The two aptamer domains of tandem glycine riboswitches were originally reported to be joined by a single-stranded linker. A recently discovered paired element formed between the linker and a conserved 5′ leader conforms to the sequence of a canonical K-turn motif and enhances glycine binding. Here, the authors find that the K-turn is not only essential for efficient glycine binding under near-physiologic conditions, but in most cases also stabilizes the global “quaternary” fold of the RNA. Despite being distal to the glycine-binding pockets, the K-turn plays an important role in the structure and function of tandem glycine riboswitches.
Keywords: quaternary structure, K-turn, RNA aptamers, ribonucleoprotein complex, SAXS
Abstract
Most known glycine riboswitches have two homologous aptamer domains arranged in tandem and separated by a short linker. The two aptamers associate through reciprocal “quaternary” interactions that have been proposed to result in cooperative glycine binding. Recently, the interaptamer linker was found to form helix P0 with a previously unrecognized segment 5′ to the first aptamer domain. P0 was shown to increase glycine affinity, abolish cooperativity, and conform to the K-turn motif consensus. We examine the global thermodynamic and structural role of P0 using isothermal titration calorimetry (ITC) and small-angle X-ray scattering (SAXS), respectively. To evaluate the generality of P0 function, we prepared glycine riboswitch constructs lacking and including P0 from Bacillus subtilis, Fusobacterium nucleatum, and Vibrio cholerae. We find that P0 indeed folds into a K-turn, supports partial pre-folding of all three glycine-free RNAs, and is required for ITC observation of glycine binding under physiologic Mg2+ concentrations. Except for the unusually small riboswitch from F. nucleatum, the K-turn is needed for maximally compacting the glycine-bound states of the RNAs. Formation of a ribonucleoprotein complex between the B. subtilis or the F. nucleatum RNA constructs and the bacterial K-turn binding protein YbxF promotes additional folding of the free riboswitch, and enhances glycine binding. Consistent with the previously reported loss of cooperativity, P0-containing B. subtilis and V. cholerae tandem aptamers bound no more than one glycine molecule per riboswitch. Our results indicate that the P0 K-turn helps organize the quaternary structure of tandem glycine riboswitches, thereby facilitating ligand binding under physiologic conditions.
INTRODUCTION
The glycine riboswitch was first described by Mandal et al. (2004), who noted two remarkable characteristics of this bacterial gene-regulatory mRNA domain. First, most known examples of this riboswitch are comprised of two homologous tandem glycine-binding aptamer domains separated by a short linker, and upstream of a single element that interfaces with the gene expression machinery (i.e., a single expression platform). Second, in vitro analysis of glycine binding by an RNA construct comprised of two aptamer domains (derived from the 5′-untranslated region of the gcvT mRNA of Vibrio cholerae; this construct is hereafter referred to as VcI-II) suggested the presence of cooperativity, with a Hill coefficient of 1.64. In contrast, the isolated second aptamer domain from the same mRNA (VcII) bound to glycine with a Hill coefficient of 0.97. Cooperativity appears to arise from interaction of the two aptamer domains in a manner analogous to cooperative ligand binding through quaternary structure formation in oligomeric proteins (Monod et al. 1965). Among known riboswitch classes, the presence of tandem aptamer domains preceding a single-expression platform is unusual. Three additional examples have been documented for guanine-ykkC, cyclic di-GMP, and glutamine aptamers (Sudarsan et al. 2006, 2008; Ames and Breaker 2011), but only glycine riboswitches generically occur as tandem aptamers with a single expression platform. For other tandem riboswitches known, an expression platform follows each aptamer domain (Sudarsan et al. 2006; Welz and Breaker 2007). Crystallographic structure determination demonstrates that isolated glycine riboswitch aptamer domains adopt the same fold as in tandem (Huang et al. 2010; Butler et al. 2011). Moreover, both in solution and in crystals, the isolated aptamer domains show a propensity to oligomerize, utilizing what appears to be the same interface as that used by the tandem aptamers when present in a single RNA chain (Huang et al. 2010; Butler et al. 2011; Erion and Strobel 2011).
Biochemical analysis of ligand binding by the glycine riboswitch demonstrated a requirement for Mg2+ concentrations of 20–100 mM, significantly above the <5 mM concentrations considered physiologic (Alatossava et al. 1985; Romani and Scarpa 1992). In particular, isothermal titration calorimetry (ITC) could detect evidence of glycine binding only above 20 mM Mg2+ concentration (Kd ∼ 30 μM and ∼3 μM for glycine binding to the isolated VcI and VcII aptamer domains, respectively). Recently, two groups have independently provided a possible explanation for the unusually high divalent cation requirement of this RNA. Kladwang et al. (2012) and Sherman et al. (2012) found that the linker segment connecting the two aptamer domains, rather than being single-stranded, forms a hitherto unrecognized helix (P0 hereafter) with a conserved RNA segment 5′ to the first aptamer domain (Fig. 1A). These groups analyzed glycine binding using chemical probing (DMS, SHAPE, and in-line probing), and found that inclusion of P0 resulted in up to 120-fold tighter glycine binding. Remarkably, Sherman et al. (2012) found that VcI–II constructs that include P0 not only bind glycine with higher affinity, but no longer show evidence of cooperativity. Kladwang et al. (2012) and Sherman et al. (2012) noted that for at least 50% of known tandem glycine riboswitches, the sequences of the P0 helices conform to a K-turn consensus (Fig. 1A). The K-turn is an RNA structural motif comprised of two helices flanking a bulge. The motif causes the helical path to bend by ∼120° and is often the binding site for proteins (Klein et al. 2001). K-turns are present in several riboswitch classes, including the Class-I S-adenosylmethionine (SAM-I) and lysine riboswitches, where it has been shown that K-turn integrity is essential for productive folding and ligand recognition (Blouin and Lafontaine 2007; Heppell and Lafontaine 2008).
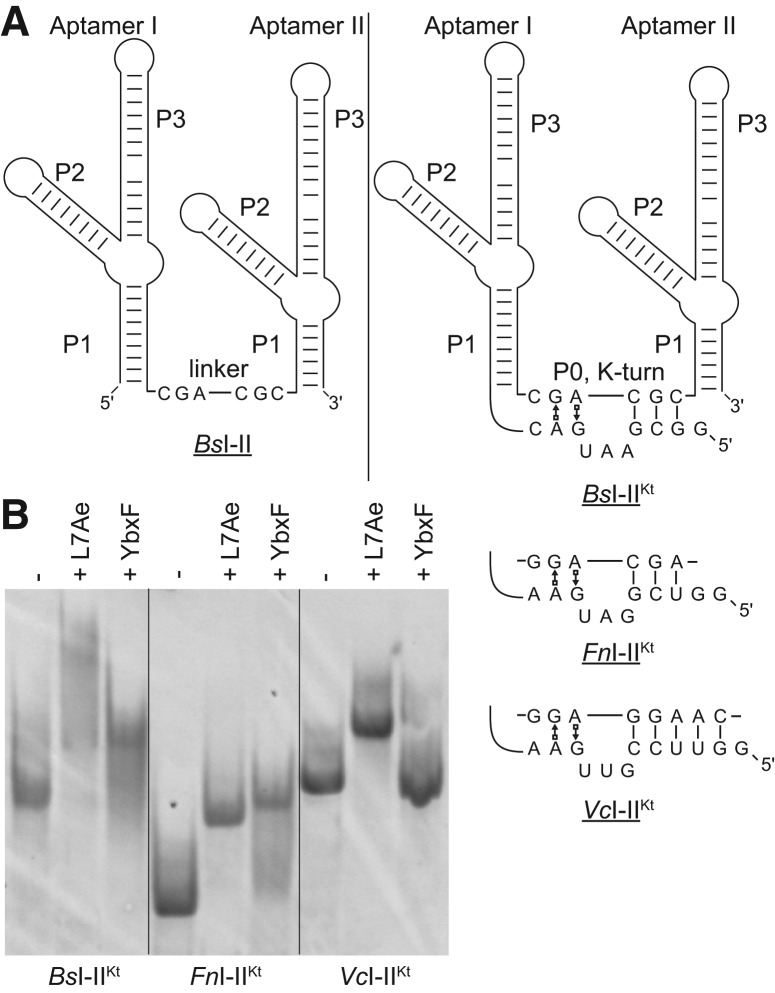
FIGURE 1.
The P0 helix of glycine riboswitches is a K-turn. (A) Tandem glycine riboswitch aptamers joined by a single-stranded linker, denoted I–II (left), and by the recently discovered (Kladwang et al. 2012; Sherman et al. 2012) K-turn, denoted I–IIKt (right). The sequence of the K-turn is given for each of the three I–IIKt examined. (B) EMSA analysis of three I–IIKt constructs using the L7Ae and YbxF K-turn binding proteins.
Kladwang et al. (2012) and Sherman et al. (2012) analyzed the effect that the presence of P0 has on the glycine riboswitch at the local, single-nucleotide level through chemical probing techniques. Because formation of a K-turn in P0 might exert its biological function by orienting the two aptamer domains and facilitating their interaction, we hypothesized that there should be a global structural change that results from inclusion of P0 in a glycine riboswitch RNA. Moreover, the K-turn might facilitate glycine binding at lower, physiologic Mg2+ concentrations. To evaluate this, we have now performed ITC and small-angle X-ray scattering (SAXS) analyses of glycine riboswitch constructs with and without the P0 K-turn (denoted I–IIKt and I–II, respectively) in the presence of physiologic divalent cation concentrations. To examine the generality of the function of the P0 K-turn, we studied three previously well-characterized tandem glycine riboswitches from V. cholerae (Vc), Bacillus subtilis (Bs), and Fusobacterium nucleatum (Fn). We find that the K-turn is required for detection of glycine binding by ITC in 5 mM Mg2+. Even in the absence of glycine, the K-turn facilitates global folding of the RNAs. In two of the three I-IIKt constructs, inclusion of the K-turn results in a more compact glycine-bound conformation relative to the construct lacking the K-turn. In addition, when the K-turn is bound specifically by the bacterial protein YbxF, a modest increase in glycine-binding affinity and additional compaction of the glycine-free RNA results. We conclude that the P0 K-turn is necessary for tandem glycine riboswitches to achieve their native (most compact) glycine-bound structures under physiologic conditions.
RESULTS
The P0 helix is a canonical K-turn
The sequences of P0 in the V. cholerae, B. subtilis, and F. nucleatum glycine riboswitches (Fig. 1A) are consistent with a K-turn motif (Kladwang et al. 2012; Sherman et al. 2012). Because of its RNA-binding specificity and high affinity, the K-turn-binding protein L7Ae has previously been used to demonstrate that RNA sequences fold into this structural motif (Schroeder et al. 2010). We used L7Ae from the archaeon Methanococcus janaschii and the homologous YbxF from the bacterium B. subtilis in electrophoretic mobility shift assays (EMSA) with the three I–IIKt constructs to demonstrate that their P0 helices indeed fold into K-turns. Each of the RNAs exhibited a gel mobility shift in the presence of L7Ae (Fig. 1B). In the presence of YbxF, only the BsI–IIKt and FnI–IIKt RNAs were shifted. That YbxF does not bind all three I–IIKt RNAs is consistent with our previous finding that YbxF is more selective regarding the K-turn motifs that it binds than L7Ae (Baird et al. 2012). ITC experiments (see below) are consistent with the binding pattern observed by EMSA for these three tandem glycine riboswitch constructs.
K-turn containing glycine riboswitches fold at physiologic Mg2+ concentration
Most structured RNAs require divalent metal ions to fold (for review, see Woodson 2005). In addition to folding, the VcI–II RNA has specific metal ion requirements for glycine binding (Huang et al. 2010; Lipfert et al. 2010; Butler et al. 2011). Many previous studies of the VcI–II RNA utilized conditions containing divalent concentrations much higher than physiologic (≥10 mM Mg2+). For example, ITC studies of glycine binding to VcI–II were performed in 20 and 40 mM Mg2+ (Huang et al. 2010). We examined the folding response of the three I–IIKt constructs to Mg2+ by titrating the divalent cation into RNA samples in buffer containing 100 mM KCl and monitored heat evolution by ITC. The response curves exhibited a transition between 3 and 4 mM Mg2+, after which the evolved heat monotonically decreased (Fig. 2A). Although this transition is at higher Mg2+ concentration than has been reported for the folding of other RNAs (e.g., <1 mM for B. subtilis RNase P specificity domain) (Baird et al. 2005), this probably is a result of the high concentrations (∼50 μM) of RNA used in these RNA folding experiments. As much as several mM Mg2+ would be expected to interact with the RNA backbone, reducing the activity of Mg2+ in solution. While the titration curves are not immediately interpretable as representing unfolded, intermediate or completely folded states, it appears that at concentrations in excess of 3–4 mM, the RNAs have folded beyond an initial transition. We therefore performed subsequent ITC experiments with samples pre-equilibrated in 5 mM Mg2+ and SAXS at various Mg2+ concentrations near 5 mM Mg2+.
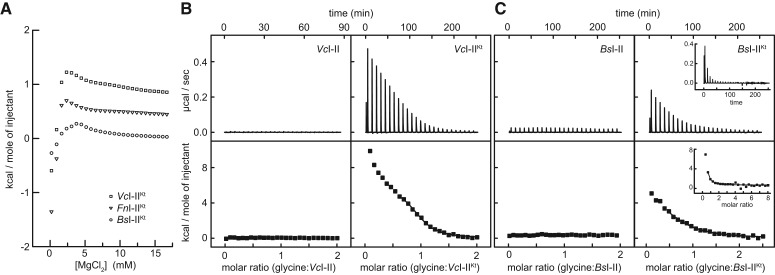
FIGURE 2.
ITC analyses of RNA folding and glycine binding. (A) Mg2+ titration of the three I–IIKt RNAs used in this study. For clarity, data for FnI–IIKt and VcI–IIKt are offset by 0.4 kcal/mol and 0.8 kcal/mol, respectively, in this panel only. (B) ITC titrations of glycine into VcI–II (left) and VcI–IIKt (right) in 5 mM Mg2+. (C) ITC titrations of glycine into BsI–II (left) and BsI–IIKt (right) in 5 mM Mg2+, including a titration in which the final molar ratio (glycine:RNA) in the solution is >8 (inset).
The K-turn is required for ITC detection of glycine binding in 5 mM Mg2+
The K-turn was reported to improve glycine-binding affinity in tandem aptamer constructs from V. cholerae, B. subtilis, and F. nucleatum (Kladwang et al. 2012; Sherman et al. 2012). We performed ITC analyses for V. cholerae and B. subtilis I–II and I–IIKt constructs under near-physiologic conditions to evaluate thermodynamically the contribution of the K-turn. In 5 mM Mg2+ and 20°C, no heat was evolved upon titration of glycine into VcI–II (Fig. 2B). This contrasts with the exothermic binding previously reported in 40 mM Mg2+ at 35°C (Huang et al. 2010). Thus, glycine binding by this RNA is sensitive to both temperature and divalent metal ion concentration. The VcI–IIKt RNA was reported to bind glycine nearly 12-fold tighter than VcI–II in 20 mM Mg2+ (Sherman et al. 2012). Under our conditions, VcI–IIKt exhibited a reproducible ITC response to glycine. The binding isotherm is endothermic; thus, the reaction is entropically driven (Fig. 2B). The shape of the isotherm is not a simple sigmoid; there appears to be a monotonic decrease in heat evolved over the concentration tested (Materials and Methods). Fitting the data as a single-site binding yields a dissociation constant, Kd, of 4.3 ± 0.75 μM and binding stoichiometry, n, of 1.05 ± 0.08 (Table 1). This Kd is similar to that observed by in-line probing in 20 mM Mg2+ (Sherman et al. 2012); the affinity for glycine of this RNA appears to be insensitive to changes in Mg2+ concentration between 5 and 20 mM.
TABLE 1.
Thermodynamic parameters for V. cholerae and B. subtilis tandem aptamers
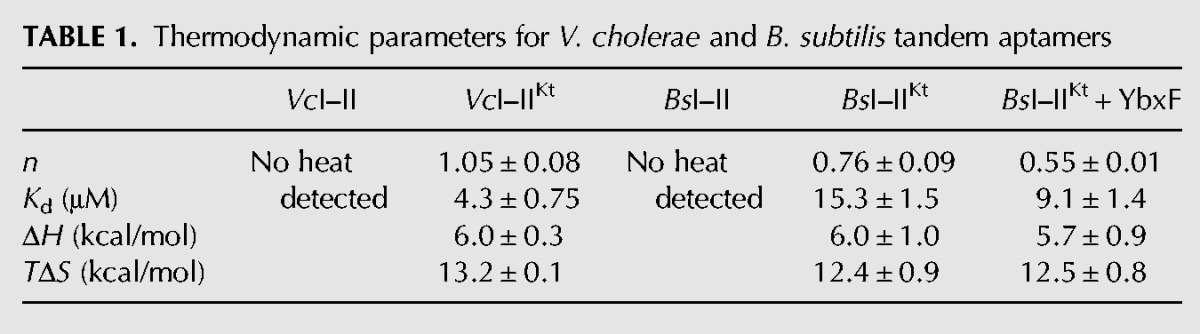
In 5 mM Mg2+, the minimal B. subtilis tandem glycine aptamer construct (BsI–II) did not yield an ITC binding isotherm (Fig. 2C), whereas BsI–IIKt did. The overall shape of the isotherm was similar to that observed for VcI–IIKt (Fig. 2B). The Kd for the BsI–IIKt-glycine complex is 15.3 μM (Table 1), about fourfold weaker than reported by in-line probing in 20 mM Mg2+ (Sherman et al. 2012). Given the identical enthalpy and entropy of glycine binding by BsI–IIKt and VcI–IIKt (ΔH ∼6 kcal/mol and TΔS ∼12 kcal/mol) (Table 1), their different Kd’s must arise from differences in stoichiometry. BsI–IIKt binds glycine with an apparent molar ratio of 0.75:1 (glycine:RNA). This subunity value for n is consistent with that recently reported (Sherman et al. 2012). No additional binding was observed by ITC even when the final molar ratio in solution exceeds 8 (Fig. 2C, inset).
The cognate K-turn binding protein YbxF enhances glycine binding to BsI–IIKt
K-turns can be stabilized by tertiary structure formation, increased Mg2+ concentrations, or by K-turn binding proteins (Schroeder et al. 2011). To test whether YbxF might improve glycine binding under physiologically relevant Mg2+ concentrations, we performed ITC as described above with the BsI–IIKt RNA and a 1.5-fold molar excess of B. subtilis YbxF in the sample cell. We found that the cognate ribonucleoprotein (RNP) complex binds glycine with ∼1.7-fold higher affinity (Kd ∼ 9 μM) than the RNA alone at 5 mM Mg2+ (Table 1). We investigated whether the protein could stabilize BsI–IIKt at even lower Mg2+ concentrations and enhance glycine binding. At 1.5 and 2.5 mM Mg2+, no heat evolution was detected upon the addition of glycine to BsI–IIKt (data not shown). Interestingly, ITC experiments in which YbxF was titrated into BsI–IIKt in 1 mM and 5 mM Mg2+ exhibited diverging properties. The signal was strongly exothermic in 5 mM Mg2+, but much weaker and endothermic in 1 mM Mg2+ (data not shown). Together, this suggests that BsI–IIKt is not well folded in lower Mg2+ concentrations and that neither YbxF nor glycine bind efficiently to the RNA under these conditions.
K-turn dependent compaction of free and bound VcI–II and VcI–IIKt
Lipfert et al. (2007, 2010) explored the contribution of various cations and glycine to the folding of VcI–II. Monovalent and divalent cations yielded partially folded states of this RNA construct lacking P0. Additional global compaction was induced by glycine. The glycine-dependent folding required the presence of certain divalent cations, implicating at least one specific metal ion in glycine binding (Lipfert et al. 2010), consistent with subsequent crystallographic structure determination (Huang et al. 2010).
K-turns often mediate long-range interactions in RNAs (Schroeder et al. 2011). The increased binding affinity we observed by ITC for VcI–IIKt over VcI–II may reflect promotion of interaptamer (“quaternary”) interactions by the K-turn. To evaluate this, we compared the global size and shape of VcI–II and VcI–IIKt by SAXS in solution at 5 and 10 mM Mg2+. For VcI–II, our experiments gave Rg values in good agreement with those previously reported both in the absence and presence of saturating glycine (hereafter “free” and “bound,” respectively) (Fig. 3A; Lipfert et al. 2007). The Kratky plot of free VcI–II presents a rise at low q, followed by a plateau at higher q, consistent with a partially folded state (Fig. 3B). The corresponding P(r) plot is broad, with a maximum intramolecular distance, Dmax (from the intercept with the abscissa) of ∼160 Å (Fig. 3C). Glycine binding to VcI–II induced a compaction (ΔRg) of ∼10 Å. The Kratky plot of bound VcI–II exhibits a peak at ∼0.05 Å−1 followed by a falloff at higher q, indicative of a better folded RNA (Fig. 3B). The P(r) plot becomes nearly bell-shaped, with Dmax ∼ 140 Å.
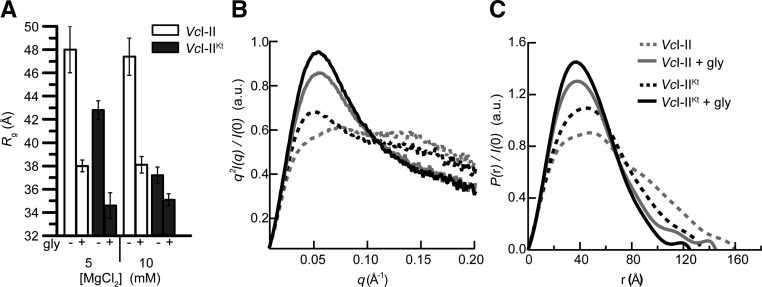
FIGURE 3.
SAXS analyses of VcI–II and VcI–IIKt. (A) Comparison of Rg in the absence and presence of saturating glycine in 5 mM and 10 mM Mg2+. (B) Kratky and (C) P(r) plots for experiments performed in 5 mM Mg2+ with VcI–II and VcI–IIKt in the absence and presence of glycine.
The Rg of free VcI–IIKt is 5 Å smaller than that of free VcI–II. This ΔRg resulting from the presence of the K-turn amounts to 50% of the glycine-binding-induced ΔRg observed for VcI–II (Fig. 3A), suggesting that P0 allows the RNA to pre-fold into a conformation more competent for glycine binding. The presence of a small peak at low q in the Kratky plot (Fig. 3B) also indicates that free VcI–IIKt is better folded than VcI–II. Glycine binding caused additional compaction of VcI–IIKt to an Rg ∼ 35 Å. This is 3 Å smaller than the Rg for bound VcI–II. The Kratky and P(r) plots also show that the bound VcI–IIKt is more compact and better folded than the bound VcI–II (Fig. 3B,C). At a Mg2+ concentration of 10 mM, the free VcI–IIKt RNA (Rg = 37.2 Å) is more compact than even the bound VcI–II (Rg = 38.1 Å).
Mg2+ and YbxF drive compaction of BsI–IIKt relative to BsI–II
Although riboswitches appear to be particularly numerous in Gram-positive bacteria (Mandal et al. 2004; Nudler and Mironov 2004), the tandem glycine aptamers of the model organism B. subtilis have not hitherto been subjected to extensive biophysical characterization. We investigated the solution structure of BsI–II and BsI–IIKt by SAXS, and compared the effect of the P0 K-turn on this RNA with those we found for the tandem aptamers from V. cholerae. The free states of both BsI–II and BsI–IIKt have Rg ∼ 46 Å in 5 mM Mg2+ (Fig. 4A). Under these conditions, the K-turn does not have an impact on the global conformation of the tandem aptamers from this species. Addition of 10 mM glycine resulted in compaction of both RNAs, although the ΔRg exhibited by BsI–IIKt was twice that of BsI–II. Comparison of the ΔRg, Kratky, and P(r) plots (Fig. 4A–C) indicates that bound BsI–IIKt is more compact than BsI–II. At 10 mM Mg2+ concentration, free BsI–IIKt collapsed to Rg ∼ 40 Å. This compaction is ∼60% of the total glycine-induced ΔRg observed in 5 mM Mg2+ (Fig. 4A). In contrast, the increased Mg2+ did not significantly alter the global conformations of free or bound BsI–II (Fig. 4A). Free BsI–IIKt in 10 mM Mg2+ is more compact than either free or bound BsI–II (Fig. 4A–D).
FIGURE 4.
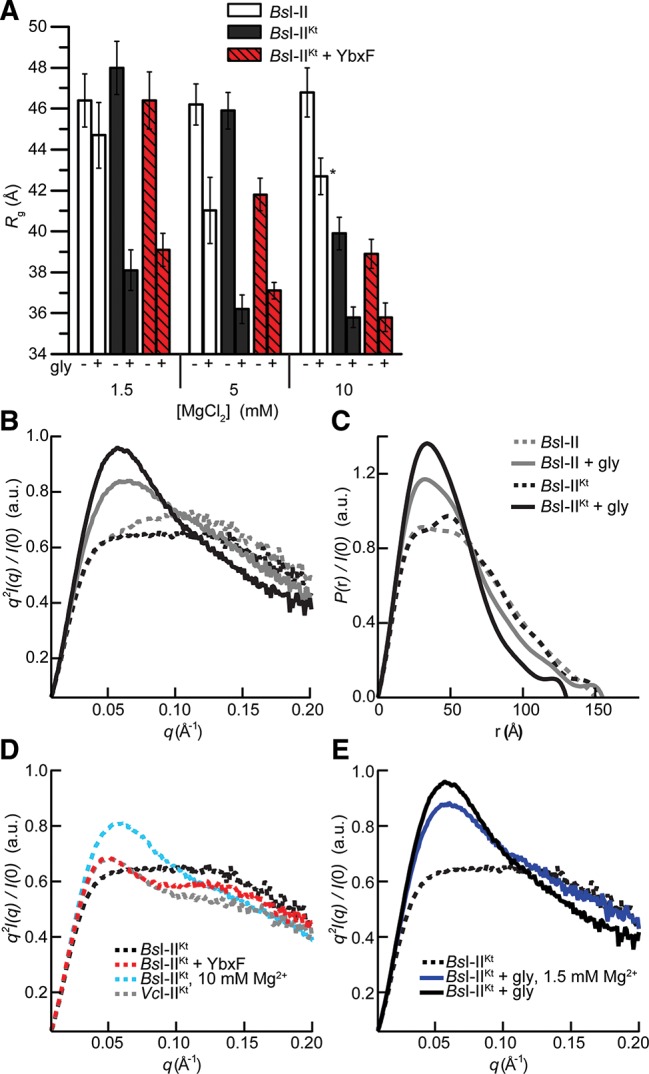
SAXS analyses of B. subtilis tandem glycine aptamers. (A) Rg in the absence and presence of saturating glycine in 1.5 mM, 5 mM, and 10 mM Mg2+ for BsI–II, BsI–IIKt, and BsI–IIKt + YbxF. Sample marked with asterisk exhibited partial oligomerization (Materials and Methods). (B) Kratky and (C) P(r) plots for experiments performed in 5 mM Mg2+ with BsI–II and BsI–IIKt in the absence and presence of glycine. (D) Kratky plots (in 5 mM Mg2+, except where noted) demonstrating the effect of Mg2+ and YbxF on the structure of BsI–IIKt in the absence of glycine. VcI–IIKt plot is reproduced from Figure 3D for additional comparison. (E) Kratky plots demonstrating the effect of glycine in 1.5 mM and 5 mM Mg2+.
We analyzed the cognate interaction between B. subtilis YbxF and BsI–IIKt and the potential contribution of the protein to the folding of this riboswitch aptamer by SAXS. Because YbxF binding to BsI–IIKt increases the mass of the scattering sample, Rg values of the complex are slightly larger than those of the RNA alone. Any observed compaction (ΔRg) of the RNA is thus under-reported in the complex. Also, the final compacted Rg of the complex is slightly larger (by ∼1 Å) than that of the protein-free RNA (Fig. 4A).
Formation of the YbxF–BsI–IIKt complex in 5 mM Mg2+ resulted in a collapse to Rg ∼ 42 Å, or ∼40% of the glycine-induced ΔRg observed for BsI–IIKt. This more compact conformation is evident as a small peak in the Kratky plot (Fig. 4D), the shape of which is reminiscent of that observed for VcI–IIKt (Figs. 3B, 4D) in the absence of protein. Neither increased Mg2+ concentration nor YbxF led to a detectable compaction of the conformation of glycine-bound BsI–IIKt (Fig. 4A).
We reasoned that since YbxF compacts free BsI–IIKt in 5 mM Mg2+, it might also confer stability to the RNA at lower Mg2+ concentrations. However, because we did not observe any glycine binding to BsI–IIKt by ITC in 1.5 mM or 2.5 mM Mg2+ (data not shown), we expected the RNA to be predominantly unfolded in the absence of YbxF. We examined the effect of YbxF on BsI–IIKt in 1.5 mM Mg2+ by SAXS. Both free and bound BsI–II exhibited Rg > 44 Å in 1.5 mM Mg2+. This suggests that the RNA requires additional Mg2+ for both folding and binding (Fig. 4A). The addition of the K-turn was insufficient to compact the RNA under these conditions. Likewise, YbxF binding did not compact the RNA (Rg ∼ 46 Å). While neither the K-turn nor YbxF induced conformational changes to the RNA in 1.5 mM Mg2+, addition of glycine did result in compaction of BsI–IIKt (Fig. 4A). This was surprising given that no heat was evolved in the glycine-binding ITC experiment under these conditions (above). Bound BsI–IIKt has Rg ∼ 38 Å, ∼2 Å larger than that observed in 5 or 10 mM Mg2+, suggesting that this RNA is not completely folded. The shape of the Kratky curve also indicates incomplete folding in 1.5 mM Mg2+ as compared with the conformation in 5 mM Mg2+ (Fig. 4E). Regardless, the glycine-induced conformational compaction of BsI–IIKt in 1.5 mM Mg2+ is K-turn dependent (Fig. 4A).
P0 results in improved ligand binding and modest compaction of glycine-free FnI-IIKt
The unusually compact F. nucleatum tandem glycine riboswitch has been the subject of multiple biochemical and structural studies (Kwon and Strobel 2008; Butler et al. 2011; Erion and Strobel 2011; Kladwang et al. 2012; Sherman et al. 2012). Indeed, the only available crystal structure of a tandem glycine riboswitch is for the RNA from this organism (Butler et al. 2011). Since the RNAs used in these crystallographic studies lacked the P0 K-turn, and in light of the significant energetic and structural contribution of the K-turn to folding of and ligand binding by the tandem glycine riboswitch aptamers from V. cholerae and B. subtilis, we investigated the effect of including P0 in the F. nucleatum tandem glycine riboswitch.
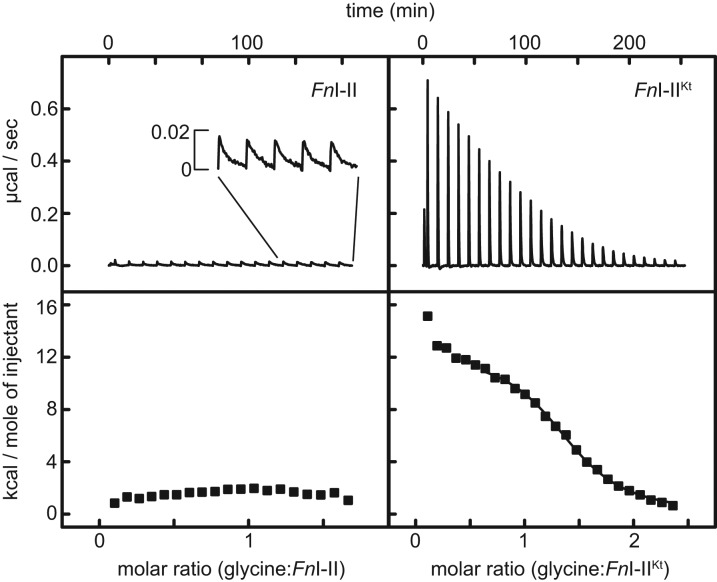
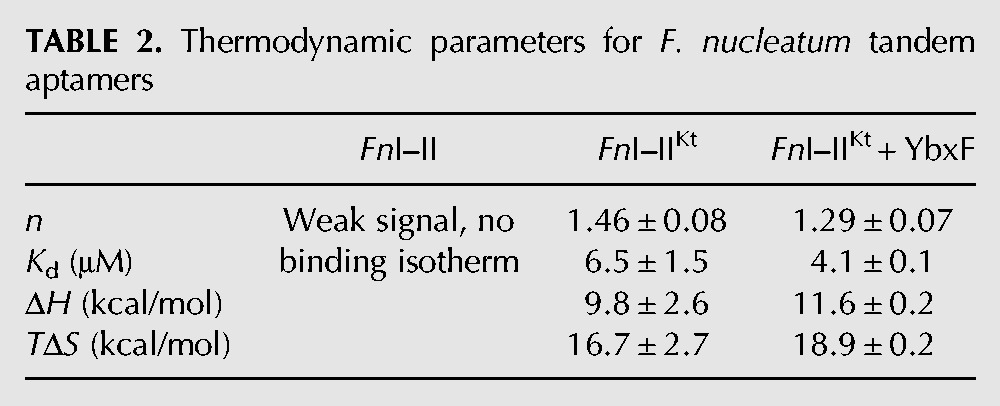
As in the cases of the VcI–II and BsI–II described above, ITC experiments in 5 mM Mg2+ did not yield an interpretable binding isotherm for the F. nucleatum tandem aptamer construct lacking P0 (Fig. 5). However, heat was evolved for each injection of glycine into FnI–II. The signal was weak and the approach to equilibrium notably slower than those of our other ITC experiments. As the experiment progressed, 10 min between injections became insufficient for re-equilibration of the sample (Fig. 5, top left inset). In contrast, FnI–IIKt yielded a reproducible binding isotherm at 5 mM Mg2+, and the reaction appeared to attain equilibrium within a few minutes of each injection of glycine (Fig. 5, top right). For FnI–IIKt, the binding isotherm is sigmoidal (Fig. 5, bottom right), unlike the isotherms for VcI–IIKt or BsI–IIKt (Fig. 2B,C, respectively). The endothermic reaction is entropically driven, with Kd = 6.5 ± 1. μM and n = 1.46 ± 0.08 (Table 2). Because YbxF binds FnI–IIKt (Fig. 1), we examined its ability to modulate glycine binding by this RNA. Titration of glycine into FnI–IIKt in the presence of 1.5-molar excess of YbxF shows that, as in the case of BsI–IIKt, YbxF slightly increased the glycine-binding affinity of FnI–IIKt (Kd = 4.0 ± 0.01 μM).
FIGURE 5.
ITC analyses of FnI–II and FnI–IIKt. (Left) Titration of glycine into FnI–II. A small heat signal is observed upon injection of glycine (inset) and is very slow to re-equilibrate (>10 min). (Right) Glycine binding by FnI–IIKt results in a binding isotherm.
TABLE 2.
Thermodynamic parameters for F. nucleatum tandem aptamers
To evaluate whether the structural compaction induced by the P0 K-turn in VcI–IIKt and BsI–IIKt is a general feature of this class of riboswitches, we examined the F. nucleatum tandem glycine riboswitch by SAXS. In 5 mM Mg2+, the Rg of free FnI–IIKt was only 1 Å smaller than that of FnI–II (Fig. 6A). The Kratky and P(r) plots of these two constructs are also very similar (Fig. 6B,C), indicating only a very modest relative compaction of FnI–IIKt. We next examined whether YbxF and Mg2+ induce folding of FnI–IIKt. In 5 mM Mg2+, the YbxF–FnI-IIKt complex was slightly more compact than the free RNA (ΔRg = 1.8 Å). Increasing Mg2+ concentration to 10 mM (in the absence of YbxF) folded FnI–IIKt to a larger extent (ΔRg = 4.7 Å) (Fig. 6A), similarly to what was observed for free VcI–IIKt and BsI–IIKt (Figs. 3, 4). Addition of YbxF to FnI–IIKt in 10 mM Mg2+ did not result in additional compaction of the Rg (Fig. 6A). It is important to note that the ΔRg is under-reported for the YbxF–FnI–IIKt complex in both 5 mM and 10 mM. The binding of YbxF to the relatively small FnI–IIKt RNA increases the mass of the scattering sample by ∼16%, and thus increases the Rg. Any compaction of the glycine-free RNA is offset by the increase in Rg of the complex. This is most readily observed as a larger Rg (>1.5 Å) for the glycine-bound conformation of the YbxF–FnI–IIKt complex relative the Rg of the RNA alone (Fig. 6A).
FIGURE 6.
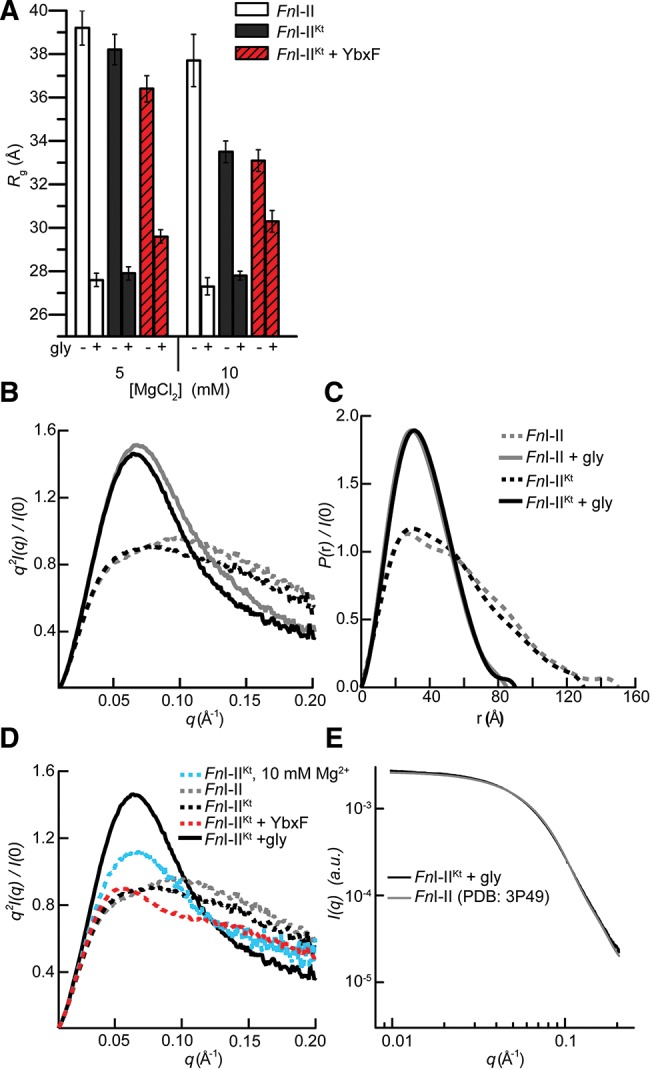
SAXS analyses of FnI–II, FnI–IIKt, and FnI-IIKt + YbxF. (A) Rg in the absence and presence of saturating glycine in 5 mM and 10 mM Mg2+. (B) Kratky and (C) P(r) plots from data collected at 5 mM Mg2+ demonstrate glycine-binding-induced compaction. (D) Kratky plots comparing the effect of Mg2+, YbxF, and glycine on folding of FnI–IIKt (in 5 mM Mg2+, except where noted). (E) Comparison of the experimental X-ray scattering profile for FnI–IIKt with the scattering profile calculated (Materials and Methods) from the crystal structure (Butler et al. 2011) of FnI–II.
In the Kratky plot (Fig. 6D), YbxF binding in 5 mM Mg2+ resulted in the emergence of a slight peak just above 0.05 Å−1, consistent with the slight reduction in Rg (Fig. 6A). Even absent YbxF, this peak became more pronounced in 10 mM Mg2+ as the RNA compacted further (ΔRg = 4.7 Å) (Fig. 6A,D). Addition of glycine to FnI–II and FnI–IIKt resulted in their maximal observed folding. Both constructs exhibited nearly identical Rg’s (∼27.7 Å), Kratky, and P(r) plots (Fig. 6A–C). Thus, at the resolution of the SAXS experiment, the presence of the P0 K-turn does not induce additional folding of the glycine-bound tandem aptamers of the F. nucleatum riboswitch. This is in marked contrast to the stabilization by the K-turn of the overall bound conformations of V. cholerae and B. subtilis tandem glycine riboswitches (Figs. 3, 4).
DISCUSSION
On the basis of in-line probing and hydroxyl radical footprinting data, Sherman et al. (2012) concluded that interaptamer interactions and glycine binding act synergistically to fold the glycine riboswitch. Moreover, they found that their data could not distinguish between the glycine-bound states of I–II and I–IIKt tandem aptamer constructs. Since chemical probing and footprinting experiments report on local reactivity of the nucleic acid, extrapolations from the results of such experiments to the global conformation of an RNA are inferences that need to be tested. SAXS experiments allow rapid comparison of the overall conformations of RNAs in various solution conditions. Our SAXS data indicate that, under physiological Mg2+ concentrations and in the absence of glycine, the K-turn does induce partial folding of the tandem aptamers of constructs from all three bacterial species tested. However, we find that VcI–IIKt and BsI–IIKt exhibit a glycine-bound global conformation that is more compact than those of their respective I–II constructs bound to glycine. Thus, under our experimental conditions, the P0 K-turns confer on these two riboswitches more stability, even in their glycine-bound states.
The global thermodynamic response of tandem glycine aptamer constructs is overall consistent with our SAXS results. For all three species tested, only constructs containing the K-turn show evidence of glycine binding by ITC at 5 mM Mg2+. At this Mg2+ concentration, constructs lacking the K-turn produced no appreciable heat when titrated with glycine. Nonetheless, SAXS experiments demonstrated glycine-induced global compaction for each I–II construct. Given the lack of heat evolution detectable by ITC, this glycine-dependent conformational change of constructs lacking P0 is likely entropically driven. Numerical fits to our ITC data are consistent with different glycine:RNA stoichiometries for the three I–IIKt constructs (Tables 1, 2). Even though the length and composition of various peripheral helical elements vary between the two aptamers of each construct, the sequences of their glycine-binding pockets are highly conserved. Thus, it was expected (Mandal et al. 2004) that two glycine molecules would bind to each dual aptamer RNA. Our ITC experiments reveal that the molar ratio (glycine:RNA) required to saturate the RNAs is <2 (Figs. 2, 5). Only FnI–IIKt exhibited a binding stoichiometry >1 (n ∼ 1.5) (Table 2). For VcI–IIKt the binding stoichiometry is unity (Table 1), while that observed for BsI–IIKt is subunity (Table 1). Intriguingly, the mean Hill coefficients previously determined by Sherman et al. (2012) from fits to their in-line probing data were subunity for all three I–IIKt constructs, which led those investigators to suggest that no glycine-binding cooperativity exists between the aptamers. In addition, structure determination of a tandem glycine aptamer construct (Butler et al. 2011) revealed only weak electron density that could be attributed to glycine. Those investigators reported that averaging of binding pocket electron density between the two aptamers and across two data sets resulted in improved glycine electron density for aptamer 1. Taken together, these results hint that glycine may not bind efficiently to both aptamers in some tandem glycine riboswitches.
Previous reports suggested that the K-turns of bacterial riboswitches containing this motif might be bound in vivo by proteins. Those studies (Blouin and Lafontaine 2007; Heppell and Lafontaine 2008) utilized the archaeal protein L7Ae. We recently found crystallographically that the B. subtilis protein YbxF is structurally homologous to L7Ae and that it binds to K-turns (Baird et al. 2012). Because the P0 K-turn is necessary for folding and glycine binding by the B. subtilis glycine riboswitch, we examined the effect of the B. subtilis YbxF on BsI–IIKt, finding that the protein provides a modest but reproducible increase in glycine-binding affinity of the RNA. To our knowledge, this is the first demonstration that formation of a cognate RNP enhances ligand binding by a riboswitch. Our SAXS experiments show that YbxF binding resulted in more compact global conformation of BsI–IIKt (Fig. 4). We speculate that YbxF binding of the K-turn of this RNA partially orients the two aptamers relative to each other. This pre-folding facilitates subsequent glycine binding. Notably, the P0 K-turn alone is insufficient to pre-fold BsI–IIKt; the K-turn binding protein YbxF is required to achieve this under physiologic Mg2+ concentrations. Therefore, the B. subtilis glycine riboswitch may function as an RNP. We have been unable to detect a YbxF homolog in the V. cholerae genome using BLAST (Altschul et al. 1997). This would be consistent with our SAXS demonstration that VcI–IIKt does not require a protein to achieve folding (Figs. 3B, 4D).
Our SAXS experiments show that the K-turn supports a partially compacted glycine-free conformation of FnI–IIKt and that this RNA is stabilized further by YbxF or increased Mg2+ concentrations. In contrast to the behavior of the RNAs from B. subtilis and V. cholerae, the K-turn does not affect the overall glycine-bound conformation of the F. nucleatum tandem aptamer constructs. To confirm this observation, we calculated the solution X-ray scattering profile expected from the crystal structure of FnI–II (Butler et al. 2011) using CRYSOL (Svergun et al. 1995). This profile agrees very closely with the experimental scattering profile of glycine-bound FnI-IIKt over the entire q-range (Fig. 6E). With the caveat that SAXS cannot resolve structural detail, it thus appears that the unusually compact glycine riboswitch from F. nucleatum can form its full complement of glycine-binding-induced quaternary interactions in the absence of P0. Most glycine riboswitches resemble the RNAs from V. cholerae and B. subtilis and contain longer or additional helices in each of their two aptamers. Additional quaternary interactions between these elements may be required for these RNAs to fold fully, and the P0 K-turn, as well as possibly a K-turn binding protein likely facilitate those. The variable requirement for P0 among glycine riboswitches is consistent with the species-to-species variation observed for riboswitches of other families (Baird and Ferré-D’Amaré 2010; Baird et al. 2010).
Structured RNAs exhibit an interesting dichotomy in that point mutations may lead to complete loss of function (Pitt and Ferré-D’Amaré 2010), yet omission of entire structural motifs may still allow limited folding and function. The previously reported ability of tandem glycine aptamers lacking the P0 K-turn to fold and bind glycine in vitro (Mandal et al. 2004; Lipfert et al. 2007; Kwon and Strobel 2008; Huang et al. 2010; Lipfert et al. 2010; Butler et al. 2011; Erion and Strobel 2011) is reminiscent of the results of truncation of the hammerhead and hairpin ribozymes. In studies of the former, efforts to generate a kinetically well-behaved minimal ribozyme yielded an RNA construct amenable to biochemical and structural characterization (for review, see Blount and Uhlenbeck 2005). However, later discovery of an important peripheral interaction (De la Peña et al. 2003; Khvorova et al. 2003) resulted in an RNA construct with faster cleavage rates (Canny et al. 2004) and a substantially altered active site conformation (Martick and Scott 2006; for review, see Ferré-D’Amaré and Scott 2010). Similarly, early studies of the hairpin ribozyme identified a minimal catalytic construct, which required only two RNA stems with appropriate internal loop sequences (Hampel and Tritz 1989; Hampel et al. 1990). Later, a hairpin ribozyme comprising the natural four-way junction was found to exhibit an increased cleavage rate and a significantly reduced requirement for divalent metal ions relative to the minimal construct (Murchie et al. 1998; for review, see Ferré-D’Amaré 2004). Here, we have shown through ITC and SAXS experiments that the P0 element recently discovered by Kladwang et al. (2012) and Sherman et al. (2012) in tandem glycine riboswitches increases the ligand binding affinity and modulates the overall quaternary arrangement of the aptamers, resulting in a considerably more compact RNA that folds under physiologic conditions.
MATERIALS AND METHODS
RNA and protein constructs
Six RNAs were used in this study. Individual plasmids containing the dual aptamer glycine riboswitches from the V. cholerae VC1422, B. subtilis gcvT, and F. nucleatum FN0328 genes were generated by PCR of overlapping synthetic oligomers (V. cholerae and F. nucleatum constructs), or PCR from genomic DNA (B. subtilis construct) followed by TOPO TA cloning (Invitrogen; Bs and Fn constructs), or ligation into pUC19 vector (Vc construct). The sequences of resulting plasmids were confirmed by Sanger sequencing. DNA templates for transcription were PCR amplified from the individual plasmids. Unique forward primers were used to generate constructs with or without the K-turn sequence upstream of the first aptamer. Sequences of the 5′ and 3′ termini the constructs are as follows: VcI–II 5′-GGGTTGAAGACTGCAG … GGACAGAG-3′; VcI–IIKt 5′-GGTTCCGTTGAAGACTG CAG … GGACAGAG-3′; BsI–II 5′-GGAGCAAGGG … GG ACAGAG-3′; BsI–IIKt 5′-GGCGAATGACAGCAAGGG … GGACAGAG-3′; FnI–II 5′-GGATATGAG … GGACGGAG-3′; FnI–IIKt 5′-GGTCGGATGAAGATATGAG … GGACG GAG-3′. All RNA constructs were transcribed from PCR products essentially as described (Xiao et al. 2008). RNAs were purified using denaturing PAGE, electroeluted (Elutrap, GE), and concentrated and exchanged with 1 M KCl once and with water three times using centrifugal ultrafiltration (Amicon Ultra, 10K MWCO, Millipore). RNAs were stored at 4°C prior to use. L7Ae and YbxF were expressed and purified as described (Hamma and Ferré-D’Amaré 2004; Baird et al. 2012).
Isothermal titration calorimetry
RNA (∼200 μM) was heated to 85°C in 20 mM HEPES-KOH (pH 7.5) and 100 mM KCl for 2 min, followed by cooling to room temperature for 5 min. Concentrated MgCl2 was added (∼1 μL) to a final concentration of 5 mM, followed by incubation at 37°C for 10 min. RNA was equilibrated in RNA folding buffer (20 mM HEPES-KOH at pH 7.5, 100 mM KCl, 5 mM MgCl2) by exchanging three times using centrifugal ultrafiltration (Amicon Ultra, Millipore). For the relevant experiments, 1.5 molar equivalents of YbxF were added prior to buffer exchange. For YbxF containing samples, the buffer was supplemented with 1 mM TCEP. Samples were diluted to a final concentration of 50–150 μM RNA in RNA folding buffer prior to placement in the ITC cell (Microcal iTC200, GE). The titration syringe contained 10–40× concentration (relative to that of the RNA in the calorimeter cell) of glycine in RNA folding buffer. All experiments were performed at 20°C. ITC experiments were analyzed using the single-site fit in the iTC200 software. Consistently, data below a molar ratio 0.5:1 (glycine:RNA) could not be analyzed. Inclusion of data points below this molar ratio yielded poor fits to the data and increased residuals to the fit. Attempts to fit the data to two sites resulted in larger residuals and overall poorer fit to the data. ITC experiments of VcI–II previously reported by Huang et al. (2010) were fitted assuming a single site after omission of initial data points. In order to facilitate comparison with the literature, we adopted this convention. ITC experiments were performed in triplicate for I–IIKt constructs, duplicate for I–IIKt–YbxF complexes, and as single experiments for I–II constructs. None of the experiments yielded a binding ratio of 2:1 (glycine:RNA) at saturation. Even when the final molar ratio (glycine:RNA) in solution was >8, no additional binding was observed. Only the fit to the FnI–IIKt data indicated n > 1. Samples for RNA folding experiments monitored by ITC were prepared as described above, except that RNA concentration was 50 μM and Mg2+ was not added prior to ITC. The titration syringe was filled with 100 mM Mg2+ in 20 mM HEPES-KOH (pH 7.5), and 100 mM KCl. Uncertainties reported in Tables 1 and 2 are the standard errors of the mean of fits to replicate experiments.
Small-angle X-ray scattering
Samples for SAXS were annealed and folded as described for ITC experiments. Then, they were subjected to size-exclusion chromatography (Superdex 200, GE) in RNA folding buffer. Each construct exhibited varying fractions of aggregated, dimeric, and monomeric RNA. Integrated peak areas monitored by absorbance at 290 nm indicate that 70%–95% of total RNA loaded was monomeric. Fractions containing the monomer were pooled and concentrated (Amicon Ultra, Millipore). Concentrated samples were then split for buffer exchange by ultrafiltration; samples were exchanged at least three times into buffers containing final experimental MgCl2 and glycine concentrations (1.5 mM Mg2+, 10 mM glycine; 5 mM Mg2+, 10 mM glycine; or 10 mM Mg2+, 5 mM glycine). Samples were diluted to a final concentration of ∼0.5, 1.0, or 2.0 g/L and shipped to the synchrotron facility overnight at 4°C. SAXS experiments were conducted at the Advanced Photon Source ID-12 BESSRC. Data were collected essentially as described elsewhere (Kulshina et al. 2009). Single experiments at a concentration of 0.5 g/L were performed for all samples in 1.5 mM and 5 mM Mg2+ except for BsI–II + glycine, as noted below. B. subtilis and F. nucleatum samples in 10 mM Mg2+ were collected in triplicate as a concentration series at 0.5, 1.0, and 2.0 g/L, except BsI–IIKt and FnI–IIKt samples including YbxF were collected as single experiments. No concentration dependence of Rg was observed for any sample in 10 mM Mg2+, except BsI–II + glycine. For the latter, samples were analyzed by native PAGE after the SAXS experiment and found to contain a small fraction of oligomer that migrated more slowly than the folded, monomeric RNA. Therefore, in an effort to remove oligomers, new samples of BsI–II + glycine in 10 mM Mg2+ were prepared and purified as described above. SAXS data collected from the new samples still exhibited concentration dependence of Rg. The Rg reported in Figure 4 was calculated from the scattering profile of the sample with the lowest concentration, even though slight oligomerization was present. As a precaution, the BsI–II + glycine in 5 mM Mg2+ measurement was performed in triplicate, given the observed concentration dependence in 10 mM Mg2+. However, no concentration dependence was observed in 5 mM Mg2+. Errors in Rg represent the variance between measurements for experiments performed in duplicate or triplicate. For single measurements, the uncertainty reported is taken from the fit to the data in the Guinier plot. SAXS data analysis was performed using Igor (Wavemetrics) and GNOM (Svergun 1992). The scattering profile of FnI–II was calculated using the atomic coordinates of Butler et al. (2011; PDB ID: 3P49) and the program CRYSOL (Svergun et al. 1995). The U1A protein and its binding site, used to facilitate crystallization (Ferré-D’Amaré 2010), were removed and the crystallographically observed water, counterions, and glycine were stripped prior to the calculation.
Electrophoretic mobility shift assay
Aliquots of monomeric VcI–IIKt, BsI–IIKt, and FnI–IIKt (∼3 μg) were taken from the samples purified by size-exclusion chromatography for SAXS experiments. Individual aliquots were mixed with no protein or more than fivefold molar excess (to ensure saturation) of L7Ae or YbxF, and were analyzed on a 8% polyacrylamide, 0.5× THE (Baird et al. 2012), 5 mM MgCl2 gel. Buffer mixing, electrophoresis, and RNA visualization were as described (Kulshina et al. 2010).
ACKNOWLEDGMENTS
We thank K. Deigan, X. Yang, and the staff of ID-12 BESSRC CAT at the Advanced Photon Source, Argonne National Laboratory, for assistance with SAXS data collection, Y.-X. Wang for generously providing beam time, and G. Piszczek for discussions on ITC data analysis. This research was supported by the Intramural Research Program of the NIH, National Heart, Lung, and Blood Institute.
REFERENCES
- Alatossava T, Jutte H, Kuhn A, Kellenberger E 1985. Manipulation of intracellular magnesium content in polymyxin B nonapeptide-sensitized Escherichia coli by ionophore A23187. J Bacteriol 162: 413–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames TD, Breaker RR 2011. Bacterial aptamers that selectively bind glutamine. RNA Biol 8: 82–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird NJ, Ferré-D’Amaré AR 2010. Idiosyncratically tuned switching behavior of riboswitch aptamer domains revealed by comparative small-angle X-ray scattering analysis. RNA 16: 598–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird NJ, Westhof E, Qin H, Pan T, Sosnick TR 2005. Structure of a folding intermediate reveals the interplay between core and peripheral elements in RNA folding. J Mol Biol 352: 712–722 [DOI] [PubMed] [Google Scholar]
- Baird NJ, Kulshina N, Ferré-D’Amaré AR 2010. Riboswitch function: Flipping the switch or tuning the dimmer? RNA Biol 7: 328–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird NJ, Zhang J, Hamma T, Ferré-D’Amaré AR 2012. YbxF and YlxQ are bacterial homologs of L7Ae and bind K-turns but not K-loops. RNA 18: 759–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blouin S, Lafontaine DA 2007. A loop–loop interaction and a K-turn motif located in the lysine aptamer domain are important for the riboswitch gene regulation control. RNA 13: 1256–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount KF, Uhlenbeck OC 2005. The structure-function dilemma of the hammerhead ribozyme. Annu Rev Biophys Biomol Struct 34: 415–440 [DOI] [PubMed] [Google Scholar]
- Butler EB, Xiong Y, Wang J, Strobel SA 2011. Structural basis of cooperative ligand binding by the glycine riboswitch. Chem Biol 18: 293–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canny MD, Jucker FM, Kellogg E, Khvorova A, Jayasena SD, Pardi A 2004. Fast cleavage kinetics of a natural hammerhead ribozyme. J Am Chem Soc 126: 10848–10849 [DOI] [PubMed] [Google Scholar]
- De la Peña M, Gago S, Flores R 2003. Peripheral regions of natural hammerhead ribozymes greatly increase their self-cleavage activity. EMBO J 22: 5561–5570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erion TV, Strobel SA 2011. Identification of a tertiary interaction important for cooperative ligand binding by the glycine riboswitch. RNA 17: 74–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferré-D’Amaré AR 2004. The hairpin ribozyme. Biopolymers 73: 71–78 [DOI] [PubMed] [Google Scholar]
- Ferré-D’Amaré AR 2010. Use of the spliceosomal protein U1A to facilitate crystallization and structure determination of complex RNAs. Methods 52: 159–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferré-D’Amaré AR, Scott WG 2010. Small self-cleaving ribozymes. Cold Spring Harb Perspect Biol 2: a003574 10.1101/cshperspect.a003574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamma T, Ferré-D’Amaré AR 2004. Structure of protein L7Ae bound to a K-turn derived from an archaeal box H/ACA sRNA at 1.8 Å resolution. Structure 12: 893–903 [DOI] [PubMed] [Google Scholar]
- Hampel A, Tritz R 1989. RNA catalytic properties of the minimum (−)sTRSV sequence. Biochemistry 28: 4929–4933 [DOI] [PubMed] [Google Scholar]
- Hampel A, Tritz R, Hicks M, Cruz P 1990. ‘Hairpin’ catalytic RNA model: Evidence for helices and sequence requirement for substrate RNA. Nucleic Acids Res 18: 299–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heppell B, Lafontaine DA 2008. Folding of the SAM aptamer is determined by the formation of a K-turn-dependent pseudoknot. Biochemistry 47: 1490–1499 [DOI] [PubMed] [Google Scholar]
- Huang L, Serganov A, Patel DJ 2010. Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch. Mol Cell 40: 774–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A, Lescoute A, Westhof E, Jayasena SD 2003. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat Struct Biol 10: 708–712 [DOI] [PubMed] [Google Scholar]
- Kladwang W, Chou FC, Das R 2012. Automated RNA structure prediction uncovers a kink-turn linker in double glycine riboswitches. J Am Chem Soc 134: 1404–1407 [DOI] [PubMed] [Google Scholar]
- Klein DJ, Schmeing TM, Moore PB, Steitz TA 2001. The kink-turn: A new RNA secondary structure motif. EMBO J 20: 4214–4221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulshina N, Baird NJ, Ferré-D’Amaré AR 2009. Recognition of the bacterial second messenger cyclic diguanylate by its cognate riboswitch. Nat Struct Mol Biol 16: 1212–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulshina N, Edwards TE, Ferré-D’Amaré AR 2010. Thermodynamic analysis of ligand binding and ligand binding-induced tertiary structure formation by the thiamine pyrophosphate riboswitch. RNA 16: 186–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon M, Strobel SA 2008. Chemical basis of glycine riboswitch cooperativity. RNA 14: 25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipfert J, Das R, Chu VB, Kudaravalli M, Boyd N, Herschlag D, Doniach S 2007. Structural transitions and thermodynamics of a glycine-dependent riboswitch from Vibrio cholerae. J Mol Biol 365: 1393–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipfert J, Sim AY, Herschlag D, Doniach S 2010. Dissecting electrostatic screening, specific ion binding, and ligand binding in an energetic model for glycine riboswitch folding. RNA 16: 708–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal M, Lee M, Barrick JE, Weinberg Z, Emilsson GM, Ruzzo WL, Breaker RR 2004. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science 306: 275–279 [DOI] [PubMed] [Google Scholar]
- Martick M, Scott WG 2006. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell 126: 309–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monod J, Wyman J, Changeux JP 1965. On the nature of allosteric transitions: A plausible model. J Mol Biol 12: 88–118 [DOI] [PubMed] [Google Scholar]
- Murchie AI, Thomson JB, Walter F, Lilley DM 1998. Folding of the hairpin ribozyme in its natural conformation achieves close physical proximity of the loops. Mol Cell 1: 873–881 [DOI] [PubMed] [Google Scholar]
- Nudler E, Mironov AS 2004. The riboswitch control of bacterial metabolism. Trends Biochem Sci 29: 11–17 [DOI] [PubMed] [Google Scholar]
- Pitt JN, Ferré-D’Amaré AR 2010. Rapid construction of empirical RNA fitness landscapes. Science 330: 376–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romani A, Scarpa A 1992. Regulation of cell magnesium. Arch Biochem Biophys 298: 1–12 [DOI] [PubMed] [Google Scholar]
- Schroeder KT, McPhee SA, Ouellet J, Lilley DMJ 2010. A structural database for k-turn motifs in RNA. RNA 16: 1463–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder KT, Daldrop P, Lilley DMJ 2011. RNA tertiary interactions in a riboswitch stabilize the structure of a kink turn. Structure 19: 1233–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman EM, Esquiaqui J, Elsayed G, Ye JD 2012. An energetically beneficial leader–linker interaction abolishes ligand-binding cooperativity in glycine riboswitches. RNA 18: 496–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsan N, Hammond MC, Block KF, Welz R, Barrick JE, Roth A, Breaker RR 2006. Tandem riboswitch architectures exhibit complex gene control functions. Science 314: 300–304 [DOI] [PubMed] [Google Scholar]
- Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR 2008. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science 321: 411–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun DI 1992. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Crystallogr 25: 495–503 [Google Scholar]
- Svergun DI, Barberato C, Koch MHJ 1995. CRYSOL – a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Crystallogr 28: 768–773 [Google Scholar]
- Welz R, Breaker R 2007. Ligand binding and gene control characteristics of tandem riboswitches in Bacillus anthracis. RNA 13: 573–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson SA 2005. Metal ions and RNA folding: A highly charged topic with a dynamic future. Curr Opin Chem Biol 9: 104–109 [DOI] [PubMed] [Google Scholar]
- Xiao H, Edwards TE, Ferré-D’Amaré AR 2008. Structural basis for specific, high-affinity tetracycline binding by an in vitro evolved aptamer and artificial riboswitch. Chem Biol 15: 1125–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]