Background: Cone snails are a rich source of α-conotoxins that target nicotinic acetylcholine receptors (nAChR).
Results: A new α-conotoxin TxIB potently blocked α6/α3β2β3 nAChRs with high selectivity.
Conclusion: TxIB is an effective inhibitor of α6/α3β2β3 nAChRs. Its structure was determined by NMR.
Significance: TxIB is a new, uniquely selective ligand for querying the structure and function of nAChRs and designing therapeutic drugs.
Keywords: Neuroscience, Neurotoxin, Nicotinic Acetylcholine Receptors, NMR, Peptide Chemical Synthesis, Peptide Conformation, α-Conotoxin, α6/α3β2β3 nAChRs, Conus, NMR
Abstract
α6β2 Nicotinic acetylcholine receptors (nAChRs) expressed by dopaminergic neurons in the CNS are potential therapeutic targets for the treatment of several neuropsychiatric diseases, including nicotine addiction and Parkinson disease. However, recent studies indicate that the α6 subunit can also associate with the β4 subunit to form α6β4 nAChRs that are difficult to pharmacologically distinguish from α6β2, α3β4, and α3β2 subtypes. The current study characterized a novel 16-amino acid α-conotoxin (α-CTx) TxIB from Conus textile whose sequence is GCCSDPPCRNKHPDLC-amide as deduced from gene cloning. The peptide and an analog with an additional C-terminal glycine were chemically synthesized and tested on rat nAChRs heterologously expressed in Xenopus laevis oocytes. α-CTx TxIB blocked α6/α3β2β3 nAChR with an IC50 of 28 nm. In contrast, the peptide showed little or no block of other tested subtypes at concentrations up to 10 μm. The three-dimensional solution structure of α-CTx TxIB was determined using NMR spectroscopy. α-CTx TxIB represents a uniquely selective ligand for probing the structure and function of α6β2 nAChRs.
Introduction
Nicotinic acetylcholine receptors (nAChRs)2 are ligand gated ion channels distributed throughout the nervous system. In addition, they are increasingly recognized as fundamental macromolecules in non-neuronal systems (1). nAChRs are assembled from α (α1-α10) and/or β (β1-β4) subunits to form a variety of subtypes that have distinct pharmacological properties and physiological functions. Expression of the α6 subunit has previously been thought to be primarily localized to catecholaminergic nuclei of the central nervous system. However, recent evidence indicates that the α6 subunit is abundantly expressed in visual pathways and is also present in peripheral tissues (2–4).
As the recognized anatomical sites of expression of α6 receptors has grown, so too has the complexity of recognized subtypes. Work to date has largely focused on α6β2* nAChRs that modulate the release of dopamine. These receptors are believed to be important in mediating tobacco addiction and are also implicated in the pathophysiology of Parkinson disease (5). Furthermore, α6* nAChRs containing the β4 rather than or in addition to the β2 subunit have recently been reported in a variety of tissues including retina (6–7), hippocampus (8), dosrsal root ganglion (3), and adrenal chromaffin cells. However, there is a paucity of ligands that can effectively discriminate between α6β2 and α6β4 nAChRs.
Toxin-producing organisms have served as a natural source of molecules that target nAChRs. Plants and animals use these toxins defensively to discourage consumption by predators or offensively to immobilize prey. Marine mollusks of the genus Conus fall into the latter category and produce an arsenal of neuroactive peptides, many of which are targeted to nAChRs. α-Conotoxins are small cysteine-rich peptide toxins that contain two or more disulfide bonds. α-Conotoxin MII from Conus magus is a signature antagonist of α6β2 nAChRs (9). However, this ligand also binds to α3β2 and α6β4 nAChRs, highlighting the need for development of further ligands (10–14). Here, we report the discovery of a novel toxin from C. textile that binds with high affinity to α6/α3β2β3 nAChRs but not α3β2 or α6β4 nAChRs. The new toxin was synthesized and we report its pharmacological profile and three-dimensional solution structure as determined by NMR.
EXPERIMENTAL PROCEDURES
Materials
Specimens of C. textile were collected from the South China Sea off Hainan Province. Living snails were frozen and stored at −80 °C. The marine animal DNA Isolation Kit, Protease K (20 mg/ml), RNase A (10 mg/ml), and 2× Taq PCR MasterMix were purchased from Tiangen Biochemistry Ltd. (Beijing, China). Oligonucleotides were synthesized and DNA clones were sequenced by Sangon Ltd. (Shanghai, China). Restriction enzymes, T4 DNA Ligase, 5-bromo-4-chloro-3-indolyl β-d-galactoside, isopropyl thio-β-d-galactoside, DL2000 DNA Marker (400 ng/5 μl), λ-EcoT14 I digest DNA marker (50 ng/μl), and pGEM-T easy vector system were purchased from TaKaRa Ltd. (Dalian, China). Agarose and Gold View DNA dye were from Amresco (USA). Acetylcholine chloride, atropine, and bovine serum albumin (BSA) were from Sigma. Reverse-phase HPLC analytical Vydac C18 (5 μm, 4.6 mm × 250 mm) and preparative C18 Vydac columns (10 μm, 22 mm × 250 mm) were obtained from Shenyue (Shanghai City, China). Reagents for peptide synthesis were from GL Biochem (Shanghai, China). Acetonitrile was from Fisher (Fisher Scientific Company L.L.C.), trifluoroacetic acid was from Tedia (Fairfield, OH). The mMessage mMachine in vitro transcription kit was from Ambion (Austin, TX). The Qiagen RNeasy kit was from Qiagen (Valencia, CA). All other chemicals used were of analytical grade. Clones of rat α2-α7 and β2-β4, as well as mouse muscle α1β1δϵ cDNAs were kindly provided by S. Heinemann (Salk Institute, San Diego, CA). Clones of β2 and β3 subunits in the high expressing pGEMHE vector were kindly provided by CW Luetje (University of Miami, Miami, FL). The α6/α3 subunit is a chimera containing the N-terminal extracellular ligand-binding portion of the α6 subunit with the remaining α3 subunit (15). This chimera was used because of poor expression of the nonchimeric form of the α6 construct as previously reported by others and us (14–16). Clones for α9 and α10 were generously provided by A.B. Elgoyen (Instituto de Investigaciones en Ingeniería Genética y Biología Molecular, Buenos Aires, Argentina).
Identification and Sequencing of a Genomic DNA Clone Encoding α-Conotoxin TxIB
Genomic DNA from the C. textile venom gland was isolated using a marine animal DNA Isolation Kit (Tiangen Biochemistry Ltd., Beijing, China). The procedure followed the manufacturer's suggested protocol for marine invertebrates as described previously (17). Briefly, frozen tissue (less than 30 mg) was placed in 200 μl of lysis buffer with 40 μl of RNase A (10 mg/ml), vortexed for 15 s, and digested for 1.5 h with 20 μl of 20 mg/ml of proteinase K solution at 56 °C. The remainder of the procedure followed the kit protocol. The resulting genomic DNA was used as a template for PCR using oligonucleotides primers, corresponding to the 3′-end of the intron preceding the toxin region of α-conotoxin prepropeptides (Primer 1) and the 3′-UTR (untranslated region) sequence of α-conotoxin prepropeptides (Primer 2). The sequence of Primer 1 was 5′-GTGGTTCTGGGTCCAGCA-3′. The sequence of Primer 2 was 5′-GTCGTGGTTCAGAGGGTC-3′. The PCR amplification was performed as described previously (17). PCR products were analyzed by electrophoresis on 1.5% agarose gel. The PCR products were purified by AxyPrep-PCR & Gel Clean up (Axygen Bio. Ltd., Hangzhou, China), which were inserted into the pGEM-T Easy vector via TA cloning (TaKaRa). Transformed colonies were screened with white-blue identification for sequence analysis. Plasmids of positive transformed colonies with conopeptide precursor DNA inserts were purified with a UNlQ-10 EZ Spin Column Plasmid Mini-Preps Kit (Yili Bioscience Ltd., Shanghai, China). Purified plasmids were sequenced by Sangon Ltd.
Chemical Synthesis
The peptide was assembled on an amide resin by solid-phase methodology on an ABI 433A peptide synthesizer using FastMoc (N-(9-fluorenyl) methoxycarbonyl) chemistry and standard side chain protection, except for cysteine residues. Cysteine residues were protected in pairs with either S-trityl on Cys2 and Cys8, or S-acetamidomethyl on Cys3 and Cys16. The peptides were removed from a solid support by treatment with reagent K (trifluoroacetic acid/water/ethanedithiol/phenol/thioanisole; 90:5:2.5:7.5:5, v/v). The released peptide was precipitated and washed several times with cold ether. A two-step oxidation protocol was used to fold the peptides selectively as described previously (18). Briefly, the disulfide bond between Cys2 and Cys8 was oxidized by adding the peptide into an equal volume of 20 mm potassium ferricyanide, 0.1 m Tris, pH 7.5. The solution was allowed to react for 45 min, and the monocyclic peptide was purified by reverse-phase HPLC. Simultaneous removal of the S-acetamidomethyl groups and oxidation of the disulfide bond between Cys3 and Cys16 were carried out by iodine oxidation as follows: the monocyclic peptide in HPLC eluent was dripped into an equal volume of iodine (10 mm) in H2O:trifluoroacetic acid:acetonitrile (73:3:24 by volume) and allowed to react for 10 min. The reaction was terminated by the addition of ascorbic acid and the bicyclic peptide was purified by HPLC on a reversed phase C18 Vydac column using a linear gradient of ACN: 0–40 min 2–42% solvent B (solvent B is 0.092% trifluoroacetic acid in 60% ACN; solvent A is 0.1% trifluoroacetic acid in H2O). Matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry was utilized to confirm the identity of the products.
cRNA Preparation and Injection
Capped cRNA for the various subunits were made using the mMessage mMachine in vitro transcription kit following linearization of the plasmid. The cRNA was purified using the Qiagen RNeasy kit. The concentration of cRNA was determined by absorbance at 260 nm. cRNAs of the various subunits were combined to give 200–500 ng/μl of each subunit cRNA. Fifty nl of this mixture was injected into each Xenopus oocyte as described previously, and incubated at 17 °C. Oocytes were injected within 1 day of harvesting and recordings were made 2–4 days post-injection.
Voltage-clamp Recording
Oocytes were voltage-clamped and exposed to ACh and peptide as described previously (10). Briefly, the oocyte chamber consisting of a cylindrical well (∼30 μl in volume) was gravity perfused at a rate of ∼2 ml/min with ND-96 buffer (96.0 mm NaCl, 2.0 mm KCl, 1.8 mm CaCl2, 1.0 mm MgCl2, 5 mm HEPES, pH 7.1–7.5) containing 1 μm atropine and 0.1 mg/ml of BSA. In the case of the α9α10, α7, and mouse muscle α1β1δϵ subtypes, the ND96 contained no atropine during recording. The membrane potential of the oocytes was clamped at −70 mV. The oocyte was subjected once a minute to a 1-s pulse of ACh. ACh concentration was 10 μm for α9α10 and α1β1δϵ subtypes, 200 μm for α7, and 100 μm for all other subtypes. Once a stable baseline was achieved, either ND-96 alone or ND-96 containing varying concentrations of the α-conotoxins were pre-applied for 5 min prior to the addition of the agonist. All recordings were done at room temperature (∼22 °C).
Data Analysis
The average of five control responses just preceding a test response was used to normalize the test response to obtain “% response.” Each data point of a dose-response curve represents the average ± S.E. of at least three oocytes. The dose-response data were fit to the equation, % response = 100/[1 + ([toxin]/IC50) × nH], where nH is the Hill coefficient, by nonlinear regression analysis using GraphPad Prism (GraphPad Software, San Diego, CA).
NMR Spectroscopy
TxIB (3.0 mg) was dissolved in 550 μl of 10% D2O, 90% H2O (∼pH 3.5) and spectra were recorded on a Bruker Avance 600 MHz NMR spectrometer. One-dimensional and two-dimensional NMR experiments including TOCSY, NOESY, and double-quantum filtered-correlated spectroscopy spectra were recorded at 280 K. Chemical shifts were referenced to internal 2,2-dimethyl-2-silapentane-5-sulfonate at 0 ppm. The TOCSY and NOESY spectra were run with mixing times of 80 and 200 ms, respectively. Two-dimensional spectra were generally collected over 4096 data points in the f2 dimension and 512 increments in the f1 dimension over a spectral width of 12 ppm. Processed spectra were analyzed and assigned using the program CCPNMR with the sequential assignment protocol (19).
Structure Calculations
Distance restraints were derived from the NOESY spectrum using peak volumes. Dihedral angle restraints were determined from 3JHN-Hα coupling constants obtained from one-dimensional 1H NMR spectrum. The φ angles were restrained to −60 ± 30° for 3JHN-Hα < 5.8 Hz and −120 ± 30° for 3JHN-Hα > 8.0 Hz. Preliminary structures were generated using CYANA (20) and the 50 final structures were generated within CNS (21) using protocols from the RECOORD database (22). The final structures were analyzed for stereochemical quality using MOLPROBITY (23) and the 20 of with lowest energy and best quality were chosen to represent the solution structure of α-CTx TxIB.
RESULTS
Gene Cloning of the Novel α-CTx TxIB
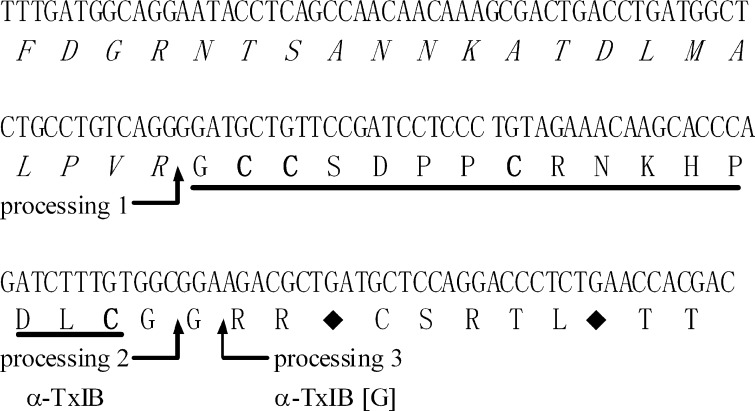
In the venom of most analyzed cone snail species there are multiple α-conotoxins targeted to nAChRs. Although the mature α-conotoxin sequences diverge considerably, the organization of α-conotoxin genes reveals substantial conservation across species. The intron immediately preceding the toxin sequence, the signal sequence within the exon, and the 3′-untranslated region show a high degree of conservation. Proteolytic processing of the precursor protein leads to the formation of the mature α-conotoxin. The coding region of the signal sequence and most of the propeptide region is separated from the toxin-coding region and 3′-untranslated region by a single intron. Primers were designed to this conserved intron sequence and used in conjunction with primers to the conserved 3′-untranslated region to PCR amplify the toxin-coding portion of the α-conotoxin gene. PCR amplification of genomic DNA from C. textile yielded the α-CTx gene product, which was cloned and sequenced. The DNA fragment was 170 bp in length and encoded an α-conotoxin precursor representing the α-conotoxin TxIB gene (tx1b). The precursor was a 47-amino acid protein (Fig. 1). This sequence displayed the characteristic structural organization of conotoxins, including a propeptide sequence at the N terminus, followed by the mature toxin region and a closing 3′-UTR (24). The predicted mature peptides exhibited the characteristic cysteine pattern of α-CTxs (CC-C–C). The EMBL accession number of the α-CTx TxIB precursor gene (tx1b) from C. textile is HE995411. The α-CTx precursor contained an N-terminal propeptide having a length of 27-amino acids and was marked by a predominance of basic residues. It was separated from the proregion by the proteolytic site-XR, which would yield a peptide with a length of 20 amino acids, and further be truncated to a length of 16 (α-CTx TxIB) or 17 (α-CTx TxIB(G)) amino acids by removal of a C-terminal glycine during C-terminal amidation (Fig. 1). The first or second glycine following the C-terminal cysteine in the mature toxin is presumed to be processed to a C-terminal amide for TxIB or TxIB(G), respectively. The deduced mature toxin sequence of TxIB is GCCSDPPCRNKHPDLC# (# = C-terminal carboxamide). TxIB(G) is an analog of α-CTx TxIB with an extra glycine residue.
FIGURE 1.
α-Conotoxin TxIB prepropeptide and encoded toxin are shown. A putative proteolytic processing site 1 (R) and amidated processing sites 2 and 3 are indicated by arrows. The pro-region is italics. The mature toxin region is underlined. The first and second glycine following the C-terminal cysteine in the mature toxin is presumed to be processed to a C-terminal amide for α-CTx TxIB and α-CTx TxIB(G), respectively. TxIB(G) is an analog of TxIB with an extra G (TxIB(G)). The cysteine residues are indicated in bold. The stop codon is indicated by the ♦. The deduced mature toxin sequence of TxIB is GCCSDPPCRNKHPDLC# (# = C-terminal carboxamide).
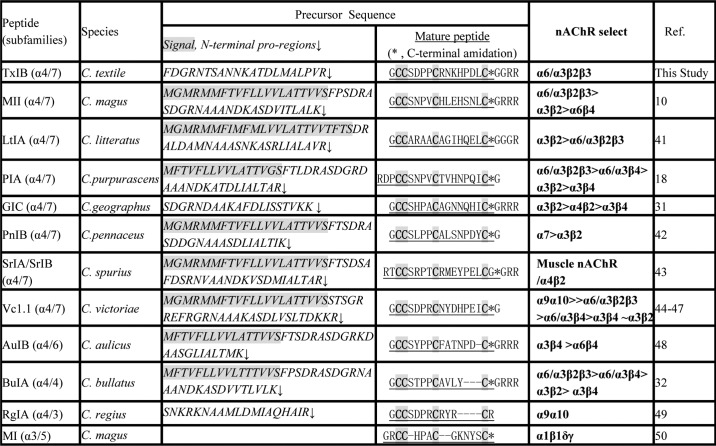
Because members of each conopeptide gene family share a highly conserved signal and propeptide sequence and a conserved arrangement of the cysteines in the sequence of the mature toxin, the new conopeptides TxIB and TxIB(G) identified in this work can be designated as members of the α-conotoxin family. Some selected α-conotoxin precursors and mature toxins from various Conus species are shown for comparison in Fig. 2 and Table 1. Note the highest sequence homology in the signal and 3′-UTR, higher sequence homology in the propeptide region and, in contrast, divergence in the mature toxin region particularly in the C-terminal portion of the peptides. Because the intron is located within the pro-region immediately preceding the toxin sequence, the α-conotoxin cloned from genomic DNA lacks the signal sequence from the toxin-containing fragment (Table 1).
FIGURE 2.
Sequence and disulfide bond connectivity of α-conotoxin TxIB (# = C-terminal carboxamide).
TABLE 1.
Protein precursor sequences of selected α-conotoxins of A-gene superfamily with CC-C-C framework
Chemical Synthesis and Oxidative Folding
Fmoc (N-(9-fluorenyl)methoxycarbonyl) chemistry was used to synthesize the linear peptides. With four cysteine residues there are three possible disulfide bond connectivities. Previously characterized α-conotoxins purified from venom typically have a disulfide bond connectivity linking cysteine I to III and II to IV, which is referred to as the “globular” form to distinguish it from the alternative “ribbon” or “beads” connectivities (25–26). We therefore synthesized both α-CTx TxIB and TxIB(G) in the globular form using a regioselective oxidation approach (Fig. 2). We protected the cysteine side chains with two orthogonal protecting groups that could be removed selectively under different conditions, allowing the formation of one disulfide bridge at a time. For this purpose, Cys2 and Cys8 were introduced as the acid labile S-trityl protected amino acids, whereas Cys3 and Cys16 were S-acetamidomethyl cysteine. The acid-labile groups were removed simultaneously with cleavage from the resin; ferricyanide was used to oxidize the first disulfide bond. Reverse-phase HPLC was used to purify the monocyclic peptide; subsequently, the acid-stable acetamidomethyl groups were removed from the second and fourth cysteines by iodine oxidation, which simultaneously formed the second disulfide bond. The two fully oxidized peptides were purified again by HPLC. Laser desorption mass spectra of synthetic α-CTx TxIB and TxIB(G) were consistent with the sequences. TxIB monoisotopic MS was: calculated 1739.70 and observed 1739.6. TxIB(G) monoisotopic MS was: calculated 1796.72 and observed 1796.6. Synthetic peptides with Cys2-Cys8; Cys3-Cys16 disulfide bond arrangement were used in all subsequently described studies.
Effect of α-CTx TxIB and TxIB(G) on ACh-evoked currents of nAChRs
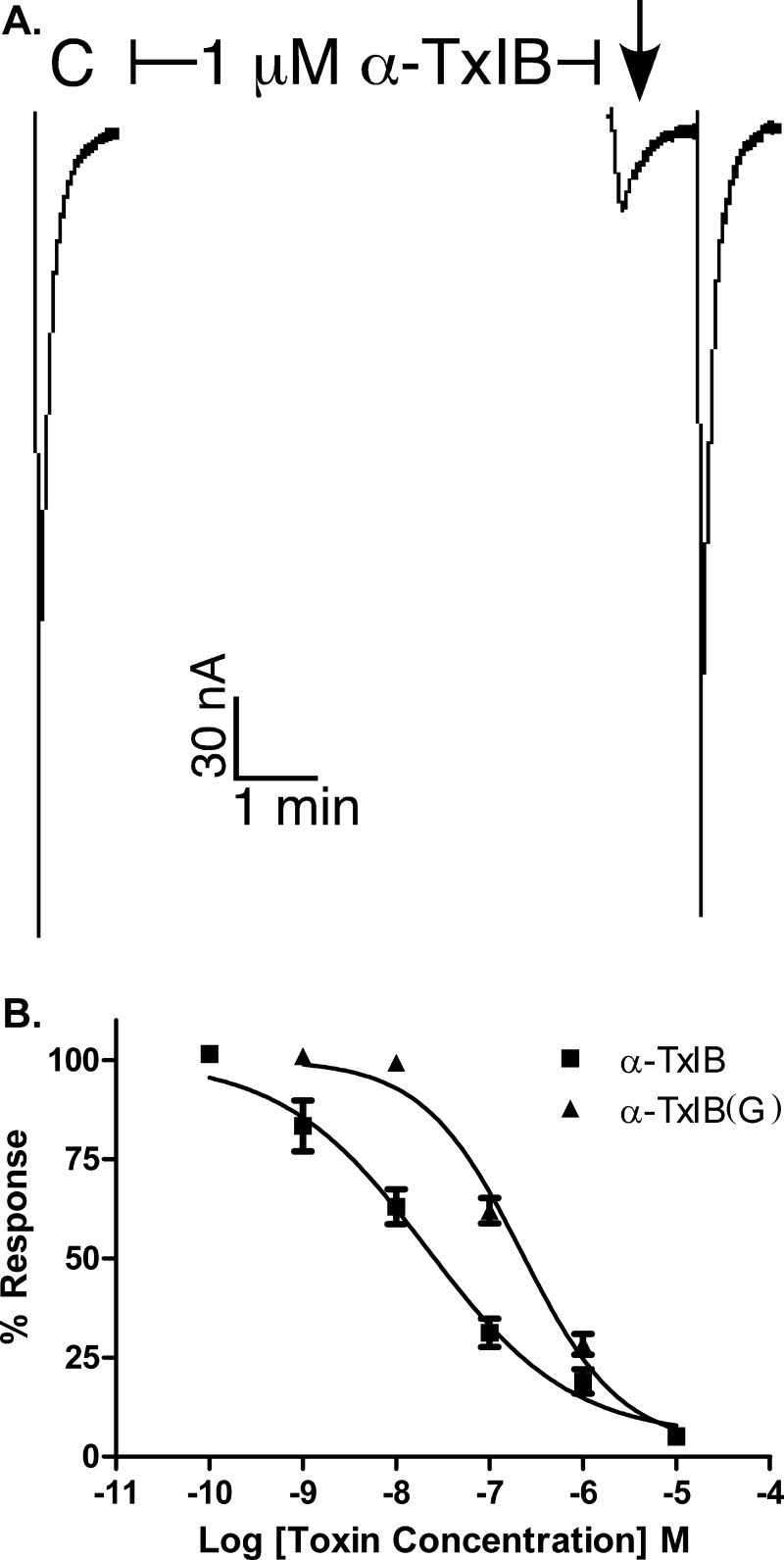
α-Conotoxins target nAChRs, in some instances with a high degree of selectivity (27). We therefore tested α-CTx TxIB and TxIB(G) on various subtypes of nAChRs, which were heterologously expressed in Xenopus oocytes. The toxins were individually tested on these subtypes for the ability to antagonize the response elicited by ACh. The toxins potently blocked α6/α3β2β3 nAChRs. Fig. 3A shows representative responses to ACh of α6/α3β2β3 nAChR in the presence and absence of 1 μm α-CTx TxIB. The block of α6/α3β2β3 nAChR by both α-CTx TxIB and TxIB(G) was rapidly reversible. The IC50 of α-CTx TxIB at the α6/α3β2β3 nAChR subtype was 28.4 (18.6–43.4) nm. TxIB(G), which differs only by an extra glycine at the C terminus, was 10-fold less potent with an IC50 of 247 (186 −329) nm (Table 2).
FIGURE 3.
α-Conotoxin TxIB blocks α6/α3β2β3 nAChRs. A, Xenopus oocytes expressing α6/α3β2β3 nAChRs were voltage clamped at −70 mV and subjected to a 1-s pulse of ACh every minute as described under “Experimental Procedures.” A representative response in a single oocyte is shown. After control responses to ACh, the oocyte was exposed to 1 μm α-CTx TxIB for 5 min. Toxin was then washed out and the response to ACh was again measured (arrow). B, concentration response of α6/α3β2β3 nAChRs exposed to α-CTx TxIB and α-CTx TxIB(G). Values shown in the graph are mean ± S.E. from 3–10 separate oocytes.
TABLE 2.
IC50 for block of nAChR subtypes by α-CTx TxIB and TxIB(G)
| Peptide | Subtype | IC50a |
|---|---|---|
| nm | ||
| TxIB | α6/α3β2β3 | 28.4 (18.6–43.4) |
| TxIB(G) | α6/α3β2β3 | 247 (186–329) |
| TxIB TxIB(G) | α6/α3β4 | >10,000b |
| α7 | >10,000b | |
| α9α10 | >10,000b | |
| Mα1β1δϵ | >10,000b | |
| α2β2 | >10,000b | |
| α2β4 | >10,000b | |
| α3β2 | >10,000b | |
| α3β4 | >10,000b | |
| α4β2 | >10,000b | |
| α4β4 | >10,000b |
a Numbers in parentheses are 95% confidence intervals.
b Less than 50% block at 10−5 m. All receptors are rat except for α1β1δϵ, which is mouse.
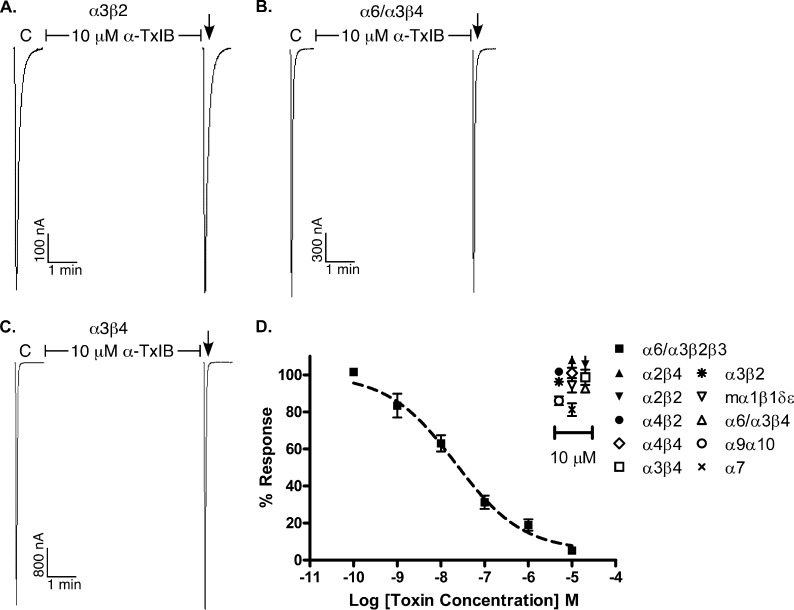
The concentration responses for α-CTx TxIB and TxIB(G) were subsequently assessed on each of the other expressed nAChR subtypes (Fig. 3C and Table 2). Fig. 4 shows representative responses to ACh of α6/α3β4, α3β2, and α3β4 nAChRs in the presence and absence of α-CTx TxIB. Whereas there was substantial block of ACh-evoked currents obtained with 1 μm α-CTx TxIB on α6/α3β2β3 nAChR (Fig. 3), there was little or no block of α6/α3β4, α3β2, and α3β4 nAChRs by 10 μm α-CTx TxIB (Fig. 4). When the β4 rather than β2 nAChR subunit was co-expressed with the α6/α3 subunit, the IC50 for α-CTx TxIB was >400-fold higher indicating that amino acid residue differences between the homologous β subunits significantly influence toxin potency. Similarly, when the α3 rather than α6/α3 nAChR subunit was co-expressed with the β2 subunit, the IC50 for α-CTx TxIB was >400-fold higher, thus indicating that amino acid residue differences between the highly homologous α6 and α3 subunits significantly influence toxin potency. In addition, there was little or no block by TxIB at 10 μm on other nAChR subtypes, including α1β1δϵ, α2β2,α2β4, α4β2, α4β4,α7, and α9α10 (Fig. 4). Thus, α-CTx TxIB was specific for the α6/α3β2β3 nAChR subtype (Table 2).
FIGURE 4.
α-Conotoxin TxIB has little or no activity at non-α6/α3β2β3 nAChR subtypes. nAChR subtypes were expressed as described under “Experimental Preocedures.” C indicates control responses to ACh. Oocytes were then exposed to 10 μm peptide for 5 min, followed by application of ACh. The peptide failed to block α3β2 (A), α6/α3β4 (B), and α3β4 (C) nAChRs. A representative response in a single oocyte is shown. D, Concentration-response of α-CTx TxIB on α6/α3β2β3 nAChRs and effect of 10 μM α-CTx TxIB on the other various nAChR subtypes. Values shown in the graph are mean ± S.E. from 3–5 separate oocytes. Dashed line shows concentration response curve of α6/α3β2β3 nAChRs for comparison.
NMR Spectroscopy
For NMR analysis, one- and two-dimensional spectra were recorded at 280 K and assigned using well established techniques (19). The fingerprint region in the NOESY spectrum of the peptide shows a complete cycle of Hα–NHi+1 sequential connectivities with the exception of the three proline residues (Pro6, Pro7, and Pro13). However, as expected, NOEs were observed from the Hα–Hδi+1 protons of the Pro6 and Pro13 residues and their preceding residues. Residue Pro7 was assigned based on a NOE peak between the Hβs of Pro6 and Hδ of Pro7. Chemical shifts for all residues are given in Table 3.
TABLE 3.
1H chemical shifts for α-CTx TxIB in H2O/D2O, pH 3.5, at 280 K
| Residue | NH | Hα | Hβ | Others |
|---|---|---|---|---|
| Gly1 | 3.90 | |||
| Cys2 | 8.94 | 4.51 | 3.24, 2.65 | |
| Cys3 | 8.75 | 4.47 | 3.33, 2.79 | |
| Ser4 | 7.91 | 4.62 | 4.04, 3.96 | |
| Asp5 | 7.98 | 5.36 | 3.16, 2.77 | |
| Pro6 | 4.22 | 2.32, 2.17 | Hγ 2.30, 2.05, Hδ 4.34, 3.86 | |
| Pro7 | 4.38 | 2.28, 1.86 | Hγ 2.06, 1.99, Hδ 3.63 | |
| Cys8 | 7.64 | 4.31 | 3.35 | |
| Arg9 | 8.75 | 4.00 | 2.03, 1.91 | Hγ 1.73, 1.53, Hδ 3.30, Hϵ 7.25, HH1 7.07 |
| Asn10 | 8.52 | 4.39 | 2.81 | Hδ 7.62, 7.03 |
| Lys11 | 7.32 | 4.25 | 1.81, 1.68 | Hδ 1.52, 1.37 |
| His12 | 7.66 | 5.19 | 3.34, 3.06 | Hδ2 7.67, Hϵ1 8.78 |
| Pro13 | 4.45 | 2.33 | Hγ 2.04, Hδ 3.63, 3.36 | |
| Asp14 | 8.81 | 4.45 | 2.85, 2.75 | |
| Leu15 | 7.64 | 4.37 | 1.83 | Hγ 1.56, Hδ 0.85, 0.77 |
| Cys16 | 7.74 | 4.94 | 3.33, 2.65 |
Structure Determination of α-CTx TxIB
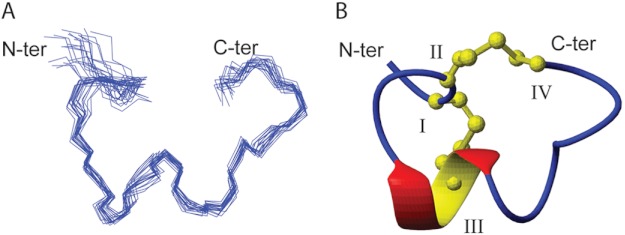
A set of 50 structures was calculated using a simulated annealing protocol in the CNS based on 133 distance restraints derived from NOESY cross-peaks, including 46 intraresidue, 49 sequential, 32 medium range (1< |i—j| < 5) and 6 long range (|i—j| ≥ 5) NOEs, and 7 dihedral restraints based on coupling constants. The calculated structures were consistent with the experimental data, with no NOE violations greater than 0.2 Å and no dihedral violations greater than 3.0°. The 20 lowest energy structures (Fig. 5) were selected based on the highest overall MOLPROBITY scores and had a global root mean square deviation of 0.70 ± 0.21 Å across backbone residues 2–16. A summary of the energy and MOLPROBITY statistics is given in Table 4. As illustrated in Fig. 5B, the secondary structure of TxIB comprises an α-helix between residues 6 and 9, as is very common in α-conotoxins. The PDB accession number of the α-CTx TxIB structure is 2LZ5.
FIGURE 5.
The three-dimensional structure of α-conotoxin TxIB. A, a stereoview of a family of the 20 lowest energy structures of α-CTx TxIB. Structures are superimposed over all the backbone atoms between residues 2–16. B, ribbon representation of the mean structure of α-CTx TxIB. The termini are labeled with N-terminal and C-terminal, and the cysteine residues are labeled with their residue numbers. The α-helical region between residues Pro6 and Arg9 is shown in red and yellow, and disulfide bonds are shown in yellow in ball-and-stick representation. The structures were generated using MOLMOL.
TABLE 4.
Energies and structural statistics for the 20 lowest energy TxIB structures
Based on structures with highest overall MOLPROBITY score (23).
| Structure data | |
|---|---|
| Energies (kcal/mol) | |
| Overall | −422.22 ± 29.70 |
| Bonds | 6.24 ± 0.50 |
| Angles | 23.36 ± 1.94 |
| Improper | 6.91 ± 1.32 |
| Van der Waals | −42.05 ± 3.80 |
| NOE | 0.09 ± 0.01 |
| cDih | 0.14 ± 0.18 |
| Dihedral | 68.65 ± 0.95 |
| Electrostatic | −485.57 ± 29.72 |
| MolProbity statistics | |
| Clashes (>0.4 Å/1000 atoms) | 12.22 ± 6.10 |
| Poor rotamers | 0.67 ± 2.05 |
| Ramachandran outliers (%) | 0.00 ± 0.00 |
| Ramachandran favored (%) | 100 ± 0 |
| MolProbity score | 1.59 ± 0.20 |
| MolProbity score percentilea | 91.45 ± 5.34 |
| Residues with bad bonds | 0.00 ± 0.00 |
| Residues with bad angles | 0.00 ± 0.00 |
| Atomic root mean square deviation (Å) | |
| Mean global backbone (residues 2–16) | 0.70 ± 0.21 |
| Mean global heavy (residues 2–16) | 1.29 ± 0.25 |
| Experimental data | |
| Distance restraints | 133 |
| Dihedral restraints | 7 |
| Total NOE violations exceeding 0.2 Å | 0 |
| Total dihedral violations exceeding 2.0° | 2 (highest 2.271) |
a 100th percentile is the best among structures of comparable resolution; 0th percentile is the worst.
DISCUSSION
C. textile hunts snails and its prey includes numerous species of prosobranch gastropods. When faced with starvation, C. textile will practice cannibalism. The venom is toxic to vertebrates and is reported to have caused human fatalities (28). The Conus genus as a whole hunts a wide variety of prey from five different phyla. These diverse feeding habits are mirrored by the diversity and complexity of the venom components of individual species. In contrast, the coding region of the signal sequence and the propeptide region of the α-CTxs are separated from the toxin-coding region and 3′-untranslated region by a single intron. In this study, by using a conserved intron sequence found within the genes that encode α-conotoxin precursors, a PCR based technique was used to identify a novel peptide from the snail hunting species C. textile. We note that native α-CTx TxIB has not been isolated from venom. Therefore, both α-CTx TxIB and α-CTx TxIB(G) peptides were chemically synthesized using the disulfide bond configuration of previously characterized α-conotoxins, that is, with a I-III, II-IV disulfide connectivity (29).
The residues between Cys-II and Cys-III and Cys-III and Cys-IV of α-conotoxins are commonly referred to as loops 1 and 2, respectively. The number of residues in each of these loops is used to further classify the α-conotoxins. For example, TxIB is classified as a 4/7 α-conotoxin, whereas the muscle nAChR selective α-CTx MI has a 3/5 spacing (Table 1). Although TxIB has a highly conserved Ser-Xaa-Pro motif in loop 1 that is crucial for potent nAChR interaction, the amino acids “RNKH” in loop 2 are distinct, suggesting that amino acids in the C-terminal half of α-CTx TxIB might be responsible for its selectivity.
α-CTx TxIB potently blocked α6/α3β2β3 nAChRS but had little or no activity at other tested subtypes. The ability of α-CTx TxIB to discriminate between α6/α3β2β3 and the other nAChR receptors is unique. No small molecules have this selectivity profile. Previously described α-conotoxins that potently block α6/α3β2β3 nAChRs also block α6/α3β4 nAChRs, α3β2 nAChRs, and/or other nAChRs subtypes. For instance, another 4/7 conotoxin, α-CTx MII from the fish-eating C. magus, potently blocks α6/α3β2β3 and α3β2 nAChRs, with weaker activity on α6/α3β4 nAChRs. A second generation analog of α-CTx MII, α-CTx MII(H9A,L15A) loses activity at the α3β2 nAChR subtype, but retains activity on the α6β4 subtype (14). Another 4/7 α-CTx, PIA, is an 18-amino acid peptide from the fish eating snail, Cyclamen purpurascens. PIA is most potent on α6/α3β2β3 nAChR but also potently blocks α6/α3β4 nAChRs (18). α-CTx BuIA is a 13-amino acid 4/4 peptide from the fish eating snail, Conus bullatus. Although it potently blocks α6/α3β2β3 nAChRs, it is also active against a broad spectrum of nAChR subtypes, including α6/α3β4, α3β2, and α3β4 (30). The kinetics of toxin unblock for the latter three toxins are slow. In contrast, the TxIB block is rapidly reversed after toxin washout, and in this way is more similar to the peptide α-CTx GIC that potently blocks both α6/α3β2β3 and α3β2 nAChRs (31).
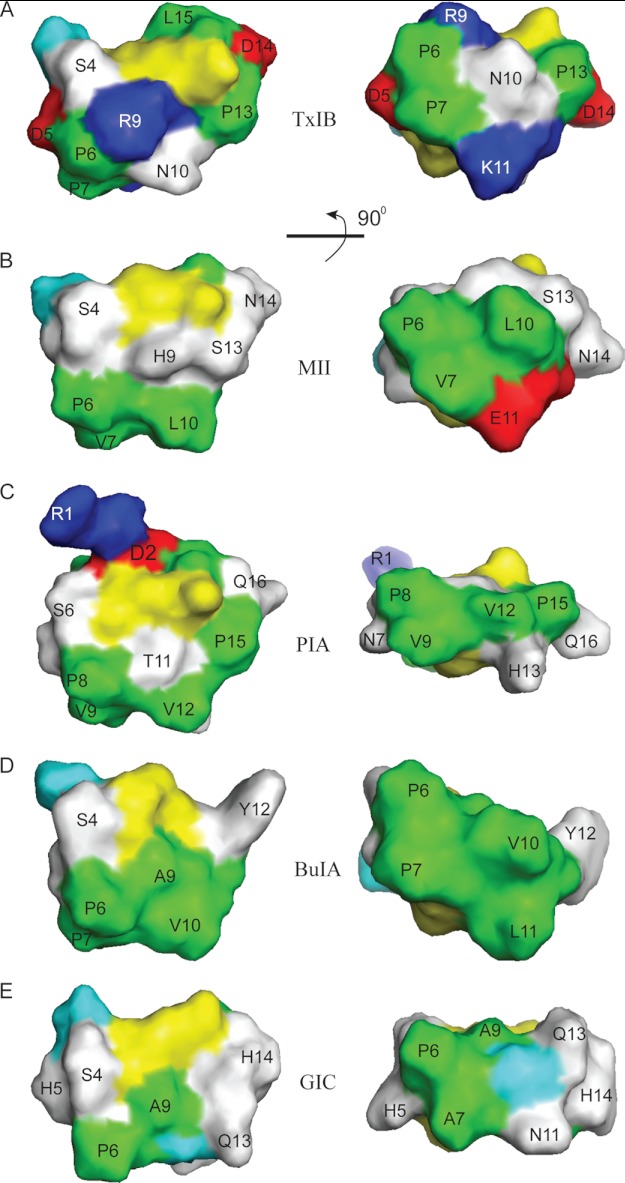
NMR and structural analyses showed that TxIB has a similar fold to other α-CTxs. As illustrated in Fig. 6, even though they have the same peptide fold, conotoxins MII, PIA, BuIA, GIC, and TxIB have different surface features in terms of the charged, polar, and hydrophobic side chains of the residues. It is clear that all of the peptides have a hydrophobic patch in loop 1. However, a discriminating feature of TxIB is that the hydrophobic patch is smaller than in MII, PIA, or BuIA, suggesting the possibility that this feature might be responsible for the higher selectivity of TxIB. However, the hydrophobic patch in GIC is also rather small, suggesting that this cannot be the only factor. A main difference between GIC and TxIB is the presence of two positive charges flanking the hydrophobic patch in TxIB. In the absence of a detailed molecular model defining the orientation of the toxins bound to the receptor it is difficult to be definitive about the reasons for the trends in specificity, but it appears likely the combination of smaller hydrophobic patch with flanking positively charged residues helps to enhance the selectivity of TxIB. Note that the presence of positive charge alone is not sufficient to confer selectivity as conotoxin PIA, which has a positive charge at R1, is nonspecific. In future studies this hypothesis could be further tested by the synthesis of derivatives of BuIA or PIA where mutations are made in the hydrophobic patches and in flanking residues.
FIGURE 6.
Surface representation of selected α-conotoxins. A, α-CTx TxIB (PDB code 2LZ5); B, α-CTx PIA (PDB code 1ZLC); C, α-CTx BuIA (PDB code 2I28); D, α-CTx GIC (PDB code 1UL2); and E, α-CTx MII (PDB code 1MII). Positively charged residues (Arg and Lys) are shown in blue, negative residues (Asp and Glu) are red, polar residues (Asn, Gln, His, Ser, Thr and Tyr) are white, and hydrophobic residues (Ala, Ile, Leu, Pro, and Val) are green, cystines are yellow, and glycines are cyan. The surface images of the peptides on the right are the images after the 90° rotation of the left images around the horizontal axis.
The nAChR α6 subunit is not widely expressed in the brain, but it is abundant in midbrain dopaminergic regions associated with pleasure, reward, and mood control (32–35), suggesting that α6*–nAChRs might play critical roles in nicotinic reward and in the regulation of mood by nicotine (36). Modulation of striatal dopamine neurotransmission plays a key role in the reinforcing effects of addictive drugs (35, 37). Striatal dopamine axon terminals express α6β2 and α4(non-α6)β2* nAChRs. Pharmacological block of the α6β2 nAChR function is associated with decreased self-administration of nicotine as well as ethanol in rat models of substance abuse (38–40). In addition, genetic deletion of the nAChR α6 subunit abolishes nicotine self-administration and lentiviral re-expression of the α6 subunit leads to restoration of nicotine self-administration (35).
Thus, α6β2* nAChR represents a potentially attractive drug target to treat substance dependence. Although TxIB itself is unlikely to cross the blood-brain barrier in significant quantity, the discovery of α-CTx TxIB represents proof-of-concept that it is possible to develop compounds that selectively block α6β2 nAChRs. α-CTx TxIB might therefore represent an initial scaffold from which to design therapeutic drugs.
In conclusion, this study describes a novel α-conotoxin TxIB that is a subtype-specific blocker of the α6/α3β2β3 nAChR. The unique selectivity of this peptide will allow probing of nAChR function in tissues where both the α6* and other nAChR subtypes occur. In addition, structural insights derived from this ligand might facilitate the development of novel therapeutics for diseases involving α6* nAChRs, including addiction.
Acknowledgments
We thank Layla Azam, Cheryl Dowell, Baldomero Olivera, and Doju Yoshikami for advice and help.
This work was supported, in whole or in part, by the Program for International Science & Technology Cooperation Program of China Grant 2011DFR31210, National Natural Science Foundation of China Grant 81160503, State High-Tech Research and Development Project (863) of the Ministry of Science and Technology of China Grant 2012AA021706, Changjiang Scholars and Innovative Research Team in University Grant PCSIRT, IRT1123. This work was also supported by National Institutes of Health Grants GM103801 and GM48677, and Australian Research Council Grant 1093115. D.J.C. acknowledges the support of a National Health & Medical Research Council Professorial Fellowship. A preliminary account of some of this work was presented in the patent literature (51).
The atomic coordinates and structure factors (code 2LZ5) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- nAChR
- nicotinic acetylcholine receptor
- CTx
- conotoxin
- TOCSY
- total correlation spectroscopy
- NOESY
- nuclear Overhauser effect spectroscopy
- α6β2*
- asterisk indicates the possible presence of additional subunits.
REFERENCES
- 1. Albuquerque E. X., Pereira E. F., Alkondon M., Rogers S. W. (2009) Mammalian nicotinic acetylcholine receptors. From structure to function. Physiol. Rev. 89, 73–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mackey E. D., Engle S. E., Kim M. R., O'Neill H. C., Wageman C. R., Patzlaff N. E., Wang Y., Grady S. R., McIntosh J. M., Marks M. J., Lester H. A., Drenan R. M. (2012) α6* Nicotinic acetylcholine receptor expression and function in a visual salience circuit. J. Neurosci. 32, 10226–10237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hone A. J., Meyer E. L., McIntyre M., McIntosh J. M. (2012) Nicotinic acetylcholine receptors in dorsal root ganglion neurons include the α6β4* subtype. FASEB J. 26, 917–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liu J., McGlinn A. M., Fernandes A., Milam A. H., Strang C. E., Andison M. E., Lindstrom J. M., Keyser K. T., Stone R. A. (2009) Nicotinic acetylcholine receptor subunits in rhesus monkey retina. Invest. Ophthalmol. Vis. Sci. 50, 1408–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Quik M., Perez X. A., Grady S. R. (2011) Role of α6 nicotinic receptors in CNS dopaminergic function. Relevance to addiction and neurological disorders. Biochem. Pharmacol. 82, 873–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Marritt A. M., Cox B. C., Yasuda R. P., McIntosh J. M., Xiao Y., Wolfe B. B., Kellar K. J. (2005) Nicotinic cholinergic receptors in the rat retina. Simple and mixed heteromeric subtypes. Mol. Pharmacol. 68, 1656–1668 [DOI] [PubMed] [Google Scholar]
- 7. Vailati S., Hanke W., Bejan A., Barabino B., Longhi R., Balestra B., Moretti M., Clementi F., Gotti C. (1999) Functional α6-containing nicotinic receptors are present in chick retina. Mol. Pharmacol. 56, 11–19 [DOI] [PubMed] [Google Scholar]
- 8. Azam L., Maskos U., Changeux J. P., Dowell C. D., Christensen S., De Biasi M., McIntosh J. M. (2010) α-Conotoxin BuIA[T5A;P6O]. A novel ligand that discriminates between α6β4 and α6β2 nicotinic acetylcholine receptors and blocks nicotine-stimulated norepinephrine release. FASEB J. 24, 5113–5123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Grady S. R., Salminen O., Laverty D. C., Whiteaker P., McIntosh J. M., Collins A. C., Marks M. J. (2007) The subtypes of nicotinic acetylcholine receptors on dopaminergic terminals of mouse striatum. Biochem. Pharmacol. 74, 1235–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cartier G. E., Yoshikami D., Gray W. R., Luo S., Olivera B. M., McIntosh J. M. (1996) A new α-conotoxin which targets α6β2 nicotinic acetylcholine receptors. J. Biol. Chem. 271, 7522–7528 [DOI] [PubMed] [Google Scholar]
- 11. Dutertre S., Nicke A., Lewis R. J. (2005) β2 Subunit contribution to 4/7 α-conotoxin binding to the nicotinic acetylcholine receptor. J. Biol. Chem. 280, 30460–30468 [DOI] [PubMed] [Google Scholar]
- 12. Harvey S. C., McIntosh J. M., Cartier G. E., Maddox F. N., Luetje C. W. (1997) Determinants of specificity for α-conotoxin MII on α6β2 neuronal nicotinic receptors. Mol. Pharmacol. 51, 336–342 [DOI] [PubMed] [Google Scholar]
- 13. Kaiser S. A., Soliakov L., Harvey S. C., Luetje C. W., Wonnacott S. (1998) Differential inhibition by α-conotoxin-MII of the nicotinic stimulation of [3H]dopamine release from rat striatal synaptosomes and slices. J. Neurochem. 70, 1069–1076 [DOI] [PubMed] [Google Scholar]
- 14. McIntosh J. M., Azam L., Staheli S., Dowell C., Lindstrom J. M., Kuryatov A., Garrett J. E., Marks M. J., Whiteaker P. (2004) Analogs of α-conotoxin MII are selective for α6-containing nicotinic acetylcholine receptors. Mol. Pharmacol. 65, 944–952 [DOI] [PubMed] [Google Scholar]
- 15. Kuryatov A., Olale F., Cooper J., Choi C., Lindstrom J. (2000) Human α6 AChR subtypes. Subunit composition, assembly, and pharmacological responses. Neuropharmacology 39, 2570–2590 [DOI] [PubMed] [Google Scholar]
- 16. Papke R. L., Dwoskin L. P., Crooks P. A., Zheng G., Zhang Z., McIntosh J. M., Stokes C. (2008) Extending the analysis of nicotinic receptor antagonists with the study of alpha6 nicotinic receptor subunit chimeras. Neuropharmacology 54, 1189–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zheng X., Gao B., Li B., Peng C., Wu A., Zhu X., Chen X., Zhangsun D., Luo S. (2011) Primer screening for new α-conotoxin gene cloning. Biotechnology 21, 40–44 [Google Scholar]
- 18. Dowell C., Olivera B. M., Garrett J. E., Staheli S. T., Watkins M., Kuryatov A., Yoshikami D., Lindstrom J. M., McIntosh J. M. (2003) α-Conotoxin PIA is selective for α6 subunit-containing nicotinic acetylcholine receptors. J. Neurosci. 23, 8445–8452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wüthrich K. (1986) NMR of Proteins and Nucleic Acids, Wiley–Interscience, New York [Google Scholar]
- 20. Güntert P., Mumenthaler C., Wüthrich K. (1997) Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 273, 283–298 [DOI] [PubMed] [Google Scholar]
- 21. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Crystallography & NMR system. A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 22. Nederveen A. J., Doreleijers J. F., Vranken W., Miller Z., Spronk C. A., Nabuurs S. B., Güntert P., Livny M., Markley J. L., Nilges M., Ulrich E. L., Kaptein R., Bonvin A. M. (2005) RECOORD. A recalculated coordinate database of 500+ proteins from the PDB using restraints from the BioMagResBank. Proteins 59, 662–672 [DOI] [PubMed] [Google Scholar]
- 23. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) MolProbity. All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Santos A. D., McIntosh J. M., Hillyard D. R., Cruz L. J., Olivera B. M. (2004) The A-superfamily of conotoxins. Structural and functional divergence. J. Biol. Chem. 279, 17596–17606 [DOI] [PubMed] [Google Scholar]
- 25. Gehrmann J., Alewood P. F., Craik D. J. (1998) Structure determination of the three disulfide bond isomers of α-conotoxin GI. A model for the role of disulfide bonds in structural stability. J. Mol. Biol. 278, 401–415 [DOI] [PubMed] [Google Scholar]
- 26. Armishaw C. J., Dutton J. L., Craik D. J., Alewood P. F. (2010) Establishing regiocontrol of disulfide bond isomers of α-conotoxin ImI via the synthesis of N-to-C cyclic analogs. Biopolymers 94, 307–313 [DOI] [PubMed] [Google Scholar]
- 27. Azam L., McIntosh J. M. (2009) α-Conotoxins as pharmacological probes of nicotinic acetylcholine receptors. Acta Pharmacol. Sin. 30, 771–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Röckel D., Korn W., Kohn A. J. (1995) Manual of the Living Conidae, pp. 308–312, Verlag Christa Hemmen, Germany [Google Scholar]
- 29. Muttenthaler M., Akondi K. B., Alewood P. F. (2011) Structure-activity studies on α-conotoxins. Curr. Pharm. Des. 17, 4226–4241 [DOI] [PubMed] [Google Scholar]
- 30. Azam L., Dowell C., Watkins M., Stitzel J. A., Olivera B. M., McIntosh J. M. (2005) α-Conotoxin BuIA, a novel peptide from Conus bullatus, distinguishes among neuronal nicotinic acetylcholine receptors. J. Biol. Chem. 280, 80–87 [DOI] [PubMed] [Google Scholar]
- 31. McIntosh J. M., Dowell C., Watkins M., Garrett J. E., Yoshikami D., Olivera B. M. (2002) α-Conotoxin GIC from Conus geographus, a novel peptide antagonist of nicotinic acetylcholine receptors. J. Biol. Chem. 277, 33610–33615 [DOI] [PubMed] [Google Scholar]
- 32. Klink R., de Kerchove d'Exaerde A., Zoli M., Changeux J. P. (2001) Molecular and physiological diversity of nicotinic acetylcholine receptors in the midbrain dopaminergic nuclei. J. Neurosci. 21, 1452–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Azam L., Winzer-Serhan U. H., Chen Y., Leslie F. M. (2002) Expression of neuronal nicotinic acetylcholine receptor subunit mRNAs within midbrain dopamine neurons. J. Comp. Neurol. 444, 260–274 [DOI] [PubMed] [Google Scholar]
- 34. Champtiaux N., Gotti C., Cordero-Erausquin M., David D. J., Przybylski C., Léna C., Clementi F., Moretti M., Rossi F. M., Le Novère N., McIntosh J. M., Gardier A. M., Changeux J. P. (2003) Subunit composition of functional nicotinic receptors in dopaminergic neurons investigated with knock-out mice. J. Neurosci. 23, 7820–7829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pons S., Fattore L., Cossu G., Tolu S., Porcu E., McIntosh J. M., Changeux J. P., Maskos U., Fratta W. (2008) Crucial role of α4 and α6 nicotinic acetylcholine receptor subunits from ventral tegmental area in systemic nicotine self-administration. J. Neurosci. 28, 12318–12327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yang K. C., Jin G. Z., Wu J. (2009) Mysterious α6-containing nAChRs. Function, pharmacology, and pathophysiology. Acta Pharmacol. Sin. 30, 740–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Exley R., Clements M. A., Hartung H., McIntosh J. M., Cragg S. J. (2008) α6 Containing nicotinic acetylcholine receptors dominate the nicotine control of dopamine neurotransmission in nucleus accumbens. Neuropsychopharmacology 33, 2158–2166 [DOI] [PubMed] [Google Scholar]
- 38. Jackson K. J., McIntosh J. M., Brunzell D. H., Sanjakdar S. S., Damaj M. I. (2009) The role of α6-containing nicotinic acetylcholine receptors in nicotine reward and withdrawal. J. Pharmacol. Exp. Ther. 331, 547–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Brunzell D. H., Boschen K. E., Hendrick E. S., Beardsley P. M., McIntosh J. M. (2010) α-Conotoxin MII-sensitive nicotinic acetylcholine receptors in the nucleus accumbens shell regulate progressive ratio responding maintained by nicotine. Neuropsychopharmacology 35, 665–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Löf E., Olausson P., deBejczy A., Stomberg R., McIntosh J. M., Taylor J. R., Söderpalm B. (2007) Nicotinic acetylcholine receptors in the ventral tegmental area mediate the dopamine activating and reinforcing properties of ethanol cues. Psychopharmacology 195, 333–343 [DOI] [PubMed] [Google Scholar]
- 41. Luo S., Akondi K. B., Zhangsun D., Wu Y., Zhu X., Hu Y., Christensen S., Dowell C., Daly N. L., Craik D. J., Wang C. I., Lewis R. J., Alewood P. F., Michael McIntosh J. (2010) α-Conotoxin LtIA from Conus litteratus targets a novel microsite of the α3β2 nicotinic receptor. J. Biol. Chem. 285, 12355–12366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fainzilber M., Hasson A., Oren R., Burlingame A. L., Gordon D., Spira M. E., Zlotkin E. (1994) New mollusc-specific α-conotoxins block Aplysia neuronal acetylcholine receptors. Biochemistry 33, 9523–9529 [DOI] [PubMed] [Google Scholar]
- 43. López-Vera E., Aguilar M. B., Schiavon E., Marinzi C., Ortiz E., Restano Cassulini R., Batista C. V., Possani L. D., Heimer de la Cotera E. P., Peri F., Becerril B., Wanke E. (2007) Novel α-conotoxins from Conus spurius and the α-conotoxin EI share high-affinity potentiation and low-affinity inhibition of nicotinic acetylcholine receptors. FEBS J. 274, 3972–3985 [DOI] [PubMed] [Google Scholar]
- 44. Sandall D. W., Satkunanathan N., Keays D. A., Polidano M. A., Liping X., Pham V., Down J. G., Khalil Z., Livett B. G., Gayler K. R. (2003) A novel α-conotoxin identified by gene sequencing is active in suppressing the vascular response to selective stimulation of sensory nerves in vivo. Biochemistry 42, 6904–6911 [DOI] [PubMed] [Google Scholar]
- 45. Satkunanathan N., Livett B., Gayler K., Sandall D., Down J., Khalil Z. (2005) α-Conotoxin Vc1.1 alleviates neuropathic pain and accelerates functional recovery of injured neurones. Brain Res. 1059, 149–158 [DOI] [PubMed] [Google Scholar]
- 46. Vincler M., Wittenauer S., Parker R., Ellison M., Olivera B. M., McIntosh J. M. (2006) Molecular mechanism for analgesia involving specific antagonism of α9α10 nicotinic acetylcholine receptors. Proc. Natl. Acad. Sci. U.S.A. 103, 17880–17884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Klimis H., Adams D. J., Callaghan B., Nevin S., Alewood P. F., Vaughan C. W., Mozar C. A., Christie M. J. (2011) A novel mechanism of inhibition of high-voltage activated calcium channels by α-conotoxins contributes to relief of nerve injury-induced neuropathic pain. Pain 152, 259–266 [DOI] [PubMed] [Google Scholar]
- 48. Luo S., Kulak J. M., Cartier G. E., Jacobsen R. B., Yoshikami D., Olivera B. M., McIntosh J. M. (1998) α-Conotoxin AuIB selectively blocks α3β4 nicotinic acetylcholine receptors and nicotine-evoked norepinephrine release. α-RgIA. J. Neurosci. 18, 8571–8579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ellison M., Haberlandt C., Gomez-Casati M. E., Watkins M., Elgoyhen A. B., McIntosh J. M., Olivera B. M. (2006) α-RgIA. A novel conotoxin that specifically and potently blocks the α9α10 nAChR. Biochemistry 45, 1511–1517 [DOI] [PubMed] [Google Scholar]
- 50. McIntosh M., Cruz L. J., Hunkapiller M. W., Gray W. R., Olivera B. M. (1982) Isolation and structure of a peptide toxin from the marine snail Conus magus. Arch. Biochem. Biophys. 218, 329–334 [DOI] [PubMed] [Google Scholar]
- 51. Luo S., Zhangsun D., Wu Y., Zhu X., Hu Y., McIntosh J. M. (2012) Chinese patent literature. CN(201210277619.8)-A [Google Scholar]