Background: Apoptosis and autophagy are two closely related systems that induce cell death.
Results: X-box-binding protein 1 (XBP1) mRNA splicing regulates BECLIN-1 transcriptional activation, a fundamental player in the initiation of autophagy.
Conclusion: XBP1 splicing induces an autophagic response in endothelial cells.
Significance: XBP1 could be used as an important pharmacological target that can regulate the autophagic machinery and endothelial cell death.
Keywords: Alternative Splicing, Atherosclerosis, Autophagy, Cell Death, Electron Microscopy (EM), Endothelial Cell, BECLIN-1, XBP1
Abstract
Sustained activation of X-box-binding protein 1 (XBP1) results in endothelial cell (EC) apoptosis and atherosclerosis development. The present study provides evidence that XBP1 mRNA splicing triggered an autophagic response in ECs by inducing autophagic vesicle formation and markers of autophagy BECLIN-1 and microtubule-associated protein 1 light chain 3β (LC3-βII). Endostatin activated autophagic gene expression through XBP1 mRNA splicing in an inositol-requiring enzyme 1α (IRE1α)-dependent manner. Knockdown of XBP1 or IRE1α by shRNA in ECs ablated endostatin-induced autophagosome formation. Importantly, data from arterial vessels from XBP1 EC conditional knock-out (XBP1eko) mice demonstrated that XBP1 deficiency in ECs reduced the basal level of LC3β expression and ablated response to endostatin. Chromatin immunoprecipitation assays further revealed that the spliced XBP1 isoform bound directly to the BECLIN-1 promoter at the region from nt −537 to −755. BECLIN-1 deficiency in ECs abolished the XBP1-induced autophagy response, whereas spliced XBP1 did not induce transcriptional activation of a truncated BECLIN-1 promoter. These results suggest that XBP1 mRNA splicing triggers an autophagic signal pathway through transcriptional regulation of BECLIN-1.
Introduction
Although tremendous recent advances have been made in the field, the functional significance of autophagy in human disease remains incompletely understood. Diseases such as atherosclerosis show evidence of cellular senescence characterized by reduced cell proliferation, irreversible growth arrest, and apoptosis. Nonetheless, a growing body of evidence suggests that elucidation of the signaling pathways regulating autophagy may provide novel therapeutic approaches in the prevention or treatment of atherosclerosis (1). Recent studies have reported that autophagy becomes dysfunctional in atherosclerosis, and its deficiency promotes atherosclerosis in part through inflammasome hyperactivation (2). Moreover, it has also been shown that macrophage autophagy plays a protective role in advanced atherosclerosis (3), whereas transmission electron microscopy of smooth muscle cells in the fibrous cap of advanced plaques revealed ultrastructural features of autophagy, such as vacuolization and formation of myelin figures (4). Endothelial cell (EC)3 dysfunction or death are also key events in the pathophysiology of the vessel wall and the development of atherosclerosis (5, 6). However, the regulation of autophagy in ECs in atherosclerotic plaques has not been fully elucidated yet. Autophagy is a multistep catabolic process, which has been shown to be involved in a variety of pathophysiological conditions (1). During the autophagic process, long-lived proteins and organelles are sequestered in a double membrane-bound autophagosome (1) and degraded via lysosome (7–11). Under physiological conditions, autophagy is a reparative and life-maintaining process. In cases of excessive induction of autophagy by environmental or intracellular stress, it becomes a cell death pathway. However, the mechanisms that are involved in the regulation of autophagy are largely unknown. We have previously shown that sustained activation of X-box-binding protein 1 (XBP1) mRNA splicing, a crucial signal transducer in endoplasmic reticulum stress response, leads to EC apoptosis and atherosclerosis development (12). XBP1 is a transcription factor that belongs to the basic region/leucine zipper family (13, 14), and it has an unspliced and a spliced form. The unspliced form has a molecular mass of 29 kDa and acts as a dominant negative of spliced XBP1 (15, 16). In response to endoplasmic reticulum (ER) stress, XBP1 mRNA undergoes unconventional splicing through the inositol-requiring enzyme 1α (IRE1α), giving rise to a 56-kDa spliced form with intact transcriptional activity (17, 18). XBP1 mRNA splicing is implicated in affecting plasma cell differentiation (19), fetal survival, and liver development (20, 21). In the present study, we provide strong evidence that XBP1 mRNA splicing is involved in the regulation of autophagy in ECs through BECLIN-1 transcriptional activation. These findings provide insights into the cell death machinery of ECs, which is a key event in atherosclerotic diseases.
EXPERIMENTAL PROCEDURES
Cell culture media and serum were purchased from Invitrogen, and cell culture supplements were purchased from Sigma. Antibodies against XBP1 (M-186), CD31, and GAPDH were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), whereas antibody against FLAG was from Abcam, and anti-LC3 (M186-3) was from MBL. Antibodies against IRE1α, pIRE1α, and BECLIN-1 (Autophagy Kit-4445) were purchased from Cell Signaling. Endostatin was from Calbiochem. Chloroquine was from Sigma. All secondary antibodies were from DAKOCytomation, and all other chemicals were from Sigma.
Cell Culture
Human umbilical vein endothelial cells were isolated from the human umbilical cord and cultured on collagen I-coated flasks in EGM-2 medium (Clonetics) or in M199 medium supplemented with 1 ng/ml β-EC growth factor (Sigma), 3 μg/ml EC growth supplement from bovine neural tissue (Sigma), 10 units/ml heparin, 1.25 μg/ml thymidine, 10% fetal bovine serum (FBS, PAA, A15-108) (12), and 100 μg/ml penicillin and streptomycin, as described previously (22). 293T cells were maintained in DMEM supplemented with 10% FBS and penicillin/streptomycin. Living cell images were assessed by a Nikon Eclipse TS100 microscope with Ph1 ADL ×10/0.25 objective lenses and Nikon DS-Fil camera at room temperature and processed by Adobe Photoshop software.
Adenoviral Gene Transfer
Ad-FLAG-XBP1s and Ad-FLAG-XBP1u were created and amplified as described previously (12). The FLAG tag is located in the N terminus. Empty vector-derived Ad-null virus was used as control virus. For adenoviral gene transfer, confluent human umbilical vein endothelial cells were infected with Ad-XBP1s or Ad-XBP1u at a multiplicity of infection (MOI) of 5 for 6 h followed by replacement with complete growth medium and further culture for the time indicated.
Transmission Electron Microscopy
Human umbilical vein endothelial cells were infected with Ad-null, Ad-XBP1u, and Ad-XBP1s viruses at 5 MOI for 48 h, or ECs were infected with non-target (shNT) or XBP1-targeting (shXBP1) shRNA lentiviruses for 48 h following by 24 h of endostatin. The cells were grown to confluence on 13-mm diameter glass coverslips. For transmission electron microscopy, cells on coverslips were fixed by removing the medium and replacing it immediately with 2.5% (v/v) glutaraldehyde in 0.1 m phosphate buffer. The cells were fixed for 2 h at 4 °C. After fixation, the coverslips were rinsed several times with 0.1 m phosphate buffer, followed by postfixation with 1% (w/v) osmium tetroxide in 0.1 m phosphate buffer (pH 7.3) for 20 min at 4 °C. The coverslips were rinsed (10 min) in phosphate buffer and dehydrated in a graded series of ethanol to 100%. The coverslips were then flooded with TAAB epoxy resin and left to infiltrate for 4 h at room temperature. To section in the plane of the monolayers, the coverslips were embedded by inverting them (monolayer side down) onto a capsule (TAAB) overfilled with resin and then polymerized for 24 h at 70 °C. Ultrathin sections (70–90 nm) were prepared using a Reichert-Jung Ultracut E ultramicrotome, mounted on 150-mesh copper grids, contrasted using uranyl acetate and lead citrate, and examined on an FEI Tecnai 12 transmission microscope operated at 120 kV. Images were acquired with an AMT 16000M digital camera.
RNA Extraction and Reverse Transcription-Polymerase Chain Reaction (RT-PCR) and Real-time PCR
RNA extraction, RT-PCR, and real-time PCR were performed as described previously (23). One μg of RNA was reverse transcribed into cDNA with random primer by MMLV reverse transcriptase (Promega). 20–50 ng of cDNA (relative to RNA amount) was amplified by standard PCR with TaqDNA polymerase (Invitrogen) or real-time PCR. The primers for each gene are as follows: for LC3β, 5′-TGTCCGACTTATTCGAGAGCAGCA-3′ and 5′-TTCACCAACAGGAAGAAGGCCTGA-3′; for XBP1, 5′-CCTTGTAGTTGAGAACCAGGAG-3′ and 5′-GGTCCAAGTTGTCCAGAATGC-3′; for BECLIN-1, 5′-AGGTTGAGAAAGGCGAGACA3′ and 5′-TTTTGATGGAATAGGAGCCG-3′; for IRE1α, 5′-GAAGATCCAGTCCTGCAGGTC-3′ and 5′-AGAAGAGAGGTTGATGGGCAG-3′; for 14-3-3T, 5′-CCACGGTGCTGGAATTGTTGGATA-3′ and 5′-CATCACCACACGCAACTTCAGCAA-3′; and for E2F1, 5′-ACGTGTCAGGACCTTCGTAGCATT-3′ and 5′-AAACATCGATCGGGCCTTGTTTGC-3′. PCR primers for real-time PCR were designed using Primer Express software (Applied Biosystems).
Indirect Immunofluorescence Assay
ECs were seeded on collagen I-coated slides 24 h before infection with Ad-XBP1s or Ad-XBP1u viruses at 5 MOI. Three days later, the slides were fixed with 4% paraformaldehyde, PBS for 15 min and permeabilized with 0.1% Triton X-100, PBS for 10 min, followed by incubation with primary and secondary antibodies, as described previously (22). Briefly, incubation with the primary antibodies, FLAG (goat), BECLIN-1 (rabbit), LC3β (rabbit), and XBP1 (rabbit), was performed for 1 h at 37 °C. The bound primary antibodies were revealed by incubation with an ALEXA 546-conjugated donkey anti-goat IgG and an FITC-conjugated swine anti-rabbit IgG at 37 °C for 30 min. Cells were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (Sigma), mounted with Floromount-G (DAKOCytomation, Glostrup, Denmark), and examined under an SP5 confocal microscope (Leica, Germany). Magnification in the figures is indicated by the scale bars.
Immunoblotting
Cells were harvested and washed with cold PBS, resuspended in lysis buffer (25 mm Tris-Cl, pH 7.5, 120 mm NaCl, 1 mm EDTA, pH 8.0, 0.5% Triton X-100) supplemented with protease inhibitors (Roche Applied Science), and lysed by ultrasonication (twice, 6 s each) (Branson Sonifier 150), as described previously (22). The protein concentration was determined using the Bio-Rad protein assay reagent. 50 μg of whole lysate was applied to SDS-PAGE and transferred to Hybond PVDF membrane (GE Healthcare), followed by a standard Western blot procedure. The bound primary antibodies were detected by the use of horseradish peroxidase (HRP)-conjugated secondary antibody and the ECL detection system (GE Healthcare).
Lentiviral Particle Transduction
Lentiviral particles were produced using MISSION shXBP1, shIRE1α and shBECLIN-1 plasmids DNA (Sigma) according to the protocol provided. The shRNA non-targeting vector was used as a negative control. Briefly, 293T cells were transfected with the lentiviral vector and the packaging plasmids, pCMV-dR8.2 and pCMV-VSV-G (both obtained from Addgene), with Fugene 6. The supernatant containing the lentivirus was harvested 48 h later and filtered, and the transducing unit was calculated, as described previously. For lentiviral infection, ECs were seeded overnight on collagen I-coated plates or flasks. The next day, cells were incubated with shXBP1 or shIRE1α or non-targeting control (1 × 107 transducing units/ml) in complete medium supplemented with 10 μg/ml Polybrene for 24 h. Subsequently, fresh medium in the absence or presence of endostatin (20 ng/ml) was added, and the cells were harvested 48 h later.
Chromatin Immunoprecipitation Assay
ECs (1 × 106 cells) were infected with Ad-XBP1s or Ad-XBP1u at 5 MOI for 6 h and further cultured for 48 h, or normal ECs were used for endogenous assays. The chromatin immunoprecipitation assay was performed with a commercial kit (EZ ChIP, Millipore) according to the protocol provided and as described previously (12, 24). In brief, cells were treated with 1% (v/v) formaldehyde at room temperature for 10 min and then quenched with glycine at room temperature. The medium was removed, and cells were harvested in lysis buffer. Following a short incubation on ice, chromatin was sheared by sonication. The sheared samples were diluted into 0.9 ml of ChIP dilution buffer and precleared with Protein G-agarose/salmon sperm DNA beads for 1 h. Subsequently, immunoprecipitation was conducted with anti-FLAG antibody (to precipitate XBP1s and XBP1u and their bound chromatin) or XBP1 antibody (Santa Cruz Biotechnology, Inc.) to precipitate the endogenous XBP1. Normal IgG was used as a control. Immunocomplexes were collected the following morning using Protein G-agarose/salmon sperm DNA beads pelleted by centrifugation and washed with low salt buffer, high salt buffer, and Tris-EDTA buffer (25 mm Tris-HCl, 150 mm NaCl, 1 mm EDTA, pH 7.2) to remove any nonspecific binding. The immunocomplexes were eluted from the beads using 200 μl of elution buffer (100 mm NaHCO3, 1% SDS), and the cross-links of the protein-DNA complexes were reversed by an overnight incubation of the elute products at 65 °C. A total of 2 μl of proteinase K (10 μg/μl) was subsequently added to the solution, and samples were incubated at 45 °C for 1 h. DNA was then purified using the spin columns provided. Aliquots of chromatin were also analyzed before immunoprecipitation and served as an input control. BECLIN-1 gene promoter sequences were amplified by PCR with primer sets 1–4: set 1, 5′-AACCCTCACGGCTCTTATTGGAGT-3′ and 5′-AGAGGCAACATTAGGGAGAAGCGA-3′; set 2, 5′-TTGTTCATCCGCTGAAGCCCGCTG-3′ and 5′-ACTCCCTTCTAAGGTCCCACCTC-3′; set 3, 5′-ACCACGCCCGGCTAGTTT-3′ and 5′-TTACCACGGGAAGTGTAGGAGCCAA-3′; set 4, 5′-TGGTCTCGAACTCCTGACCTTGTGAT-3′ and 5′-ATCCCAGCTTAATCCCAGCTACTC-3′. The PCR products were analyzed in 2% agarose gel, and images were assessed with the BioSpectrum AC imaging system and Vision- WorksLS software.
Luciferase Activity Assay
The BECLIN-1 promoter (bp −206 to −910) region was amplified by PCR with a primer set of 5′-CAGCATCTCGAGTGTTGTTCTGAGATGGAG-3′ and 5′-GCCGGTAAGCTTACTCCAATAAGAGCCGTG-3′ and cloned into the XhoI/HindIII sites of pGL3-Luc reporter vector and verified by DNA sequencing. Additionally, pGL3-Luc-BECLIN-1 plasmid was digested with the restriction enzymes PmlI and StuI, and a truncated construct lacking the XBP1s-specific binding site was generated. In brief, ECs were transfected with full-length or truncated pGL3-Luc-BECLIN-1 (0.33 μg/well) together with expression plasmid (0.16 μg/well) encoded from XBP1s and XBP1u. pGL3-Luc Renilla (0.1 μg/well) was included in all transfection assays as an internal control, and pShuttle2 vector was also used as a mock control. Luciferase and Renilla (Promega) activity assays were run 24 and 48 h after transfection using a standard protocol. A relative luciferase unit was defined as the ratio of luciferase activity to Renilla activity, with that of control set as 1.0.
Ex Vivo Experiments
For the generation of XBP1 EC conditional knock-out (XBP1eko) mice, an 11.5-kb DNA fragment encompassing XBP1 gene exons 1–5 was isolated from the mouse genome. Two LoxP sites were inserted into the promoter and intron 2 regions, respectively. An FRT-flanked Neo cassette was inserted upstream of the second LoxP site in the intron 2 for positive clone selection. Recombinant 129Sv/Pas ES cell clones were injected into recipient blastocysts isolated from pregnant C57BL/6J females. The injected blastocysts were then reimplanted into OF1 pseudopregnant females and allowed to develop to term. The F1 generation of chimeras was cross-bred with CAG-Cre deleter to create global knock-out XBP1 mice, whereas they were cross-bred with FLPe transgenic mice to create XBP1loxP/+ mice (25, 26). All of the gene cloning, ES clone selection and injection, and the XBP+/− and XBP1loxP/+ mouse creation were performed by genOway. The following breeding procedures were performed in the King's College London animal facility. All animal experiments in this study were performed according to protocols approved by the Institutional Committee for Use and Care of Laboratory Animals. Arterial vessels from aortas were isolated from wild type and XBP1eko mice and cut into 2 × 2-mm sections. The sections were incubated with M199 supplemented with 10% FBS in the presence or absence of endostatin (20 ng/ml) for 24 h. The segments were fixed with 4% paraformaldehyde, followed by immunofluorescence staining with anti-XBP1, anti-CD31, and anti-LC3β antibodies, as described in the supplemental material. The segments were mounted on a slide with the vessel lumen facing up. Images were taken under an SP5 confocal microscope (Leica). Magnification is indicated in the figures by scale bars.
Statistical Analysis
Data are expressed as the means ± S.E. and were analyzed with a two-tailed Student's t test for two groups or pairwise comparisons. A value of p < 0.05 was considered to be significant.
RESULTS
Sustained Activation of XBP1 mRNA Splicing Induces Autophagy in ECs
In the present study, we tested the hypothesis that XBP1 splicing induced autophagy in ECs. The study was initiated by infected ECs with XBP1 spliced form adenovirus (Ad-XBP1s), where a number of structures that could be either droplets or autophagic vacuoles were observed (Fig. 1, A–C). No such structures or very few of them were observed in ECs infected with unspliced XBP1 form (Ad-XBP1u) or null control (Ad-null) (Fig. 1, A–C). Oil Red O staining demonstrated that these structures were not oil droplets. To explore whether these structures had an autophagic phenotype, ECs were infected with Ad-XBP1s, Ad-XBP1u, or Ad-null viruses for 48 h, and samples were prepared and observed using transmission electron microscopy. The results confirmed the presence of autophagosomes in ECs infected with Ad-XBP1s (Fig. 1, D–I). The overexpression of the unspliced and spliced XBP1 is confirmed by conventional and real-time PCR in Fig. 1, J and K, respectively. These results indicate that XBP1s may trigger an autophagic response in ECs.
FIGURE 1.
Sustained activation of XBP1 splicing induces autophagy in ECs. A–C, ECs were infected with Ad-null, Ad-XBP1u, and Ad-XBP1s viruses at 5 MOI for 6 h and cultured for 48 h, followed by hematoxylin staining (scale bars (A–C), 25 μm). D–I, ECs were infected with Ad-null, Ad-XBP1u, and Ad-XBP1s viruses at 5 MOI for 48 h and grown to confluence on 13-mm diameter glass coverslips. The cells were fixed with 2.5% (v/v) glutaraldehyde in 0.1 m phosphate buffer, and ultrathin sections (70–90 nm) were prepared and examined on an FEI Tecnai 12 transmission microscope showing the presence of autophagosomes in ECs infected with Ad-XBP1s (scale bars, 2 μm (D–F) and 0.5 μm (G–I)). The overexpression of the unspliced and spliced XBP1 is shown by conventional (J) and real-time PCR. *, p < 0.05 (K). Data presented are representative of or an average from three independent experiments. Error bars, S.E.
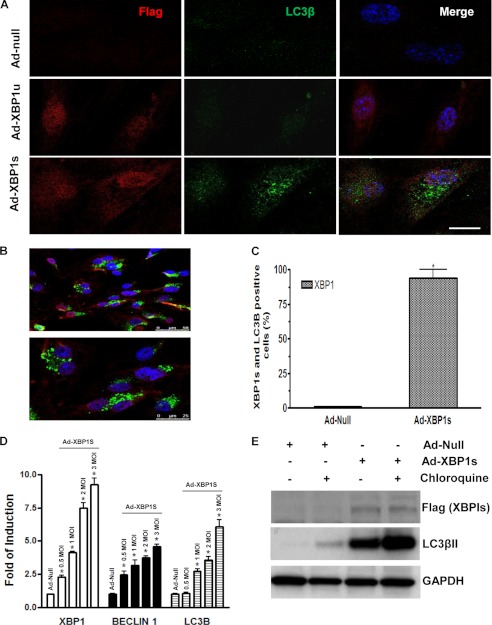
Ad-XBP1s-infected ECs Show a Typical “Punctate Staining” for the LC3β Marker
Immunofluorescence staining of LC3β showed a typical punctate staining for the LC3β marker in Ad-XBP1s-infected cells (Fig. 2A). In order to answer the question of whether XBP1 and LC3β proteins are found in the same cell, more fields have been examined, as shown in Fig. 2B, and the percentage of the XBP1 and LC3β double-positive cells has been calculated and found to be around 95% (Fig. 2C). Additional experiments have revealed that low levels of XBP1s overexpression are sufficient to trigger an autophagic response in ECs by increasing the expression of BECLIN-1 and LC3β (Fig. 2D). Importantly, when ECs were infected with Ad-XBP1s and treated with chloroquine, an inhibitor of the fusion of autophagosomes with lysosomes, a further increase in LC3βII expression level was observed (Fig. 2E). These results further confirm that XBP1s triggers an autophagic response in ECs.
FIGURE 2.
Ad-XBP1s-infected ECs showed a typical “punctate staining” for the LC3β marker. ECs were infected with Ad-XBP1u or Ad-XBP1s at 5 MOI and stained with LC3β antibody and anti-FLAG antibody to detect exogenous XBP1s. A, immunofluorescence staining of LC3β showed a typical punctate staining for the LC3β marker in Ad-XBP1s-infected cells. B, more fields were examined and confirmed that XBP1 and LC3β proteins are found in the same cell. C, the percentage of the XBP1- and LC3β-positive cells has been calculated and found to be around 95%. *, p < 0.05. D, ECs were infected with Ad-null or Ad-XBP1s at 0.5–3 MOI, and real time quantitative RT-PCR was performed for XBP1, BECLIN-1, and LC3βII. *, p < 0.05. E, ECs were infected with Ad-null or Ad-XBP1s and treated with chloroquine for 6 h, and Western blot for LC3βII, an inhibitor of the fusion of autophagosomes with lysosomes, is shown. Data presented are representative of or an average from three independent experiments. Error bars, S.E.
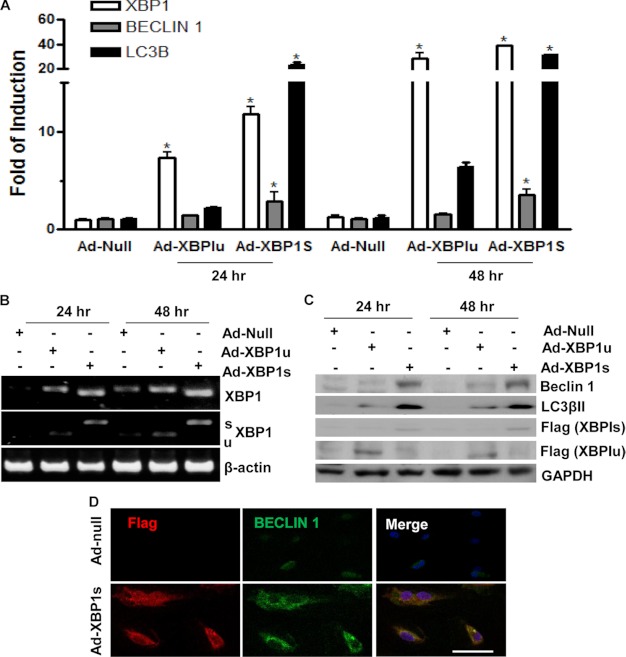
Overexpression of XBP1s Increases Autophagy Gene Expression
To verify that XBP1 mRNA splicing induces autophagy, XBP1s and XBP1u were overexpressed in ECs by adenoviral gene transfer, and the expression of autophagy genes was assessed at the mRNA and protein levels. Real-time quantitative RT-PCR showed that XBP1s increases BECLIN-1 and LC3β autophagy genes when compared with control virus-infected cells or Ad-XBP1u-infected cells in a time course experiment (Fig. 3A). Routine RT-PCR confirmed the overexpression of the two isoforms of XBP1 in ECs (Fig. 3B). XBP1 mRNA splicing was detected by routine RT-PCR, followed by PstI digestion. There is a PstI site in the 26-nucleotide intron of XBP1u but not in XBP1s mRNA. Digestion of the RT-PCR product with a PstI restriction enzyme allows XBP1s (not digested) and XBP1u (digested into two smaller bands) to be distinguished. Western blot analysis showed induced expression of BECLIN-1 and autophagosome-specific isoform LC3βII, indicating activation of autophagy (Fig. 3C). Furthermore, immunofluorescence staining revealed that Ad-XBP1s increased BECLIN-1 expression in ECs (Fig. 3D). These results suggest that XBP1 splicing is involved in the induction of the autophagy gene transcription in ECs.
FIGURE 3.
Overexpression of XBP1 increases autophagic gene expression. A-C, ECs were infected with Ad-null, Ad-XBP1u, and Ad-XBP1s viruses at 5 MOI. Total RNA and proteins were harvested at 24 and 48 h postinfection, followed by real-time quantitative RT-PCR (A), routine RT-PCR (B), and Western blot (C) analysis. *, p < 0.05. D, ECs were infected with Ad-null and Ad-XBP1s viruses at 5 MOI for 48 h, followed by immunofluorescence staining with BECLIN-1 antibody. FLAG antibody was included to detect the exogenous XBP1s. Scale bar, 25 μm. Data presented are representative of or an average from three independent experiments. Error bars, S.E.
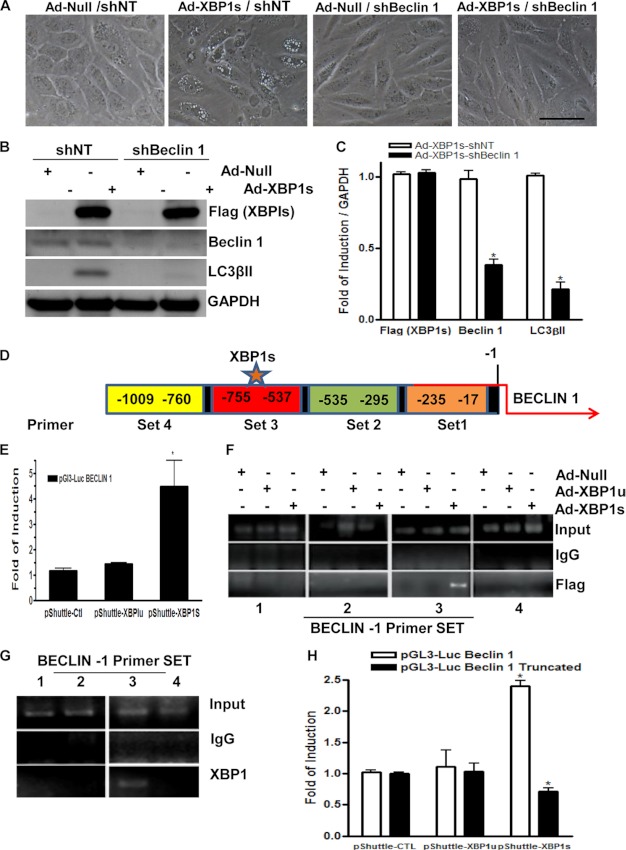
BECLIN-1 Deficiency in ECs Abolishes the XBP1-induced Autophagy Response
To shed light on the mechanisms involved in XBP1s-induced autophagy, further experiments were performed to explore the effect of XBP1s on BECLIN-1 expression. BECLIN-1 has been shown to play an important role in the vesicle nucleation during the initial steps of autophagy (27). Importantly, when BECLIN-1 was knocked down by shRNA in ECs, the induction of autophagy mediated by XBP1 was ablated because autophagic vacuole structures were not observed (Fig. 4A). Further experiments revealed that knockdown of BECLIN-1 abolished XBP1-induced LC3βII expression at the protein level (Fig. 4, B and C, quantification). Recent reports have shown that 14-3-3T and E2F1 regulate the BECLIN-1 gene and initiate autophagy (28). Therefore, we explored whether XBP1s could regulate BECLIN-1 through 14-3-3T and E2F1. However, neither XBP1s nor XBP1u could regulate 14-3-3T and E2F1 gene expression (supplemental Fig. S1), suggesting that XBP1s-regulated BECLIN-1 expression is not through these two genes. Because XBP1s is a transcription factor, we hypothesized that it may directly regulate BECLIN-1 gene expression at the transcriptional level. Luciferase assays demonstrated that overexpression of XBP1s induced the expression of the BECLIN-1 promoter in ECs (Fig. 4E). This transcriptional regulation of BECLIN-1 was specific for the XBP1s and not for the XBP1u (Fig. 4E). These results demonstrate that knockdown of BECLIN-1 abolished XBP1-induced autophagy, highlighting the importance of BECLIN-1 in induction of autophagy mediated by XBP1s.
FIGURE 4.
XBP1 spliced isoform regulates BECLIN-1 through transcriptional activation. BECLIN-1 deficiency in ECs abolishes the XBP1-induced autophagy response. A, ECs were infected with non-target (shNT) or BECLIN-1 (shBeclin1) shRNA, and 24 h later, Ad-null of Ad-XBP1s were introduced. Images showing that knockdown of BECLIN-1 abolished XBP1-induced autophagy were taken at 48 h upon XBP1s overexpression. Scale bar, 25 μm. B, knockdown of BECLIN-1 abolished XBP1-induced LC3βII gene expression, as shown by Western blot analysis (C) and quantification. *, p < 0.05. D, a map of the BECLIN-1 promoter showing the relative regions covered by the primer sets 1–4. E, XBP1s increased BECLIN-1-Luc reporter expression in luciferase activity assays. *, p < 0.05. F, ChIP assays demonstrated that XBP1s bound directly on the BECLIN-1 gene promoter. Anti-FLAG antibody was used to pull down exogenous XBP1s and its bound DNA fragments. To define the XBP1 binding site to the BECLIN-1 gene promoter, four sets of primers covering the −17 to −1009 nt promoter region were used to amplify the precipitated DNA fragments. G, ChIP assays were performed with an anti-XBP1 antibody to pull down the endogenous XBP1s and its bound DNA fragments in normal ECs. H, when ECs were co-transfected with plasmids encoded by XBPIu, XBP1s, or control empty vector together with a truncated construct of pGL3-Luc-Beclin promoter lacking the −537 to −755 region, luciferase activity assays demonstrated an ablated transcriptional activation of BECLIN-1 induced by XBP1s, p < 0.05. Data presented are representative of or an average from three independent experiments. Error bars, S.E.
XBP1s Directly Binds to the BECLIN-1 Gene Promoter
To investigate whether XBP1s directly binds to the BECLIN-1 promoter region, chromatin immunoprecipitation assays were performed with anti-FLAG antibody. A map of the BECLIN-1 promoter showing the relative regions covered by the primer sets 1–4 is illustrated in Fig. 4D. As expected, a binding of XBP1s to the BECLIN-1 gene promoter was detected, which locates at region −537 to −755 nt upstream of the transcription initiation site (Fig. 4F). Moreover, the binding of XBP1 to the BECLIN-1 gene promoter was verified in experiments with antibody against endogenous XBP1 (Fig. 4G). Further experiments were performed to explore whether the direct binding of XBP1s to the BECLIN-1 promoter is essential for BECLIN-1 transcription. A truncated construct of the BECLIN-1 promoter lacking the −537 to −755 region was created. Transient transfections with the intact and the truncated pGL3-Luc-BECLIN-1 promoter constructs were performed. As shown in Fig. 4H, deletion of the −537 to −755 region abolished XBP1s-induced BECLIN-1 reporter gene expression. These results suggest that XBP1 mRNA splicing triggers an autophagic response in ECs through BECLIN-1 transcriptional activation by direct binding to the −537 to −755 nt region in BECLIN-1 gene promoter.
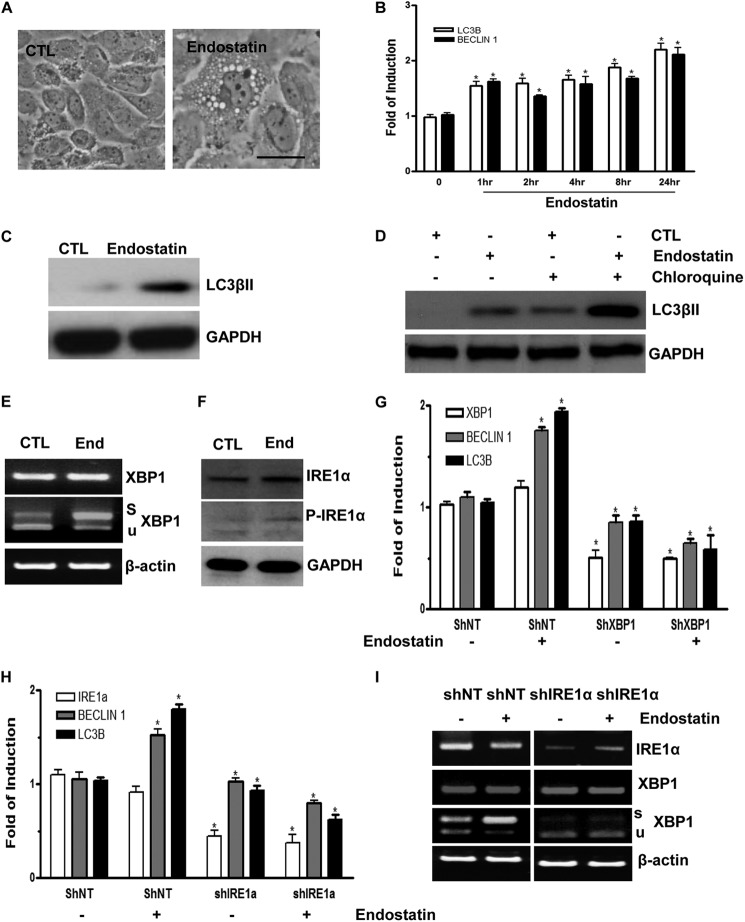
Endostatin Induces Autophagy in ECs through IRE1α-mediated XBP1 mRNA Splicing
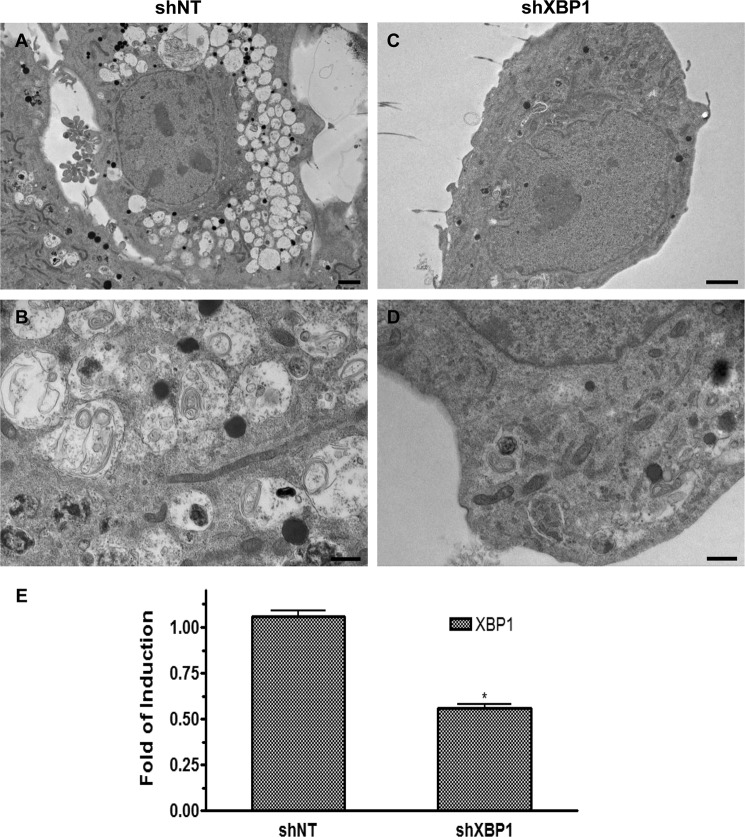
Angiogenesis inhibitors, such as endostatin, are reported to induce autophagy in ECs (29). In this study, ECs treated with endostatin induced autophagy, as demonstrated by the appearance of autophagic vacuole structures (Fig. 5A). Further experiments revealed that endostatin induced the expression levels of genes related to autophagy at both the RNA level in a time course experiment (Fig. 5B) and at the protein level (Fig. 5C) after 24 h upon endostatin stimulation. Importantly, when ECs were treated with endostatin in the presence of chloroquine, an accumulation of LC3βII was observed (Fig. 5D). To determine whether XBP1 mRNA splicing is involved in the endostatin-induced autophagy process, the effect of endostatin on XBP1 mRNA splicing was assessed. Endostatin treatment robustly increased XBP1 splicing but had no effect on the total XBP1 mRNA level (Fig. 5E). XBP1 mRNA undergoes unconventional splicing through the phosphorylation of IRE1α (17, 18). Therefore, we further tested whether endostatin could alter the phosphorylation status of IRE1α. Indeed, endostatin induced IRE1α phosphorylation, as revealed by Western blot analysis (Fig. 5F). These results suggest that endostatin can activate XBP1 mRNA splicing through IRE1α phosphorylation. To investigate whether XBP1 mRNA splicing is necessary for endostatin-induced autophagy gene expression, ECs were infected with non-target (shNT) or XBP1-targeting (shXBP1) shRNA lentiviruses for 72 h and then treated with endostatin for 4 h. Real-time PCR analysis demonstrated that knockdown of XBP1 abolished endostatin-induced autophagy gene expression (Fig. 5G). Similarly, knockdown of IRE1α also suppressed endostatin-induced autophagy-related gene expression (Fig. 5, H and I). These results suggest that IRE1α-mediated XBP1 mRNA splicing is necessary for endostatin-induced autophagy in ECs. In order to address the important question of whether knockdown of XBP1 could inhibit the induction of autophagy, ECs were infected with Non-target (shNT) or XBP1-targeting (shXBP1) shRNA lentiviruses for 48 h, followed by 24-h endostatin treatment. The cells were fixed, and samples were prepared to be observed under a transmission electron microscope. It is shown that knockdown of XBP1 by shRNA abolished the presence of autophagosomes in ECs mediated by endostatin treatment (Fig. 6, A–D). A real-time PCR assay confirmed the knockdown of XBP1 by shRNA in ECs (Fig. 6E). These results reveal that knockdown of XBP1 abolished the induction of autophagy mediated by endostatin, highlighting the important role of XBP1s in the induction of autophagy.
FIGURE 5.
Endostatin induces autophagy in ECs through IRE1α-mediated XBP1 mRNA splicing. A, endostatin-induced autophagy in ECs. Images were taken at 24 h after 20 ng/ml endostatin treatment. Scale bar, 25 μm. B, endostatin up-regulates autophagic gene expression at the mRNA level in time course experiments. C, ECs stimulated with endostatin for 24 h showing induced LC3βII expression at the protein level. D, ECs were treated with endostatin for 24 h prior to chloroquine treatment for 6 h, and Western blot for LC3βII is shown. E, endostatin-induced XBP1 mRNA splicing and IRE1α phosphorylation. ECs were treated with endostatin for 4 h, followed by RT-PCR plus PstI digestion and Western blot analysis (F). Note that endostatin induced IRE1α phosphorylation and robust XBP1 mRNA splicing. CTL, control; End, endostatin; s, spliced band; u, unspliced band. β-Actin and GAPDH were included as mRNA and protein loading control, respectively. G, knockdown of XBP1 abolished endostatin-induced autophagy gene expression. Non-target (shNT) or XBP1 (shXBP1) shRNA lentiviruses-infected ECs (for 72 h) were treated with endostatin for 4 h, followed by real-time PCR analysis of autophagy gene expression. H and I, knockdown of IRE1α abolished endostatin-induced autophagy gene expression. Non-target (shNT) or IRE1α (shIRE1α) shRNA lentivirus-infected ECs (for 72 h) were treated with endostatin for 4 h, followed by real-time PCR analysis of autophagy gene expression or routine RT-PCR. Note that treatment with endostatin even decreased autophagy gene expression in IRE1α-deficient cells. *, p < 0.05. Data presented are representative of or an average of three independent experiments. Error bars, S.E.
FIGURE 6.
Knockdown of XBP1 abolished the induction of autophagy mediated by Endostatin. ECs were infected with non-target (shNT) or XBP1-targeting (shXBP1) shRNA lentiviruses for 48 h, followed by 24-h endostatin treatment. A–D, electron micrographs show that knockdown of XBP1 abolished the presence of autophagosomes in ECs mediated by endostatin. Scale bars, 2 μm (A and B) and 0.5 μm (C and D). E, confirmation of knockdown of XBP1 in ECs by a real-time PCR assay. *, p < 0.05. Data presented are representative of or an average from three independent experiments. Error bars, S.E.
XBP1 Deficiency in ECs Abolishes the Endostatin-induced Autophagy Response
The above data have demonstrated that XBP1 mRNA splicing is involved in autophagy response in cultured ECs. To further explore the role of XBP1 in autophagy in intact endothelium, we generated a conditional null allele of XBP1 in endothelial cells. Targeting of XBP1 was performed by introducing LoxP sites into the promoter and intron 2 regions through homologous recombination (supplemental Fig. S2A). The introduction of a LoxP site into the promoter region does not affect XBP1 transcription. The mutation mediated by Cre recombinase deletes a 2.2-kb DNA fragment comprising part of the promoter and the exons 1 and 2, totally ablating XBP1 transcription. Germ line transmission was detected by Southern blot (supplemental Fig. S2B), and deletion of XBP1 was confirmed by PCR at the genomic level (supplemental Fig. S2, C and D). XBP1neo-loxP mice were bred to CAG-Cre (25) transgenic mice, which express Cre recombinase ubiquitously, allowing for the generation of XBP1+/− mice. XBP1loP/loxP mice were bred to Tie2-Cre (26) transgenic mice, which express Cre recombinase in ECs and bone marrow progenitor cells, allowing for the generation of EC conditional knock-out mice (Tie2Cre/XBP1loxP/loxP). Supplemental Fig. S2D shows the genotypes of the littermates from the cross-breeding of Tie2Cre/XBP1loxP/+ and Tie2Cre/XBP1loxP/loxP (XBP1eko) mice. The XBP1eko mice can survive, showing defects in angiogenesis.4
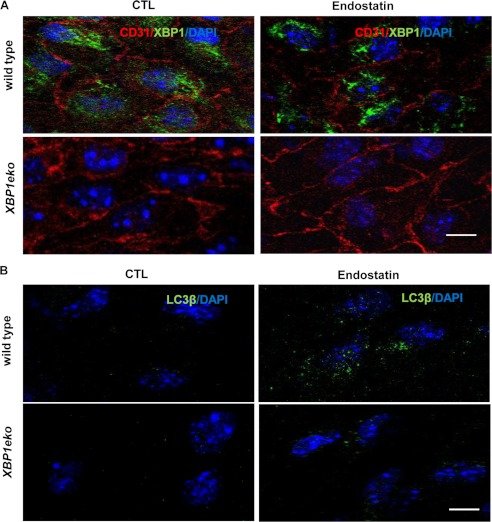
To this end, ex vivo experiments were performed on arterial vessel sections isolated from wild type and XBP1eko mice in which XBP1 is specifically disrupted in ECs. Arterial vessels were isolated from wild type and XBP1eko mice, dissected into 2 × 2-mm sections, and incubated with M199 supplemented with 10% FBS in the absence or presence of endostatin for 24 h, followed by immunofluorescence staining with anti-XBP1, anti-CD31 (Fig. 7A), and anti-LC3β (Fig. 7B) antibodies. As expected, no XBP1 can be detected in ECs on vessels from XBP1eko mice (Fig. 7A, bottom). Importantly, XBP1 deficiency not only reduced the basal level of LC3β (Fig. 7B, left panels) but also abolished the endostatin-induced LC3β expression (Fig. 7B, right panels). In XBP1eko mice, a low level of LC3β could be detected after endostatin treatment, indicating that endostatin-induced LC3β expression and autophagy response can also be activated in XBP1-independent pathways. These results confirm that XBP1 is involved in endostatin-induced autophagy.
FIGURE 7.
XBP1 deficiency in ECs ablates the endostatin-induced autophagy. Arterial vessels were isolated from wild type and XBP1eko mice and dissected into 2 × 2-mm sections. The sections were incubated with M199 supplemented with 10% FBS in the absence or presence of endostatin for 24 h, followed by immunofluorescence staining with anti-XBP1, anti-CD31 (A), and anti-LC3β (B) antibodies. Note that XBP1 deficiency reduces the basal level of LC3β expression and its response to endostatin stimulation. Data presented are representative images from 20 views. Scale bar, 25 μm.
DISCUSSION
The findings of this study reveal that XBP1 mRNA splicing is implicated in the induction of autophagy in ECs through transcriptional regulation of BECLIN-1. In line with this finding, endostatin activated autophagic gene expression through XBP1 mRNA splicing in a IRE1α-dependent manner. Targeting of XBP1 or IRE1α by shRNA in ECs ablated endostatin-induced autophagy, whereas XBP1 deficiency in ECs reduced the basal level of LC3β expression and ablated response to endostatin stimulation in XBP1 EC conditional knock-out (XBP1eko) mice.
Among the causes that stimulate autophagy are intracellular stress conditions and hypoxia (11, 30). In response to ER stress, XBP1 mRNA undergoes unconventional splicing through IRE1α, giving rise to a spliced form (17, 18). A number of previous reports have already highlighted the link of ER stress response with the induction of autophagy (31–34). For instance, Yorimitsu et al. (35) have shown that ER stress induces autophagy, which can protect against cell death. Furthermore, hypoxia has been reported to activate an autophagy-lysosomal degradation pathway, and unfolded protein response enhances the capacity of hypoxic tumor cells to carry out autophagy and induces cell survival (36). Recent evidence is also providing mechanistic insights into this concept by demonstrating that activating transcription factor 4 (ATF4) is required for ER stress and hypoxia-induced expansion of autophagy through direct binding to a cyclic AMP-response element binding site in the LC3β promoter. This results in LC3β up-regulation and implication in cell survival (37). In contrast, inhibition of ER stress by knockdown of IRE1 inhibited autophagy and adipogenesis (38). In the present study, we provide novel findings that suggest that XBP1s, a crucial signal transducer of ER stress response, is involved in the induction of autophagy in ECs, providing insight into EC survival.
Autophagy has also been activated in response to angiogenesis inhibitors (39). Endostatin, which is a well characterized inhibitor of angiogenesis that induces apoptosis (29, 40), has been shown to induce autophagy in ECs (40) and increase BECLIN-1 expression through β-catenin- and Wnt-mediated signaling pathways (29, 41). In the present study, we provide evidence that endostatin stimulation is implicated in autophagic gene expression through XBP1 mRNA splicing in an IRE1α-dependent manner. When XBP1 or IRE1α was targeted by shRNA in ECs or in XBP1eko mice, the endostatin-induced autophagic gene expression and autophagosome formation were ablated. These results further support the notion that XBP1s is involved in the regulation of autophagy in ECs.
BECLIN-1 is an essential autophagic protein, which has a crucial role in the initial stages of autophagy (27). Previous studies have shown that BECLIN-1 levels and autophagic vesicle formation were regulated by Bcl-2 and Bcl-xL (29), which are important players in a cell's decision to progress to an apoptotic or necrotic death (27, 42–44). In order to shed light on the mechanistic insights into the regulation of autophagy by XBP1 splicing in ECs, further experiments revealed that activation of XBP1 splicing triggers BECLIN-1 transcriptional induction through directly binding to the BECLIN-1 promoter at the region of nt −537 to −755. A number of potential transcription factor binding sites in the region of nt −537 to −755 have been identified using TFMATRIX software, including CREBP. Therefore, XBP1s may function as homodimer or heterodimer with another transcription factor, such as CREBP. Importantly, further experiments have demonstrated that XBP1s did not induce transcriptional activation of a truncated construct of the pGL3-Luc-BECLIN-1 promoter lacking the −537 to −755 region, providing further insights into the BECLIN-1 transcriptional activation mechanism through XBP1s. XBP1 has been reported to regulate histone H4 acetylation (45), whereas XBP1s is also a target of acetylation and deacetylation mediated by p300 and SIRT1 (sirtuin 1), respectively (46). In addition, it has been recently shown that the acetylation status of the ER is regulated by IRE1/XBP1, by controlling the influx of acetyl-CoA through the membrane transporter AT-1, which results in the regulation of autophagy (47). Thus, XBP1s may control the transcriptional activation of BECLIN-1 through recruiting other co-factors that induce acetylation and protein stability and/or inhibit deacetylation in a cell-dependent manner. Further experiments will elucidate these pathways and enhance our knowledge on this aspect.
Vidal et al. (50) have recently reported that targeting XBP1 protects against Huntington disease through the regulation of FoxO1 and autophagy. The authors also showed that XBP1 deficiency enhanced the severity of spinal cord injury in a mouse model (48), whereas it did not alter prion pathogenesis (49), which indicates that the XBP1-regulated autophagy pathway contributes only to certain diseases with distinct outputs (50). Therefore, based on those findings and taking into account our data, we can now describe important aspects in the regulation of autophagy, for example, the activation of XBP1 splicing has a precise role in the autophagy, and autophagy-mediated by XBP1 splicing in ECs is cell type-dependent. For instance, activation of XBP1 splicing in vascular smooth muscle cells does not induce autophagic response.4
Evidence is accumulating that autophagy occurs in advanced atherosclerotic plaques (51). For instance, in advanced atherosclerosis, the role of autophagy in macrophages (3) and vascular smooth muscle cells (52) has been shown. In addition, Ox-LDL can also activate autophagy (53) in ECs through the LC3β/BECLIN-1 pathway, leading to the degradation of ox-LDL through lysosomes (54), whereas induction of autophagy promotes angiogenesis by activating the vascular endothelial growth factor (55). Therefore, autophagy may be defined as “a final attempt for survival” based on an adaptive response to fight against cellular stress. However, it remains elusive whether autophagy is a harmful or a protective mechanism in atherosclerotic plaques. The key questions are “how long” and “at what levels” autophagy can be activated without causing detrimental consequences (56, 57). Recent studies have shown that caspase activation regulates the extracellular export of autophagic vacuoles, which reveals that apoptosis and autophagy are two closely related and tightly regulated processes (58). We have previously shown that sustained activation of XBP1 results in EC apoptosis and atherosclerosis development (12). In the present study, we provide evidence that XBP1s triggers an autophagy response in ECs. Thus, it seems that the tight regulation of the expression levels and duration of splicing activation of key molecules, such as XBP1s, could determine the threshold of the autophagic responses through regulation of precise mechanisms in a cell-specific manner. Cross-breeding of the mouse model for atherosclerosis apolipoprotein E with XBP1eko mice is currently in progress, and it will undoubtedly help to answer these questions and demonstrate the role of XBP1s in the regulation of autophagy in ECs and the development of atherosclerosis. Further studies will elucidate whether XBP1 mRNA splicing could be used as an important pharmacological target that can regulate the autophagic machinery.
In summary, endostatin and other stimuli activate IRE1α phosphorylation, which in turn removes the 26-nt intron from XBP1 mRNA, causing the open reading frame shift and giving rise to the spliced isoform. The spliced XBP1 translocates into the nucleus and binds as homodimer or heterodimer to the BECLIN-1 gene promoter, leading to BECLIN-1 transcription. BECLIN-1 activates autophagic response, leading to cell survival or apoptosis (Fig. 8). This study supports the notion that XBP1 induced an autophagic response in ECs through transcriptional regulation of BECLIN-1 gene expression, which mediates activation of LC3βII and in turn induces autophagy, providing key insights into the threshold of the autophagic responses.
FIGURE 8.
A schematic illustration showing that XBP1 splicing induces autophagic response in endothelial cells. Under normal conditions, ER-resident IRE1α associates with Grp78. Upon treatment with endostatin or other stimuli, IRE1α dissociates from Grp78 and oligomerizes, leading to its autophosphorylation. The activated IRE1α catalyzes the excision of a 26-nucleotide unconventional intron from XBP1 mRNA, which causes an open reading frameshift in the XBP1 mRNA, giving rise to a 371-amino acid, 54-kDa protein with an activation domain, the XBP1s isoform. The XBP1s translocates into the nucleus and binds as homodimers or heterodimers to the BECLIN-1 gene promoter region at −537 to −755 nt upstream of the transcription initiation site, leading to the activation of BECLIN-1 transcription, which mediates activation of LC3βII and in turn induces autophagic response, leading to EC survival or apoptosis.
Supplementary Material
This work was supported by Grants from the British Heart Foundation and the Oak Foundation.

This article contains supplemental Figs. S1 and S2.
L. Zeng, Q. Xiao, H. Li, A. Margariti, D. Martin, S. Alam, A. Zampetaki, Y. Hu, Q. Xu, unpublished data.
- EC
- endothelial cell
- ER
- endoplasmic reticulum
- MOI
- multiplicity of infection
- nt
- nucleotide(s)
- CREBP
- cAMP-response element-binding protein.
REFERENCES
- 1. Martinet W., De Meyer G. R. (2008) Autophagy in atherosclerosis. Curr. Atheroscler. Rep. 10, 216–223 [DOI] [PubMed] [Google Scholar]
- 2. Razani B., Feng C., Coleman T., Emanuel R., Wen H., Hwang S., Ting J. P., Virgin H. W., Kastan M. B., Semenkovich C. F. (2012) Autophagy links inflammasomes to atherosclerotic progression. Cell Metab. 15, 534–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liao X., Sluimer J. C., Wang Y., Subramanian M., Brown K., Pattison J. S., Robbins J., Martinez J., Tabas I. (2012) Macrophage autophagy plays a protective role in advanced atherosclerosis. Cell Metab. 15, 545–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Martinet W., De Meyer G. R. (2009) Autophagy in atherosclerosis. A cell survival and death phenomenon with therapeutic potential. Circ. Res. 104, 304–317 [DOI] [PubMed] [Google Scholar]
- 5. Mitra A. K., Dhume A. S., Agrawal D. K. (2004) “Vulnerable plaques.” Ticking of the time bomb. Can J. Physiol. Pharmacol. 82, 860–871 [DOI] [PubMed] [Google Scholar]
- 6. Jia G., Cheng G., Agrawal D. K. (2006) Differential effects of insulin-like growth factor-1 and atheroma-associated cytokines on cell proliferation and apoptosis in plaque smooth muscle cells of symptomatic and asymptomatic patients with carotid stenosis. Immunol. Cell Biol. 84, 422–429 [DOI] [PubMed] [Google Scholar]
- 7. Klionsky D. J., Emr S. D. (2000) Autophagy as a regulated pathway of cellular degradation. Science 290, 1717–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yoshimori T. (2004) [Autophagy as a bulk protein degradation system. It plays various roles]. Tanpakushitsu Kakusan Koso 49, 1029–1032 [PubMed] [Google Scholar]
- 9. Levine B., Klionsky D. J. (2004) Development by self-digestion. Molecular mechanisms and biological functions of autophagy. Dev. Cell 6, 463–477 [DOI] [PubMed] [Google Scholar]
- 10. Yorimitsu T., Klionsky D. J. (2005) Autophagy. Molecular machinery for self-eating. Cell Death Differ. 12, 1542–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yoshimori T. (2007) Autophagy. paying Charon's toll. Cell 128, 833–836 [DOI] [PubMed] [Google Scholar]
- 12. Zeng L., Zampetaki A., Margariti A., Pepe A. E., Alam S., Martin D., Xiao Q., Wang W., Jin Z. G., Cockerill G., Mori K., Li Y. S., Hu Y., Chien S., Xu Q. (2009) Sustained activation of XBP1 splicing leads to endothelial apoptosis and atherosclerosis development in response to disturbed flow. Proc. Natl. Acad. Sci. U.S.A. 106, 8326–8331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Clauss I. M., Chu M., Zhao J. L., Glimcher L. H. (1996) The basic domain/leucine zipper protein hXBP-1 preferentially binds to and transactivates CRE-like sequences containing an ACGT core. Nucleic Acids Res. 24, 1855–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liou H. C., Boothby M. R., Finn P. W., Davidon R., Nabavi N., Zeleznik-Le N. J., Ting J. P., Glimcher L. H. (1990) A new member of the leucine zipper class of proteins that binds to the HLA DR α promoter. Science 247, 1581–1584 [DOI] [PubMed] [Google Scholar]
- 15. Sriburi R., Jackowski S., Mori K., Brewer J. W. (2004) XBP1. A link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J. Cell Biol. 167, 35–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lee A. H., Iwakoshi N. N., Anderson K. C., Glimcher L. H. (2003) Proteasome inhibitors disrupt the unfolded protein response in myeloma cells. Proc. Natl. Acad. Sci. U.S.A. 100, 9946–9951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee K., Tirasophon W., Shen X., Michalak M., Prywes R., Okada T., Yoshida H., Mori K., Kaufman R. J. (2002) IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 16, 452–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yoshida H., Matsui T., Yamamoto A., Okada T., Mori K. (2001) XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107, 881–891 [DOI] [PubMed] [Google Scholar]
- 19. Reimold A. M., Iwakoshi N. N., Manis J., Vallabhajosyula P., Szomolanyi-Tsuda E., Gravallese E. M., Friend D., Grusby M. J., Alt F., Glimcher L. H. (2001) Plasma cell differentiation requires the transcription factor XBP-1. Nature 412, 300–307 [DOI] [PubMed] [Google Scholar]
- 20. Clauss I. M., Gravallese E. M., Darling J. M., Shapiro F., Glimcher M. J., Glimcher L. H. (1993) In situ hybridization studies suggest a role for the basic region-leucine zipper protein hXBP-1 in exocrine gland and skeletal development during mouse embryogenesis. Dev. Dyn. 197, 146–156 [DOI] [PubMed] [Google Scholar]
- 21. Reimold A. M., Etkin A., Clauss I., Perkins A., Friend D. S., Zhang J., Horton H. F., Scott A., Orkin S. H., Byrne M. C., Grusby M. J., Glimcher L. H. (2000) An essential role in liver development for transcription factor XBP-1. Genes Dev. 14, 152–157 [PMC free article] [PubMed] [Google Scholar]
- 22. Margariti A., Zampetaki A., Xiao Q., Zhou B., Karamariti E., Martin D., Yin X., Mayr M., Li H., Zhang Z., De Falco E., Hu Y., Cockerill G., Xu Q., Zeng L. (2010) Histone deacetylase 7 controls endothelial cell growth through modulation of β-catenin. Circ. Res. 106, 1202–1211 [DOI] [PubMed] [Google Scholar]
- 23. Margariti A., Xiao Q., Zampetaki A., Zhang Z., Li H., Martin D., Hu Y., Zeng L., Xu Q. (2009) Splicing of HDAC7 modulates the SRF-myocardin complex during stem-cell differentiation towards smooth muscle cells. J. Cell Sci. 122, 460–470 [DOI] [PubMed] [Google Scholar]
- 24. Margariti A., Winkler B., Karamariti E., Zampetaki A., Tsai T. N., Baban D., Ragoussis J., Huang Y., Han J. D., Zeng L., Hu Y., Xu Q. (2012) Direct reprogramming of fibroblasts into endothelial cells capable of angiogenesis and reendothelialization in tissue-engineered vessels. Proc. Natl. Acad. Sci. U.S.A. 109, 13793–13798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sakai K., Miyazaki J. (1997) A transgenic mouse line that retains Cre recombinase activity in mature oocytes irrespective of the cre transgene transmission. Biochem. Biophys. Res. Commun. 237, 318–324 [DOI] [PubMed] [Google Scholar]
- 26. Kisanuki Y. Y., Hammer R. E., Miyazaki J., Williams S. C., Richardson J. A., Yanagisawa M. (2001) Tie2-Cre transgenic mice. A new model for endothelial cell-lineage analysis in vivo. Dev. Biol. 230, 230–242 [DOI] [PubMed] [Google Scholar]
- 27. Maiuri M. C., Zalckvar E., Kimchi A., Kroemer G. (2007) Self-eating and self-killing. Cross-talk between autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 8, 741–752 [DOI] [PubMed] [Google Scholar]
- 28. Wang B., Ling S., Lin W. C. (2010) 14-3-3Tau regulates Beclin 1 and is required for autophagy. PLoS One 5, e10409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nguyen T. M., Subramanian I. V., Xiao X., Ghosh G., Nguyen P., Kelekar A., Ramakrishnan S. (2009) Endostatin induces autophagy in endothelial cells by modulating Beclin 1 and β-catenin levels. J. Cell Mol. Med. 13, 3687–3698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gozuacik D., Kimchi A. (2004) Autophagy as a cell death and tumor suppressor mechanism. Oncogene 23, 2891–2906 [DOI] [PubMed] [Google Scholar]
- 31. Fouillet A., Levet C., Virgone A., Robin M., Dourlen P., Rieusset J., Belaidi E., Ovize M., Touret M., Nataf S., Mollereau B. (2012) ER stress inhibits neuronal death by promoting autophagy. Autophagy 8, 915–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ghavami S., Yeganeh B., Stelmack G. L., Kashani H. H., Sharma P., Cunnington R., Rattan S., Bathe K., Klonisch T., Dixon I. M., Freed D. H., Halayko A. J. (2012) Apoptosis, autophagy, and ER stress in mevalonate cascade inhibition-induced cell death of human atrial fibroblasts. Cell Death Dis. 3, e330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vidal R. L., Hetz C. (2012) Crosstalk between the UPR and autophagy pathway contributes to handling cellular stress in neurodegenerative disease. Autophagy 8, 970–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rovetta F., Stacchiotti A., Consiglio A., Cadei M., Grigolato P. G., Lavazza A., Rezzani R., Aleo M. F. (2012) ER signaling regulation drives the switch between autophagy and apoptosis in NRK-52E cells exposed to cisplatin. Exp. Cell Res. 318, 238–250 [DOI] [PubMed] [Google Scholar]
- 35. Yorimitsu T., Nair U., Yang Z., Klionsky D. J. (2006) Endoplasmic reticulum stress triggers autophagy. J. Biol. Chem. 281, 30299–30304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rouschop K. M., van den Beucken T., Dubois L., Niessen H., Bussink J., Savelkouls K., Keulers T., Mujcic H., Landuyt W., Voncken J. W., Lambin P., van der Kogel A. J., Koritzinsky M., Wouters B. G. (2010) The unfolded protein response protects human tumor cells during hypoxia through regulation of the autophagy genes MAP1LC3B and ATG5. J. Clin. Invest. 120, 127–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rzymski T., Milani M., Pike L., Buffa F., Mellor H. R., Winchester L., Pires I., Hammond E., Ragoussis I., Harris A. L. (2010) Regulation of autophagy by ATF4 in response to severe hypoxia. Oncogene 29, 4424–4435 [DOI] [PubMed] [Google Scholar]
- 38. Younce C., Kolattukudy P. (2012) MCP-1-induced protein promotes adipogenesis via oxidative stress, endoplasmic reticulum stress and autophagy. Cell Physiol. Biochem. 30, 307–320 [DOI] [PubMed] [Google Scholar]
- 39. Nguyen T. M., Subramanian I. V., Kelekar A., Ramakrishnan S. (2007) Kringle 5 of human plasminogen, an angiogenesis inhibitor, induces both autophagy and apoptotic death in endothelial cells. Blood 109, 4793–4802 [DOI] [PubMed] [Google Scholar]
- 40. Chau Y. P., Lin S. Y., Chen J. H., Tai M. H. (2003) Endostatin induces autophagic cell death in EAhy926 human endothelial cells. Histol. Histopathol. 18, 715–726 [DOI] [PubMed] [Google Scholar]
- 41. Gao C., Cao W., Bao L., Zuo W., Xie G., Cai T., Fu W., Zhang J., Wu W., Zhang X., Chen Y. G. (2010) Autophagy negatively regulates Wnt signalling by promoting Dishevelled degradation. Nat. Cell Biol. 12, 781–790 [DOI] [PubMed] [Google Scholar]
- 42. Oberstein A., Jeffrey P. D., Shi Y. (2007) Crystal structure of the Bcl-XL-Beclin 1 peptide complex. Beclin 1 is a novel BH3-only protein. J. Biol. Chem. 282, 13123–13132 [DOI] [PubMed] [Google Scholar]
- 43. Pattingre S., Levine B. (2006) Bcl-2 inhibition of autophagy. A new route to cancer? Cancer Res. 66, 2885–2888 [DOI] [PubMed] [Google Scholar]
- 44. Pattingre S., Tassa A., Qu X., Garuti R., Liang X. H., Mizushima N., Packer M., Schneider M. D., Levine B. (2005) Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 122, 927–939 [DOI] [PubMed] [Google Scholar]
- 45. Tao R., Chen H., Gao C., Xue P., Yang F., Han J. D., Zhou B., Chen Y. G. (2011) Xbp1-mediated histone H4 deacetylation contributes to DNA double-strand break repair in yeast. Cell Res. 21, 1619–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang F. M., Chen Y. J., Ouyang H. J. (2011) Regulation of unfolded protein response modulator XBP1s by acetylation and deacetylation. Biochem. J. 433, 245–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pehar M., Jonas M. C., Hare T. M., Puglielli L. (2012) SLC33A1/AT-1 protein regulates the induction of autophagy downstream of IRE1/XBP1 pathway. J. Biol. Chem. 287, 29921–29930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Valenzuela V., Collyer E., Armentano D., Parsons G. B., Court F. A., Hetz C. (2012) Activation of the unfolded protein response enhances motor recovery after spinal cord injury. Cell Death Dis. 3, e272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hetz C., Lee A. H., Gonzalez-Romero D., Thielen P., Castilla J., Soto C., Glimcher L. H. (2008) Unfolded protein response transcription factor XBP-1 does not influence prion replication or pathogenesis. Proc. Natl. Acad. Sci. U.S.A. 105, 757–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vidal R. L., Figueroa A., Court F. A., Thielen P., Molina C., Wirth C., Caballero B., Kiffin R., Segura-Aguilar J., Cuervo A. M., Glimcher L. H., Hetz C. (2012) Targeting the UPR transcription factor XBP1 protects against Huntington's disease through the regulation of FoxO1 and autophagy. Hum. Mol. Genet 21, 2245–2262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schrijvers D. M., De Meyer G. R., Martinet W. (2011) Autophagy in atherosclerosis. A potential drug target for plaque stabilization. Arterioscler. Thromb. Vasc. Biol. 31, 2787–2791 [DOI] [PubMed] [Google Scholar]
- 52. Hu P., Lai D., Lu P., Gao J., He H. (2012) ERK and Akt signaling pathways are involved in advanced glycation end product-induced autophagy in rat vascular smooth muscle cells. Int. J. Mol. Med. 29, 613–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Han Q., Zhang Y. L., You S. J., Liu H. H., Cao Y. J., Chen R., Liu C. F. (2011) [Autophagy of human vascular endothelial cells by oxidized low-density lipoprotein. Involvement of oxidative stress but no oxidized low density lipoprotein-1]. Zhonghua Yi Xue Za Zhi 91, 2216–2220 [PubMed] [Google Scholar]
- 54. Zhang Y. L., Cao Y. J., Zhang X., Liu H. H., Tong T., Xiao G. D., Yang Y. P., Liu C. F. (2010) The autophagy-lysosome pathway. A novel mechanism involved in the processing of oxidized LDL in human vascular endothelial cells. Biochem. Biophys. Res. Commun. 394, 377–382 [DOI] [PubMed] [Google Scholar]
- 55. Du J., Teng R. J., Guan T., Eis A., Kaul S., Konduri G. G., Shi Y. (2012) Role of autophagy in angiogenesis in aortic endothelial cells. Am. J. Physiol. Cell Physiol. 302, C383–C391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Debnath J., Baehrecke E. H., Kroemer G. (2005) Does autophagy contribute to cell death? Autophagy 1, 66–74 [DOI] [PubMed] [Google Scholar]
- 57. Levine B., Yuan J. (2005) Autophagy in cell death. An innocent convict? J. Clin. Invest. 115, 2679–2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sirois I., Groleau J., Pallet N., Brassard N., Hamelin K., Londono I., Pshezhetsky A. V., Bendayan M., Hébert M. J. (2012) Caspase activation regulates the extracellular export of autophagic vacuoles. Autophagy 8, 927–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.