Abstract
Transmission of HIV-1 results in the establishment of a new infection, typically starting from a single virus particle. That virion replicates to generate viremia and persistent infection in all of the lymphoid tissue in the body. HIV-1 preferentially infects T cells with high levels of CD4 and those subsets of T cells that express CCR5, particularly memory T cells. Most of the replicating virus is in the lymphoid tissue, yet most of samples studied are from blood. For the most part the tissue and blood viruses represent a well-mixed population. With the onset of immunodeficiency, the virus evolves to infect new cell types. The tropism switch involves switching from using CCR5 to CXCR4 and corresponds to an expansion of infected cells to include naïve CD4+ T cells. Similarly, the virus evolves the ability to enter cells with low levels of CD4 on the surface and this potentiates the ability to infect macrophages, although the scope of sites where infection of macrophages occurs and the link to pathogenesis is only partly known and is clear only for infection of the central nervous system. A model linking viral evolution to these two pathways has been proposed. Finally, other disease states related to immunodeficiency may be the result of viral infection of additional tissues, although the evidence for a direct role for the virus is less strong. Advancing immunodeficiency creates an environment in which viral evolution results in viral variants that can target new cell types to generate yet another class of opportunistic infections (i.e., HIV-1 with altered tropism).
Over time, HIV-1 can evolve to infect new cell types. For example, it can switch from infecting T cells that express CCR5 to those that express CXCR4, or to infect cells with low levels of CD4 such as macrophages.
The viral population from the time of initiation of infection to the time of overt immunodeficiency undergoes remarkable changes. The large viral population in an infected person is usually founded by a single infected CD4+ T cell in the mucosal tissue proximal to the site of exposure. For much of the time course of the infection, viral evolution is apparent, a result of evading the humoral and cell-mediated immune responses, while the virus continues to replicate in CD4+ T cells using CCR5 as the coreceptor. Initially, T cells in the gut associated lymphoid tissue (GALT) are massively depleted even though a majority of these cells are not in the activated state, which is preferred for HIV-1 infection in cell culture. The massive loss of GALT CD4+ T cells happens early and therefore cannot be the direct cause of immunodeficiency, which occurs late. However, the GALT is likely the source for a significant fraction of the virus in the blood, although the relationship between production of virus in lymphoid tissue and its transfer to the blood is unknown.
Important insights have been gained from examining the dynamics of both the infected cell and free virus particles, especially when the system is perturbed with antiviral drugs. These lessons are summarized by Coffin and Swanstrom (2011) and they fill out the story of virus-host interactions viewed from the perspective of the virus. In most settings, the virus turns over quickly such that changes in the production of virus are readily measured, at least for 99.9% of the virus. Most of the time virus is produced from CD4+ T cells that have a short half-life. However, some cells are latently infected and present a major challenge to eradication of the virus (Siliciano and Greene 2011). Recently it has been possible to identify a variant of HIV-1 that has evolved to replicate in a new cell type with a different half-life (see below). Thus, the dynamics of virus and infected cell turnover offer important lessons into how the virus sustains itself in the host (Coffin and Swanstrom 2011).
Although the long-term persistent replication of virus leads to immunodeficiency, the damage to the host that leads to this state must be multifactorial. The early loss of most of the CD4+ T cells in the GALT results in the translocation of bacterial products beyond the gut, potentially exacerbating one of the key correlates of disease progression—immune activation (Lackner et al. 2011). Loss of the capacity to make T cells and loss of the support structure to mature and regulate T cells may also contribute to the loss of immunologic capacity.
The onset of immunodeficiency sets the stage for opportunistic infections by common microbes that are otherwise controlled by the healthy host. The virus contributes to this phenomenon as shown by the appearance of variants that allow the virus to replicate in new cell types. At any one time, the virus is limited to cell types in which it can maintain a steady-state infection that is not cleared by the immune system. Growth in alternative cells likely is a challenge for the virus because replication in suboptimal cell targets would likely result in slow replication and easier containment by the immune system. With immunodeficiency the host response to replication in alternative cell types would be slower, giving the virus a chance to adapt to the new environment. However, in the clearest cases the virus stays close to home in that it always requires CD4, but can adapt to use lower levels, and it at most swaps one chemokine coreceptor for another, CXCR4 for CCR5. Whether the virus can evolve beyond these limits in an infected host is unknown but the tools for finding such viruses are in place.
In this article we follow the virus from transmission through dissemination and the adaptation to new target cells late in infection. Some of the ideas overlap other articles, which we have cited and which can be consulted for more detail. The potential for the virus to generate a localized replicating population is well established for several tissues/compartments, and it would not be surprising if other clear examples are demonstrated in the future, although such studies are difficult given the inaccessibility of the tissues of interest. Still, the potential for the virus to participate in pathogenic processes beyond immunodeficiency make such questions relevant.
TRANSMISSION
Frequency and Mechanism
Transmission is covered at length by Shaw and Hunter (2011). There are several aspects of transmission that influence viral pathogenesis. In general, transmission rates for HIV-1 are low, with the highest rates suggested to range from 10% per exposure to 0.1% (Powers et al. 2008; Boily et al. 2009). Under these circumstances, it is understandable that the infecting dose is a single virus particle; that is, limiting rates of infection result in infection with the minimal infectious dose. This inference has largely been confirmed with the molecular analysis of HIV-1 present after acute infection. In most cases, systemic infection is established by a single genetic variant in sexual transmission, transmission by intravenous drug use, or vertical transmission, although the frequencies of single variant versus multiple variant infections may vary by route (see Shaw and Hunter 2011).
Nature of the Founder Virus
There is ongoing discussion about where the genetic bottleneck associated with transmission occurs. There may be compartmentalization and restriction of the population in the donor at the site that produces the transmitted virus, although there is little evidence in support of this (see below). Alternatively, there may be physical barriers that limit the exposure in the recipient to very small amounts of virus; such barriers might include mucus and epithelium in the genital tract, which would limit access of virus to susceptible cells. Finally, multiple cells could become infected at the site of transmission, but only one variant succeed in establishing a systemic infection. There are rare examples in which the initially observed virus is not the same as the transmitted virus (Kim et al. 2005; Russell et al. 2011). However, in the setting of low frequency infection in humans, transmission of a single variant appears to be the norm (Shaw and Hunter 2011).
An early observation important for understanding viral pathogenesis was that some viruses isolated from people late in infection were able to grow and cause syncytia in transformed T-cell lines (Asjo et al. 1986), earning the name syncytium-inducing (SI). The remaining viruses were dubbed nonsyncytium-inducing (NSI). These NSI viruses were found early in infection and had some capacity to infect macrophages. This supported the initial dichotomy of less pathogenic/macrophage-tropic viruses being transmitted (van’t Wout et al. 1994), which evolved into more pathogenic SI viruses. This picture has been supplanted with the understanding that SI viruses use CXCR4, which is expressed on transformed T-cell lines, in which the normal HIV-1 coreceptor CCR5 typically is absent, accounting for the apparent pathogenicity in T-cell lines of late viral isolates that have evolved to use CXCR4 (see Wilen et al. 2011). More recently, it was discovered that only a fraction of viruses that use CCR5 can infect macrophages because of the low levels of surface CD4 (see below) and these are not the transmitted viruses. Indeed, viruses with macrophage-tropic Env proteins appear only late in infection, calling into question the role of macrophage infection in systemic infection. Thus the transmitted virus typically uses CCR5 and replicates in activated T cells as evidenced by its need for high levels of surface CD4 (Isaacman-Beck et al. 2009; Salazar-Gonzalez et al. 2009; Alexander et al. 2010).
Early Events in Replication
It is now clear that CD4+ T cells in or near the epithelial layer must represent the initial target cells for mucosal transmission. In vitro, dendritic cells (DCs) can efficiently capture virus and transmit it to target CD4+ T cells in the absence of viral replication (potentially by the high mannose glycans on the viral Env protein [Doores et al. 2010] binding to lectins on the surface of DCs [Wu and KewalRamani 2006]), but the importance of DCs facilitating the initial infection of CD4+ T cells in vivo is still an open question (Martin et al. 2004; Boily-Larouche et al. 2009). In any case, there is little evidence to suggest that viral replication in DCs is an important part of the mechanism of transmission. The initial focal replication in both activated and resting CD4+ T cells in the proximal tissue has been documented in the SIV/macaque model (Zhang et al. 2004; Miller et al. 2005), and this early replication is followed by movement of the virus to proximal lymphoid organs and then to a systemic infection (see Lackner et al. 2011).
There are several potential mechanisms for the virus to move from a localized infection to a systemic infection. The simplest model is that the concentration of virus is sufficiently high in the extracellular space to diffuse to adjacent target cells and tissues. Alternatively, virus could be transported by dendritic cells to proximal lymphoid tissue. Similarly, infected T cells could migrate to different body compartments to deliver virus. Finally, it has been suggested that the viral Env protein in the transmitted virus acts as a mimic of α4β7 integrin to target virus to CD4+ T cells in the GALT (Nawaz et al. 2011); a corollary of this model is that viruses without this feature might initiate a local mucosal infection but not establish a systemic infection because of failure to traffic to the GALT. If stable infections were failing because of an absence of the ability to mimic α4β7 integrin to target GALT one might expect to see seroconversion based on transient replication in the proximal mucosal tissues, as is seen for HSV-2 infection, but this is not observed for putative abortive HIV-1 infections. It will be important to place this binding property of Env in the context of a large sampling of transmitted viruses to determine the frequency of this property.
TARGET CELLS
T-Cell Subsets
CD4 is required for natural isolates of HIV-1 to infect cells. Thus, robust infection of cells is limited to those expressing CD4. The normal function of CD4 is to act as a coreceptor along with the T-cell receptor in binding to Class II MHC, which is on antigen presenting cells and has the role of presenting heterologous peptides to the CD4+ T helper cell. Other cell types can express lower levels of CD4, for example monocytes and macrophages, and it has been reported that CD4 plays an alternative role as the receptor for IL-16 (Liu et al. 1999).
CD4+ T cells are heterogeneous in the expression of the CCR5 coreceptor. In the peripheral blood the memory cell subset and not the naive cell subset expresses significant levels of CCR5, whereas CXCR4 is expressed at relatively high levels on both memory and naive CD4+ T cells (Bleul et al. 1997; Lee et al. 1999; Nicholson et al. 2001). This pattern of CCR5 expression is consistent with memory CD4+ T cells being the predominant cell that is infected in vivo (Sleasman et al. 1996; Douek et al. 2002; Brenchley et al. 2004a), with a minor population of CD8+ T cells that express a low level of CD4 also infected (Cochrane et al. 2004). However, these studies have relied on linking the presence of viral DNA to cell surface markers, as opposed to using a marker of active viral replication. Because of down regulation of CD4 by HIV-1, cells actively producing virus are seen as CD4/CD8 double-negative T cells (Kaiser et al. 2007). In cell culture HIV-1 infects activated cells with much greater efficiency than quiescent cells (Korin and Zack 1998), with central and effector memory cells as the primary targets (Pfaff et al. 2010). However, in vivo there appears to be a type of CD4+ T cell that does not express surface activation markers but supports significant levels of infection, particularly in the gut mucosa (Veazey et al. 2000; Brenchley et al. 2004b; Li et al. 2005; Mattapallil et al. 2005; Mehandru et al. 2007).
As immunodeficiency progresses the virus can evolve to enter cells using CXCR4 (Wilen et al. 2011). The appearance of CXCR4-using viruses (X4 viruses) is correlated with more rapid progression of disease, but it is still unclear if the evolution of these variants is the cause or a marker of rapid disease progression (Schuitemaker et al. 1992; Brumme et al. 2005; Hunt et al. 2006). Both may be true. X4 viruses are rarely transmitted, and it is curious why the virus primarily uses CCR5 when more CD4+ T cells in the blood express CXCR4 than CCR5. However, CCR5 is generally up-regulated with infection and immune activation and CXCR4 is down-regulated (Ostrowski et al. 1998), and CCR5-expressing cells are significantly enriched in lymphoid tissue such as the GALT (Agace et al. 2000; Anton et al. 2000; Veazey et al. 2003; Brenchley et al. 2004b). Naïve CD4+ T cells become more extensively infected either by infection with the X4 virus (Blaak et al. 2000; van Rij et al. 2000) or by expansion of infection into these cells by both the X4 and R5 viruses (Heeregrave et al. 2009). Although R5-to-X4 evolution is common, it is not essential for progression to disease. A large fraction of untreated HIV-infected individuals progress to AIDS and die with no evidence of X4 virus.
There are distinctive sequence features of the Env protein that allow use of CXCR4. An accumulation of basic amino acid substitutions at specific positions in the V3 loop of SU is associated with the coreceptor switch, which is presumed to increase specificity or affinity for CXCR4 (Wilen et al. 2011). There are other positions in Env but outside of V3 that also contribute to the X4 phenotype, but the specific contribution of these other sites is not clear (Hoffman et al. 2002; Pastore et al. 2006; Huang et al. 2008, 2011). There also seem to be differences among the subtypes for their propensity to evolve X4 variants, with X4 variants being more common in subtype D isolates (Tscherning et al. 1998; Huang et al. 2007). The reason for this difference is unclear, although the evolutionary distance between R5 and X4 variants of each subtype might be different. Conversely, X4 variants of subtype C appear to evolve less frequently and often include more dramatic sequence changes in V3, including deletions, compared to the sequence changes seen in subtype B (Coetzer et al. 2006, 2011).
Monocytes, Macrophages, and NK Cells
Monocytes are found in the blood and migrate to tissue where they differentiate into macrophages. This can be mimicked in cell culture by differentiating isolated blood monocytes into macrophages (monocyte-derived macrophages MDM) by exposure to cytokines. Monocytes and dendritic cells isolated from blood express very low levels of CD4 (Lee et al. 1999). Peripheral monocytes have been reported to be infected in vivo (Zhu et al. 2002; Fulcher et al. 2004; Ellery et al. 2007; Xu et al. 2008), along with complex collections of other blood myeloid cells (Centlivre et al. 2011). However, a recent analysis of viral DNA in blood cell subsets failed to detect a significant amount of viral DNA in the monocyte pool (Joseffson et al. 2011). Infection of monocytes in vitro is limited by the low levels of surface CD4 and blocks to entry (Arfi et al. 2008), reduced viral DNA synthesis (Sonza et al. 1996; Triques and Stevenson 2004; Arfi et al. 2008), and reduced viral gene expression (Dong et al. 2009).
Macrophage-tropic viruses infect cells with low levels of surface CD4 (Gorry et al. 2002; Peters et al. 2004; Martin-Garcia et al. 2006; Thomas et al. 2007). The only place where there is clear evidence for the evolution of these viruses is in the brain, where at least a fraction of the cases of HIV-associated dementia involve the presence of macrophage-tropic virus (see Spudich and Gonzalez-Scarano 2011). There are two potential cell targets in the brain, microglia cells (macrophage-like cells in the parenchyma), and perivascular macrophages that migrate into the brain as part of an inflammatory response. It is not known which cell type supports the evolution and replication of macrophage-tropic viruses in the central nervous system (CNS). In addition to detecting these viruses in brain tissue at autopsy, it has now been possible to link the slow decay of virus in the CSF during therapy (Ellis et al. 2000; Haas et al. 2000; Cinque et al. 2001; Schnell et al. 2009), which is distinct from the rapid decay seen in the blood (Ho et al. 1995; Wei et al. 1995), with the presence of macrophage-tropic virus in the CSF, thus indicating replication in a long-lived cell (Schnell et al. 2011).
It has been possible to isolate viruses from blood that can enter macrophages (Li et al. 1999). However, these are not the viruses that evolve in the CNS, which are absent from the blood (Schnell et al. 2011) and the peripheral tissue (Peters et al. 2006). Understanding the range of infection in the body by macrophage-tropic viruses will require sampling different tissues but under conditions of more extensive disease progression to look for these late-evolving variants. In this regard, macrophages appear to be preferentially infected in macaque tissues after extensive depletion of CD4+ T cells in lymphoid tissue by SIV (Veazey et al. 1998; Igarashi et al. 2001).
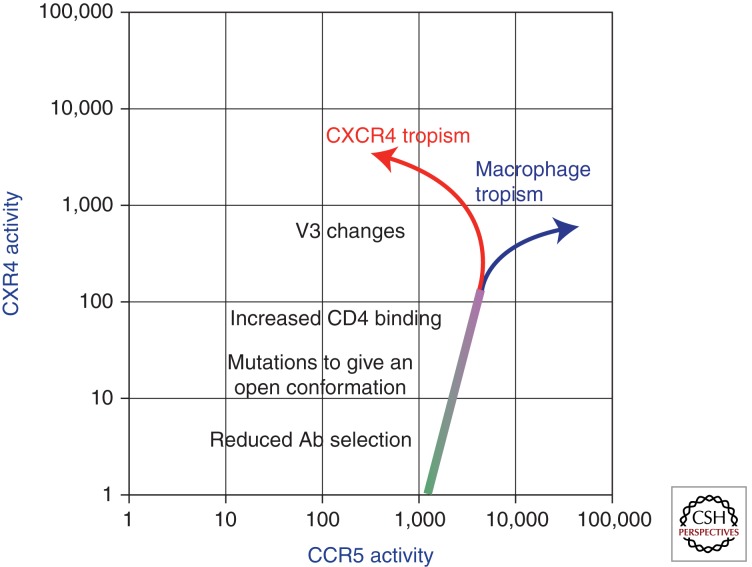
Both X4 viruses and macrophage-tropic viruses evolve from R5 progenitors, which require high levels of CD4, as found on activated T cells. Both of these variants appear late in the disease course and are therefore linked to increasing immunodeficiency of the host. Neither is efficiently transmitted. Thus, they both appear to be evolutionary dead-ends that evolve anew in each host, representing the extension of infection into new cell types (Fig. 1). One evolutionary model is based on adaptation of the Env protein structure in response to a changing host environment (Cheng-Mayer et al. 2009; Ince et al. 2010). This model suggests that in the presence of a weakened antibody response, the Env protein can evolve to be in a more open conformation rather than the closed, neutralization-resistant conformation that avoids sensitivity to antibodies targeting the coreceptor binding face. This concept is shown in Figure 2, in which the evolution of the open conformation may allow faster entry into T cells but also potentiate interaction with CXCR4 and the ability to enter cells with lower levels of CD4, setting the stage for further evolution.
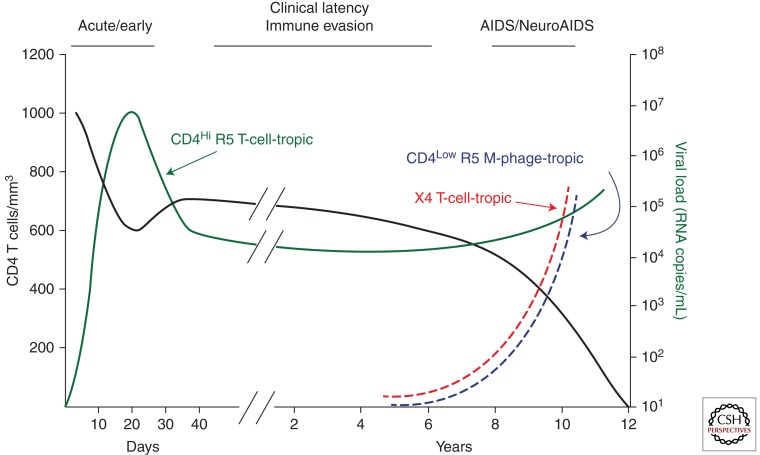
Figure 1.
Time course of a typical HIV-1 infection with the appearance of host range variants late. The time course for different components of the infection are shown. There is an initial loss of CD4+ T cells during acute infection followed by a partial recovery and then a slow decay during the period of clinical latency (black line). There is an initial viremia of the transmitted virus that uses CCR5 (R5) and requires high levels of CD4 to enter cells, and this virus establishes a set point during the period of clinical latency (CD4Hi R5 T-cell-tropic, green line). With the loss of CD4+ T cells there is increasing immunodeficiency (AIDS, including NeuroAIDS), a trend toward increasing viral load, and in a subset of subjects the appearance of host range variants that evolve to use a different coreceptor (X4 T-cell-tropic, red line) or evolve the ability to infect cells, presumably macrophages, with low levels of CD4 (CD4Low R5 M-phage-tropic, blue line).
Figure 2.
Evolution of host range variants. In this model the variant of HIV-1 that is replicating in memory T cells (requiring high levels of CD4 on the surface of the cell and using CCR5 as the coreceptor, shown in green) is exposed to reduced host surveillance in the form of reduced selective pressure from antibodies. This allows the virus to evolve such that the Env protein assumes a more open conformation that allows increased binding to CD4. The open conformation may also expose a latent low level tropism for CXCR4. These changes potentiate the subsequent evolution to use CXCR4 efficiently (X4 virus—red) or to use CD4 more efficiently (macrophage tropism—blue).
NK cells can also be infected in vitro, although with a limited range of viruses (Robinson et al. 1988; Valentin et al. 2002; Harada et al. 2007; Bernstein et al. 2009). Viral DNA can also be found in this cell lineage in vivo (Valentin et al. 2002).
DISSEMINATION AND PERSISTENCE IN TARGET TISSUES
HIV-1 has been isolated from every bodily fluid and it can be assumed the virus will replicate in activated CD4+ T cells virtually anywhere in the body. Thus, the virus is broadly disseminated in the body. Given this wide distribution, it is not clear what the source of virus is in the blood, from which viral samples have been most extensively studied. CD4+ T cells in the blood represent just a few percent of the CD4+ T cells in the body, making it likely that most viral replication is taking place in lymphoid tissues rich in CD4+ T cells. It is not clear whether the virus in the blood is produced by infected cells in tissue or in blood, or if all virus-producing tissues shunt virus into the blood with equal efficiency. Studies of tissues require biopsy or analysis of autopsy material, preventing careful time course studies, and the analysis of viral sequences is usually of DNA, which can include archival and defective viral DNA that may not represent the currently replicating virus. Also, free virus, the most sensitive and reliable instantaneous indicator of the state of infection, is not accessible from solid tissue samples. For this reason, more accessible fluids such as semen, cervico-vaginal mucus, and cerebral spinal fluid are often used as surrogates for the corresponding tissue.
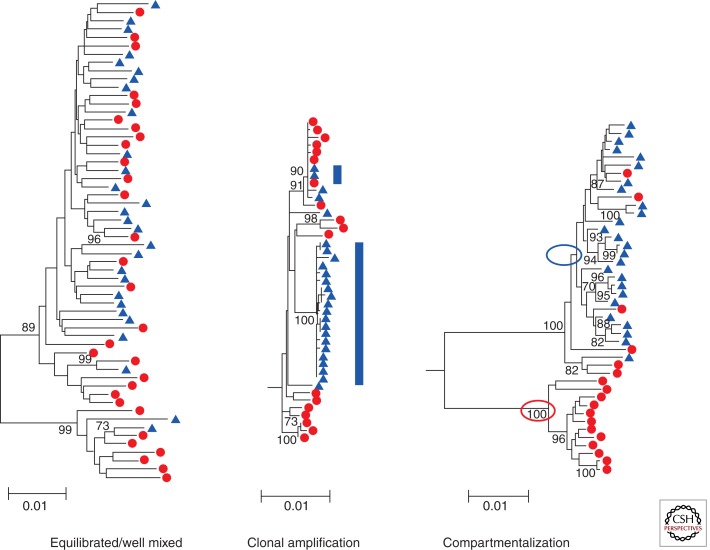
Genetic analysis of virus from tissues (or their liquid surrogates) has generated several unifying observations. In many cases the genetic diversity of the viral population in the blood overlaps the population in the tissue, suggesting a well-mixed relationship in which the virus in the blood is derived from that tissue or imported into the tissue, and if imported into the tissue the virus must undergo little replication given its similarity to the virus in the blood. A variation on the mixing includes a specific lineage of the virus in the compartment disproportionately and transiently expanding, resulting in clonal expansion or amplification of a subset of viral sequences. In a third scenario, the viral population in a tissue can be distinct from the virus in the blood, indicating an independently replicating population that is not exchanging between the compartments. Examples of these types of genetic relationships are shown in Figure 3 (comparing blood and semen, taken from Anderson et al. 2010). One unifying model is that for many tissues virus is imported by some mechanism from the blood compartment at a low level, giving the appearance of equilibrated populations. Only when local viral replication reaches a level in which it significantly increases the local viral RNA load does it become apparent that there is an independently replicating population that can be recognized as genetically distinct.
Figure 3.
Phylogenetic trees demonstrating different relationships between viral populations. In these trees are examples of viral populations in blood (red) and semen (blue). On the left is an example of well-mixed populations with the sequences derived from blood and semen intermingled. In the middle is an example of clonal amplification (blue bars) in which a nearly homogeneous set of sequences appears only in the semen creating a population that is distinct from the blood. On the right is an example of compartmentalization in which the sequences in the semen are distinct from the bulk of the sequences in the blood. These two lineages are indicated by the circles. In addition, sequences present in the seminal tract appear to have migrated back into the blood compartment. (These trees were originally published in Anderson et al. 2010; reprinted with permission from the author.)
Studies of compartmentalization can be confounded by the use of a bulk PCR strategy to generate material for cloning and/or sequencing. The use of cDNA end point dilution followed by multiple PCR amplifications (also called single genome sequencing or single genome amplification) avoids both PCR resampling (which results in artifactual population homogeneity) and PCR-mediated recombination (which confounds the genetic structure of the population) and therefore provides the most reliable source of sequences for the analysis of issues dealing with viral population structure.
Blood and Lymph Nodes
Viral populations in the blood tend to be complex genetically, consistent with a large and diverse population. The rapid decay of viral loads with the initiation of therapy is consistent with 99% of the virus in the blood being produced from short lived cells, presumably activated CD4+ T cells (Ho et al. 1995; Wei et al. 1995). Genetic variants within this population decay at similar rates, including both X4 and R5 variants, indicating that the virus-producing cells have mostly similar half-lives (Ince et al. 2009). One exception is a small percentage of infected cells that appear to be slow to integrate viral DNA, so that when an integrase inhibitor is used an even larger fraction of the viral load is accounted for in cells with a short half-life (Murray et al. 2007). A small fraction of the virus in the blood decays with much slower kinetics. The significance of these populations is discussed by Coffin and Swanstrom (2011) and Siliciano and Greene (2011).
Within the lymph node the infected CD4+ T cells are found outside of the germinal centers within the paracortex (Haase 2011). In addition, there is a diffuse distribution of virus trapped on follicular dendritic cells throughout the lymph node (Lackner et al. 2011). The high efficiency of virus spread between cells has the potential to generate overlapping foci of clonal infection, resulting in local spread of a genetically homogeneous population (Gratton et al. 2000). In this view the virus creates many independent sites of replication in lymphoid tissue driven by high local concentrations of virus, with the complex population seen in the blood the sum of production from all of these independent foci. This effect could limit the chance for recombination to sites of overlapping foci, or require virus produced at distal sites to colonize new tissues to allow encounter of new recombination partners. However, the findings of recombinants in vivo (discussed in Brown et al. 2011), and the ease with which the entire population turns over during development of drug resistance, indicate there must be significant mixing of viral populations between tissues (Shriner et al. 2006). The similarity of the viral sequences at blood and tissue sites implies that they represent the same well-mixed population, consistent with the observation that CTL escape mutations sequentially move through these compartments (Vanderford et al. 2011).
Ongoing viral replication eventually results in irreversible damage in the capacity to replenish CD4+ T cells. The extent of the damage is related to the length of time of infection (or more appropriately to the nadir of CD4+ T-cell count). This damage is seen by an inability to completely replace the CD4+ T-cell compartment after therapy in initiated (Kelley et al. 2009), which may be due in part to depletion of the host capacity to generate CD4+ T cells over time, or to damage of the architecture of the lymphoid tissue that results in reduced capacity to support CD4+ T-cell function (Lackner et al. 2011).
Gut-Associated Lymphoid Tissue (GALT)
The GALT is the largest lymphoid tissue in the body, containing on the order of one-half or more of the lymphoid cells. The CD4+ T cells in this tissue are rapidly depleted in primary infection, more so than in the blood or other lymphoid sites. Curiously, the cells that are depleted are in more of a resting state rather than an activated state, but none-the-less able to support robust viral replication (Li et al. 1999; Veazey et al. 2000; Mehandru et al. 2007). Massive and early damage to the GALT is proposed to contribute to damage to the intestinal lining, which results in the translocation of bacterial products to the blood where they have the potential to enhance the generalized immune activation that is a central feature of HIV disease (see Lackner et al. 2011). Exposure of an epithelial cell monolayer to virus or purified gp120 can impair the integrity of the monolayer through the induction of inflammatory cytokines, as one potential mechanism of damage (Nazli et al. 2010). As noted above, there is little evidence for compartmentalization of viral sequences between blood and GALT, implying frequent exchange of virus or infected cells between these sites (Vanderford et al. 2011). Viral RNA can be detected in rectal secretions (Zuckerman et al. 2004), however, this virus has not been extensively studied in terms of compartmentalization or equilibration with the blood population of virus.
CNS
Virus can be detected in the CSF early after infection and in many subjects throughout infection (Spudich and Gonzalez-Scarano 2011). However, it is not clear if low levels of virus can move (by an unknown mechanism) into the CSF, or if detection of virus in CSF always means there is ongoing viral replication in the CNS itself. Compartmentalized virus, distinct from that in blood, can be detected in the CSF (and in brain tissue at autopsy) late in disease associated with dementia, providing clear evidence for independent replication in the CNS. The compartmentalized virus can involve either macrophage-tropic or T-cell-tropic (R5) virus, which suggests a complex interaction between HIV-1 and CNS tissue (Schnell et al. 2011). An important question in understanding HIV-1 pathogenesis is determining when independent replication can occur in the CNS because such replication is likely to have an associated pathogenic outcome. A compartmentalized CSF population has been detected early in infection associated with a diagnosis of aseptic meningitis, indicating the potential for HIV-1 to establish an independently replicating population of virus associated with a pathogenic outcome even early after infection (Schnell et al. 2010). Compartmentalization has been detected in chronic infection in the presence or absence of neurocognitive impairment (Harrington et al. 2009). Viral replication in the CNS will remain an important consideration of viral evolution given the role of this tissue in pathogenesis.
Genital Tract
There is special interest in virus in the male genital tract (MGT) and in the female genital tract (FGT) as these are the sites involved in sexual transmission. Virus is present in both seminal plasma and FGT mucus in the absence of blood contamination, indicating there must be mechanisms for introduction of virus into genital secretions. A key question is whether genital tract virus is equilibrated with the virus in the blood or is genetically distinct, indicating local sites of viral production.
The MGT contains potential target cells at several anatomical sites (Le Tortorec et al. 2008). It has been reported that viral load does not change after vasectomy, suggesting that virus is not produced in large amounts in the testes (Anderson et al. 1991; Krieger et al. 1998). However, this observation has not been tied to subjects who had evidence of local production of virus in the MGT so the lack of involvement of the testes is clearest for virus that is likely equilibrated with the blood. Virus can be produced locally within the MGT because in a subset of men the virus in semen can be genetically distinct from the virus in the blood (Delwart et al. 1998; Paranjpe et al. 2002; Diem et al. 2008; Anderson et al. 2010). There appear to be two related mechanisms at work in local production of virus. In one there is sustained independent replication to generate a population that is distinct from that in the blood. In the other, there is a rapid short-term expansion of a relatively homogeneous variant that becomes a significant portion of the local population. This pattern of clonal amplification has been observed in the background of virus that is otherwise similar to that in blood or of virus compartmentalized within the MGT (Anderson et al. 2010). Given the limited complexity of the clonally amplified population, it is assumed that this expansion occurs over a short period of time but is not sustained, although the longitudinal relationship between clonal amplification and the long term compartmentalization of a genetically distinct population is not clear.
Virus production in the FGT occurs in the context of a thick layer of squamous epithelium in the vagina and a single cell layer of columnar epithelium starting at the cervix. There is no obvious single tissue mass (equivalent to the prostate, for example) that could provide a source of localized virus production. Several studies have reported compartmentalization of viral sequences in the FMT within at least a subset of subjects (Poss et al. 1998; Kemal et al. 2003; Adal et al. 2005; Andreoletti et al. 2007). More recently, this compartmentalization was interpreted to be largely the result of clonal amplification (Bull et al. 2009). There is a need for both longitudinal analysis of viral populations within the genital tract, and an examination of the impact of concurrent localized bacterial infections with attendant inflammation.
Other Cell and Tissue Types
Long-term infection with HIV-1 can have a variety of clinical manifestations in additional organ systems. The ongoing debate is over the extent to which they reflect infection of cells in the affected organ, dysfunction of cells in the organ caused by interaction with soluble viral proteins, or indirect pathogenesis because of concurrent systemic changes such as constitutive immune activation. As discussed above, HIV-1 evolves to infect cells using a different coreceptor (X4 tropism) and cells with low levels of CD4 (macrophage tropism). Phenotypic and genotypic evidence for viral evolution to infect additional cell types is much less convincing compared to CXCR4 tropism and macrophage tropism. It is possible that alternative cell types are fortuitously infected at a low level in vivo, but it is not clear that infection in these cells can be independently sustained, which would likely be accompanied by some type of cell-specific viral evolution. It is also possible that such evolution goes on in one or more tissue types but signatures of this process have thus far evaded detection. Similarly, the potential pathogenic role of soluble viral proteins is limited by the level of protein in vivo that reaches the target cell or tissue. However, given the difficulty is sampling tissues beyond blood, much of the work addressing these questions has been performed in cell culture systems either by infection or by exposure to purified viral proteins; in some cases this approach has been complemented with work using mice transgenic for HIV-1 genes.
Kidney: HIV-associated nephropathy can play a significant role in end-stage renal disease in infected people (Medapalli et al. 2011). Infection of renal epithelial cells has been frequently invoked to explain this condition. HIV-1 sequences have been detected in these cells (Bruggeman et al. 2000; Marras et al. 2002). The viral Nef protein has been implicated in dysregulation of cell function through interaction with cellular signaling cascades (Husain et al. 2005).
Liver: Another potential site where HIV-1 could have a pathogenic effect is the liver, in which Kupffer cells represent at least 10% of the total cells and are of the macrophage lineage. It is clear that HIV-1 infection can substantially accelerate liver disease in patients coinfected with HIV-1 and HCV (Macias et al. 2009), although it is not known if this is because of general immunodeficiency or local HIV-1 replication. Viral DNA, RNA, and protein have been detected in liver samples from HIV-1-infected subjects (Housset et al. 1990; Cao et al. 1992; van’t Wout et al. 1998). In cell culture systems, HIV-1 has been reported to infect Kupffer cells (Schmitt et al. 1990) and stellate cells (Tuyama et al. 2010). HIV-1 infection of stellate cells may contribute to liver fibrosis by promoting collagen I expression and secretion of the proinflammatory cytokine monocyte chemoattractant protein-1 (Tuyama et al. 2010). Another proposed pathogenic mechanism is induction of apoptosis in hepatocytes after exposure to HCV E2 and HIV gp120 (Munshi et al. 2003; Vlahakis et al. 2003).
Lung: The possibility of local infection in the lung is of interest given the importance of the lung as the site of opportunistic infections and pulmonary TB. Both CD4+ T cells and alveolar macrophages are potential target cells in the lung. The isolation of the macrophage-tropic strain Ba-L from a lavage sample (Gartner et al. 1986) is consistent with the potential for viral replication and evolution in this compartment. Initial reports suggested that independent replication can occur in the lung (Itescu et al. 1994; Nakata et al. 1995). However, more recent analysis of viral populations in blood and lung showed only modest evidence for compartmentalization (including local clonal expansion), representing a relationship that suggests mixing between the R5 T-cell-tropic viruses in the blood and the lung compartment (Heath et al. 2009). One limitation in the work to date in defining the genetics of virus in the lung is that it has not focused on later stage infections when expansion of virus into new cell types, particularly macrophages, is more likely to be occurring, such as during severe immunodeficiency (Jeffrey et al. 1991).
Hematopoietic stem cells: The potential for infection of HSC was shown many years ago (Folks et al. 1988; Stanley et al. 1992), although they are relatively difficult to infect in culture (Shen et al. 1999; Zhang et al. 2007). More recently, interest in this phenomenon has focused on potential pathogenic outcomes such as anemia (Redd et al. 2007), or as a source of long-lived virus making up part of the HIV-1 latent reservoir (Carter et al. 2010). For subtype B HIV-1 the ability to infect a multipotent cell appears to be restricted to X4 viruses (Carter et al. 2011).
Breast: Viral dissemination into breast tissue is relevant in the context of transmission during breast feeding. Recent work comparing virus in blood and breast milk has indicated that compartmentalization is largely limited to clonal amplification (Gantt et al. 2010; Salazar-Gonzalez et al. 2011). It is possible that milk production results in a flushing effect of this compartment that limits the opportunity for sustained localized replication.
MECHANISM OF CELL KILLING
The mechanisms by which an infected cell dies remain controversial. In culture, infected CD4+ T cells die in a matter of days, whereas infected monocyte-derived macrophages can produce virus over a period of weeks, indicating there is nothing inherent about viral replication that leads to cell death. Indeed many retroviruses infect cells without killing them. There must be specific features of the virus and its interaction with the host cell that are responsible for cell killing. The ability to kill CD4+ T cells in culture is consistent with the short half-life of virus in the blood after the initiation of therapy (Ho et al. 1995; Wei et al. 1995). That this is in large part a virologic effect is seen by the fact that the decay of virus in the blood goes on at a similar rate in the presence or absence of CD8+ cytotoxic T cells (Klatt et al. 2010). In addition to the direct killing of infected cells, a number of mechanisms have been proposed that result in indirect killing of uninfected cells because of their proximity to infected cells and the general state of immune activation (Lackner et al. 2011).
Many viral proteins have been implicated as participating in cellular apoptotic (and antiapoptotic) pathways. Expression of Env protein is toxic to the infected cell and surface expression can mediate syncytium formation or interaction with chemokine receptors on adjacent cells. However, other viral proteins have also been implicated in several cellular apoptotic pathways (Varbanov et al. 2006). Recently, a proteolytic fragment of Caspase 8, generated by the viral protease, has been shown to induce apoptosis through Caspase 9 activation (Sainski et al. 2011). Also, partial viral DNA products have been implicated in a cell killing mechanism of abortive infection that may contribute to apparent indirect killing (Zhou et al. 2008; Doitsh et al. 2010). It is clear that during acute infection massive loss of CD4+ T cells is largely the result of direct killing of infected cells (Li et al. 2005; Mattapallil et al. 2005). However, the mix of direct killing versus indirect killing over the course of the infection and the range of mechanisms involved remain under discussion.
SUMMARY
Viral dissemination and evolution are central to HIV pathogenesis and the constantly changing virus-host interaction. Evidence for evolution to infect new cell types is strongest for X4 viruses and macrophage-tropic viruses, but even in these cases their contributions to viral pathogenesis are incompletely understood. Many additional mechanisms of viral pathogenesis have been proposed and will continue to be explored. The evolution of virus to form distinct replicating populations, immune escape, or distinct entry phenotypes has provided the most compelling evidence for the virus responding to a changing virus-host environment. Linking viral biology to genetic markers of viral evolution will continue to provide a strong experimental paradigm for viral pathogenesis.
ACKNOWLEDGMENTS
We would like to thank Elena Dukhovlinova and Sarah Joseph for assistance with specific sections. We also thank Elizabeth Pollom for Figure 1 and William Ince for Figure 2. The authors’ own research has been supported by funding from the National Institutes of Health.
Footnotes
Editors: Frederic D. Bushman, Gary J. Nabel, and Ronald Swanstrom
Additional Perspectives on HIV available at www.perspectivesinmedicine.org
REFERENCES
*Reference is also in this collection.
- Adal M, Ayele W, Wolday D, Dagne K, Messele T, Tilahun T, Berkhout B, Mayaan S, Pollakis G, Dorigo-Zetsma W 2005. Evidence of genetic variability of human immunodeficiency virus type 1 in plasma and cervicovaginal lavage in Ethiopian women seeking care for sexually transmitted infections. AIDS Res Hum Retroviruses 21: 649–653 [DOI] [PubMed] [Google Scholar]
- Agace WW, Roberts AI, Wu L, Greineder C, Ebert EC, Parker CM 2000. Human intestinal lamina propria and intraepithelial lymphocytes express receptors specific for chemokines induced by inflammation. Eur J Immunol 30: 819–826 [DOI] [PubMed] [Google Scholar]
- Alexander M, Lynch R, Mulenga J, Allen S, Derdeyn CA, Hunter E 2010. Donor and recipient envs from heterosexual human immunodeficiency virus subtype C transmission pairs require high receptor levels for entry. J Virol 84: 4100–4104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DJ, Politch JA, Martinez A, Van Voorhis BJ, Padian NS, O’Brien TR 1991. White blood cells and HIV-1 in semen from vasectomised seropositive men. Lancet 338: 573–574 [DOI] [PubMed] [Google Scholar]
- Anderson JA, Ping LH, Dibben O, Jabara CB, Arney L, Kincer L, Tang Y, Hobbs M, Hoffman I, Kazembe P, et al. 2010. HIV-1 Populations in Semen Arise through Multiple Mechanisms. PLoS Pathog 6: e1001053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreoletti L, Skrabal K, Perrin V, Chomont N, Saragosti S, Gresenguet G, Moret H, Jacques J, Longo Jde D, Matta M, et al. 2007. Genetic and phenotypic features of blood and genital viral populations of clinically asymptomatic and antiretroviral-treatment-naive clade a human immunodeficiency virus type 1-infected women. J Clin Microbiol 45: 1838–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anton PA, Elliott J, Poles MA, McGowan IM, Matud J, Hultin LE, Grovit-Ferbas K, Mackay CR, Chen ISY, Giorgi JV 2000. Enhanced levels of functional HIV-1 co-receptors on human mucosal T cells demonstrated using intestinal biopsy tissue. AIDS 14: 1761–1765 [DOI] [PubMed] [Google Scholar]
- Arfi V, Riviere L, Jarrosson-Wuilleme L, Goujon C, Rigal D, Darlix JL, Cimarelli A 2008. Characterization of the early steps of infection of primary blood monocytes by human immunodeficiency virus type 1. J Virol 82: 6557–6565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asjo B, Morfeldt-Manson L, Albert J, Biberfeld G, Karlsson A, Lidman K, Fenyo EM 1986. Replicative capacity of human immunodeficiency virus from patients with varying severity of HIV infection. Lancet 2: 660–662 [PubMed] [Google Scholar]
- Bernstein HB, Wang G, Plasterer MC, Zack JA, Ramasastry P, Mumenthaler SM, Kitchen CM 2009. CD4+ NK cells can be productively infected with HIV, leading to downregulation of CD4 expression and changes in function. Virology 387: 59–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaak H, van’t Wout AB, Brouwer M, Hooibrink B, Hovenkamp E, Schuitemaker H 2000. In vivo HIV-1 infection of CD45RA+CD4+ T cells is established primarily by syncytium-inducing variants and correlates with the rate of CD4+ T cell decline. Proc Natl Acad Sci 97: 1269–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleul CC, Wu L, Hoxie JA, Springer TA, Mackay CR 1997. The HIV coreceptors CXCR4 and CCR5 are differentially expressed and regulated on human T lymphocytes. Proc Natl Acad Sci 94: 1925–1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boily MC, Baggaley RF, Wang L, Masse B, White RG, Hayes RJ, Alary M 2009. Heterosexual risk of HIV-1 infection per sexual act: Systematic review and meta-analysis of observational studies. Lancet Infect Dis 9: 118–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boily-Larouche G, Iscache AL, Zijenah LS, Humphrey JH, Mouland AJ, Ward BJ, Roger M 2009. Functional genetic variants in DC-SIGNR are associated with mother-to-child transmission of HIV-1. PLoS One 4: e7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenchley JM, Hill BJ, Ambrozak DR, Price DA, Guenaga FJ, Casazza JP, Kuruppu J, Yazdani J, Migueles SA, Connors M, et al. 2004a. T-cell subsets that harbor human immunodeficiency virus (HIV) in vivo: Implications for HIV pathogenesis. J Virol 78: 1160–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenchley JM, Schacker TW, Ruff LE, Price DA, Taylor JH, Beilman GJ, Nguyen PL, Khoruts A, Larson M, Haase AT, et al. 2004b. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J Exp Med 200: 749–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RJ, Peters PJ, Caron C, Gonzalez-Perez MP, Stones L, Ankghuambom C, Pondei K, McClure CP, Alemnji G, Taylor S, et al. 2011. Intercompartmental recombination of HIV-1 contributes to env intrahost diversity and modulates viral tropism and sensitivity to entry inhibitors. J Virol 85: 6024–6037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggeman LA, Ross MD, Tanji N, Cara A, Dikman S, Gordon RE, Burns GC, D’Agati VD, Winston JA, Klotman ME, et al. 2000. Renal epithelium is a previously unrecognized site of HIV-1 infection. J Am Soc Nephrol 11: 2079–2087 [DOI] [PubMed] [Google Scholar]
- Brumme ZL, Goodrich J, Mayer HB, Brumme CJ, Henrick BM, Wynhoven B, Asselin JJ, Cheung PK, Hogg RS, Montaner JS, et al. 2005. Molecular and clinical epidemiology of CXCR4-using HIV-1 in a large population of antiretroviral-naive individuals. J Infect Dis 192: 466–474 [DOI] [PubMed] [Google Scholar]
- Bull M, Learn G, Genowati I, McKernan J, Hitti J, Lockhart D, Tapia K, Holte S, Dragavon J, Coombs R, et al. 2009. Compartmentalization of HIV-1 within the female genital tract is due to monotypic and low-diversity variants not distinct viral populations. PLoS One 4: e7122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao YZ, Dieterich D, Thomas PA, Huang YX, Mirabile M, Ho DD 1992. Identification and quantitation of HIV-1 in the liver of patients with AIDS. AIDS 6: 65–70 [DOI] [PubMed] [Google Scholar]
- Carter CC, Onafuwa-Nuga A, McNamara LA, Riddell JT, Bixby D, Savona MR, Collins KL 2010. HIV-1 infects multipotent progenitor cells causing cell death and establishing latent cellular reservoirs. Nat Med 16: 446–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter CC, McNamara LA, Onafuwa-Nuga A, Shackleton M, Riddell J 4th, Bixby D, Savona MR, Morrison SJ, Collins KL 2011. HIV-1 utilizes the CXCR4 chemokine receptor to infect multipotent hematopoietic stem and progenitor cells. Cell Host Microbe 9: 223–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centlivre M, Legrand N, Steingrover R, van der Sluis R, Grijsen ML, Bakker M, Jurriaans S, Berkhout B, Paxton WA, Prins JM, et al. 2011. Altered dynamics and differential infection profiles of lymphoid and myeloid cell subsets during acute and chronic HIV-1 infection. J Leukoc Biol 89: 785–795 [DOI] [PubMed] [Google Scholar]
- Cheng-Mayer C, Tasca S, Ho SH 2009. Coreceptor switch in infection of nonhuman primates. Curr HIV Res 7: 30–38 [DOI] [PubMed] [Google Scholar]
- Cinque P, Presi S, Bestetti A, Pierotti C, Racca S, Boeri E, Morelli P, Carrera P, Ferrari M, Lazzarin A 2001. Effect of genotypic resistance on the virological response to highly active antiretroviral therapy in cerebrospinal fluid. AIDS Res Hum Retroviruses 17: 377–383 [DOI] [PubMed] [Google Scholar]
- Cochrane A, Imlach S, Leen C, Scott G, Kennedy D, Simmonds P 2004. High levels of human immunodeficiency virus infection of CD8 lymphocytes expressing CD4 in vivo. J Virol 78: 9862–9871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coetzer M, Cilliers T, Ping LH, Swanstrom R, Morris L 2006. Genetic characteristics of the V3 region associated with CXCR4 usage in HIV-1 subtype C isolates. Virology 356: 95–105 [DOI] [PubMed] [Google Scholar]
- Coetzer M, Nedellec R, Cilliers T, Meyers T, Morris L, Mosier DE 2011. Extreme genetic divergence is required for coreceptor switching in HIV-1 subtype C. J Acquir Immune Defic Syndr 56: 9–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Coffin L, Swanstrom R 2011. HIV pathogenesis: Dynamics and genetics of viral populations and infected cells. Cold Spring Harb Perspect Med 10.1101/cshperspect.a012526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delwart EL, Mullins JI, Gupta P, Learn GH Jr, Holodniy M, Katzenstein D, Walker BD, Singh MK 1998. Human immunodeficiency virus type 1 populations in blood and semen. J Virol 72: 617–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diem K, Nickle DC, Motoshige A, Fox A, Ross S, Mullins JI, Corey L, Coombs RW, Krieger JN 2008. Male genital tract compartmentalization of human immunodeficiency virus type 1 (HIV). AIDS Res Hum Retroviruses 24: 561–571 [DOI] [PubMed] [Google Scholar]
- Doitsh G, Cavrois M, Lassen KG, Zepeda O, Yang Z, Santiago ML, Hebbeler AM, Greene WC 2010. Abortive HIV infection mediates CD4 T cell depletion and inflammation in human lymphoid tissue. Cell 143: 789–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Kwas C, Wu L 2009. Transcriptional restriction of human immunodeficiency virus type 1 gene expression in undifferentiated primary monocytes. J Virol 83: 3518–3527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doores KJ, Bonomelli C, Harvey DJ, Vasiljevic S, Dwek RA, Burton DR, Crispin M, Scanlan CN 2010. Envelope glycans of immunodeficiency virions are almost entirely oligomannose antigens. Proc Natl Acad Sci 107: 13800–13805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douek DC, Brenchley JM, Betts MR, Ambrozak DR, Hill BJ, Okamoto Y, Casazza JP, Kuruppu J, Kunstman K, Wolinsky S, et al. 2002. HIV preferentially infects HIV-specific CD4+ T cells. Nature 417: 95–98 [DOI] [PubMed] [Google Scholar]
- Ellery PJ, Tippett E, Chiu YL, Paukovics G, Cameron PU, Solomon A, Lewin SR, Gorry PR, Jaworowski A, Greene WC, et al. 2007. The CD16+ monocyte subset is more permissive to infection and preferentially harbors HIV-1 in vivo. J Immunol 178: 6581–6589 [DOI] [PubMed] [Google Scholar]
- Ellis RJ, Gamst AC, Capparelli E, Spector SA, Hsia K, Wolfson T, Abramson I, Grant I, McCutchan JA 2000. Cerebrospinal fluid HIV RNA originates from both local CNS and systemic sources. Neurology 54: 927–936 [DOI] [PubMed] [Google Scholar]
- Folks TM, Kessler SW, Orenstein JM, Justement JS, Jaffe ES, Fauci AS 1988. Infection and replication of HIV-1 in purified progenitor cells of normal human bone marrow. Science 242: 919–922 [DOI] [PubMed] [Google Scholar]
- Fulcher JA, Hwangbo Y, Zioni R, Nickle D, Lin X, Heath L, Mullins JI, Corey L, Zhu T 2004. Compartmentalization of human immunodeficiency virus type 1 between blood monocytes and CD4+ T cells during infection. J Virol 78: 7883–7893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantt S, Carlsson J, Heath L, Bull ME, Shetty AK, Mutsvangwa J, Musingwini G, Woelk G, Zijenah LS, Katzenstein DA, et al. 2010. Genetic analyses of HIV-1 env sequences demonstrate limited compartmentalization in breast milk and suggest viral replication within the breast that increases with mastitis. J Virol 84: 10812–10819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner S, Markovits P, Markovitz DM, Kaplan MH, Gallo RC, Popovic M 1986. The role of mononuclear phagocytes in HTLV-III/LAV infection. Science 233: 215–219 [DOI] [PubMed] [Google Scholar]
- Gorry PR, Taylor J, Holm GH, Mehle A, Morgan T, Cayabyab M, Farzan M, Wang H, Bell JE, Kunstman K, et al. 2002. Increased CCR5 affinity and reduced CCR5/CD4 dependence of a neurovirulent primary human immunodeficiency virus type 1 isolate. J Virol 76: 6277–6292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratton S, Cheynier R, Dumaurier MJ, Oksenhendler E, Wain-Hobson S 2000. Highly restricted spread of HIV-1 and multiply infected cells within splenic germinal centers. Proc Natl Acad Sci 97: 14566–14571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas DW, Clough LA, Johnson BW, Harris VL, Spearman P, Wilkinson GR, Fletcher CV, Fiscus S, Raffanti S, Donlon R, et al. 2000. Evidence of a source of HIV type 1 within the central nervous system by ultraintensive sampling of cerebrospinal fluid and plasma. AIDS Res Hum Retroviruses 16: 1491–1502 [DOI] [PubMed] [Google Scholar]
- Haase AT 2011. Early events in sexual transmission of HIV and SIV and opportunities for interventions. Annu Rev Med 62: 127–139 [DOI] [PubMed] [Google Scholar]
- Harada H, Goto Y, Ohno T, Suzu S, Okada S 2007. Proliferative activation up-regulates expression of CD4 and HIV-1 co-receptors on NK cells and induces their infection with HIV-1. Eur J Immunol 37: 2148–2155 [DOI] [PubMed] [Google Scholar]
- Harrington PR, Schnell G, Letendre SL, Ritola K, Robertson K, Hall C, Burch CL, Jabara CB, Moore DT, Ellis RJ, et al. 2009. Cross-sectional characterization of HIV-1 env compartmentalization in cerebrospinal fluid over the full disease course. AIDS 23: 907–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath L, Fox A, McClure J, Diem K, van 't Wout AB, Zhao H, Park DR, Schouten JT, Twigg HL, Corey L, Mullins JI, Mittler JE 2009. Evidence for limited genetic compartmentalization of HIV-1 between lung and blood. PLoS One 4: e6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heeregrave EJ, Geels MJ, Brenchley JM, Baan E, Ambrozak DR, van der Sluis RM, Bennemeer R, Douek DC, Goudsmit J, Pollakis G, et al. 2009. Lack of in vivo compartmentalization among HIV-1 infected naive and memory CD4+ T cell subsets. Virology 393: 24–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho DD, Neumann AU, Perelson AS, Chen W, Leonard JM, Markowitz M 1995. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 373: 123–126 [DOI] [PubMed] [Google Scholar]
- Hoffman NG, Seillier-Moiseiwitsch F, Ahn J, Walker JM, Swanstrom R 2002. Variability in the human immunodeficiency virus type 1 gp120 Env protein linked to phenotype-associated changes in the V3 loop. J Virol 76: 3852–3864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Housset C, Lamas E, Brechot C 1990. Detection of HIV1 RNA and pp24 antigen in HIV1-infected human liver. Res Virol 141: 153–159 [DOI] [PubMed] [Google Scholar]
- Huang W, Eshleman SH, Toma J, Fransen S, Stawiski E, Paxinos EE, Whitcomb JM, Young AM, Donnell D, Mmiro F, et al. 2007. Coreceptor tropism in human immunodeficiency virus type 1 subtype D: High prevalence of CXCR4 tropism and heterogeneous composition of viral populations. J Virol 81: 7885–7893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Toma J, Fransen S, Stawiski E, Reeves JD, Whitcomb JM, Parkin N, Petropoulos CJ 2008. Coreceptor tropism can be influenced by amino acid substitutions in the gp41 transmembrane subunit of human immunodeficiency virus type 1 envelope protein. J Virol 82: 5584–5593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Frantzell A, Toma J, Fransen S, Whitcomb JM, Stawiski E, Petropoulos CJ 2011. Mutational pathways and genetic barriers to CXCR4-mediated entry by human immunodeficiency virus type 1. Virology 409: 308–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt PW, Harrigan PR, Huang W, Bates M, Williamson DW, McCune JM, Price RW, Spudich SS, Lampiris H, Hoh R, et al. 2006. Prevalence of CXCR4 tropism among antiretroviral-treated HIV-1-infected patients with detectable viremia. J Infect Dis 194: 926–930 [DOI] [PubMed] [Google Scholar]
- Husain M, D’Agati VD, He JC, Klotman ME, Klotman PE 2005. HIV-1 Nef induces dedifferentiation of podocytes in vivo: A characteristic feature of HIVAN. AIDS 19: 1975–1980 [DOI] [PubMed] [Google Scholar]
- Igarashi T, Brown CR, Endo Y, Buckler-White A, Plishka R, Bischofberger N, Hirsch V, Martin MA 2001. Macrophage are the principal reservoir and sustain high virus loads in rhesus macaques after the depletion of CD4+ T cells by a highly pathogenic simian immunodeficiency virus/HIV type 1 chimera (SHIV): Implications for HIV-1 infections of humans. Proc Natl Acad Sci 98: 658–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ince WL, Harrington PR, Schnell GL, Patel-Chhabra M, Burch CL, Menezes P, Price RW, Eron JJ Jr, Swanstrom RI 2009. Major coexisting human immunodeficiency virus type 1 env gene subpopulations in the peripheral blood are produced by cells with similar turnover rates and show little evidence of genetic compartmentalization. J Virol 83: 4068–4080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ince WL, Zhang L, Jiang Q, Arrildt K, Su L, Swanstrom R 2010. Evolution of the HIV-1 env gene in the Raγ2−/−γC−/− humanized mouse model. J Virol 84: 2740–2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacman-Beck J, Hermann EA, Yi Y, Ratcliffe SJ, Mulenga J, Allen S, Hunter E, Derdeyn CA, Collman RG 2009. Heterosexual transmission of human immunodeficiency virus type 1 subtype C: Macrophage tropism, alternative coreceptor use, and the molecular anatomy of CCR5 utilization. J Virol 83: 8208–8220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itescu S, Simonelli PF, Winchester RJ, Ginsberg HS 1994. Human immunodeficiency virus type 1 strains in the lungs of infected individuals evolve independently from those in peripheral blood and are highly conserved in the C-terminal region of the envelope V3 loop. Proc Natl Acad Sci 91: 11378–11382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey AA, Israel-Biet D, Andrieu JM, Even P, Venet A 1991. HIV isolation from pulmonary cells derived from bronchoalveolar lavage. Clin Exp Immunol 84: 488–492 [PMC free article] [PubMed] [Google Scholar]
- Josefsson L, King MS, Makitalo B, Brönnström H, Shao W, Maldarelli F, Kearney MF, Hu WS, Chen J, Gaines H, et al. 2011. Majority of CD4+ T cells from peripheral blood of HIV-1-infected individuals contain only one HIV DNA molecule. Proc Natl Acad Sci 108: 11199–11204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser P, Joos B, Niederost B, Weber R, Gunthard HF, Fischer M 2007. Productive human immunodeficiency virus type 1 infection in peripheral blood predominantly takes place in CD4/CD8 double-negative T lymphocytes. J Virol 81: 9693–9706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley CF, Kitchen CM, Hunt PW, Rodriguez B, Hecht FM, Kitahata M, Crane HM, Willig J, Mugavero M, Saag M, et al. 2009. Incomplete peripheral CD4+ cell count restoration in HIV-infected patients receiving long-term antiretroviral treatment. Clin Infect Dis 48: 787–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemal KS, Foley B, Burger H, Anastos K, Minkoff H, Kitchen C, Philpott SM, Gao W, Robison E, Holman S, et al. 2003. HIV-1 in genital tract and plasma of women: Compartmentalization of viral sequences, coreceptor usage, and glycosylation. Proc Natl Acad Sci 100: 12972–12977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EY, Busch M, Abel K, Fritts L, Bustamante P, Stanton J, Lu D, Wu S, Glowczwskie J, Rourke T, et al. 2005. Retroviral recombination in vivo: Viral replication patterns and genetic structure of simian immunodeficiency virus (SIV) populations in rhesus macaques after simultaneous or sequential intravaginal inoculation with SIVmac239δvpx/δvpr and SIVmac239δnef. J Virol 79: 4886–4895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klatt NR, Shudo E, Ortiz AM, Engram JC, Paiardini M, Lawson B, Miller MD, Else J, Pandrea I, Estes JD, et al. 2010. CD8+ lymphocytes control viral replication in SIVmac239-infected rhesus macaques without decreasing the lifespan of productively infected cells. PLoS Pathog 6: e1000747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korin YD, Zack JA 1998. Progression to the G1b phase of the cell cycle is required for completion of human immunodeficiency virus type 1 reverse transcription in T cells. J Virol 72: 3161–3168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger JN, Nirapathpongporn A, Chaiyaporn M, Peterson G, Nikolaeva I, Akridge R, Ross SO, Coombs RW 1998. Vasectomy and human immunodeficiency virus type 1 in semen. J Urol 159: 820–825; discussion 825–826 [PubMed] [Google Scholar]
- *.Lackner AA, Lederman MM, Rodriguez B HIV pathogenesis—The host. Cold Spring Harb Perspect Med 10.1101/cshperspect.a007005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Tortorec A, Satie AP, Denis H, Rioux-Leclercq N, Havard L, Ruffault A, Jegou B, Dejucq-Rainsford N 2008. Human prostate supports more efficient replication of HIV-1 R5 than X4 strains ex vivo. Retrovirology 5: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B, Sharron M, Montaner LJ, Weissman D, Doms RW 1999. Quantification of CD4, CCR5, and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages. Proc Natl Acad Sci 96: 5215–5220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Juarez J, Alali M, Dwyer D, Collman R, Cunningham A, Naif HM 1999. Persistent CCR5 utilization and enhanced macrophage tropism by primary blood human immunodeficiency virus type 1 isolates from advanced stages of disease and comparison to tissue-derived isolates. J Virol 73: 9741–9755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Duan L, Estes JD, Ma ZM, Rourke T, Wang Y, Reilly C, Carlis J, Miller CJ, Haase AT 2005. Peak SIV replication in resting memory CD4+ T cells depletes gut lamina propria CD4+ T cells. Nature 434: 1148–1152 [DOI] [PubMed] [Google Scholar]
- Liu Y, Cruikshank WW, O’Loughlin T, O’Reilly P, Center DM, Kornfeld H 1999. Identification of a CD4 domain required for interleukin-16 binding and lymphocyte activation. J Biol Chem 274: 23387–23395 [DOI] [PubMed] [Google Scholar]
- Macias J, Berenguer J, Japon MA, Giron JA, Rivero A, Lopez-Cortes LF, Moreno A, Gonzalez-Serrano M, Iribarren JA, Ortega E, et al. 2009. Fast fibrosis progression between repeated liver biopsies in patients coinfected with human immunodeficiency virus/hepatitis C virus. Hepatology 50: 1056–1063 [DOI] [PubMed] [Google Scholar]
- Marras D, Bruggeman LA, Gao F, Tanji N, Mansukhani MM, Cara A, Ross MD, Gusella GL, Benson G, D’Agati VD, et al. 2002. Replication and compartmentalization of HIV-1 in kidney epithelium of patients with HIV-associated nephropathy. Nat Med 8: 522–526 [DOI] [PubMed] [Google Scholar]
- Martin MP, Lederman MM, Hutcheson HB, Goedert JJ, Nelson GW, van Kooyk Y, Detels R, Buchbinder S, Hoots K, Vlahov D, et al. 2004. Association of DC-SIGN promoter polymorphism with increased risk for parenteral, but not mucosal, acquisition of human immunodeficiency virus type 1 infection. J Virol 78: 14053–14056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Garcia J, Cao W, Varela-Rohena A, Plassmeyer ML, Gonzalez-Scarano F 2006. HIV-1 tropism for the central nervous system: Brain-derived envelope glycoproteins with lower CD4 dependence and reduced sensitivity to a fusion inhibitor. Virology 346: 169–179 [DOI] [PubMed] [Google Scholar]
- Mattapallil JJ, Douek DC, Hill B, Nishimura Y, Martin M, Roederer M 2005. Massive infection and loss of memory CD4+ T cells in multiple tissues during acute SIV infection. Nature 434: 1093–1097 [DOI] [PubMed] [Google Scholar]
- Medapalli RK, He JC, Klotman PE 2011. HIV-associated nephropathy: Pathogenesis. Curr Opin Nephrol Hypertens 20: 306–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehandru S, Poles MA, Tenner-Racz K, Manuelli V, Jean-Pierre P, Lopez P, Shet A, Low A, Mohri H, Boden D, et al. 2007. Mechanisms of gastrointestinal CD4+ T-cell depletion during acute and early human immunodeficiency virus type 1 infection. J Virol 81: 599–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CJ, Li Q, Abel K, Kim EY, Ma ZM, Wietgrefe S, La Franco-Scheuch L, Compton L, Duan L, Shore MD, et al. 2005. Propagation and dissemination of infection after vaginal transmission of simian immunodeficiency virus. J Virol 79: 9217–9227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munshi N, Balasubramanian A, Koziel M, Ganju RK, Groopman JE 2003. Hepatitis C and human immunodeficiency virus envelope proteins cooperatively induce hepatocytic apoptosis via an innocent bystander mechanism. J Infect Dis 188: 1192–1204 [DOI] [PubMed] [Google Scholar]
- Murray JM, Emery S, Kelleher AD, Law M, Chen J, Hazuda DJ, Nguyen BY, Teppler H, Cooper DA 2007. Antiretroviral therapy with the integrase inhibitor raltegravir alters decay kinetics of HIV, significantly reducing the second phase. AIDS 21: 2315–2321 [DOI] [PubMed] [Google Scholar]
- Nakata K, Weiden M, Harkin T, Ho D, Rom WN 1995. Low copy number and limited variability of proviral DNA in alveolar macrophages from HIV-1-infected patients: Evidence for genetic differences in HIV-1 between lung and blood macrophage populations. Mol Med 1: 744–757 [PMC free article] [PubMed] [Google Scholar]
- Nawaz F, Cicala C, Van Ryk D, Block KE, Jelicic K, McNally JP, Ogundare O, Pascuccio M, Patel N, Wei D, et al. 2011. The genotype of early-transmitting HIV gp120s promotes αβ-reactivity, revealing αβCD4+ T cells as key targets in mucosal transmission. PLoS Pathog 7: e1001301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazli A, Chan O, Dobson-Belaire WN, Ouellet M, Tremblay MJ, Gray-Owen SD, Arsenault AL, Kaushic C 2010. Exposure to HIV-1 directly impairs mucosal epithelial barrier integrity allowing microbial translocation. PLoS Pathog 6: e1000852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson JK, Browning SW, Hengel RL, Lew E, Gallagher LE, Rimland D, McDougal JS 2001. CCR5 and CXCR4 expression on memory and naive T cells in HIV-1 infection and response to highly active antiretroviral therapy. J Acquir Immune Defic Syndr 27: 105–115 [DOI] [PubMed] [Google Scholar]
- Ostrowski MA, Justement SJ, Catanzaro A, Hallahan CA, Ehler LA, Mizell SB, Kumar PN, Mican JA, Chun TW, Fauci AS 1998. Expression of chemokine receptors CXCR4 and CCR5 in HIV-1-infected and uninfected individuals. J Immunol 161: 3195–3201 [PubMed] [Google Scholar]
- Paranjpe S, Craigo J, Patterson B, Ding M, Barroso P, Harrison L, Montelaro R, Gupta P 2002. Subcompartmentalization of HIV-1 quasispecies between seminal cells and seminal plasma indicates their origin in distinct genital tissues. AIDS Res Hum Retroviruses 18: 1271–1280 [DOI] [PubMed] [Google Scholar]
- Pastore C, Nedellec R, Ramos A, Pontow S, Ratner L, Mosier DE 2006. Human immunodeficiency virus type 1 coreceptor switching: V1/V2 gain-of-fitness mutations compensate for V3 loss-of-fitness mutations. J Virol 80: 750–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters PJ, Bhattacharya J, Hibbitts S, Dittmar MT, Simmons G, Bell J, Simmonds P, Clapham PR 2004. Biological analysis of human immunodeficiency virus type 1 R5 envelopes amplified from brain and lymph node tissues of AIDS patients with neuropathology reveals two distinct tropism phenotypes and identifies envelopes in the brain that confer an enhanced tropism and fusigenicity for macrophages. J Virol 78: 6915–6926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters PJ, Sullivan WM, Duenas-Decamp MJ, Bhattacharya J, Ankghuambom C, Brown R, Luzuriaga K, Bell J, Simmonds P, Ball J, et al. 2006. Non-macrophage-tropic human immunodeficiency virus type 1 R5 envelopes predominate in blood, lymph nodes, and semen: Implications for transmission and pathogenesis. J Virol 80: 6324–6332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaff JM, Wilen CB, Harrison JE, Demarest JF, Lee B, Doms RW, Tilton JC 2010. HIV-1 resistance to CCR5 antagonists associated with highly efficient use of CCR5 and altered tropism on primary CD4+ T cells. J Virol 84: 6505–6514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss M, Rodrigo AG, Gosink JJ, Learn GH, de Vange Panteleeff D, Martin HL Jr, Bwayo J, Kreiss JK, Overbaugh J 1998. Evolution of envelope sequences from the genital tract and peripheral blood of women infected with clade A human immunodeficiency virus type 1. J Virol 72: 8240–8251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers KA, Poole C, Pettifor AE, Cohen MS 2008. Rethinking the heterosexual infectivity of HIV-1: A systematic review and meta-analysis. Lancet Infect Dis 8: 553–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redd AD, Avalos A, Essex M 2007. Infection of hematopoietic progenitor cells by HIV-1 subtype C, and its association with anemia in southern Africa. Blood 110: 3143–3149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson WE Jr, Mitchell WM, Chambers WH, Schuffman SS, Montefiori DC, Oeltmann TN 1988. Natural killer cell infection and inactivation in vitro by the human immunodeficiency virus. Hum Pathol 19: 535–540 [DOI] [PubMed] [Google Scholar]
- Russell ES, Kwiek JJ, Keys J, Barton K, Mwapasa V, Montefiori DC, Meshnick SR, Swanstrom R 2011. The genetic bottleneck in vertical transmission of subtype C HIV-1 is not driven by selection of especially neutralization resistant virus from the maternal viral population. J Virol 85: 8253–8262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainski AM, Natesampillai S, Cummins NW, Bren GD, Taylor J, Saenz DT, Poeschla EM, Badley AD 2011. The HIV-1 specific protein Casp8p41, induces death of infected cells through Bax/Bak. J Virol 85: 7965–7975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar-Gonzalez JF, Salazar MG, Keele BF, Learn GH, Giorgi EE, Li H, Decker JM, Wang S, Baalwa J, Kraus MH, et al. 2009. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J Exp Med 206: 1273–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar-Gonzalez JF, Salazar MG, Learn GH, Fouda GG, Kang HH, Mahlokozera T, Wilks AB, Lovingood RV, Stacey A, Kalilani L, et al. 2011. Origin and evolution of HIV-1 in breast milk determined by single-genome amplification and sequencing. J Virol 85: 2751–2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt MP, Gendrault JL, Schweitzer C, Steffan AM, Beyer C, Royer C, Jaeck D, Pasquali JL, Kirn A, Aubertin AM 1990. Permissivity of primary cultures of human Kupffer cells for HIV-1. AIDS Res Hum Retroviruses 6: 987–991 [DOI] [PubMed] [Google Scholar]
- Schnell G, Spudich S, Harrington P, Price RW, Swanstrom R 2009. Compartmentalized human immunodeficiency virus type 1 originates from long-lived cells in some subjects with HIV-1-associated dementia. PLoS Pathog 5: e1000395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell G, Price RW, Swanstrom R, Spudich S 2010. Compartmentalization and clonal amplification of HIV-1 variants in the cerebrospinal fluid during primary infection. J Virol 84: 2395–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell G, Joseph S, Spudich S, Price RW, Swanstrom R 2011. HIV-1 replication in the central nervous system occurs in two distinct cell types. PLoS Pathogens (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuitemaker H, Koot M, Kootstra NA, Dercksen MW, de Goede RE, van Steenwijk RP, Lange JM, Schattenkerk JK, Miedema F, Tersmette M 1992. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: Progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J Virol 66: 1354–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Shaw GM, Hunter E 2011. HIV transmission. Cold Spring Harb Perspect Med 10.1101/cshperspect.a006965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Cheng T, Preffer FI, Dombkowski D, Tomasson MH, Golan DE, Yang O, Hofmann W, Sodroski JG, Luster AD, et al. 1999. Intrinsic human immunodeficiency virus type 1 resistance of hematopoietic stem cells despite coreceptor expression. J Virol 73: 728–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shriner D, Liu Y, Nickle DC, Mullins JI 2006. Evolution of intrahost HIV-1 genetic diversity during chronic infection. Evolution 60: 1165–1176 [PubMed] [Google Scholar]
- *.Siliciano RF, Greene WC 2001. HIV latency. Cold Spring Harb Perspect Med 10.1101/cshperspect.a007096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleasman JW, Aleixo LF, Morton A, Skoda-Smith S, Goodenow MM 1996. CD4+ memory T cells are the predominant population of HIV-1-infected lymphocytes in neonates and children. AIDS 10: 1477–1484 [DOI] [PubMed] [Google Scholar]
- Sonza S, Maerz A, Deacon N, Meanger J, Mills J, Crowe S 1996. Human immunodeficiency virus type 1 replication is blocked prior to reverse transcription and integration in freshly isolated peripheral blood monocytes. J Virol 70: 3863–3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Spudich S, González-Scarano F 2011. HIV-1-related CNS disease: Current issues in pathogenesis, diagnosis, and treatment. Cold Spring Harb Perspect Med 10.1101/cshperspect.a007120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley SK, Kessler SW, Justement JS, Schnittman SM, Greenhouse JJ, Brown CC, Musongela L, Musey K, Kapita B, Fauci AS 1992. CD34+ bone marrow cells are infected with HIV in a subset of seropositive individuals. J Immunol 149: 689–697 [PubMed] [Google Scholar]
- Thomas ER, Dunfee RL, Stanton J, Bogdan D, Taylor J, Kunstman K, Bell JE, Wolinsky SM, Gabuzda D 2007. Macrophage entry mediated by HIV Envs from brain and lymphoid tissues is determined by the capacity to use low CD4 levels and overall efficiency of fusion. Virology 360: 105–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triques K, Stevenson M 2004. Characterization of restrictions to human immunodeficiency virus type 1 infection of monocytes. J Virol 78: 5523–5527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tscherning C, Alaeus A, Fredriksson R, Bjorndal A, Deng H, Littman DR, Fenyo EM, Albert J 1998. Differences in chemokine coreceptor usage between genetic subtypes of HIV-1. Virology 241: 181–188 [DOI] [PubMed] [Google Scholar]
- Tuyama AC, Hong F, Saiman Y, Wang C, Ozkok D, Mosoian A, Chen P, Chen BK, Klotman ME, Bansal MB 2010. Human immunodeficiency virus (HIV)-1 infects human hepatic stellate cells and promotes collagen I and monocyte chemoattractant protein-1 expression: Implications for the pathogenesis of HIV/hepatitis C virus-induced liver fibrosis. Hepatology 52: 612–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin A, Rosati M, Patenaude DJ, Hatzakis A, Kostrikis LG, Lazanas M, Wyvill KM, Yarchoan R, Pavlakis GN 2002. Persistent HIV-1 infection of natural killer cells in patients receiving highly active antiretroviral therapy. Proc Natl Acad Sci 99: 7015–7020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rij RP, Blaak H, Visser JA, Brouwer M, Rientsma R, Broersen S, de Roda Husman AM, Schuitemaker H 2000. Differential coreceptor expression allows for independent evolution of non-syncytium-inducing and syncytium-inducing HIV-1. J Clin Invest 106: 1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van’t Wout AB, Kootstra NA, Mulder-Kampinga GA, Albrecht-van Lent N, Scherpbier HJ, Veenstra J, Boer K, Coutinho RA, Miedema F, Schuitemaker H 1994. Macrophage-tropic variants initiate human immunodeficiency virus type 1 infection after sexual, parenteral, and vertical transmission. J Clin Invest 94: 2060–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van’t Wout AB, Ran LJ, Kuiken CL, Kootstra NA, Pals ST, Schuitemaker H 1998. Analysis of the temporal relationship between human immunodeficiency virus type 1 quasispecies in sequential blood samples and various organs obtained at autopsy. J Virol 72: 488–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderford TH, Bleckwehl C, Engram JC, Dunham RM, Klatt NR, Feinberg MB, Garber DA, Betts MR, Silvestri G 2011. Viral CTL escape mutants are generated in lymph nodes and subsequently become fixed in plasma and rectal mucosa during acute SIV infection of macaques. PLoS Pathog 7: e1002048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varbanov M, Espert L, Biard-Piechaczyk M 2006. Mechanisms of CD4 T-cell depletion triggered by HIV-1 viral proteins. AIDS Rev 8: 221–236 [PubMed] [Google Scholar]
- Veazey RS, DeMaria M, Chalifoux LV, Shvetz DE, Pauley DR, Knight HL, Rosenzweig M, Johnson RP, Desrosiers RC, Lackner AA 1998. Gastrointestinal tract as a major site of CD4+ T cell depletion and viral replication in SIV infection. Science 280: 427–431 [DOI] [PubMed] [Google Scholar]
- Veazey RS, Mansfield KG, Tham IC, Carville AC, Shvetz DE, Forand AE, Lackner AA 2000. Dynamics of CCR5 expression by CD4+ T cells in lymphoid tissues during simian immunodeficiency virus infection. J Virol 74: 11001–11007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veazey RS, Marx PA, Lackner AA 2003. Vaginal CD4+ T cells express high levels of CCR5 and are rapidly depleted in simian immunodeficiency virus infection. J Infect Dis 187: 769–776 [DOI] [PubMed] [Google Scholar]
- Vlahakis SR, Villasis-Keever A, Gomez TS, Bren GD, Paya CV 2003. Human immunodeficiency virus-induced apoptosis of human hepatocytes via CXCR4. J Infect Dis 188: 1455–1460 [DOI] [PubMed] [Google Scholar]
- Wei X, Ghosh SK, Taylor ME, Johnson VA, Emini EA, Deutsch P, Lifson JD, Bonhoeffer S, Nowak MA, Hahn BH, et al. 1995. Viral dynamics in human immunodeficiency virus type 1 infection. Nature 373: 117–122 [DOI] [PubMed] [Google Scholar]
- *.Wilen CB, Tilton JC, Doms RW 2011. HIV: Cell binding and entry. Cold Spring Harb Perspect Med 10.1101/cshperspect.a006866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, KewalRamani VN 2006. Dendritic-cell interactions with HIV: Infection and viral dissemination. Nat Rev Immunol 6: 859–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Zhu H, Wilcox CK, van’t Wout A, Andrus T, Llewellyn N, Stamatatos L, Mullins JI, Corey L, Zhu T 2008. Blood monocytes harbor HIV type 1 strains with diversified phenotypes including macrophage-specific CCR5 virus. J Infect Dis 197: 309–318 [DOI] [PubMed] [Google Scholar]
- Zhang ZQ, Wietgrefe SW, Li Q, Shore MD, Duan L, Reilly C, Lifson JD, Haase AT 2004. Roles of substrate availability and infection of resting and activated CD4+ T cells in transmission and acute simian immunodeficiency virus infection. Proc Natl Acad Sci 101: 5640–5645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Scadden DT, Crumpacker CS 2007. Primitive hematopoietic cells resist HIV-1 infection via p21. J Clin Invest 117: 473–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Shen L, Yang HC, Siliciano RF 2008. Preferential cytolysis of peripheral memory CD4+ T cells by in vitro X4-tropic human immunodeficiency virus type 1 infection before the completion of reverse transcription. J Virol 82: 9154–9163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T, Muthui D, Holte S, Nickle D, Feng F, Brodie S, Hwangbo Y, Mullins JI, Corey L 2002. Evidence for human immunodeficiency virus type 1 replication in vivo in CD14+ monocytes and its potential role as a source of virus in patients on highly active antiretroviral therapy. J Virol 76: 707–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerman RA, Whittington WL, Celum CL, Collis TK, Lucchetti AJ, Sanchez JL, Hughes JP, Coombs RW 2004. Higher concentration of HIV RNA in rectal mucosa secretions than in blood and seminal plasma, among men who have sex with men, independent of antiretroviral therapy. J Infect Dis 190: 156–161 [DOI] [PubMed] [Google Scholar]