Background: High resolution structural information only covers 15% of the full-length sequence of RyR.
Results: Pseudo-atomic structures are generated for RyR1 fragments 850–1,056 and RyR2 861–1,067, docked into cryo-EM maps, and supported by FRET experiments.
Conclusion: The binding interface between RyR and FKBP consists of electrostatic contacts and contains mutations.
Significance: The fragments docked into a domain that forms an intersubunit interaction with a phosphorylation domain.
Keywords: Calcium Channels, Calcium Intracellular Release, FRET, Molecular Modeling, Ryanodine Receptor, Cryo-EM Docking, Intersubunit Interaction
Abstract
Ryanodine receptors (RyRs) form a class of intracellular calcium release channels in various excitable tissues and cells such as muscles and neurons. They are the major cellular mediators of the release of calcium ions from the sarcoplasmic reticulum, an essential step in muscle excitation-contraction coupling. Several crystal structures of skeletal muscle RyR1 peptide fragments have been solved, but these cover less than 15% of the full-length RyR1 sequence. In this study, by combining modeling techniques with sub-nanometer resolution cryo-electron microscopy (cryo-EM) maps, we obtained pseudo-atomic models for RyR fragments consisting of residues 850–1,056 in rabbit RyR1 or residues 861–1,067 in mouse RyR2. These fragments are docked into a domain that connects the central vestibule and corner clamp region of RyR, resulting in a good match of the secondary structure elements in the cryo-EM map and the pseudo-atomic models, which is also consistent with our previous mappings of GFP insertions by cryo-EM and with FRET measurements involving RyR and FK506-binding protein (FKBP). A combined model of the RyR fragment and FKBP docked into the cryo-EM map suggests that the fragment is positioned adjacent to the FKBP-binding site. Its predicted binding interface with FKBP consists primarily of electrostatic contacts and contains several disease-associated mutations. A dynamic interaction between the fragment and an RyR phosphorylation domain, characterized by FRET experiments, also supports the structural predictions of the pseudo-atomic models.
Introduction
Ryanodine receptors (RyRs)2 are a class of intracellular calcium release channels existing in various excitable tissues and cells such as muscles and neurons. RyRs play an important role in regulating cytoplasmic calcium, which acts as a secondary messenger in cells. In striated muscle, RyRs release calcium from the sarcoplasmic reticulum in response to membrane depolarization. Mammalian RyRs exist in three isoforms, RyR1, RyR2, and RyR3, which distribute predominantly in skeletal muscle, cardiac muscle, and various tissues, respectively (1, 2), and which share ∼65% identity over their full-length sequences (3).
All three isoforms of RyR have been cloned, and each primary sequence contains ∼5,000 amino acids (3–5). The N-terminal ∼4,300 amino acids form a large cytoplasmic assembly that interacts with numerous intracellular modulators, and the C-terminal ∼700 residues form a trans-membrane assembly that acts as an ion-conducting pore (6). Functional RyR molecules are homotetramers of 2.3 MDa (565 kDa per monomer), making RyRs the largest ion channels known to date and contributing to the difficulty in obtaining crystals suitable for atomic structure determination by x-ray crystallography. In the past decades, several groups have determined three-dimensional reconstructions of intact RyRs at intermediate resolutions by cryo-electron microscopy (cryo-EM) (7–12). Recently, crystal structures of RyR1 fragments, mainly from the N-terminal region, have been solved to atomic resolution (13–16). The largest fragment contains the first 559 residues and covers ∼11% of the entire RyR1 sequence (16). It includes three separate domains, domain A (residues 1–205), domain B (206–394), and domain C (395–532). Docking the crystal structure of the ABC domains into a full-length RyR1 cryo-EM map has placed the domains in the cytoplasmic portion of RyR1, forming a central rim in the cytoplasmic assembly (16). More recently, crystal structures of a phosphorylation domain in RyR1 (residues 2,733/2734–2,940) and its corresponding domains in RyR2 (residues 2,699–2,904) and in RyR3 (residues 2,597–2,800) were reported by two independent research groups. These fragments were docked into a region near each corner of the cytoplasmic assembly (17, 18). Nevertheless, further progress by this approach toward a high resolution structure for RyR requires improvement in the cryo-EM resolution of structurally intact RyR and determination of additional fragment crystal structures to fit into the cryo-EM maps.
RyRs are modulated by numerous natural and pharmacological ligands and by covalent modifications such as phosphorylation, nitrosylation, and oxidation/reduction of cysteine sulfhydryl moieties (19–21). Most of these bound ligands or modifications occur at specific sites on the cytoplasmic assembly, a fact that perhaps offers one explanation for the unusually large size of the cytoplasmic region. Cryo-EM has been utilized to determine the physical locations of some RyR protein binding partners, including calmodulin (22–24), a 12- or 12.6-kDa FK506-binding protein (FKBP12 or FKBP12.6) (22, 25, 26), chloride intracellular channel 2 (27), imperatoxin A from scorpion venom (28), and natrin snake venom toxin (29).
FKBP is a protein prolyl isomerase that is widely expressed and involved in many cell functions. Two isoforms, FKBP12 and FKBP12.6, bind tightly to RyR1 and RyR2, respectively, and may be considered as integral subunits of RyR (30, 31). FKBP12 and FKBP12.6 share a high degree of sequence homology (85%), and it has been demonstrated by a site-directed mutagenesis study that three residues specific to FKBP12.6 (Gln-31, Asn-32, and Phe-59) account for the selective binding to cardiac RyR2 (32). However, sequences in RyR that participate in FKBP binding remain controversial. The valine 2461 to proline 2462 motif in RyR1 (corresponding to isoleucine 2427 to proline 2428 in RyR2) has been identified as being required for FKBP binding by site-directed mutagenesis (33). However, the corresponding isoleucine 2427 to proline 2428 motif in RyR2 is not required for FKBP12.6 binding for RyR2 (34). Protein kinase A (PKA) phosphorylation of RyR2 dissociates FKBP12.6 and affects the RyR2 channel open probability, and phosphorylation of RyR2 at a single residue serine 2808 (serine 2843 in RyR1) activates the channel by releasing FKBP (35, 36). However, this proposed mechanism is not consistent with other findings (37–40). In addition, C-terminal truncation analysis demonstrated that a fragment between residues 1,815 and 1,855 in RyR2 is essential for GST-FKBP12.6 binding (41). It is likely that more than one sequence in RyR participates in FKBP binding, as multiple sequences may form a binding pocket for FKBP when RyR folds and four subunits assemble into a homotetramer. In support of this hypothesis, we have mapped two residues, Tyr-846 and Thr-1874, that are widely separated in RyR2's primary sequence onto the three-dimensional structure by cryo-EM, and we found that the structural domains bearing the two residues are both adjacent to the FKBP-binding site (41, 42).
In this study, sequence homology was detected between the phosphorylation domain residues 2,733–2,940 in RyR1, for which a 2.2-Å crystal structure exists (18), and residues 850–1,056 in rabbit RyR1 (henceforth called “RyR1 fragment”) or residues 861–1,067 in mouse RyR2 (henceforth called “RyR2 fragment”). Pseudo-atomic models were generated for these fragments based on the available crystal structure, and these models were fitted into sub-nanometer resolution cryo-EM maps of RyR1 in both closed and open conformations using rigid-body docking and flexible fitting. The fragments docked optimally into a domain that connects the central vestibule and corner clamp region of RyR, which is consistent with our previous results from RyR-GFP mappings by three-dimensional cryo-EM and from FRET measurement of RyR-FKBP interactions (42). The docked location was further supported by detection of secondary structure features in the cryo-EM map, which matched the fitted models. The fragments were located adjacent to the FKBP-binding site, and docking of an FKBP crystal structure revealed that a largely electrostatic type of interaction between the fragment and FKBP is likely. Several positions of disease-associated mutations were mapped into the modeled structure, and they are mainly located either in the binding interface with FKBP or near an interface that interacts with another RyR structural domain in the corner of the clamp region. We also designed a FRET pair to test for the existence of a dynamic interaction between the fragment in the modeled structure and a RyR phosphorylation domain. The FRET data demonstrate that these two structural domains belong to two neighboring RyR subunits, and they form an intersubunit interaction that is sensitive to RyR channel activation. Taken together, these data suggest that the structural domain formed by the RyR1/RyR2 fragment plays a role in mediating a dynamic domain-domain interaction within RyR and in channel modulation by FKBP.
EXPERIMENTAL PROCEDURES
BLAST Sequence Analysis and Comparative Modeling
To identify sequences in RyR whose structures could be modeled using structural templates from the PDB, we performed protein BLAST against PDB protein sequences for every 1200 residues with a small overlap of 200 residues for each alignment group. All five resulting sequence groups were aligned using the protein blast plug-in of UCSF Chimera (43). A sequence portion of the third group (amino acids 2,001–3,200) matched the sequence of an RyR protein fragment (amino acids 2,733–2,940) whose crystal structure was known (PDB code 3RQR). In addition, residues 850–1,056 shared a high sequence homology with the 3RQR crystal structure sequence. The corresponding sequence in mouse RyR2 was identified by multiple sequence alignment using ClustalW2 (44). Next, we chose the two isoforms as the target sequences with the 3RQR chain A as the template to complete comparative modeling using the program MODELLER (45). Five models were generated that differed to a slight degree mainly in the loop regions. We chose the model with the lowest estimated root mean square deviation to perform the fitting in the next step. The differences between the modeling template and the homology model mainly exist in loop 3, helix 3, loop 4, and loop 5. We also submitted the sequences for the two isoforms to the I-TASSER server (46), an on-line platform developed for protein structure predictions by an ab initio modeling protocol. Three models were generated for each isoform, and we chose the first model because of its highest C-score value, which is used for estimating the accuracy of the I-TASSER prediction (46). When we superimposed models obtained from these two different modeling methods for individual isoforms by using Chimera, they were almost the same except for minor differences at the middle portion of loop 3, the C-terminal portion of helix 4 and loop 4, and there were no β-sheets modeled in both isoforms by I-TASSER (comparisons for each isoform are shown in supplemental Figs. S1 and S2).
Rigid-body Docking of Template and Simulated Atomic Models
Rigid-body docking was performed by using the FFT-accelerated 6D exhaustive search of the Situs package (47). The modeling template 3RQR deposited in the PDB (rabbit RyR1 amino acids 2,733–2,940), two comparatively modeled structures of rabbit RyR1 fragment 850–1,056, and mouse RyR2 fragment 861–1,067 were docked into RyR1 cryo-EM maps, including a 13.6-Å resolution map for the closed state (EMDB entry code 5014 (10)), a 9.6-Å resolution map for the closed state (EMDB entry code 1275 (12)), and two 10.2-Å resolution maps for both closed and open conformations (EMDB entry codes 1606 and 1607 (11)). Laplacian filter and standard linear cross-correlation criteria were both applied.
Molecular Dynamics Flexible Fitting into Cryo-EM Maps
We performed Molecular Dynamics Flexible Fitting by using VMD (48) and NAMD (49), two program used for molecular visualization and molecular dynamics simulations, respectively. The top ranked solution for each monomer previously obtained by rigid-body docking was used as the initial structure. We segmented a sub-volume from the overall homotetramer three-dimensional reconstruction. The rigid-body docking model is located inside the sub-volume, and we used the sub-volume as the target map. To prevent over-fitting, we increased simulation steps gradually with varying energy minimization steps and inspected the cross-correlation coefficient between each simulated map created from the MDFF trajectories and the segmented target map. The scaling factor was set to 1.0 for both the first 200 minimization steps and the subsequent minimization steps. The cross-correlation coefficient reached a plateau rapidly after thousands of simulated steps for models created by MODELLER or tens of thousands for models generated by the I-TASSER server in both the closed and the open state maps (see parameter and results in supplemental Table S1).
Secondary Structural Elements Detection in RyR1 Cryo-EM Map
We used program VolTrac from the Situs package (50) to predict the distribution of potential α-helices in regions of cryo-EM maps where we docked the modeled structures.
cDNA Construction and FRET Measurement
cDNAs encoding RyR2Y846-CFP and RyR2Y2801-YFP were constructed according to the previously described procedure (42, 51). FRET measurements in the HEK293 cells co-expressing RyR2Y846-CFP and RyR2Y2801-YFP were performed as described previously (52).
RESULTS
Homology Detection and Pseudo-atomic Model Generation
The full-length rabbit RyR1 subunit consisted of 5,037 amino acid residues (4), and the three-dimensional architectures of RyR that have been determined by cryo-EM revealed that the native homotetrameric RyR1 consisted of a transmembrane region and a large cytoplasmic assembly that is composed of at least 15 structural domains (10). Most of the solved crystal structures of RyR fragments are in the N-terminal region (13–16). The largest peptide fragment, consisting of the first 559 residues, was docked in three structural domains in the center of cytoplasmic assembly (16). Crystal structures of the phosphorylation domain in RyR1 (residues 2,733/2734–2,940), as well as the corresponding domains in RyR2 (residues 2,699–2,904) and in RyR3 (residues 2,597–2,800), were recently reported, and they were docked into a region near the corners of the square-shaped cytoplasmic assembly, also known as “clamp” structures (17, 18).
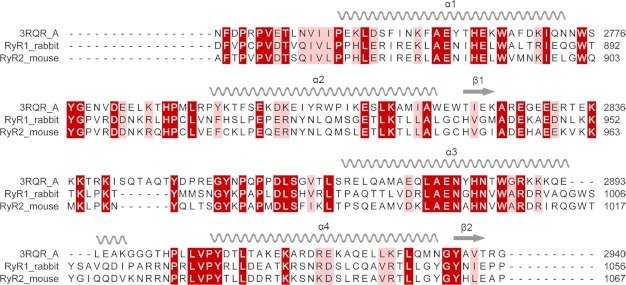
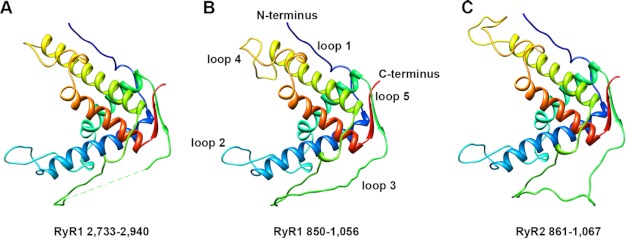
We performed a protein alignment search of the Protein Data Bank for the rabbit RyR1 sequence starting from residue 533, rather than 559, because the last 27 residues of the crystallized fragment (1–559) are not visible in the electron density (16). Multiple sequence alignments of this sequence in RyR1 using NCBI BLAST, I-TASSER, and Swiss Model Workspace protocols all found significant homology between sequence 850–1,056 in RyR1 and sequence 2,733–2,940 in RyR1, for which the x-ray-crystallographic structure is known (PDB entry code 3RQR) (18). Sequence identity between the two fragments ranged between 28.3% (Swiss Model) to 32.3% (NCBI BLAST and I-TASSER). We used ClustalW2 to align the corresponding sequence in mouse RyR2 (Fig. 1) (44). Pseudo-atomic models were generated by alignment of RyR1(850–1,056) (or RyR2(861–1,067)) with sequence 2,733–2,940, using the crystal structure of fragment 2,733–2,940 as the template. Two separate modeling procedures were used as follows: homology modeling using MODELLER (45) and ab initio modeling using the I-TASSER server (46). Modeled structures generated from the two procedures are very similar and remain close to the available crystal structure (Fig. 2, supplemental Figs. S1 and S2). They all have four α-helices linked by three loops and one loop each at the N and C termini. The only substantial differences between the modeled structures and the template structure are in the loops and in helix 3.
FIGURE 1.
Sequence alignment and secondary structural elements. A protein BLAST alignment of rabbit RyR1 and mouse RyR2 sequences against the sequences for all structures deposited in the PDB was performed. The result shows that one portion of RyR1, residues 850–1,056 (or for RyR2, 861–1,067), has been aligned with the sequence of 3RQR chain A (a crystal structure of a fragment of rabbit RyR1, residues 2,733–2,940) with a highly statistically significant alignment score. Sequence identity between 850–1,056 and 2,733–2,940 is 32.3%. Corresponding secondary structural elements are indicated above the sequences. α-Helices are indicated by coils and β-strands by arrows. Regions with identical amino acids are highlighted in dark red and similar residues in light red.
FIGURE 2.
Molecular modeling of RyR1 and RyR2 fragments. Pseudo-atomic models of rabbit RyR1 fragment (residues 850–1,056) (B) and mouse RyR2 fragment (residues 861–1,067) (C) generated by homology modeling based on the template crystal structure 3RQR (A) using the program MODELLER. The sequence order shown in the figure is as follows: N terminus, loop 1, helix 1 (blue); loop 2, helix 2 (green); loop 3, helix 3 (yellow); loop 4, helix 4 (red); loop 5, C terminus.
Docking Pseudo-atomic Models into Cryo-EM Maps
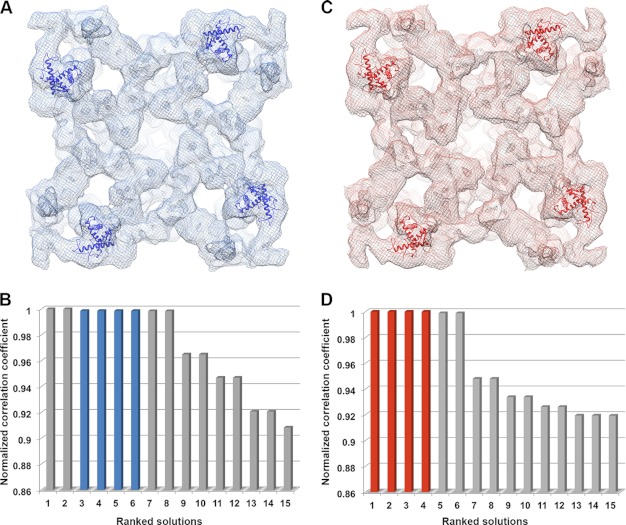
To determine the likely location of the RyR1 and RyR2 fragments in intact RyR, we first performed rigid-body docking using the Colores program from Situs package (47). We used four cryo-EM maps of RyR1 as follows: a 13.6-Å resolution map for the closed state (EMDB entry code 5014, (10)), a 9.6-Å resolution map for closed state (EMDB entry code 1275, (12)), and two 10.2-Å resolution maps for both closed and open conformations (EMDB entry codes 1606 and 1607 (11)). The resolution of the available RyR2 cryo-EM maps is poorer than that of the best RyR1 maps, but RyR1 and RyR2 share a high degree of sequence identity (66%) (53). Intact RyR1 and RyR2 structures determined by cryo-EM (7, 8) or fragment structures determined by x-ray crystallography (14) are also very similar to each other. We therefore estimated the location of the RyR2 model by docking it into RyR1 cryo-EM maps.
It was suggested by Tung et al. (16) that applying a Laplacian filter during the rigid-body docking was necessary for accuracy. Application of a Laplacian filter while docking our two pseudo-atomic models localized them both to the central vestibule region of the cryo-EM maps of RyR1 closed state, even though credible evidence indicates that this region contains the first 559 amino acid residues (16). For the RyR1 open state cryo-EM map, the RyR1 fragment model docked in a region that contained both domains 5 and 9, whereas the RyR2 fragment model docked into domain 10, where the phosphorylation domain 2,733/2734–2,940 was previously docked (supplemental Fig. S3) (17, 18). Using the standard linear cross-correlation fitting criterion instead of the Laplacian filter in the rigid-body docking localized both models to the region between domains 5 and 9, the same docking position that was obtained for the RyR1 fragment in the RyR1 open state conformation with the Laplacian filter applied. Given that there are four repeats, one in each subunit of the tetrameric structure, these were the four top ranked solutions in the open state cryo-EM maps, and solutions ranked 3–6 in the closed state maps (Fig. 3). Surprisingly, the top two solutions in the closed state did not show four repeats for the tetrameric RyR structure, but docked the models in two diagonally related subunits in a mirror orientation to the solutions ranked 3–6. These two mirrored positions were similar to those for the phosphorylation domain in structural domain 10 described by Yuchi et al. (17), as well as those we found for the crystal structure 3RQR in open conformation RyR1 map (supplemental Fig. S3). The solutions ranked seven and eight for the closed state or five and six for the open state, also repeated in only two diagonal subunits, with a slight rotation as compared with the top two solutions in the closed state. All other rigid-body docking solutions have much lower correlation coefficients.
FIGURE 3.
Rigid-body docking of the RyR1 fragment model into cryo-EM maps of RyR1. A, pseudo-atomic model of RyR1 fragment (residues 850–1,056, shown as blue ribbons) docked into the cryo-EM map of a closed conformation RyR1 (EMDB 1606, shown as blue mesh). B, top ranked docking solutions of RyR1 fragment in the closed state RyR1, solutions ranked 3–6 (blue bars) are the four positions shown as blue ribbons in A. C, RyR1 fragment pseudo-atomic model (shown as red ribbons) docked into the cryo-EM map of the open conformation of RyR1 (EMDB 1607, shown as red mesh). D, top ranked docking solutions of RyR1 fragment in the open state RyR1, solutions ranked 1–4 (red bars) are the positions shown as red ribbons in C.
Next, we docked the pseudo-atomic model for the RyR2 fragment into cryo-EM maps of RyR1. Interestingly, this pseudo-atomic model demonstrated a better fit, with the four top ranked solutions in both the closed and open states assuming the same location and orientation (supplemental Fig. S4). This location is the same as the top four solutions in the open state and solutions ranked 3–6 in the closed state for the pseudo-atomic model of RyR1 residues 850–1,056. We favor this docking position because it is highly ranked for both RyR1 and RyR2 fragments, and it has the requisite 4-fold symmetry for the tetrameric RyR architecture. More importantly, the N terminus (loop 1) of both models approaches the upper (cytoplasmic facing) surface of RyR1 in this position, which is consistent with our previous cryo-EM mapping of residue Tyr-846 in RyR2 (42). The N terminus of the models in other top ranked docking positions (ranked 5 or 6 for open state or ranked 1, 2, 7, or 8 for closed state) all point to the bottom or interior, which does not agree with the cryo-EM location of Tyr-846. In addition, this docking position is adjacent to the FKBP12-binding site (22, 26), which is consistent with our previous FRET characterization of two FRET pairs as follows: one pair between RyR2-YFP (YFP inserted after residue Tyr-846) and CFP-FKBP12.6 (CFP fused to the N terminus of FKBP12.6) and another pair between RyR2-GFP (GFP inserted after residue Tyr-846) and Alexa Fluor555-FKBP12.6 (labeled at residue Cys-14) (42).
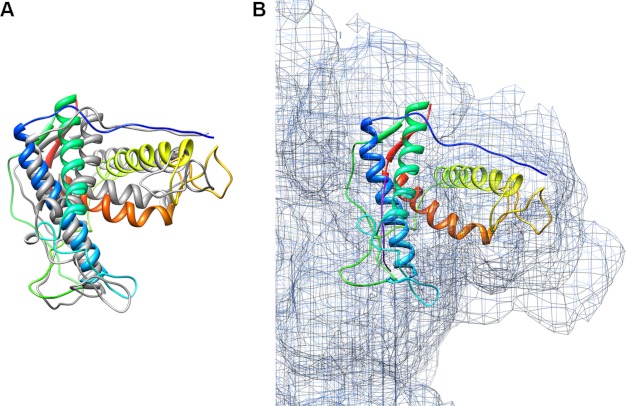
To obtain a better fit of the models into the cryo-EM maps, we used the MDFF program (54, 55) implemented in NAMD (49) to perform flexible fitting of the rigid-body fitted pseudo-atomic models. This program optimizes the fit of the pseudo-atomic models into the cryo-EM maps using restrained molecular dynamics simulations. The stereochemical properties and the secondary structure elements in the pseudo-atomic models are maintained through restraints, with the cryo-EM map as a guiding potential for model optimization in addition to the standard force field. Forces proportional to the gradient of the cryo-EM map are applied to drive atoms into high density regions of the map, resulting in a new structure that more closely conforms to the presumably native conformation of RyR captured by the cryo-EM data (54). The resulting flexibly fitted pseudo-atomic models reveal slight changes compared with their initial rigid-body docked conformations (Fig. 4A). The MDFF protocol was optimized to achieve the best cross-correlation coefficient and to minimize over-fitting (supplemental Table S1). The cross-correlation coefficients were improved from 0.67 to 0.70 by MDFF optimization for the RyR1 fragment, and from 0.66 to 0.70 for the RyR2 fragment, indicating slightly better fittings for both modeled fragments.
FIGURE 4.
Flexible fitting of RyR1 fragment model into RyR1 cryo-EM maps. A, MDFF of rigid-body docked RyR1 fragment model into the closed state RyR1 cryo-EM map. The starting rigid-body fit (gray ribbons) was obtained using the Colores program in the Situs package. The cross-correlation coefficient was improved from 0.67 to 0.70 after MDFF (shown as colored ribbons). B, close-up view of MDFF optimized RyR1 fragment model in the clamp region of RyR1 cryo-EM map in the closed conformation. The fine purple line next to helix 1 is the helical density traced by the secondary structure element detection program VolTrac in the Situs package.
Because both of our models contain four characteristic α-helices, these should match densities resembling α-helices in the region of cryo-EM maps where the modeled structures are positioned. We used the program VolTrac from the Situs package to trace α-helical densities in the cryo-EM map (50). It predicted one helix (represented as a fine purple line in Fig. 4B), which matched helix 1, the longest α-helix in each model. Other rod-like densities, which could correspond to α-helices, are visually apparent in the cryo-EM maps and are also located where the other three α-helices are positioned in the modeled fragments. VolTrac presumably assigned low confidence scores to these densities because they appear discontinuous in the current intermediate resolution (∼10 Å) cryo-EM maps.
Interaction between Modeled Fragments and FKBP
In a previous study, Samso et al. (26) determined the binding interface between FKBP12 and RyR1 and found a unique orientation for FKBP12 when fitted into a 16-Å resolution three-dimensional cryo-EM map. Two RyR1 cryo-EM maps used in this study were reconstructed with bound FKBP12 (11). Both pseudo-atomic models, for RyR1 residues 850–1,056 and for RyR2 residues 861–1,067, are docked adjacent to the FKBP-binding site. To gain further insight into the interaction between RyR and FKBP, we docked the crystal structures of FKBP12 (PDB entry code 2PPN) and FKBP12.6 (PDB entry code 1C9H) into the 10.2-Å resolution RyR1 cryo-EM maps. The resulting docked model shows a direct interaction between the pseudo-atomic model for RyR1 residues 850–1,056 and FKBP12 and between the pseudo-atomic model for RyR2 residues 861–1,067 and FKBP12.6, respectively (Fig. 5 and supplemental Fig. S5).
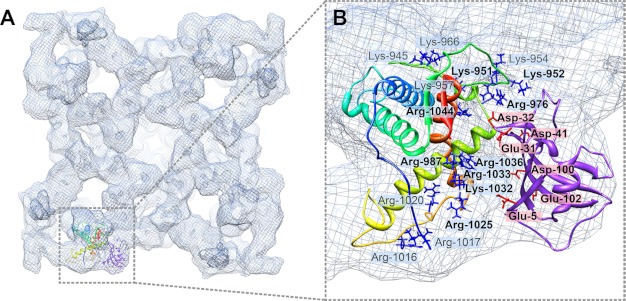
FIGURE 5.
Interaction between modeled structure RyR1(850–1,056) and FKBP12. A, flexibly fitted RyR1 fragment model and rigid-body fitted FKBP12 in the closed conformation RyR1 cryo-EM map, with docked structures shown in only one RyR1 subunit for clarity. B, close-up view of interface between the RyR1 fragment and FKBP12. Positively charged residues in RyR1 are depicted as blue stick structures, and negatively charged residues in FKBP12 are indicated as red stick structures. Residues with numbers in boldface are located at the direct interface. Residue Arg-976 in RyR1 is close enough to residue Asp-32 in FKBP12 to form an intermolecular salt bridge.
Analysis of the interface between FKBP12 and the pseudo-atomic model for the RyR1 fragment reveals a high occurrence of charged residues. There are six negatively charged residues in FKBP12 (Glu-5, Glu-31, Asp-32, Asp-40, Asp-100, and Glu-102), which are all oriented toward RyR1. Correspondingly, there are 16 positively charged residues in RyR1, among which nine residues are located in the interface that directly contacts FKBP12 (Fig. 5B). These nine residues (Lys-951, Lys-952, Arg-976, Arg-987, Arg-1025, Arg-1032, Arg-1033, Arg-1036, and Arg-1044) are located mainly in the C-terminal portion of loop 3, the N-terminal portion of helix 3, and the C-terminal portions of both loop 4 and helix 4. A total of 11 positively charged residues are either in loop 3 (Lys-945, Lys-951, Lys-952, Lys-954, Lys-957, Lys-966, and Arg-976) or in loop 4 (Arg-1016, Arg-1017, Arg-1020, and Arg-1025). Both loops are likely to be flexible and can probably adjust to interact more favorably with FKBP12. One noticeable residue in loop 3, Arg-976, is less than 4 Å away from the residue Asp-32 in FKBP12, which suggests that they could participate in a salt bridge between RyR1 and FKBP12. Such complementary salt bridges could explain why FKBP12 is one of the strongest binding partners for RyR1 (32, 56). Many charged residues are also clustered near the interface between the pseudo-atomic model of RyR2 residues 861–1,067 and FKBP12.6. These include 17 positively charged residues in RyR2 and four negatively charged residues in FKBP12.6 (supplemental Fig. S5). However, there are no residue pairs close enough to form an obvious salt bridge connecting RyR2 and FKBP12.6. Thus, our two models suggest that electrostatic interactions may play an important role in FKBP binding to RyR.
Disease-causing Mutations in the Modeled Structures
Multiple mutations in both RyR1 and RyR2 have been identified and linked to skeletal myopathies and cardiovascular disease. Mutations in RyR1 are associated with malignant hyperthermia and central core disease (57). In human skeletal muscles, the R1043C mutation in RyR1 causes malignant hyperthermia, a rare life-threatening condition that is usually triggered by exposure to certain drugs used for general anesthesia (57). Mutations in RyR2 are now linked to two genetic forms of cardiac arrhythmia: catecholaminergic polymorphic ventricular tachycardia and arrhythmogenic right ventricular cardiomyopathy (58–60). Catecholaminergic polymorphic ventricular tachycardia is an autosomal dominant arrhythmic syndrome characterized by exercise-induced polymorphic ventricular tachycardia, and it has been linked to two missense mutations in human cardiac muscle RyR2, R1013Q, and R1051P (61, 62).
Three of these disease-causing mutations are located in the sequence range of the modeled RyR1 and RyR2 fragments. These correspond to Ala-1002 (hRyR2 Arg-1013), Cys-1040 (hRyR2 Arg-1051), and Arg-1044 (hRyR1 Arg-1043) in the rabbit RyR1 sequence. Their locations in the pseudo-atomic model for the RyR1 fragment are shown in Fig. 6A. The corresponding locations of these residues (mouse RyR2 Arg-1013, Arg-1051, and Arg-1055) in the pseudo-atomic model for the docked RyR2 fragment are shown in supplemental Fig. S6A. Both Cys-1040 and Arg-1044 in RyR1 (or Arg-1051 and Arg-1055 in RyR2) are located in helix 4 and could directly interact with FKBP. Future experiments are needed to determine whether these mutations alter the RyR binding affinity for FKBP, a mechanism that was previously proposed for RyR channel dysfunction (35, 36). The third residue, Ala-1002 in RyR1 (or Arg-1013 in RyR2) is located in the C-terminal portion of helix 3. It is not close to the RyR/FKBP interface but is in an interface with the phosphorylation domain (see below).
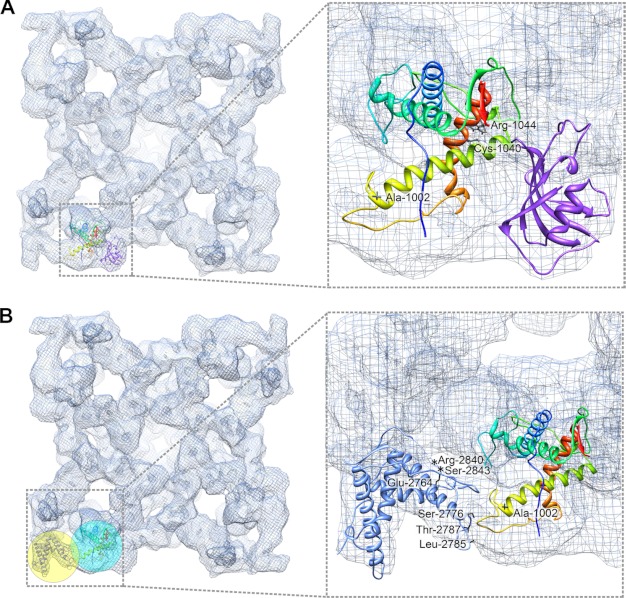
FIGURE 6.
Mapping disease-causing mutations in RyR1 fragment model. A, in the flexibly fitted RyR1 fragment model, mutant residues are depicted as black stick structures. Two residues, Cys-1040 and Arg-1044, are located in helix 4 (orange-red), which is at RyR1/FKBP12 interface, and possibly involved in the RyR-FKBP interaction. B, one residue Ala-1002 is located at interface between domain 9 (cyan sphere) and domain 10 (yellow sphere), where the RyR1 fragment interacts with docked crystal structure 3RQR (residues 2,733–2,940), which is a phosphorylation domain in RyR1. Seven mutant residues in this phosphorylation domain that show direct contacts with the RyR1 fragment model are highlighted with asterisks (in the unstructured loop) or black stick structures.
Domain-Domain Interaction between the RyR Fragment and the Phosphorylation Domain
The crystal structure 3RQR, the template for our RyR1 and RyR2 fragment models, contains RyR1 residues 2,733–2,940 (18). This fragment is highly enriched with serine residues, making it a hot spot for RyR phosphorylation by PKA or Ca2+/calmodulin-dependent protein kinase II. An independent study reported a crystal structure for an almost identical fragment (2,734–2,940, PDB entry code 4ERT) (17). Both crystal structures have been docked into domain 10, in the corner of the clamp region in the RyR three-dimensional cryo-EM map (17, 18). Notably, domain 10 is next to the structural domains 9 and 5, where our RyR1 and RyR2 fragments were docked (Fig. 6B and supplemental Fig. S6B). This suggests the likelihood of a direct domain-domain interaction between the phosphorylation domain and the RyR1 or RyR2 fragment. The mutant residue, Ala-1002 in RyR1 (or Arg-1013 in RyR2) is located on the interface (see close-up view in Fig. 6B and supplemental Fig. S6B). At least 11 disease-causing mutations were also found in the phosphorylation domain (RyR1 residues 2,733–2,940), among which seven residues, E2764K, S2776F, S2776M, L2785V, T2787S, R2840W, and S2843P, have a direct contact with the loop 4 in the RyR1 fragment model, which contains mutant residue Ala-1002 (close-up view in Fig. 6B). In addition, the loop that contains mutations R2840W and S2843P is also termed a phosphorylation loop because it has multiple serines and threonines, which are the major phosphorylation targets for PKA and CaMKII (17). In human RyR2, mutant residues Ser-2808, Thr-2810, Ser-2811, Ser-2814, Thr-2820, and Ser-2821, are all located in the phosphorylation loop and directly contact the RyR2 fragment model (close-up view in supplemental Fig. S6B). Taken together, the mutations and residues that can be phosphorylated may alter the domain-domain interaction between the RyR fragment and the phosphorylation domain, causing improper conformational changes that lead to RyR channel dysfunction (see under “Discussion”).
FRET Characterization of Dynamic Domain-Domain Interaction
Based on the docking results, we hypothesized a direct interaction between the phosphorylation domain and the RyR1 and RyR2 fragment models. To test this hypothesis experimentally, we designed a FRET pair to assess the dynamics of such a domain-domain interaction. FRET is a biophysical technique to quantify dynamical behavior in molecules, such as protein-protein interactions, protein-DNA interactions, and protein conformational changes. To monitor conformational changes in a large protein complex such as RyR, one structural domain can be labeled with a donor and the other with an acceptor. The FRET signals can then be measured and used to identify interactions between the labeled structural domains. The efficiency of this energy transfer is inversely proportional to the sixth power of the distance between donor and acceptor making FRET extremely sensitive to small distance changes. Previously, we mapped a residue in the phosphorylation domain and a residue close to the N terminus of the modeled structure in RyR2 by three-dimensional cryo-EM with GFP inserted as a structural marker (42, 51). In this study, we replaced the GFP that was inserted after Tyr-846 with a CFP and replaced the GFP after Tyr-2801 with a YFP. When the two cDNAs (RyR2Y846-CFP and RyR2Y2801-YFP) are co-expressed in HEK293 cells, different hybrid tetrameric RyRs could be formed (see models in Fig. 7A). FRET pairs will be formed only when the following two criteria are both satisfied as follows: 1) the donor (CFP) and acceptor (YFP) are within a certain distance of each other (normally below 100 Å); 2) the two domains containing CFP and YFP must belong to two neighboring RyR2 subunits. Models displayed in Fig. 7A highlight FRET pairs formed only when the two domains form an intersubunit interaction. No FRET will be detected in the case of an intrasubunit interaction. Based on our previous cryo-EM mappings, the CFP and YFP are close to each other (center-to-center distance is about 65 Å), and according to a previous FRET study, the two domains likely belong to different RyR subunits (52). FRET signals were detected in the HEK293 cells that co-expressed cDNAs of RyR2Y846-CFP and RyR2Y2801-YFP (Fig. 7, B–D). These FRET signals were also altered by caffeine, which is a RyR channel activator. Both of these results provide an experimental support for our docked models. The conformational change also seems to be sensitive to the dose of caffeine and can be reversed by removal of caffeine (Fig. 7D). The decreases in FRET efficiency (Fig. 7C) and in acceptor/donor emission ratio (Fig. 7D) upon caffeine treatment indicate that the two structural domains separate from one another when the RyR channel switches from the closed to the open conformation.
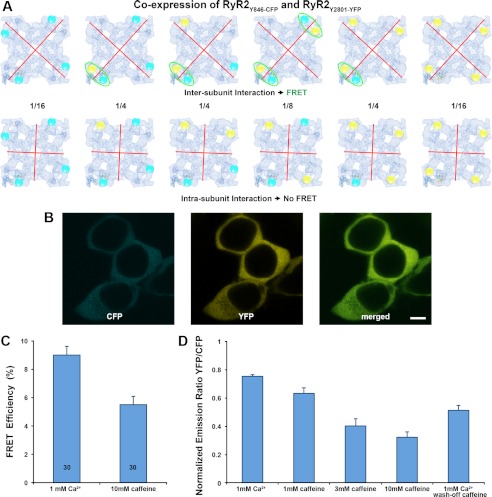
FIGURE 7.
FRET analysis in HEK293 cells that co-express RyR2Y846-CFP and RyR2Y2801-YFP. A, models of six possible hybrid RyR2 tetramer molecules when two cDNAs that encode RyR2Y846-CFP and RyR2Y2801-YFP are co-expressed. The cyan and yellow spheres represent the CFP and YFP. Red lines represent the possible boundary between RyR subunits. The top row shows six RyR2 tetramer structures for the situation in which two structural domains bearing Tyr-846-CFP and Tyr-2801-YFP belong to two different subunits (i.e. an intersubunit interaction). In this case, four out of six structures have at least one FRET pair within one corner of the RyR cytoplasmic assembly (highlighted by green ellipses). The bottom row shows six RyR2 tetramer structures for the case in which the two structural domains bearing Tyr-846-CFP and Tyr-2801-YFP are contained within one RyR subunit (i.e. an intrasubunit interaction). In this case, no FRET signal will be detected, because the distance between CFP and YFP in two separate corners is over 200 Å. Numbers between two rows are the mathematical probability of a ratio among six possible structures when RyR2 tetramers are formed randomly. B, images of live HEK293 cells co-transfected with cDNAs of RyR2Y846-CFP and RyR2Y2801-YFP showing co-localization of CFP and YFP; scale bar, 5 μm. C, FRET efficiency determined by photo-bleaching of acceptor. Data are mean ± S.E., with the number of cells indicated in the bars. D, change of FRET signal determined by monitoring acceptor/donor emission ratio. Data are mean ± S.E., averaged from seven separate experiments.
DISCUSSION
Multiple Sequences in RyR Involved in FKBP Binding
Marx et al. (35) hypothesized that PKA hyper-phosphorylates Ser-2808 in RyR2 (or Ser-2843 in RyR1) causing FKBP12.6 (FKBP12) dissociation from RyR2 (or RyR1), which directly enhances sarcoplasmic reticulum Ca2+ leak and systolic dysfunction in heart failure (36). However, this proposed mechanism has been challenged by other laboratory findings; Li et al. (37) did not find any effect of PKA phosphorylation of the RyR2 on calcium sparks in mouse ventricular myocytes. Jiang et al. (38)did not observe dissociation of FKBP12.6 from RyR2 in cardiac microsomal membranes treated with PKA. Stange et al. (39) made site-directed substitutions of RyR1 at Ser-2843 and RyR2 at Ser-2809 (rabbit RyR2, corresponding to Ser-2808 in mouse) and showed that mutant RyR2s neither abolished FKBP12.6 binding nor substantially changed channel functional properties (21). Xiao et al. (40) found that FKBP12.6 can bind to both the Ser-2808-phosphorylated and -nonphosphorylated forms of RyR2 and that an S2808D phosphomimetic mutant retained the ability to bind FKBP12.6. Furthermore, complete phosphorylation at Ser-2808 by exogenous PKA did not disrupt the FKBP12.6-RyR2 complex. Using three-dimensional cryo-EM, we have mapped the phosphorylation site by GFP insertion and by antibody binding, and we found neither of these labels close to the FKBP-binding site (51). This structural evidence is further supported by the docked positions of crystal structures of the RyR phosphorylation domain (17, 18), which suggest that Ser-2808 does not directly interact with FKBP. If phosphorylated Ser-2808 does cause a decrease in RyR's affinity for FKBP, it could do so through an indirect mechanism, such as through changes in interaction with the RyR1 or RyR2 fragments modeled in this study.
Besides Ser-2808, other motifs or sequences in RyR have been proposed to be important for FKBP binding, including the Val-2461–Pro-2462 motif in RyR1 (Ile-2427–Pro-2428 in RyR2) (33), a region contained within the sequence 1,815–1,855 in RyR2 (41), and a C-terminal fragment spanning RyR2 residues 3,788–4,765 (63). The FKBP12- and FKBP12.6-binding sites were mapped to the same position in the three-dimensional structure of RyR1 and RyR2 by cryo-EM, and the position is at the junction of RyR structural domains 3, 5, and 9 (22, 25, 26). These findings suggest that there could be more than one sequence in RyR that participates in FKBP binding, as multiple sequences may form a binding pocket for FKBP when RyR folds to its native structure and assembles into a homotetramer. In support of this hypothesis, we have mapped two residues in the three-dimensional structure of RyR2, Tyr-846 and Thr-1874, and we found the structural domains bearing the two residues are both adjacent to the FKBP-binding site (41, 42). Here, our docking of the RyR1 and RyR2 fragment models into an RyR region comprising a portion of structural domain 5 and the upper portion of domain 9 is consistent with the three-dimensional location of Tyr-846. Other FKBP-binding motifs or sequences may fold into the lower portion of domain 9 and in domain 3, and altogether form a binding pocket for FKBP.
Potential Impact of Certain RyR Mutations on FKBP Binding
Mutations in RyR2 in patients with catecholaminergic polymorphic ventricular tachycardia reduce the binding affinity of FKBP12.6 for RyR2 and cause increased channel activity under conditions that simulate exercise, resulting in exercise-induced sudden cardiac death (64). In this study, the RyR1 and RyR2 fragment models docked into RyR cryo-EM maps indicate a direct interaction with FKBP. Three disease-causing mutations have been identified in the region encompassed by the fragments, including two catecholaminergic polymorphic ventricular tachycardia mutations in human RyR2 and one malignant hyperthermia mutation in human RyR1. We have located their corresponding residues in the models; the catecholaminergic polymorphic ventricular tachycardia mutation R1051P and the malignant hyperthermia mutation R1043C are at the interface with FKBP. Intriguingly, both mutations convert a positively charged residue, arginine, to neutral residues, proline or cysteine. Also in this work, we presented evidence that the interaction between RyR and FKBP is mainly electrostatic, which suggests that both of these mutations might reduce the binding affinity for FKBP through alterations in electrostatic interactions.
Certain Mutations in RyR Alter the Interaction between RyR Structural Domains
Another potential underlying mechanism for mutant RyR channel dysfunction involves abnormal domain-domain interactions (65). According to this hypothesis, several RyR structural domains work in a coordinated manner to perform necessary conformational changes that control the Ca2+ channel gating. An example of such a domain-domain interaction, for which many disruptive disease-causing mutations have been discovered, involves an N-terminal region and a sequence from the central region of the RyR sequence. Several lines of experimental evidence support the hypothesis that mutations in either domain weaken the normal interdomain interaction and lead to enhance RyR Ca2+ channel opening (66–68). Recently, some disease-causing mutations were mapped in the crystal structure of the RyR1 fragment containing the first 559 residues, which include 33 mutations in RyR1 and 23 mutations in RyR2 (positions in RyR1 that correspond to mutated residues in RyR2). Among them, only six mutations (three in RyR1 and three in RyR2) were buried within a folding domain, and the other 50 mutations were all distributed at domain-domain interfaces either between the N-terminal ABC domains or between ABC domains and other neighboring domains (16). One notable interface occurs between domains A and B across two subunits (an intersubunit interaction), with 19 mutations clustering at this interface, suggesting an intersubunit contact plays an important role in RyR function (16). Ikemoto and co-workers (52) hypothesized a domain-domain interaction that involves N-terminal and central mutation domains, and it has also been identified as an intersubunit interaction. In this work, the domain-domain interaction between the RyR fragment models and the phosphorylation domain is also characterized as an intersubunit interaction. Similar to many potassium channels, a functional RyR is a homotetramer that is composed of four identical subunits that are arranged symmetrically around the pore. In the KcsA potassium channel, there is evidence to suggest that direct interaction between the four subunits leads to a cooperative opening and closing of the ion-conducting pore (69), which is structurally similar to the pore region of RyR (10, 11). Here, we have identified a third intersubunit domain-domain pair involving two structurally homologous domains derived from amino acid residues 850–1,056 in one subunit and 2,733–2,940 in an adjacent subunit. Based on the present results and previous studies, we suggest that mutations in either domain may weaken the normal subunit-subunit interactions, thus altering the stability of the RyR channel.
Supplementary Material
Acknowledgments
We thank Terence Wagenknecht for critically reviewing this manuscript. We gratefully acknowledge the Advanced Light Microscopy and Image Analysis Core Facilities at the Wadsworth Center.
This work was supported, in whole or in part, by National Institutes of Health Grant R01HL095541 (to Z. L.). This work was also supported by a Canadian Institutes of Health Research grant (to S. R. W. C.).

This article contains supplemental Figs. S1–S6 and Table S1.
- RyR
- ryanodine receptor
- cryo-EM
- cryo-electron microscopy
- RyR1
- ryanodine receptor type 1 (the skeletal muscle isoform)
- RyR2
- ryanodine receptor type 2 (the cardiac muscle isoform)
- MDFF
- molecular dynamics flexible fitting
- PDB
- Protein Data Bank
- FKBP
- FK506-binding protein
- h
- human
- CFP
- cyan fluorescent protein
- YFP
- yellow fluorescent protein
- GFP
- green fluorescent protein
- FRET
- fluorescence resonance energy transfer.
REFERENCES
- 1. Coronado R., Morrissette J., Sukhareva M., Vaughan D. M. (1994) Structure and function of ryanodine receptors. Am. J. Physiol. Cell Physiol. 266, C1485–C1504 [DOI] [PubMed] [Google Scholar]
- 2. Lanner J. T., Georgiou D. K., Joshi A. D., Hamilton S. L. (2010) Ryanodine receptors. Structure, expression, molecular details, and function in calcium release. Cold Spring Harbor Perspect. Biol. 2, a003996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hakamata Y., Nakai J., Takeshima H., Imoto K. (1992) Primary structure and distribution of a novel ryanodine receptor/calcium release channel from rabbit brain. FEBS Lett. 312, 229–235 [DOI] [PubMed] [Google Scholar]
- 4. Takeshima H., Nishimura S., Matsumoto T., Ishida H., Kangawa K., Minamino N., Matsuo H., Ueda M., Hanaoka M., Hirose T. (1989) Primary structure and expression from complementary DNA of skeletal muscle ryanodine receptor. Nature 339, 439–445 [DOI] [PubMed] [Google Scholar]
- 5. Zorzato F., Fujii J., Otsu K., Phillips M., Green N. M., Lai F. A., Meissner G., MacLennan D. H. (1990) Molecular cloning of cDNA encoding human and rabbit forms of the Ca2+ release channel (ryanodine receptor) of skeletal muscle sarcoplasmic reticulum. J. Biol. Chem. 265, 2244–2256 [PubMed] [Google Scholar]
- 6. Du G. G., MacLennan D. H. (2005) in Ryanodine Receptors: Structure, Function and Dysfunction in Clinical Disease (Wehrens X. H. T., Marks A. R., eds) pp. 9–23, Springer-Verlag, New York [Google Scholar]
- 7. Radermacher M., Rao V., Grassucci R., Frank J., Timerman A. P., Fleischer S., Wagenknecht T. (1994) Cryo-electron microscopy and three-dimensional reconstruction of the calcium release channel/ryanodine receptor from skeletal muscle. J. Cell Biol. 127, 411–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sharma M. R., Penczek P., Grassucci R., Xin H.-B., Fleischer S., Wagenknecht T. (1998) Cryoelectron microscopy and image analysis of the cardiac ryanodine receptor. J. Biol. Chem. 273, 18429–18434 [DOI] [PubMed] [Google Scholar]
- 9. Sharma M. R., Jeyakumar L. H., Fleischer S., Wagenknecht T. (2000) Three-dimensional structure of ryanodine receptor isoform three in two conformational states as visualized by cryo-electron microscopy. J. Biol. Chem. 275, 9485–9491 [DOI] [PubMed] [Google Scholar]
- 10. Samsó M., Wagenknecht T., Allen P. D. (2005) Internal structure and visualization of transmembrane domains of the RyR1 calcium release channel by cryo-EM. Nat. Struct. Mol. Biol. 12, 539–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Samso M., Feng W., Pessah I. N., Allen P. D. (2009) Coordinated movement of cytoplasmic and transmembrane domains of RyR1 upon gating. PLoS Biol. 7, e1000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ludtke S. J., Serysheva I. I., Hamilton S. L., Chiu W. (2005) The pore structure of the closed RyR1 channel. Structure 13, 1203–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Amador F. J., Liu S., Ishiyama N., Plevin M. J., Wilson A., MacLennan D. H., Ikura M. (2009) Crystal structure of type I ryanodine receptor amino-terminal β-trefoil domain reveals a disease-associated mutation “hot spot” loop. Proc. Natl. Acad. Sci. U.S.A. 106, 11040–11044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lobo P. A., Van Petegem F. (2009) Crystal structures of the N-terminal domains of cardiac and skeletal muscle ryanodine receptors: insights into disease mutations. Structure 17, 1505–1514 [DOI] [PubMed] [Google Scholar]
- 15. Lobo P.-A., Kimlicka L., Tung C. C., Van Petegem F. (2011) The deletion of exon 3 in the cardiac ryanodine receptor is rescued by β strand switching. Structure 19, 790–798 [DOI] [PubMed] [Google Scholar]
- 16. Tung C. C., Lobo P. A., Kimlicka L., Van Petegem F. (2010) The amino-terminal disease hot spot of ryanodine receptors forms a cytoplasmic vestibule. Nature 468, 585–588 [DOI] [PubMed] [Google Scholar]
- 17. Yuchi Z., Lau K., Van Petegem F. (2012) Disease mutations in the ryanodine receptor central region. Crystal structures of a phosphorylation hot spot domain. Structure 20, 1201–1211 [DOI] [PubMed] [Google Scholar]
- 18. Sharma P., Ishiyama N., Nair U., Li W., Dong A., Miyake T., Wilson A., Ryan T., MacLennan D. H., Kislinger T., Ikura M., Dhe-Paganon S., Gramolini A. O. (2012) Structural determination of the phosphorylation domain of the ryanodine receptor. FEBS J. 279, 3952–3964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wehrens X. H., Lehnart S. E., Marks A. R. (2005) in Ryanodine Receptors: Structure, Function and Dysfunction in Clinical Disease (Wehrens X. H. T., Marks A. R., eds) pp. 151–161, Springer-Verlag, New York [Google Scholar]
- 20. Laver D. R. (2010) in Structure and Function of Calcium Release Channels (Serysheva I. I., ed) pp. 69–89, Academic Press, Burlington, MA [Google Scholar]
- 21. Meissner G. (2010) in Structure and Function of Calcium Release Channels (Serysheva I. I., ed) pp. 91–113, Academic Press, Burlington, MA [Google Scholar]
- 22. Wagenknecht T., Radermacher M., Grassucci R., Berkowitz J., Xin H.-B., Fleischer S. (1997) Locations of calmodulin and FK506-binding protein on the three-dimensional architecture of the skeletal muscle ryanodine receptor. J. Biol. Chem. 272, 32463–32471 [DOI] [PubMed] [Google Scholar]
- 23. Samsó M., Wagenknecht T. (2002) Apocalmodulin and Ca2+-calmodulin bind to neighboring locations on the ryanodine receptor. J. Biol. Chem. 277, 1349–1353 [DOI] [PubMed] [Google Scholar]
- 24. Huang X., Fruen B., Farrington D. T., Wagenknecht T., Liu Z. (2012) Calmodulin binding locations on the skeletal and cardiac ryanodine receptors. J. Biol. Chem. 287, 30328–30335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sharma M. R., Jeyakumar L. H., Fleischer S., Wagenknecht T. (2006) Three-dimensional visualization of FKBP12.6 binding to an open conformation of cardiac ryanodine receptor. Biophys. J. 90, 164–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Samsó M., Shen X., Allen P. D. (2006) Structural characterization of the RyR1-KBP12 interaction. J. Mol. Biol. 356, 917–927 [DOI] [PubMed] [Google Scholar]
- 27. Meng X., Wang G., Viero C., Wang Q., Mi W., Su X.-D., Wagenknecht T., Williams A. J., Liu Z., Yin C.-C. (2009) CLIC2-RyR1 interaction and structural characterization by cryo-electron microscopy. J. Mol. Biol. 387, 320–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Samsó M., Trujillo R., Gurrola G. B., Valdivia H. H., Wagenknecht T. (1999) Three-dimensional location of the imperatoxin a binding site on the ryanodine receptor. J. Cell Biol. 146, 493–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhou Q., Wang Q.-L., Meng X., Shu Y., Jiang T., Wagenknecht T., Yin C.-C., Sui S.-F., Liu Z. (2008) Structural and functional characterization of ryanodine receptor-natrin toxin interaction. Biophys. J. 95, 4289–4299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jeyakumar L. H., Ballester L., Cheng D. S., McIntyre J. O., Chang P., Olivey H. E., Rollins-Smith L., Barnett J. V., Murray K., Xin H.-B., Fleischer S. (2001) FKBP binding characteristics of cardiac microsomes from diverse vertebrates. Biochem. Biophys. Res. Commun. 281, 979–986 [DOI] [PubMed] [Google Scholar]
- 31. Jayaraman T., Brillantes A. M., Timerman A. P., Fleischer S., Erdjument-Bromage H., Tempst P., Marks A. R. (1992) FK506-binding protein associated with the calcium release channel (ryanodine receptor). J. Biol. Chem. 267, 9474–9477 [PubMed] [Google Scholar]
- 32. Xin H.-B., Rogers K., Qi Y., Kanematsu T., Fleischer S. (1999) Three amino acid residues determine selective binding of FK506-binding protein 12.6 to the cardiac ryanodine receptor. J. Biol. Chem. 274, 15315–15319 [DOI] [PubMed] [Google Scholar]
- 33. Gaburjakova M., Gaburjakova J., Reiken S., Huang F., Marx S. O., Rosemblit N., Marks A. R. (2001) FKBP12 binding modulates ryanodine receptor channel gating. J. Biol. Chem. 276, 16931–16935 [DOI] [PubMed] [Google Scholar]
- 34. Masumiya H., Wang R., Zhang J., Xiao B., Chen S. R. (2003) Localization of the 12.6-kDa FK506-binding protein (FKBP12.6) binding site to the NH2-terminal domain of the cardiac Ca2+ release channel (ryanodine receptor). J. Biol. Chem. 278, 3786–3792 [DOI] [PubMed] [Google Scholar]
- 35. Marx S. O., Reiken S., Hisamatsu Y., Jayaraman T., Burkhoff D., Rosemblit N., Marks A. R. (2000) PKA phosphorylation dissociates FKBP12.6 from the calcium release channel (ryanodine receptor). Defective regulation in failing hearts. Cell 101, 365–376 [DOI] [PubMed] [Google Scholar]
- 36. Reiken S., Lacampagne A., Zhou H., Kherani A., Lehnart S. E., Ward C., Huang F., Gaburjakova M., Gaburjakova J., Rosemblit N., Warren M. S., He K.-L., Yi G.-H., Wang J., Burkhoff D., Vassort G., Marks A. R. (2003) PKA phosphorylation activates the calcium release channel (ryanodine receptor) in skeletal muscle. J. Cell Biol. 160, 919–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li Y., Kranias E. G., Mignery G. A., Bers D. M. (2002) Protein kinase A phosphorylation of the ryanodine receptor does not affect calcium sparks in mouse ventricular myocytes. Circ. Res. 90, 309–316 [DOI] [PubMed] [Google Scholar]
- 38. Jiang M. T., Lokuta A. J., Farrell E. F., Wolff M. R., Haworth R. A., Valdivia H. H. (2002) Abnormal Ca2+ release, but normal ryanodine receptors, in canine and human heart failure. Circ. Res. 91, 1015–1022 [DOI] [PubMed] [Google Scholar]
- 39. Stange M., Xu L., Balshaw D., Yamaguchi N., Meissner G. (2003) Characterization of recombinant skeletal muscle (Ser-2843) and cardiac muscle (Ser-2809) ryanodine receptor phosphorylation mutants. J. Biol. Chem. 278, 51693–51702 [DOI] [PubMed] [Google Scholar]
- 40. Xiao B., Jiang M. T., Zhao M., Yang D., Sutherland C., Lai F. A., Walsh M. P., Warltier D. C., Cheng H., Chen S. R. (2005) Characterization of a novel PKA phosphorylation site, serine-2030, reveals no PKA hyperphosphorylation of the cardiac ryanodine receptor in canine heart failure. Circ. Res. 96, 847–855 [DOI] [PubMed] [Google Scholar]
- 41. Zhang J., Liu Z., Masumiya H., Wang R., Jiang D., Li F., Wagenknecht T., Chen S. R. (2003) Three-dimensional localization of divergent region 3 of the ryanodine receptor to the clamp-shaped structures adjacent to the FKBP-binding sites. J. Biol. Chem. 278, 14211–14218 [DOI] [PubMed] [Google Scholar]
- 42. Wang R., Zhong X., Meng X., Koop A., Tian X., Jones P. P., Fruen B. R., Wagenknecht T., Liu Z., Chen S. R. (2011) Localization of the dantrolene-binding sequence near the FK506-binding protein-binding site in the three-dimensional structure of the ryanodine receptor. J. Biol. Chem. 286, 12202–12212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 44. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 45. Sali A., Blundell T. L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 46. Zhang Y. (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chacón P., Wriggers W. (2002) Multi-resolution contour-based fitting of macromolecular structures. J. Mol. Biol. 317, 375–384 [DOI] [PubMed] [Google Scholar]
- 48. Humphrey W., Dalke A., Schulten K. (1996) VMD. Visual molecular dynamics. J. Mol. Graph. 14, 33–38 [DOI] [PubMed] [Google Scholar]
- 49. Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kalé L., Schulten K. (2005) Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rusu M., Wriggers W. (2012) Evolutionary bidirectional expansion for the tracing of α-helices in cryo-electron microscopy reconstructions. J. Struct. Biol. 177, 410–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Meng X., Xiao B., Cai S., Huang X., Li F., Bolstad J., Trujillo R., Airey J., Chen S. R., Wagenknecht T., Liu Z. (2007) Three-dimensional localization of serine 2808, a phosphorylation site in cardiac ryanodine receptor. J. Biol. Chem. 282, 25929–25939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liu Z., Wang R., Tian X., Zhong X., Gangopadhyay J., Cole R., Ikemoto N., Chen S. R., Wagenknecht T. (2010) Dynamic, intersubunit interactions between the N-terminal and central mutation regions of cardiac ryanodine receptor. J. Cell Sci. 123, 1775–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Otsu K., Willard H. F., Khanna V. K., Zorzato F., Green N. M., MacLennan D. H. (1990) Molecular cloning of cDNA encoding the Ca2+ release channel (ryanodine receptor) of rabbit cardiac muscle sarcoplasmic reticulum. J. Biol. Chem. 265, 13472–13483 [PubMed] [Google Scholar]
- 54. Trabuco L. G., Villa E., Mitra K., Frank J., Schulten K. (2008) Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure 16, 673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Trabuco L. G., Villa E., Schreiner E., Harrison C. B., Schulten K. (2009) Molecular dynamics flexible fitting. A practical guide to combine cryo-electron microscopy and x-ray crystallography. Methods 49, 174–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lee E. H., Rho S.-H., Kwon S.-J., Eom S. H., Allen P. D., Kim D. H. (2004) N-terminal region of FKBP12 is essential for binding to the skeletal ryanodine receptor. J. Biol. Chem. 279, 26481–26488 [DOI] [PubMed] [Google Scholar]
- 57. Robinson R., Carpenter D., Shaw M.-A., Halsall J., Hopkins P. (2006) Mutations in RYR1 in malignant hyperthermia and central core disease. Hum. Mutat. 27, 977–989 [DOI] [PubMed] [Google Scholar]
- 58. Herren T., Gerber P. A., Duru F. (2009) Arrhythmogenic right ventricular cardiomyopathy/dysplasia: a not so rare “disease of the desmosome” with multiple clinical presentations. Clin. Res. Cardiol. 98, 141–158 [DOI] [PubMed] [Google Scholar]
- 59. Liu N., Ruan Y., Priori S. G. (2008) Catecholaminergic polymorphic ventricular tachycardia. Prog. Cardiovasc. Dis. 51, 23–30 [DOI] [PubMed] [Google Scholar]
- 60. Priori S. G., Chen S. R. (2011) Inherited dysfunction of sarcoplasmic reticulum Ca2+ handling and arrhythmogenesis. Circ. Res. 108, 871–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Marjamaa A., Laitinen-Forsblom P., Lahtinen A. M., Viitasalo M., Toivonen L., Kontula K., Swan H. (2009) Search for cardiac calcium cycling gene mutations in familial ventricular arrhythmias resembling catecholaminergic polymorphic ventricular tachycardia. BMC Med. Genet. 10, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Medeiros-Domingo A., Bhuiyan Z. A., Tester D. J., Hofman N., Bikker H., van Tintelen J. P., Mannens M. M., Wilde A. A., Ackerman M. J. (2009) The RYR2-encoded ryanodine receptor/calcium release channel in patients diagnosed previously with either catecholaminergic polymorphic ventricular tachycardia or genotype negative, exercise-induced long QT syndrome. A comprehensive open reading frame mutational analysis. J. Am. Coll. Cardiol. 54, 2065–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zissimopoulos S., Lai F. A. (2005) Interaction of FKBP12.6 with the cardiac ryanodine receptor C-terminal domain. J. Biol. Chem. 280, 5475–5485 [DOI] [PubMed] [Google Scholar]
- 64. Wehrens X. H., Lehnart S. E., Huang F., Vest J. A., Reiken S. R., Mohler P. J., Sun J., Guatimosim S., Song L.-S., Rosemblit N., D'Armiento J. M., Napolitano C., Memmi M., Priori S. G., Lederer W. J., Marks A. R. (2003) FKBP12.6 deficiency and defective calcium release channel (ryanodine receptor) function linked to exercise-induced sudden cardiac death. Cell 113, 829–840 [DOI] [PubMed] [Google Scholar]
- 65. Ikemoto N. (2005) in Ryanodine Receptors: Structure, Function and Dysfunction in Clinical Disease (Wehrens X. H. T., Marks A. R., eds) pp. 53–65, Springer-Verlag, New York [Google Scholar]
- 66. Yamamoto T., Ikemoto N. (2002) Peptide probe study of the critical regulatory domain of the cardiac ryanodine receptor. Biochem. Biophys. Res. Commun. 291, 1102–1108 [DOI] [PubMed] [Google Scholar]
- 67. Oda T., Yano M., Yamamoto T., Tokuhisa T., Okuda S., Doi M., Ohkusa T., Ikeda Y., Kobayashi S., Ikemoto N., Matsuzaki M. (2005) Defective regulation of interdomain interactions within the ryanodine receptor plays a key role in the pathogenesis of heart failure. Circulation 111, 3400–3410 [DOI] [PubMed] [Google Scholar]
- 68. Yang Z., Ikemoto N., Lamb G. D., Steele D. S. (2006) The RyR2 central domain peptide DPc10 lowers the threshold for spontaneous Ca2+ release in permeabilized cardiomyocytes. Cardiovasc. Res. 70, 475–485 [DOI] [PubMed] [Google Scholar]
- 69. Blunck R., McGuire H., Hyde H. C., Bezanilla F. (2008) Fluorescence detection of the movement of single KcsA subunits reveals cooperativity. Proc. Natl. Acad. Sci. U.S.A. 105, 20263–20268 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.