Abstract
Insights into the evolution of hemoglobins and their genes are an abundant source of ideas regarding hemoglobin function and regulation of globin gene expression. This article presents the multiple genes and gene families encoding human globins, summarizes major events in the evolution of the hemoglobin gene clusters, and discusses how these studies provide insights into regulation of globin genes. Although the genes in and around the α-like globin gene complex are relatively stable, the β-like globin gene clusters are more dynamic, showing evidence of transposition to a new locus and frequent lineage-specific expansions and deletions. The cis-regulatory modules controlling levels and timing of gene expression are a mix of conserved and lineage-specific DNA, perhaps reflecting evolutionary constraint on core regulatory functions shared broadly in mammals and adaptive fine-tuning in different orders of mammals.
The α-like globin gene complex is relatively stable, but the β-like globin gene clusters show evidence of transposition and lineage-specific expansions and deletions.
A wide range of animals, vertebrate and invertebrate, use hemoglobins to transport oxygen, carrying it from lungs, gills, or other respiratory organs to peripheral tissues that need the oxygen for efficient metabolism. Hence it is natural to compare the structure and function of hemoglobin proteins between species both to explore adaptation and to discover aspects of biochemistry and physiology that are conserved. Comparative studies also have been conducted on the genes and gene clusters that encode the hemoglobins, revealing a rich history of gene duplications and losses as well as translocations. One motivation for comparative studies has been to use the insights from the evolutionary analyses to better understand mechanisms of gene regulation. Many human hemoglobinopathies result from inadequate expression of globin genes, and attempts to modulate globin gene expression are a fundamental approach to seek novel avenues to therapy. This article summarizes broad aspects of the evolution of hemoglobins and related globins, starting with the five basic types of globins found in humans, and then progressively focuses more specifically on hemoglobin gene clusters in vertebrates and eutherian mammals. Finally, the impact of these and related studies on hemoglobin gene regulation is discussed.
HEMOGLOBINS WITHIN THE FAMILY OF GLOBIN HEME PROTEINS
Hemoglobins were originally discovered as abundant proteins in red blood cells of mammals and other jawed vertebrates (gnathostomes) that bind and release oxygen reversibly. The major hemoglobin in adult humans, hemoglobin A, is a heterotetramer composed of two α-globin and two β-globin polypeptides, each with an associated heme group. These are encoded by the duplicated HBA1 and HBA2 genes and by the HBB gene, respectively (Fig. 1). Hemoglobins are produced only in erythroid cells, where they are the major protein. The multi-subunit hemoglobin binds oxygen cooperatively in the lungs, transports the oxygen through the blood, and releases it in peripheral tissues to support oxidative metabolism. The heme molecule contains an iron atom (Fe) in the reduced state (ferrous or +2 oxidation state), and five of the six coordination sites of the Fe are occupied, four by the porphyrin ring and one by the “proximal” histidine (His) of the surrounding globin polypeptide. The sixth coordination site is bound reversibly by oxygen. Oxygen is loaded onto hemoglobin, transported, and unloaded with no chemical (covalent) change to either the oxygen or the heme groups, and the iron in the heme group stays in the reduced state. This contrasts with the familiar role of many heme proteins, such as cytochromes, which catalyze redox reactions involving changes in the oxidation state of the heme iron.
Figure 1.
Model for evolution of vertebrate globin genes. The deduced times of duplication and divergence are shown along the horizontal axis, and contemporary human globin genes are shown at the right. Major duplication events in globin gene evolution are noted along the tree, and time of origin of some major animal groups is indicated along the horizontal axis. For simplicity, only the gene tree is shown. For more information on the inferred times of these duplications relative to speciation events, refer to figures in the primary references (Burmester et al. 2002; Opazo et al. 2008a,b; Hoffmann et al. 2010). Times of major separations used in creation of the figure are taken from TimeTree (Hedges et al. 2006).
Myoglobin (encoded by the MB gene) is a related, monomeric heme-bound globin protein found predominantly in skeletal and heart muscle. The Fe in the heme is pentacoordinate. It has long been described as an oxygen storage protein, and it facilitates diffusion of oxygen to the mitochondria (Wittenberg and Wittenberg 1987).
Additional heme-containing globins were discovered by mining the wealth of information in the sequence of the human genome and transcripts produced from it. Cytoglobin, encoded by the CYGB gene (Fig. 1), is found in many tissues (Burmester et al. 2002; Trent and Hargrove 2002), in sharp contrast to the stringently tissue-specific expression pattern of hemoglobin and myoglobin genes. The most distantly related globin found in the human genome is neuroglobin, encoded by NGB (Burmester et al. 2000). Its mRNA is abundant in brain tissue but also is present in many other tissues. It is related to invertebrate nerve globins, indicating that an ancestral gene was present before the divergence of vertebrates and invertebrates more than 800 million years ago (Fig. 1). In contrast to the pentacoordinate heme complex in hemoglobins and MB, heme forms a hexacoordinate complex with both NGB and CYGB, having two His residues, termed proximal and distal, coordinated with the Fe. Ligands such as oxygen and nitric oxide compete with the distal His for binding, but despite this, NGB and CYBG still have high affinity for the ligands. Both these hexacoordinate heme globins have been implicated in nitric oxide metabolism, with CYGB showing nitric oxide dioxygenase activity, converting nitric oxide to nitrate (Oleksiewicz et al. 2011), and NGB showing nitrite reductase activity to form nitric oxide (Tiso et al. 2011). The latter activity also has been shown for myoglobin (Hendgen-Cotta et al. 2008) and deoxy-hemoglobin (Gladwin and Kim-Shapiro 2008). Physiologically, the nitrite reductase activity could provide a means to produce nitric oxide under hypoxic conditions, signaling from which could regulate mitochondrial respiration and protect tissues (nerves by NGB, heart muscle by MB) from damage under ischemic conditions (Dietz 2011). A role for CYGB in oxygen-requiring reactions, such as hydroxylation, has not been ruled out (Fago et al. 2004). These proposed enzymatic roles in nitric oxide and other metabolism may harken back to functions performed by ancestral hemoglobins in primordial life (Hardison 1998, 1999; Tiso et al. 2011).
The five types of globin genes listed in Figure 1 are located on five different chromosomes: HBA1 and HBA2 at chromosomal position 16p13.3, HBB at 11p15.4, MB at 22q12.3, CYGB at 17q25.1, and NGB at 14q24.3. MB, CYGB, and NGB are present as single-copy genes, whereas HBB and HBAs are in clusters with multiple related genes. All of the genes consist of at least three exons separated by two introns. Although the introns differ dramatically in size, they are in homologous locations. The CYGB and NGB genes each have an additional exon. The conservation of intron position in vertebrate globin genes has been proposed to facilitate the shuffling of exons during protein evolution (Gilbert 1978). However, intron positions differ considerably in globin genes outside vertebrates, suggesting that the conservation of intron position could simply reflect an ancestral state that has not changed over vertebrate evolution (Hardison 1998).
Species in an early diverging branch of vertebrates, the cyclostomes (represented by hagfish and lampreys), also use a heme-containing globin for oxygen transport, but surprisingly, it is more closely related to CYGB than to the gnathostome hemoglobins (Fig. 1) (Hoffmann et al. 2010). This suggests that the oxygen transport function of heme-containing globins arose by independent, convergent evolution in the two major branches of vertebrates. For cyclostomes, it appears that oxygen transport is derived by cooption of the widely expressed CYGB gene.
HEMOGLOBIN GENE CLUSTERS IN JAWED VERTEBRATES
Developmental Regulation of Expression within α-Globin and β-Globin Gene Clusters
In all jawed vertebrates, erythrocytes produced at distinct developmental stages contain different forms of hemoglobin. All species examined make embryonic-specific hemoglobins in primitive erythroid cells derived from the yolk sac, some species make a fetal-specific form in the liver, and all species produce an “adult” hemoglobin in erythroid cells produced in the bone marrow (Maniatis et al. 1980; Karlsson and Nienhuis 1985). Like the major adult hemoglobin A, each of these is a heterotetramer of two α-like globins and two β-like globins, each bound by heme. The α-like globins are paralogous, meaning that they are homologous genes generated by gene duplication. ζ-globin is made in embryonic red cells, and α-globin is produced in fetal and adult red cells (Fig. 2) (Higgs et al. 2005). Likewise, the paralogous β-like globin genes are also expressed at progressive stages of gestation. In humans, ε-globin is made in embryonic erythrocytes, γ-globins are produced in fetal liver erythroid cells, and the δ- and β-globins are made in erythroid cells from adult bone marrow (Grosveld et al. 1993). The hemoglobins produced at distinct developmental stages have different affinities for oxygen and are subject to complex regulation by cofactors, favoring an overall movement of oxygen from the maternal bloodstream to that of the fetus or embryo.
Figure 2.
Models for evolution of hemoglobin gene complexes in jawed vertebrates. Gene clusters in contemporary species are diagrammed at the top of the figure, and inferred gene arrangements in the last common ancestor to jawed vertebrates are diagrammed at the bottom. Genes in brackets with a question mark could either be present in the LCA and lost in one or more descendant lineages, or they could be absent in the LCA but acquired via transposition in some descendant lineages. (Thick gray lines) The major bifurcations during evolution of the lineages exemplified by species at the top. Maps of the gene clusters were derived from a combination of viewing annotations in the UCSC Genome Browser (Kent et al. 2002) of assembled genomes of human (Lander et al. 2001), platypus (Warren et al. 2008), chicken (Hillier et al. 2004), frog Xenopus tropicalis (Hellsten et al. 2010), and fish Medaka (Kasahara et al. 2007), and from recent publications (Fuchs et al. 2006; Opazo et al. 2008b; Patel et al. 2008). The genes are represented by boxes, with those above the line transcribed from left to right and those below the line in the opposite orientation. (Red) β-like globin genes; (yellow) α-like globin genes; (light blue) OR genes; others have colors distinctive for each locus (e.g., shades of green for nonglobin genes at the LA locus). (Small orange circles) Major regulatory regions. The gene maps are not complete, nor are they to scale; the genes shown here were chosen to illustrate logic for the proposed ancestral arrangements and transpositions in two clades. The number to the left of each cluster specifies the chromosome on which it is located; for the frog gene clusters, the scaffold identifier is given. The Greek letter name is specified for hemoglobin genes in human, platypus, and chicken, but generic “α-globin” or “β-globin” is used for frog and fish because the genes are less well characterized. (This diagram is adapted from Hardison 2008.)
The multiple, developmentally regulated genes in the gnathostome α-globin gene clusters are derived from a common ancestral gene cluster (Flint et al. 2001). However, the β-like globin genes in mammals are more similar to each other than they are to the multiple β-like globin genes in birds (Hardison and Miller 1993; Reitman et al. 1993). This implies that the β-like globin gene clusters were generated by independent gene duplications in the bird and mammal lineages. Because differential regulation during development is a consistent property of these independently derived gene clusters, either an ancestral developmental regulatory mechanism was enforced on the newly duplicated genes or the mechanism evolved by convergence. Regulatory mechanisms are complex, and as is discussed in the last section, the current mechanisms are combinations of conserved and acquired features.
Coordinated Regulation between α-Globin and β-Globin Gene Clusters
Expression of α-like and β-like globin genes must be strictly coordinated. A balanced production of α-globin and β-globin in erythroid cells is required for the efficient formation of hemoglobin, and an imbalance leads to the pathological phenotypes of inherited anemias called thalassemias (Weatherall and Clegg 2001). The separation of α-like and β-like globin gene clusters in amniotes requires coordination of expression between different chromosomes.
Fish species show an interesting contrast, in that the gene cluster orthologous (homologous genes generated by a speciation event) to that of the mammalian α-globin gene cluster contains both α-like and β-like globin genes (Fig. 2). Some of the genes in the larger globin gene cluster in fish are expressed in larvae, and others are expressed in adults (Chan et al. 1997). Thus within this fish globin gene cluster, genes are regulated coordinately (balancing α-globin and β-globin synthesis) and differentially during development (larval vs. adult).
Evolution of Multiple Globin Gene Clusters in Vertebrates
As just discussed, differential expression of paralogous globin genes within a cluster and coordinated regulation between gene clusters on different chromosomes are consistent properties in amniotes (birds and mammals). The unfolding story regarding how this arose during vertebrate evolution is dynamic and complex. Analysis of the maps of globin genes and surrounding genes in contemporary vertebrate species suggests a model featuring movement to new locations or differential retention of globin genes but still leading to multiple hemoglobin gene clusters in most if not all vertebrates examined (Fig. 2).
Genes diagnostic for a particular genomic region can be found flanking hemoglobin gene clusters (Bulger et al. 1999; Flint et al. 2001; Gillemans et al. 2003). The diagram in Figure 2 focuses on single-copy, flanking diagnostic genes for clarity (Hardison 2008). One globin gene cluster is found in all gnathostomes examined; it is flanked on one side by the genes MPG and NPRL3 (Flint et al. 2001), and the locus can be called “MN,” the acronym for these two genes (Fig. 2). The major DNA region regulating expression of the globin genes (MRE) is located in an intron of NPRL3 (Higgs et al. 1990). Frequently, the gene RHBDF1 is adjacent to the MPG gene. In contrast to placental mammals and chickens, which have only α-like globin genes at the MN locus, the orthologous loci in the monotreme platypus and in marsupials have a set of α-like globin genes plus a globin gene related to β-globin, the ω-globin gene (Wheeler et al. 2004; Patel et al. 2008). In addition, the platypus MN locus contains a homolog to the globin Y gene (GBY), a globin discovered in amphibians. Direct molecular cloning from the genome of the frog Xenopus laevis (Jeffreys et al. 1980) and examination of the genome assembly of Xenopus tropicalis (Fuchs et al. 2006; Hellsten et al. 2010) reveal a different β-globin gene linked to several α-globin genes at the MN locus. Given the presence of globin genes at this locus in all gnathostomes examined, one can infer with considerable confidence that the MN locus contained globin genes in the last common ancestor (LCA) of vertebrates (Fig. 2).
A second locus contains α- and β-globin genes in the pufferfish Fugu rubripes (Gillemans et al. 2003), and examination of the genome assemblies of zebrafish and Medaka shows a similar arrangement (Fig. 2). The globin genes in this locus are flanked by the genes LCMT1 and AQP8, and the locus can be called “LA.” The gene ARHGAP17 is also part of this locus in many species. These three nonglobin genes are in the same arrangement and order in the tetrapods (human, platypus, chicken, and frog), but the LA locus is devoid of globin genes in these species. This suggests two different models for this locus in the LCA of jawed vertebrates. One model posits that the LCA had globin genes at the LA locus (Gillemans et al. 2003), and these globin genes were retained in fish but lost in tetrapods. The converse posits that the globin genes were not present at the LA locus of the LCA, but moved into it during the lineage to fish.
A third locus contains only β-like globin genes in amniotes. The β-like globin genes in amniotes are flanked by olfactory receptor (OR) genes (Bulger et al. 1999; Patel et al. 2008). In placental mammals, hundreds of OR genes are in this locus, along with additional multigene families such as TRIM genes. Thus one has to look several megabases away from the β-like globin genes to find single-copy genes that are distinctive for this locus, which are DCHS1 on one side and STIM1 on the other. Hence this locus can be called DS (Fig. 2); the RRM1 gene is adjacent to STIM1 in many species. The presence of β-like globin genes in the DS locus in amniotes but absence in both fish and amphibians is most easily explained by transposition of the β-like globin genes into the DS locus in the stem amniote (Fig. 2) (Patel et al. 2008). A proposal that they were present at the DS in the LCA of jawed vertebrates also requires independent deletions in the fish and amphibian lineages; thus, parsimony favors the transposition model. One possible source for the β-like globin genes could be the MN locus (Patel et al. 2008), but it also could be from the LA locus (Hardison 2008).
No globin genes have been mapped to the LA or DS loci in the current assembly of X. tropicalis, but one contig covers a cluster of β-like globin genes linked to RHBDF1 (Fig. 2). Further work is needed to ascertain whether this cluster is linked to the MN locus (Fuchs et al. 2006) or if they are on different chromosomes.
In summary, the history of the gene clusters encoding hemoglobins is dynamic and complex. The MN locus now contains only α-globin genes in eutherians; it retained these and non-globin flanking genes since the gnathostome LCA, while losing β-globin genes in many vertebrate lineages. β-like globin genes were acquired at the DS locus in the stem amniote, and subsequently they duplicated and acquired differential developmental expression independently in the avian and mammalian lineages. The LA locus has undergone dramatic losses or gains of globin genes.
Relative Stability of the α-Like Globin Gene Cluster
The consistent location of α-like globin genes in the MN locus in gnathostomes indicates a more stable history than that of the β-like globin genes. This greater stability is also seen in the composition and expression patterns of the α-like globin genes. Extensive phylogenetic comparisons indicate that this gene cluster in the LCA of tetrapods contained orthologs to ζ-globin, μ-globin (also called αD), and α-globin (or αA) genes (Hoffmann and Storz 2007; Hoffmann et al. 2008), and this arrangement is still seen in chickens (Fig. 3). Before the divergence of the three major subclasses of mammals (monotremes, marsupials, and placentals), both the ζ-globin and α-globin genes duplicated. Most contemporary mammals retain at least two copies of these genes (in some cases, they are pseudogenes). The θ-globin gene appears to have been generated by a duplication of an α-globin gene after the divergence of monotremes from the other mammals (Hoffmann et al. 2008). The μ-globin and θ-globin genes each are present in only a single copy, and although they are transcribed, no evidence has been found for polypeptide products of either in mammals (Clegg 1987; Hsu et al. 1988; Leung et al. 1989; Goh et al. 2005; Cooper et al. 2006). The ortholog of the μ-globin gene is expressed in adult erythroid cells in birds, producing αD-globin.
Figure 3.
Maps of orthologous α-like globin genes and expression timing in amniotes. Each gene is shown as a rectangle, colored by the orthology relationship to the human genes, labeled by the Greek letters at the top. The timing of expression is indicated by distinctive background shading as indicated in the legend. The letter “p” denotes a pseudogene. Sizes of and spacing between genes are not to scale. The gene clusters or parts of them are duplicated or triplicated in rabbits, mouse, rat, and tenrec, indicated by the parentheses and subscripts. The ω-globin gene encodes a β-like globin that has been identified in marsupials and monotremes. The assignments of orthologous relationships are based on grouping within phylogenetic comparisons of coding sequences and flanking regions (Hardison and Gelinas 1986; Cheng et al. 1987; Hoffmann et al. 2008) and automated determination of orthologs using a method that recognizes gene conversions (Song et al. 2012). The “junction” sequence of the rabbit HBA cluster, associated with recombination breakpoints, is assigned to the μ position in this diagram, but it contains only a remnant of a globin gene.
The expression timing of the genes encoding α-like globins is remarkably consistent. In all species examined, including birds, the active orthologs to the ζ-globin gene are expressed in embryonic erythroid cells, and the orthologs of the α-globin gene are expressed in fetal and adult erythroid cells (Fig. 3) (Higgs et al. 1989; Whitelaw et al. 1990).
The α-like globin gene cluster does show some dynamic features. Genes are lost and gained in specific lineages (Hoffmann et al. 2008), and some of the genes have undergone multiple conversion events during mammalian evolution (Hess et al. 1984; Song et al. 2011, 2012). However, the genomic context, that is, the MN locus, has been a constant across gnathostome evolution, and the expression patterns of the ζ- and α-globin genes are strikingly consistent in amniotes.
Lineage-Specific Gains and Losses of β-Like Globin Genes
Within the three major subclasses of mammals, the β-like globin genes at the DS locus have been duplicated and lost in specific lineages (Fig. 4). Both monotremes and marsupials have two β-like globin genes. In marsupials the ε-globin ortholog is expressed in embryonic erythrocytes, whereas the β-globin ortholog is expressed in fetal and adult erythroid cells (Koop and Goodman 1988). The two β-globin genes in platypus are more similar to each other than to other β-like globin genes, consistent with either a gene conversion event (Patel et al. 2008) or a gene duplication independent of the one that established therian ε-globin and β-globin genes (Opazo et al. 2008b). Both the β-globin genes in platypus are expressed in adults (Patel et al. 2008), but no information is available currently on whether the leftmost β-globin gene in platypus (in the orientation in Fig. 4) is also expressed in embryonic erythroid cells, as is expected based on its position.
Figure 4.
Maps of orthologous β-like globin genes and expression timing in mammals. The symbols and backgrounds are similar to those in Figure 3. When the timing of expression is predicted rather than experimentally determined (or highly likely, as in the case of embryonic expression of ε-globin gene orthologs), the background shading is outlined with a dashed line. The gene clusters are triplicated in goats and duplicated in cows, indicated by the parentheses and subscripts. The orthology and expression assignments for these are based largely on the first copy of the segmental duplication; the three β-globin orthologs are expressed in juvenile, adult, and fetal goats (Townes et al. 1984). The assignments of orthologous relationships are based on grouping within phylogenetic comparisons of coding sequences (Koop and Goodman 1988; Opazo et al. 2008a,b, 2009; Patel et al. 2008) and automated determination of orthologs using a method that recognizes gene conversions (Song et al. 2012). The timing of expression is based on multiple reports in the literature (Stockell et al. 1961; LeCrone 1970; Efstratiadis et al. 1980; Rohrbaugh and Hardison 1983; Shapiro et al. 1983; Townes et al. 1984; Schimenti and Duncan 1985; Koop and Goodman 1988; Tagle et al. 1988; Whitelaw et al. 1990; Johnson et al. 1996, 2000; Satoh et al. 1999; Patel et al. 2008).
A proposed cluster of five β-like globin genes, in the orientation 5′-ε-γ-η-δ-β-3′, in the stem eutherian is consistent with the gene arrangements in contemporary species (Goodman et al. 1984; Hardies et al. 1984; Hardison 1984). The relative similarities among orthologous genes indicate that this gene cluster was formed by a series of duplications, first to make the ancestor to β- and δ-globin genes and the ancestor to ε-, γ-, and η-globin genes, followed by duplications to generate the proposed five-gene cluster (Hardison and Miller 1993). The initial duplication established two major lineages of β-like globin genes that differ in their positions in the gene clusters and in their timing of expression (Fig. 4). Genes in the β- and δ-globin lineage are located to the right in the gene clusters and, if active, are expressed in fetal and/or adult erythroid cells. Genes in the ε-, γ-, and η-globin gene lineage are toward the left in the gene clusters and are expressed in embryonic erythroid cells, except for the γ-globin genes in anthropoid primates, which were coopted for fetal-specific expression.
The full set of five β-like globin genes is not used in any extant mammal examined. At least one pseudogene is found in this gene cluster for almost every eutherian species (Fig. 4), and any exceptions to this could reflect a lack of detailed characterization of the genes. Pseudogenes are DNA segments with sequences homologous to those of actively expressed globin genes, but they harbor mutations, such as frameshifts or chain terminators, that preclude expression to form a globin protein.
Deletion can completely inactivate a gene, and gene loss has also occurred widely in the β-like globin gene clusters of eutherians. Some gene losses tend to be consistent across the members of each eutherian order (Fig. 4). No ortholog for the η-globin gene is found in the species sampled from the order Glires (rodents and lagomorphs), but it is present in the sister clades Primates and Laurasiatherians (represented by dog, horse, bat, goat, and cow). This strongly suggests that the η-globin gene was lost in the LCA for Glires (Opazo et al. 2008a). The η-globin gene is also absent from sampled members of the superorders Xenarthra or Afrotheria, which can be explained either by gene loss in the LCA, or perhaps the duplication to form η-globin occurred after these superorders diverged from the other eutherians. No active γ-globin gene has been identified in Laurasiatherians, with the gene either being absent, partially deleted, or harboring inactivating mutations. Note that the loss of the γ-globin gene was not in the stem Laurasiatherian, but rather different losses and inactivations have occurred in the lineages to each species.
All species examined within the therians (marsupial and placental mammals) have an ortholog of the ε-globin gene. This gene has the most consistent features across species of any of the paralogous β-like globin genes. It is always present at the left end of the gene cluster, it is almost always a single gene, and in all species examined, it is expressed only in embryonic erythroid cells (Fig. 4).
The γ-globin genes of both the prosimian primate galago and species in order Glires (rabbit, mouse, and rat) are expressed in embryonic erythroid cells (Rohrbaugh and Hardison 1983; Whitelaw et al. 1990; Satoh et al. 1999), whereas the γ-globin genes of anthropoid primates (monkeys, apes, and humans) are expressed only in fetal erythroid cells (Fig. 4) (Johnson et al. 1996, 2000). One common interpretation is that the embryonic expression pattern was ancestral, and the recruitment to fetal expression was an adaptation in the anthropoids, coinciding with a duplication of the γ-globin gene (Johnson et al. 1996). γ-globin genes are also present in Afrotherians, but the developmental timing of their expression has not been reported.
The η-globin gene homolog in goats is expressed embryonically (Shapiro et al. 1983). Currently, this is the only example of an active η-globin gene, but studies of expression in other Laurasiatherians would reveal whether it is active in other species, and if the timing of expression is embryonic. Fetal and adult hemoglobins were found to be identical for horse (Stockell et al. 1961) and dog (LeCrone 1970), and based on the absence of evidence for a fetal-specific hemoglobin, the η-globin homologs are predicted to be expressed embryonically in Figure 4. The η-globin gene is a pseudogene in all primates.
The δ-globin gene is present in almost all eutherian species examined, but it is frequently a pseudogene (Fig. 4). In every case examined in sufficient detail, the δ-globin gene has been involved in a gene conversion, with sequence from the paralogous β-globin gene copied into the δ-globin gene locus (Spritz et al. 1980; Martin et al. 1983; Hardies et al. 1984; Hardison and Margot 1984; Song et al. 2012). The boundaries of the conversions are different in each species, indicating that these are independent gene conversions. The structural and mechanistic bases for this propensity for conversion are not understood. In galago, the replacement with β-globin gene sequences extends into the promoter region, leading to high-level expression from this gene (Tagle et al. 1991). That, in turn, led to efforts to engineer a form of the δ-globin gene that would express at sufficiently high levels to provide potential therapy (Tang et al. 1997).
In most eutherian species, the β-globin gene is expressed in fetal and adult erythroid cells (Fig. 4). Concomitantly with the recruitment of γ-globin genes to fetal expression in anthropoid primates, the onset of expression of the β-globin gene was delayed to shortly before birth in catarrhine primates (Old World monkeys, apes, and humans) (Johnson et al. 2000). The onset of β-globin gene expression is earlier in fetal life in the New World monkeys (Johnson et al. 1996), perhaps representing a transitional state intermediate between the fetal onset seen in most eutherians and the prenatal onset observed in humans.
This overview of the evolution of β-globin genes illustrates the diversity of events that have been inferred, including duplications, deletions, inactivations, and reactivations. It shows that the ε-globin genes have been stable over eutherian evolution, whereas the γ-, η-, and δ-globin genes have been gained and lost frequently, sometimes in entire orders of mammals. Furthermore, the timing of expression can change dramatically between clades, notably the delay in γ-globin (fetal) and β-globin (adult) gene expression in anthropoid primates. Strategies being pursued to reactivate γ-globin gene expression in adult erythroid cells, either pharmacologically or by gene therapy, in a sense are attempts to modulate expression patterns in humans that recapitulate expression changes that have occurred during eutherian evolution.
INSIGHTS INTO GENE REGULATION
The previous discussion shows that expression of globin genes is tightly regulated. Hemoglobin gene expression is restricted to erythroid cells. The genes are expressed at extremely high levels late in erythroid differentiation, with balanced production of α-globin and β-globin. Paralogous globin genes are expressed at progressive developmental stages. This exquisite regulation is exerted, at least in part, by the binding of specific transcription factors to DNA sequences that serve as cis-regulatory modules (CRMs), such as promoters and enhancers (Maniatis et al. 1987).
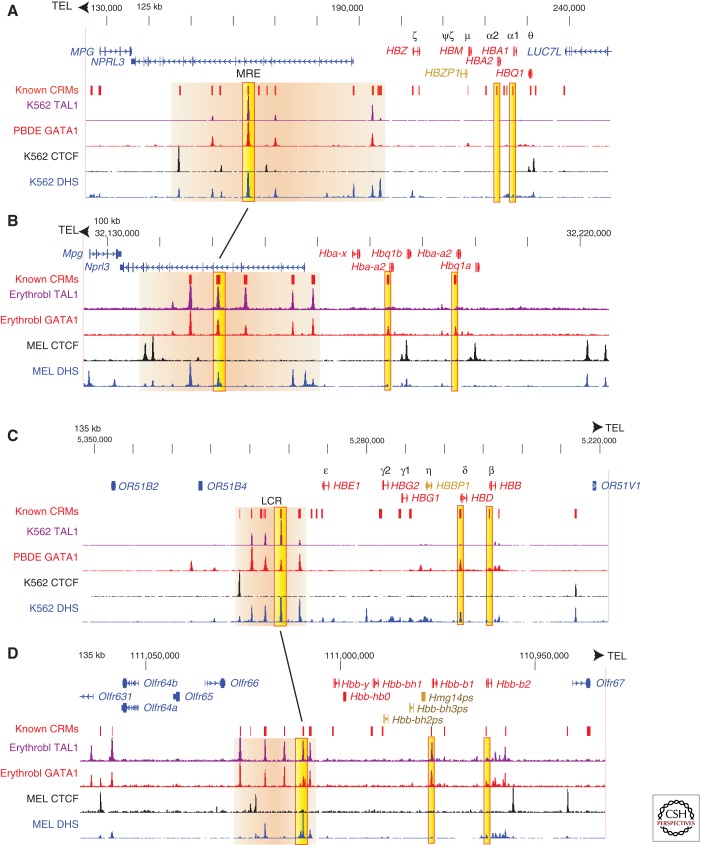
Detailed studies over the past three decades have led to the discovery of numerous CRMs in both the α-globin gene (HBA) and β-globin gene (HBB) clusters (Fig. 5). Some are located proximal to and within the genes, such as promoters and internal enhancers (Mellon et al. 1981; Wright et al. 1984; Myers et al. 1986; Antoniou et al. 1988; Wall et al. 1988), and others are located distal to the genes (Grosveld et al. 1987; Talbot et al. 1989; Higgs et al. 1990). For instance, the major regulatory element (MRE) of the HBA gene complex is located distal to the adult HBA genes (∼60 kb upstream in human), residing in an intron of the large NPRL3 gene (Fig. 5A). Several additional CRMs are present around the MRE (Anguita et al. 2004) both in human and mouse (Fig. 5A,B). A cluster of CRMs called the locus control region (LCR) is found 50–70 kb upstream of the HBB gene (Grosveld et al. 1987; Talbot et al. 1989; Moon and Ley 1990) in human and mouse (Fig. 5C,D). These distal regulatory regions are enhancers (Tuan et al. 1989; Ney et al. 1990; Pondel et al. 1992) required for high-level expression of the globin genes (Grosveld et al. 1987; Talbot et al. 1989; Higgs et al. 1990; Bender et al. 2000a; Anguita et al. 2002). They are in regions of open chromatin marked by DNase hypersensitive sites (Forrester et al. 1986, 1990; Vyas et al. 1992; Gourdon et al. 1995), and they can protect against some repressive position effects (Grosveld et al. 1987; Caterina et al. 1991; Milot et al. 1996). They are bound by key transcription factors active in erythroid cells, such as GATA1 and TAL1 (Johnson et al. 2002; Anguita et al. 2004; Grass et al. 2006). The protein CTCF is bound at specific sites in the gene clusters, some of which serve as insulators that localize the effects of distal enhancers on target genes (Bulger et al. 2003). The detailed information gleaned from decades of work on these gene clusters is recapitulated with high sensitivity and specificity in recent genome-wide analyses (Cheng et al. 2009; Fujiwara et al. 2009; Yu et al. 2009; Kassouf et al. 2010; Soler et al. 2010; Wilson et al. 2010; Wu et al. 2011), as illustrated in Figure 5.
Figure 5.
Genes and regulatory modules in the human and mouse hemoglobin gene clusters. Each panel shows chromosomal positions (human genome assembly GRCh37/hg19 and mouse genome assembly NCBI37/mm9), the direction to the telomere (TEL), genes (from UCSC genes unless otherwise indicated) with rectangles for exons and lines with the direction of transcription shown as arrowheads, known cis-regulatory modules (CRMs), and signals for occupancy of TAL1, GATA1, and CTCF based on genome-wide ChIP-seq and the signal for DNase hypersensitive sites (DHSs) based on genome-wide DNase-seq. Strong enhancers (MRE in the HBA clusters and HS2 in the HBB clusters) and adult globin gene promoters are in gold, and groups of distal CRMs (such as the LCR) are in tan. (A) The human α-globin gene (HBA) cluster, showing chr16:125,001–250,000. The known CRMs are from Hughes et al. (2005). (B) The mouse α-globin gene (Hba) cluster, showing chr11:32,125,501–32,225,500. The known CRMs are from Anguita et al. (2004). (C) The human β-globin gene (HBB) cluster, showing chr11:5,218,001–5,353,000. The known CRMs are from King et al. (2005). (D) The mouse β-globin gene (Hbb) cluster, showing chr7:110,932,001–111,067,000. The known CRMs are from Zhang et al. (2009). The orientation of the gene clusters is the reverse of the genome assembly in panels C and D, so that globin genes in all panels are arranged with transcription from left to right. The factor occupancy and DNase sensitivity tracks show data from the human (ENCODE Project Consortium et al. 2011, 2012) and mouse ENCODE projects (Wu et al. 2011; Mouse ENCODE Project Consortium 2012). The images are from tracks displayed at the UCSC Genome Browser (Kent et al. 2002). K562 is a human tumor-derived cell line with some erythroid properties. PBDE, human peripheral blood-derived erythroblasts; Erythrobl, mouse Ter119-positive erythroblasts purified from fetal liver; MEL, murine erythroleukemia cells.
Given the large number of diverse regulatory modules in the globin gene clusters and the multiple modes of regulation, it is not surprising that the mechanisms of gene regulation are complex and not fully understood. A few themes have emerged. Many of the distal CRMs interact in some manner, as shown by phenotypes of mutations (Bungert et al. 1995; Jackson et al. 1996; Molete et al. 2001) and by direct physical mapping using chromosome conformation capture (Dostie et al. 2006; Bau et al. 2011). One proposal is that they form a discrete structure called an active chromatin hub (de Laat and Grosveld 2003; Palstra et al. 2003; Zhou et al. 2006), whereas other experiments indicate that the distal CRMs act more independently (Bender et al. 2000b, 2001). Regardless of the structure(s) formed by the distal CRMs, activation of target globin genes involves a direct interaction between the distal LCR (or equivalent) and the promoter of the activated genes, presumably through a looping mechanism (Carter et al. 2002; Tolhuis et al. 2002; Vakoc et al. 2005; Ragoczy et al. 2006; Vernimmen et al. 2009). Activation of globin genes and other erythroid genes occurs with relocation of the chromatin domain to transcriptionally active regions of the nucleus (Schubeler et al. 2000; Osborne et al. 2004; Schoenfelder et al. 2010). Stabilization of the transcription complex through LCR–promoter interactions, perhaps at transcription factories, is one mechanism for gene activation (Wijgerde et al. 1995).
The general similarities in arrangement of CRMs, transcription factor occupancy, and mechanisms of regulation in globin gene clusters suggest that cross-species evolutionary comparisons of genomic DNA could be effective for finding regulatory regions (Hardison et al. 1993, 1997a; Gumucio et al. 1996; Hardison 2000). This approach has been successful in many studies (e.g., Gumucio et al. 1992; Elnitski et al. 1997; Hardison et al. 1997b; Flint et al. 2001; Hughes et al. 2005), but it has some limitations. The approach is most sensitive for finding CRMs under evolutionary constraint, and although this is true for many CRMs (Pennacchio et al. 2006), a large fraction is species specific (King et al. 2007; Bourque et al. 2008; Hardison and Taylor 2012). Only a small fraction of CRMs are preserved throughout vertebrate evolution (King et al. 2007), and comparisons of noncoding genomic DNA between human and fish have not been a rich resource for discovery of CRMs in globin gene clusters (Flint et al. 2001). Furthermore, critical aspects of the expression patterns or mechanisms can differ, even between mouse and human. For example, the phenotypes differ between mouse and human for deletions of the homologous distal MREs of the HBA gene complexes (Anguita et al. 2002). This showed that some aspects of regulation differ between the two species, and further studies were pursued by engineering a homologous replacement of the mouse Hba gene complex with that of human (Wallace et al. 2007). The change in developmental timing of expression of γ-globin orthologs between mouse and human (Fig. 4) is driven by interspecies differences in the expression of the transcription factor BCL11A, which is a repressor of γ-globin gene expression (Sankaran et al. 2009). This latter study illustrates the larger point that hypotheses can be derived from evolutionary signatures of either constraint or adaptation. Comparative analyses continue to be a rich source of insights, but they must be performed in a context that embraces both conservation and lineage-specific innovation (Hardison and Taylor 2012).
One useful model for interpreting the differences in phylogenetic depth of preservation of CRMs is that those involved in core regulatory functions may be conserved across a wide group of species and show evidence of evolutionary constraint. The CRMs found in one or a limited range of species could be involved in fine-tuning those core regulatory processes, perhaps helping that species adapt to a unique niche. For example, a strong distal enhancer is required for high-level expression of globin genes. Both the HBA and HBB gene complexes have such enhancers (Fig. 5), and interspecies DNA alignments show that both are preserved across mammals and have been under strong constraint (Elnitski et al. 1997; Flint et al. 2001). These strong enhancers could be a key component of the distal regulatory structure, perhaps an active chromatin hub. In contrast, some of the other distal CRMs are present only in mouse, such as the one in the first intron of Nrpl3 and the most distal CRM in the Hbb LCR (Fig. 5). These could contribute to functional differences between the species, perhaps by modifying the distal regulatory structure.
CONCLUDING REMARKS
The diversity of hemoglobins, their critical functions, their exquisite regulation, and the pathological consequences of some mutations make this a fascinating family of proteins and genes. Exploration of these genes in many different species continues to illuminate some and challenge other evolutionary models. Production of different forms of hemoglobin at progressive developmental stages is widespread in vertebrates and beyond, and studies of hemoglobin switching are pursued in several non-human species as models of the process in humans. The evolutionary comparisons summarized here illustrate the power of this approach, but they also remind us that such studies are best done while embracing both interspecies conservation of some elements and lineage-specific changes for others. Indeed, this can lead to important insights, such as the impact of differences in expression pattern of a key transcription factor driving a change in developmental timing of expression in humans.
ACKNOWLEDGMENTS
This work is supported by National Institutes of Health grants from NIDDK (R01 DK65806) and NHGRI (RC2 HG005573).
Footnotes
Editors: David Weatherall, Alan N. Schechter, and David G. Nathan
Additional Perspectives on Hemoglobin and Its Diseases available at www.perspectivesinmedicine.org
REFERENCES
- Anguita E, Sharpe JA, Sloane-Stanley JA, Tufarelli C, Higgs DR, Wood WG 2002. Deletion of the mouse α-globin regulatory element (HS-26) has an unexpectedly mild phenotype. Blood 100: 3450–3456 [DOI] [PubMed] [Google Scholar]
- Anguita E, Hughes J, Heyworth C, Blobel GA, Wood WG, Higgs DR 2004. Globin gene activation during haemopoiesis is driven by protein complexes nucleated by GATA-1 and GATA-2. EMBO J 23: 2841–2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniou M, deBoer E, Habets G, Grosveld F 1988. The human β-globin gene contains multiple regulatory regions: Identification of one promoter and two downstream enhancers. EMBO J 7: 377–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bau D, Sanyal A, Lajoie BR, Capriotti E, Byron M, Lawrence JB, Dekker J, Marti-Renom MA 2011. The three-dimensional folding of the α-globin gene domain reveals formation of chromatin globules. Nat Struct Mol Biol 18: 107–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender MA, Bulger M, Close J, Groudine M 2000a. β-globin gene switching and DNase I sensitivity of the endogenous β-globin locus in mice do not require the locus control region. Mol Cell 5: 387–393 [DOI] [PubMed] [Google Scholar]
- Bender MA, Mehaffey MG, Telling A, Hug B, Ley TJ, Groudine M, Fiering S 2000b. Independent formation of DnaseI hypersensitive sites in the murine β-globin locus control region. Blood 95: 3600–3604 [PubMed] [Google Scholar]
- Bender MA, Roach JN, Halow J, Close J, Alami R, Bouhassira EE, Groudine M, Fiering SN 2001. Targeted deletion of 5′HS1 and 5′HS4 of the β-globin locus control region reveals additive activity of the DNaseI hypersensitive sites. Blood 98: 2022–2027 [DOI] [PubMed] [Google Scholar]
- Bourque G, Leong B, Vega VB, Chen X, Lee YL, Srinivasan KG, Chew JL, Ruan Y, Wei CL, Ng HH, et al. 2008. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res 18: 1752–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulger M, von Doorninck JH, Saitoh N, Telling A, Farrell C, Bender MA, Felsenfeld G, Axel R, Groudine M 1999. Conservation of sequence and structure flanking the mouse and human β-globin loci: The β-globin genes are embedded within an array of odorant receptor genes. Proc Natl Acad Sci 96: 5129–5134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulger M, Schubeler D, Bender MA, Hamilton J, Farrell CM, Hardison RC, Groudine M 2003. A complex chromatin “landscape” revealed by patterns of nuclease sensitivity and histone modification within the mouse β-globin locus. Mol Cell Biol 23: 5234–5244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bungert J, Dave U, Lim K-C, Kieuw KH, Shavit JA, Liu Q, Engel JD 1995. Synergistic regulation of human β-globin gene switching by locus control region elements HS3 and HS4. Genes Dev 9: 3083–3096 [DOI] [PubMed] [Google Scholar]
- Burmester T, Weich B, Reinhardt S, Hankeln T 2000. A vertebrate globin expressed in the brain. Nature 407: 520–523 [DOI] [PubMed] [Google Scholar]
- Burmester T, Ebner B, Weich B, Hankeln T 2002. Cytoglobin: A novel globin type ubiquitously expressed in vertebrate tissues. Mol Biol Evol 19: 416–421 [DOI] [PubMed] [Google Scholar]
- Carter D, Chakalova L, Osborne CS, Dai YF, Fraser P 2002. Long-range chromatin regulatory interactions in vivo. Nat Genet 32: 623–626 [DOI] [PubMed] [Google Scholar]
- Caterina JJ, Ryan TM, Pawlik KM, Palmiter RD, Brinster RL, Behringer RR, Townes TM 1991. Human β-globin locus control region: Analysis of the 5′ DNaseI hypersensitive site HS2 in transgenic mice. Proc Natl Acad Sci 88: 1626–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan F, Robinson J, Brownlie A, Shivdasani R, Donovan A, Brugnara C, Kim J, Lau B, Witkowska H, Zon L 1997. Characterization of adult α- and β-globin genes in the zebrafish. Blood 89: 688–700 [PubMed] [Google Scholar]
- Cheng J-F, Raid L, Hardison RC 1987. Block duplications of α ζ–ζ–α–τ gene set in the rabbit α-like globin gene cluster. J Biol Chem 262: 5414–5421 [PubMed] [Google Scholar]
- Cheng Y, Wu W, Kumar SA, Yu D, Deng W, Tripic T, King DC, Chen KB, Zhang Y, Drautz D, et al. 2009. Erythroid GATA1 function revealed by genome-wide analysis of transcription factor occupancy, histone modifications, and mRNA expression. Genome Res 19: 2172–2184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegg JB 1987. Can the product of the θ gene be a real globin? Nature 329: 465–466 [DOI] [PubMed] [Google Scholar]
- Cooper SJ, Wheeler D, De Leo A, Cheng JF, Holland RA, Marshall Graves JA, Hope RM 2006. The mammalian αD-globin gene lineage and a new model for the molecular evolution of α-globin gene clusters at the stem of the mammalian radiation. Mol Phylogenet Evol 38: 439–448 [DOI] [PubMed] [Google Scholar]
- de Laat W, Grosveld F 2003. Spatial organization of gene expression: The active chromatin hub. Chromosome Res 11: 447–459 [DOI] [PubMed] [Google Scholar]
- Dietz GP 2011. Protection by neuroglobin and cell-penetrating peptide-mediated delivery in vivo: A decade of research. Comment on Cai et al.: TAT-mediated delivery of neuroglobin protects against focal cerebral ischemia in mice. Exp Neurol. 2011; 227(1): 224–31. Exp Neurol 231: 1–10 [DOI] [PubMed] [Google Scholar]
- Dostie J, Richmond TA, Arnaout RA, Selzer RR, Lee WL, Honan TA, Rubio ED, Krumm A, Lamb J, Nusbaum C, et al. 2006. Chromosome Conformation Capture Carbon Copy (5C): A massively parallel solution for mapping interactions between genomic elements. Genome Res 16: 1299–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efstratiadis A, Posakony JW, Maniatis T, Lawn RM, O'Connell C, Spritz RA, DeRiel JK, Forget BG, Weissman SM, Slightom JL, et al. 1980. The structure and evolution of the human β-globin gene family. Cell 21: 653–668 [DOI] [PubMed] [Google Scholar]
- Elnitski L, Miller W, Hardison R 1997. Conserved E boxes function as part of the enhancer in hypersensitive site 2 of the β-globin locus control region: Role of basic helix–loop–helix proteins. J Biol Chem 272: 369–378 [DOI] [PubMed] [Google Scholar]
- ENCODE Project Consortium, Myers RM, Stamatoyannopoulos J, Snyder M, Dunham I, Hardison RC, Bernstein BE, Gingeras TR, Kent WJ, Birney E, et al. 2011. A user’s guide to the Encyclopedia of DNA Elements (ENCODE). PLoS Biol 9: e1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fago A, Hundahl C, Dewilde S, Gilany K, Moens L, Weber RE 2004. Allosteric regulation and temperature dependence of oxygen binding in human neuroglobin and cytoglobin. Molecular mechanisms and physiological significance. J Biol Chem 279: 44417–44426 [DOI] [PubMed] [Google Scholar]
- Flint J, Tufarelli C, Peden J, Clark K, Daniels RJ, Hardison R, Miller W, Philipsen S, Tan-Un KC, McMorrow T, et al. 2001. Comparative genome analysis delimits a chromosomal domain and identifies key regulatory elements in the α globin cluster. Hum Mol Genet 10: 371–382 [DOI] [PubMed] [Google Scholar]
- Forrester WC, Thompson C, Elder JT, Groudine M 1986. A developmentally stable chromatin structure in the human β-globin gene cluster. Proc Natl Acad Sci 83: 1359–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester WC, Epner E, Driscoll MC, Enver T, Brice M, Papayannopoulou T, Groudine M 1990. A deletion of the human β-globin locus activation region causes a major alteration in chromatin structure and replication across the entire β-globin locus. Genes Dev 4: 1637–1649 [DOI] [PubMed] [Google Scholar]
- Fuchs C, Burmester T, Hankeln T 2006. The amphibian globin gene repertoire as revealed by the Xenopus genome. Cytogenet Genome Res 112: 296–306 [DOI] [PubMed] [Google Scholar]
- Fujiwara T, O’Geen H, Keles S, Blahnik K, Linnemann AK, Kang YA, Choi K, Farnham PJ, Bresnick EH 2009. Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy. Mol Cell 36: 667–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W 1978. Why genes in pieces? Nature 271: 501. [DOI] [PubMed] [Google Scholar]
- Gillemans N, McMorrow T, Tewari R, Wai AW, Burgtorf C, Drabek D, Ventress N, Langeveld A, Higgs D, Tan-Un K, et al. 2003. A functional and comparative analysis of globin loci in pufferfish and man. Blood 101: 2842–2849 [DOI] [PubMed] [Google Scholar]
- Gladwin MT, Kim-Shapiro DB 2008. The functional nitrite reductase activity of the heme-globins. Blood 112: 2636–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh SH, Lee YT, Bhanu NV, Cam MC, Desper R, Martin BM, Moharram R, Gherman RB, Miller JL 2005. A newly discovered human α-globin gene. Blood 106: 1466–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman M, Koop BF, Czelusniak J, Weiss ML 1984. The η-globin gene. Its long evolutionary history in the β-globin gene family of mammals. J Mol Biol 180: 803–823 [DOI] [PubMed] [Google Scholar]
- Gourdon G, Sharpe JA, Higgs DR, Wood WG 1995. The mouse α-globin locus regulatory element. Blood 86: 766–775 [PubMed] [Google Scholar]
- Grass JA, Jing H, Kim SI, Martowicz ML, Pal S, Blobel GA, Bresnick EH 2006. Distinct functions of dispersed GATA factor complexes at an endogenous gene locus. Mol Cell Biol 26: 7056–7067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosveld F, van Assendelft GB, Greaves D, Kollias G 1987. Position-independent, high-level expression of the human β-globin gene in transgenic mice. Cell 51: 975–985 [DOI] [PubMed] [Google Scholar]
- Grosveld F, Antoniou M, Berry M, de Boer E, Dillon N, Ellis J, Fraser P, Hanscombe O, Hurst J, Imam A, et al. 1993. The regulation of human globin gene switching. Philos Trans R Soc Lond 339: 183–191 [DOI] [PubMed] [Google Scholar]
- Gumucio DL, Heilstedt-Williamson H, Gray TA, Tarle SA, Shelton DA, Tagle D, Slightom J, Goodman M, Collins FS 1992. Phylogenetic footprinting reveals a nuclear protein which binds to silencer sequences in the human γ and ε globin genes. Mol Cell Biol 12: 4919–4929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumucio D, Shelton D, Zhu W, Millinoff D, Gray T, Bock J, Slightom J, Goodman M 1996. Evolutionary strategies for the elucidation of cis and trans factors that regulate the developmental switching programs of the β-like globin genes. Mol Phylog Evol 5: 18–32 [DOI] [PubMed] [Google Scholar]
- Hardies SC, Edgell MH, Hutchison CA III 1984. Evolution of the mammalian β-globin gene cluster. J Biol Chem 259: 3748–3756 [PubMed] [Google Scholar]
- Hardison RC 1984. Comparison of the β-like globin gene families of rabbits and humans indicates that the gene cluster 5′ ε-γ-δ-β 3′ predates the mammalian radiation. Mol Biol Evol 1: 390–410 [DOI] [PubMed] [Google Scholar]
- Hardison R 1998. Hemoglobins from bacteria to man: Evolution of different patterns of gene expression. J Exp Biol 201: 1099–1117 [DOI] [PubMed] [Google Scholar]
- Hardison R 1999. The evolution of hemoglobin: Studies of a very ancient protein suggest that changes in gene regulation are an important part of the evolutionary story. Am Sci 87: 126–137 [Google Scholar]
- Hardison RC 2000. Conserved noncoding sequences are reliable guides to regulatory elements. Trends Genet 16: 369–372 [DOI] [PubMed] [Google Scholar]
- Hardison RC 2008. Globin genes on the move. J Biol 7: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardison RC, Gelinas R 1986. Assignment of orthologous relationships among mammalian α-globin genes by examining flanking regions reveals a rapid rate of evolution. Mol Biol Evol 3: 243–261 [DOI] [PubMed] [Google Scholar]
- Hardison RC, Margot JB 1984. Rabbit globin pseudogene pseudoβ2 is a hybrid of δ- and β-globin gene sequences. Mol Biol Evol 1: 302–316 [DOI] [PubMed] [Google Scholar]
- Hardison R, Miller W 1993. Use of long sequence alignments to study the evolution and regulation of mammalian globin gene clusters. Mol Biol Evol 10: 73–102 [DOI] [PubMed] [Google Scholar]
- Hardison RC, Taylor J 2012. Genomic approaches towards finding cis-regulatory modules in animals. Nat Rev Genet 13: 469–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardison R, Xu J, Jackson J, Mansberger J, Selifonova O, Grotch B, Biesecker J, Petrykowska H, Miller W 1993. Comparative analysis of the locus control region of the rabbit β-like globin gene cluster: HS3 increases transient expression of an embryonic ε-globin gene. Nucl Acids Res 21: 1265–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardison R, Oeltjen J, Miller W 1997a. Long human–mouse sequence alignments reveal novel regulatory elements: A reason to sequence the mouse genome. Genome Res 7: 959–966 [DOI] [PubMed] [Google Scholar]
- Hardison R, Slightom JL, Gumucio DL, Goodman M, Stojanovic N, Miller W 1997b. Locus control regions of mammalian β-globin gene clusters: Combining phylogenetic analyses and experimental results to gain functional insights. Gene 205: 73–94 [DOI] [PubMed] [Google Scholar]
- Hedges SB, Dudley J, Kumar S 2006. TimeTree: A public knowledge-base of divergence times among organisms. Bioinformatics 22: 2971–2972 [DOI] [PubMed] [Google Scholar]
- Hellsten U, Harland RM, Gilchrist MJ, Hendrix D, Jurka J, Kapitonov V, Ovcharenko I, Putnam NH, Shu S, Taher L, et al. 2010. The genome of the Western clawed frog Xenopus tropicalis. Science 328: 633–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendgen-Cotta UB, Merx MW, Shiva S, Schmitz J, Becher S, Klare JP, Steinhoff HJ, Goedecke A, Schrader J, Gladwin MT, et al. 2008. Nitrite reductase activity of myoglobin regulates respiration and cellular viability in myocardial ischemia-reperfusion injury. Proc Natl Acad Sci 105: 10256–10261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess J, Schmid C, Shen C 1984. A gradient of sequence divergence in the human adult α-globin duplication units. Science 226: 67–70 [DOI] [PubMed] [Google Scholar]
- Higgs DR, Vickers MA, Wilkie AO, Pretorius IM, Jarman AP, Weatherall DJ 1989. A review of the molecular genetics of the human α-globin gene cluster. Blood 73: 1081–1104 [PubMed] [Google Scholar]
- Higgs D, Wood W, Jarman A, Sharpe J, Lida J, Pretorious IM, Ayyub H 1990. A major positive regulatory region located far upstream of the human α-globin gene locus. Genes Dev 4: 1588–1601 [DOI] [PubMed] [Google Scholar]
- Higgs DR, Garrick D, Anguita E, De Gobbi M, Hughes J, Muers M, Vernimmen D, Lower K, Law M, Argentaro A, et al. 2005. Understanding α-globin gene regulation: Aiming to improve the management of thalassemia. Ann NY Acad Sci 1054: 92–102 [DOI] [PubMed] [Google Scholar]
- Hillier LW, Miller W, Birney E, Warren W, Hardison RC, Ponting CP, Bork P, Burt DW, Groenen MA, Delany ME, et al. 2004. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432: 695–716 [DOI] [PubMed] [Google Scholar]
- Hoffmann FG, Storz JF 2007. The αD-globin gene originated via duplication of an embryonic α-like globin gene in the ancestor of tetrapod vertebrates. Mol Biol Evol 24: 1982–1990 [DOI] [PubMed] [Google Scholar]
- Hoffmann FG, Opazo JC, Storz JF 2008. Rapid rates of lineage-specific gene duplication and deletion in the α-globin gene family. Mol Biol Evol 25: 591–602 [DOI] [PubMed] [Google Scholar]
- Hoffmann FG, Opazo JC, Storz JF 2010. Gene cooption and convergent evolution of oxygen transport hemoglobins in jawed and jawless vertebrates. Proc Natl Acad Sci 107: 14274–14279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu S, Marks J, Shaw J, Tam M, Higgs D, Shen C, Shen C 1988. Structure and expression of the human θ1 globin gene. Nature 331: 94–96 [DOI] [PubMed] [Google Scholar]
- Hughes JR, Cheng JF, Ventress N, Prabhakar S, Clark K, Anguita E, De Gobbi M, de Jong P, Rubin E, Higgs DR 2005. Annotation of cis-regulatory elements by identification, subclassification, and functional assessment of multispecies conserved sequences. Proc Natl Acad Sci 102: 9830–9835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson JD, Petrykowska H, Philipsen S, Miller W, Hardison R 1996. Role of DNA sequences outside the cores of DNase hypersensitive sites (HSs) in functions of the β-globin locus control region: Domain opening and synergism between HS2 and HS3. J Biol Chem 271: 11871–11878 [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Wilson V, Wood D, Simons JP, Kay RM, Williams JG 1980. Linkage of adult α- and β-globin genes in X. laevis and gene duplication by tetraploidization. Cell 21: 555–564 [DOI] [PubMed] [Google Scholar]
- Johnson RM, Buck S, Chiu C, Schneider H, Sampaio I, Gage DA, Shen TL, Schneider MP, Muniz JA, Gumucio DL, et al. 1996. Fetal globin expression in New World monkeys. J Biol Chem 271: 14684–14691 [DOI] [PubMed] [Google Scholar]
- Johnson RM, Buck S, Chiu CH, Gage DA, Shen TL, Hendrickx AG, Gumucio DL, Goodman M 2000. Humans and Old World monkeys have similar patterns of fetal globin expression. J Exp Zool 288: 318–326 [DOI] [PubMed] [Google Scholar]
- Johnson KD, Grass JA, Boyer ME, Keikhaefer CM, Blobel GA, Weiss MJ, Bresnick EH 2002. Cooperative activities of hematopoietic regulators recruit RNA polymerase II to a tissue-specific chromatin domain. Proc Natl Acad Sci 99: 11760–11765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson S, Nienhuis AW 1985. Developmental regulation of human globin genes. Annu Rev Biochem 54: 1071–1108 [DOI] [PubMed] [Google Scholar]
- Kasahara M, Naruse K, Sasaki S, Nakatani Y, Qu W, Ahsan B, Yamada T, Nagayasu Y, Doi K, Kasai Y, et al. 2007. The medaka draft genome and insights into vertebrate genome evolution. Nature 447: 714–719 [DOI] [PubMed] [Google Scholar]
- Kassouf MT, Hughes JR, Taylor S, McGowan SJ, Soneji S, Green AL, Vyas P, Porcher C 2010. Genome-wide identification of TAL1’s functional targets: Insights into its mechanisms of action in primary erythroid cells. Genome Res 20: 1064–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D 2002. The human genome browser at UCSC. Genome Res 12: 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King DC, Taylor J, Elnitski L, Chiaromonte F, Miller W, Hardison RC 2005. Evaluation of regulatory potential and conservation scores for detecting cis-regulatory modules in aligned mammalian genome sequences. Genome Res 15: 1051–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King DC, Taylor J, Zhang Y, Cheng Y, Lawson HA, Martin J, ENCODE Groups for Transcriptional Regulation and Multispecies Sequence Analysis, Chiaromonte F, Miller W, Hardison RC 2007. Finding cis-regulatory elements using comparative genomics: Some lessons from ENCODE data. Genome Res 17: 775–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koop B, Goodman M 1988. Evolutionary and developmental aspects of two hemoglobin β-chain genes (εM and βM) of opossum. Proc Natl Acad Sci 85: 3893–3897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- LeCrone CN 1970. Absence of special fetal hemoglobin in beagle dogs. Blood 35: 451–452 [PubMed] [Google Scholar]
- Leung S, Whitelaw E, Proudfoot N 1989. Transcriptional and translational analysis of the human θ globin gene. Nucleic Acids Res 17: 8283–8300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Fritsch EF, Lauer J, Lawn RM 1980. The molecular genetics of human hemoglobins. Annu Rev Genet 14: 145–178 [DOI] [PubMed] [Google Scholar]
- Maniatis T, Goodbourn S, Fischer JA 1987. Regulation of inducible and tissue-specific gene expression. Science 236: 1237–1245 [DOI] [PubMed] [Google Scholar]
- Martin SL, Vincent KA, Wilson AC 1983. Rise and fall of the δ globin gene. J Mol Biol 164: 513–528 [DOI] [PubMed] [Google Scholar]
- Mellon P, Parker V, Gluzman Y, Maniatis T 1981. Identification of DNA sequences required for transcription of the human α1-globin gene in a new SV40 host-vector system. Cell 27: 279–288 [DOI] [PubMed] [Google Scholar]
- Milot E, Strouboulis J, Trimborn T, Wijgerde M, de Boer E, Langeveld A, Tan-Un K, Vergeer W, Yannoutsos N, Grosveld F, et al. 1996. Heterochromatin effects on the frequency and duration of LCR-mediated gene transcription. Cell 87: 105–114 [DOI] [PubMed] [Google Scholar]
- Molete JM, Petrykowska H, Bouhassira EE, Feng YQ, Miller W, Hardison RC 2001. Sequences flanking hypersensitive sites of the β-globin locus control region are required for synergistic enhancement. Mol Cell Biol 21: 2969–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon AM, Ley TJ 1990. Conservation of the primary structure, organization, and function of the human and mouse β-globin locus-activating regions. Proc Natl Acad Sci 87: 7693–7697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse ENCODE Project Consortium 2012. An encyclopedia of mouse DNA elements (Mouse ENCODE). Genome Biol 13: 418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers RM, Tilly K, Maniatis T 1986. Fine structure genetic analysis of a β-globin promoter. Science 232: 613–618 [DOI] [PubMed] [Google Scholar]
- Ney P, Sorrentino B, McDonagh K, Nienhuis A 1990. Tandem AP-1-binding sites within the human β-globin dominant control region function as an inducible enhancer in erythroid cells. Genes Dev 4: 993–1006 [DOI] [PubMed] [Google Scholar]
- Oleksiewicz U, Liloglou T, Field JK, Xinarianos G 2011. Cytoglobin: Biochemical, functional and clinical perspective of the newest member of the globin family. Cell Mol Life Sci 68: 3869–3883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opazo JC, Hoffmann FG, Storz JF 2008a. Differential loss of embryonic globin genes during the radiation of placental mammals. Proc Natl Acad Sci 105: 12950–12955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opazo JC, Hoffmann FG, Storz JF 2008b. Genomic evidence for independent origins of β-like globin genes in monotremes and therian mammals. Proc Natl Acad Sci 105: 1590–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opazo JC, Sloan AM, Campbell KL, Storz JF 2009. Origin and ascendancy of a chimeric fusion gene: The β/δ-globin gene of paenungulate mammals. Mol Biol Evol 26: 1469–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, et al. 2004. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet 36: 1065–1071 [DOI] [PubMed] [Google Scholar]
- Palstra RJ, Tolhuis B, Splinter E, Nijmeijer R, Grosveld F, de Laat W 2003. The β-globin nuclear compartment in development and erythroid differentiation. Nat Genet 35: 190–194 [DOI] [PubMed] [Google Scholar]
- Patel VS, Cooper SJ, Deakin JE, Fulton B, Graves T, Warren WC, Wilson RK, Graves JA 2008. Platypus globin genes and flanking loci suggest a new insertional model for β-globin evolution in birds and mammals. BMC Biol 6: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, et al. 2006. In vivo enhancer analysis of human conserved non-coding sequences. Nature 444: 499–502 [DOI] [PubMed] [Google Scholar]
- Pondel MD, George M, Proudfoot NJ 1992. The LCR-like α-globin positive regulatory element functions as an enhancer in transiently transfected cells during erythroid differentiation. Nucleic Acids Res 20: 237–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragoczy T, Bender MA, Telling A, Byron R, Groudine M 2006. The locus control region is required for association of the murine β-globin locus with engaged transcription factories during erythroid maturation. Genes Dev 20: 1447–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitman M, Grasso JA, Blumentahl R, Lewit P 1993. Primary sequence, evolution and repetitive elements of the G. gallus (chicken) β-globin cluster. Genomics 18: 616–626 [DOI] [PubMed] [Google Scholar]
- Rohrbaugh ML, Hardison RC 1983. Analysis of rabbit β-like globin gene transcripts during development. J Mol Biol 164: 395–417 [DOI] [PubMed] [Google Scholar]
- Sankaran VG, Xu J, Ragoczy T, Ippolito GC, Walkley CR, Maika SD, Fujiwara Y, Ito M, Groudine M, Bender MA, et al. 2009. Developmental and species-divergent globin switching are driven by BCL11A. Nature 460: 1093–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh H, Inokuchi N, Nagae Y, Okazaki T 1999. Organization, structure, and evolution of the nonadult rat β-globin gene cluster. J Mol Evol 49: 122–129 [DOI] [PubMed] [Google Scholar]
- Schimenti JC, Duncan CH 1985. Structure and organization of the bovine β-globin genes. Mol Biol Evol 2: 514–525 [DOI] [PubMed] [Google Scholar]
- Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, et al. 2010. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet 42: 53–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubeler D, Francastel C, Cimbora DM, Reik A, Martin DI, Groudine M 2000. Nuclear localization and histone acetylation: A pathway for chromatin opening and transcriptional activation of the human β-globin locus. Genes Dev 14: 940–950 [PMC free article] [PubMed] [Google Scholar]
- Shapiro SG, Schon EA, Townes TM, Lingrel JB 1983. Sequence and linkage of the goat εI and εII β-globin genes. J Mol Biol 169: 31–52 [DOI] [PubMed] [Google Scholar]
- Soler E, Andrieu-Soler C, de Boer E, Bryne JC, Thongjuea S, Stadhouders R, Palstra RJ, Stevens M, Kockx C, van Ijcken W, et al. 2010. The genome-wide dynamics of the binding of Ldb1 complexes during erythroid differentiation. Genes Dev 24: 277–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song G, Hsu CH, Riemer C, Zhang Y, Kim HL, Hoffmann F, Zhang L, Hardison RC, Green ED, Miller W 2011. Conversion events in gene clusters. BMC Evol Biol 11: 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song G, Riemer C, Dickins B, Kim HL, Zhang L, Zhang Y, Hsu CH, Hardison RC, NISC Comparative Sequencing Program, Green ED, et al. 2012. Revealing mammalian evolutionary relationships by comparative analysis of gene clusters. Genome Biol Evol 4: 586–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spritz R, DeRiel J, Forget B, Weissman S 1980. Complete nucleotide sequence of the human δ-globin gene. Cell 21: 639–646 [DOI] [PubMed] [Google Scholar]
- Stockell A, Perutz MF, Muirhead H, Glauser SC 1961. A comparison of adult and foetal horse haemoglobins. J Mol Biol 3: 112–116 [Google Scholar]
- Tagle DA, Koop BF, Goodman M, Slightom J, Hess DL, Jones RT 1988. Embryonic ε and γ globin genes of a prosimian primate (Galago crassicaudatus): Nucleotide and amino acid sequences, developmental regulation and phylogenetic footprints. J Mol Biol 203: 7469–7480 [DOI] [PubMed] [Google Scholar]
- Tagle D, Slightom J, Jones R, Goodman M 1991. Concerted evolution led to high expression of a prosimian primate δ globin gene locus. J Biol Chem 266: 7469–7480 [PubMed] [Google Scholar]
- Talbot D, Collis P, Antoniou M, Vidal M, Grosveld F, Greaves DR 1989. A dominant control region from the human β-globin locus conferring integration site-independent gene expression. Nature 338: 352–355 [DOI] [PubMed] [Google Scholar]
- Tang DC, Ebb D, Hardison RC, Rodgers GP 1997. Restoration of the CCAAT box and insertion of the CACCC motif activate δ-globin gene expression. Blood 90: 421–427 [PubMed] [Google Scholar]
- Tiso M, Tejero J, Basu S, Azarov I, Wang X, Simplaceanu V, Frizzell S, Jayaraman T, Geary L, Shapiro C, et al. 2011. Human neuroglobin functions as a redox-regulated nitrite reductase. J Biol Chem 286: 18277–18289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W 2002. Looping and interaction between hypersensitive sites in the active β-globin locus. Mol Cell 10: 1453–1465 [DOI] [PubMed] [Google Scholar]
- Townes TM, Fitzgerald MC, Lingrel JB 1984. Triplication of a four-gene set during evolution of the goat β-globin locus produced three genes now expressed differentially during development. Proc Natl Acad Sci 81: 6589–6593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trent JT III, Hargrove MS 2002. A ubiquitously expressed human hexacoordinate hemoglobin. J Biol Chem 277: 19538–19545 [DOI] [PubMed] [Google Scholar]
- Tuan D, Solomon W, London I, Lee D 1989. An erythroid-specific, developmental-stage-independent enhancer far upstream of the human “β-like globin” genes. Proc Natl Acad Sci 86: 2554–2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakoc CR, Letting DL, Gheldof N, Sawado T, Bender MA, Groudine M, Weiss MJ, Dekker J, Blobel GA 2005. Proximity among distant regulatory elements at the β-globin locus requires GATA-1 and FOG-1. Mol Cell 17: 453–462 [DOI] [PubMed] [Google Scholar]
- Vernimmen D, Marques-Kranc F, Sharpe JA, Sloane-Stanley JA, Wood WG, Wallace HA, Smith AJ, Higgs DR 2009. Chromosome looping at the human α-globin locus is mediated via the major upstream regulatory element (HS-40). Blood 114: 4253–4260 [DOI] [PubMed] [Google Scholar]
- Vyas P, Vickers MA, Simmons DL, Ayyub H, Craddock CF, Higgs DR 1992. Cis-acting sequences regulating expression of the human α-globin cluster lie within constitutively open chromatin. Cell 69: 781–793 [DOI] [PubMed] [Google Scholar]
- Wall L, deBoer E, Grosveld F 1988. The human β-globin gene 3′ enhancer contains multiple binding sites for an erythroid-specific protein. Genes Dev 2: 1089–1100 [DOI] [PubMed] [Google Scholar]
- Wallace HA, Marques-Kranc F, Richardson M, Luna-Crespo F, Sharpe JA, Hughes J, Wood WG, Higgs DR, Smith AJ 2007. Manipulating the mouse genome to engineer precise functional syntenic replacements with human sequence. Cell 128: 197–209 [DOI] [PubMed] [Google Scholar]
- Warren WC, Hillier LW, Marshall Graves JA, Birney E, Ponting CP, Grutzner F, Belov K, Miller W, Clarke L, Chinwalla AT, et al. 2008. Genome analysis of the platypus reveals unique signatures of evolution. Nature 453: 175–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatherall DJ, Clegg JB 2001. Thalassemia syndromes. Blackwell Science, Oxford [Google Scholar]
- Wheeler D, Hope RM, Cooper SJ, Gooley AA, Holland RA 2004. Linkage of the β-like ω-globin gene to α-like globin genes in an Australian marsupial supports the chromosome duplication model for separation of globin gene clusters. J Mol Evol 58: 642–652 [DOI] [PubMed] [Google Scholar]
- Whitelaw E, Tsai SF, Hogben P, Orkin SH 1990. Regulated expression of globin chains and the erythroid transcription factor GATA-1 during erythropoiesis in the developing mouse. Mol Cell Biol 10: 6596–6606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijgerde M, Grosveld F, Fraser P 1995. Transcription complex stability and chromatin dynamics in vivo. Nature 377: 209–213 [DOI] [PubMed] [Google Scholar]
- Wilson NK, Foster SD, Wang X, Knezevic K, Schutte J, Kaimakis P, Chilarska PM, Kinston S, Ouwehand WH, Dzierzak E, et al. 2010. Combinatorial transcriptional control in blood stem/progenitor cells: Genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell 7: 532–544 [DOI] [PubMed] [Google Scholar]
- Wittenberg BA, Wittenberg JB 1987. Myoglobin-mediated oxygen delivery to mitochondria of isolated cardiac myocytes. Proc Natl Acad Sci 84: 7503–7507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S, Rosenthal A, Flavell R, Grosveld F 1984. DNA sequences required for regulated expression of β-globin genes in murine erythroleukemia cells. Cell 38: 265–273 [DOI] [PubMed] [Google Scholar]
- Wu W, Cheng Y, Keller CA, Ernst J, Kumar SA, Mishra T, Morrissey C, Dorman CM, Chen KB, Drautz D, et al. 2011. Dynamics of the epigenetic landscape during erythroid differentiation after GATA1 restoration. Genome Res 21: 1659–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Riva L, Xie H, Schindler Y, Moran TB, Cheng Y, Yu D, Hardison R, Weiss MJ, Orkin SH, et al. 2009. Insights into GATA-1-mediated gene activation versus repression via genome-wide chromatin occupancy analysis. Mol Cell 36: 682–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wu W, Cheng Y, King DC, Harris RS, Taylor J, Chiaromonte F, Hardison RC 2009. Primary sequence and epigenetic determinants of in vivo occupancy of genomic DNA by GATA1. Nucleic Acids Res 37: 7024–7038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou GL, Xin L, Song W, Di LJ, Liu G, Wu XS, Liu DP, Liang CC 2006. Active chromatin hub of the mouse α-globin locus forms in a transcription factory of clustered housekeeping genes. Mol Cell Biol 26: 5096–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]