Abstract
Technological advances in genetics have made feasible and affordable large studies to identify genetic variants that cause or modify a trait. Genetic studies have been carried out to assess variants in candidate genes, as well as polymorphisms throughout the genome, for their associations with heritable clinical outcomes of cystic fibrosis (CF), such as lung disease, meconium ileus, and CF-related diabetes. The candidate gene approach has identified some predicted relationships, while genome-wide surveys have identified several genes that would not have been obvious disease-modifying candidates, such as a methionine sulfoxide transferase gene that influences intestinal obstruction, or a region on chromosome 11 proximate to genes encoding a transcription factor and an apoptosis controller that associates with lung function. These unforeseen associations thus provide novel insight into disease pathophysiology, as well as suggesting new therapeutic strategies for CF.
Non-CFTR gene modifiers strongly influence the age of onset and the organ systems affected in cystic fibrosis patients. Studies have examined genes associated with lung and liver disease, meconium ileus, and diabetes.
Cystic fibrosis (CF) is a Mendelian “monogenic” recessive genetic disorder caused by mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene (Welsh et al. 2001). There is a broad range of age-of-onset and disease activity for different organ systems in CF, including lung disease, meconium ileus, diabetes, and liver disease, even for CF patients who are homozygous for the most common mutation, F508del (Cutting 2010). Therefore, non-CFTR genetic variation and/or environmental influences must contribute to the variability of clinical phenotypes.
To enhance understanding of the pathogenesis of organ-system disease in CF and identify novel therapies, it is key to determine the magnitude of environmental and genetic effects of non-CFTR gene modifiers on clinical phenotypes. If variability in disease largely reflects environmental influences, there should be intensified focus on these factors, which might include the intensity of medical care, treatment of different types of lung infections, socioeconomic status, and effect of geographical temperature (Schechter 2004; Collaco et al. 2010, 2011). If the variability in clinical phenotypes largely reflects genetic variation in the genome, it is key to determine which genes are involved, and the mechanism of biological effects. Recent evidence indicates that there is a strong influence of non-CFTR genetic variants on clinical phenotype in CF, but most of the genetic variation has not yet been defined (Mekus et al. 2000; Vanscoy et al. 2007).
Recent advances in genetic technology and the formation of The International CF Gene Modifier Consortium put CF at the cutting edge of gene-modifier research in “monogenic” disorders. This combination of events provides a critical opportunity for CF, because identification of the most important gene modifiers will provide insight into the mechanisms of disease pathogenesis, and offer the potential for novel prognostic approaches and therapies. This article provides an overview and update on the status of gene modifier research in CF.
HETEROGENEITY OF CF CLINICAL PHENOTYPES RELATED TO CFTR MUTATIONS
Overview
CF is the most common autosomal recessive disorder among Caucasians, and the median age of survival is only ∼39 years, despite improved treatments. The cystic fibrosis transmembrane regulator (CFTR) gene encodes a 1480-amino acid protein that functions as a cAMP-mediated Cl− channel, which plays a key role in hydrating airway secretions and regulating other cellular functions, including Na+ transport in airway epithelia (Welsh et al. 2001; Cutting 2010). Because clinical disease in CF results primarily from mutations in CFTR, CF is termed a “monogenic” disorder. There are >1500 mutations in CFTR (see Cystic Fibrosis Gene Analysis Consortium, www.genet.sickkids.on.ca/cftr/) and the most prevalent mutation results in deletion of phenylalanine at position 508 (F508del), which occurs on 70% of CF chromosomes in the U.S.
Heterogeneity of Disease Related to Mutations in CFTR
There is strong correlation between the general type of CFTR mutation and disease phenotype. Specifically, those mutations without residual function, such as F508del, are associated with pancreatic exocrine insufficiency, whereas ∼10% of CFTR mutations retain some residual function and are associated with pancreatic exocrine sufficiency. Indeed, patients with at least one copy of a mutant CFTR allele with residual function have better nutritional status and milder lung disease, even though there is broad heterogeneity of disease severity (Mickle and Cutting 1993, 2000; The Cystic Fibrosis Genotype-Phenotype Consortium 1993). The development of meconium ileus, CF related diabetes (CFRD), and severe CF liver disease with portal hypertension (CFLD) is largely confined to patients with CFTR mutations with no residual function (Blackman et al. 2006, 2009b; Sontag et al. 2006; Bartlett et al. 2009; Moran et al. 2009). However, among patients carrying two mutations with no residual function, there is also a very broad range of lung disease severity, and there are strikingly different prevalences of meconium ileus, diabetes, and liver disease.
HERITABILITY OF CLINICAL PHENOTYPES
Overview
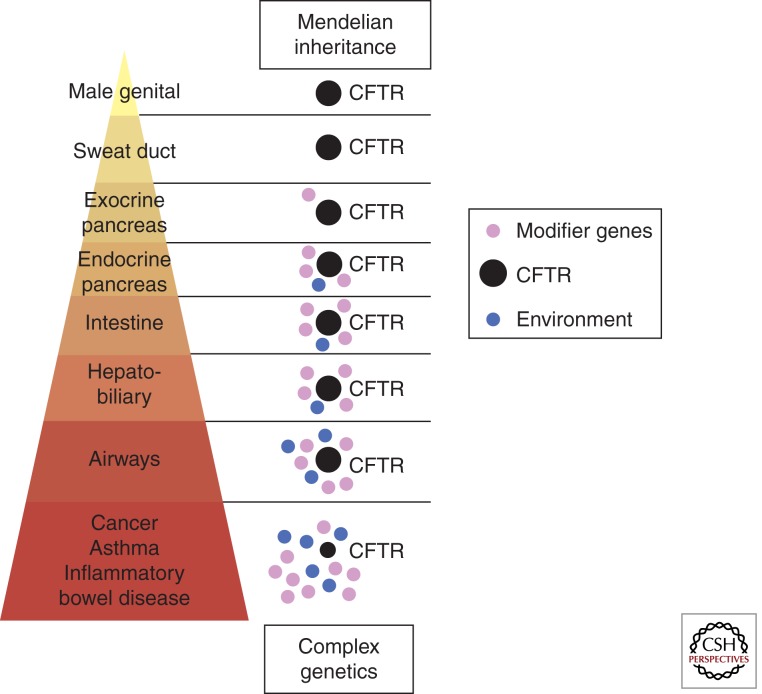
Before extensive genetic data was available, the prevailing concept was that the heterogeneity of clinical phenotypes reflected differing magnitudes of genetic versus environmental influences (Fig. 1) (Castaldo et al. 2001; Drumm 2001; Davies et al. 2005). For example, obstructive azoospermia (congenital bilateral absence of the vas deferens, CBAVD), occurs in nearly all CF males, regardless of whether the CFTR mutation has some residual function, or not; therefore, CBAVD is determined by genetics underlying mutations in CFTR (Cutting 2010). CF-related diabetes (CFRD) and severe CF liver disease with portal hypertension (CFLD) is limited to patients with two CFTR mutations without residual function (Bartlett et al. 2009; Blackman et al. 2009b; Moran et al. 2009). CFLD occurs before adulthood in the vast majority of CF patients who develop CFLD. The prevalence of CFRD is age-dependent, and occurs in ∼25%–35% of CF adults. Lung disease occurs in patients regardless of whether CFTR mutations have residual function, or not, but the range of disease severity is very great; thus, there are likely major contributions from both non-CFTR genetic modifiers and environmental influences (Mekus et al. 2000; Vanscoy et al. 2007).
Figure 1.
Schematic display of the magnitude of the effect of mutant CFTR versus environmental influences and modifier genes. (Image courtesy of Peter Durie, Toronto.)
Formal studies of genetic heritability of different clinical phenotypes have largely confirmed previous speculations (see above), and clearly show major roles for non-CFTR genetic variation in the different clinical phenotypes (Table 1). Genes that cause many “monogenic” disorders have been described, but there is little knowledge of the other specific factors that contribute to variability of the clinical phenotype. Investigations of inbred mice provide evidence that overall genetic background has a major effect on clinical phenotype in monogenic disorders, which is a concept that relates to non-CFTR genetic influences on disease phenotype in CF (Nadeau 2001). There are also CFTR-related “mono-organ” disorders, such as male infertility, pancreatitis and sinusitis, which implicate complex genetic variation as important in mono-organ disease. Therefore, the study of modifier genes in CF is well-grounded from a conceptual perspective.
Table 1.
Heritability estimates for CF-related phenotypes
| Phenotype | Lung function | Persistent infection | Diabetes | Meconium ileus | BMI | Weight for height | References |
|---|---|---|---|---|---|---|---|
| Heritability, all subjects | 0.54–1.0 | 0.76–0.85 | ≤1 | ≤1 | ∼0.6 | ∼0.8 | Blackman et al. 2006, 2008, 2009b; Vanscoy et al. 2007; Stanke et al. 2011; Green et al. 2012 |
| Heritability, F508del only | 0.56–0.86 | No estimate | No estimate | No estimate | No estimate | ∼0.6 | Vanscoy et al. 2007; Stanke et al. 2011 |
Lung
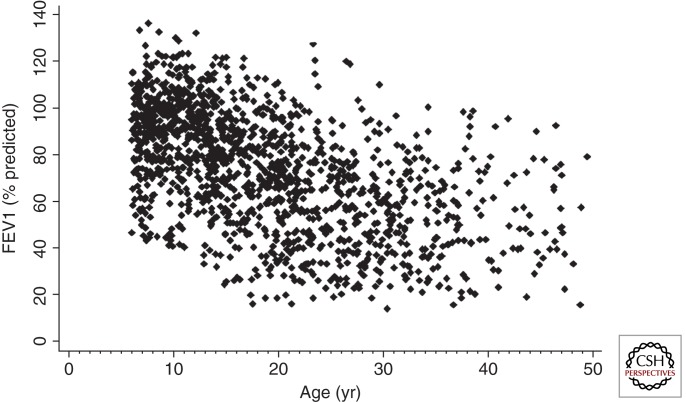
There is a broad range of lung disease severity, even for patients who are F508del homozygotes. This is illustrated in a representative population of Canadian CF patients by great variability in forced expired volume in 1 sec (FEV1) (Fig. 2), which is a measure of airflow obstruction that correlates with lung disease severity and survival in CF. Two twin/sibling studies have concluded that there is a strong non-CFTR genetic contribution (0.54–1.0) to the CF lung phenotype (Table 1—“Heritability”). A key feature for studying non-CFTR genotype-phenotype relationships is the ability to define a meaningful clinical phenotype in quantitative, or clear-cut binary, terms. Successful studies involve substantial effort to define the pertinent phenotype (see section below, “Challenges: Phenotypes”).
Figure 2.
Lung disease severity assessed by FEV1 versus age in 1357 Canadian CF patients with biallelic loss-of-function (pancreatic insufficient) mutations. (Image courtesy of Peter Durie, Toronto.)
Meconium Ileus
About 15%–20% of CF neonates are afflicted by an intestinal blockage that develops in utero, a concretion composed of meconium and mucus, and characteristically forms in the distal ileum. Evidence of this condition, known as meconium ileus (MI), is often detected before birth by ultrasound as a dense region on the sonogram. Histologically, there is an increase in the number and size of goblet cells, and elongated villi and distended crypts are seen as well. MI appears to be under nearly complete control of genetic factors, as heritability of this trait approaches 100% (Table 1).
CF-Related Diabetes
CF-related diabetes (CFRD) is a common comorbidity in CF, with about a fifth of adolescents affected and nearly half of adults (Moran et al. 2009). As a clinical entity, CFRD is distinct, although sharing properties of type 1 and type 2 diabetes (T1DM and T2DM, respectively). Type 1 features include reduced β-cell mass and diminished insulin secretion, but the etiology is not one of autoimmunity. Similar to type 2 diabetes, the pancreases of CFRD patients display amyloid deposition (Couce et al. 1996). Also, insulin resistance is common and ketosis is rare, consistent with insulin secretion being reduced but not absent. Unlike type 2 diabetes, obesity is not a risk factor, nor are the dyslipidemia profiles characteristic of type 2 diabetes.
Whereas the morphometric and biochemical risk factors for T2DM do not appear to be risk factors for CFRD, the genetics point to a relationship between T2DM and CFRD. For example, having a relative with T2DM increases the risk of a CF patient for developing CFRD (Blackman et al. 2009a). In fact, the heritability of CFRD is high, approaching 1 (Table 1), substantially higher than for T2DM, suggesting that CFRD is much less influenced by environmental factors, such as diet, than T2DM with which it appears to share genetic etiology.
CF Liver Disease
CF liver disease begins with loss of CFTR (Cl− channel) function on the apical membrane of cholangiocytes. This dysfunction leads to defective (sluggish) bile flow and an inflammatory response with activation and proliferation of hepatic stellate cells, which results in cholangitis and fibrosis in focal portal tracts (Rowland and Bourke 2011). However, only a small subset (∼5%) of patients with CF develops severe liver disease with portal hypertension (CFLD). Moreover, CFLD occurs at a median age of only 10 years, and >90% of CFLD occurs by age 20 years; thus, non-CFTR genetic variations likely contribute to risk for CFLD (Bartlett et al. 2009), but there are no formal estimates of heritability. Finally, there is also an increased prevalence of CFLD in males (2:1) over females, suggesting a role for gender and/or male/female hormones in this disorder.
MODIFIER GENETICS AND RECESSIVE MENDELIAN DISORDERS
Approaches to Study Genetic Modifiers in CF
The concept of gene modifiers in a Mendelian disorder such as cystic fibrosis shares some features, but is fundamentally different from the concept of genetic variants in non-Mendelian, complex genetic disorders, such as asthma. In complex genetic disorders, genetic variants interact with each other and the environment to cause the disorder (Van Heyningen and Yeyati 2004). In contrast, cystic fibrosis results from mutations in CFTR, and genetic variants that affect pathogenesis of mutant CFTR modify the severity of the phenotype (Drumm 2001; Cutting 2010). Genetic variation that has little effect in normal subjects may modify disease in cystic fibrosis.
Two different types of genetic studies have been used to discover gene modifiers in cystic fibrosis, i.e., case-control association studies and family-based (linkage) studies (Cutting 2010). The case-control (association) study is usually more robust for detecting common genetic variants, in part because of the larger number of subjects that can be enrolled in association studies, as compared with family-based studies. Enrollment into family-based twin/sibling linkage studies is more challenging, but it allows better definition of environmental versus genetic effects on the phenotype. Thus, complementary studies employing both the association and family-based methodologies would be optimal, and this strategy has been undertaken in cystic fibrosis by the International CF Gene Modifier Consortium.
Challenges: Phenotypes
Studies of genetic effects on phenotype require a meaningful and accurate definition of phenotype. For lung disease, FEV1 relates to clinical disease severity and outcome; therefore, the development of a quantitative lung phenotype has been developed for CF, using multiple measures of FEV1 and adjusting for mortality attrition (Taylor et al. 2011). This rigorous, quantitative lung phenotype has been used in a GWAS study of ∼3500 CF patients (Wright et al. 2011). For meconium ileus, a binary (yes/no) approach is used to define phenotype, but requires careful review of source documents to be accurate (Wood et al. 2007; Sun et al. 2012). A recent publication has also determined that the heritability of chronic respiratory infection with Pseudomonas aeruginosa is very strong (0.76–0.85), which provides another lung phenotype that can be studied for genetic modifiers (Green et al. 2012). For CF-related diabetes there is an increasing prevalence with age; therefore, genetic studies use age-of-onset (“survival”) type analyses (Blackman et al. 2009b). For liver disease, a binary (yes/no) approach is used, but this requires extensive review of source documents to accurately determine which patients have severe liver disease with portal hypertension (CFLD) (Bartlett et al. 2009). Many genetic studies have been flawed by nonmeaningful or imprecise definition of phenotype, but the CF community has focused on establishing meaningful and accurate phenotypes.
IDENTIFIED MODIFIERS OF DISEASE PHENOTYPES BY CANDIDATE GENE APPROACH
Concept
Initial searches for non-CFTR genetic variants that influence pulmonary phenotype used the candidate gene approach, which is based on biological plausibility. This approach is attractive because testing a small number of carefully selected candidates avoids severe penalties from multiple comparisons during statistical analysis. However, the candidate gene approach has substantial bias, because of incomplete understanding of the pathophysiology of CF lung disease. Because an association between a genetic variant and disease phenotype can reflect relatively small differences in allele frequency, it is critical to replicate any association in a separate population, preferably several times. In CF, many candidate genes have been tested, but only a few candidates have been tested in >1000 CF patients, and only a few have been replicated (Table 2). A comprehensive review of all gene modifier studies in CF is beyond the scope of this article, so we report a summary of candidate genes that have been replicated in at least one organ in ≥2 populations entailing at least 500 individuals total (Table 2; see text below). Several recent publications review all candidate genes that have been tested in CF (Cutting 2005; Davies et al. 2005; Knowles 2006; Collaco and Cutting 2008).
Table 2.
Candidate gene modifiers of clinical phenotypes in cystic fibrosis
| Gene | Lung function (FEV1) | Ps. a. acquisition | Meconium ileus | CF-related diabetes | CF liver disease | References |
|---|---|---|---|---|---|---|
| MBL2 | REPLICATEDa | REPLICATED | Gabolde et al. 1999; Garred et al. 1999; Davies et al. 2004; Yarden et al. 2004; Trevisiol et al. 2005; Dorfman et al. 2008; McDougal et al. 2010 | |||
| EDNRA | REPLICATED | Darrah et al. 2010 | ||||
| TGF-β1 | REPLICATED | Negativec | Probably negatived | Arkwright et al. 2000, 2003; Drumm et al. 2005; Brazova et al., 2006; Bremer et al. 2008; Corvol et al. 2008; Dorfman et al. 2008; Bartlett et al. 2009; Faria et al. 2009 | ||
| IFRD1 | Replicatedb | Gu et al. 2009 | ||||
| IL8 | Replicated | Hillian et al. 2008 | ||||
| MSRA | REPLICATED | Henderson et al. 2012 | ||||
| ADIPOR2 | Replicated | Dorfman et al. 2009 | ||||
| TCF7L2 | REPLICATED | Blackman et al. 2009a | ||||
| SERPINA1 | Negative | Probably negative | Replicated | Arkwright et al. 2000, 2003; Drumm et al. 2005; Brazova et al. 2006; Bremer et al. 2008; Corvol et al. 2008 Dorfman et al. 2008; Bartlett et al. 2009; Faria et al. 2009 |
aREPLICATED: seen in ≥3 populations with total ≥1000 individuals.
bReplicated: seen in ≥2 populations with total ≥500 individuals.
cNegative: negative in ≥3 populations with total ≥1000 individuals.
dProbably negative: negative in ≥2 populations with total ≥500 individuals.
Lung Disease
Lung disease in CF is linked to defective ion transport, which leads to inadequate hydration of airway mucus and results in defective mucociliary and cough clearance. Acute and chronic airway infection occurs secondary to defective mucociliary clearance, which contributes to an inflammatory response and adversely affects the phenotype. These defects of ion transport, mucus clearance, and infection and inflammation leads to the development of bronchiectasis. Therefore, genetic variation in many genes in these pathways could modify the clinical phenotype, and be associated with more severe (or milder) lung disease, and affect survival.
Multiple studies have tested whether functional variants in mannose binding lectin (MBL2) associate with lung disease severity in CF. MBL2 is biologically plausible, because of its role in innate immunity and the predilection of MBL2 deficient patients to bacterial and viral infection. Six studies involving >2000 CF patients have shown that MBL-deficient genotypes (OO or AO) are associated with worse lung function (Table 2) (Gabolde et al. 1999; Garred et al. 1999; Davies et al. 2004; Yarden et al. 2004; Trevisiol et al. 2005; Dorfman et al. 2008), and three studies reported earlier age of infection with Pseudomonas aeruginosa (Ps. a.) (Table 2) (Trevisiol et al. 2005; Dorfman et al. 2008; McDougal et al. 2010). Because severity of lung disease, Ps. a. infection, and age are correlated, a modeling framework addressed these confounding variables, and concluded that deficiency in MBL predisposes to earlier infection with Ps. a. and thereby leads to worse lung disease (McDougal et al. 2010). Several studies did not report an association of MBL deficient genotypes with CF lung disease, which might reflect large numbers of older patients and/or other factors related to bacterial infection (Cutting 2010).
Genetic variation in the endothelial receptor type A (EDNRA) has been associated with CF lung disease severity (Table 2), and replicated in three additional populations of CF patients (1478 total) (Darrah et al. 2010). The variant EDNRA (rs5335) is in the 3′ UTR, and the risk allele for worse lung disease is associated with increased expression of EDNRA in cultured human tracheal smooth muscle cells, as well as increased proliferation of smooth muscle cells that are homozygous for the risk variant. Thus, the mechanism for adversely modifying CF lung disease likely reflects increased smooth muscle activity in airways and/or vasculature.
Transforming growth factor β1 (TGF-β1) modifies risk for airways disease in asthma and COPD, and has a key role in regulating airway inflammation and remodeling; hence, TGF-β1 has been repeatedly studied as a modifier of CF lung disease (Table 2). Alleles in the promotor (-509) and first exon (codon 10) of TGF-β1 are associated with worse lung function, and was replicated in the Genetic Modifier Study (GMS) of >1300 CF patients (Drumm et al. 2005). This discovery was independently replicated when haplotypes of alleles at -509 and codon 10 were analyzed in a separate population (Bremer et al. 2008). Further, other gene and environmental interactions are relevant to TGF-β1, because interactions with variants in MBL2 and second-hand smoke exposure modify TGF-β1 association with lung disease. A few small studies did not report the same association of TGF-β1 and CF lung disease (Cutting 2010).
Genetic variation in the interferon-related developmental regulator 1 (IFRD1) gene was identified as one of six candidate regions, based on study of 320 CF patients in the GMS study with “mild” (n = 160) versus “severe” (n = 160) lung disease (Table 2). Polymorphisms in IFRD1 replicated in the larger GMS sample, and then showed association in a family-based (twin and siblings) population (Gu et al. 2009). In vitro and studies in mice indicated that IFRD1 acts to modify neutrophil function during bacterial infection, via transcriptional mechanisms, but this gene has not been further replicated in additional CF populations.
IL-8 is a cytokine that is well-recognized to be associated with airways inflammation in CF, and known to mediate neutrophil chemotaxis. The association of IL-8 variants and CF lung disease was first noted in 737 CF patients and was most strongly shown in males (Table 2). The replication was verified for males in an additional 385 patients. The mechanism appears to reflect differential gene expression through IL-8 promoter variants.
Meconium Ileus
The heritability of MI (Table 1) clearly indicates that genes other than CFTR are involved in its etiology, but the candidate gene approach has not been applied to this CF trait. A novel candidate region approach was used however, in which a segment of mouse chromosome 7 that was found to influence intestinal obstruction in CF mice (Rozmahel et al. 1996) was examined for the corresponding region of the human genome for its effect on MI. Initial reports indicated that variants in this region did, in fact, associate with MI (Zielenski et al. 1999), but a causative gene was never identified and subsequent studies failed to replicate this finding (Blackman et al. 2006). Although the genetic contribution to MI seems clear, the genes responsible remained elusive.
CF-Related Diabetes
The observation that family history of T2DM increases risk for CFRD, as described above, made obvious the need to test specific genes associated with T2DM for similar associations with CFRD. Thus, a candidate gene approach was initiated and showed that single nucleotide polymorphisms near the transcription factor 7-like 2 (TCF7L2) gene showed an association with type 2 diabetes with an estimated odds ratio of ∼1.5 per allele (Blackman et al. 2009b).
In addition to showing a specific genetic relationship between T2DM and CFRD, this study showed an important concept regarding the confounding effects that nongenetic, or environmental factors may have on genetic studies, sometimes obscuring them. In this case, the nongenetic influences appear to be pharmacology, specifically steroidal anti-inflammatory drugs. Glucocorticoids are commonly used as an anti-inflammatory treatment for CF, and it is well established that a side effect of glucocorticoids is hyperglycemia, albeit transiently. Thus, this important pharmacotherapy for the lungs has the potential to mask differences in glycemic levels that would be caused by genetic variation. When controlling for the effect of this steroidal treatment by examining patients not recently using glucocorticoids, the risk associated with TCF7L2 alleles increased to 2.9 per allele (Blackman et al. 2009b).
Other evidence of similarities to T2DM is emerging as well. A screen of six genes associated with T2DM found that a polymorphism in Calpain-10 associated nominally with response to a glucose tolerance test (Derbel et al. 2006). Other T2DM genes are sure to be tested in the future, but as the effect of many of them is quite small it remains to be seen whether the cohort sizes available for CF studies will have the power to detect them.
CF Liver Disease
CF liver disease with portal hypertension (CFLD) occurs early in life in CF patients with biallelic loss-of-function (pancreatic insufficient) mutations in CFTR. The largest two-stage study of CFLD focused on five candidate genes that had previously been studied in CF “liver disease,” but not all patients in previous studies had portal hypertension (Bartlett et al. 2009). The two-stage study showed strong association (and replication) of the Z-allele of the α1-antiprotease (SERPINA1) gene with CFLD in a total of 260 patients with CFLD versus 1931 CF patients without CFLD (Table 2). The mechanism is unclear, but probably does not reflect cumulative efforts of the folding mutations in CFTR (F508del) and SERPINA1 (Z-allele), because these two genes are predominately expressed in two different cell types (cholangiocytes and hepatocytes, respectively) in the liver. Variants in TGF-β1 were associated with CFLD in stage 1, but did not replicate in stage 2 of the CFLD study (Bartlett et al. 2009).
IDENTIFIED MODIFIERS OF DISEASE PHENOTYPE BY GWAS ASSOCIATION AND LINKAGE
The ability to perform unbiased study of relatively common genetic polymorphisms across the entire human genome in a cost-effective fashion has recently come-of-age. To maximize power to detect novel modifiers of CF phenotypes using genome-wide approaches, three different study groups have joined forces and created a consortium with >3500 well-phenotyped CF patients to study modifiers in CF. This consortium includes three different study designs: (1) Gene Modifier Study (GMS), which enrolled F508del homozygotes with extremes-of-lung phenotype (Drumm et al. 2005); (2) Canadian Genetic Study (CGS), which enrolled population-based CF patients; and (3) family-based, twins and siblings (TSS), which enrolled patients from families with more than one CF child. The GMS and CGS are best-suited for association analysis of unrelated patients, whereas the TSS (family-based) study can be used for both linkage and association analyses. Using this complementary study design allows for discovery by association (or by linkage) and then testing for replication in the alternate population(s).
Lung Disease
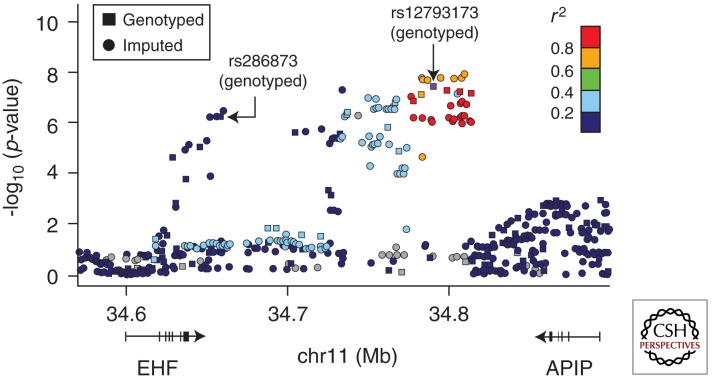
The combined genome-wide association and linkage study (GWALS) genotyped 3467 CF patients (Illumina 610 Quad platform) to identify genomic regions associated with CF lung disease severity. The strongest association signal (p = 3 × 10−8) was located at chromosome11p13, intergenic to EHF and APIP and replicated in the TSS (family-based) patients (Fig. 3) (Wright et al. 2011). EHF is an epithelia-specific Ets transcription factor, and may be an important regulator of differentiation under stress conditions and inflammation. APIP is a known inhibitor of apoptosis, and eQTLs for APIP in lymphocytes suggests the risk SNP in this region may be associated with increased expression of APIP, implying that inhibition of apoptosis worsens CF lung disease. This is congruent with the notion that inhibition of apoptosis in airways delays clearance of neutrophils and leads to a hyper-inflammatory state (Dibbert et al. 1999; Harris et al. 2005). Inhibition of apoptosis may also contribute to goblet cell metaplasia, which is a feature of CF airways disease (Dibbert et al. 1999; Harris et al. 2005). Polymorphisms in the intergenic region of chr11p13 may also affect regulatory domains, which could alter expression of EHF and APIP and other genes in the region, including ELF5, PDHX, and CD44. Studies are ongoing to elucidate the mechanism of disease pathogenesis from genomic polymorphisms at chromosome11p13.
Figure 3.
A locus-zoom plot of association in the GMS and CGS F508del homozygotes at chr11p13 EHF/APIP region, showing both genotyped and HapMap-imputed SNPs. Colors represent HapMap CEU linkage disequilibrium r2 with the most significant genotyped SNP (rs12793173; p = 3 × 10−8). The secondary genotyped peak (rs286873) has low r2 with the primary peak. Addition of the imputed SNPs to genotyped SNPs illustrates the coverage and LD blocks across the region.
There was also a near-significant genome-wide association at the chromosome 6p21.3 HLA class II DR locus. Genetic variation at this locus has been widely recognized to play a role in multiple infections and inflammatory disorders, which would be relevant to the infections and inflammatory nature of CF airway disease (Handunnetthi et al. 2010). This locus is very genetically heterogeneous, and studies are underway to define the specific DR allele that is associated with severity of CF lung disease.
There were five other regions of suggestive association (Wright et al. 2011), which will be further analyzed by an ongoing GWAS in an additional ∼3500 CF patients. Two previously reported modifiers of CF lung disease, TGF-β1 and IFRD1, did not achieve genome-wide significance. However, TGF-β1 achieved p-values of 10−3 to 10−4 in GMS patients, depending on the covariates used in the analyses.
Linkage analysis of 486 sibling pairs from the family-based study identified a robust linkage signal at chr20q13.2 (LOD = 5.03). A region below the linkage peak was analyzed for association in the GMS and CGS patients, and replicated under a region-wide analytical approach (p = 1 × 10−4). The chr20p13.2 linkage region contains five genes (CBLN4; MC3R; CASS4; CSTF1; AURKA) that are expressed in fetal or adult lung, or bronchial epithelia. MC3R is of particular interest, because it has been implicated in regulation of energy balance and modulating neutrophil accumulation in lung inflammation, which are both relevant to CF lung disease (Savastano et al. 2009). Studies are underway to define the mechanism of lung disease in CF that reflects the linkage signal at chr20p13.2.
Meconium Ileus
The high heritability of MI certainly suggested that genes other than CFTR must be involved, and the availability of high density markers throughout the genome provided the opportunity to carry out linkage studies at high resolution. Using this approach, Blackman and colleagues found by genome-wide analyses several regions of suggestive linkage (logarithm of the odds of linkage >2.0) for contributing to, or protecting from development of, meconium ileus. These regions were found on chromosomes 4q35.1, 8p23.1, and 11q25 (MI contributing loci) and chromosomes 20p11.22 and 21q22.3 (protective loci) (Blackman et al. 2006).
The locus on chromosome 8 is a gene-dense region and was followed up by haplotype analysis and identified a three polymorphism haplotype that most strongly associated and corresponded to a gene, MSRA, encoding an enzyme, methionine sulfoxide reductase A, that maintains the amino acid methonine in a reduced state. This candidate was then examined functionally by assessing its effect on obstruction in a CF mouse model lacking functional MsrA, and found to dramatically decrease the incidence of obstruction compared to CF mice with functional MsrA (Henderson et al. 2012).
Another linkage study provided evidence for MI-influencing loci on chromosome 12p13.3. Using densely spaced markers in the 12p13.3 region in a cohort of pancreatic insufficient Canadian CF patients with and without MI, one gene, ADIPOR2 in MI (p = 0.002), was implicated. The frequency of alleles in this gene was distorted from Hardy-Weinberg proportions, but only in the MI group. The deviation from expected proportions suggests a causative effect and the association was replicated in an independent sample of CF families. A second locus, but this one protective, was found on chromosome 4q13.3 and one candidate gene in the region, SLC4A4, a member of the solute transport family of proteins, showed evidence of association (p = 0.002) (Dorfman et al. 2009).
Association studies can also be used to identify genes involved in meconium ileus, and a novel modification of the association approach has identified several candidate genes. This approach, termed “informed GWAS” incorporates functional annotation into the analyses, giving higher import to those genes with a particular function attributed to them. Using a model in which genes influencing epithelial function were the focus, polymorphisms near SLC6A14 on the X chromosome and SLC26A9 on chromosome 1 accounted for ∼5% of variance in MI and these associations were replicated in an independent sample of affected individuals (Sun et al. 2012).
Other
Genome-wide association data have not yet been reported for CFRD or CFLD, although the Consortium is analyzing data from GWAS1, and awaiting data from GWAS2.
CONCLUDING REMARKS
The ability to identify genes, or genomic regions, that influence disease processes hold enormous promise for better understanding those processes and for manipulating them for therapeutic gain. However, an important caveat of genetic screens is that they may identify genes involved in a process, but as such they are only markers; they do not provide information about the mechanisms by which the variants exert their phenotypic effects. Thus, for the results of any genetic study to have more than prognostic value, the genes must be followed up by functional studies to have clinical utility.
The potential for clinical application is not only great, it is near. There are already approved drugs that target the protein product encoded by several of the genes identified (Table 2), including antagonists to the endothelin and angiotensin receptors, drugs that alter TGF-β1 signaling and small molecule screening provides the potential to rapidly identify new drugs that will target these gene products or the pathways in which they act.
ACKNOWLEDGMENTS
This work is supported in part by the U.S. National Institutes of Health grants R01HL068890 and P30DK27651, and Cystic Fibrosis Foundation grants KNOWLE00A0 and DRUMM0A00. The authors thank Fred A. Wright, Lisa J. Strug, Scott M. Blackman, Rhonda G. Pace, Johanna M. Rommens, Jaclyn R. Stonebraker, Wanda K. O’Neal, Peter R. Durie, Garry R. Cutting, and Tony T. Dang for contributions; plus Syanne D. Olson for editorial contributions and Elizabeth A. Godwin for administrative support.
Footnotes
Editors: John R. Riordan, Richard C. Boucher, and Paul M. Quinton
Additional Perspectives on Cystic Fibrosis available at www.perspectivesinmedicine.org
REFERENCES
- Arkwright PD, Laurie S, Super M, Pravica V, Schwarz MJ, Webb AK, Hutchinson IV 2000. TGF-β1 genotype and accelerated decline in lung function of patients with cystic fibrosis. Thorax 55: 459–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkwright PD, Pravica V, Geraghty PJ, Super M, Webb AK, Schwarz M, Hutchinson IV 2003. End-organ dysfunction in cystic fibrosis: Association with angiotensin I converting enzyme and cytokine gene polymorphisms. Am J Respir Crit Care Med 167: 384–389 [DOI] [PubMed] [Google Scholar]
- Bartlett JR, Friedman KJ, Ling SC, Pace RG, Bell SC, Bourke B, Castaldo G, Castellani C, Cipolli M, Colombo C, et al. 2009. Genetic modifiers of liver disease in cystic fibrosis. JAMA 302: 1076–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackman SM, Deering-Brose R, McWilliams R, Naughton K, Coleman B, Lai T, Algire M, Beck S, Hoover-Fong J, Hamosh A, et al. 2006. Relative contribution of genetic and nongenetic modifiers to intestinal obstruction in cystic fibrosis. Gastroenterology 131: 1030–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackman SM, Vanscoy LL, Collaco JM, Naughton KM, Cutting GR 2008. Variability in body mass index in cystic fibrosis is determined partly by a genetic locus on chromosome 5. Pediatr Pulmonol Suppl 31: 271 [Google Scholar]
- Blackman SM, Hsu S, Ritter SE, Naughton KM, Wright FA, Drumm ML, Knowles MR, Cutting GR 2009a. A susceptibility gene for type 2 diabetes confers substantial risk for diabetes complicating cystic fibrosis. Diabetologia 52: 1858–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackman SM, Hsu S, Vanscoy LL, Collaco JM, Ritter SE, Naughton K, Cutting GR 2009b. Genetic modifiers play a substantial role in diabetes complicating cystic fibrosis. J Clin Endocrinol Metab 94: 1302–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazova J, Sismova K, Vavrova V, Bartosova J, Macek M Jr, Lauschman H, Sediva A 2006. Polymorphisms of TGF-β1 in cystic fibrosis patients. Clin Immunol 121: 350–357 [DOI] [PubMed] [Google Scholar]
- Bremer LA, Blackman SM, Vanscoy LL, McDougal KE, Bowers A, Naughton KM, Cutler DJ, Cutting GR 2008. Interaction between a novel TGF-β1 haplotype and CFTR genotype is associated with improved lung function in cystic fibrosis. Hum Mol Genet 17: 2228–2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaldo G, Fuccio A, Salvatore D, Raia V, Santostasi T, Leonardi S, Lizzi N, La Rosa M, Rigillo N, Salvatore F 2001. Liver expression in cystic fibrosis could be modulated by genetic factors different from the cystic fibrosis transmembrane regulator genotype. Am J Med Genet 98: 294–297 [DOI] [PubMed] [Google Scholar]
- Collaco JM, Cutting GR 2008. Update on gene modifiers in cystic fibrosis. Curr Opin Pulm Med 14: 559–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaco JM, Blackman SM, McGready J, Naughton KM, Cutting GR 2010. Quantification of the relative contribution of environmental and genetic factors to variation in cystic fibrosis lung function. J Pediatr 157: 802–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaco JM, McGready J, Green DM, Naughton KM, Watson CP, Shields T, Bell SC, Wainwright CE, Cutting GR 2011. Effect of temperature on cystic fibrosis lung disease and infections: A replicated cohort study. PLoS ONE 6: e27784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corvol H, Boelle PY, Brouard J, Knauer N, Chadelat K, Henrion-Caude A, Flamant C, Muselet-Charlier C, Boule M, Fauroux B, et al. 2008. Genetic variations in inflammatory mediators influence lung disease progression in cystic fibrosis. Pediatr Pulmonol 43: 1224–1232 [DOI] [PubMed] [Google Scholar]
- Couce M, O’Brien TD, Moran A, Roche PC, Butler PC 1996. Diabetes mellitus in cystic fibrosis is characterized by islet amyloidosis. J Clin Endocrinol Metab 81: 1267–1272 [DOI] [PubMed] [Google Scholar]
- Cutting GR 2005. Modifier genetics: Cystic fibrosis. Annu Rev Genomics Hum Genet 6: 237–260 [DOI] [PubMed] [Google Scholar]
- Cutting GR 2010. Modifier genes in Mendelian disorders: The example of cystic fibrosis. Ann NY Acad Sci 1214: 57–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darrah R, McKone E, O’Connor C, Rodgers C, Genatossio A, McNamara S, Gibson R, Stuart EJ, Ennis M, Gallagher CG, et al. 2010. EDNRA variants associate with smooth muscle mRNA levels, cell proliferation rates, and cystic fibrosis pulmonary disease severity. Physiol Genomics 41: 71–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JC, Turner MW, Klein N, London MBL CF Study Group 2004. Impaired pulmonary status in cystic fibrosis adults with two mutated MBL-2 alleles. Eur Respir J 24: 298–804 [DOI] [PubMed] [Google Scholar]
- Davies JC, Griesenbach U, Alton E 2005. Modifier genes in cystic fibrosis. Pediatr Pulmonol 39: 383–391 [DOI] [PubMed] [Google Scholar]
- Derbel S, Doumaguet C, Hubert D, Mosnier-Pudar H, Grabar S, Chelly J, Bienvenu T 2006. Calpain 10 and development of diabetes mellitus in cystic fibrosis. J Cyst Fibros 5: 47–51 [DOI] [PubMed] [Google Scholar]
- Dibbert B, Weber M, Nikolaizik WH, Vogt P, Schoni MH, Blaser K, Simon HU 1999. Cytokine-mediated Bax deficiency and consequent delayed neutrophil apoptosis: A general mechanism to accumulate effector cells in inflammation. Proc Natl Acad Sci 96: 13330–13335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorfman R, Sandford A, Taylor C, Huang B, Frangolias D, Wang Y, Sang R, Pereira L, Sun L, Berthiaume Y, et al. 2008. Complex two-gene modulation of lung disease severity in children with cystic fibrosis. J Clin Invest 118: 1040–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorfman R, Li W, Sun L, Lin F, Wang Y, Sandford A, Pare PD, McKay K, Kayserova H, Piskackova T, et al. 2009. Modifier gene study of meconium ileus in cystic fibrosis: Statistical considerations and gene mapping results. Hum Genet 126: 763–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drumm ML 2001. Modifier genes and variation in cystic fibrosis. Respir Res 2: 125–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drumm ML, Konstan MW, Schluchter MD, Handler A, Pace R, Zou F, Zariwala M, Fargo D, Xu A, Dunn JM, et al. 2005. Gene modifiers of lung disease in cystic fibrosis. N Engl J Med 353: 1443–1453 [DOI] [PubMed] [Google Scholar]
- Faria EJ, Faria IC, Ribeiro JD, Ribeiro AF, Hessel G, Bertuzzo CS 2009. Association of MBL2, TGF-β1 and CD14 gene polymorphisms with lung disease severity in cystic fibrosis. J Bras Pneumol 35: 334–342 [DOI] [PubMed] [Google Scholar]
- Gabolde M, Guilloud-Bataille M, Feingold J, Besmond C 1999. Association of variant alleles of mannose binding lectin with severity of pulmonary disease in cystic fibrosis: Cohort study. Br Med J 319: 1166–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garred P, Pressler T, Madsen HO, Frederiksen B, Svejgaard A, Hoiby N, Schwartz M, Koch C 1999. Association of mannose-binding lectin gene heterogeneity with severity of lung disease and survival in cystic fibrosis [see comments]. J Clin Invest 104: 431–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green DM, Collaco JM, McDougal KE, Naughton KM, Blackman SM, Cutting GR 2012. Heritability of respiratory infection with Pseudomonas aeruginosa in cystic fibrosis. J Pediatr 10.1016/j.jpeds.2012.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Harley IT, Henderson LB, Aronow BJ, Vietor I, Huber LA, Harley JB, Kilpatrick JR, Langefeld CD, Williams AH, et al. 2009. Identification of IFRD1 as a modifier gene for cystic fibrosis lung disease. Nature 458: 1039–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handunnetthi L, Ramagopalan SV, Ebers GC, Knight JC 2010. Regulation of major histocompatibility complex class II gene expression, genetic variation and disease. Genes Immun 11: 99–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris JF, Fischer MJ, Hotchkiss JR, Monia BP, Randell SH, Harkema JR, Tesfaigzi Y 2005. Bcl-2 sustains increased mucous and epithelial cell numbers in metaplastic airway epithelium. Am J Respir Crit Care Med 171: 764–772 [DOI] [PubMed] [Google Scholar]
- Henderson LB, Doshi VK, Blackman SM, Naughton KM, Pace RG, Moskovitz J, Knowles MR, Durie PR, Drumm ML, Cutting GR 2012. Variation in MSRA modifies risk of neonatal intestinal obstruction in cystic fibrosis. PLoS Genet 8: e1002580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillian AD, Londono D, Dunn JM, Goddard KAB, Pace RG, Knowles MR, Drumm ML, CF Gene Modifier Study Group 2008. Modulation of cystic fibrosis lung disease by variants in interleukin-8. Genes Immun 9: 501–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles MR 2006. Gene modifiers of lung disease. Curr Opin Pulm Med 12: 416–421 [DOI] [PubMed] [Google Scholar]
- McDougal KE, Green DM, Vanscoy LL, Fallin MD, Grow M, Cheng S, Blackman SM, Collaco JM, Henderson LB, Naughton K, et al. 2010. Use of a modeling framework to evaluate the effect of a modifier gene (MBL2) on variation in cystic fibrosis. Eur J Hum Genet 18: 680–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekus F, Ballmann M, Bronsveld I, Bijman J, Veeze H, Tummler B 2000. Categories of deltaF508 homozygous cystic fibrosis twin and sibling pairs with distinct phenotypic characteristics. Twin Res 3: 277–293 [DOI] [PubMed] [Google Scholar]
- Mickle JE, Cutting GR 2000. Genotype-phenotype relationships in cystic fibrosis. Med Clin North Am 84: 197–202 [DOI] [PubMed] [Google Scholar]
- Moran A, Dunitz J, Nathan B, Saeed A, Holme B, Thomas W 2009. Cystic fibrosis-related diabetes: Current trends in prevalence, incidence, and mortality. Diabetes Care 32: 1626–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau JH 2001. Modifier genes in mice and humans. Nat Rev Genet 2: 165–174 [DOI] [PubMed] [Google Scholar]
- Rowland M, Bourke B 2011. Liver disease in cystic fibrosis. Curr Opin Pulm Med 17: 461–466 [DOI] [PubMed] [Google Scholar]
- Rozmahel R, Wilschanski M, Matin A, Plyte S, Oliver M, Auerbach W, Moore A, Forstner J, Durie P, Nadeau J, et al. 1996. Modulation of disease severity in cystic fibrosis transmembrane conductance regulator deficient mice by a secondary genetic factor. Nat Genet 12: 280–287 [DOI] [PubMed] [Google Scholar]
- Savastano DM, Tanofsky-Kraff M, Han JC, Ning C, Sorg RA, Roza CA, Wolkoff LE, Anandalingam K, Jefferson-George KS, Figueroa RE, et al. 2009. Energy intake and energy expenditure among children with polymorphisms of the melanocortin-3 receptor. Am J Clin Nutr 90: 912–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schechter MS 2004. Non-genetic influences on CF lung disease: The role of sociodemographic characteristics, environmental exposures and healthcare interventions. Pediatr Pulmonol Suppl 26: 82–85 [DOI] [PubMed] [Google Scholar]
- Sontag MK, Corey M, Hokanson JE, Marshall JA, Sommer SS, Zerbe GO, Accurso FJ 2006. Genetic and physiologic correlates of longitudinal immunoreactive trypsinogen decline in infants with cystic fibrosis identified through newborn screening. J Pediatr 149: 650–657 [DOI] [PubMed] [Google Scholar]
- Stanke F, Becker T, Kumar V, Hedtfeld S, Becker C, Cuppens H, Tamm S, Yarden J, Laabs U, Siebert B, et al. 2011. Genes that determine immunology and inflammation modify the basic defect of impaired ion conductance in cystic fibrosis epithelia. J Med Genet 48: 24–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Rommens JM, Corvol H, Li W, Li X, Chiang TA, Lin F, Dorfman R, Busson PF, Parekh RV, et al. 2012. Multiple apical plasma membrane constituents are associated with susceptibility to meconium ileus in individuals with cystic fibrosis. Nat Genet 44: 562–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor C, Commander CW, Collaco JM, Strug LJ, Li W, Wright FA, Webel AD, Pace RG, Stonebraker JR, Naughton K, et al. 2011. A novel lung disease phenotype adjusted for mortality attrition for cystic fibrosis genetic modifier studies. Pediatr Pulmonol 46: 857–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cystic Fibrosis Genotype-Phenotype Consortium 1993. Correlation between genotype and phenotype in patients with cystic fibrosis. N Engl J Med 329: 1308–1313 [DOI] [PubMed] [Google Scholar]
- Trevisiol C, Boniotto M, Giglio L, Poli F, Morgutti M, Crovella S 2005. MBL2 polymorphisms screening in a regional Italian CF Center. J Cyst Fibros 4: 189–191 [DOI] [PubMed] [Google Scholar]
- Van Heyningen V, Yeyati PL 2004. Mechanisms of non-Mendelian inheritance in genetic disease. Hum Mol Genet 13: R225–R233 [DOI] [PubMed] [Google Scholar]
- Vanscoy LL, Blackman SM, Collaco JM, Bowers A, Lai T, Naughton K, Algire M, McWilliams R, Beck S, Hoover-Fong J, et al. 2007. Heritability of lung disease severity in cystic fibrosis. Am J Respir Crit Care Med 175: 1036–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh MJ, Ramsey BW, Accurso FJ, Cutting GR 2001. Cystic fibrosis. In The metabolic and molecular bases of inherited disease (ed. CR Scriver, et al. ), pp. 5121–5188 McGraw-Hill, New York [Google Scholar]
- Wood SD, Adams S, Dunn J, Baler R, Xu A, Pace RG, Kohl A, Yeatts J, Stonebraker JR, Konstan MW, et al. 2007. Inaccuracy of reporting of meconium ileus on case report forms of the gene modifier study. Pediatr Pulmonol Suppl 30: 272 [Google Scholar]
- Wright FA, Strug LJ, Doshi VK, Commander CW, Blackman SM, Sun L, Berthiaume Y, Cutler D, Cojocaru A, Collaco JM, et al. 2011. Genome-wide association and linkage identify modifier loci of lung disease severity in cystic fibrosis at 11p13 and 20q13.2. Nat Genet 43: 539–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarden J, Radojkovic D, De Boeck K, Macek M Jr, Zemkova D, Vavrova V, Vlietinck R, Cassiman JJ, Cuppens H 2004. Polymorphisms in the mannose binding lectin gene affect the cystic fibrosis pulmonary phenotype. J Med Genet 41: 629–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielenski J, Corey M, Rozmahel R, Markiewicz D, Aznarez I, Casals T, Larriba S, Mercier B, Cutting GR, Krebsova A, et al. 1999. Detection of a cystic fibrosis modifier locus for meconium ileus on human chromosome 19q13. Nat Genet 22: 128–129 [DOI] [PubMed] [Google Scholar]