Background: Bcl6 is a potent inhibitor of cell senescence, and somatic mutations accumulate in senescent cells.
Results: Mutation frequencies in senescent Bcl6-deficient cells were higher than senescent wild-type cells, and ADAR1 is overexpressed in Bcl6-deficient cells.
Conclusion: Bcl6 negatively regulates ADAR1 expression, and ADAR1 overexpression induces adenosine-targeted DNA mutations.
Significance: Bcl6 may protect senescent cells from accumulation of somatic mutations.
Keywords: Cellular Senescence, Gene Knock-out, Gene Regulation, Somatic Cell Genetics, Transcription Repressor, Transgenic Mice
Abstract
Somatic mutations accumulate in senescent cells. Bcl6, which functions as a transcriptional repressor, has been identified as a potent inhibitor of cell senescence, but a role of Bcl6 in the accumulation of somatic mutations has remained unclear. Ig class-switch recombination simultaneously induces somatic mutations in an IgM class-switch (Ig-Sμ) region of IgG B cells. Surprisingly, mutations were detected in the Ig-Sμ region of Bcl6-deficient IgM B cells without class-switch recombination, and these mutations were mainly generated by conversion of adenosine to guanosine, suggesting a novel DNA mutator in the B cells. The ADAR1 (adenosine deaminase acting on RNA1) gene was overexpressed in Bcl6-deficient cells, and its promoter analysis revealed that ADAR1 is a molecular target of Bcl6. Exogenous ADAR1 induced adenosine-targeted DNA mutations in IgM B cells from ADAR1-transgenic mice and in wild-type mouse embryonic fibroblasts (MEFs). These mutations accumulated in senescent MEFs accompanied with endogenous ADAR1 expression, and the frequency in senescent Bcl6-deficient MEFs was higher than senescent wild-type MEFs. Thus, Bcl6 protects senescent cells from accumulation of adenosine-targeted DNA mutations induced by ADAR1.
Introduction
Somatic mutations accumulate in mammals with age (1) and have been proposed to contribute to aging (2). Protection of DNA against somatic mutations may lead to prevention of aging. Bcl6 has been identified as a potent inhibitor of cell senescence by a senescence rescue screen (3). Bcl6 overexpression efficiently immortalizes primary mouse embryonic fibroblasts (MEFs)2 by blocking anti-proliferative p19ARF-p53 signaling. The Bcl6 gene encodes a nuclear phosphoprotein that binds to silencer regions of target genes to repress expression of these genes as a sequence-specific transcriptional repressor (4), and thus Bcl6 was suggested to repress expression of its target genes on the p19ARF-p53 signaling. Indeed, a recent report has demonstrated that the p53 gene is a molecular target of Bcl6 (5). However, a role for Bcl6 in accumulation of somatic mutations in senescent cells has remained unclear.
There are many sources of somatic mutations (6). In addition to external sources, there are cell intrinsic sources such as reactive oxygen species. Activation-induced cytidine deaminase (AID) (7, 8) is one of cell-intrinsic sources. AID belongs to an RNA-editing cytidine deaminase family (9), and there are several models to explain the molecular mechanism of AID-mediated somatic mutations (10). A significant body of data on the mechanism has been reported (11), and thus a consensus of the mechanism is a “DNA deamination” model. AID deaminates cytosine (C) to uracil (U) directly on ssDNA of transcription bubbles (12–17) and creates U-guanine (G) base pair mismatches that recruit DNA repair machinery. Attempted error-prone repair of these lesions triggers somatic mutations (18).
AID-induced somatic mutations are mainly restricted to variable regions of rearranged Ig genes in germinal center (GC) B cells. The Bcl6 gene is ubiquitously expressed and predominantly in GC B cells. However, we cannot examine a role for Bcl6 in accumulation of somatic mutations in GC B cells using Bcl6-deficient (Bcl6-KO) mice because GC formation was impaired in Bcl6-KO mice (19–21). Recent reports demonstrated that Bcl6 is required for survival and proliferation of GC B cells by regulating expression of DNA damage response and checkpoint genes (22, 23). AID-induced somatic mutations are also detected in an unrearranged IgM class-switch (Ig-Sμ) region and the c-myc gene of Ig class-switched B cells (24–26). Naive IgM B cells from Bcl6-KO mice normally differentiate into class-switched IgG1 B cells after immunization (27), and thus we examined frequencies of somatic mutations in the Ig-Sμ region and the c-myc gene of Bcl6-KO IgG1 B cells. The frequencies in Bcl6-KO IgG1 B cells were higher than wild-type (WT) IgG1 B cells, suggesting an inhibitory role of Bcl6 in accumulation of somatic mutations.
We simultaneously examined the frequencies in Bcl6-KO IgM B cells without Ig class-switch recombination. Surprisingly, a frequency in the Ig-Sμ region of Bcl6-KO IgM B cells was significantly higher than WT IgM B cells. These mutations were mainly generated by conversion of adenosine to guanosine, suggesting a novel cell-intrinsic inducer of somatic mutations other than AID in Bcl6-KO IgM B cells. Then, we looked for adenosine-targeted RNA-editing genes as the novel inducer. An ADAR (adenosine deaminase acting on RNA) family (28) converts adenosine of pre-mRNA into inosine (29), which is subsequently translated as guanosine, and thus we examined expression of ADAR family genes in various cells from Bcl6-KO mice. Here, we show that the ADAR1 gene among the ADAR family genes was overexpressed in various cells from Bcl6-KO mice and that the ADAR1 gene is a molecular target of Bcl6. ADAR1 overexpression induced adenosine-targeted DNA mutations in the Ig-Sμ region of IgM B cells from spleens of ADAR1 transgenic (Tg) mice and in the Ig-Sμ region and the c-myc gene of WT MEFs without induction of AID. These mutations accumulated in MEFs at a senescent stage accompanied with endogenous ADAR1 expression, and the frequencies in senescent Bcl6-KO MEFs were higher than senescent WT MEFs. We discuss a physiologic role of Bcl6 in protection of senescent cells against accumulation of adenosine-targeted DNA mutations induced by ADAR1.
EXPERIMENTAL PROCEDURES
Mice
C57BL/6 mice were purchased from Japan SLC (Hamamatsu, Japan). Bcl6-KO mice (20) and Bcl6-Tg mice with the exogenous Bcl6 gene under the control of the Lck distal promoter (30) were as described. Murine ADAR1 cDNAs (31) subcloned into the BamH1 site of the Lck distal promoter expression cassette (pLck(d)-ADAR1-hGH) (32) were microinjected into a male pronuclear of fertilized eggs from (C57BL/6 × DBA2) F1 mice to generate ADAR1-Tg mice. Conditional ADAR1-deficient (ADAR1-cKO) mice were developed by crossing ADAR1flox/flox mice (33) with CD21-cre Tg mice (34). These mice were backcrossed with C57BL/6 mice more than five generations and maintained under specific pathogen-free conditions in the animal center of Graduate School of Medicine, Chiba University (Chiba, Japan). All procedures conformed to the Chiba University Resolution on use of animals in research and were approved by the Institutional Animal Care and Use Committee at Chiba University, Graduate School of Medicine.
B Cell Culture
Single cell suspensions were prepared from spleens, and cells were incubated with a mixture of biotinylated Abs against CD43 and IgG1 (Pharmingen). IgM B cells were purified from spleen cells by depleting CD43+ and IgG1+ cells with the magnetic cell sorting system (Miltenyi Biotec). Purity of IgM B cells was 89–95%. Purified IgM B cells were cultured, as described (27). Briefly, purified IgM B cells (5.0 × 105/ml) were cultured in RPMI 1640 medium (Sigma) supplemented with 10% fetal calf serum (Intergen), 50 μm 2-Mercaptoethanol, 100 μg/ml of streptomycin (Wako Chemical Co., Osaka, Japan), and 100 units/ml of penicillin G potassium (Banyu Pharmaceutical Co., Tokyo, Japan). For B cell activation, IgM B cells were cultured with lipopolysaccharide (LPS) (4 μg/ml; Sigma) and interleukin-4 (rIL-4, 1000 units/ml) for 4 days in a humidified atmosphere at 37 °C with 5% CO2. In some experiments, IgM B cells and IgG1 B cells were sorted by FACSVantage (Becton Dickinson). Purity of IgG1 B cells was >95%.
MEF Culture
MEFs were established by harvesting embryonic day 13.5 embryos. Head and liver were removed from embryos, and the remaining embryonic tissues were trypsinized at 37 °C for 30 min. The disrupted tissues were plated in DMEM (Sigma) supplemented with 10% fetal calf serum (Sigma) and cultured at 37 °C in 5% CO2. The 3T3 type serial MEF cultivation was done as described (35). Briefly, 3 × 105 cells were cultured on a 6-cm well for 3 days (passage 1; P1), the total cell number was counted, then 3 × 105 cells were cultured on a separate well for 3 days (P2). In some experiments, MEFs were cultured until P5 and then treated with 10–100 μm of H2O2 for 2 h.
Isolation of CD4 T Cells and GC B Cells
Spleen cells were blocked with 20 μg/ml of anti-Fc receptor II/III (2.4G2; Pharmingen) Abs. These cells were incubated with phycoerythrin-labeled anti-CD4 (Pharmingen) Abs, and CD4 T cells were sorted by FACSVantage. Purity of these sorted cells was >95%. For isolation of GC B cells, mice were immunized intraperitoneally with 50 μg of (4-hydroxy-3-nitrophenyl)acetyl-chicken γ globulin (NP25-CG, Biosearch Technologies, Novato, CA) precipitated in alum plus 106 pertussis. GC B cells were isolated from spleens of these mice 10 days after immunization. Spleen cells after blocking were incubated with PE-labeled anti-B220 (eBioscience, San Diego, CA) and fluorescein isothiocyanate-labeled peanut agglutinin (PNA, Vector Laboratories, Burlingame, CA). GC (B220+PNA+) B cells were sorted by FACSVantage (Becton Dickinson). Purity of those cells was >95%.
PCR and Sequencing
DNA isolation methods were as described (36). Briefly, high molecular weight nuclear DNA was extracted with SDS/proteinase K lysis, followed by phenol/chloroform extraction. PCR was done with the primers shown below using LA Taq polymerase (TaKaRa). After purification, the PCR fragments were ligated into the pGEM-T Easy Vector System (Promega). The ligation mixture was used for transformation and the library was plated without preculturing to avoid amplification of sister clones. Nucleotide sequences were determined using an ABI PRISM 310 genetic analyzer (PE Biosystems). The 400 base pairs of the Ig-Sμ region (the first base corresponding to position 4800 in GenBankTM/EBI accession number J00440) and the 300 base pairs of the c-myc gene (the first base corresponding to the position 23103197 in GenBankTM/EBI accession number NT039621) were sequenced. The primers used were as follows: Ig-Sμ (25), 5′-AATGGATACCTCAGTGGTTTTTAAT-3′ and 5′-TCTCGGTTAAGCCTAGTTTA-3′; c-myc (26), 5′-ACCTAAGAAGGCAGCTCTGGAGTG-3′ and 5′-TCATCTTGACAAGTCGCTCTACCC-3′. Common primers for SP6 or T7 were used for sequencing PCR products in the pGEM-T Easy Vector.
Real-time Quantitative Reverse Transcription (qRT)-PCR
Real-time qRT-PCR was performed as described elsewhere (37). Total RNA was extracted from various organs and cells with the TRIzol reagent (Invitrogen). Total RNA was reverse-transcribed using SuperScript III Reverse Transcriptase (Invitrogen) and oligo(dT) (Pharmacia), and the cDNAs were used for PCR. Real-time PCR with cDNAs was conducted using a SYBR Green PCR Master Mix (Perkin-Elmer Applied Biosystems) and run on an ABI Prism 7000 Sequence Detection System (Perkin-Elmer Applied Biosystems). GAPDH mRNA was used as an internal control for the amount of mRNA. Primers for the cDNA amplification were as follows: ADAR1, 5′-AAGGTCGGCCGAGTCAGTGTTAT-3′ and 5′-ACAACTCTTTCCCTGGTCCATC-3′; ADAR2, 5′-TGTCAAAGATGCCAAGGTGA-3′ and 5′-AGGTGGAACTGCACGGTATC-3′; ADAR3, 5′-AGTCCAGACGTGGGATGGCATCC-3′ and 5′-CTCACCCGGGTGCCAAGACCGGC-3′; ADAT1, 5′-AAGAAGACTGTGGAGGCTCA-3′ and 5′-GGACAGCAAGGCTGTTCTTC-3′; AID, 5′-CTGCCAAACCTGATGTCTTGATTTT-3′ and 5′-CAACGTGGCGTCCAAACAGGCACTT-3′; and GAPDH, 5′-TGTGTCCGTCGTGGATCTGA-3′ and 5′-TTGCTGTTGAAGTCGCAGGAG-3′.
Somatic Hypermutation (SHM) in the VH186.2 Gene
NP-binding IgG1 B cells were sorted from spleens of mice immunized with NP25-CG by FACSVantage (27), and RNA was prepared from these isolated IgG1 B cells. RT-PCR was done, and the VH186.2 gene followed by D and J regions and a proximal segment of the Cγ1 gene was amplified by two rounds of nested PCR with high-fidelity Pfu-Ultra DNA polymerase (Stratagene, La Jolla, CA). After purification, the PCR fragments were ligated into the pGEM-T Easy Vector System (Promega). The forward and reverse primers for the first-round PCR were 5′-CATGCTCTTCTTGGCAGCAACAGC-3′ and 5′-GTGCACACCGCTGGACAGGGATCC-3′, and for second-round PCR, primers were 5′-CAGGTCCAACTGCAGCCAG-3′ and 5′-AGTTTGGGCAGCAGA-3′. Nucleotide sequences of the cloned VH186.2 gene were determined by automated sequencing using an ABI PRISM 3100 DNA sequencer (PE Biosystems, Foster City, CA).
Chromatin Immunoprecipitation (ChIP) Assay
ChIP assay was done using the ChIP assay kit (UpState Biotechnology) and then conducted according to the manufacturer's instructions. The following Abs were used: Bcl6-specific rabbit polyclonal Abs (Santa Cruz Biotechnology) and rabbit IgG (Santa Cruz Biotechnology). Primers used for analysis in the promoter region and intron 1A of the ADAR1 gene were as follows: region 1, 5′-TGGTGGCACACACCTGTAATCC-3′ and 5′-ACCATGATGCTGTCTCTTACTGTCC-3′; region 2, 5′-GCCCATGCTACACAGAGAAACTGTC-3′ and 5′-CCTAGCTCCTGCAAGACAATGG-3′; region 3, 5′-AATAGCTCCCAAGCTCTCAGAGAAC-3′ and 5′-TGACCCTCGTAATTCACACTACGC-3′; region 4, 5′-GCAGAGGCAGGTATTTCTGAGTCC-3′ and 5′-CCATTAAGAGCTGGGGAGATGATGAC-3′.
Preparation of Nuclear Extracts and EMSA
Nuclear extracts were prepared from splenic IgM B cells and activated B cells and rapidly stored at −70 °C after determination of the protein amount using a Bio-Rad protein assay (Bio-Rad). Oligonucleotides were labeled with biotin using the biotin 3′ end DNA labeling kit (Pierce). The binding reaction was done using the LightShift chemiluminescent EMSA kit (Pierce). The following oligonucleotide sequences were used for DNA probes or competitor fragments in EMSA: region 1, 5′-TGGAATCTGTTTCCAAGAAAATAAAGACT-3′; region 2, 5′-TCCACCTGTTTTCCAAGAAATGCTGCAGCA-3′; region 3, 5′-GGGGAAGCGTTTTCAAGGAAACGAACTGGA-3′; region 4, 5′-GGCCCTCAGGCTTCCAGGAATTTTTCTTCT-3′; WT Bcl6, 5′-CATTTAACTTTCCTTGAAAAACTAGTAAAA-3′; and Mut Bcl6, 5′-CATTTAACTTTCCTTTTAAAACTAGTAAA-3′. The supershift assay was performed using 2 μg of Bcl6-specific rabbit polyclonal Abs or rabbit IgG (Santa Cruz Biotechnology).
Reporter Constructs with the ADAR1 Promoter Region
Genomic DNA fragments of the ADAR1 promoter region (38) were inserted into cloning sites of a pGL3-Basic vector carrying the SV40 enhancer (pADAR1 Luc) and the firefly luciferase reporter gene (Promega). Mutant luciferase constructs (pADAR1 Luc (mutBcl6)) were generated by PCR using oligonucleotide primers carrying point mutations (QuikChangeXL site-directed mutagenesis kit, Stratagene). The primer used for the mutation on a Bcl6-binding sequence of region 3 (MutBcl6): 5′-CATTTAACTTTCCTTTTAAAACTAGTAAA-3′. The Bcl6 expression vector (pcDNA3-Bcl6) was constructed, as described (39).
Transient Transfection and Luciferase Assay
Transfection of genes was conducted by electroporation using the Bio-Rad Gene PulserII (Bio-Rad). Cells (1 × 107) were incubated with DNA (total 20 μg) for 10 min on ice and then electroporated at 960 microfarads with 220 mV for Ramos cells (Human Burkitt's lymphoma cell line). These cells were harvested 24 h after electroporation. Luciferase activity in cell extracts was determined using a luciferase assay kit (Promega).
The ADAR1-EGFP Fusion Gene and Its Transfection
Purified IgM B cells or MEFs were transfected with the pEGFP-ADAR1(La) vector as described (31). RetroNectin (TaKaRa) was used for the transfection. EGFP+ IgG1 B cells or MEFs were isolated using FACSVantage.
Statistical Analysis
Statistical analysis was made using unpaired t test. p values of less than 0.05 were considered to be significant.
RESULTS
Somatic Mutations in the Ig-Sμ Region and the c-myc Gene of Bcl6-KO B Cells
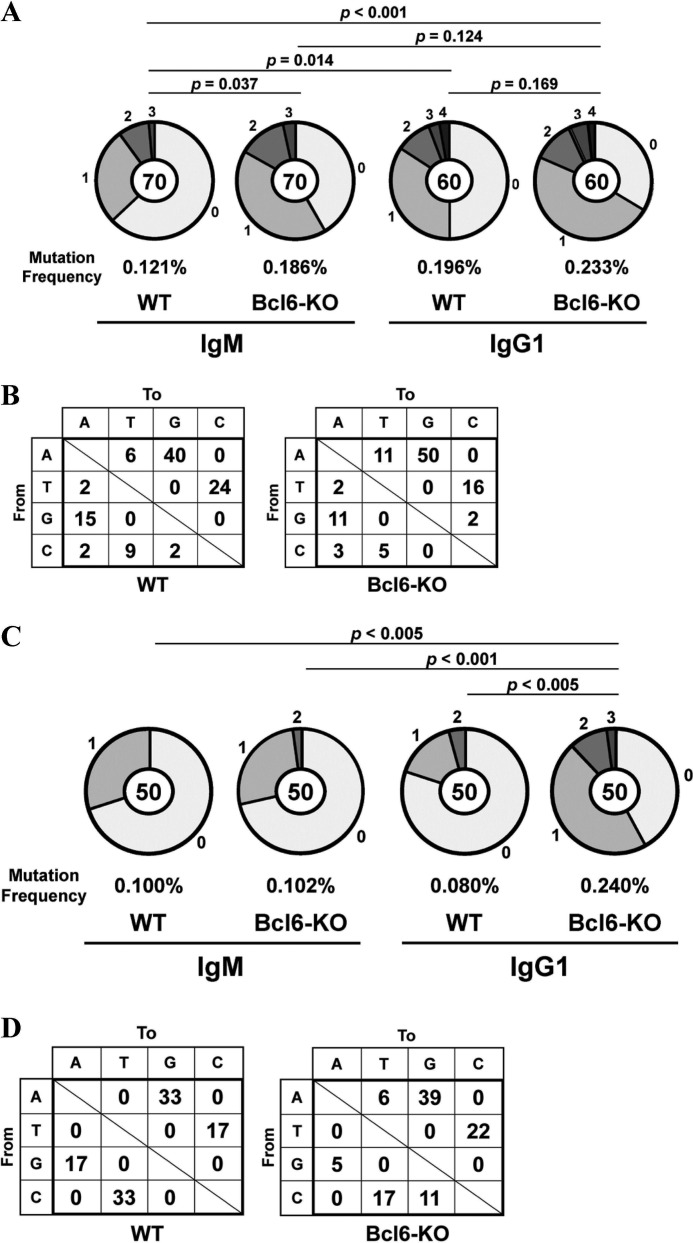
Naive IgM B cells from spleens of WT and Bcl6-KO mice were stimulated with LPS and IL-4 for 4 days, and IgM B cells and IgG1 B cells were isolated by fluorescence activated cell sorter (FACS). Mutation frequencies in the Ig-Sμ region of IgG1 B cells were compared with IgM B cells. As we expected, frequencies in the Ig-Sμ region of WT IgG1 B cells (p = 0.014) and Bcl6-KO IgG1 B cells (p < 0.001) were higher than WT IgM B cells (Fig. 1A). Surprisingly, the frequency in Bcl6-KO IgM B cells was also higher than WT IgM B cells (p = 0.037). The frequency in Bcl6-KO IgG1 B cells was slightly higher than Bcl6-KO IgM B cells (p = 0.124) and WT IgG1 B cells (p = 0.169), although the difference was not significant. Percentages of mutations from adenine (A)/thymine (T) to G/C among total mutations were very high in WT (74% = 25/34) and Bcl6-KO (63% = 33/52) IgM B cells (data not shown) and in WT (64%) and Bcl6-KO (66%) IgG1 B cells (Fig. 1B). Percentages of mutations from C/G to T/A were also high in the WT (18% = 6/34) and the Bcl6-KO (12% = 6/52) IgM B cells and in the WT (24%) and the Bcl6-KO (16%) IgG1 B cells.
FIGURE 1.
Somatic mutations in the Ig-Sμ region and the c-myc gene of Bcl6-KO B cells. Splenic IgM B cells from WT and Bcl6-KO mice were stimulated with LPS and IL-4 for 4 days. The Ig-Sμ region and the c-myc gene of IgM B cells and IgG1 B cells were sequenced. A and C, pie charts indicate mutations in the Ig-Sμ region (A) and the c-myc gene (C) of IgM B cells and IgG1 B cells. Sectors of the pie are proportional to the number of sequences with a given number of mutations. The number of sequences is shown in the center of the pie chart. A frequency of mutations is described below the pie chart. B and D, types of nucleotide substitutions in the Ig-Sμ region (B) and the c-myc gene (D) of IgG1 B cells are shown as percentages of total mutations.
AID-induced somatic mutations are also detected in the c-myc gene of class-switched B cells, and thus, somatic mutations in the c-myc gene of these IgG1 B cells were analyzed. The frequency in the Bcl6-KO IgG1 B cells was significantly higher than the WT IgM B cells (p < 0.005), the Bcl6-KO IgM B cells (p < 0.001) and the WT IgG1 B cells (p < 0.005) (Fig. 1C). However, the frequency in the Bcl6-KO IgM B cells was not higher than the WT IgM B cells (p = 0.475). We observed very high percentages of mutations from A/T to G/C among total mutations in the WT (50%) and the Bcl6-KO (61%) IgG1 B cells (Fig. 1D). Percentages of mutations from C/G to T/A were also high in the WT (50%) and the Bcl6-KO (22%) IgG1 B cells.
Overexpression of ADAR1 in Various Cells from Bcl6-KO Mice
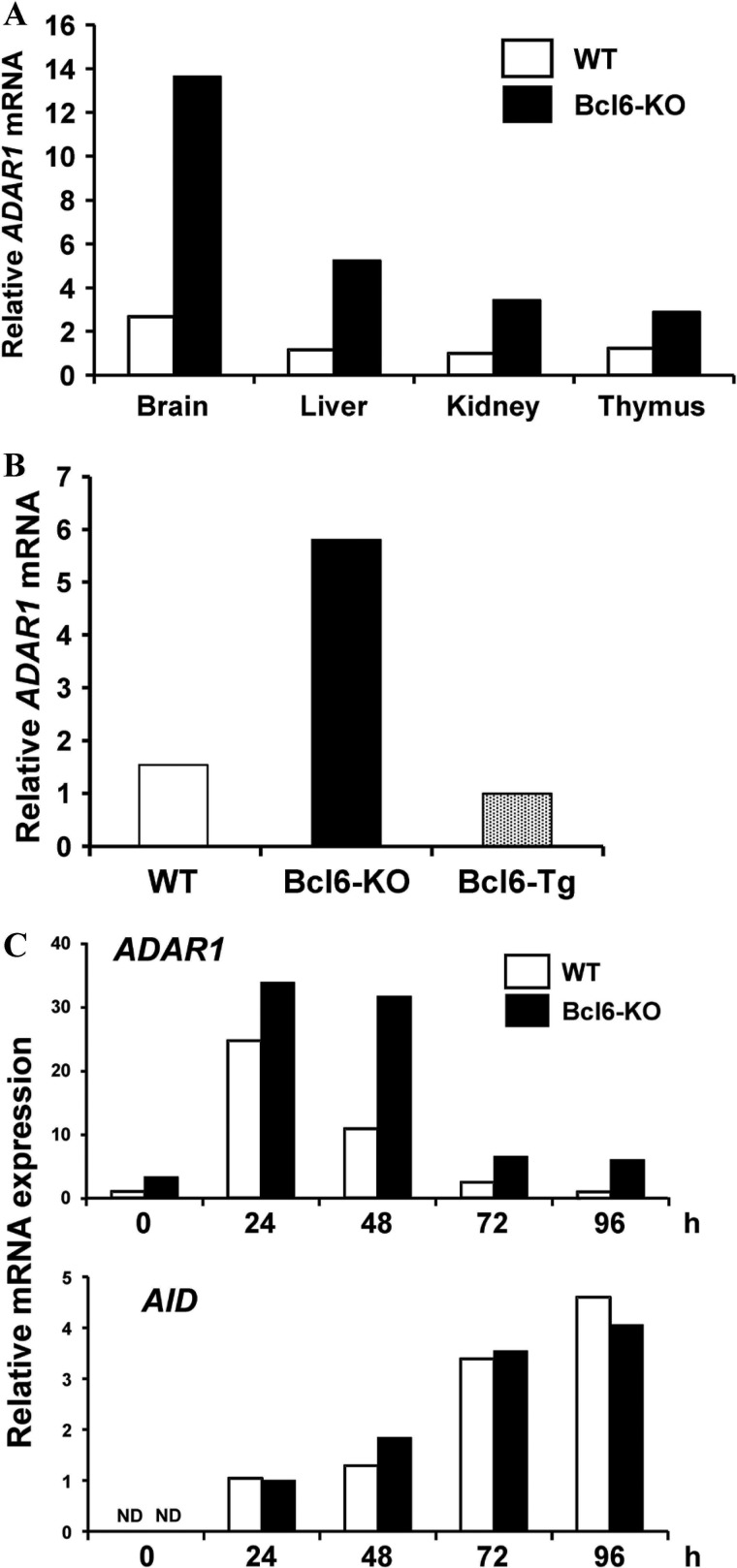
Somatic mutations from A/T to G/C were major mutations in the Ig-Sμ region of Bcl6-KO IgM B cells, suggesting induction of a novel cell-intrinsic inducer other than AID in the B cells. Because RNA-editing adenosine deaminase such as ADAR converts adenosine of pre-mRNA into inosine (29), which is subsequently translated as guanosine, we speculated that ADAR could be responsible for the DNA mutations. We then examined expression of ADAR family genes in various organs from WT and Bcl6-KO mice by real-time qRT-PCR. ADAR1 expression was detected in brain in which ADAR1 edits pre-mRNA of serotonin-2C receptor (40) but not in other organs examined from WT mice and augmented in various organs from Bcl6-KO mice (Fig. 2A). Expression of other ADAR family (ADAR2, ADAR3, and ADAT1) genes (41) in various organs from WT mice was similar to that in Bcl6-KO mice (data not shown). ADAR1 expression was also augmented in splenic CD4 T cells from Bcl6-KO mice and suppressed in those from lck-Bcl6 Tg (Bcl6-Tg) mice (Fig. 2B). The ADAR1 gene was expressed in Bcl6-KO IgM B cells but not in WT IgM B cells (Fig. 2C). When WT and Bcl6-KO IgM B cells were activated with LPS and IL-4, ADAR1 expression was transiently induced in both B cells and detected longer time in activated Bcl6-KO B cells, whereas AID expression in the Bcl6-KO B cells was similar to the WT B cells. Expression of other ADAR family genes was not detected in naive IgM B cells and activated IgG1 B cells from both mice (data not shown). These results suggested that ADAR1 was involved in induction of somatic mutations in the Ig-Sμ region of Bcl6-KO IgM B cells.
FIGURE 2.
ADAR1 mRNA expression in various organs and cells from WT, Bcl6-KO, and Bcl6-Tg mice. A and B, amounts of ADAR1 mRNA in various organs (A) and CD4 T cells (B) from WT, Bcl6-KO, and Bcl6-Tg mice were analyzed by real-time qRT-PCR. Data presented are as a representative of three independent experiments. C, ADAR1 and AID mRNA levels in activated B cells. Splenic IgM B cells from WT and Bcl6-KO mice were stimulated with LPS and IL-4 for 4 days. ADAR1 and AID mRNA levels were analyzed by real-time qRT-PCR. Data are presented as a representative of three independent experiments. ND, not detected.
ADAR1 as a Molecular Target of Bcl6
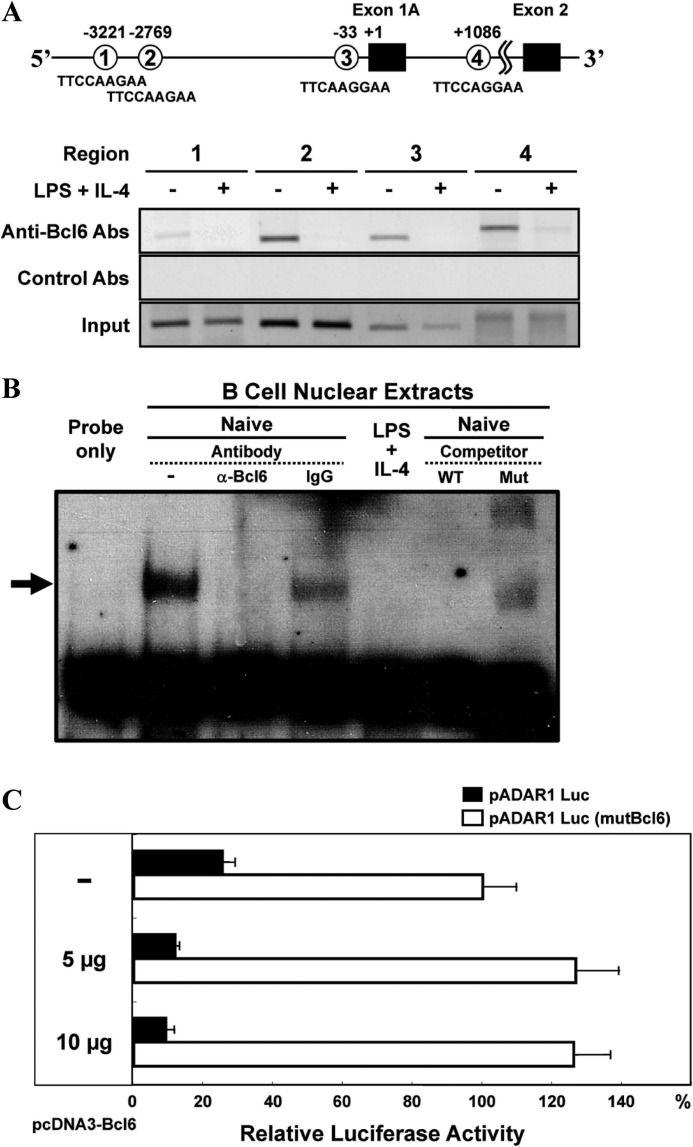
To examine whether the ADAR1 gene is a molecular target of Bcl6, we looked for Bcl6-binding sequences in the murine ADAR1 gene (GenBankTM/EBI accession no. AAK16102) and found four putative Bcl6-binding sequences in the promoter region (upstream region of the exon 1A) and the intron 1A (Fig. 3A). ChIP assay demonstrated Bcl6 binding to these regions in naive IgM B cells but not in B cells activated with LPS and IL-4. Specific binding of Bcl6 to Bcl6-binding sequences at regions 2 and 3 was confirmed by EMSA with a cold competitor. Nuclear extracts from naive IgM B cells but not those from activated B cells contained binding molecules to regions 2 (data not shown) and 3 (Fig. 3B), and the binding complex was removed by the addition of anti-Bcl6 Abs. The cold competitor but not that with mutations in the Bcl6-binding sequence blocked the binding. Furthermore, we confirmed a potential of exogenous Bcl6 for repressing ADAR1 promoter activity in Ramos cells by co-transfection of the reporter (pADAR1 Luc) gene under the control of the ADAR1 promoter containing region 3 and a Bcl6 expression vector (pcDNA3-Bcl6). Co-transfection of various amounts (5–10 μg) of pcDNA3-Bcl6 repressed the promoter activity in Ramos cells (Fig. 3C). Bcl6-dependent repression was confirmed by co-transfection of the reporter gene with mutations in the Bcl6-binding sequence of region 3 (pADAR1 Luc (mutBcl6)) and pcDNA3-Bcl6. These results indicated that the ADAR1 gene is a molecular target of Bcl6.
FIGURE 3.
Bcl6-binding to putative Bcl6-binding sequences in the ADAR1 promoter region. A and B, splenic IgM B cells from WT mice were stimulated with LPS and IL-4 for 6 h. A, ChIP analysis for Bcl6 binding to the ADAR1 promoter region of B cells. DNA samples were analyzed by PCR with primers specific for each indicated region, and 10% of input DNA (bottom) was analyzed by PCR. Data presented are as a representative of three independent experiments. B, EMSA for Bcl6-binding to region 3. An arrow indicates the retarded bands. For cold probe competition experiments, 50-fold molar of unlabeled competitor oligonucleotides was included in binding reactions with nuclear extracts from naive IgM B cells. Data are presented as a representative of three independent experiments. C, repression of ADAR1 promoter activity by Bcl6. Ramos cells were transfected with pADAR1 Luc (filled bar) or pADAR1 Luc (mutBcl6) (open bar) and serial dilution of pcDNA3-Bcl6. Luciferase activity of these cells transfected with pADAR1 Luc (mutBcl6) was as 100%. Data are presented as the mean ± S.D. from three independent experiments.
Effect of ADAR1 on Induction of Somatic Mutations in B Cells
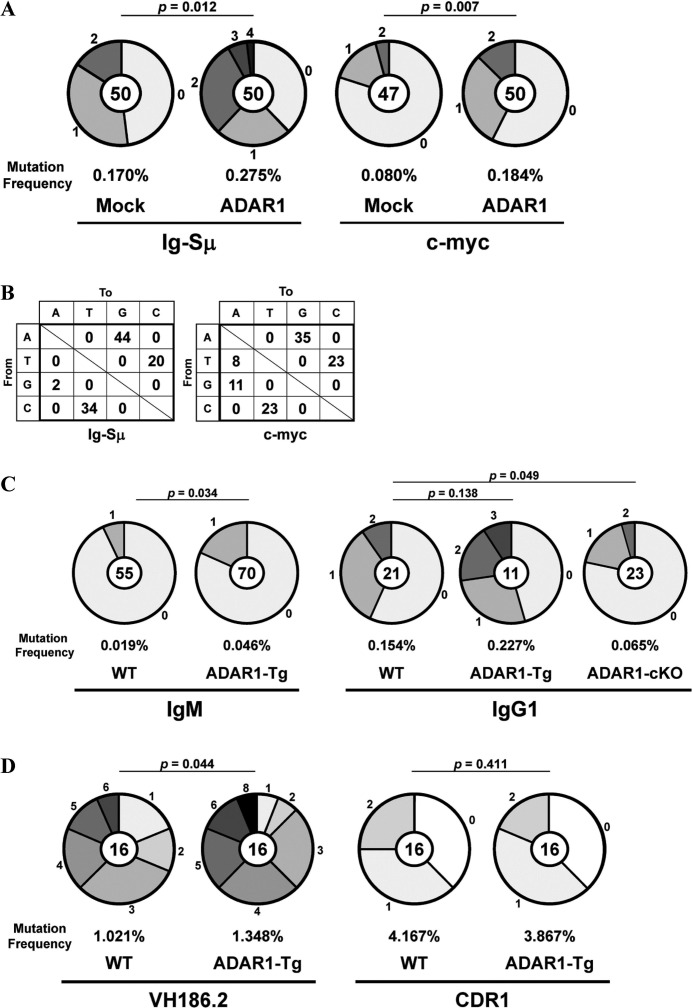
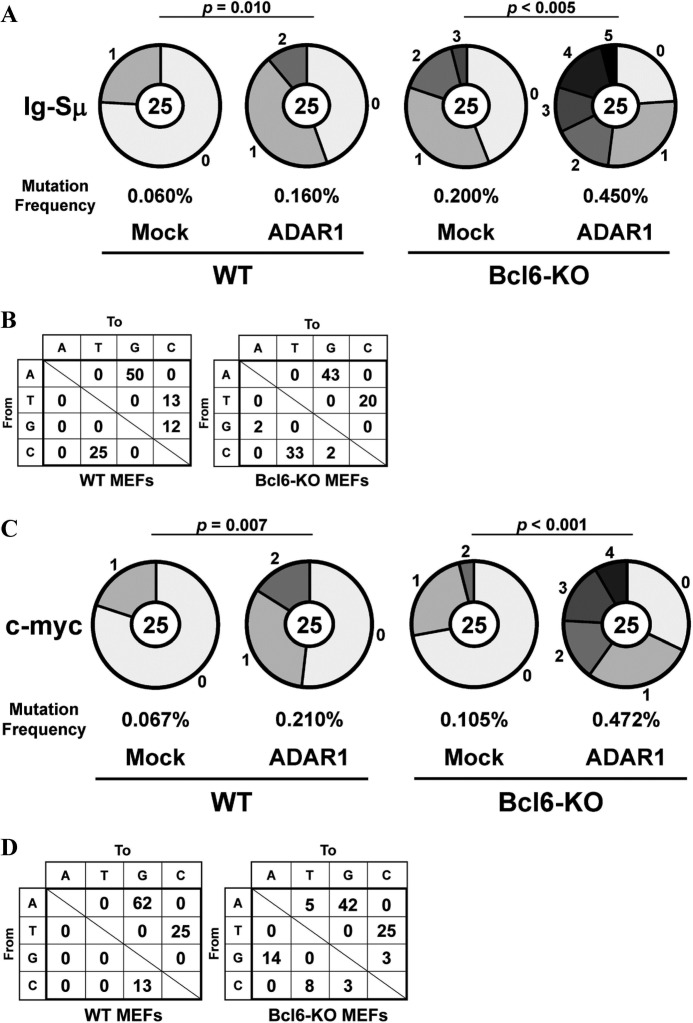
We transfected the ADAR1-EGFP fusion gene into IgM B cells from spleens of WT mice. These IgM B cells were stimulated with LPS and IL-4 for 4 days, and EGFP+ IgG1 B cells were isolated by FACS. The Ig-Sμ region and the c-myc gene of EGFP+ IgG1 B cells were sequenced. Mutation frequencies in these regions of ADAR1-transfected IgG1 B cells were significantly higher than mock-transfected IgG1 B cells (Ig-Sμ, p = 0.012; c-myc, p = 0.007) (Fig. 4A). Percentages of mutations from A/T to G/C among total mutations were very high in the Ig-Sμ region (64%) and the c-myc gene (58%) of ADAR1-transfected IgG1 B cells (Fig. 4B). Percentages of mutations from C/G to T/A were also high in both regions (Ig-Sμ, 36%; c-myc, 34%) of the IgG1 B cells.
FIGURE 4.
Somatic mutations in the Ig-Sμ region and the c-myc gene of B cells with the exogenous ADAR1 gene. A and B, naive WT IgM B cells transfected with the ADAR1-EGFP fusion (ADAR1) gene or the Mock-EGFP (Mock) gene were stimulated with LPS and IL-4 for 4 days. EGFP+ IgG1 B cells were isolated by FACS. The Ig-Sμ region and the c-myc gene of EGFP+ IgG1 B cells were sequenced. C, naive IgM B cells were stimulated with LPS and IL-4 for 4 days. The Ig-Sμ region of naive IgM B cells and IgG1 B cells from WT, ADAR1-Tg, and ADAR1-cKO mice was sequenced. D, ADAR1-Tg mice were immunized with NP-CG, and NP-binding IgG1 B cells were isolated by FACS. The VH186.2 gene of isolated IgG1 B cells was sequenced. A, C, and D, pie charts indicate mutations in the Ig-Sμ region and the c-myc gene of ADAR1-transfected IgG1 B cells (A), in the Ig-Sμ region of IgM B cells and IgG1 B cells (C) or in the VH186.2 gene of NP-binding IgG1 B cells (D). Sectors of the pie are proportional to the number of sequences with a given number of mutations. The number of sequences is shown in the center of the pie chart. A frequency of mutations is described below the pie chart. B, types of nucleotide substitutions in the Ig-Sμ region and the c-myc gene of ADAR1-transfected IgG1 B cells are shown as percentages of total mutations.
We generated Lckd-ADAR1 Tg (ADAR1-Tg) mice to examine whether exogenous ADAR1 induces somatic mutations in the Ig-Sμ region of IgM B cells without class-switch recombination. A mutation frequency in the Ig-Sμ region of IgM B cells from spleens of ADAR1-Tg mice was higher than WT IgM B cells (p = 0.034) (Fig. 4C). Mutations from A/T to G/C were major in the Ig-Sμ region of ADAR1-Tg IgM B cells (62% = 8/13). Mutations from C/G to T/A were also detected in the B cells (15% = 2/13). IgM B cells from ADAR1-Tg or ADAR1-cKO mice were stimulated with LPS and IL-4 for 4 days, and IgG1 B cells were isolated by FACS. The Ig-Sμ region of IgG1 B cells was sequenced. Mutation frequencies in the region of ADAR1-Tg and ADAR1-cKO IgG1 B cells were higher (p = 0.138) and lower (p = 0.049) than WT IgG1 B cells, respectively. Furthermore, we examined effect of the exogenous ADAR1 on induction of SHM in GC B cells because the amount of ADAR1 mRNA in GC B cells from spleens of ADAR1-Tg mice was ∼6-fold more than GC B cells from WT mice 10 days after immunization (data not shown). We immunized ADAR1-Tg mice with NP-CG in alum and examined SHM of the rearranged VH186.2 gene (42) in NP-binding IgG1 B cells from spleens of ADAR1-Tg mice 10 days after immunization. A mutation frequency in the VH186.2 gene of ADAR1-Tg IgG1 B cells was higher than WT IgG1 B cells (p = 0.044), whereas the frequency in the CDR1 region of the VH186.2 gene of the ADAR1-Tg IgG1 B cells was similar to the WT IgG1 B cells (p = 0.411) (Fig. 4D). The percentage of mutations from A/T to G/C among total mutations in the VH186.2 gene of the ADAR1-Tg IgG1 B cells (30% = 20/66) was slightly higher than the WT IgG1 B cells (20% = 10/50). Mutations from C/G to T/A in the VH186.2 gene of the ADAR1-Tg IgG1 B cells (32% = 21/66) were similar to the WT IgG1 B cells (32% = 16/50).
Effect of Exogenous ADAR1 on Induction of Somatic Mutations in MEFs
Because AID expression is restricted to activated B cells (9), we examined whether exogenous ADAR1 induces somatic mutations in the Ig-Sμ region and the c-myc gene of cells other than B cells without AID expression. We transfected the ADAR1-EGFP gene into MEFs from WT and Bcl6-KO mice at P5. EGFP+ MEFs were isolated 3 days after transfection by FACS. Higher mutation frequencies were observed in the Ig-Sμ region of ADAR1-transfected MEFs as compared with mock-transfected MEFs (WT, p = 0.010; Bcl6-KO, p < 0.005) (Fig. 5A). Mutations from A/T to G/C were major in these WT (63%) and Bcl6-KO (62%) MEFs (Fig. 5B). Percentages of mutations from C/G to T/A among total mutations were also high in these WT (25%) and Bcl6-KO (35%) MEFs. ADAR1-induced DNA mutations were detected in the c-myc gene, and these mutation frequencies in the ADAR1-transfected WT and Bcl6-KO MEFs were higher than the mock-transfected MEFs (WT, p = 0.007; Bcl6-KO, p < 0.001) (Fig. 5C). Mutations from A/T to G/C were major in these WT (87%) and Bcl6-KO (67%) MEFs (Fig. 5D). Percentages of mutations from C/G to T/A were high in the Bcl6-KO MEFs (22%) but not in the WT MEFs (0%).
FIGURE 5.
Somatic mutations in the Ig-Sμ region and the c-myc gene of MEFs transfected with the ADAR1 gene. WT and Bcl6-KO MEFs at P5 transfected with the ADAR1-EGFP fusion (ADAR1) gene or the Mock-EGFP (Mock) gene were cultured for 3 days. EGFP+ MEFs were isolated by FACS, and the Ig-Sμ region and the c-myc gene were sequenced. A and C, pie charts indicate mutations in the Ig-Sμ region (A) and the c-myc gene (C) of ADAR1-transfected MEFs. Sectors of the pie are proportional to the number of sequences with a given number of mutations. The number of sequences is shown in the center of the pie chart. A frequency of mutations is described below the pie chart. B and D, types of nucleotide substitutions in the Ig-Sμ region (B) and the c-myc gene (D) of ADAR1-transfected MEFs are shown as percentages of total mutations.
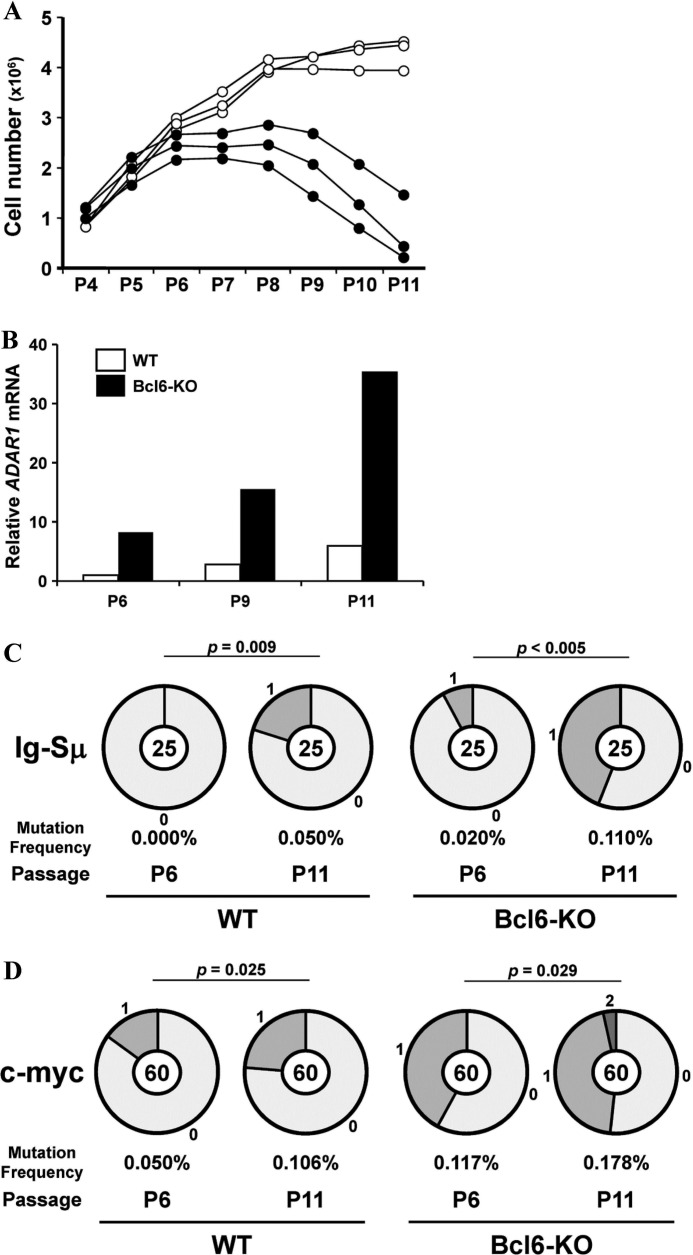
Accumulation of Somatic Mutations in Senescent MEFs
To examine a role for Bcl6 in accumulation of somatic mutations in senescent MEFs, MEFs from WT and Bcl6-KO mice were cultured by the 3T3 protocol. Bcl6-KO MEFs entered a senescent stage after P6, whereas WT MEFs did after P9 (Fig. 6A). We confirmed expression of senescence-related (p16, p21, and p53) genes in Bcl6-KO MEFs at P9 by RT-PCR (data not shown). These results confirmed the inhibitory effect of Bcl6 on induction of cell senescence (3). We examined expression of ADAR1 and AID mRNA in WT and Bcl6-KO MEFs by real-time qRT-PCR. ADAR1 expression was detected in WT and Bcl6-KO MEFs after P11 and P6, respectively (Fig. 6B). However, AID expression was not detected at all in these MEFs until P11 (data not shown). Then, we examined somatic mutations in the Ig-Sμ region and the c-myc gene of WT and Bcl6-KO MEFs at P6 and P11. Mutation frequencies significantly increased in the Ig-Sμ region of MEFs at P11 as compared with those at P6 (WT, p = 0.009; Bcl6-KO, p < 0.005) (Fig. 6C). Frequencies also increased in the c-myc gene of these MEFs at P11 (WT, p = 0.025; Bcl6-KO, p = 0.029) (Fig. 6D). Mutations from A/T to G/C were major in the Ig-Sμ region (WT, 80% = 4/5; Bcl6-KO, 82% = 9/11) and the c-myc gene (WT, 71% = 5/7; Bcl6-KO, 46% = 6/13) of these MEFs at P11.
FIGURE 6.
Somatic mutations in the Ig-Sμ region and the c-myc gene of MEFs at the senescent stage. MEFs from WT and Bcl6-KO mice were cultured by the 3T3 protocol. A, cell numbers of WT (open circle) and Bcl6-KO (filled circle) MEFs were demonstrated. B, amounts of ADAR1 mRNA in MEFs were analyzed by real-time qRT-PCR. Data are presented as representative of three independent experiments. C and D, pie charts indicate mutations in the Ig-Sμ region (C) and the c-myc gene (D) of MEFs. Sectors of the pie are proportional to the number of sequences with a given number of mutations. The number of sequences is shown in the center of the pie chart. A frequency of mutations is described below the pie chart.
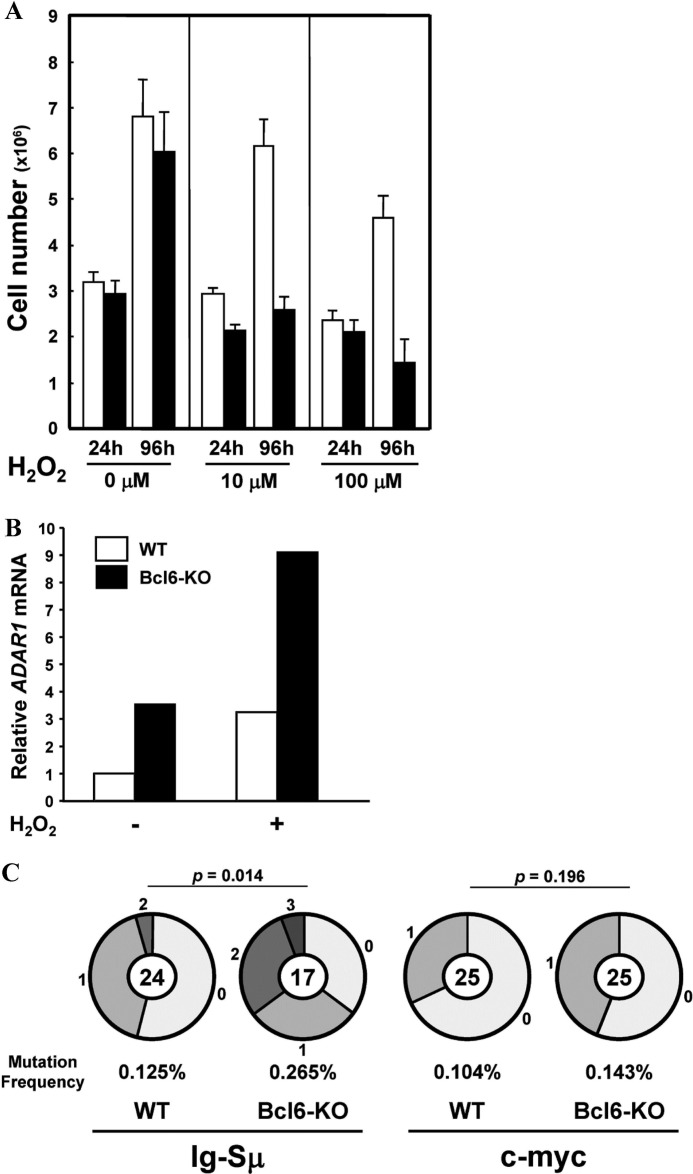
Oxidative stress induces MEFs to enter the premature senescent stage (43), and thus, MEFs at P5 were treated with an oxidative stress inducer, H2O2 (44), for 2 h and then cultured for 4 days. Proliferation of WT MEFs was partly inhibited after treatment with 100 μm H2O2, whereas Bcl6-KO MEFs stopped proliferation after treatment with 10 μm H2O2 (Fig. 7A). ADAR1 mRNA was detected in Bcl6-KO MEFs but not in WT MEFs before treatment and induced in both MEFs after treatment with 100 μm H2O2 (Fig. 7B). Frequencies of somatic mutations in the Ig-Sμ region (p = 0.014) and the c-myc gene (p = 0.196) of H2O2-treated Bcl6-KO MEFs were higher than H2O2-treated WT MEFs, although difference of frequencies in the c-myc gene between them was not significant (Fig. 7C). Percentages of mutations from A/T to G/C among total mutations were very high in the Ig-Sμ region (WT, 92% = 11/12; Bcl6-KO, 71% = 12/17) and high in the c-myc gene (WT, 50% = 4/8; Bcl6-KO, 55% = 6/11) of these MEFs.
FIGURE 7.
Somatic mutations in the Ig-Sμ region and the c-myc gene of MEFs after treatment with H2O2. Bcl6-KO and WT MEFs at P5 were treated with various doses of H2O2 for 2 h and then cultured for 96 h. A, cell numbers of WT (open bar) and Bcl6-KO (filled bar) MEFs were calculated 24 and 96 h after treatment. Data are presented as a representative of three independent experiments. B, amounts of ADAR1 mRNA in MEFs after treatment with 100 μm H2O2 were analyzed by real-time qRT-PCR. Data are presented as a representative of three independent experiments. C, pie charts indicate mutations in the Ig-Sμ region and the c-myc gene of MEFs after treatment with 100 μm H2O2. Sectors of the pie are proportional to the number of sequences with a given number of mutations. The number of sequences is shown in the center of the pie chart. A frequency of mutations is described below the pie chart.
DISCUSSION
Frequencies of somatic mutations in the Ig-Sμ region of IgM B cells and CD4 T cells from spleens of naive Bcl6-KO mice were higher than IgM B cells and CD4 T cells (p = 0.031) from naive WT mice, respectively. Ectopic AID induces somatic mutations in various genes of T cells from AID-Tg mice (26), but we could not detect AID mRNA in these cells from Bcl6-KO mice by RT-PCR (data not shown). Expression of ADAR1 among ADAR family genes was detected in these Bcl6-KO cells, and ADAR1 overexpression increased mutation frequencies in the Ig-Sμ region and the c-myc gene of IgG1 B cells. Exogenous ADAR1 induced somatic mutations in the Ig-Sμ region of IgM B cells from ADAR1-Tg mice and in the Ig-Sμ region and the c-myc gene of WT MEFs at P5 without AID expression. Furthermore, endogenous ADAR1 expression was observed in senescent WT and Bcl6-KO MEFs, and mutation frequencies in the Ig-Sμ region and the c-myc gene of these senescent MEFs increased. Mutations from A/T to G/C were major among those mutations. Thus, the large amount of ADAR1 can induce adenosine-targeted DNA mutations in murine cells without AID expression.
ADAR1 selectively edits in vivo two adenosines to inosines in the serotonin 2C receptor pre-mRNA of nervous tissue (45), and the RNA edition is essential for embryogenesis (46). A nuclear localization signal and a Z-DNA binding domain are present near the N-terminal region of ADAR1, and endogenous ADAR1 is localized in both nucleus and cytoplasm (31, 47). These results suggest that the molecular mechanism of somatic mutations induced by exogenous ADAR1 is explained by a DNA deamination model similar to the core process of AID-induced DNA mutations. ADAR1 may induce adenosine deamination on genomic DNA at replication. This deaminating adenosine as inosine will be subsequently transcribed as guanosine by replication and eventually become fixed in one of the daughter cells with inosine-C base pair mismatches. These mismatches recruit DNA repair machinery, and attempted repair of these lesions results in induction of somatic mutations. However, it is still possible that ADAR1 functions as a catalytic subunit of a RNA-editing holoenzyme for a novel inducer of somatic mutations. Further study is required to elucidate mechanisms of ADAR1-induced DNA mutations.
Variable regions of rearranged Ig genes in GC B cells are physiologically mutated by AID (8), and the state of open chromatin including histone acetylation is required for AID-induced SHM (48, 49). A constant region in the same transcription unit is protected from AID-induced mutations. Induction of mutations at the variable region but not the constant-region of the rearranged Ig gene in a BL2 cell line was associated with hyperacetylation of histone at chromatin of the Ig gene, and AID overexpression induced somatic mutations in both variable and constant regions (49). Overexpression of AID lacking a functional nuclear export signal caused more mutations in a nonphysiologic target gene in fibroblasts (50). These results suggest that chromatin structure affects induction of AID-induced somatic mutations and that the state of chromatin associated with high levels of transcription may provide sufficient accessibility for physiologic AID (12, 15). Although the Ig-Sμ region is not transcribed in MEFs, ADAR1 overexpression induced adenosine-targeted DNA mutations in the Ig-Sμ region of WT MEFs at P5, and the mutations, albeit a low frequency, were induced in WT MEFs at a senescent stage accompanied with endogenous ADAR1 expression. ChIP assay demonstrated that chromatin histone at the Ig-Sμ region was not acetylated in WT MEFs until P6 but was acetylated in senescent WT MEFs at P11 (data not shown). These results suggest that chromatin structure with histone acetylation is required for induction of somatic mutations by a physiologic level of ADAR1.
Cell senescence was accompanied with expression of endogenous ADAR1 in MEFs, and adenosine-targeted DNA mutations were accumulated in senescent MEFs. Because frequencies of mutations in the Ig-Sμ region and the c-myc gene of senescent Bcl6-KO MEFs were higher than senescent WT MEFs, Bcl6 may protect senescent cells from accumulation of somatic mutations. IgM B cells, CD4 T cells, and kidney cells (data not shown) from naive Bcl6-KO mice also accumulated adenosine-targeted DNA mutations in the Ig-Sμ region with overexpression of the endogenous ADAR1 gene. Bcl6 binds to the Bcl6-binding sequences (regions 2 and 3) in the ADAR1 promoter region and represses its promoter activity by recruiting the histone deacetylase complex (4). Bcl6 is ubiquitously expressed, and thus, Bcl6 suppresses endogenous ADAR1 expression to avoid unexpected DNA mutations in various cells. Frequencies of somatic mutations in the Ig-Sμ region and the c-myc gene of Bcl6-KO MEFs transfected with the ADAR1 gene were higher than ADAR1-transfected WT MEFs, suggesting that Bcl6-dependent protection against somatic mutations cannot be explained only by repression of ADAR1 expression. The chromatin histone at the Ig-Sμ region of Bcl6-KO MEFs was acetylated at a senescent stage (data not shown). There is a Bcl6-binding sequence at ∼500 bp upstream of the Ig-Sμ region and Bcl6 in nuclear extracts from IgM B cells specifically bound to the sequence (data not shown). These results suggest that Bcl6 protects its target genes from somatic mutations by deacetylating chromatin histone at its target sites. At physiologic condition, Bcl6 may protect senescent cells from accumulation of adenosine-targeted DNA mutations by repressing ADAR1 expression and by deacetylating chromatin histone at its target sites.
The exogenous ADAR1 increased a frequency of SHM from A/T to G/C but not from C/G to T/A in IgG1 B cells from spleens of ADAR1-Tg mice 10 days after immunization. The endogenous ADAR1 expression was suppressed in WT GC B cells with a large amount of Bcl6 until day 7 after immunization but became detectable in WT GC B cells on day 10 after immunization (data not shown). Thus, this detectable ADAR1 in GC B cells may be involved in A-T targeted SHM, including a suite of DNA repair polymerases and enzymes led by an A-T mutator, DNA polymerase-η (51, 52). ADAR1 selectively edits two adenosines to inosines in serotonin 2C receptor pre-mRNA (45), and this substrate specificity is the same as for polymerase-η copying off a DNA template, which mutates adenosine preferentially when preceded by adenosine or thymidine. Furthermore, the recent report demonstrated the strong statistical association between quantitative patterns of dsRNA secondary structure and adenosine to guanosine mutation spectrum in variable regions of rearranged Ig genes, providing evidence of a link between SHM and adenosine to inosine RNA editing (53). This implies that an RNA editing step could play a significant role in induction of SHM and that a physiologic level of ADAR1 in association with AID may be one of mutators for A-T targeted SHM in GC B cells at their late differentiation phase. Further study is required for elucidating a role of ADAR1 in induction of A-T targeted SHM using GC B cells from ADAR1-cKO mice.
In summary, ADAR1 is a molecular target of Bcl6 and ADAR1 overexpression increased frequencies of adenosine-targeted DNA mutations in the Ig-Sμ region and the c-myc gene of IgG B cells and MEFs. Endogenous ADAR1 expression was observed in senescent MEFs, and mutation frequencies in the Ig-Sμ region and the c-myc gene of senescent Bcl6-KO MEFs were higher than senescent WT MEFs. Thus, ADAR1 induces adenosine-targeted DNA mutations in senescent cells, and Bcl6 may protect senescent cells from accumulation of the somatic mutations at physiologic condition.
Acknowledgments
We thank Dr. K. Rajewsky (Harvard University) for CD21-cre Tg mice and S. Nakamura for secretarial assistance.
This work was supported by Grants-in-aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan and Global COE Program (Global Center for Education and Research in Immune System Regulation and Treatment), MEXT, Japan. The work was also supported by grants from the National Institutes of Health, the Ellison Medical Foundation, and the Commonwealth Universal Research Enhancement Program, Pennsylvania Department of Health (to K. N.).
- MEF
- mouse embryonic fibroblast
- AID
- activation-induced cytidine deaminase
- GC
- germinal center
- Ig-Sμ
- IgM class-switch
- ADAR1
- adenosine deaminase acting on RNA1
- Tg
- transgenic
- ADAR1-cKO
- ADAR1-conditional deficient
- P
- passage
- NP-CG
- (4-hydroxy-3-nitrophenyl)acetyl-chicken γ globulin
- SHM
- somatic hypermutation
- Ab
- antibody
- qRT
- real-time quantitative RT.
REFERENCES
- 1. Vijg J. (2000) Somatic mutations and aging: a re-evaluation. Mutat. Res. 447, 117–135 [DOI] [PubMed] [Google Scholar]
- 2. Karanjawala Z. E., Lieber M. R. (2004) DNA damage and aging. Mech. Ageing Dev. 125, 405–416 [DOI] [PubMed] [Google Scholar]
- 3. Shvarts A., Brummelkamp T. R., Scheeren F., Koh E., Daley G. Q., Spits H., Bernards R. (2002) A senescence rescue screen identifies BCL6 as an inhibitor of anti-proliferative p19(ARF)-p53 signaling. Genes Dev. 16, 681–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wong C. W., Privalsky M. L. (1998) Components of the SMRT corepressor complex exhibit distinctive interactions with the POZ domain oncoproteins PLZF, PLZF-RARα, and BCL-6. J. Biol. Chem. 273, 27695–27702 [DOI] [PubMed] [Google Scholar]
- 5. Phan R. T., Dalla-Favera R. (2004) The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature 432, 635–639 [DOI] [PubMed] [Google Scholar]
- 6. Lombard D. B., Chua K. F., Mostoslavsky R., Franco S., Gostissa M., Alt F. W. (2005) DNA repair, genome stability, and aging. Cell 120, 497–512 [DOI] [PubMed] [Google Scholar]
- 7. Revy P., Muto T., Levy Y., Geissmann F., Plebani A., Sanal O., Catalan N., Forveille M., Dufourcq-Labelouse R., Gennery A., Tezcan I., Ersoy F., Kayserili H., Ugazio A. G., Brousse N., Muramatsu M., Notarangelo L. D., Kinoshita K., Honjo T., Fischer A., Durandy A. (2000) Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2). Cell 102, 565–575 [DOI] [PubMed] [Google Scholar]
- 8. Muramatsu M., Kinoshita K., Fagarasan S., Yamada S., Shinkai Y., Honjo T. (2000) Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 102, 553–563 [DOI] [PubMed] [Google Scholar]
- 9. Muramatsu M., Sankaranand V. S., Anant S., Sugai M., Kinoshita K., Davidson N. O., Honjo T. (1999) Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J. Biol. Chem. 274, 18470–18476 [DOI] [PubMed] [Google Scholar]
- 10. Muramatsu M., Nagaoka H., Shinkura R., Begum N. A., Honjo T. (2007) Discovery of activation-induced cytidine deaminase, the engraver of antibody memory. Adv. Immunol. 94, 1–36 [DOI] [PubMed] [Google Scholar]
- 11. Goodman M. F., Scharff M. D., Romesberg F. E. (2007) AID-initiated purposeful mutations in immunoglobulin genes. Adv. Immunol. 94, 127–155 [DOI] [PubMed] [Google Scholar]
- 12. Bransteitter R., Pham P., Scharff M. D., Goodman M. F. (2003) Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc. Natl. Acad. Sci. U.S.A. 100, 4102–4107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chaudhuri J., Tian M., Khuong C., Chua K., Pinaud E., Alt F. W. (2003) Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature 422, 726–730 [DOI] [PubMed] [Google Scholar]
- 14. Dickerson S. K., Market E., Besmer E., Papavasiliou F. N. (2003) AID mediates hypermutation by deaminating single stranded DNA. J. Exp. Med. 197, 1291–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pham P., Bransteitter R., Petruska J., Goodman M. F. (2003) Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature 424, 103–107 [DOI] [PubMed] [Google Scholar]
- 16. Ramiro A. R., Stavropoulos P., Jankovic M., Nussenzweig M. C. (2003) Transcription enhances AID-mediated cytidine deamination by exposing single-stranded DNA on the nontemplate strand. Nat. Immunol. 4, 452–456 [DOI] [PubMed] [Google Scholar]
- 17. Sohail A., Klapacz J., Samaranayake M., Ullah A., Bhagwat A. S. (2003) Human activation-induced cytidine deaminase causes transcription-dependent, strand-biased C to U deaminations. Nucleic Acids Res. 31, 2990–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rada C., Di Noia J. M., Neuberger M. S. (2004) Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol. Cell 16, 163–171 [DOI] [PubMed] [Google Scholar]
- 19. Dent A. L., Shaffer A. L., Yu X., Allman D., Staudt L. M. (1997) Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science 276, 589–592 [DOI] [PubMed] [Google Scholar]
- 20. Fukuda T., Yoshida T., Okada S., Hatano M., Miki T., Ishibashi K., Okabe S., Koseki H., Hirosawa S., Taniguchi M., Miyasaka N., Tokuhisa T. (1997) Disruption of the Bcl6 gene results in an impaired germinal center formation. J. Exp. Med. 186, 439–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ye B. H., Cattoretti G., Shen Q., Zhang J., Hawe N., de Waard R., Leung C., Nouri-Shirazi M., Orazi A., Chaganti R. S., Rothman P., Stall A. M., Pandolfi P. P., Dalla-Favera R. (1997) The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nat. Genet. 16, 161–170 [DOI] [PubMed] [Google Scholar]
- 22. Phan R. T., Saito M., Basso K., Niu H., Dalla-Favera R. (2005) BCL6 interacts with the transcription factor Miz-1 to suppress the cyclin-dependent kinase inhibitor p21 and cell cycle arrest in germinal center B cells. Nat. Immunol. 6, 1054–1060 [DOI] [PubMed] [Google Scholar]
- 23. Shaffer A. L., Yu X., He Y., Boldrick J., Chan E. P., Staudt L. M. (2000) BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity 13, 199–212 [DOI] [PubMed] [Google Scholar]
- 24. Dunnick W., Hertz G. Z., Scappino L., Gritzmacher C. (1993) DNA sequences at immunoglobulin switch region recombination sites. Nucleic Acids Res. 21, 365–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nagaoka H., Muramatsu M., Yamamura N., Kinoshita K., Honjo T. (2002) Activation-induced deaminase (AID)-directed hypermutation in the immunoglobulin Smu region: implication of AID involvement in a common step of class switch recombination and somatic hypermutation. J. Exp. Med. 195, 529–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Okazaki I. M., Hiai H., Kakazu N., Yamada S., Muramatsu M., Kinoshita K., Honjo T. (2003) Constitutive expression of AID leads to tumorigenesis. J. Exp. Med. 197, 1173–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Toyama H., Okada S., Hatano M., Takahashi Y., Takeda N., Ichii H., Takemori T., Kuroda Y., Tokuhisa T. (2002) Memory B cells without somatic hypermutation are generated from Bcl6-deficient B cells. Immunity 17, 329–339 [DOI] [PubMed] [Google Scholar]
- 28. Nishikura K. (2010) Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 79, 321–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Seeburg P. H. (2002) A-to-I editing: new and old sites, functions and speculations. Neuron 35, 17–20 [DOI] [PubMed] [Google Scholar]
- 30. Ichii H., Sakamoto A., Hatano M., Okada S., Toyama H., Taki S., Arima M., Kuroda Y., Tokuhisa T. (2002) Role for Bcl-6 in the generation and maintenance of memory CD8+ T cells. Nat. Immunol. 3, 558–563 [DOI] [PubMed] [Google Scholar]
- 31. Yang J. H., Nie Y., Zhao Q., Su Y., Pypaert M., Su H., Rabinovici R. (2003) Intracellular localization of differentially regulated RNA-specific adenosine deaminase isoforms in inflammation. J. Biol. Chem. 278, 45833–45842 [DOI] [PubMed] [Google Scholar]
- 32. Wildin R. S., Wang H. U., Forbush K. A., Perlmutter R. M. (1995) Functional dissection of the murine lck distal promoter. J. Immunol. 155, 1286–1295 [PubMed] [Google Scholar]
- 33. Wang Q., Miyakoda M., Yang W., Khillan J., Stachura D. L., Weiss M. J., Nishikura K. (2004) Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J. Biol. Chem. 279, 4952–4961 [DOI] [PubMed] [Google Scholar]
- 34. Kraus M., Alimzhanov M. B., Rajewsky N., Rajewsky K. (2004) Survival of resting mature B lymphocytes depends on BCR signaling via the Igα/β heterodimer. Cell 117, 787–800 [DOI] [PubMed] [Google Scholar]
- 35. Todaro G. J., Green H. (1963) Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 17, 299–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Petersen S., Casellas R., Reina-San-Martin B., Chen H. T., Difilippantonio M. J., Wilson P. C., Hanitsch L., Celeste A., Muramatsu M., Pilch D. R., Redon C., Ried T., Bonner W. M., Honjo T., Nussenzweig M. C., Nussenzweig A. (2001) AID is required to initiate Nbs1/γ-H2AX focus formation and mutations at sites of class switching. Nature 414, 660–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yoshida K., Sakamoto A., Yamashita K., Arguni E., Horigome S., Arima M., Hatano M., Seki N., Ichikawa T., Tokuhisa T. (2006) Bcl6 controls granzyme B expression in effector CD8+ T cells. Eur. J. Immunol. 36, 3146–3156 [DOI] [PubMed] [Google Scholar]
- 38. George C. X., Samuel C. E. (1999) Characterization of the 5′-flanking region of the human RNA-specific adenosine deaminase ADAR1 gene and identification of an interferon-inducible ADAR1 promoter. Gene 229, 203–213 [DOI] [PubMed] [Google Scholar]
- 39. Takeda N., Arima M., Tsuruoka N., Okada S., Hatano M., Sakamoto A., Kohno Y., Tokuhisa T. (2003) Bcl6 is a transcriptional repressor for the IL-18 gene. J. Immunol. 171, 426–431 [DOI] [PubMed] [Google Scholar]
- 40. Liu Y., Emeson R. B., Samuel C. E. (1999) Serotonin-2C receptor pre-mRNA editing in rat brain and in vitro by splice site variants of the interferon-inducible double-stranded RNA-specific adenosine deaminase ADAR1. J. Biol. Chem. 274, 18351–18358 [DOI] [PubMed] [Google Scholar]
- 41. Maas S., Rich A., Nishikura K. (2003) A-to-I RNA editing: recent news and residual mysteries. J. Biol. Chem. 278, 1391–1394 [DOI] [PubMed] [Google Scholar]
- 42. Takahashi Y., Dutta P. R., Cerasoli D. M., Kelsoe G. (1998) In situ studies of the primary immune response to (4-hydroxy-3-nitrophenyl)acetyl. V. Affinity maturation develops in two stages of clonal selection. J. Exp. Med. 187, 885–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dasari A., Bartholomew J. N., Volonte D., Galbiati F. (2006) Oxidative stress induces premature senescence by stimulating caveolin-1 gene transcription through p38 mitogen-activated protein kinase/Sp1-mediated activation of two GC-rich promoter elements. Cancer Res. 66, 10805–10814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wolf F. I., Torsello A., Covacci V., Fasanella S., Montanari M., Boninsegna A., Cittadini A. (2002) Oxidative DNA damage as a marker of aging in WI-38 human fibroblasts. Exp. Gerontol. 37, 647–656 [DOI] [PubMed] [Google Scholar]
- 45. Burns C. M., Chu H., Rueter S. M., Hutchinson L. K., Canton H., Sanders-Bush E., Emeson R. B. (1997) Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature 387, 303–308 [DOI] [PubMed] [Google Scholar]
- 46. Wang Q., Khillan J., Gadue P., Nishikura K. (2000) Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science 290, 1765–1768 [DOI] [PubMed] [Google Scholar]
- 47. Eckmann C. R., Neunteufl A., Pfaffstetter L., Jantsch M. F. (2001) The human but not the Xenopus RNA-editing enzyme ADAR1 has an atypical nuclear localization signal and displays the characteristics of a shuttling protein. Mol. Biol. Cell 12, 1911–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Odegard V. H., Kim S. T., Anderson S. M., Shlomchik M. J., Schatz D. G. (2005) Histone modifications associated with somatic hypermutation. Immunity 23, 101–110 [DOI] [PubMed] [Google Scholar]
- 49. Woo C. J., Martin A., Scharff M. D. (2003) Induction of somatic hypermutation is associated with modifications in immunoglobulin variable region chromatin. Immunity 19, 479–489 [DOI] [PubMed] [Google Scholar]
- 50. McBride K. M., Barreto V., Ramiro A. R., Stavropoulos P., Nussenzweig M. C. (2004) Somatic hypermutation is limited by CRM1-dependent nuclear export of activation-induced deaminase. J. Exp. Med. 199, 1235–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rogozin I. B., Pavlov Y. I., Bebenek K., Matsuda T., Kunkel T. A. (2001) Somatic mutation hotspots correlate with DNA polymerase η error spectrum. Nat. Immunol. 2, 530–536 [DOI] [PubMed] [Google Scholar]
- 52. Zeng X., Winter D. B., Kasmer C., Kraemer K. H., Lehmann A. R., Gearhart P. J. (2001) DNA polymerase η is an A-T mutator in somatic hypermutation of immunoglobulin variable genes. Nat. Immunol. 2, 537–541 [DOI] [PubMed] [Google Scholar]
- 53. Steele E. J., Lindley R. A., Wen J., Weiller G. F. (2006) Computational analyses show A-to-G mutations correlate with nascent mRNA hairpins at somatic hypermutation hotspots. DNA Repair 5, 1346–1363 [DOI] [PubMed] [Google Scholar]