORF), 5′-CTGGTTACACCCACGACGGTTCTT-3′ and 5′-CAGACTTCAAAGCCTTGTAGACG3′; RPS5 (Promoter), 5′-GGCCAACTTCTACGCTCACGTTAG-3′ and 5′-CGGTGTCAGACATCTTTGGAATGGTC-3′; CTT1 (Promoter), 5′-GGCTGCAGGCTAGCCTAGCCGAT-3′ and 5′-GGAATAGAGGTAAAGCAACGACTTC-3′; CTT1 (ORF), 5′-TGCTGAACTGTCAGGCTCCCACC-3′ and 5′-GGGGAATTCCTTGTGTGGCCATATT-3′; STL1 (Promoter), 5′-ACCTTTGATAGGGCTTTCATTGGGGC-3′ and 5′-TCTAAGGCCAAGCAGCGTTGAAG-3′; STL1 (ORF), 5′-ACACTAGACGCGGATCCAAAT-3′ and 5′-AGCGTTACAACCAGTAAATTGCTGG-3′; CUP1 (Promoter), 5′-TCTTCTAGAAGCAAAAAGAGCGATG-3′ and 5′-CGCTGAACATTTTATGTGATGATTG3′; CUP1 (ORF), 5′-ACTTCCAAAATGAAGGTCATGAGTG-3′ and 5′-AGCAGCATGACTTCTTGGTTTCTTC-3′; PGK1 (Promoter), 5′-GAATCGTGTGACAACAACAGCCTG-3′ and 5′-CTTGCATTGACCAATTTATGC-3′; PGK1 (ORF), 5′-GACGAAGTTGTCAAGAGCTCTGC-3′ and 5′-GAAAGCAACACCTGGCAATTCCT-3′; PYK1 (Promoter), 5′-CTCGCCATCAAAACGATATTCG-3′ and 5′-TAACTTTGAAAGGGGACCATG-3′; PYK1 (ORF), 5′-AAGTTTCCGATGTCGGTAACGCTAT-3′ and 5′-TTGGCAAGTAAGCGATAGCTTGTTC-3′. UAS is the upstream activating sequence, and “promoter” is the core promoter.
Autoradiograms were scanned and quantitated by the National Institutes of Health Image 1.62 program. The ChIP signal is the ratio of immunoprecipitate over the input in the autoradiogram. The experiments were carried out multiple times. These experiments are biologically independent. The average ChIP signal of the biologically independent experiments is reported with standard deviation.
Both the wild type and Δrad14 mutant strains grew similarly in YPR up to an A600 of 0.9 before transfer to YPG for 90 min of transcriptional induction of GAL genes before cross-linking. Likewise, both the wild type and Δrad14 mutant strains grew similarly in YPD before cross-linking. Whole cell extract preparation and sonication for both the wild type and Δrad14 strains were carried out in parallel. Furthermore, the wild type and Δrad14 mutant strains had equal amounts of DNA after whole cell extract preparation and sonication as evident by equal input signals of the wild type and mutant strains.
Total RNA Preparation
The total RNA was prepared from yeast cell culture as described by Peterson et al. (60). Briefly, 10 ml of yeast culture was harvested and then suspended in 100 μl of RNA preparation buffer (500 mm NaCl, 200 mm Tris-HCl, 100 mm Na2EDTA, and 1% SDS) along with 100 μl of phenol/chloroform/isoamyl alcohol and 100 μl of a volume equivalent of glass beads (acid-washed; Sigma). Subsequently, yeast cell suspension was vortexed with a maximum speed (10 in VWR Mini-vortexer; catalog no. 58816-121) 5 times (30 s each). Cells suspension was put in ice for 30 s between pulses. After vortexing, 150 μl of RNA preparation buffer and 150 μl of phenol/chloroform/isoamyl alcohol were added to the yeast cell suspension followed by vortexing for 15 s with a maximum speed on VWR Mini-vortexer. The aqueous phase was collected after 5 min of centrifugation at a maximum speed in microcentrifuge machine. The total RNA was isolated from the aqueous phase by ethanol precipitation.
Reverse Transcriptase (RT)-PCR Analysis
RT-PCR analysis was performed according to the standard protocols (61). Briefly, total RNA was prepared from 10 ml of yeast culture. Ten micrograms of total RNA was used in the reverse transcription assay for both the wild type and mutant strains. RNA was treated with RNase-free DNase (M610A, Promega) and then reverse-transcribed into cDNA using oligo(dT) as described in the protocol supplied by Promega (A3800, Promega). PCR was performed using synthesized first strand as template and the primer pairs targeted to the GAL1, GAL7, GAL10, ADH1, PGK1, PYK1, RPS5, CTT1, STL1, and CUP1 ORFs. RT-PCR products were separated by 2.2% agarose gel electrophoresis and visualized by ethidium bromide staining. The primer pairs used in the PCR analysis were as follows: GAL1, 5′-CAGAGGGCTAAGCATGTGTATTCT-3′ and 5′-GTCAATCTCTGGACAAGAACATTC-3′; GAL7, 5′-TGAGACCTTGGTCATTTCAAAGAAG-3′ and 5′-ATGGATACCCATTGAGTATGGGAAA-3′; GAL10, 5′-TTAATGCGAATCATAGTAGTATCGG-3′ and 5′-TTACCAATAGATCACCTGGAAATTC-3′; ADH1, 5′-CGGTAACAGAGCTGACACCAGAGA-3′ and 5′-ACGTATCTACCAACGATTTGACCC-3′; RPS5, 5′-AGGCTCAATGTCCAATCATTGAAAG-3′ and 5′-CAACAACTTGGATTGGGTTTTGGTC-3′; CUP1, 5′-ACTTCCAAAATGAAGGTCATGAGTG-3′ and 5′-AGCAGCATGACTTCTTGGTTTCTTC-3′; CTT1, 5′-TGCTGAACTGTCAGGCTCCCACC-3′ and 5′-GGGGAATTCCTTGTGTGGCCATATT-3′; STL1, 5′-ACACTAGACGCGGATCCAAAT-3′ and 5′-AGCGTTACAACCAGTAAATTGCTGG-3′; PGK1, 5′-GACGAAGTTGTCAAGAGCTCTGC-3′ and 5′-GAAAGCAACACCTGGCAATTCCT-3′; PYK1, 5′-AAGTTTCCGATGTCGGTAACGCTAT-3′ and 5′-TTGGCAAGTAAGCGATAGCTTGTTC-3′.
Whole Cell Extract Preparation and Western Blot Analysis
For analysis of global levels of TBP, Rpb1p, Rad3p, and actin in the RAD14 deletion mutant and its isogenic wild type equivalent, the yeast cells were grown in YPR up to an A600 of 0.9 and then induced for 90 min in YPG. The harvested cells were lysed and sonicated to prepare the whole cell extract with solubilized chromatin following the protocol as described previously for the ChIP assay (55–58). The whole cell extract was run on SDS-polyacrylamide gel and then analyzed by Western blot. The anti-TBP (obtained from Michael R. Green, University of Massachusetts Medical School), anti-Rpb1p (8WG16; Covance), anti-myc (9E10; Santa Cruz Biotechnology), and anti-actin (A2066; Sigma) antibodies against TBP, Rpb1p, myc-tagged Rpb3p, and actin, respectively, were used for Western blot analysis.
RESULTS
Rad14p Associates with the Promoter and Coding Sequence of the Transcriptionally Active, but Not Inactive, GAL1 Gene
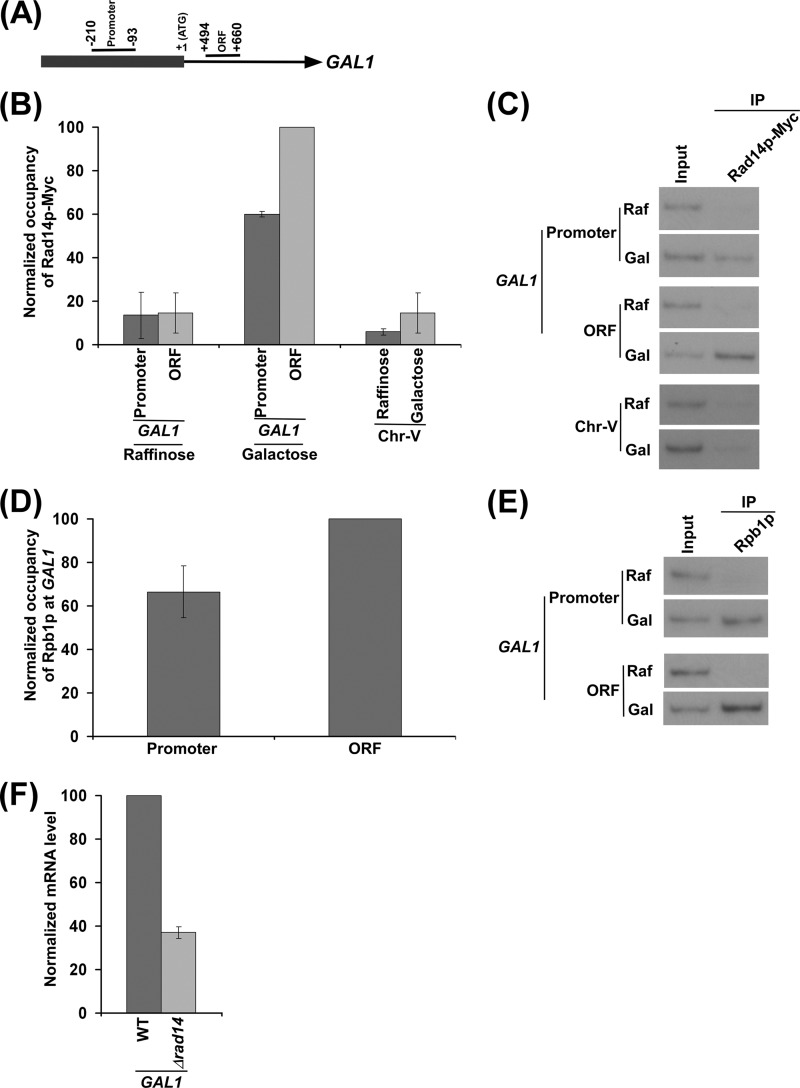
To determine whether Rad14p associates with active gene in the absence of lesion, we employed the ChIP assay to analyze the level of Rad14p at the promoter and coding sequence of a galactose-regulated gene, GAL1, under inducible (galactose-containing growth medium) and non-inducible (raffinose-containing growth medium) conditions. For the ChIP experiments, we tagged Rad14p by Myc epitope at the C terminus in its chromosomal locus in the W303a strain. The generated yeast strain expressing Myc epitope-tagged Rad14p (ZDY3) was grown in either raffinose- or galactose-containing growth medium up to an A600 of 1.0 before formaldehyde-based in vivo cross-linking. Subsequently, immunoprecipitation was performed using an anti-Myc antibody against Myc epitope-tagged Rad14p. Immunoprecipitated DNA was analyzed by PCR using the primer pairs targeted to the promoter and coding sequence (ORF) of GAL1 (Fig. 1A). A primer pair targeted to the transcriptionally inactive region of chromosome V was also used in the PCR analysis as a nonspecific DNA control (62). Our ChIP analysis revealed that Rad14p was associated with the promoter and coding sequence of GAL1 under transcriptionally inducible conditions in galactose-containing growth medium (Fig. 1, B and C). However, Rad14p did not associate with the GAL1 promoter and coding sequence (Fig. 1, B and C) when transcription of GAL1 was off in raffinose-containing growth medium (58, 63). Furthermore, Rad14p was not associated with the transcriptionally inactive region of chromosome V in both raffinose and galactose-containing growth media (Fig. 1, B and C). Together, these results support that Rad14p associates with the promoter and coding sequence of the transcriptionally active, but not inactive, GAL1 gene in the absence of DNA lesion.
FIGURE 1.
Rad14p is associated with the promoter and coding sequence of the GAL1 gene in a transcription-dependent manner. A, a schematic diagram shows the PCR primer pairs located at the promoter and coding sequence (or ORF) of the GAL1 gene in the ChIP assay. The numbers are presented with respect to the position of the first nucleotide of the initiation codon (+1). B, Rad14p is associated with the promoter and coding sequence of the active GAL1 gene. The yeast strain expressing Myc epitope-tagged Rad14p (ZDY3) was grown in raffinose (YPR)- or galactose (YPG)-containing growth medium up to an A600 of 1.0 before cross-linking. Immunoprecipitation was performed using a mouse monoclonal antibody against the c-myc epitope-tag (9E10; Santa Cruz Biotechnology). Primer pairs targeted to the promoter and coding sequence of the GAL1 gene and an inactive region of the chromosome-V (Chr-V) were used for PCR analysis of the immunoprecipitated DNA samples. The maximum ChIP signal was set to 100, and other ChIP signals were normalized with respect to 100. The normalized ChIP signals (represented as normalized occupancy) were plotted in the form of a histogram. C, the autoradiograms for the data presented in the panel B are shown. IP, immunoprecipitate. D, RNA polymerase II is associated with the promoter and coding sequence of the active GAL1 gene. The yeast strain was grown in YPR up to an A600 of 0.9 and then switched to YPG for 90 min before cross-linking. Immunoprecipitation was performed using a mouse monoclonal antibody 8WG16 (Covance) against the C-terminal domain of the largest subunit (Rpb1p) of RNA polymerase II. The maximum ChIP signal was set to 100, and another ChIP signal was normalized with respect to 100. The normalized ChIP signal was plotted in the form of a histogram. E, the autoradiograms for the data presented in the panel D. F, RT-PCR analysis. Both the wild type and Δrad14 strains were grown as in panel D. The Δrad14 strain (PCY25) was generated in the W303a wild type background. GAL1 mRNA level in the wild type strain was set to 100, and the mRNA level in the Δrad14 strain was normalized with respect to 100. The normalized mRNA level was plotted in the form of a histogram.
How does Rad14p associate with the transcriptionally active, but not inactive, GAL1 gene? It is quite likely that Rad14p interacts with RNA polymerase II, and such interaction plays an important role to bring down Rad14p to the active gene. In fact, a previous biochemical study demonstrated the interaction of Rad14p with RNA polymerase II (51). Furthermore, RNA polymerase II (Rpb1p) is associated with the promoter and coding sequence of GAL1 under inducible conditions (Fig. 1, D and E), consistent with previous studies (58, 63). However, RNA polymerase II does not associate with the GAL1 promoter and coding sequence under non-inducible conditions (Fig. 1E; Refs. 58 and 63). Thus, the interaction of RNA polymerase II with Rad14p is attributed for the association of Rad14p with the active gene. However, we observed a higher level of Rad14p association with the GAL1 coding sequence than the promoter. This could be due to a high level of RNA polymerase II at the GAL1 coding sequence in comparison to the promoter. Indeed, a high level of RNA polymerase II was observed at the GAL1 coding sequence when compared with the promoter (Fig. 1, D and E). Furthermore, the association of Rad14p with the active GAL1 gene can also be attributed to be mediated via RPA, as a previous biochemical study (51) demonstrated the interaction of Rad14p with RPA and a recent study (52) showed the association of RPA with active gene via its interaction with ssDNA at the transcription bubble.
Rad14p Promotes Transcription of GAL1
Because Rad14p is associated with the transcriptionally active GAL1 gene, it may play an important role to promote GAL1 transcription. To test this possibility, we analyzed GAL1 transcription in the Δrad14 strain (PCY25) using the RT-PCR assay. In this direction, we isolated total RNA from the wild type and Δrad14 strains after a 90-min transcriptional induction in galactose-containing growth medium. Subsequently, we analyzed GAL1 mRNA levels in these strains. Interestingly, we find that the absence of Rad14p significantly lowered the level of GAL1 mRNA (Fig. 1F). Thus, our results support the role of Rad14p in stimulation of GAL1 transcription.
Rad14p Enhances the Association of RNA Polymerase II with the Promoter and Coding Sequence of GAL1
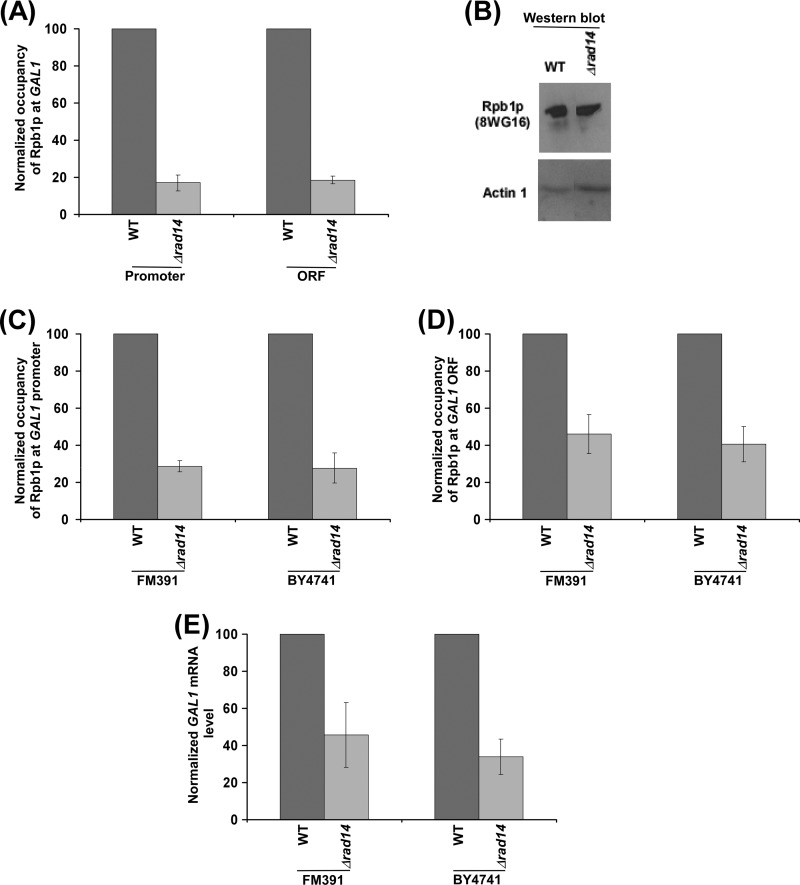
Because transcription of GAL1 was significantly reduced in the absence of Rad14p, it is likely that the association of RNA polymerase II with active GAL1 would be decreased in the Δrad14 strain in comparison to the wild type equivalent. To test this, we analyzed the association of RNA polymerase II with the GAL1 promoter and coding sequence in the wild type and Δrad14 (PCY25) strains using the ChIP assay. We find that the association of RNA polymerase II (Rpb1p) with the GAL1 promoter and coding sequence was significantly decreased in the Δrad14 strain (Fig. 2A). Such a reduction in the association of RNA polymerase II with GAL1 in the Δrad14 strain could be due to the decreased stability of Rpb1p. However, our Western blot analysis revealed that the global level of Rpb1p was not altered in the Δrad14 strain (Fig. 2B). The level of actin was monitored as a loading control (Fig. 2B). Thus, our results demonstrate that Rad14p promotes the association of RNA polymerase II with GAL1 (Fig. 2, A and B), and hence, the transcription of GAL1 was significantly decreased in the Δrad14 strain as compared with the wild type equivalent (Fig. 1F). However, there is a possibility that a hidden mutation, but not null, mutation of RAD14 in our strain reduced GAL1 transcription. To address this issue we have performed similar experiments using commercial Δrad14 strain (YMR201C; Open Biosystems) and its isogenic wild type equivalent (BY4741; Open Biosystems). We find that the association of RNA polymerase II with GAL1 was significantly impaired in this commercial Δrad14 strain as compared with the wild type equivalent (Fig. 2, C and D). Consistently, transcription of GAL1 was significantly decreased in this Δrad14 strain (Fig. 2E). Furthermore, we have generated a RAD14 deletion mutant strain (PCY35) in the FM391 wild type background and performed similar experiments. Again, using this new Δrad14 strain in the FM391 wild type background, we found that Rad14p promotes the association of RNA polymerase II with GAL1 (Fig. 2, C and D) and, hence, transcription (Fig. 2E). Collectively, we find a significant impairment of RNA polymerase II association with GAL1 and hence transcription in the Δrad14 strains generated from three different parental backgrounds (W303a, BY4741, and FM391), thus supporting the role of Rad14p in promoting transcription.
FIGURE 2.
Rad14p promotes the association of RNA polymerase II with GAL1. A, analysis of Rpb1p association with the GAL1 promoter and coding sequence in the Δrad14 strain (PCY25) and its isogenic wild type equivalent is shown. The wild type and Δrad14 strains were grown, cross-linked, and immunoprecipitated as in Fig. 1D. The maximum ChIP signal for the wild type strain was set to 100, and the ChIP signal of the Δrad14 strain was normalized with respect to 100. The normalized ChIP signal was plotted in the form of a histogram. B, Western blot analysis. C and D, analysis of Rpb1p association with GAL1 in the Δrad14 strains (PCY35 and YMR201C) derived from the FM391 and BY4741 wild type backgrounds is shown. Yeast strains were grown, cross-linked, and immunoprecipitated as in panel A. E, analysis of GAL1 transcription in the Δrad14 strains (PCY35 and YMR201C) is shown. Yeast strains were grown as in panel A.
Rad14p Promotes the Recruitment of TBP, TFIIH, and RNA Polymerase II to the GAL1 Promoter and Hence Transcriptional Initiation
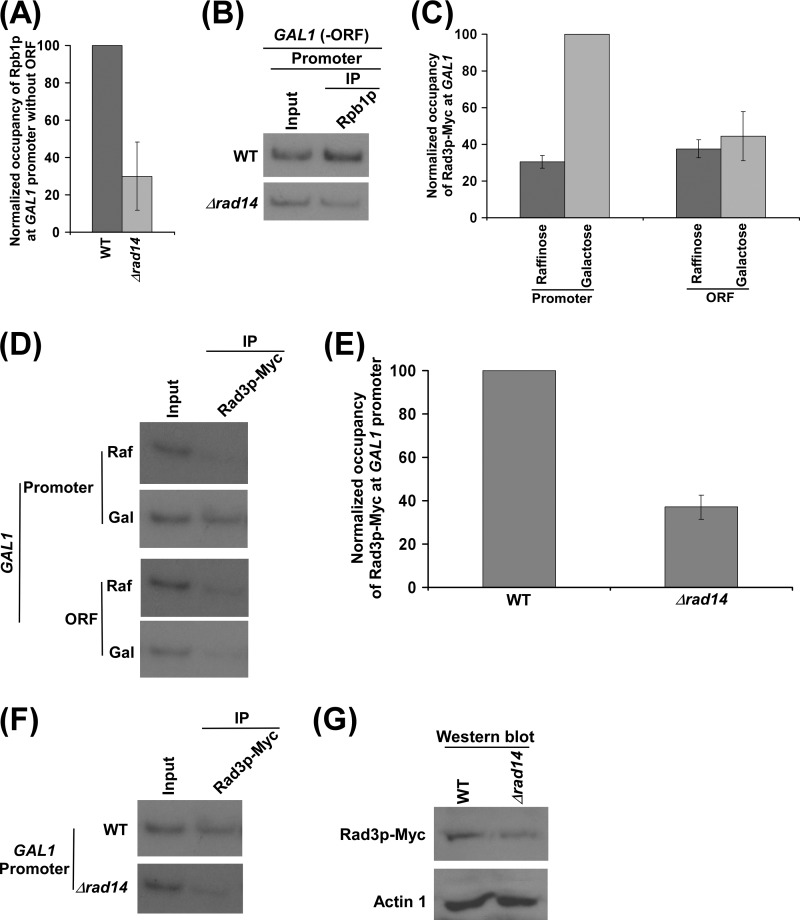
So far we find that the absence of Rad14p decreases the association of RNA polymerase II with the GAL1 promoter and coding sequence and hence transcription. Next, we asked whether the decrease in the association of RNA polymerase II with the GAL1 coding sequence in the Δrad14 strain is a result of the defect of RNA polymerase II recruitment to the promoter. To address this question, we analyzed the role of Rad14p in recruitment of initiating RNA polymerase II to the GAL1 promoter. In this direction, we deleted the coding sequence of GAL1 from its chromosomal locus and then analyzed the recruitment of RNA polymerase II to the GAL1 promoter in the presence and absence of Rad14p. We found that the recruitment of RNA polymerase II (Rpb1p) to the GAL1 promoter was significantly decreased in the absence of Rad14p, when the coding sequence was deleted (Fig. 3, A and B). This observation supports that Rad14p promotes the recruitment of RNA polymerase II to the GAL1 promoter (and hence transcriptional initiation). Otherwise, the recruitment of RNA polymerase II to the GAL1 promoter would not have been altered in the Δrad14 strain, when the coding sequence was deleted, consistent with our recent studies (64) demonstrating the role of Rad26p in promoting the association of elongating but not initiating RNA polymerase II with GAL1.
FIGURE 3.
Rad14p promotes the formation of the PIC assembly. A, regulation of Rpb1p occupancy at the GAL1 promoter by Rad14p in the absence of the coding sequence. Both the wild type and Δrad14 strains without GAL1 ORF (PCY31 and PCY32) were grown, cross-linked, and immunoprecipitated as in Fig. 1D. B, the autoradiograms for the data presented in panel A. C, Rad3p is predominantly associated with the GAL1 promoter. The yeast strain expressing Myc epitope-tagged Rad3p (PCY2) was grown, cross-linked, and immunoprecipitated as in Fig. 1B. The maximum ChIP signal was set to 100, and other ChIP signals were normalized with respect to 100. The normalized ChIP signals were plotted in the form of a histogram. D, the autoradiograms of the data presented in the panel C. E, Rad14p promotes the recruitment of Rad3p. Both the wild type and Δrad14 strains expressing myc-tagged Rad3p (PCY2 and PCY35) were grown and cross-linked as in Fig. 1D. F, the autoradiograms for the data presented in the panel E. G, Western blot analysis is shown.
How does Rad14p promote the association of RNA polymerase II with the GAL1 promoter? A previous biochemical study (51) demonstrated the interaction of Rad14p with RNA polymerase II and TFIIH. TFIIH is known to be recruited to the promoter after RNA polymerase II during the formation of the pre-initiation complex (PIC) (65–67). Because Rad14p interacts with both TFIIH and RNA polymerase II (51), it is likely to enhance their recruitment and/or stabilize the PIC. Furthermore, the decreased association of TFIIH with the promoter would weaken the transition of the initiating RNA polymerase II to the elongating form, leading to the low level of RNA polymerase II at the coding sequence, as previous studies (67–71) implicated TFIIH-mediated phosphorylation of RNA polymerase II for entering into the elongation phase. Thus, Rad14p is likely to promote/stabilize the PIC formation for transcriptional initiation. Therefore, we observed a significant decrease in the recruitment of RNA polymerase II to the GAL1 promoter in the Δrad14 strain in the absence of the coding sequence (Fig. 3, A and B). Because TFIIH joins the PIC assembly after RNA polymerase II, we would also expect to observe a decrease in the recruitment of TFIIH in the absence of Rad14p. To test this, we tagged the Rad3p component of TFIIH by Myc epitope in its chromosomal locus in the wild type and Δrad14 strains and subsequently performed the ChIP analysis at the GAL1 promoter. Our ChIP analysis revealed that the absence of Rad14p significantly lowered the recruitment of Rad3p to the GAL1 promoter (Fig. 3, C–F). However, such a decreased level of Rad3p to the GAL1 promoter was not due to an altered stability of Rad3p in the Δrad14 strain as evident by the Western blot analysis (Fig. 3G). Thus, our data support that the recruitment of TFIIH to the GAL1 promoter is significantly impaired in the absence of Rad14p.
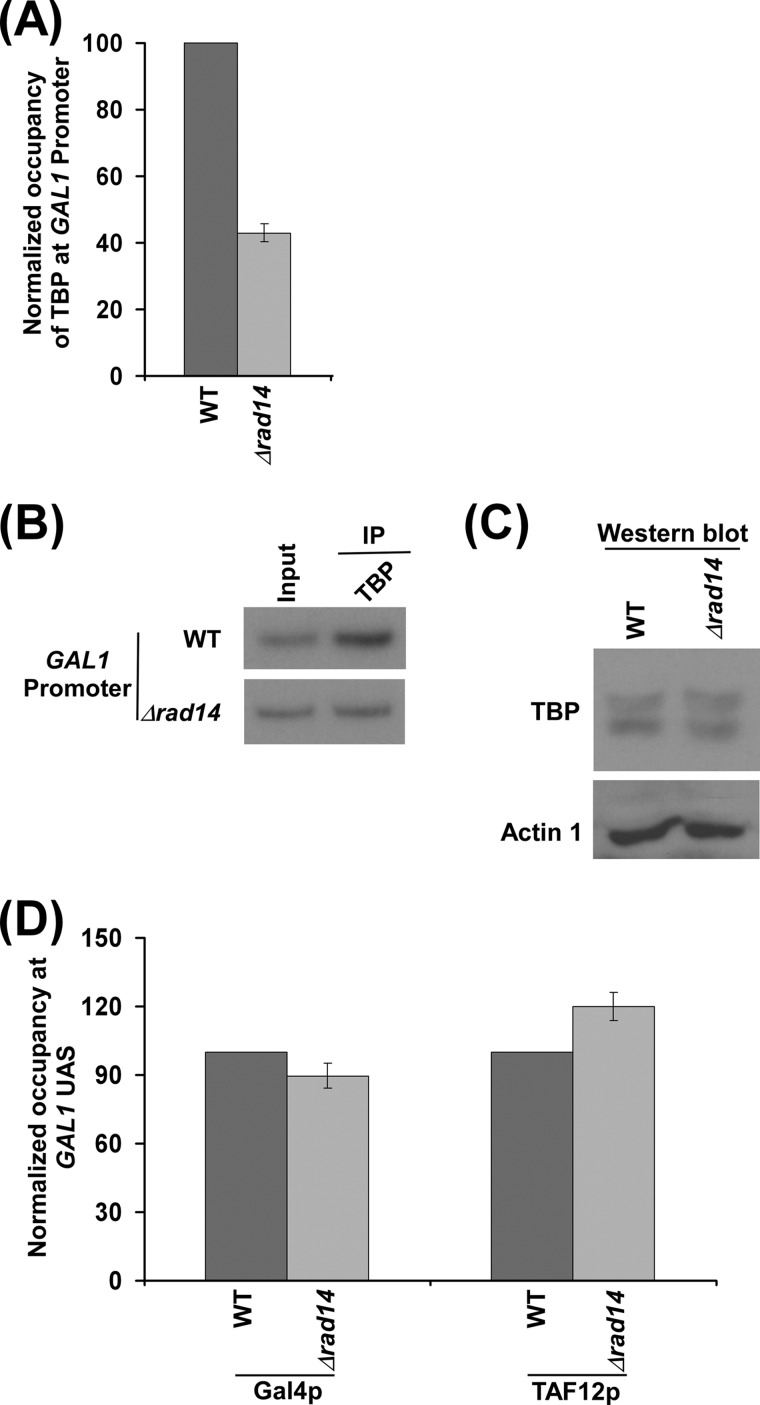
TBP nucleates the PIC formation, and RNA polymerase II and TFIIH join toward the end of the PIC assembly (65–67). Thus, the defect in the recruitment of RNA polymerase II and TFIIH during PIC formation may also have an effect on the recruitment of TBP due to the loss of reciprocal cooperativity. Indeed, we observe a decrease in the recruitment of TBP to the GAL1 promoter in the absence of Rad14p (Fig. 4, A and B). However, such a decreased level of TBP recruitment in the Δrad14 strain is not mediated via an altered stability of TBP as evident by the Western blot analysis (Fig. 4C). Taken together, our results reveal that Rad14p promotes the formation of the PIC, and hence a significantly decreased level of RNA polymerase II was observed at the GAL1 coding sequence in the absence of Rad14p (Fig. 2A). However, the decreased association of RNA polymerase II with the GAL1 coding sequence may also be due to the defect in transcriptional elongation in addition to an altered transcriptional initiation in the Δrad14 strain. This possibility remains to be further elucidated.
FIGURE 4.
Rad14p regulates the recruitment of TBP, but not Gal4p and SAGA, to GAL1. A, Rad14p facilitates the recruitment of TBP. Both the wild type and Δrad14 strains were grown and cross-linked as in Fig. 3E. Immunoprecipitation was performed using an anti-TBP antibody against TBP. B, the autoradiograms for the data presented in the panel A. C, Western blot analysis. D, Rad14p does not regulate the recruitment of Gal4p and SAGA (TAF12p) to the GAL1 UAS. Both the wild type and Δrad14 strains were grown and cross-linked as in Fig. 3E. Immunoprecipitations were performed using anti-Gal4p (Santa Cruz Biotechnology) and anti-TAF12p (Michael R. Green, University of Massachusetts Medical School) antibodies against Gal4p and TAF12p, respectively. Immunoprecipitated DNAs were analyzed by PCR using the primer pair targeted to the GAL1 UAS.
Next, we asked whether an impairment of the PIC formation at the GAL1 promoter in the absence of Rad14p is mediated via activator Gal4p or co-activator SAGA. To address this question, we analyzed the recruitment of Gal4p and SAGA (TAF12p) to the GAL1 UAS in the Δrad14 strain and its isogenic wild type equivalent. Our ChIP analysis revealed that the absence of Rad14p did not alter the recruitment of Gal4p and SAGA to the GAL1 UAS (Fig. 4D). Thus, Rad14p appears to promote the PIC formation directly (and hence transcription initiation) without altering the recruitment of activator or co-activator.
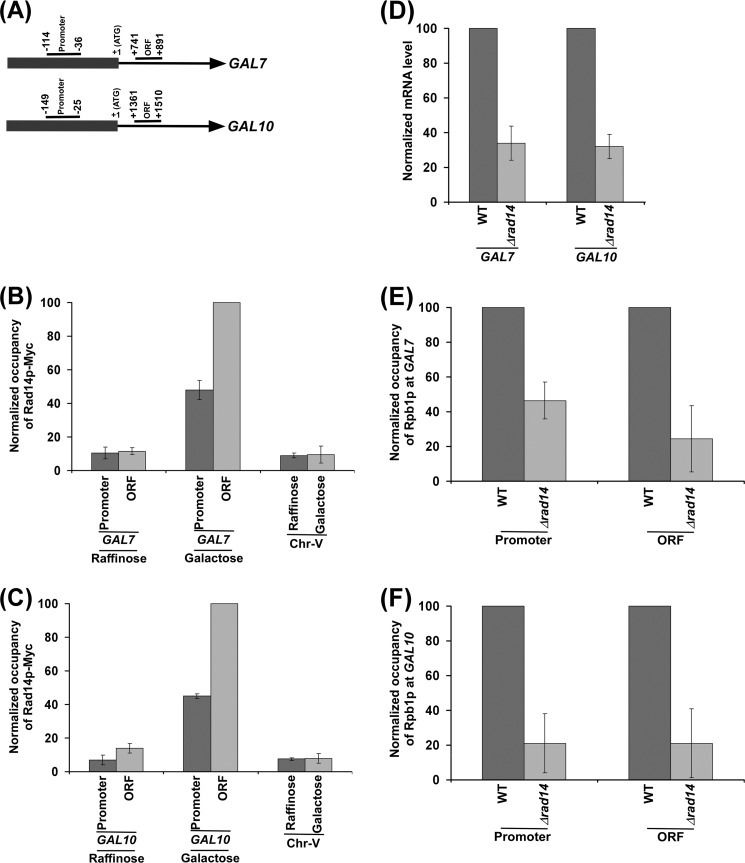
Rad14p Associates with the Active GAL7 and GAL10 Genes to Promote Transcription
We find above that Rad14p associates with the promoter and coding sequence of GAL1 and stimulates its transcription. To determine whether Rad14p also associates with other active genes and subsequently promotes their transcription, we analyzed the association of Rad14p with the promoters and coding sequences of other GAL genes such as GAL7 and GAL10 (Fig. 5A) under transcriptionally inducible and non-inducible conditions. Similar to the results at GAL1, we found that Rad14p associated with the promoters and coding sequences of GAL7 and GAL10 under transcriptionally inducible conditions (Fig. 5, B and C). However, Rad14p did not associate with these genes under non-inducible conditions (Fig. 5, B and C). Thus, Rad14p associates with active GAL7 and GAL10 genes in the absence of lesion. Next, we analyzed the role of Rad14p in regulation of transcription of GAL7 and GAL10 genes. In this direction, we isolated total RNA from the wild type and Δrad14 strains and then analyzed the levels of GAL7 and GAL10 mRNAs. We found that the levels of GAL7 and GAL10 mRNAs were significantly decreased in the absence of Rad14p when compared with the wild type equivalent (Fig. 5D). Thus, similar to the results at GAL1, we find that Rad14p promotes the transcription of GAL7 and GAL10. Consistently, the association of RNA polymerase II with the active GAL7 and GAL10 genes was significantly decreased in the absence of Rad14p (Fig. 5, E and F). Taken together, our results demonstrate that Rad14p associates with active GAL7 and GAL10 genes and promotes their transcription, similar to the results obtained at GAL1.
FIGURE 5.
Rad14p associates with the promoters and coding sequences of the GAL7 and GAL10 genes in a transcription-dependent manner. A, the schematic diagrams show the PCR primer pairs located at the promoters and ORFs of the GAL7 and GAL10 genes in the ChIP assay. The numbers are presented with respect to the position of the first nucleotide of the initiation codon (+1). B and C, Rad14p associates with the promoters and coding sequences of the active GAL7 and GAL10 genes. The yeast strain expressing Myc epitope-tagged Rad14p (ZDY3) was grown, cross-linked, and immunoprecipitated as in Fig. 1B. D, RT-PCR analysis is shown. Both the wild type and Δrad14 strains (ZDY2 and PCY25) were grown for analysis of GAL7 and GAL10 transcriptions as in Fig. 1F. E and F, analysis of Rpb1p association with the promoters and coding sequences of the GAL7 and GAL10 genes in the Δrad14 strain (PCY25) and its isogenic wild type equivalent. The wild type and Δrad14 strains were grown, cross-linked, and immunoprecipitated as in Fig. 2A.
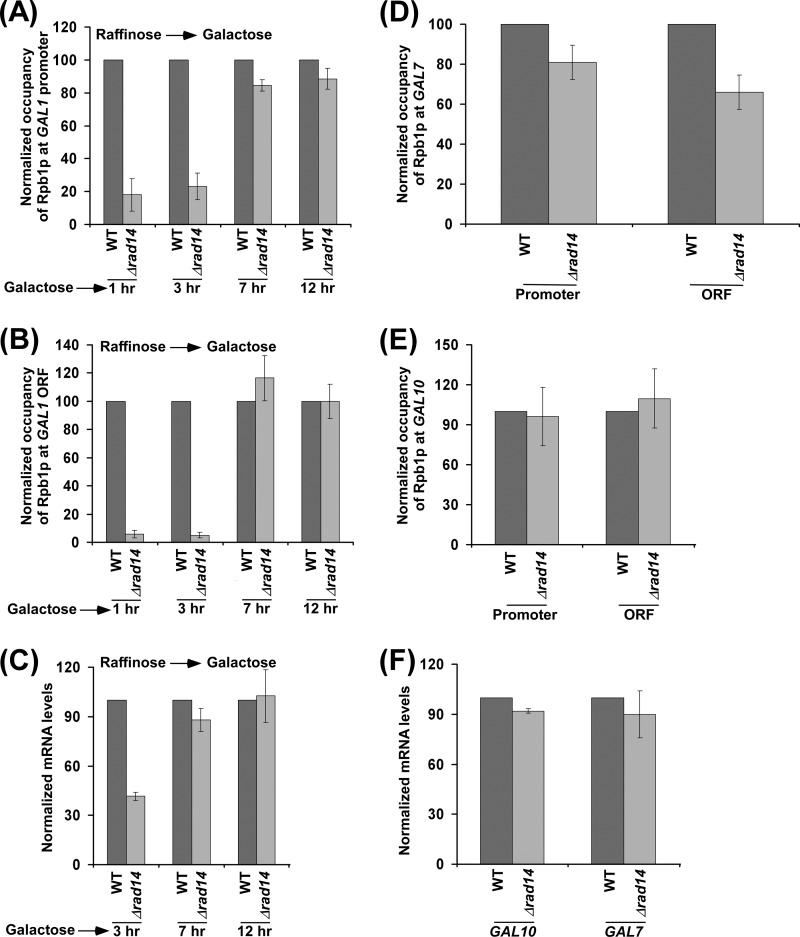
Effect of Rad14p on the Steady-state Level of Transcription
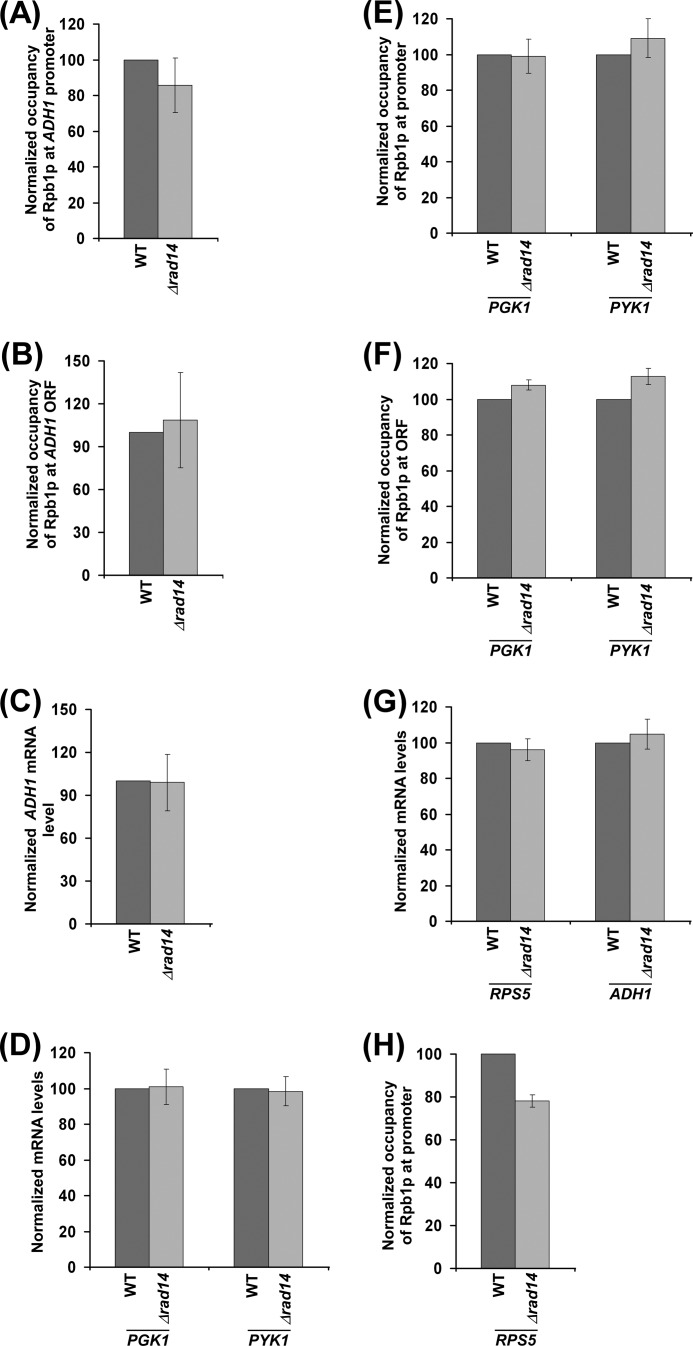
We next asked whether the occupancy of RNA polymerase II at the GAL gene in the Δrad14 strain could reach to the wild type level if given enough time for attaining the steady state. To address this question, we analyzed the association of RNA polymerase II with GAL1 at different times (1, 3, 7, and 12 h) after transcriptional induction. Fig. 6, A and B, show that the association of RNA polymerase II with GAL1 in the Δrad14 strain reached to the wild type level after long transcriptional induction (e.g. 7 and 12 h). Consistently, transcription of GAL1 in the Δrad14 strain became the same as the wild type level after longer transcriptional induction (Fig. 6C). Likewise, we found that the effect of Rad14p on the association of RNA polymerase II with the GAL7 and GAL10 promoters and coding sequences (and hence transcription) was minimal (or absent) after a long induction in galactose-containing growth medium (Fig. 6, D–F). Therefore, it is expected that there would not be a dramatic effect of Rad14p on the association of RNA polymerase II (and hence transcription) with constitutively active genes. Indeed, we found that Rad14p did not alter the association of RNA polymerase II with a constitutively active gene, ADH1, in dextrose-containing growth medium (Fig. 7, A and B). Therefore, transcription of ADH1 was not changed in the absence of Rad14p (Fig. 7C). Likewise, we found that the transcription of other constitutively active genes such as PGK1 and PYK1 was not changed in the absence of Rad14p (Fig. 7D). Therefore, the association of RNA polymerase II with these genes was not changed in the Δrad14 strain (Fig. 7, E and F). Similar results were also obtained at another constitutively active gene, RPS5 (Fig. 7, G and H). Thus, Rad14p appears to promote the initial rate of transcription. However, its effect is minimal or absent on the steady-state level or constitutive transcription.
FIGURE 6.
Effect of Rad14p on the association of RNA polymerase II with GAL genes under steady-state conditions. A and B, kinetic analysis for the association of Rpb1p with the GAL1 promoter and coding sequence following transcriptional induction. Yeast cells were initially grown in YPR at 30 °C and then switched to YPG for different induction time periods before cross-linking. C, RT-PCR analysis is shown. D and E, analysis of Rpb1p association with the GAL7 and GAL10 promoters and coding sequences in the Δrad14 strain (PCY25) and its isogenic wild type equivalent after continuous growth in YPG up to an A600 of 1.0 before cross-linking. F, RT-PCR analysis is shown.
FIGURE 7.
Effect of Rad14p on transcription of constitutively active genes. A and B, analysis of Rpb1p association with the ADH1 promoter and coding sequence in the Δrad14 strain (PCY25) and its isogenic wild type equivalent following continuous growth in YPD up to an A600 of 1.0 before cross-linking. C, RT-PCR analysis of ADH1 is shown. D, RT-PCR analysis of PGK1 and PYK1. E and F, analysis of Rpb1p association with the promoters and coding sequences of PGK1 and PYK1 in the wild type and Δrad14 strains. G, RT-PCR analysis of RPS5. H, analysis of Rpb1p association with RPS5 in the wild type and Δrad14 strains.
Rad14p Promotes Transcriptional Induction of Non-GAL Genes
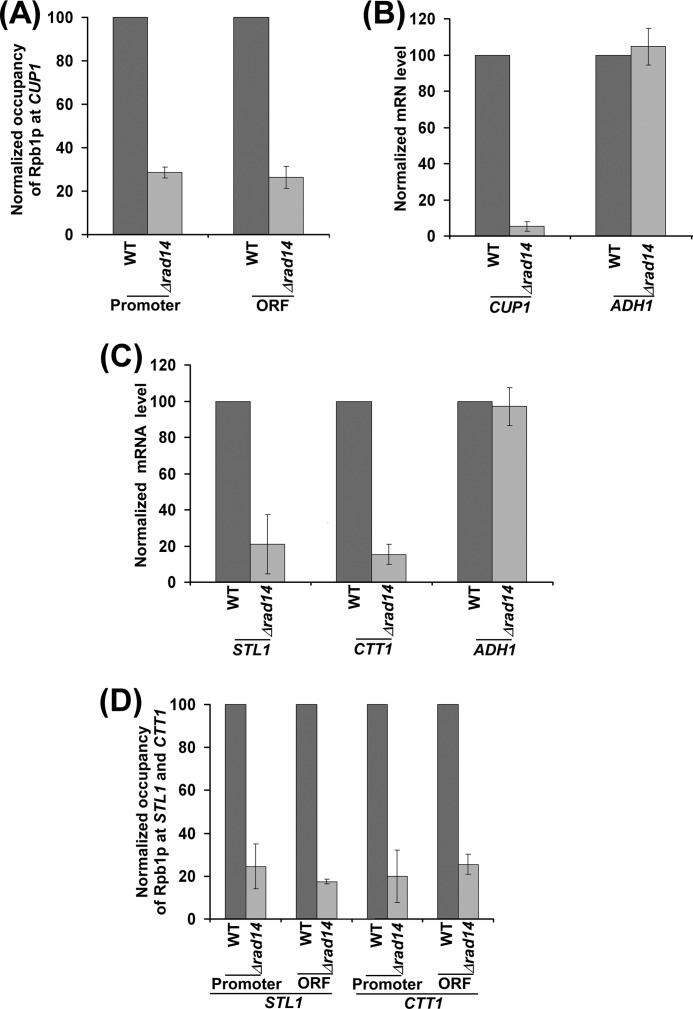
So far, we have demonstrated that Rad14p promotes transcriptional induction of GAL genes but not the steady-state level or constitutive transcription. To determine whether Rad14p can also promote transcriptional induction of other non-GAL gene, we analyzed the association of RNA polymerase II with an inducible CUP1 gene in the presence of CuSO4 in the wild type and Δrad14 strains. Fig. 8A shows that the occupancy of RNA polymerase II at CUP1 was significantly impaired in the Δrad14 strain. Consistently, transcription of CUP1 was also dramatically reduced after transcriptional induction in the absence of Rad14p (Fig. 8B). ADH1 transcript levels in the wild type and Δrad14 strains were monitored under the similar inducible growth conditions as controls. As expected, transcription of the constitutively active ADH1 gene was not altered in the absence of Rad14p (Fig. 8B). Thus, Rad14p promotes transcription of CUP1 after transcriptional induction. Likewise, Rad14p promotes transcription of STL1 and CTT1 after transcriptional induction by NaCl (Fig. 8C). Consistently, the association of RNA polymerase II with these genes was also significantly impaired in the absence of Rad14p (Fig. 8D). Together, our data support that Rad14p promotes transcriptional induction of GAL as well as non-GAL genes.
FIGURE 8.
Rad14p promotes transcriptional induction of CUP1, STL1, and CTT1. A, Rad14p promotes the association of RNA polymerase II with CUP1. B, RT-PCR analysis of CUP1. C, RT-PCR analysis of STL1 and CTT1 in the wild type and Δrad14 strains following transcriptional inductions by NaCl is shown. D, analysis of Rpb1p association with STL1 and CTT1 in the wild type and Δrad14 strains.
DISCUSSION
Previous studies have implicated Rad14p in recognizing DNA lesions (21, 22), and subsequently Rad14p interacts with DNA repair factors for NER (1, 18, 20–23). Like Rad14p, Rad26p recognizes the DNA lesion at the active gene (59) and promotes repair (72). Intriguingly, Rad26p also associates with active gene in a transcription or elongating RNA polymerase II-dependent manner in the absence of lesion and promotes transcriptional elongation (59, 64). However, it is not clearly known whether, like Rad26p, the DNA lesion recognition factor Rad14p can also associate with active gene in the absence of lesion and promote transcription. Here, we demonstrate that like Rad26p, Rad14p associates with active gene independently of lesion. Such an association of Rad14p with active genes promotes transcription. Thus, this study demonstrates a new role of Rad14p in promoting transcription in addition to its well known function in NER.
Although Rad14p has been implicated to recognize DNA lesion (21, 22), it has not been investigated whether Rad14p can also interact with active genomic region in the absence of lesion. Here, we show that Rad14p is not recruited to the promoters and coding sequences of several GAL genes when these genes are transcriptionally silent or not induced in raffinose-containing growth medium (Figs. 1, B and C, and 5, B and C). Intriguingly, we find that Rad14p associates with the promoters and coding sequences of several GAL genes when these genes are transcriptionally active in galactose-containing growth medium (Figs. 1, B and C, and 5, B and C). These observations support that Rad14p associates with active GAL genes even in the absence of lesion.
How does Rad14p associate with the promoter and coding sequence of the active gene? Because Rad14p, like RNA polymerase II, associates with the promoter and coding sequence of the active gene, it is likely that Rad14p interacts with RNA polymerase II and gets recruited to the active gene. In support of this possibility, a previous biochemical study (51) revealed the interaction of Rad14p with the largest subunit (Rpb1p) of RNA polymerase II. Thus, the interaction of Rad14p with RNA polymerase II (51) is attributed for the association of Rad14p with the active gene in the absence of lesion. Likewise, RNA polymerase II has been recently shown to play an important role to bring down Rad26p to the coding sequence of active gene (59, 64). However, unlike Rad14p, Rad26p does not associate with the promoter (59). Thus, Rad26p does not interact with initiating RNA polymerase II but rather with elongating RNA polymerase II (59). Therefore, Rad26p associates with the active gene in an elongating RNA polymerase II-dependent manner (59). On the other hand, Rad14p appears to associate with the active gene in initiating and elongating RNA polymerase II-dependent manner. Furthermore, Rad14p has been previously shown to interact with RPA (51), which associates with active gene (52). Thus, the interaction of RPA with Rad14p is also likely to play an important role to bring down Rad14p to the active gene in the absence of lesion.
Because Rad14p associates with the active gene, it is likely to enhance transcription. Indeed, our data demonstrate that the transcription of several GAL genes is significantly decreased in the absence of Rad14p after 90 min of transcriptional induction in galactose-containing growth medium (Figs. 1F and 5D). Similarly, Rad14p promotes transcriptional induction of non-GAL genes such as CUP1, CTT1, and STL1 (Fig. 8, B and C). These results support the role of Rad14p in facilitation of transcription. Likewise, human homologue of Rad14p has recently been shown to associate with the RARβ2 promoter to facilitate transcription after all-trans retinoic acid induction in HeLa cell lines (73). However, in a striking contrast to our results and recent study (73) in HeLa cell lines, Prakash and co-workers (74, 75) did not find a change in GAL transcription in the absence of Rad14p in their Northern blot analysis. Such contradictory results raise the possibility that either our strain or the strain used in the Prakash laboratory contained a hidden mutation that impaired transcription in our study or suppressed Δrad14 phenotype in the previous studies of Prakash and co-workers (74, 75). To address this issue, we have carried out experiments using three Δrad14 mutants generated from three different parental backgrounds (W303a, BY4741, and FM391). We find that Rad14p promotes GAL1 transcription (Figs. 1F and 2, A, C, D, and E). Thus, the role of Rad14p in promoting transcription does not appear to be due to a hidden mutation in our strain. Furthermore, the growth conditions in the previous studies of Prakash and co-workers (74, 75) were different from our study. Moreover, the strong signals (or exposures) in the reported northern blots of Prakash and co-workers (74, 75) may not adequately show up a change of transcriptional defect. These factors might have led to the discrepancy in our results and previous studies of Prakash and co-workers (74, 75). Nonetheless, our ChIP and RT-PCR data support the function of Rad14p in promoting transcription, consistent with the recently reported role of its homologue in HeLa cell lines (73).
How does Rad14p regulate transcription? A previous biochemical study (51) demonstrated the interaction of Rad14p with RNA polymerase II and TFIIH. Both RNA polymerase II and TFIIH are involved in transcriptional initiation (65–67). TFIIH has a kinase activity that phosphorylates serine 5 at the C-terminal domain of the Rpb1p subunit of RNA polymerase II. Such phosphorylation is essential for RNA polymerase II to enter into the elongation phase of transcription (68–71). Transcription is initiated via the formation of PIC by RNA polymerase II and a set of general transcription factors (GTFs) such as TBP, TFIIB, TFIIE, TFIIF, and TFIIH. TBP nucleates the assembly of GTFs to form the PIC. RNA polymerase II and TFIIH join the PIC toward the end to initiate transcription. The fact that Rad14p interacts with TFIIH and RNA polymerase II suggests that the absence of Rad14p would lower the formation of the PIC and, hence, transcriptional initiation. Indeed, we find that the absence of Rad14p significantly lowers the association of RNA polymerase II, TFIIH, and TBP with the GAL1 promoter (Figs. 2A, 3, E and F, and 4, A and B). Furthermore, we show that the recruitment of RNA polymerase II to the GAL1 promoter is significantly decreased in the Δrad14 strain, when the coding sequence is deleted (Fig. 3, A and B). These results support the role of Rad14p in promoting the PIC formation (and hence transcriptional initiation). Furthermore, we find that Rad14p associates with the active coding sequence (Figs. 1, B and C, and 5, B and C). Hence, Rad14p may have a hidden role in transcriptional elongation, and such function may be mediated via RPA as RPA interacts with Rad14p (51) and regulates transcriptional elongation (52). This possibility remains to be further elucidated. Nonetheless, our results demonstrate the role of a DNA repair factor, Rad14p, in stimulation of transcription in addition to its well known function in NER, thus implicating Rad14p as an important regulator of gene expression.
Acknowledgments
We thank Michael R. Green for anti-TBP and anti-TAF12p antibodies and Ali Shilatifard and Judy Davie for yeast strains.
This work was supported, in whole or in part, by National Institutes of Health Grant 1R15GM088798-01. This work was also supported by American Heart Association (Greater Midwest Affiliate) Grant-in-aid 10GRNT4300059, a Mallinckrodt Foundation grant, and Excellence in Academic Medicine (EAM) grants from Southern Illinois University School of Medicine.
- NER
- Nucleotide excision repair
- XP
- xeroderma pigmentosum
- XPA
- XP group A
- RPA
- replication protein A
- PIC
- pre-initiation complex
- UAS
- upstream activating sequence
- TBP
- TATA-box binding protein.
REFERENCES
- 1. Prakash S., Prakash L. (2000) Nucleotide excision repair in yeast. Mutat. Res. 451, 13–24 [DOI] [PubMed] [Google Scholar]
- 2. Guzder S. N., Sung P., Prakash L., Prakash S. (1996) Nucleotide excision repair in yeast is mediated by sequential assembly of repair factors and not by a pre-assembled repairosome. J. Biol. Chem. 271, 8903–8910 [DOI] [PubMed] [Google Scholar]
- 3. de Laat W. L., Jaspers N. G., Hoeijmakers J. H. (1999) Molecular mechanism of nucleotide excision repair. Genes Dev. 13, 768–785 [DOI] [PubMed] [Google Scholar]
- 4. Vermeulen W. (2011) Dynamics of mammalian NER proteins. DNA Repair 10, 760–771 [DOI] [PubMed] [Google Scholar]
- 5. Bergoglio V., Magnaldo T. (2006) Nucleotide excision repair and related human diseases. Genome Dyn. 1, 35–52 [DOI] [PubMed] [Google Scholar]
- 6. Nouspikel T. (2008) Nucleotide excision repair and neurological diseases. DNA Repair 7, 1155–1167 [DOI] [PubMed] [Google Scholar]
- 7. Shuck S. C., Short E. A., Turchi J. J. (2008) Eukaryotic nucleotide excision repair. From understanding mechanisms to influencing biology. Cell Res. 18, 64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Costa R. M., Chiganças V., Galhardo Rda S., Carvalho H., Menck C. F. (2003) The eukaryotic nucleotide excision repair pathway. Biochimie 85, 1083–1099 [DOI] [PubMed] [Google Scholar]
- 9. Leibeling D., Laspe P., Emmert S. (2006) Nucleotide excision repair and cancer. J. Mol. Histol. 37, 225–238 [DOI] [PubMed] [Google Scholar]
- 10. Andressoo J. O., Hoeijmakers J. H., Mitchell J. R. (2006) Nucleotide excision repair disorders and the balance between cancer and aging. Cell Cycle 5, 2886–2888 [DOI] [PubMed] [Google Scholar]
- 11. Lichon V., Khachemoune A. (2007) Xeroderma pigmentosum. Beyond skin cancer. J. Drugs Dermatol. 6, 281–288 [PubMed] [Google Scholar]
- 12. Lockett K. L., Snowhite I. V., Hu J. J. (2005) Nucleotide-excision repair and prostate cancer risk. Cancer Lett. 220, 125–135 [DOI] [PubMed] [Google Scholar]
- 13. Magnaldo T., Sarasin A. (2004) Xeroderma pigmentosum. From symptoms and genetics to gene-based skin therapy. Cells Tissues Organs 177, 189–198 [DOI] [PubMed] [Google Scholar]
- 14. Lagerwerf S., Vrouwe M. G., Overmeer R. M., Fousteri M. I., Mullenders L. H. (2011) DNA damage response and transcription. DNA Repair 10, 743–750 [DOI] [PubMed] [Google Scholar]
- 15. Guzder S. N., Habraken Y., Sung P., Prakash L., Prakash S. (1995) Reconstitution of yeast nucleotide excision repair with purified Rad proteins, replication protein A, and transcription factor TFIIH. J. Biol. Chem. 270, 12973–12976 [DOI] [PubMed] [Google Scholar]
- 16. Habraken Y., Sung P., Prakash L., Prakash S. (1993) Yeast excision repair gene RAD2 encodes a single-stranded DNA endonuclease. Nature 366, 365–368 [DOI] [PubMed] [Google Scholar]
- 17. Sung P., Reynolds P., Prakash L., Prakash S. (1993) Purification and characterization of the Saccharomyces cerevisiae RAD1/RAD10 endonuclease. J. Biol. Chem. 268, 26391–26399 [PubMed] [Google Scholar]
- 18. Guzder S. N., Sommers C. H., Prakash L., Prakash S. (2006) Complex formation with damage recognition protein Rad14 is essential for Saccharomyces cerevisiae Rad1-Rad10 nuclease to perform its function in nucleotide excision repair in vivo. Mol. Cell. Biol. 26, 1135–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Habraken Y., Sung P., Prakash S., Prakash L. (1996) Transcription factor TFIIH and DNA endonuclease Rad2 constitute yeast nucleotide excision repair factor 3. Implications for nucleotide excision repair and Cockayne syndrome. Proc. Natl. Acad. Sci. U.S.A. 93, 10718–10722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mardiros A., Benoun J. M., Haughton R., Baxter K., Kelson E. P., Fischhaber P. L. (2011) Rad10-YFP focus induction in response to UV depends on RAD14 in yeast. Acta Histochem. 113, 409–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Guzder S. N., Sung P., Prakash L., Prakash S. (1993) Yeast DNA-repair gene RAD14 encodes a zinc metalloprotein with affinity for ultraviolet-damaged DNA. Proc. Natl. Acad. Sci. U.S.A. 90, 5433–5437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jones G. W., Reed S. H., Waters R. (1997) Characterization of the rad14–2 mutant of Saccharomyces cerevisiae. Implications for the recognition of UV photoproducts by the Rad14 protein. Yeast 13, 31–36 [DOI] [PubMed] [Google Scholar]
- 23. Aboussekhra A., Biggerstaff M., Shivji M. K., Vilpo J. A., Moncollin V., Podust V. N., Proti M., Hübscher U., Egly J.-M., Wood R. D. (1995) Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell 80, 859–868 [DOI] [PubMed] [Google Scholar]
- 24. Rademakers S., Volker M., Hoogstraten D., Nigg A. L., Moné M. J., Van Zeeland A. A., Hoeijmakers J. H., Houtsmuller A. B., Vermeulen W. (2003) Xeroderma pigmentosum group A protein loads as a separate factor onto DNA lesions. Mol. Cell. Biol. 23, 5755–5767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wu X., Zhao H., Wei Q., Amos C. I., Zhang K., Guo Z., Qiao Y., Hong W. K., Spitz M. R. (2003) XPA polymorphism associated with reduced lung cancer risk and a modulating effect on nucleotide excision repair capacity. Carcinogenesis 24, 505–509 [DOI] [PubMed] [Google Scholar]
- 26. Bankmann M., Prakash L., Prakash S. (1992) Yeast RAD14 and human xeroderma pigmentosum group A DNA-repair genes encode homologous proteins. Nature 355, 555–558 [DOI] [PubMed] [Google Scholar]
- 27. Reardon J. T., Sancar A. (2002) Molecular anatomy of the human excision nuclease assembled at sites of DNA damage. Mol. Cell. Biol. 22, 5938–5945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Reardon J. T., Sancar A. (2003) Recognition and repair of the cyclobutane thymine dimer, a major cause of skin cancers, by the human excision nuclease. Genes Dev. 17, 2539–2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu J., Zhang Z., Cao X. L., Lei D. P., Wang Z. Q., Jin T., Pan X. L. (2012) XPA A23G polymorphism and susceptibility to cancer. A meta-analysis. Mol. Biol. Rep. 39, 6791–6799 [DOI] [PubMed] [Google Scholar]
- 30. Park J. Y., Park S. H., Choi J. E., Lee S. Y., Jeon H. S., Cha S. I., Kim C. H., Park J. H., Kam S., Park R. W., Kim I. S., Jung T. H. (2002) Polymorphisms of the DNA repair gene xeroderma pigmentosum group A and risk of primary lung cancer. Cancer Epidemiol. Biomarkers Prev. 11, 993–997 [PubMed] [Google Scholar]
- 31. Butkiewicz D., Popanda O., Risch A., Edler L., Dienemann H., Schulz V., Kayser K., Drings P., Bartsch H., Schmezer P. (2004) Association between the risk for lung adenocarcinoma and a (-4) G-to-A polymorphism in the XPA gene. Cancer Epidemiol. Biomarkers Prev. 13, 2242–2246 [PubMed] [Google Scholar]
- 32. Vogel U., Overvad K., Wallin H., Tjønneland A., Nexø B. A., Raaschou-Nielsen O. (2005) Combinations of polymorphisms in XPD, XPC and XPA in relation to risk of lung cancer. Cancer Lett. 222, 67–74 [DOI] [PubMed] [Google Scholar]
- 33. Zienolddiny S., Campa D., Lind H., Ryberg D., Skaug V., Stangeland L., Phillips D. H., Canzian F., Haugen A. (2006) Polymorphisms of DNA repair genes and risk of non-small cell lung cancer. Carcinogenesis 27, 560–567 [DOI] [PubMed] [Google Scholar]
- 34. De Ruyck K., Szaumkessel M., De Rudder I., Dehoorne A., Vral A., Claes K., Velghe A., Van Meerbeeck J., Thierens H. (2007) Polymorphisms in base-excision repair and nucleotide-excision repair genes in relation to lung cancer risk. Mutat. Res. 631, 101–110 [DOI] [PubMed] [Google Scholar]
- 35. Raaschou-Nielsen O., Sørensen M., Overvad K., Tjønneland A., Vogel U. (2008) Polymorphisms in nucleotide excision repair genes, smoking, and intake of fruit and vegetables in relation to lung cancer. Lung Cancer 59, 171–179 [DOI] [PubMed] [Google Scholar]
- 36. Qian B., Zhang H., Zhang L., Zhou X., Yu H., Chen K. (2011) Association of genetic polymorphisms in DNA repair pathway genes with non-small cell lung cancer risk. Lung Cancer 73, 138–146 [DOI] [PubMed] [Google Scholar]
- 37. Sugimura T., Kumimoto H., Tohnai I., Fukui T., Matsuo K., Tsurusako S., Mitsudo K., Ueda M., Tajima K., Ishizaki K. (2006) Gene-environment interaction involved in oral carcinogenesis. Molecular epidemiological study for metabolic and DNA repair gene polymorphisms. J. Oral. Pathol. Med. 35, 11–18 [DOI] [PubMed] [Google Scholar]
- 38. Bau D. T., Tsai M. H., Huang C. Y., Lee C. C., Tseng H. C., Lo Y. L., Tsai Y., Tsai F. J. (2007) Relationship between polymorphisms of nucleotide excision repair genes and oral cancer risk in Taiwan. Evidence for modification of smoking habit. Chin J. Physiol. 50, 294–300 [PubMed] [Google Scholar]
- 39. Abbasi R., Ramroth H., Becher H., Dietz A., Schmezer P., Popanda O. (2009) Laryngeal cancer risk associated with smoking and alcohol consumption is modified by genetic polymorphisms in ERCC5, ERCC6, and RAD23B but not by polymorphisms in five other nucleotide excision repair genes. Int. J. Cancer 125, 1431–1439 [DOI] [PubMed] [Google Scholar]
- 40. Jelonek K., Gdowicz-Klosok A., Pietrowska M., Borkowska M., Korfanty J., Rzeszowska-Wolny J., Widlak P. (2010) Association between single-nucleotide polymorphisms of selected genes involved in the response to DNA damage and risk of colon, head and neck, and breast cancers in a Polish population. J. Appl. Genet. 51, 343–352 [DOI] [PubMed] [Google Scholar]
- 41. Guo W., Zhou R. M., Wan L. L., Wang N., Li Y., Zhang X. J., Dong X. J. (2008) Polymorphisms of the DNA repair gene xeroderma pigmentosum groups A and C and risk of esophageal squamous cell carcinoma in a population of high incidence region of North China. J Cancer Res. Clin. Oncol. 134, 263–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pan J., Lin J., Izzo J. G., Liu Y., Xing J., Huang M., Ajani J. A., Wu X. (2009) Genetic susceptibility to esophageal cancer. The role of the nucleotide excision repair pathway. Carcinogenesis 30, 785–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Crew K. D., Gammon M. D., Terry M. B., Zhang F. F., Zablotska L. B., Agrawal M., Shen J., Long C. M., Eng S. M., Sagiv S. K., Teitelbaum S. L., Neugut A. I., Santella R. M. (2007) Polymorphisms in nucleotide excision repair genes, polycyclic aromatic hydrocarbon-DNA adducts, and breast cancer risk. Cancer Epidemiol. Biomarkers Prev. 16, 2033–2041 [DOI] [PubMed] [Google Scholar]
- 44. Dong Z., Guo W., Zhou R., Wan L., Li Y., Wang N., Kuang G., Wang S. (2008) Polymorphisms of the DNA repair gene XPA and XPC and its correlation with gastric cardiac adenocarcinoma in a high incidence population in North China. J. Clin. Gastroenterol. 42, 910–915 [DOI] [PubMed] [Google Scholar]
- 45. Palli D., Polidoro S., D'Errico M., Saieva C., Guarrera S., Calcagnile A. S., Sera F., Allione A., Gemma S., Zanna I., Filomena A., Testai E., Caini S., Moretti R., Gomez-Miguel M. J., Nesi G., Luzzi I., Ottini L., Masala G., Matullo G., Dogliotti E. (2010) Polymorphic DNA repair and metabolic genes. A multigenic study on gastric cancer. Mutagenesis 25, 569–575 [DOI] [PubMed] [Google Scholar]
- 46. Miller K. L., Karagas M. R., Kraft P., Hunter D. J., Catalano P. J., Byler S. H., Nelson H. H. (2006) XPA, haplotypes, and risk of basal and squamous cell carcinoma. Carcinogenesis 27, 1670–1675 [DOI] [PubMed] [Google Scholar]
- 47. Lin J., Pu X., Wang W., Matin S., Tannir N. M., Wood C. G., Wu X. (2008) Case-control analysis of nucleotide excision repair pathway and the risk of renal cell carcinoma. Carcinogenesis 29, 2112–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hansen R. D., Sørensen M., Tjønneland A., Overvad K., Wallin H., Raaschou-Nielsen O., Vogel U. (2007) XPA A23G, XPC K939Q, XPD K751Q, and XPD D312N polymorphisms, interactions with smoking, alcohol, and dietary factors, and risk of colorectal cancer. Mutat. Res. 619, 68–80 [DOI] [PubMed] [Google Scholar]
- 49. Weiss J. M., Weiss N. S., Ulrich C. M., Doherty J. A., Voigt L. F., Chen C. (2005) Interindividual variation in nucleotide excision repair genes and risk of endometrial cancer. Cancer Epidemiol. Biomarkers Prev. 14, 2524–2530 [DOI] [PubMed] [Google Scholar]
- 50. Hall J., Hashibe M., Boffetta P., Gaborieau V., Moullan N., Chabrier A., Zaridze D., Shangina O., Szeszenia-Dabrowska N., Mates D., Janout V., Fabiánová E., Holcatova I., Hung R. J., McKay J., Canzian F., Brennan P. (2007) The association of sequence variants in DNA repair and cell cycle genes with cancers of the upper aerodigestive tract. Carcinogenesis 28, 665–671 [DOI] [PubMed] [Google Scholar]
- 51. Rodriguez K., Talamantez J., Huang W., Reed S. H., Wang Z., Chen L., Feaver W. J., Friedberg E. C., Tomkinson A. E. (1998) Affinity purification and partial characterization of a yeast multiprotein complex for nucleotide excision repair using histidine-tagged Rad14 protein. J. Biol. Chem. 273, 34180–34189 [DOI] [PubMed] [Google Scholar]
- 52. Sikorski T. W., Ficarro S. B., Holik J., Kim T., Rando O. J., Marto J. A., Buratowski S. (2011) Sub1 and RPA associate with RNA polymerase II at different stages of transcription. Mol. Cell 44, 397–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sikorski R. S., Hieter P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 55. Bhaumik S. R., Green M. R. (2002) Differential requirement of SAGA components for recruitment of TATA-box-binding protein to promoters in vivo. Mol. Cell. Biol. 22, 7365–7371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bhaumik S. R., Green M. R. (2003) Interaction of Gal4p with components of transcription machinery in vivo. Methods Enzymol. 370, 445–454 [DOI] [PubMed] [Google Scholar]
- 57. Shukla A., Stanojevic N., Duan Z., Sen P., Bhaumik S. R. (2006) Ubp8p, a histone deubiquitinase whose association with SAGA is mediated by Sgf11p, differentially regulates lysine 4 methylation of histone H3 in vivo. Mol. Cell. Biol. 26, 3339–3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bhaumik S. R., Raha T., Aiello D. P., Green M. R. (2004) in vivo target of a transcriptional activator revealed by fluorescence resonance energy transfer. Genes Dev. 18, 333–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Malik S., Chaurasia P., Lahudkar S., Durairaj G., Shukla A., Bhaumik S. R. (2010) Rad26p, a transcription-coupled repair factor, is recruited to the site of DNA lesion in an elongating RNA polymerase II-dependent manner in vivo. Nucleic Acids Res. 38, 1461–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Peterson C. L., Kruger W., Herskowitz I. (1991) A functional interaction between the C-terminal domain of RNA polymerase II and the negative regulator SIN1. Cell 64, 1135–1143 [DOI] [PubMed] [Google Scholar]
- 61. Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Struhl K. (2001) Current Protocols in Molecular Biology, John Wiley & Sons, Inc., New York [Google Scholar]
- 62. Lee J. S., Shukla A., Schneider J., Swanson S. K., Washburn M. P., Florens L., Bhaumik S. R., Shilatifard A. (2007) Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell 131, 1084–1096 [DOI] [PubMed] [Google Scholar]
- 63. Bhaumik S. R., Green M. R. (2001) SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev. 15, 1935–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Malik S., Chaurasia P., Lahudkar S., Uprety B., Bhaumik S. R. (2012) Rad26p regulates the occupancy of histone H2A-H2B dimer at the active genes in vivo. Nucleic Acids Res. 40, 3348–3363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bhaumik S. R. (2011) Distinct regulatory mechanisms of eukaryotic transcriptional activation by SAGA and TFIID. Biochim. Biophys. Acta 1809, 97–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bhaumik S. R., Malik S. (2008) Diverse regulatory mechanisms of eukaryotic transcriptional activation by the proteasome complex. Crit. Rev. Biochem. Mol. Biol. 43, 419–433 [DOI] [PubMed] [Google Scholar]
- 67. Roeder R. G. (1996) The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci. 21, 327–335 [PubMed] [Google Scholar]
- 68. Cismowski M. J., Laff G. M., Solomon M. J., Reed S. I. (1995) KIN28 encodes a C-terminal domain kinase that controls mRNA transcription in Saccharomyces cerevisiae but lacks cyclin-dependent kinase-activating kinase (CAK) activity. Mol. Cell. Biol. 15, 2983–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Rodriguez C. R., Cho E.-J., Keogh M.-C., Moore C. L., Greenleaf A. L., Buratowski S. (2000) Kin28, the TFIIH-associated carboxy-terminal domain kinase, facilitates the recruitment of mRNA processing machinery to RNA polymerase II. Mol. Cell. Biol. 20, 104–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Valay J. G., Simon M., Dubois M. F., Bensaude O., Facca C., Faye G. (1995) The KIN28 gene is required for both RNA polymerase II mediated transcription and phosphorylation of the Rbb1p CTD. J. Mol. Biol. 249, 535–544 [DOI] [PubMed] [Google Scholar]
- 71. Keogh M. C., Cho E. J., Podolny V., Buratowski S. (2002) Kin28 is found within TFIIH and a Kin28-Ccl1-Tfb3 trimer complex with differential sensitivities to T-loop phosphorylation. Mol. Cell. Biol. 22, 1288–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. van Gool A. J., Verhage R., Swagemakers S. M., van de Putte P., Brouwer J., Troelstra C., Bootsma D., Hoeijmakers J. H. (1994) RAD26, the functional S. cerevisiae homolog of the Cockayne syndrome B gene ERCC6. EMBO J. 13, 5361–5369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Le May N., Mota-Fernandes D., Vélez-Cruz R., Iltis I., Biard D., Egly J. M. (2010) NER factors are recruited to active promoters and facilitate chromatin modification for transcription in the absence of exogenous genotoxic attack. Mol. Cell 38, 54–66 [DOI] [PubMed] [Google Scholar]
- 74. Lee S. K., Yu S. L., Prakash L., Prakash S. (2002) Requirement of yeast RAD2, a homolog of human XPG gene, for efficient RNA polymerase II transcription. Implications for Cockayne syndrome. Cell 109, 823–834 [DOI] [PubMed] [Google Scholar]
- 75. Lee S. K., Yu S. L., Prakash L., Prakash S. (2002) Yeast RAD26, a homolog of the human CSB gene, functions independently of nucleotide excision repair and base excision repair in promoting transcription through damaged bases. Mol. Cell. Biol. 22, 4383–4389 [DOI] [PMC free article] [PubMed] [Google Scholar]