Background: Glycolytic enzymes (GEs) are membrane-bound in oxygenated erythrocytes, but some GEs do not bind to the NH2 terminus of band 3.
Results: Additional GE binding sites are identified on erythrocyte membrane proteins that associate with band 3.
Conclusion: Complexes of GEs exist on the membrane in areas where ATP is consumed.
Significance: The architecture of the GE complex is defined in greater detail.
Keywords: Enzymes, Erythrocyte, Glycolysis, Membrane Proteins, Membrane Structure, Ankyrin Complex, Erythrocyte Band 3, Glycolysis Metabolon, Glycolytic Enzyme Complex, Junctional Complex
Abstract
Glycolytic enzymes (GEs) have been shown to exist in multienzyme complexes on the inner surface of the human erythrocyte membrane. Because no protein other than band 3 has been found to interact with GEs, and because several GEs do not bind band 3, we decided to identify the additional membrane proteins that serve as docking sites for GE on the membrane. For this purpose, a method known as “label transfer” that employs a photoactivatable trifunctional cross-linking reagent to deliver a biotin from a derivatized GE to its binding partner on the membrane was used. Mass spectrometry analysis of membrane proteins that were biotinylated following rebinding and photoactivation of labeled GAPDH, aldolase, lactate dehydrogenase, and pyruvate kinase revealed not only the anticipated binding partner, band 3, but also the association of GEs with specific peptides in α- and β-spectrin, ankyrin, actin, p55, and protein 4.2. More importantly, the labeled GEs were also found to transfer biotin to other GEs in the complex, demonstrating for the first time that GEs also associate with each other in their membrane complexes. Surprisingly, a new GE binding site was repeatedly identified near the junction of the membrane-spanning and cytoplasmic domains of band 3, and this binding site was confirmed by direct binding studies. These results not only identify new components of the membrane-associated GE complexes but also provide molecular details on the specific peptides that form the interfacial contacts within each interaction.
Introduction
As our understanding of subcellular structures in eukaryotes improves, a conclusion is emerging that proteins in common pathways often organize into complexes that facilitate the performance of the pathway's metabolic/signaling function (1, 2). In the case of the human erythrocyte, complexes have been suggested to exist for gas transport (3–6), cation efflux pumping (7–9), and glycolysis (10). Determination of whether additional complexes also exist to respond to oxidative stress (11), mechanical deformation (12), or hemostatic stimuli (13) may require further characterization.
It is now generally accepted that human erythrocyte band 3 constitutes the center of organization of a glycolytic enzyme (GE)2 complex on the red cell membrane (10). Binding sites of glyceraldehyde-3-phosphate dehydrogenase (GAPDH), aldolase, and phosphofructokinase have been carefully mapped to sequences near the NH2 terminus of band 3, and association of GEs with this site has been shown to lead to enzyme inhibition (14–17). Moreover, assembly of the GE complex on band 3 has been shown to be regulated by both the oxygenation state of hemoglobin and the phosphorylation state of band 3 (10). Recently, Su et al. (18) have reported the binding of GAPDH to the COOH terminus of both erythrocyte and kidney band 3, suggesting that additional GE docking sites may exist elsewhere on the erythrocyte membrane. This hypothesis has, in fact, been strengthened by observations showing that LDH and pyruvate kinase isoform M1/M2 (PK) do not bind to the common GE site at the NH2 terminus of band 3, yet their membrane association is regulated by hemoglobin oxygenation and band 3 phosphorylation similarly to GEs that dock at the NH2-terminal site (17). Moreover, an analogous band 3-centered complex of GEs has been found in murine erythrocytes (19), where defects or deletions in band 3-associated proteins have been observed to exert profound effects on GE complex stability, suggesting the involvement of additional membrane proteins in the architecture of the GE complex.
In an effort to identify the other major components of the erythrocyte membrane GE complex, a method known as “label transfer” was employed to detect membrane proteins with which the major GEs interact. The method involves the labeling of a desired “bait” protein (i.e. a glycolytic enzyme) with sulfo-SBED (a photoactivatable trifunctional biotin transfer reagent) and then allowing the labeled GE to reassemble on the membrane. After exposure to UV light (which triggers biotin transfer to any proximal membrane protein), the newly biotinylated membrane proteins are identified by Western blot analysis and/or mass spectrometry. Using this method, we report that GEs associate with highly specific peptides on β-spectrin, ankyrin, actin, protein 4.2, and other GEs as well as a second prominent site on band 3.
EXPERIMENTAL PROCEDURES
GAPDH, aldolase, lactate dehydrogenase (LDH), PK, all rabbit muscle enzymes, and endoproteinase Lys-C (sequencing grade) were purchased from Roche Applied Science as suspensions in ammonium sulfate. Streptavidin HRP was purchased from Jackson ImmunoResearch. Sulfo-SBED, micro-BCA assay kit, SuperSignal West Pico chemiluminescent substrate, and CL-Xposure film were purchased from Pierce. Softlink Soft Release Avidin resin and sequencing grade trypsin were purchased from Promega. d-Biotin was from Supelco. RapiGest was purchased from Waters. dl-Dithiothreitol, phenylmethylsulfonyl fluoride (PMSF), and iodoacetamide were obtained from Sigma. Molecular weight markers (SeeBlue prestained standard) were purchased from Invitrogen.
Labeling of the GE with SulfoSBED
Enzymes were dialyzed overnight at 4 °C against a buffer containing 50 mm Hepes, 150 mm NaCl, 0.5 mm PMSF, and pH 7.3. Sulfo-SBED (1 mg), a photocross-linking reagent that reacts in the dark with primary amines (e.g. lysine side chains), was dissolved in 100 μl of N,N-dimethylformamide in the dark and used as indicated by the manufacturer at a 1:1 molar ratio to label the GEs. Labeling was performed in the dark at room temperature for 30 min under constant stirring. Samples were then dialyzed overnight at 4 °C in the dark against Hepes buffer to remove all unreacted cross-linker and then either used immediately or stored frozen in liquid nitrogen for later use.
Preparation of Erythrocyte Ghosts
Blood was obtained from normal volunteers and sedimented immediately at 1000 × g for 10 min. Plasma and buffy coat were removed, and cells were carefully washed three times in 0.9% NaCl, each time discarding the upper layer of red cells to assure complete elimination of white cells. After washing, each red cell suspension was inspected for contaminating leukocytes using a hemocytometer and washed again until residual leukocytes constituted <0.01% of the cells. When desired, the washed erythrocytes were treated with chymotrypsin to cleave band 3 at an extracellular site (20).
Membranes were prepared from washed erythrocytes following established protocols (21). Membranes were invariably used within 48 h of preparation.
Identification of GE Binding Partners on Human Erythrocyte Membranes
Freshly prepared leaky membranes and enzymes were separately dialyzed against incubation buffer (5 mm PO4, 0.5 mm EDTA, 0.5 mm PMSF, pH 7.0), after which their protein concentrations were determined by a BCA assay. For GE reconstitution purposes, the band 3 content of the dialyzed membranes was assumed to be ∼29% of the total membrane protein (22, 23), and based on this calculation, a 2–3-fold molar excess of the labeled GE over band 3 was added to the membrane suspension. Following incubation of the mixture for 90 min in the dark at 4 °C under agitation, the suspension was washed twice in the dark with incubation buffer to remove unbound GE. The washed pellet was carefully resuspended by gentle vortexing and then exposed to UV light (model UVM-57, UVP (Upland, CA)) for 15 min on ice at a distance of 5 cm from the membrane pellet. The resulting photolabeled membranes were then incubated for 15 min in 5 mm sodium phosphate buffer, pH 8, containing 0.5 mm EDTA and 100 mm DTT to reduce the disulfide bond linking the photolabeling reagent to the GE. After washing the sample again in the same DTT-containing buffer and then finally in the buffer lacking DTT, the samples were incubated for 15 min in 50 mm iodoacetamide to block reformation of disulfide bonds. The sample was then washed in the same buffer lacking iodoacetamide and divided into two aliquots; one was mixed with SDS sample buffer for analysis by SDS-PAGE, and the other was frozen at −80 °C for subsequent trypsin digestion and characterization by mass spectrometry.
Electrophoresis and Immunoblotting
Samples were allowed to stand at 50 °C for 15 min before electrophoretic separation in 10% polyacrylamide gels. After SDS-PAGE, samples were electrophoretically transferred to nitrocellulose membranes according to Bio-Rad protocols, and proteins were visualized with Ponceau S stain (24) and photographed. Specific visualization of those proteins that were biotinylated during the photocross-linking process was achieved by washing the membranes in Tris-buffered saline (TBS) (20 mm Tris-HCl, pH 7.5, 0.5 m NaCl, and 0.05% Tween 20) and blocking nonspecific sites overnight at 4 °C with 5% powdered milk in the same buffer. Then, after 30 min of incubation with streptavidin-HRP (1:60,000), the membranes were washed for 60 min in TBS buffer and reacted with SuperSignal West Pico chemiluminescent substrate for 1 min. Finally, the membranes were exposed to a CL-Xposure film for 1 min, and the film was developed using standard procedures.
Sample Preparation for Mass Spectrometry
Biotinylated membrane samples were centrifuged at 4 °C and, after discarding the supernatant, were resuspended at a final concentration of 1% RapiGest (Waters) containing 100 mm DTT. Solubilized samples were incubated for 10 min at 40 °C to promote maximal membrane dissolution and then diluted 10× with warm 20 mm Tris buffer, pH 8. Samples were then sonicated for 1 h and digested overnight at 37 °C with endoproteinase Lys-C (0.1 μg per 90 μg of protein) to unfold the proteins and generate large peptide fragments. To create smaller peptides appropriate for flight in the mass spectrometer, the samples were again digested for 24 h at 37 °C with trypsin at a protease/protein ratio of 1:50. RapiGest detergent was then removed by incubation for 40 min at 37 °C in 5% acetic acid followed by centrifugation for 10 min at 23,000 × g. The supernatant was collected and neutralized to pH 7 with 1 m NH4HCO3, pH 8.3, in preparation for affinity purification.
Isolation of biotinylated peptides by avidin affinity chromatography was accomplished with monomeric avidin-derivatized beads (Promega) according to established protocols (25). Samples containing the eluted affinity-purified peptides were dried using a Labconco vacuum concentrator and stored frozen until mass spectrometry analysis.
Nanoflow LC-MS/MS
Eight microliters of sample were injected into an Agilent 1100 HPLC system linked to an in-house reverse phase C18 capillary column packed with 5-μm C18 Magic beads (Michrom; 75-μm inner diameter and 12-cm bed length). Peptides were eluted with a shallow linear gradient over 60 min of 0.1% formic acid in water to 0.1% formic acid in 100% acetonitrile at a flow rate of 0.3 μl/min. The electrospray ionization emitter tip was generated on the prepacked column with a laser puller (model P-2000, Sutter Instrument Co.). The Agilent 1100 HPLC system was coupled online with an LTQ linear ion trap mass spectrometer (Thermo Electron, San Jose, CA). The mass spectrometer was operated in the data-dependent mode, in which a full scan MS was followed by MS/MS scans of the 10 most abundant ions with +2 to +3 charge states. The mass exclusion time was 180 s.
Peptide and Protein Identification
The MS/MS data were converted to an mzXML format using the open source Trans-Peptide Pipeline (TPP) software (version 2.9.4), and the resulting mzXML files were searched against the International Protein Index human database (Version 3.68 with 87,061 protein entries) using the SEQUEST algorithm on the Sorcerer IDA server (software version 2.5.6; SageN, Inc., San Jose, CA). Peptide mass tolerance was set at 3.0 atomic mass units, and MS/MS tolerance was set internally by the software, with values varying from 0 to 1 atomic mass unit. Search criteria allowed detection of methionine oxidation (15.9949 Da), cysteine alkylation (+57.0214 Da), and semitryptic digestion, where a maximum of 2 missed cleavages in any peptide was permitted. Minimum protein and peptide probabilities were set at 0.8 and 0.9, respectively (26, 27).
Generation of a Fragment of the Cytoplasmic Domain of Band 3 Comprising Residues 330–391
An expression vector for residues 330–391 of band 3 was generated using the ThioFusion Expression Kit from Invitrogen according to the manufacturer's instructions. Briefly, the cDNA for residues 330–391 of band 3 was amplified by PCR using the following primers: forward, 5′-caggcactgctcagtct-3′; reverse, 5′-gtagcggcgccggatatc-3′. The resulting PCR product was inserted into the pBAD/Thio-TOPO vector. After sequencing to assure proper insertion, the plasmid was transformed into TOP10 E. coli cells, and the band 3 peptide comprising residues 330–391 was expressed as a thioredoxin fusion protein containing a His tag at the COOH terminus and thioredoxin at its NH2 terminus. The fusion protein was purified by nickel-NTA bead chromatography (Qiagen) and analyzed by SDS-PAGE and Western blotting using a polyclonal antibody against CDB3. When desired, bovine enterokinase (Invitrogen) was used to remove the NH2-terminal thioredoxin.
Analysis of GE Binding to the Peptide Fragment Comprising Residues 330–391 of CDB3
The peptide fragment described above was dialyzed into binding buffer (7.5 mm NaH2PO4, 90 mm KCl, 20 mm imidazole, 10% sucrose, 0.4 mm PMSF, and 0.1% BSA, pH 7.3) and mixed with increasing concentrations of GEs that had previously been dialyzed against the same buffer. Binding was allowed to proceed overnight at 4 °C with rotation, after which the samples and appropriate controls were allowed to interact for 30 min at 4 °C with nickel beads that had been equilibrated in the same buffer containing 1% BSA. Beads were pelleted and washed three times with binding buffer and again with the same buffer lacking BSA prior to dot blot analysis.
Nitrocellulose membranes were spotted with the above samples/controls using a guided plastic template. When required, a set of known dilutions of pure GEs were also applied to obtain a calibration curve. Upon drying, membranes were allowed to incubate in TBS containing 5% milk to block nonspecific sites. Membranes were then incubated with the appropriate primary antibody for 60 min, washed several times to remove unbound antibody, and incubated with secondary antibody conjugated to HRP. Protein spots were analyzed and quantified using an Eastman Kodak Co. molecular imaging system.
Entrapment of Antibodies and Fusion Peptides into Erythrocytes
BRIC 170, nonspecific mouse IgG, or a fusion protein containing residues 330–391 of band 3 linked to thioredoxin (or buffer) were entrapped at 50% hematocrit in resealed ghosts according to published protocols (28). The resealed erythrocytes were washed in PBS containing 5 mm glucose and 0.1% BSA until the supernatant was cleared of hemoglobin, and the washed resealed ghosts were incubated at room temperature for 30 min before fixation with freshly prepared 0.5% acrolein in PBS. The resealed ghosts were then processed for immunostaining as described previously (10).
RESULTS
Interaction of GEs with Erythrocyte Membrane Proteins Is Specific
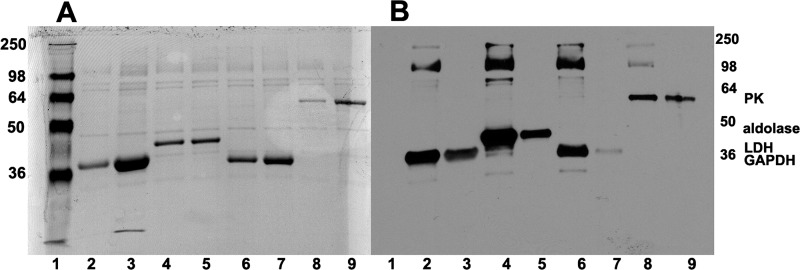
As described under “Experimental Procedures,” the glycolytic enzymes GAPDH, aldolase, LDH, and PK were each labeled with sulfo-SBED (a photoactivatable reagent that transfers biotin to adjacent proteins upon illumination with UV light) and incubated with freshly prepared erythrocyte ghosts either in the presence (lanes 3, 5, 7, and 9) or absence (lanes 2, 4, 6, and 8) of an excess of the unlabeled GE counterpart. After allowing time for the labeled GEs to exchange with the endogenous GEs (29, 30) and washing to remove unbound GEs, biotin transfer was photoactivated, and membrane proteins were separated by SDS-PAGE. Following transfer of separated proteins onto nitrocellulose, blots were analyzed for both total protein (Fig. 1A) and biotinylated protein (Fig. 1B). As seen in the Ponceau stain of Fig. 1A, all lanes showed transfer of the major erythrocyte membrane proteins to the nitrocellulose paper, with prominent transfer of the GEs that were added in excess to saturate all membrane sites. Moreover, after removal of the reversible Ponceau S dye and restaining of the same blots with streptavidin-horseradish peroxidase, analysis revealed multiple biotinylated proteins, suggesting association of labeled GEs with multiple proteins on the membrane. More specifically, reconstituted GAPDH transferred biotin not only to itself (Fig. 1B, lane 2, ∼36 kDa) but also to proteins that migrate with molecular masses near those of band 3 (101 kDa) and spectrin (∼240 kDa). Similarly, aldolase (lane 4, ∼40 kDa) and LDH (lane 6, ∼35 kDa) transferred biotin not only to themselves but also to polypeptides that migrate near protein 4.1 and/or 4.2 (∼78 kDa), band 3, and spectrin. Pyruvate kinase, in contrast, primarily transferred label to itself (lane 8, ∼57 kDa), band 3, and spectrin and/or ankyrin.
FIGURE 1.
Identification of membrane proteins labeled by photoactivated biotin transfer from sulfo-SBED-derivatized glycolytic enzymes. Sulfo-SBED-labeled enzymes were incubated with fresh erythrocyte ghosts as described under “Experimental Procedures.” Samples were separated on 10% SDS-polyacrylamide gels, transferred to nitrocellulose, and stained first with Ponceau S stain (A) to reveal all transferred proteins and, after removal of Ponceau stain, with streptavidin HRP (B) to selectively reveal only biotinylated proteins. Lane 1, molecular weight markers; lane 2, sulfo-SBED-GAPDH incubated with ghosts; lane 3, same as lane 2 but containing a 10-fold excess of unlabeled GAPDH; lane 4, sulfo-SBED-aldolase incubated with ghosts; lane 5, same as lane 4 but containing a 6-fold excess of unlabeled aldolase; lane 6, sulfo-SBED-LDH incubated with ghosts; lane 7, same as lane 6 but containing a 10-fold excess of unlabeled LDH; lane 8, sulfo-SBED-PK incubated with ghosts; lane 9, same as lane 7 but containing a 10-fold excess of unlabeled PK. Molecular weights are indicated in the right and left margins. Positions of the enzymes are labeled.
In order to determine if the above label transfer events reflect specific interactions, parallel experiments were performed precisely as above except that a ∼10-fold excess of each unlabeled GE was included in the binding and photolabeling steps to compete for specific interactions. As seen in lanes 3, 5, 7, and 9, competition with excess GE not only prevented biotin transfer to each membrane protein but also significantly reduced biotin transfer to the labeling GE. These data suggest that the binding partners identified in the photolabeling reactions, with the exception of some autologous GE labeling, associate in a specific manner with each GE. Moreover, the inability to quantitatively block GE self-labeling was expected, because intramolecular biotin transfer always occurs with this methodology (31).
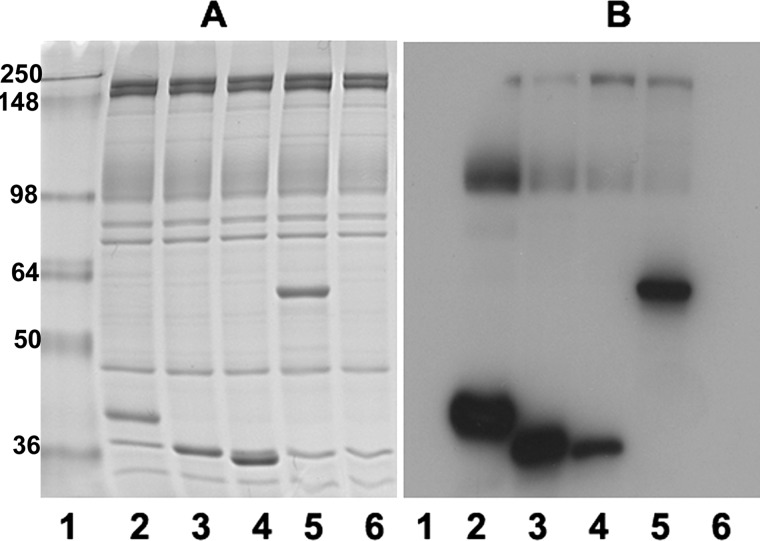
To further confirm the specificity of the sulfo-SBED label transfer reaction, we repeated the above studies except that sulfo-SBED-ovalbumin was also incubated with the leaky ghosts instead of a labeled GE (negative control). As shown in Fig. 2, sulfo-SBED-ovalbumin failed to transfer biotin to any membrane protein, although sulfo-SBED-labeled GEs transferred biotin to the usual membrane proteins in the same experiment. Together with the competition data described above, these results demonstrate that biotin transfer from sulfo-SBED-labeled GEs arises from physiological GE-protein interactions and not from transient nonspecific collisions.
FIGURE 2.
Binding of aldolase, GAPDH, LDH, PK, and ovalbumin to fresh porous ghosts. Sulfo-SBED-labeled aldolase, GAPDH, LDH, PK, and ovalbumin were incubated with fresh ghosts, as described under “Experimental Procedures,” and after washing and photoactivating, bound proteins were separated by SDS-PAGE and stained with Coomassie Blue (A) or transferred to nitrocellulose and incubated with streptavidin-HRP to reveal biotinylated proteins (B). Because ovalbumin does not bind to erythrocyte membranes, washing to remove unbound protein prevents labeling of any membrane proteins with sulfo-SBED-ovalbumin. Lane 1, molecular weight markers; lanes 2–6, ghosts that were incubated with sulfo-SBED-aldolase (lane 2), sulfo-SBED-GAPDH (lane 3), sulfo-SBED-LDH (lane 4), sulfo-SBED-PK (lane 5), and sulfo-SBED-ovalbumin (lane 6).
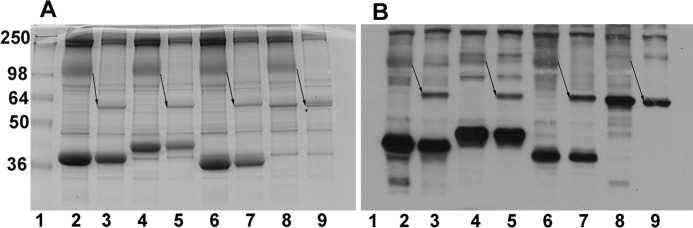
Confirmation That Band 3 Constitutes the Major Biotinylated Membrane Protein
To confirm that the biotinylated protein of ∼100 kDa is indeed band 3, intact erythrocytes were divided into two aliquots. One aliquot was treated with chymotrypsin prior to processing and analysis, and the other was left unmodified (see “Experimental Procedures”). As shown in Fig. 3, the biotinylated band at ∼100 kDa decreases to a molecular mass of ∼55 kDa (see arrows), as expected for band 3 (20), demonstrating that the anion transporter is indeed a major binding partner of each GE. Interestingly, digestion of band 3 also reveals other biotinylated proteins that are occluded by the more intense staining of band 3 in undigested cells. Whether these minor biotinylated proteins correspond to adducin or a fragment of ankyrin, both of which are labeled by this methodology (Table 1 and supplemental Table 1), cannot be ascertained from the blots.
FIGURE 3.
Treatment of intact erythrocytes with chymotrypsin shifts the ∼100 kDa band to ∼55 kDa, as expected for band 3. Ghosts prepared from control cells (lanes 2, 4, 6, and 8) or chymotrypsin-digested whole cells (lanes 3, 5, 7, and 9) were incubated with sulfo-SBED-labeled enzymes and then processed for SDS-PAGE as described under “Experimental Procedures.” Proteins were separated in 10% SDS-polyacrylamide gels and stained with Coomassie (A) or transferred to nitrocellulose and visualized with streptavidin HRP (B). The NH2-terminal half of band 3 containing the full cytoplasmic domain and part of the membrane domain migrates at ∼55 kDa following chymotrypsin digestion of intact erythrocytes (see arrows). This new 55 kDa band is labeled by each of the GE lanes (3, 5, 7, and 9). GAPDH (lanes 2 and 3), aldolase (lanes 4 and 5), LDH (lanes 6 and 7), and PK (lanes 8 and 9).
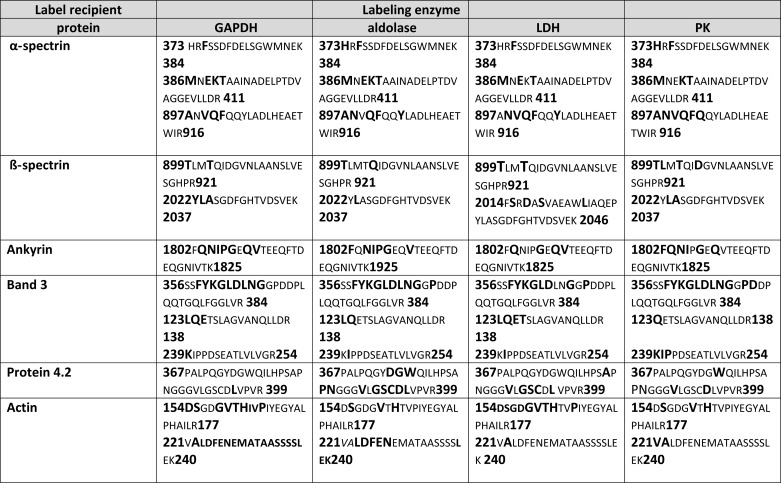
TABLE 1.
Common erythrocyte membrane peptides labeled by all glycolytic enzymes derivatized with a photoactivatable cross-linking reagent
The first column shows the identified membrane protein. The second, third, fourth, and fifth columns list the peptides tagged by GAPDH, aldolase, lactate dehydrogenase, and pyruvate kinase, respectively. The number at the beginning and end of each peptide shows where the peptide is located in the protein sequence. The boldface characters in larger font constitute the biotin-tagged amino acids.
Identification of Biotinylated Proteins and Their Labeled Peptides
Although the proteolysis results of Fig. 3 confirm the involvement of band 3 in GE binding, the complex banding patterns of the other biotinylated polypeptides prevent unequivocal identification of the remaining membrane binding partners of the GEs. To obtain more detailed information on the identities of these additional biotinylated proteins, membrane proteins were trypsin-digested, and biotinylated peptides were isolated by avidin affinity chromatography and then processed for mass spectrometry analysis. Four separate label transfer experiments using blood from three different donors were independently processed and analyzed. Mass spectrometry results were then interrogated using the International Protein Index human database, where peptides with an extra mass of 620 atomic mass units (amu), corresponding to the molecular weight of the attached biotin, were identified (32). Based on the fragmentation pattern, the specific amino acid in each labeled peptide was also identified. Moreover, in order to eliminate any false positives, only peptides with probability scores of ≥0.9 were considered. For ease of analysis, red blood cell membrane peptides that were labeled by all sulfo-SBED-GEs are listed in Table 1, whereas peptides that were labeled by only some of the derivatized GEs are shown in supplemental Table 1, with labeled amino acids always shown in boldface type.
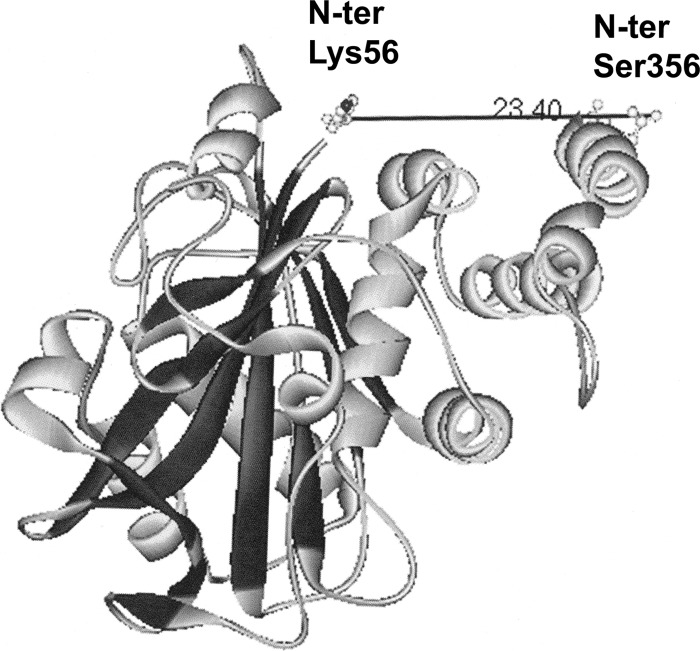
Several conclusions can be drawn from the data. First, relatively few peptides are labeled in each membrane protein, and in many cases, the same peptide is labeled by all four GEs. Moreover, labeling is generally not limited to a single amino acid but rather extends to multiple amino acids within the same peptide (Table 1 and supplemental Table 1). For example, the peptide comprising residues 356–384 of band 3 is strongly labeled by GAPDH, aldolase, LDH, and PK (see labeling map in Fig. 3A). Furthermore, this peptide is biotinylated on 9–11 different residues, depending on which GE is used for labeling. This result is perhaps not surprising, because analysis of the crystal structure of CDB3 suggests that residues 356–384 must lie adjacent to the well established GE binding site at the NH2 terminus of band 3 (Fig. 5) (33). As will be shown below, this newly identified GE binding peptide also displays high affinity for the various GEs in direct binding assays (Fig. 6) and is able to displace GEs when entrapped inside resealed red blood cells (Fig. 8B).
FIGURE 5.
Crystal structure of CDB3 (Protein Data Bank entry 1HYN). The distance in angstroms from the first resolved amino acid at the NH2 terminus of band 3 (Lys-56) to the last resolved amino acid near the COOH terminus of CDB3 (Ser-356) is shown.
FIGURE 6.
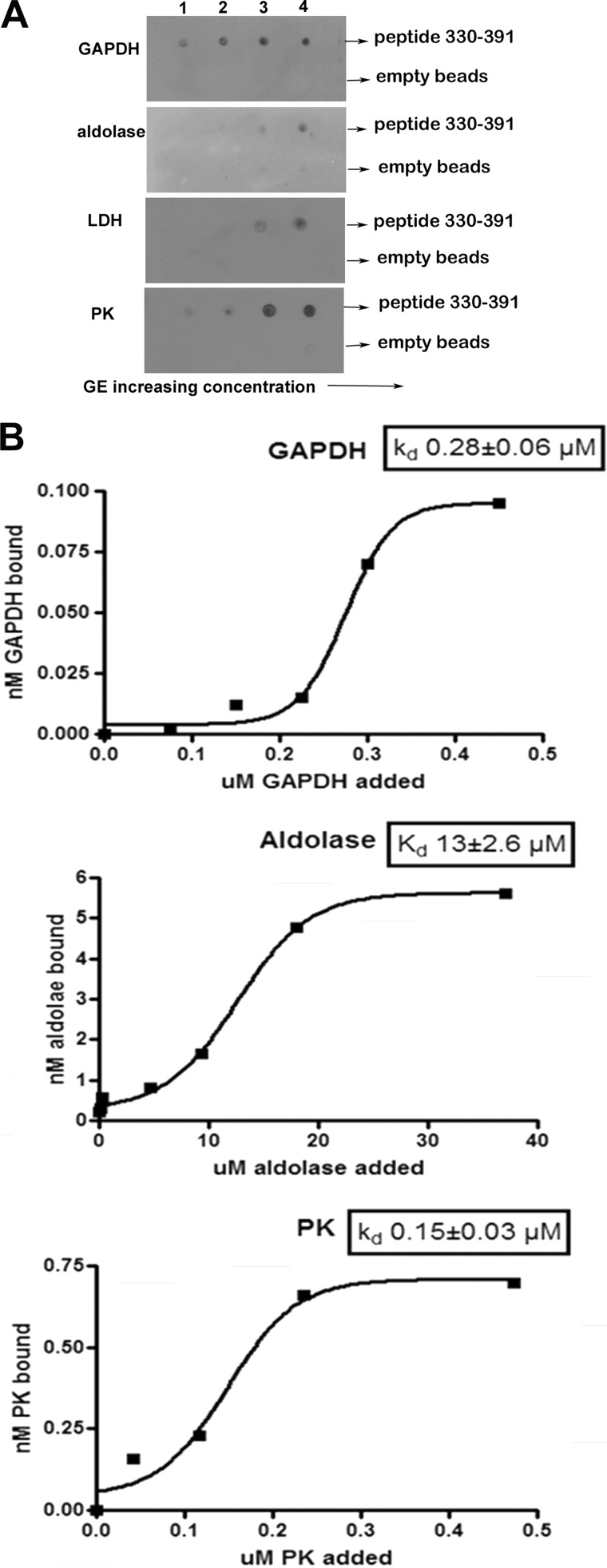
Effect of increasing GE concentration on the binding of GEs to the peptide comprising residues 330–391 of band 3. A, evaluation of the affinity of GEs for the His-tagged fusion protein containing residues 330–391 of band 3 by nickel bead pull-down assay. Fusion protein (0.17 μmol) was incubated with increasing amounts (5–50 nmol) of GE in a total volume of 100 μl overnight at 4 °C. Nickel beads blocked with binding buffer containing 5% BSA were added, and the His-tagged fragment with bound GE was collected and washed before analysis. A similar sample containing only GE was run in parallel and dot-blotted as a control for nonspecific binding to the nickel beads. The concentrations of GE tetramers applied to the blot in each lane were as follows. Lane 1, GAPDH, 54 nm; aldolase, 51 nm; LDH, 50 nm; PK, 51 nm. Lane 2, GAPDH, 134 nm; aldolase, 126 nm; LDH, 126 nm; PK, 123 nm. Lane 3, GAPDH, 268 nm; aldolase, 252 nm; LDH, 246 nm; PK, 249 nm. Lane 4, GAPDH, 536 nm; aldolase, 504 nm; LDH, 492 nm; PK, 497 nm. B, determination of the Kd of each GE for the peptide 330–391 of band 3. Data are plotted using GraphPad Prism 4 and fitted to a Boltzman sigmoidal equation.
FIGURE 8.
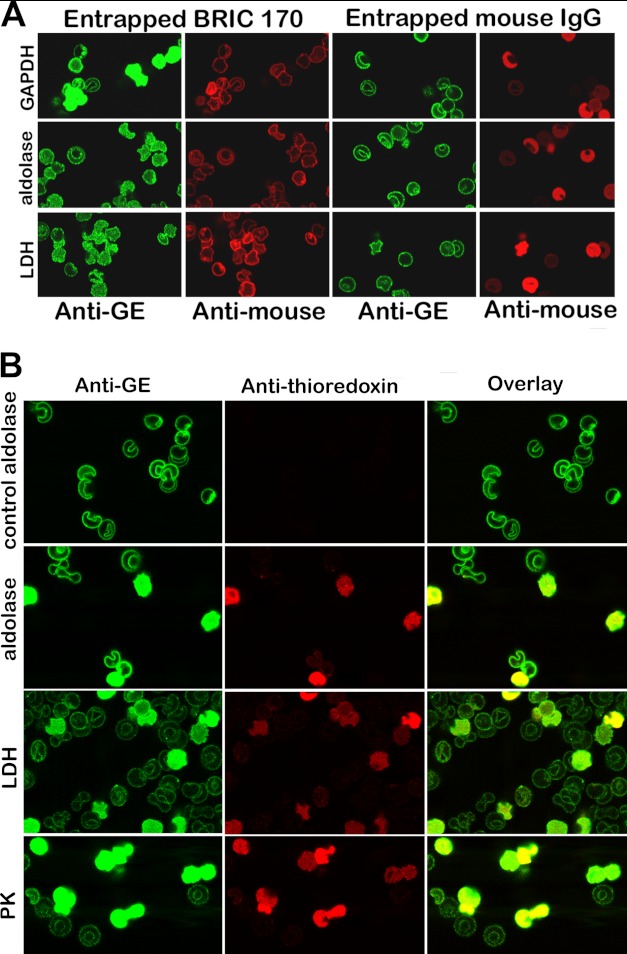
Displacement of GEs from erythrocyte membranes by entrapment of a monoclonal antibody to residues 368–382 of band 3 (BRIC 170) (A) or a fusion protein comprising residues 330–391 linked to thioredoxin (B). A, the first and second columns show the location of endogenous GEs (green stain) and the entrapment of BRIC 170 (red stain). Parallel control experiments following entrapment of a nonspecific mouse IgG are shown in columns 3 and 4. Quantitation of the intensity of GE staining in the cytosol using an Olympus Fluo View microscope version 2.1 reveals that the cytosolic GE staining for GAPDH, aldolase, and LDH is 20, 4, and 8 times greater on average in BRIC 170-entrapped cells than in isotype control-entrapped cells. B, a fusion protein containing peptide 330–391 linked to thioredoxin was entrapped into resealed ghosts, and its effect on GE binding was evaluated by confocal microscopy. A control in which no peptide fusion protein was entrapped during the incubation step with leaky ghosts was run in parallel and is shown in the first row (labeled control aldolase). The first column shows the staining of the GEs; the second column identifies the “ghosts” that have successfully entrapped the fusion protein (red stain of the anti-thioredoxin antibody); and the third column shows the overlay of both stains.
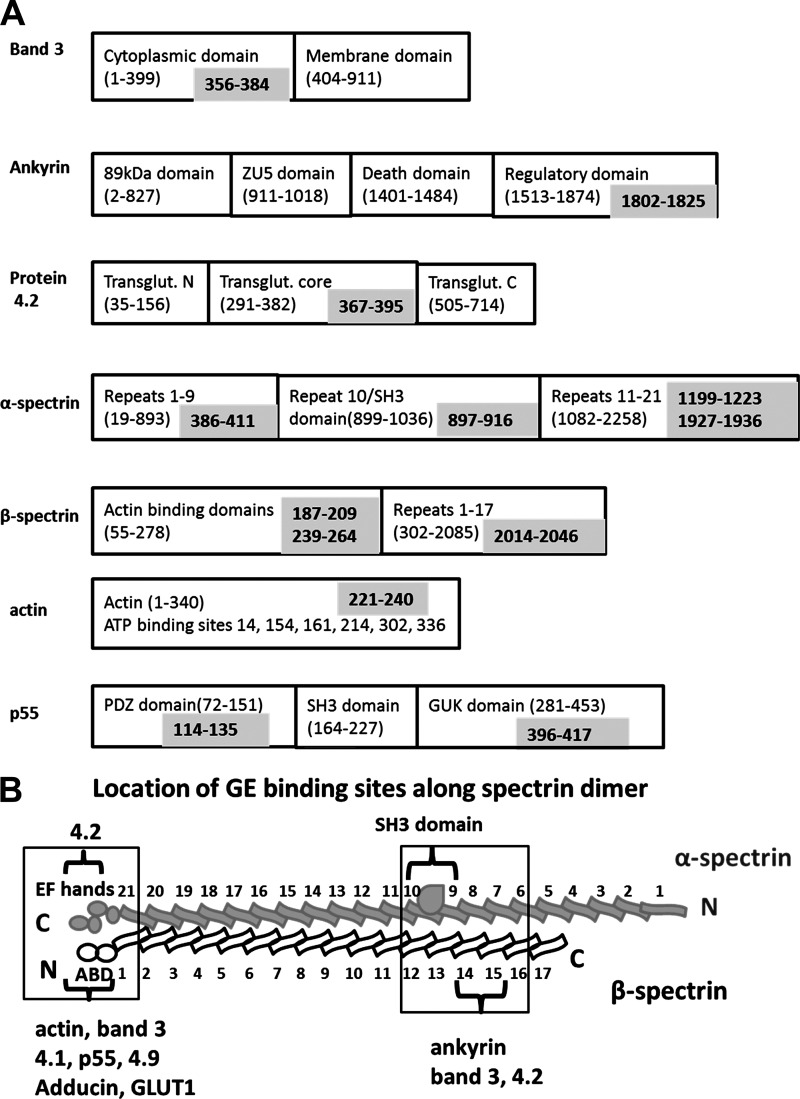
A second major conclusion is that the GE complexes must exist at multiple sites on the erythrocyte membrane. For example, amino acids 1802–1825 of ankyrin, residues 367–395 of protein 4.2, amino acids 373–384 and 386–411 of α-spectrin, and residues 2014–2046 of β-spectrin are all multiply labeled by all four GEs (see labeling map in Fig. 4A). Because these peptides reside within or near the ankyrin-band 3 complex, it can be suggested that a major GE nucleation site must exist at this ankyrin complex. Importantly, a similar argument can be made for a GE cluster near the junctional complex. Thus, common GE labeling sites are found near the COOH terminus of α-spectrin (residues 1927–1936), the NH2 terminus of β-spectrin (residues 187–209 and 239–264), on actin (residues 154–177 and 221–240), and on p55 (residues 114–135 and 396–417 (see labeling maps in Fig. 4A, Table 1, and supplemental Table 1). Because each of these peptides is thought to be located at the junctional complex, a second GE nucleation site probably also exists at this nexus of membrane interactions. This latter contention is, in fact, supported by the labeling of other junctional complex proteins by at least one (but not all) GEs (e.g. adducin, protein 4.1, stomatin, dematin, and GLUT1; see supplemental Table 1).
FIGURE 4.
Schematic representation of the prominent labeling sites of GEs on major membrane proteins. A, the domain structure of each major membrane protein is shown in the black boxes, with the component amino acid sequences provided in parentheses. The specific peptide in each protein that is photolabeled during the biotin transfer reaction is indicated in the gray box. B, a more detailed map of the labeling of spectrin.
Third, a few membrane sites not thought to belong to either the ankyrin or junctional complex are also labeled, including α-spectrin repeat domain 10 (peptides 897–916 and 969–981), which also contains the polypeptide's SH3 domain (Table 1, Fig. 4, A and B, and supplemental Table 1). Whether these labeled sequences constitute independent GE docking sites or fortuitous transient interactions cannot be ascertained from the data.
Fourth, the data strongly suggest that several GEs associate with each other. For example, GAPDH labels aldolase on peptide 1–22, and aldolase labels GAPDH on peptide 82–107 (see supplemental Table 1). This mutual cross-labeling should perhaps have been anticipated, because Ovádi and Keleti (34) have previously shown that GAPDH and aldolase display direct affinities for each other. In fact, our observation that each GE cross-labels at least one additional GE is consistent with evidence from many laboratories that specific associations among multiple GEs exist (35) and that these interactions have probably arisen to enhance substrate flow between GEs in their macromolecular assemblies (36).
Finally, we were surprised that the long established GE binding site at the NH2 terminus of band 3 was not labeled by any GE. Thus, many laboratories (including our own) have shown that the peptide comprising residues 1–23 of band 3 binds multiple GEs avidly and thereby inhibits their catalytic activities (14–16). This interaction, in fact, has been shown to be critical for both the assembly of GE complexes on the membrane (10) and the reversible shift in glucose metabolism between glycolysis and the pentose phosphate pathway during erythrocyte deoxygenation and oxygenation, respectively (37). Because the NH2-terminal peptide has also been found to be essential for regulation of glycolysis by erythrocyte tyrosine kinases (38, 39), our inability to detect this peptide in the mass spectrometry analysis was perplexing. However, upon further investigation, we noted that the first trypsin cleavage site in band 3 lies at lysine 56, yielding an NH2-terminal peptide that would be difficult to observe by mass spectrometry for two reasons. First, the unusually large peptide lies above the high molecular weight end (Mr = 6413) of the resolution range of our mass spectrometer, and second, the peptide is too acidic to fly in the positive mode used in all of our mass spectrometry analyses (i.e. the NH2-terminal 56 residues include an acetylated NH2-terminal methionine, 19 acidic residues, and only one basic residue (Lys-56)). Indeed, we have been unable to detect even the unlabeled NH2-terminal peptide of band 3 in mass spectrometry analyses of total red cell membrane proteins, although most of the other band 3 peptides are easily observed by this method.
GEs Dock at the Junction between the Cytoplasmic and Membrane-spanning Domains of Band 3
The most prominently biotinylated peptide in the entire erythrocyte membrane was found to reside at the junction between the cytoplasmic and membrane-spanning domains of band 3 (i.e. the bridging sequence comprising residues 356–384). In fact, all four GEs labeled this peptide at multiple sites (Table 1). Because a second GE binding site on band 3 has been frequently hypothesized (18, 29, 40), and because this bridging peptide probably locates near the well known GE binding site at the NH2 terminus of CDB3 (Fig. 5), we elected to directly determine whether the bridging peptide might also associate with GEs. For this purpose, we cloned and expressed a fusion protein composed of thioredoxin linked to the NH2 terminus of residues 330–391 and a His tag at the COOH terminus of the same peptide. As seen in the nickel bead pull-down assays shown in Fig. 6A, the bridging peptide was observed to pull down all four GEs examined. More detailed binding studies revealed half-saturation values of 0.28 ± 0.06, 13 ± 2.6, and 0.15 ± 0.03 μm for GAPDH, aldolase, and PK, respectively (Fig. 6B), in agreement with the lower affinity site for GAPDH found by McDaniels et al. (29). Because empty nickel beads lacking the fusion peptide failed to pull down any GEs, we conclude that the bridging peptide constitutes a second GE docking site on band 3. This result is significant, because PK and LDH do not display affinity for the well characterized GE site at the NH2 terminus of CDB3, although they respond to all regulatory signals that control the binding of GAPDH, aldolase, and phosphofructokinase at this NH2-terminal site (10).
Binding of GEs to the Secondary Site on CDB3 Does Not Inhibit GE Activity but Contributes to the Stability of the GE Complex
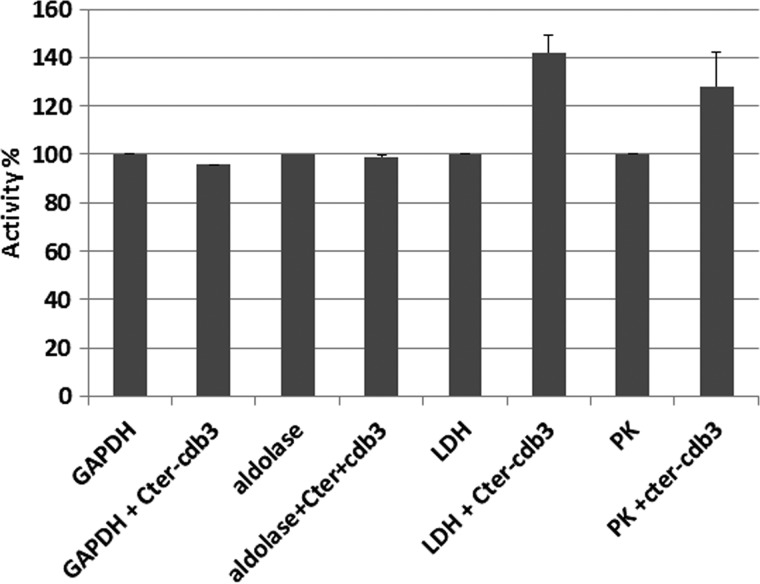
Because all GEs known to associate with the NH2 terminus of CDB3 are inhibited upon binding, the question naturally arose whether GE association with this secondary site might also suppress catalytic activity. To examine this question, standard enzyme assays of all four GEs were performed in the presence and absence of the fusion peptide (17). As seen in Fig. 7, no decrease in enzyme activity was observed upon binding. These data suggest that, similar to the association of GAPDH with the COOH terminus of erythrocyte or kidney band 3 (18), the interaction of GEs with the COOH of CDB3 is not inhibitory.
FIGURE 7.
Effect of the band 3 peptide comprising residues 330–391 on the catalytic activities of GEs. GEs were incubated without or with the above band 3 peptide for 5 min prior to assaying the activities of the indicated enzymes as described by Chu and Low (17). Error bars, S.E.
To demonstrate that GEs associate specifically with residues 330–391 of CDB3 in situ, the impact of entrapping a mouse monoclonal antibody to residues 366–382 (BRIC 170) on GE binding was assessed by confocal immunofluorescence microscopy in resealed ghosts. As seen in Fig. 8A, resealed ghosts containing entrapped BRIC 170, a monoclonal antibody that binds to CDB3 residues 368–382 (red labeling, second column), display GEs (green labeling, first column) that are at least partially distributed throughout the cytoplasm, whereas resealed ghosts containing entrapped nonspecific control mouse IgG (third and fourth columns) show no GE displacement.
Finally, a similar result was obtained when the peptide 330–391 containing the thioredoxin tag was entrapped in the resealed ghosts. As seen in Fig. 8B, resealed ghosts that enclosed the fusion peptide (indicated by the red labeling of anti-thioredoxin) display strong cytoplasmic GE staining, suggesting GE displacement from the membrane, whereas resealed ghosts that failed to entrap the fusion peptide do not. These results confirm that although the binding of GEs to this new site on band 3 is not inhibitory, the new site still contributes to the stability of the GE complex.
DISCUSSION
A variety of experiments have previously established that most if not all GEs are membrane-associated in oxygenated human erythrocytes, with band 3 (AE1, the anion transporter) serving as their primary docking site (10, 17, 19). Questions remained, however, regarding which population of band 3 might be interacting with the GEs and whether these interactions were specific or simply driven by nonselective interactions (41). To explore the nature of these interactions further, we used a trifunctional photocross-linking reagent (sulfo-SBED) that has been employed successfully by many laboratories, including our own (42), to identify nearest neighbor proteins in complex macromolecular structures and organelles (43–45). The photocross-linking reagent has a length of 1.4 nm and generates a nitrene upon photoactivation, enabling insertion into any proximal peptide (Pierce). Although the nitrene will admittedly prefer to react with nucleophiles, such as lysine, threonine, serine, and cysteine side chains, it will also insert into virtually any peptide bond and even label adjacent aliphatic amino acid side chains. The fact that every amino acid was labeled in the GE binding partners except arginine (see Table 1 and supplemental Table 1) suggests that the photocross-linking reaction was determined predominantly by protein proximity and not chemical reactivity.
Although most proteins in both the ankyrin and junctional complexes were labeled, several lines of evidence suggest that this labeling was specific and not a consequence of random GE collisions. First, photolabel transfer from GEs to their membrane docking sites could be blocked by the addition of unlabeled GEs. Second, when ovalbumin was substituted for GEs as the photolabel donor, no membrane proteins were labeled. Third, any unbound GEs were invariably removed during the washing steps prior to photoactivation, leaving only membrane-bound GEs to transfer label to their nearest neighbors. Fourth, only specific peptides in each multidomain binding partner were photolabeled, as demonstrated in four independent photolabeling experiments, suggesting that the labeling was selective for the most proximal peptides. Although a variety of polypeptides were indeed labeled, this diversity of “nearest neighbors” was not unexpected, because band 3 is located at both the junctional and ankyrin complexes, and the peptides that were labeled are also primarily located in these two complexes.
A major finding of this study was that GEs bind not only to the NH2 terminus of band 3 but also to a peptide comprising residues 356–384 that resides adjacent to the NH2 terminus of CDB3 in the crystal structure of the polypeptide (33). Although the exact location of this auxiliary site was previously unknown, a number of prior observations had clearly pointed to its existence. Thus, both deoxyhemoglobin (which binds to residues 12–23 of band 3) and band 3 tyrosine phosphorylation on Tyr-8 and Tyr-21 had been known to displace LDH and PK from the membrane (i.e. similar to GEs that dock at the NH2 terminus of band 3), although neither LDH nor PK had been found to exhibit affinity for this NH2-terminal sequence. Second, Steck and colleagues (16) had reported that an NH2-terminal 22-kDa fragment of band 3 displays a potency in competitive GE displacement assays that is “inexplicably reduced 30-fold” compared with intact band 3. Similarly, Alper and colleagues (40) had noted that “a peptide comprising residues 1–56 from the human AEl cytoplasmic domain displaces GAPDH from erythrocyte membranes with l00-fold lower potency than does intact band 3, indicating the existence of other binding sites.” Thus, although no information had been previously available on the location of this auxiliary GE site, it was clearly anticipated based on data in the literature. The important conclusion resulting from this study is that we not only know where this second GE site resides, but we also know much more about its function. Thus, a picture emerges in which this second GE binding site not only serves to provide a primary binding site for LDH and PK, neither of which were observed to bind the NH2 terminus of band 3, but also appears to augment the affinity of GAPDH (and probably other GEs) for their primary site at the NH2 terminus of band 3. The observation that GEs are competitively displaced from the membrane by both this peptide and a monoclonal antibody directed against its sequence (BRIC 170) suggests that this auxiliary GE site is also important for GE assembly in vivo.
The fact that both tyrosine phosphorylation and deoxyhemoglobin binding to the NH2 terminus of band 3 displace all GEs from the membrane (10, 19) suggests that most, if not all, GE binding sites reside on or near band 3. From these observations, it naturally follows that the majority of biotinylated proteins reported in Table 1 and supplemental Table 1 must reside near one of the major populations of band 3. This assumption presents no problem for most of the peptides identified in the above tables, because the most prominently labeled peptides are components of ankyrin, protein 4.1, protein 4.2, adducin, Rh, p55, dematin, actin, or spectrin (i.e. proteins already known to assemble into one of the major band 3-containing protein complexes) (6, 12, 42, 46–51). However, the labeling of peptides in repeat domain 10 of α-spectrin (peptides 897–916 and 969–981) by the GEs cannot be easily explained by standard models of red cell membrane architecture. Because this sequence contains α-spectrin's SH3 domain, we suggest that either the SH3 domain of α-spectrin somehow folds into a position proximal to one of the band 3 complexes (Fig. 4B), or an independent GE binding site exists near the SH3 domain of α-spectrin. Regarding the latter possibility, it is interesting that a proline-rich sequence fitting the criteria of an SH3 domain binding peptide (residues 815PPKYHPDVP823) is present in the largest cytoplasmic loop of the membrane-spanning domain of band 3, where it could tether the SH3 domain of α-spectrin to the freely diffusing subpopulation of band 3. It will be interesting in the future to evaluate whether the SH3 domain of α-spectrin actually displays direct affinity for this sequence in the membrane-spanning domain of band 3.
Finally, based on the crystal structure of CDB3, the newly identified GE site resides ∼2.4 nm from the well known GE site at the NH2 terminus of band 3 (Fig. 5). Because the diameters of the GEs average ∼7.5 ± 2 nm (measurements obtained from the Protein Data Bank using WebLab ViewerPro 2.0 (MSI, San Diego, CA)), it seems reasonable to speculate that some GEs might associate with only one site on band 3, whereas others could bind both simultaneously. Based on studies in other cell types, GEs are also known to assemble on actin filaments, microtubules, myofibrils, and intracellular organelles (i.e. in general at sites where the ATP they produce is needed) (52–55). In the red cell, the only source of ATP is glycolysis, and the major consumers of ATP are in the membrane, where ATP is used to fuel ion pumps, maintain phospholipid asymmetry, control actin filament dynamics, facilitate kinase signaling, and regulate vascular tone via release of ATP into the vasculature (13, 56, 57). Evolutionarily, it would also make sense for the erythrocyte to localize an ATP-producing metabolon near the site of ATP consumption on the membrane (6, 42, 46, 58–60). Consistent with this expectation, we have recently found that the ATP pool that fuels the Na/K− and Ca− pumps resides on the membrane within or near the band 3-ankyrin complex (9).
Supplementary Material
Acknowledgments
We thank Dr. Joseph F. Hoffman for helpful discussions and critical review of the manuscript and Dr. Anton Iliuk for assistance with the mass spectrometry.
This work was supported, in whole or in part, by National Institutes of Health Grants R01GM24417-33 and R01GM088317.

This article contains supplemental Table 1.
- GE
- glycolytic enzyme
- LDH
- lactate dehydrogenase
- PK
- pyruvate kinase isoform M1/M2
- SH3
- Src homology 3.
REFERENCES
- 1. Vonck J., Schäfer E. (2009) Supramolecular organization of protein complexes in the mitochondrial inner membrane. Biochim. Biophys. Acta 1793, 117–124 [DOI] [PubMed] [Google Scholar]
- 2. Dai S., Hall D. D., Hell J. W. (2009) Supramolecular assemblies and localized regulation of voltage-gated ion channels. Physiol. Rev. 89, 411–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sterling D., Reithmeier R. A., Casey J. R. (2001) A transport metabolon. J. Biol. Chem. 276, 47886–47894 [DOI] [PubMed] [Google Scholar]
- 4. Sowah D., Casey J. R. (2011) An intramolecular transport metabolon. Fusion of carbonic anhydrase II to the COOH terminus of the Cl−/HCO3− exchanger, AE1. Am. J. Physiol. Cell Physiol. 301, C336–C346 [DOI] [PubMed] [Google Scholar]
- 5. Endeward V., Cartron J.-P., Ripoche P., Gros G. (2008) RhAG protein of the Rhesus complex is a CO2 channel in the human red cell membrane. FASEB J. 22, 64–73 [DOI] [PubMed] [Google Scholar]
- 6. Bruce L. J., Beckmann R., Ribeiro M. L., Peters L. L., Chasis J. A., Delaunay J., Mohandas N., Anstee D. J., Tanner M. J. (2003) A band 3-based macrocomplex of integral and peripheral proteins in the RBC membrane. Blood 101, 4180–4188 [DOI] [PubMed] [Google Scholar]
- 7. Hoffman J. F., Dodson A., Proverbio F. (2009) On the functional use of the membrane compartmentalized pool of ATP by the Na+ and Ca2+ pumps in human red blood cell ghosts. J. Gen. Physiol. 134, 351–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tiffert T., Lew V. L. (2011) Elevated intracellular Ca2+ reveals a functional membrane nucleotide pool in intact human red blood cells. J. Gen. Physiol. 138, 381–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chu H., Puchulu-Campanella E., Galan J. A., Tao W. A., Low P. S., Hoffman J. F. (2012) Identification of cytoskeletal elements enclosing the ATP pools that fuel human red blood cell membrane cation pumps. Proc. Natl. Acad. Sci. 109, 12794–12799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Campanella M. E., Chu H., Low P. S. (2005) Assembly and regulation of a glycolytic enzyme complex on the human erythrocyte membrane. Proc. Natl. Acad. Sci. U.S.A. 102, 2402–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li B. (2009) An antioxidant metabolon at the red blood cell membrane. M.S. thesis, Concordia University, Montreal, Canada [Google Scholar]
- 12. Salomao M., Zhang X., Yang Y., Lee S., Hartwig J. H., Chasis J. A., Mohandas N., An X. (2008) Protein 4.1R-dependent multiprotein complex. New insights into the structural organization of the red blood cell membrane. Proc. Natl. Acad. Sci. U.S.A. 105, 8026–8031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sprague R. S., Stephenson A. H., Ellsworth M. L. (2007) Red not dead. Signaling in and from erythrocytes. Trends Endocrinol. Metab. 18, 350–355 [DOI] [PubMed] [Google Scholar]
- 14. Strapazon E., Steck T. L. (1977) Interaction of aldolase and membrane of human erythrocytes. Biochemistry 16, 2966–2971 [DOI] [PubMed] [Google Scholar]
- 15. Murthy S. N., Liu T., Kaul R. K., Köhler H., Steck T. L. (1981) The aldolase-binding site of the human erythrocyte membrane is at the NH2 terminus of band 3. J. Biol. Chem. 256, 11203–11208 [PubMed] [Google Scholar]
- 16. Tsai I. H., Murthy S. N., Steck T. L. (1982) Effect of red cell membrane binding on the catalytic activity of glyceraldehyde-3-phosphate dehydrogenase. J. Biol. Chem. 257, 1438–1442 [PubMed] [Google Scholar]
- 17. Chu H., Low P. S. (2006) Mapping of glycolytic enzyme-binding sites on human erythrocyte band 3. Biochem. J. 400, 143–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Su Y., Blake-Palmer K. G., Fry A. C., Best A., Brown A. C., Hiemstra T. F., Horita S., Zhou A., Toye A. M., Karet F. E. (2011) Glyceraldehyde 3-phosphate dehydrogenase is required for band 3 (anion exchanger 1) membrane residency in the mammalian kidney. Am. J. Physiol. Renal Physiol. 300, F157–F166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Campanella M. E., Chu H., Wandersee N. J., Peters L. L., Mohandas N., Gilligan D. M., Low P. S. (2008) Characterization of glycolytic enzyme interactions with murine erythrocyte membranes in wild-type and membrane protein knockout mice. Blood 112, 3900–3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Steck T. L., Ramos B., Strapazon E. (1976) Proteolytic dissection of band 3, predominant transmembrane polypeptide of human erythrocyte-membrane. Biochemistry 15, 1153–1161 [DOI] [PubMed] [Google Scholar]
- 21. Bennett V. (1983) Proteins involved in membrane-cytoskeleton association in human erythrocytes. Spectrin, ankyrin, and band 3. Methods Enzymol. 96, 313–324 [DOI] [PubMed] [Google Scholar]
- 22. Hassoun H., Wang Y., Vassiliadis J., Lutchman M., Palek J., Aish L., Aish I. S., Liu S. C., Chishti A. H. (1998) Targeted inactivation of murine band 3 (AE1) gene produces a hypercoagulable state causing widespread thrombosis in vivo. Blood 92, 1785–1792 [PubMed] [Google Scholar]
- 23. Gallagher P. G., Lux S. E. (eds) (2003) Disorders of the Erythrocyte Membrane. in Hematology of Infancy and Children (Nathan D., Orkin S. H., Oski F. A., eds) pp. 560–684, W.B. Saunders, Philadelphia [Google Scholar]
- 24. Stochaj W. R., Berkelman T., Laird N. (2006) Staining membrane-bound proteins with Ponceau S. Cold Spring Harb. Protoc. 2006, doi: 10.1101/pdb.prot4543 [DOI] [PubMed] [Google Scholar]
- 25. Simpson R. J. (2003) Stage 3. Selection of tagged proteins using an avidin column. in Proteins and Proteomics: A Laboratory Manual (Simpson R. J., ed) pp. 529–531, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 26. Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 27. Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 28. Murphy S. C., Harrison T., Hamm H. E., Lomasney J. W., Mohandas N., Haldar K. (2006) Erythrocyte G protein as a novel target for malarial chemotherapy. PLoS Med. 3, e528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McDaniel C. F., Kirtley M. E., Tanner M. J. (1974) The interaction of glyceraldehyde-3-phosphate dehydrogenase with human erythrocyte membranes. J. Biol. Chem. 249, 6478–6485 [PubMed] [Google Scholar]
- 30. Kant J. A., Steck T. L. (1973) Specificity in the association of glyceraldehyde 3-phosphate dehydrogenase with isolated human erythrocyte membranes. J. Biol. Chem. 248, 8457–8464 [PubMed] [Google Scholar]
- 31. Hurst G. B., Lankford T. K., Kennel S. J. (2004) Mass spectrometric detection of affinity purified crosslinked peptides. J. Am. Soc. Mass Spectrom. 15, 832–839 [DOI] [PubMed] [Google Scholar]
- 32. Kovacic L., Sribar J., Krizaj I. (2007) A new photoprobe for studying biological activities of secreted phospholipases A2. Bioorg. Chem. 35, 295–305 [DOI] [PubMed] [Google Scholar]
- 33. Zhang D., Kiyatkin A., Bolin J. T., Low P. S. (2000) Crystallographic structure and functional interpretation of the cytoplasmic domain of erythrocyte membrane band 3. Blood 96, 2925–2933 [PubMed] [Google Scholar]
- 34. Ovádi J., Keleti T., Salerno C., Fasella P. (1978) Physico-chemical evidence for the interaction between aldolase and glyceraldehyde-3-phosphate dehydrogenase. Eur. J. Biochem. 90, 499–503 [DOI] [PubMed] [Google Scholar]
- 35. Ovádi J. (1988) Old pathway-new concept. Control of glycolysis by metabolite-modulated dynamic enzyme associations. Trends Biochem. Sci. 13, 486–490 [DOI] [PubMed] [Google Scholar]
- 36. Srere P. A., Ovadi J. (1990) Enzyme-enzyme interactions and their metabolic role. FEBS Lett. 268, 360–364 [DOI] [PubMed] [Google Scholar]
- 37. Lewis I. A., Campanella M. E., Markley J. L., Low P. S. (2009) Role of band 3 in regulating metabolic flux of red blood cells. Proc. Natl. Acad. Sci. 106, 18515–18520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Harrison M. L., Rathinavelu P., Arese P., Geahlen R. L., Low P. S. (1991) Role of band 3 tyrosine phosphorylation in the regulation of erythrocyte glycolysis. J. Biol. Chem. 266, 4106–4111 [PubMed] [Google Scholar]
- 39. Low P. S., Geahlen R. L., Mehler E., Harrison M. L. (1990) Extracellular control of erythrocyte metabolism mediated by a cytoplasmic tyrosine kinase. Biomed. Biochim. Acta 49, S135–S140 [PubMed] [Google Scholar]
- 40. Ercolani L., Brown D., Stuart-Tilley A., Alper S. L. (1992) Colocalization of GAPDH and band 3 (AE1) proteins in rat erythrocytes and kidney intercalated cell membranes. Am. J. Physiol. Renal Physiol. 262, F892–F896 [DOI] [PubMed] [Google Scholar]
- 41. Kelley G. E., Winzor D. J. (1984) Quantitative characterization of the interactions of aldolase and glyceraldehyde-3-phosphate dehydrogenase with erythrocyte membranes. Biochim. Biophys. Acta 778, 67–73 [DOI] [PubMed] [Google Scholar]
- 42. Anong W. A., Franco T., Chu H., Weis T. L., Devlin E. E., Bodine D. M., An X., Mohandas N., Low P. S. (2009) Adducin forms a bridge between the erythrocyte membrane and its cytoskeleton and regulates membrane cohesion. Blood 114, 1904–1912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mitola S., Ravelli C., Moroni E., Salvi V., Leali D., Ballmer-Hofer K., Zammataro L., Presta M. (2010) Gremlin is a novel agonist of the major proangiogenic receptor VEGFR2. Blood 116, 3677–3680 [DOI] [PubMed] [Google Scholar]
- 44. Emara M., Royer P.-J., Mahdavi J., Shakib F., Ghaemmaghami A. M. (2012) Retagging identifies dendritic cell-specific intercellular adhesion molecule-3 (ICAM3)-grabbing non-integrin (DC-SIGN) protein as a novel receptor for a major allergen from house dust mite. J. Biol. Chem. 287, 5756–5763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang Q., Yu L., Yu C.-A. (2010) Cross-talk between mitochondrial malate dehydrogenase and the cytochrome bc1 complex. J. Biol. Chem. 285, 10408–10414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. van den Akker E., Satchwell T. J., Williamson R. C., Toye A. M. (2010) Band 3 multiprotein complexes in the red cell membrane. Of mice and men. Blood Cells Mol. Dis. 45, 1–8 [DOI] [PubMed] [Google Scholar]
- 47. Su Y., Ding Y., Jiang M., Jiang W., Hu X., Zhang Z. (2006) Associations of protein 4.2 with band 3 and ankyrin. Mol. Cell. Biochem. 289, 159–166 [DOI] [PubMed] [Google Scholar]
- 48. Satchwell T. J., Shoemark D. K., Sessions R. B., Toye A. M. (2009) Protein 4.2. A complex linker. Blood Cells Mol. Dis. 42, 201–210 [DOI] [PubMed] [Google Scholar]
- 49. Satchwell T. J., Bell A. J., Pellegrin S., Kupzig S., Ridgwell K., Daniels G., Anstee D. J., van den Akker E., Toye A. M. (2011) Critical band 3 multiprotein complex interactions establish early during human erythropoiesis. Blood 118, 182–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Marfatia S. M., Leu R. A., Branton D., Chishti A. H. (1995) Identification of the protein 4.1 binding interface on glycophorin C and p55, a homologue of the Drosophila discs-large tumor suppressor protein. J. Biol. Chem. 270, 715–719 [DOI] [PubMed] [Google Scholar]
- 51. Mankelow T. J., Satchwell T. J., Burton N. M. (2012) Refined views of multi-protein complexes in the erythrocyte membrane. Blood Cells Mol. Dis. 49, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kim Y.-H., Haidl G., Schaefer M., Egner U., Mandal A., Herr J. C. (2007) Compartmentalization of a unique ADP/ATP carrier protein SFEC (sperm flagellar energy carrier, AAC4) with glycolytic enzymes in the fibrous sheath of the human sperm flagellar principal piece. Dev. Biol. 302, 463–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dhar-Chowdhury P., Harrell M. D., Han S. Y., Jankowska D., Parachuru L., Morrissey A., Srivastava S., Liu W., Malester B., Yoshida H., Coetzee W. A. (2005) The glycolytic enzymes, glyceraldehyde-3-phosphate dehydrogenase, triose-phosphate isomerase, and pyruvate kinase are components of the KATP channel macromolecular complex and regulate its function. J. Biol. Chem. 280, 38464–38470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Raikar L. S., Vallejo J., Lloyd P. G., Hardin C. D. (2006) Overexpression of caveolin-1 results in increased plasma membrane targeting of glycolytic enzymes. The structural basis for a membrane associated metabolic compartment. J. Cell. Biochem. 98, 861–871 [DOI] [PubMed] [Google Scholar]
- 55. Pomel S., Luk F. C., Beckers C. J. (2008) Host cell egress and invasion induce marked relocations of glycolytic enzymes in Toxoplasma gondii tachyzoites. PLoS Pathog. 4, e1000188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bergfeld G. R., Forrester T. (1992) Release of ATP from human erythrocytes in response to a brief period of hypoxia and hypercapnia. Cardiovasc. Res. 26, 40–47 [DOI] [PubMed] [Google Scholar]
- 57. Sprague R. S., Ellsworth M. L., Stephenson A. H., Lonigro A. J. (2001) Participation of cAMP in a signal-transduction pathway relating erythrocyte deformation to ATP release. Am. J. Physiol. Cell Physiol. 281, C1158–C1164 [DOI] [PubMed] [Google Scholar]
- 58. Franco T., Low P. S. (2010) Erythrocyte adducin. A structural regulator of the red blood cell membrane. Transfus. Clin. Biol. 17, 87–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kim S., Brandon S., Zhou Z., Cobb C. E., Edwards S. J., Moth C. W., Parry C. S., Smith J. A., Lybrand T. P., Hustedt E. J., Beth A. H. (2011) Determination of structural models of the complex between the cytoplasmic domain of erythrocyte band 3 and ankyrin-R repeats 13–24. J. Biol. Chem. 286, 20746–20757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Harper S. L., Li D., Maksimova Y., Gallagher P. G., Speicher D. W. (2010) A fused α-β “mini-spectrin” mimics the intact erythrocyte spectrin head-to-head tetramer. J. Biol. Chem. 285, 11003–11012 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.