Background: Laboratory of genetics and physiology 2 (LGP2) is a cytoplasmic RNA receptor required for innate antiviral signaling.
Results: LGP2 uses ATP hydrolysis to diversify RNA recognition and enhance antiviral signaling.
Conclusion: LGP2 mediates antiviral responses by ATP-enhanced RNA recognition.
Significance: This study reveals a novel property of LGP2 providing a mechanistic basis for its positive role in antiviral signaling.
Keywords: Innate Immunity, Interferon, Pattern Recognition Receptor, Rig-like Receptors (RLR), RNA-binding Protein, RNA-Protein Interaction, LGP2, RNA Recognition
Abstract
Laboratory of genetics and physiology 2 (LGP2) is a member of the RIG-I-like receptor family of cytoplasmic pattern recognition receptors that detect molecular signatures of virus infection and initiate antiviral signal transduction cascades. The ATP hydrolysis activity of LGP2 is essential for antiviral signaling, but it has been unclear how the enzymatic properties of LGP2 regulate its biological response. Quantitative analysis of the dsRNA binding and enzymatic activities of LGP2 revealed high dsRNA-independent ATP hydrolysis activity. Biochemical assays and single-molecule analysis of LGP2 and mutant variants that dissociate basal from dsRNA-stimulated ATP hydrolysis demonstrate that LGP2 utilizes basal ATP hydrolysis to enhance and diversify its RNA recognition capacity, enabling the protein to associate with intrinsically poor substrates. This property is required for LGP2 to synergize with another RIG-I-like receptor, MDA5, to potentiate IFNβ transcription in vivo during infection with encephalomyocarditis virus or transfection with poly(I:C). These results demonstrate previously unrecognized properties of LGP2 ATP hydrolysis and RNA interaction and provide a mechanistic basis for a positive regulatory role for LGP2 in antiviral signaling.
Introduction
The mammalian type I interferon (IFN) response is a powerful antiviral system that directly interferes with virus replication and contributes to both innate and adaptive immune responses. Intracellular accumulation of virus replication intermediates such as double-stranded RNA (dsRNA) or RNAs with triphosphorylated 5′-ends are detected by cytosolic pattern recognition receptors, including the CARD-helicase proteins, RIG-I-like receptor (RIG-I)3 and MDA5. RIG-I and MDA5 recognize non-self RNAs with their C-terminal DEXD/H-box helicase domains and regulatory domains (RD) and use their N-terminal CARD regions for interactions with downstream signaling components, leading to transcriptional activation of IFN and other immediate antiviral response genes (1, 2).
A third member of the RIG-I-like receptor (RLR) family, LGP2, has significant sequence identity within the helicase domain and RD but lacks a CARD region. The functions of LGP2 in antiviral signaling have been controversial, as different experimental strategies have demonstrated seemingly antithetic biological activities. The intracellular LGP2 concentration appears to regulate a switch between positive and negative functions in antiviral signal transduction. When the protein is expressed in cells from plasmid expression vectors, LGP2 can function as a negative regulator of RLR signaling (3–5). This finding together with the observation that LGP2 expression is stimulated by virus infection led to the initial characterization of LGP2 as a feedback inhibitor of antiviral responses. In contrast, mice with a targeted disruption in the LGP2 locus are more susceptible to virus infections and have defects in antiviral IFNβ and cytokine responses. LGP2 deficiency reduces host responses to several RNA viruses including the picornaviruses encephalomyocarditis virus and poliovirus that had been previously linked to detection by MDA5 (6). The effect of LGP2 deficiency extends to innate cytokine responses triggered by cytosolic dsDNA and DNA-genome pathogens, which are impaired in cells lacking LGP2 (7). Experiments in LGP2-deficient cells also revealed a synergistic signal transduction activity resulting from co-expression of LGP2 with MDA5 (8, 9). This led to the suggestion that LGP2 may promote more efficient RNA detection by RLRs, possibly by inducing conformational alteration of viral RNAs to enable downstream recognition or by facilitating interaction between the RLRs and additional signaling machinery. Replacement of LGP2 with an ATP hydrolysis-inactivating helicase domain mutant results in a phenotypic copy of the LGP2 null mice, indicating that enzymatic activity is essential for LGP2 to function in the antiviral system (8). A positive antiviral role for LGP2 is also highlighted by the discovery that interferon antagonist proteins encoded by paramyxoviruses can target the LGP2 helicase domain, disrupting its ATP hydrolysis activity (10). Together, these findings are consistent with LGP2 acting as an upstream mediator of non-self RNA recognition and antiviral signaling.
ATP hydrolysis is critical for proper antiviral function of all three RLR proteins, but the role for energy utilization in their biological responses is poorly understood. Data suggesting ATP-dependent dsRNA unwinding has been reported for RIG-I (11, 12), but FRET assays failed to detect strand displacement (13); there are no reports of MDA5- or LGP2-mediated helicase activity. This uncertainty regarding the ability of RLRs to function as true RNA modifying proteins evokes a question about the exact role(s) for ATP hydrolysis in their biological functions.
A single-molecule study has demonstrated that purified RIG-I is able to translocate along RNA substrates (13). RIG-I translocation requires ATP, and the rate is increased when the dsRNA substrate contains a 5′-triphosphate (5′-PPP) modification. The ability of RIG-I to translocate along dsRNA without unwinding the duplex has been suggested to be an exclusive function of RLR proteins (14). However, the generality of dsRNA translocation as a property of MDA5 and LGP2, and the important question of the biological significance of RNA translocation in RLR-mediated antiviral signaling have not yet been addressed.
In this study biochemical and biophysical approaches were used to conduct a quantitative evaluation of the enzymatic and RNA binding properties of LGP2. The results reveal that LGP2 possesses high basal ATP hydrolysis activity that can be mutagenically distinguished from dsRNA-stimulated ATP hydrolysis. Moreover, experimental evidence demonstrates that LGP2 utilizes this basal ATP hydrolysis to enhance and diversify its RNA recognition capacity. The consequence of this ATP-dependent phenomenon is revealed in antiviral signaling assays demonstrating that synergistic activation of MDA5-mediated IFNβ production by LGP2 requires ATP hydrolysis. These findings define a previously unrecognized ATP-dependent RNA sensing property of LGP2 and provide a mechanistic basis for its upstream antiviral signal transduction activity.
EXPERIMENTAL PROCEDURES
Plasmids and Recombinant Baculovirus
FLAG-tagged RIG-I and RIG-I-C cDNA was cloned into the baculovirus transfer vector pBac2cp (Novagen). FLAG-tagged LGP2 cDNA was cloned into the pVL-1393 transfer vector (BD Biosciences). Site-directed mutagenesis was carried out using QuikChangeXL mutagenesis (Stratagene). Mutations introduced to generate the LGP2 motif mutants were confirmed by DNA sequencing. The baculovirus transfer vector containing the cDNA of interest was co-transfected with linearized baculovirus DNA (BD Biosciences) into Sf9 insect cells grown in Sf900-II (Invitrogen) supplemented with 1% penicillin/streptomycin (Invitrogen) and 10% fetal bovine serum (Invitrogen). Homologous recombination generated a baculovirus expressing RIG-I, RIG-I-C, LGP2, or LGP2 mutant proteins. These baculoviruses were amplified in Sf9 cells three times, for 3 days each, reaching a titer of ∼1 × 108 pfu/ml quantified by plaque assay. Virus stocks were stored in the dark at 4 °C.
Protein Expression and Purification
Sf9 cells were infected with recombinant baculovirus (1 pfu/cell) for 4 days, then cell lysates were prepared in whole cell extract buffer (50 mm Tris, pH 8.0, 280 mm NaCl, 0.5% IGEPAL, 0.2 mm EDTA, 10% glycerol, 1 mm DTT) supplemented with protease inhibitor mixture (Roche Applied Science) and 200 mm Na2VO3. Lysates were pre-cleared with Sepharose 6B beads (Sigma), and the FLAG-tagged proteins were then immunoaffinity purified with anti-FLAG M2 affinity beads (Sigma). Immunoprecipitated proteins were eluted with 150 μg/ml of 3× FLAG peptide, and quantified by Bradford assay (27). The purity of the proteins was assessed by SDS-PAGE and Coomassie Blue staining. Spectrophotometric measurement of the 260:280 nm absorbance ratios revealed no nucleic acid contamination with values ranging from 0.47 to 0.65.
ATP Hydrolysis Assays
ATP hydrolysis assays were completed using the Enz-Chek Phosphate Assay Kit (Molecular Probes) according to the manufacturer's protocol with 100 nm protein concentrations and 500 μm ATP, unless otherwise noted. The ATP hydrolysis rate is calculated based on the slope of a standard curve generated in parallel for each assay and is expressed in nanomoles of ATP hydrolyzed per minute per milligram of protein (nmol of ATP min−1 mg−1). The nonhydrolyzable ATP analogue AMP-PNP was from Sigma (A2647), and ADP-AlF4 was made by incubating 10 mm ADP (Sigma A2754), 50 mm NaF (Sigma S7920), and 10 mm AlCl3 (Sigma 563919). Vmax, Km, and kcat were calculated according to standard Michaelis-Menten enzyme kinetics, using the initial rates of ATP hydrolysis for the indicated protein at ATP concentrations between 50 and 1000 μm. All ATP hydrolysis assays completed to obtain enzymatic efficiency were conducted using 150 nm protein concentration.
Flow Cell Preparation and Single Molecule Microscopy
Flow cells for single molecule observations were created by coating a microscope slide with polyethylene glycol (PEG, Mr ≈ 5000, Laysan Bio) and a coverslip with 98% PEG and 2% PEG-biotin (Laysan Bio). The coverslip and slide are assembled to create individual chambers incubated with neutravidin followed by the biotinylated dsRNA substrate. RNA substrates were imaged in T-50 buffer (10 mm Tris-HCl, pH 7.5, 3 mm MgCl2, and 50 mm NaCl) containing an oxygen scavenging system (1 mg/ml of glucose oxidase (Sigma), 0.04 mg/ml of catalase (Sigma), and 0.4% dextrose) to reduce photobleaching, and 1% β-mercaptoethanol (Acros) serving as a triplet state quencher. The prepared flow cell was placed on a heated objective at 37 °C for imaging using a Olympus IX81 TIRF microscope (×100/1.45 NA objective), ImageEM EMCCD camera (Hammamatsu), and 561 nm (75 milliwatts) laser (Melles Griot). Image acquisition was done using Slidebook software (Olympus) capturing one frame per 100 ms for 2 min. Protein/ATP was diluted as indicated in imaging buffer and added to the flow cell immediately before image capture.
RNA Substrates
All RNA substrates were ordered from Integrated DNA Technology (IDT) modified with a 3′ Cy3 fluorophore, or a 3′ biotin tag. The fluorescently labeled strand (RNA 3′-Cy3) was used for all substrates. The biotinylated strand was either completely complementary or contained additional nucleotides to create the series of bulged substrates depicted in Fig. 3. Each single molecule experiment with LGP2 ± ATP and the imperfect dsRNA substrates was done in parallel with the standard dsRNA substrate within different chambers of the same flow cell. This enables all four conditions to be tested (bulged dsRNA with 80 nm LGP2, bulged dsRNA with 80 nm LGP2 and 500 μm ATP, dsRNA with 80 nm LGP2, and dsRNA with 80 nm LGP2 and 500 μm ATP) with minimized concern for effects due to batch-to-batch variability in flow cell preparation. The RNA sequences used to create all duplexes were: RNA 3′-Cy3, 5′-GCAGAGGGUGGCGCUCCCGACAAGC-3′ Cy3; dsRNA, biotin-3′CGUCUCCCACCGCGAGGGCUGUUCG-5′; 3 top bulge, biotin-3′-CGUCUCCCACCGCGAGUAGGGCUGUUCG-5′; 3 bottom bulge, biotin-3′-CGUCUCCUAGCACCGCGAGGGCUGUUCG-5′; 4 bulge, biotin-3′-CGUCUCCCACCGCGAGUGAGGGCUGUUCG-5′; 5 bulge, biotin-3′-CGUCUCCCACCGCGAGUGUAGGGCUGUUCG-5′; 6 bulge, biotin-3′-CGUCUCCCACCGCGAGUGGUAGGGCUGUUCG-5′; 7 bulge, biotin-3′-CGUCUCCCACCGCGAGUGAGUAGGGCUGUUCG-5′; double bulge, biotin-3′-CGUCUCCCACCUGAGCGAGGGUAGCUGUUCG-5′;triple bulge, biotin-3′-CGUCUCCUGACACCGCUGAGAGGGCUAGUGUUCG-5′.
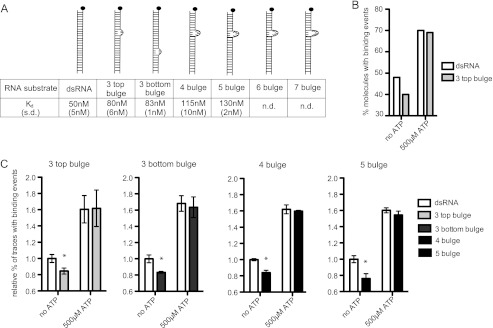
FIGURE 3.
ATP enables LGP2 to associate with diverse imperfect dsRNAs. A, diagram illustrates the standard dsRNA and a series of increasingly noncomplementary dsRNA substrates tested. The Kd measured in the absence of ATP at 80 nm protein concentration is indicated below with S.D. in parentheses. B, representative experimental data demonstrating the percent of dsRNA molecules with binding events at 80 nm LGP2 in the absence and presence of 500 μm ATP. The dsRNA substrate and 3 top bulge substrate were evaluated in parallel chambers of the same flow cell. C, analysis of ATP-enhanced RNA recognition by 80 nm LGP2 in the presence of 500 μm ATP. Bulged substrate recognition is greatly enhanced by ATP. For each experiment, the percentage of dsRNA molecules recognized by LGP2 in the absence of ATP is set equal to 1 for comparison. Fewer bulged substrates are recognized in the absence of ATP, but in the presence of ATP similar numbers of bulged and perfectly complementary substrates are bound by LGP2. * = p < 0.05.
Data Analysis
Following data acquisition, a custom written spot-identifying computer program was used to select fluorescence “spots” (i.e. RNA substrates) based on their individual fluorescence intensity, and their separation from other fluorophores (supplemental Fig. S1a). This program then generates fluorescence intensity time trajectories (“traces”) for individual molecules and classifies fluorescence intensities as a function of time as level 1, RNA alone; level 2, RNA with protein bound; or level 0, bleached fluorophore. The accuracy of this spot-identifying program was verified by manual classification of binding and unbinding events (supplemental Fig. S1). The results of this first analysis are processed by a second custom-written program to determine the percent of total traces that have at least one binding event, as well as the number of binding events per trace.
A third custom-written program was then used to histogram the dwell times in the unbound or bound states before a binding or unbinding event occurs, respectively (supplemental Fig. S1c). These histograms are fit to exponential decay forms y = y0 e−kx, determining k as on or off rates. Kd values are calculated in the absence of ATP using these rates where Kd = (koff/kon)[protein]. All Kd measurements were taken at 80 nm protein concentration, unless otherwise indicated.
Luciferase Reporter Gene Assays
Human embryonic kidney cell line HEK293T were seeded in 24-well tissue culture plates, and cotransfected using Lipofectamine 2000 (Invitrogen) with the reporter gene and expression vectors for Renilla luciferase along with a plasmid containing the indicated helicase. The reporter gene contains the firefly luciferase open reading frame under the control of the −110 IFNβ promoter. The concentration of MDA5 plasmid was held constant at 50 ng/well, and the amount of LGP2 or LGP2 mutant plasmid was varied at 5, 10, and 50 ng/well. The following day cells were infected with EMCV Mengo strain at multiplicity of infection = 3, or transfected with 5 μg/ml of poly(I:C) using Lipofectamine 2000 for 8 h before harvesting. Relative luciferase activity was measured using Dual LuciferaseTM reporter assay (Promega). Data are plotted as the average of triplicate samples with error bars representing S.D. The data presented are representative of three independent experiments.
RESULTS
LGP2 Has Both Basal and dsRNA-activated ATP Hydrolysis Activity
The ATP hydrolysis kinetics were quantitatively compared for both LGP2 and RIG-I (Fig. 1A). RIG-I exhibits very low ATP hydrolysis activity in the absence of dsRNA, and is activated by the addition of poly(I:C), reaching an ATP hydrolysis rate of 1,173 ± 175 nmol of ATP min−1 mg−1 (Table 1). Addition of poly(I:C) stimulates the enzyme efficiency of RIG-I (kcat/Km) from 0.08 ± 0.1 to 0.64 ± 0.6 min−1 μm−1 (Table 2). In contrast to the dsRNA-inducible enzymatic activity of RIG-I, LGP2 exhibits a high ATP hydrolysis activity even in the absence of poly(I:C), with a basal rate of 393 ± 48 nmol of ATP min−1 mg−1, and a kcat/Km of 0.27 ± 0.03 min−1 μm−1. The addition of poly(I:C) increases the rate of LGP2 ATP hydrolysis to 766 ± 31 nmol of ATP min−1 mg−1, whereas the kcat/Km does not change significantly at 0.23 ± 0.02 min−1 μm−1 (Tables 1 and 2). This indicates that whereas the catalytic efficiency of RIG-I is greatly enhanced by poly(I:C) stimulation, the efficiency of LGP2 is similar in the presence and absence of poly(I:C). This analysis also defines two enzymatic activities of LGP2: basal dsRNA-independent ATP hydrolysis and dsRNA-stimulated ATP hydrolysis.
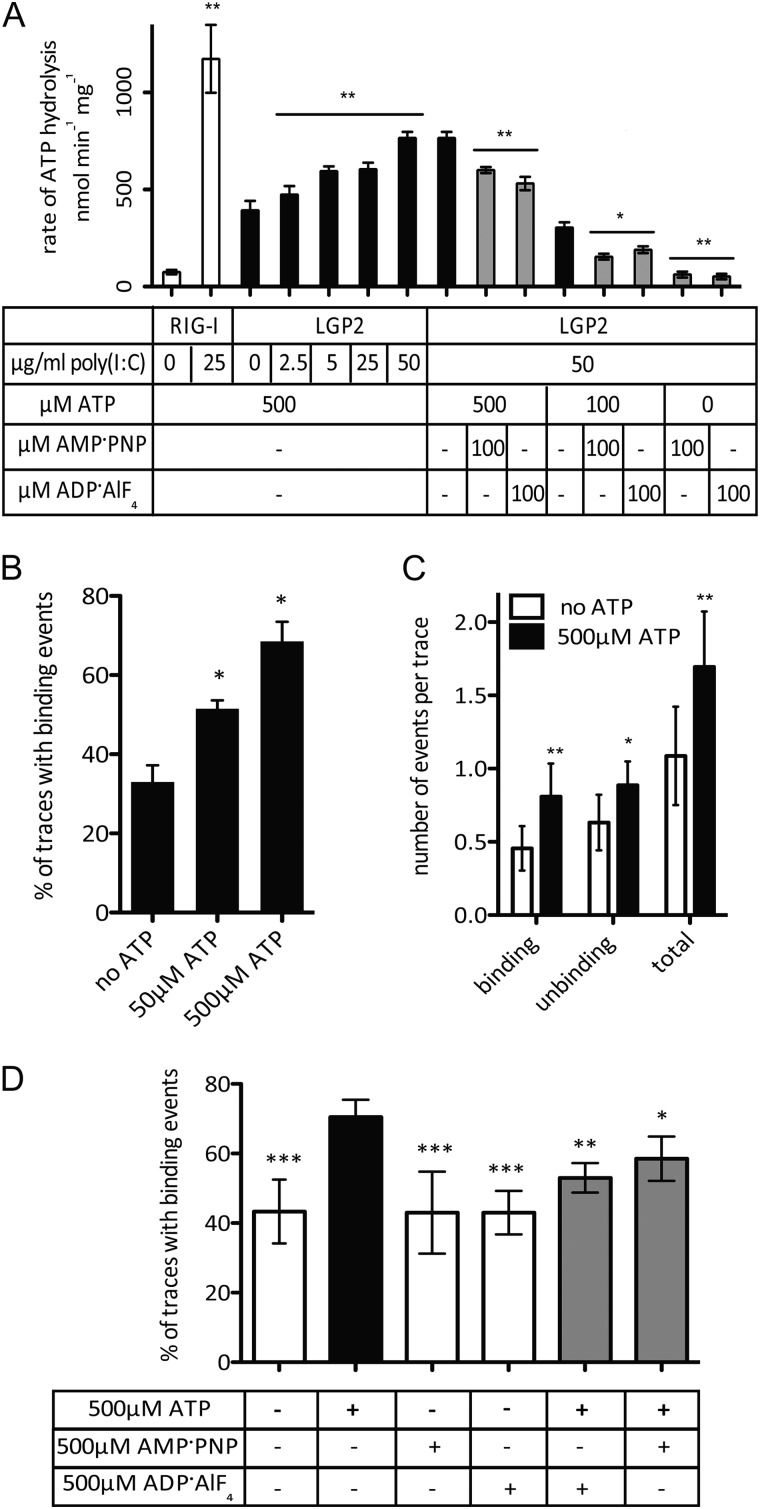
FIGURE 1.
ATP enhances LGP2 interaction with dsRNA. A, the ATP hydrolysis assay demonstrates dsRNA-induced activity for RIG-I (white bars), but basal and dsRNA-induced activity for LGP2 at varying concentrations of poly(I:C) (black bars). The addition of the nonhydrolyzable analogues AMP-PNP and ADP-AlF4 to the assay inhibits the ATP hydrolysis activity of LGP2 (gray bars). * = p < 0.05. B, quantitation of dsRNA molecules bound by LGP2 (80 nm) in the absence and presence of 50 or 500 μm ATP. ATP increases the percentage of dsRNA substrates (traces) that exhibit binding by LGP2. * = p < 0.05; ** = p < 0.008. C, quantification of the number of binding and unbinding events by LGP2 per dsRNA molecule (trace) in the absence and presence of 500 μm ATP over a 120-s (1200 frame) image capture. ATP increases the number of events per RNA molecule. * = p < 0.05; ** = p < 0.02; *** = p < 0.0003. D, percent of dsRNA molecules bound by LGP2 (80 nm) in the absence and presence of ATP (black bar), AMP-PNP, ADP-AlF4, (white bars) or equimolar mixtures of ATP and analogue (gray bars). Values presented are the average of at least two independent experiments, and error bars represent S.D. * = p < 0.02; ** = p < 0.003; *** = p < 0.0003.
TABLE 1.
Rates of ATP hydrolysis by LGP2 and RIG-I
| Protein | Poly(I:C) | ATP | Non-hydrolyzable ATP analogue | Rate of ATP hydrolysis (±S.D.)a |
|---|---|---|---|---|
| LGP2 | 0 μg/ml | 500 μm | 393 ± 48 nmol min−1 mg−1 | |
| 2.5 μg/ml | 474 ± 44 nmol min−1 mg−1 | |||
| 5 μg/ml | 592 ± 25 nmol min−1 mg−1 | |||
| 25 μg/ml | 605 ± 33 nmol min−1 mg−1 | |||
| 50 μg/ml | 766 ± 31 nmol min−1 mg−1 | |||
| LGP2 | 50 μg/ml | 500 μm | 100 μm AMP-PNP | 600 ± 16 nmol min−1 mg−1 |
| 100 μm ADP-AIF4 | 531 ± 34 nmol min−1 mg−1 | |||
| LGP2 | 50 μg/ml | 100 μm | 304 ± 27 nmol min−1 mg−1 | |
| 100 μm AMP-PNP | 154 ± 15 nmol min−1 mg−1 | |||
| 100 μm ADP-AIF4 | 190 ± 17 nmol min−1 mg−1 | |||
| LGP2 | 50 μg/ml | - | 100 μm AMP-PNP | 65 ± 15 nmol min−1 mg−1 |
| - | 100 μm ADP-AIF4 | 52 ± 14 nmol min−1 mg−1 | ||
| RIG-I | 0 μg/ml | 500 μm | 75 ± 11 nmol min−1 mg−1 | |
| 25 μg/ml | 1173 ± 175 nmol min−1 mg−1 | |||
| No protein | 0 μg/ml | 500 μm | 41 ± 13 nmol min−1 mg−1 | |
| 5 μg/ml | 38 ± 15 nmol min−1 mg−1 |
a Values are the average of at least two independent experiments shown ±S.D.
TABLE 2.
Enzymatic properties of RLRs and mutant proteins
| Substratea | Vmaxb | Km | kcat | kcat/Km | |
|---|---|---|---|---|---|
| μm/min | μm | min−1 | min−1 μm−1 | ||
| LGP2 | 3.2 ± 0.5 | 78 ± 6.0 | 21.5 ± 2.1 | 0.27 ± 0.03 | |
| Poly(I:C) | 5.6 ± 0.6 | 160 ± 8.5 | 37.4 ± 3.7 | 0.23 ± 0.02 | |
| LGP2 MIIa | - | NAc | NA | NA | NA |
| Poly(I:C) | 6 ± 0.5 | 145 ± 4.3 | 40 ± 2.2 | 0.27 ± 0.03 | |
| LGP2 K651E | - | NA | NA | NA | NA |
| Poly(I:C) | 3.5 ± 0.5 | 155 ± 7.2 | 23.7 ± 2.1 | 0.15 ± 0.04 | |
| RIG-I | - | 1.5 ± 0.4 | 121 ± 9.0 | 10.1 ± 1.9 | 0.08 ± 0.01 |
| Poly(I:C) | 18 ± 0.9 | 187 ± 8.2 | 120 ± 4.8 | 0.64 ± 0.06 | |
| RIG-I-C | - | 7.8 ± 0.7 | 102 ± 6.6 | 52 ± 4.1 | 0.51 ± 0.05 |
| Poly(I:C) | 14 ± 1.0 | 235 ± 9.4 | 94 ± 5.0 | 0.40 ± 0.06 |
a Poly(I:C) was used at 50 μg/ml.
b Values are the average of at least two independent experiments shown ±S.D.
c NA, not applicable. LGP2 MIIa and LGP2 K651E did not display significant basal ATP hydrolysis (−) above the background level for the no protein control, so measurements of catalytic efficiency are not applicable.
Single Molecule Analysis of LGP2 RNA Binding
A single molecule imaging assay was used to quantitatively evaluate the RNA binding properties of LGP2 (13). Fluorescently labeled dsRNA substrates are tethered to a glass slide and incubated with purified RLR proteins. Protein binding increases the fluorescence intensity of the bound dsRNA molecule via a phenomenon referred to as protein-induced fluorescence enhancement, or PIFE (13, 15). RNA association, dissociation, and translocation can be observed and measured using TIRF microscopy to monitor the intensity of a single fluorophore over time. The data output consists of individual traces corresponding to the fluorescence intensity of a single RNA molecule over time (supplemental Figs. S1 and S2). Measurements were made in parallel chambers of a dsRNA-coated slide to minimize effects due to variability in slide preparation. The interactions of an RLR with RNA substrates can thereby be analyzed with or without ATP, at different concentrations, or under distinct buffer conditions.
Analysis of these experiments revealed two apparent RNA populations. One population of RNA molecules featured LGP2 binding and unbinding, whereas the other did not exhibit binding by LGP2. The population of RNA molecules that interacted with LGP2 displayed highly reproducible dwell times in the bound or unbound state. Due to their consistent behavior, only traces containing at least one binding or unbinding event were used to calculate binding kinetics, whereas the non-interacting population was not included in the calculation. From the bound RNAs, histograms of the dwell times in the bound or unbound state were generated. These histograms were then fit to the exponential decay form y = y0 e−kx, determining k as an on rate for histograms of unbound dwell times, or off rate for histograms of bound dwell times. These rates were then used to calculate the Kd as (koff/kon) × [protein] (supplemental Fig. S1).
As a positive control, the binding and translocation of a RIG-I protein lacking the N-terminal CARDs (RIG-I-C) was evaluated. In the absence of ATP, RIG-I-C bound to the 25-bp dsRNA with a Kd of 98 ± 13 nm, similar to the previously reported 76.3 nm (13). In the presence of ATP RIG-I-C translocation was observed with an average period (Δt) of 4.0 s at 50 μm ATP in agreement with the reported Δt of 3.7 s at 60 μm ATP (13) (supplemental Fig. S2).
In the absence of ATP, LGP2 was calculated to have a Kd of 50 ± 5 nm (Table 3, data analysis as described in supplemental Fig. S1), but in the presence of ATP, RNA translocation by LGP2 was not observed under conditions permissive for RIG-I-C translocation. RNA binding experiments using various LGP2 preparations, ATP concentrations, or buffer conditions did not reveal LGP2 translocation. Instead, a different behavior of LGP2 was observed in single-molecule analysis in the presence of ATP. Analysis of 80 nm LGP2 protein without ATP resulted in only 30 ± 3% of all dsRNAs with at least one binding event, leaving the majority of RNA molecules in the unbound population. In parallel analysis, 50 μm ATP combined with 80 nm LGP2 increased the number of LGP2-associated dsRNAs to 51.5 ± 2%, whereas 500 μm ATP resulted in 69 ± 3.5% of RNA molecules bound by LGP2 (Fig. 1b, n = 3). Higher concentrations of ATP did not result in a significant increase in the fraction of RNA molecules able to be recognized by LGP2. To rule out effects resulting from batch-to-batch variation in flow cell preparation, adjacent chambers of the same flow cell were used to compare samples with and without ATP side-by-side. Over the course of eight separate experiments with 80 nm LGP2, the presence of 500 μm ATP resulted in an average 30 ± 7% increase in the number of RNA molecules able to be bound, compared with parallel experiments conducted in the absence of ATP.
TABLE 3.
RNA binding and ATP hydrolysis rates of LGP2 and mutants
| Protein | Kd with dsRNA ± S.D.a | Basal ATP hydrolysis (0 μg/ml) of poly(I:C) ± S.D.b | dsRNA-stimulated ATP hydrolysis (5 μg/ml) of poly(I:C) ± S.D. |
|---|---|---|---|
| LGP2 | 50 ± 5 nm | 393 ± 48 nmol min−1 mg−1 | 592 ± 25 nmol min−1 mg−1 |
| LGP2 MI | 57 ± 11 nnm | 48 ± 17 nmol min−1 mg−1 | 55 ± 11 nmol min−1 mg−1 |
| LGP2 MIIa | 335 ± 70 nnm | 33 ± 10 nmol min−1 mg−1 | 530 ± 33 nmol min−1 mg−1 |
| LGP2 MIII | NDc | 62 ± 13 nmol min−1 mg−1 | 37 ± 9 nmol min−1 mg−1 |
| LGP2 K651E | 462 ± 28 nnm | 41 ± 12 nmol min−1 mg−1 | 543 ± 20 nmol min−1 mg−1 |
| LGP2 N461I | 53 ± 8 nnm | 50 ± 7 nmol min−1 mg−1 | 43 ± 14 nmol min−1 mg−1 |
| No protein | ND | 41 ± 13 nmol min−1 mg−1 | 38 ± 15 nmol min−1 mg−1 |
a Values are the average of at least two independent experiments shown ±S.D.
b All experiments were conducted at 500 μM ATP.
c ND, not detected.
In one representative experiment, in the absence of ATP 1,005 total traces were analyzed from five separate fields of view and only 374 (37%) had observable binding events, but in the presence of 500 μm ATP 926 total analyzed traces contained 646 with observable binding events (70%). Moreover, the presence of ATP resulted in more robust and dynamic LGP2-dsRNA interactions, increasing the number of binding events per molecule from 1.1 ± 0.3 to 1.7 ± 0.34 events per 120-s (1200 frame) trace (Fig. 1c). These results indicate that ATP dramatically enhances the ability of LGP2 to recognize RNA.
ATP Hydrolysis Is Required for Enhanced RNA Recognition by LGP2
Nonhydrolyzable ATP analogues were used in both single molecule and biochemical assays to determine whether the enhanced recognition requires ATP hydrolysis. Both ADP-AlF4 and AMP-PNP reduced the rate of ATP hydrolysis when used in a competition experiment (Fig. 1d, Table 1). When added to 100 μm ATP, 100 μm AMP-PNP decreased the rate of ATP hydrolysis from 304 ± 27 to 154 ± 15 nmol of ATP min−1 mg−1, and 100 μm ADP-AlF4 decreased the rate to 190 ± 17 nmol of ATP min−1 mg−1 (Table 1). In single molecule analysis, neither nonhydrolyzable analogue was sufficient to enhance LGP2 dsRNA binding, and the addition of equimolar ATP and ATP analogues decreased the level of enhanced RNA recognition accordingly (Fig. 1d). These results demonstrate that the energy from ATP hydrolysis is required to enhance LGP2 interactions with dsRNA.
Mutagenesis of LGP2 Distinguishes Basal and dsRNA-stimulated ATP Hydrolysis
To further investigate the relationship between enzymatic activity and RNA binding, LGP2 mutations were designed to target key amino acids involved in ATP binding, ATP hydrolysis, or RNA binding (Fig. 2A). The LGP2 mutant MI targets the Walker A motif and lacks ATP hydrolysis but retains RNA binding (16–19). Mutant MIII is defective for both ATP hydrolysis and RNA binding (16). Three other LGP2 mutations were designed to target residues that available structural information indicated might be important for biological activity (20–23). LGP2 mutant MIIa targets a lysine and a tyrosine (K138E, Y142F) conserved in RIG-I and LGP2 motif IIa (21–23). LGP2 mutant N461I targets a conserved asparagine that mediates dsRNA binding interactions in the RIG-I crystal structure (23). LGP2 mutant K651E was based on the observation that mutation of LGP2 lysine 651 to glutamic acid completely disrupts RNA recognition by the isolated C-terminal RD (20).
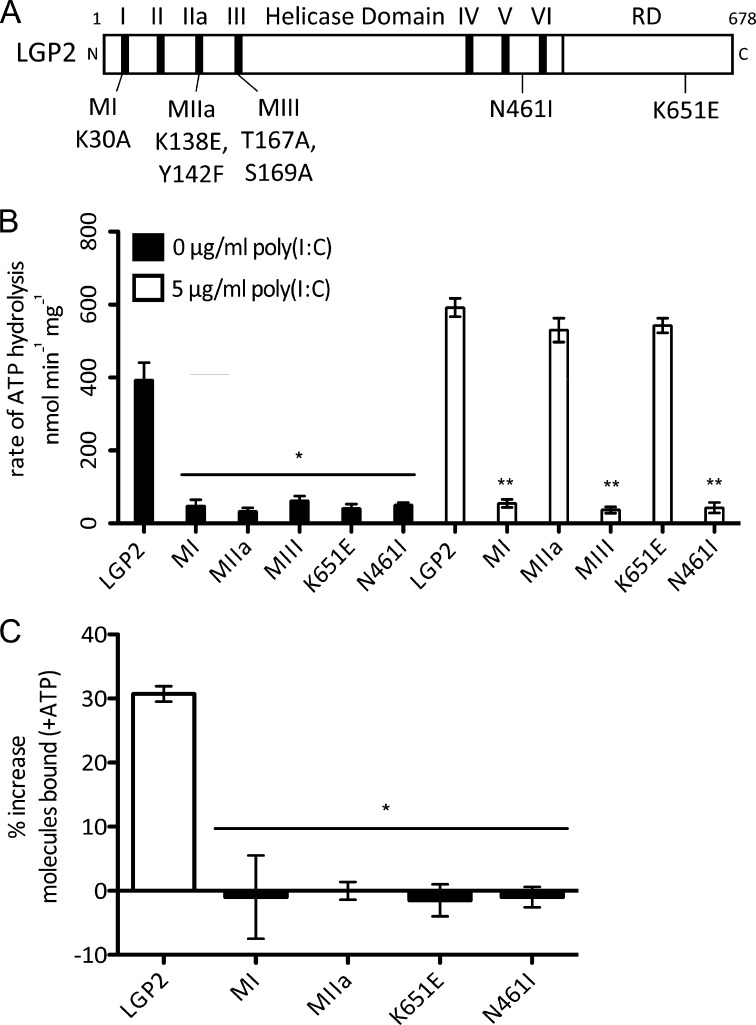
FIGURE 2.
LGP2 basal ATP hydrolysis, but not dsRNA-stimulated ATP hydrolysis, is required for enhanced dsRNA recognition. A, diagram representing the domain structure of LGP2 and illustrating mutations. B, rate of ATP hydrolysis by LGP2 and mutants in the absence (black bars) or presence of 5 μg/ml of poly(I:C) (white bars). All mutants lack basal ATP hydrolysis activity, however, the presence of poly(I:C) stimulates LGP2 MIIa and K651E to hydrolyze ATP near wild type levels. * = p < 0.003; ** = p < 0.0001. C, analysis of ATP-enhanced RNA recognition by LGP2 and mutants, represented as the percent increase in the number of dsRNA molecules with binding events after addition of 500 μm ATP, compared with protein without ATP added. LGP2 displays an average 30% increase in the number of molecules bound with the addition of ATP, but no mutants display increased recognition in the presence of ATP. * = p < 0.003.
The enzymatic activity and RNA binding properties of these five mutants were measured and compared with wild type LGP2 (Fig. 2, Table 3). In agreement with previous observations (16), LGP2 mutant MI was defective for ATP hydrolysis but bound dsRNA with an affinity similar to wild type, whereas mutant MIII was defective for both ATP hydrolysis and RNA binding. Both mutant MIIa and K651E displayed measurable but decreased dsRNA affinity (335 ± 70 and 462 ± 28 nm, respectively). LGP2 mutant N461I was able to bind dsRNA similarly to wild type (Kd = 53 ± 8 nm).
Analysis of the ATP hydrolysis activities revealed that none of the mutants retained basal ATP hydrolysis (Fig. 2B, black bars), but addition of poly(I:C) stimulated ATP hydrolysis by two of the LGP2 mutants to nearly wild type levels (MIIa to 530 ± 33 nmol of ATP min−1 mg−1 and K651E to 543 ± 20 nmol of ATP min−1 mg−1; see Fig. 2B and Table 3). The addition of poly(I:C) also significantly increased their catalytic efficiency (Table 2). These mutant proteins display only dsRNA-stimulated ATP hydrolysis and not basal ATP hydrolysis, providing tools to investigate the importance of basal ATP hydrolysis in LGP2 RNA binding.
Basal ATP Hydrolysis Enhances LGP2 RNA Binding
Single-molecule experiments were performed to test the LGP2 mutants' RNA binding properties in the absence or presence of ATP (Fig. 2c). In these experiments 500 μm ATP induced a 30% average increase in the number of molecules bound by wild type LGP2, but all of the mutants showed no increased RNA recognition in the presence of ATP, irrespective of their ability to hydrolyze ATP. Although LGP2 mutants MIIa and K651E retained their dsRNA-stimulated ATP hydrolysis activity, they failed to exhibit ATP-enhanced RNA binding (Fig. 2C). These results indicate that basal, rather than dsRNA-stimulated, enzymatic activity is required for enhanced RNA recognition by LGP2.
ATP Increases Substrate Recognition Capacity of LGP2
Although the role of LGP2 in antiviral defense is not completely resolved, studies of LGP2-deficient mice have been interpreted to indicate it may function upstream of MDA5 and RIG-I to remodel RNA or enhance RNA recognition (8, 10, 24). Analysis of the single-molecule RNA binding data suggested that ATP hydrolysis may allow LGP2 increased access to heterogeneous dsRNA species within the population. To directly test this hypothesis, a series of imperfect dsRNA substrates were designed that contain 3–7 nucleotide noncomplementary bulges at defined positions (Fig. 3A). These bulged RNAs are progressively poorer substrates for LGP2 recognition when compared with the standard complementary dsRNA (Fig. 3A). The RNAs with the greatest modification (6 bulge, 7 bulge) displayed no detectable binding above background (Fig. 3A, ND).
When these modified substrates were tested in the presence and absence of ATP, the observed effect of ATP hydrolysis-mediated enhancement of RNA recognition by LGP2 was even more pronounced. A single example from a representative experiment with the RNA substrate referred to as the “3 top bulge” is displayed in Fig. 3B. In the absence of ATP, significantly fewer of the modified substrates are recognized by LGP2 in comparison to the standard dsRNA substrate. However, in the presence of ATP the percentage of modified substrates recognized by LGP2 is almost identical to the observed recognition of the standard dsRNA substrates.
To compare the ATP-dependent increase in the number of dsRNA molecules recognized without concern for slight differences in flow cell preparation or general experimental variation, each bulged substrate was analyzed in individual chambers of the same flow cell with or without ATP in parallel with the dsRNA substrate. For each individual experiment the percent of standard dsRNA molecules recognized by LGP2 in the absence of ATP is set equal to 1 (Fig. 3C). Each bulged substrate displays reduced recognition by LGP2 in the absence of ATP, producing values lower than 1. In the presence of ATP, LGP2 recognizes the bulge-modified substrates and the complementary dsRNA substrates similarly. These results indicate that ATP hydrolysis increases LGP2 interactions with intrinsically poor dsRNA substrates, and accommodates heterogeneity that is present in a population of dsRNAs.
LGP2 Requires ATP Hydrolysis to Potentiate MDA5-mediated IFNβ Production
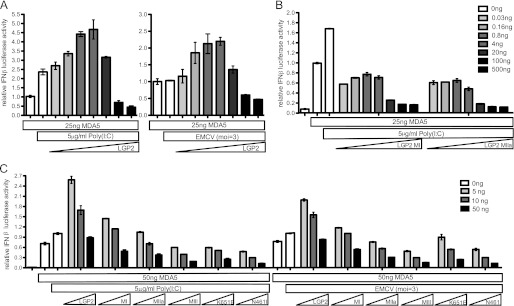
Previous studies have suggested that LGP2 may work in conjunction with MDA5 to mediate antiviral signaling and IFNβ production (8, 24). The single-molecule experiments presented here indicate that LGP2 uses the energy from ATP hydrolysis to increase its interactions with a more diverse range of dsRNA substrates. The biological impact of this ATP-dependent RNA recognition was tested in an antiviral signaling assay in which LGP2 or mutant LGP2 was titrated into MDA5-dependent IFNβ promoter transcription assays. MDA5 is constitutively active in this assay and can activate the IFNβ promoter when expressed, irrespective of infection with encephalomyocarditis virus (EMCV) or transfection with poly(I:C) (Fig. 4) (16, 25). Introducing low levels of wild type LGP2 is synergistic with MDA5 and results in a 2-fold increase in IFNβ promoter activity (Fig. 4A and supplemental Fig. S3). Raising the level of LGP2 expression reduces the MDA5-dependent IFNβ promoter activity, consistent with the reported feedback inhibition effect of LGP2 expression (3–5). To test if ATP hydrolysis is required for synergistic MDA5-mediated IFNβ promoter activity, two LGP2 mutants were co-transfected at a range of plasmid concentrations. LGP2 MI is defective for both basal and RNA-stimulated ATP hydrolysis, whereas LGP2 MIIa lacks basal ATP hydrolysis, but retains RNA-stimulated ATP hydrolysis. Unlike wild type LGP2, neither of these mutants is able to enhance MDA5-mediated signaling at low expression levels, but both display the inhibitory activity of LGP2 at higher expression levels (Fig. 4B and supplemental Fig. S3). Parallel analysis of all the LGP2 mutants defective for ATP hydrolysis verified that none of the mutant proteins were able to enhance MDA5-mediated signaling (Fig. 4C), demonstrating that intact ATP hydrolysis is critical for LGP2 to stimulate MDA5 signal transduction responses.
FIGURE 4.
LGP2 ATP hydrolysis is required for enhanced MDA5-mediated IFNβ signaling. A, HEK293T cells were transfected with a −110 IFNβ-luciferase reporter gene, control Renilla luciferase plasmid, and expression vectors for the indicated helicase proteins MDA5 or LGP2. MDA5 was transfected at a constant 25 ng of plasmid/well, whereas the amount of LGP2 transfected varied at 0.03, 0.16, 0.8, 4, 20, 100, and 500 ng. Following a 24-h transfection, cells were transfected with 5 μg/ml of poly(I:C) (left) or infected with 3 pfu/cell EMCV (right) for 8 h before harvesting. At low concentrations LGP2 enhances MDA5-mediated IFNβ signaling, but at higher concentrations LGP2 functions as a negative regulator. B, HEK293T cells were transfected with a −110 IFNβ-luciferase reporter gene, control Renilla luciferase plasmid, and expression vectors for the indicated helicase proteins MDA5, LGP2 MI, or LGP2 MIIa mutants. MDA5 was transfected at a constant 25 ng of plasmid/well, whereas the amount of LGP2 transfected varied at 0.03, 0.16, 0.8, 4, 20, 100, and 500 ng. Following a 24-h transfection, cells were transfected with 5 μg/ml of poly(I:C) for 8 h before harvesting. Neither LGP2 MI nor LGP2 MIIa are able to enhance MDA5-mediated signaling. C, HEK293T cells were transfected with a −110 IFNβ-luciferase reporter gene, control Renilla luciferase plasmid, and expression vectors for the indicated helicase proteins MDA5, LGP2, or LGP2 mutants. MDA5 was transfected at a constant 50 ng of plasmid/well, whereas the amount of LGP2 transfected was varied at 5 (white), 10 (gray), and 50 ng (black) of plasmid. Following a 24-h transfection, cells were infected with 3 pfu/cell EMCV or transfected with 5 μg/ml of poly(I:C) for 8 h before harvesting. None of the ATP hydrolysis defective mutants are able to enhance MDA5-mediated signaling.
DISCUSSION
The cytosolic innate pattern recognition receptors, RIG-I and MDA5, are thought to scan the cytoplasm for RNA signatures indicative of virus infection. Identification of non-self RNA triggers activation of downstream antiviral responses through protein interactions mediated by their CARD regions. Due to the absence of these signaling domains in LGP2, its role in upstream activation of antiviral signaling was not initially apparent, but analysis of mice lacking LGP2 expression indicated that LGP2 was required for the optimal detection of and response to RNA virus infections in vivo (8, 9). The present studies demonstrate that LGP2 is capable of using energy derived from ATP hydrolysis to enhance its ability to associate with diverse dsRNA species, enabling it to act in concert with MDA5 to maximize antiviral signal transduction. These results provide a unique mechanistic basis for an ATP-dependent function of LGP2 in promoting signal transduction in response to intracellular virus infections.
All of the RLRs require ATP hydrolysis for their biological activity in the antiviral system. LGP2 uses basal ATP hydrolysis to enhance association with the dsRNA species but RIG-I, the archetypal RLR, does not hydrolyze ATP in the absence of dsRNA. When bound to dsRNA, RIG-I can use ATP to power translocation, presumably to scan the molecule for its preferred molecular signature, 5′-end triphosphorylation (13), which locks it in an active signaling configuration (26). The exact roles for ATP hydrolysis in MDA5 biological activity remain to be determined, but like RIG-I, it does not hydrolyze ATP in the absence of dsRNA (16). The results presented here demonstrate that LGP2 is distinct in its ability to hydrolyze ATP in the absence of dsRNA, and that dsRNA-independent basal ATP hydrolysis and dsRNA-dependent hydrolysis can be separated by site-directed mutagenesis. Moreover, analysis of these mutant proteins indicates that basal, but not dsRNA-dependent, ATP hydrolysis is required for enhanced recognition of dsRNA species by LGP2.
A role for LGP2 in the initial phase of RNA virus detection is consistent with reports that it is essential for responses to diverse pathogen-associated molecular patterns and pathogens including dsRNA and RNA viruses (8, 9), dsDNA and DNA viruses, and the intracellular bacterium, Listeria monocytogenes (7). Cellular responses to many virus types that have been linked previously to RIG-I, such as vesicular stomatitis virus and Sendai virus, were impaired in the absence of LGP2, and picornaviruses including EMCV, poliovirus, and Mengo virus, were highly sensitive to LGP2 deficiency (8). This led to the conclusion that LGP2 may function upstream of other RLRs to facilitate pattern recognition and subsequent antiviral signal transduction (8, 10, 24). The observed ATP-enhanced dsRNA interaction mediated by LGP2 provides a biochemical means to explain the effects of LGP2 on RLR RNA recognition. This is supported by the inability of ATP hydrolysis-defective mutants to enhance MDA5-mediated IFNβ gene expression. This phenomenon also accounts for the fact that an ATPase-defective LGP2 protein fails to compensate for LGP2 deficiency in mice (8). Furthermore, naturally occurring paramyxovirus-encoded immune evasion proteins target both MDA5 and LGP2, but not RIG-I. These LGP2 inhibitors bind to the helicase domain and interfere specifically with ATP hydrolysis activity (10), underscoring the importance of enzymatic activity in LGP2 function.
Clearly ATP hydrolysis by LGP2 is required for antiviral responses in vivo, but this does not exclude the possibility of a feedback inhibitory role for LGP2 that is regulated by its intracellular concentration. At low levels, LGP2 was found to be a positive regulator of IFNβ signaling, and this is consistent with observed low levels of LGP2 protein in mouse and human cells and tissues at steady state. As the concentration of LGP2 is increased, its inhibitory properties are revealed, consistent with increased LGP2 abundance in response to virus infections. Notably, LGP2 mutants with defective ATP hydrolysis display only the inhibitory properties, even at the lowest concentrations tested. These observations may resolve the seemingly contradictory properties of LGP2 by revealing a concentration-regulated dual role for LGP2 in antiviral signaling.
In summary, these data indicate that LGP2 uses intrinsic RNA-independent ATP hydrolysis activity to increase its RNA recognition repertoire and enhance cooperative activation of RLR signaling. The exact nature of this cooperation is currently a matter of speculation (24), but may involve several mechanisms including but not limited to allowing more efficient engagement of RLRs with diverse substrates that are present during virus infections, alteration of RNA secondary and tertiary structures, displacement of proteins from ribonucleoprotein complexes, or alteration in the subcellular localization of RNAs and RLRs. It is predicted that the ATP hydrolysis-mediated enhanced RNA recognition by LGP2 will form a common basis for functions of this RLR protein in mediating antiviral defenses to specific pathogens in distinct physiological or cellular contexts in vivo.
Supplementary Material
Acknowledgments
We are grateful to members of the Horvath and Marko laboratories for critical comments and helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants R01AI073919 and U01AI082984 (to C. M. H.) and 1U54CA143869–01 (NU-PS-OC) and National Science Foundation Grant MCB-1022117 (to J. M.), and an Immune Mechanisms of Virus Control Pilot Award (to C. M. H. and S. M.).

This article contains supplemental Figs. S1–S4.
- RIG-I
- RIG-I-like receptor
- RD
- regulatory domain
- RLR
- RIG-I-like receptor
- EMCV
- encephalomyocarditis virus
- LGP2
- laboratory of genetics and physiology 2
- AMP-PNP
- adenosine 5′-(β,γ-imino)triphosphate
- CARD
- caspase activation and recruitment domain.
REFERENCES
- 1. Yoneyama M., Fujita T. (2009) RNA recognition and signal transduction by RIG-I-like receptors. Immunol. Rev. 227, 54–65 [DOI] [PubMed] [Google Scholar]
- 2. Bruns A. M., Horvath C. M. (2012) Activation of RIG-I-like receptor signal transduction. Crit. Rev. Biochem. Mol. Biol. 47, 194–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Saito T., Hirai R., Loo Y. M., Owen D., Johnson C. L., Sinha S. C., Akira S., Fujita T., Gale M., Jr. (2007) Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc. Natl. Acad. Sci. U.S.A. 104, 582–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Komuro A., Horvath C. M. (2006) RNA- and virus-independent inhibition of antiviral signaling by RNA helicase LGP2. J. Virol. 80, 12332–12342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rothenfusser S., Goutagny N., DiPerna G., Gong M., Monks B. G., Schoenemeyer A., Yamamoto M., Akira S., Fitzgerald K. A. (2005) The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J. Immunol. 175, 5260–5268 [DOI] [PubMed] [Google Scholar]
- 6. Yoneyama M., Kikuchi M., Matsumoto K., Imaizumi T., Miyagishi M., Taira K., Foy E., Loo Y. M., Gale M., Jr., Akira S., Yonehara S., Kato A., Fujita T. (2005) Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 175, 2851–2858 [DOI] [PubMed] [Google Scholar]
- 7. Pollpeter D., Komuro A., Barber G. N., Horvath C. M. (2011) Impaired cellular responses to cytosolic DNA or infection with Listeria monocytogenes and vaccinia virus in the absence of the murine LGP2 protein. PLoS ONE 6, e18842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Satoh T., Kato H., Kumagai Y., Yoneyama M., Sato S., Matsushita K., Tsujimura T., Fujita T., Akira S., Takeuchi O. (2010) LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc. Natl. Acad. Sci. U.S.A. 107, 1512–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Venkataraman T., Valdes M., Elsby R., Kakuta S., Caceres G., Saijo S., Iwakura Y., Barber G. N. (2007) Loss of DEXD/H box RNA helicase LGP2 manifests disparate antiviral responses. J. Immunol. 178, 6444–6455 [DOI] [PubMed] [Google Scholar]
- 10. Parisien J. P., Bamming D., Komuro A., Ramachandran A., Rodriguez J. J., Barber G., Wojahn R. D., Horvath C. M. (2009) A shared interface mediates paramyxovirus interference with antiviral RNA helicases MDA5 and LGP2. J. Virol. 83, 7252–7260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Marques J. T., Devosse T., Wang D., Zamanian-Daryoush M., Serbinowski P., Hartmann R., Fujita T., Behlke M. A., Williams B. R. (2006) A structural basis for discriminating between self and nonself double-stranded RNAs in mammalian cells. Nat. Biotechnol. 24, 559–565 [DOI] [PubMed] [Google Scholar]
- 12. Takahasi K., Yoneyama M., Nishihori T., Hirai R., Kumeta H., Narita R., Gale M., Jr., Inagaki F., Fujita T. (2008) Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Molecular Cell 29, 428–440 [DOI] [PubMed] [Google Scholar]
- 13. Myong S., Cui S., Cornish P. V., Kirchhofer A., Gack M. U., Jung J. U., Hopfner K. P., Ha T. (2009) Cytosolic viral sensor RIG-I is a 5′-triphosphate-dependent translocase on double-stranded RNA. Science 323, 1070–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Myong S., Ha T. (2010) Stepwise translocation of nucleic acid motors. Curr. Opin. Struct. Biol. 20, 121–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hwang H., Kim H., Myong S. (2011) Protein induced fluorescence enhancement as a single molecule assay with short distance sensitivity. Proc. Natl. Acad. Sci. U.S.A. 108, 7414–7418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bamming D., Horvath C. M. (2009) Regulation of signal transduction by enzymatically inactive antiviral RNA helicase proteins MDA5, RIG-I, and LGP2. J. Biol. Chem. 284, 9700–9712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yoneyama M., Kikuchi M., Natsukawa T., Shinobu N., Imaizumi T., Miyagishi M., Taira K., Akira S., Fujita T. (2004) The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5, 730–737 [DOI] [PubMed] [Google Scholar]
- 18. Caruthers J. M., McKay D. B. (2002) Helicase structure and mechanism. Curr. Opin. Struct. Biol. 12, 123–133 [DOI] [PubMed] [Google Scholar]
- 19. Shi H., Cordin O., Minder C. M., Linder P., Xu R. M. (2004) Crystal structure of the human ATP-dependent splicing and export factor UAP56. Proc. Natl. Acad. Sci. U.S.A. 101, 17628–17633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li X., Ranjith-Kumar C. T., Brooks M. T., Dharmaiah S., Herr A. B., Kao C., Li P. (2009) The RIG-I-like receptor LGP2 recognizes the termini of double-stranded RNA. J. Biol. Chem. 284, 13881–13891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kowalinski E., Lunardi T., McCarthy A. A., Louber J., Brunel J., Grigorov B., Gerlier D., Cusack S. (2011) Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell 147, 423–435 [DOI] [PubMed] [Google Scholar]
- 22. Luo D., Ding S. C., Vela A., Kohlway A., Lindenbach B. D., Pyle A. M. (2011) Structural insights into RNA recognition by RIG-I. Cell 147, 409–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jiang F., Ramanathan A., Miller M. T., Tang G. Q., Gale M., Jr., Patel S. S., Marcotrigiano J. (2011) Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature 479, 423–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Moresco E. M., Beutler B. (2010) LGP2. Positive about viral sensing. Proc. Natl. Acad. Sci. U.S.A. 107, 1261–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gitlin L., Benoit L., Song C., Cella M., Gilfillan S., Holtzman M. J., Colonna M. (2010) Melanoma differentiation-associated gene 5 (MDA5) is involved in the innate immune response to Paramyxoviridae infection in vivo. PLoS Pathog. 6, e1000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schmidt A., Rothenfusser S., Hopfner K. P. (2012) Sensing of viral nucleic acids by RIG-I. From translocation to translation. Eur. J. Cell Biol. 91, 78–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.