In this paper, the authors compare the microRNA-suppressive capacity of different inhibitor RNAs expressed from plasmid DNA and lentiviral vectors. They demonstrate comparable miRNA suppressive activity of two decoy-type inhibitors. They conclude that RNA Pol II-transcribed hairpin-structured Tough Decoy inhibitors elicit potent miRNA suppression.
Keywords: miRNA inhibition, Tough Decoy, TuD, lentiviral vectors
Abstract
MicroRNAs (miRNAs) are key regulators of gene expression and modulators of diverse biological pathways. Analyses of miRNA function as well as therapeutic managing of miRNAs rely on cellular administration of miRNA inhibitors which may be achieved by the use of viral vehicles. This study explores the miRNA-suppressive capacity of inhibitors expressed intracellularly from lentivirus-derived gene vectors. Superior activity of two decoy-type inhibitors, a “Bulged Sponge” with eight miRNA recognition sites and a hairpin-shaped “Tough Decoy” containing two miRNA recognition sites, is demonstrated in a side-by-side comparison of seven types of miRNA inhibitors transcribed as short RNAs from an RNA Pol III promoter. We find that lentiviral vectors expressing Tough Decoy inhibitors are less vulnerable than Bulged Sponge-encoding vectors to targeting by the cognate miRNA and less prone, therefore, to reductions in transfer efficiency. Importantly, it is demonstrated that Tough Decoy inhibitors retain their miRNA suppression capacity in the context of longer RNA transcripts expressed from an RNA Pol II promoter. Such RNA Pol II-transcribed Tough Decoy inhibitors are new tools in managing of miRNAs and may have potential for temporal and spatial regulation of miRNA activity as well as for therapeutic targeting of miRNAs that are aberrantly expressed in human disease.
INTRODUCTION
MicroRNAs (miRNAs) are endogenously encoded small RNA species that regulate gene expression by mediating mRNA degradation and translational repression. As part of the large multicomponent RNA-induced silencing complex (RISC), miRNAs target mRNAs through imperfect or perfect base-pairing with recognition sites located mainly in the 3′ untranslated region (UTR). Imperfect base-pairing promotes enhanced mRNA degradation or translational suppression, whereas perfect or near-perfect base-pairing promotes mRNA cleavage. mRNA cleavage occurs at a single phosphodiester bond corresponding to nucleotides 10–11 of the paired miRNA (Elbashir et al. 2001), and base-pairing at the central part of the miRNA is imperative for RISC-mediated cleavage to occur, as it provides the required geometry at the active site to make the complex catalytically active (Ameres et al. 2007). To date, more than 2000 different mature miRNAs in humans have been identified (miRBase, August 2012, release 19.0). The expanding number of miRNAs has revealed an extensive and intricate level of gene regulation in which a single gene can be under the regulation of several different miRNAs and, vice versa, that a single miRNA can potentially regulate hundreds of genes. Aberrant miRNA activity has been associated with a wide range of pathological conditions, demonstrating the functional significance of miRNAs in disease development and their potential use as diagnostic biomarkers or therapeutic targets (McDermott et al. 2011).
With the intensifying research in miRNA target identification and miRNA gain- and loss-of-function phenotypes, molecular tools have been developed for overexpression and inhibition of miRNA function. MicroRNA overexpression is achieved by mimicking endogenous miRNA expression. Hence, the miRNA-encoding genomic region can be cloned and expressed from a vector of choice, or alternatively, chemically synthesized miRNA precursor molecules can be delivered by cellular transfection (Zeng et al. 2002, 2005; Dickins et al. 2005; Chang et al. 2006; Guimaraes-Sternberg et al. 2006). Methods of miRNA inhibition have primarily relied on transfection of synthetic antisense molecules of different chemistries that are fully complementary to the mature miRNA and thus act as decoys to stoichiometrically sequester miRNAs from their natural targets (Hutvágner et al. 2004; Davis et al. 2006). Less studied synthetic miRNA inhibition molecules include synthetic siRNAs, small molecule inhibitors, and target-protecting masks (Jing et al. 2005; Lee et al. 2005; Choi et al. 2007; Vasudevan et al. 2007; Gumireddy et al. 2008; Medina and Slack 2009; Young et al. 2010; Zhang et al. 2010). Although chemically synthesized miRNA inhibitors may effectively target miRNAs, disadvantages of this approach include the requirement for repeated administration to achieve persistent miRNA inhibition, high production costs, and low transfection efficiency in some tissues and cell types.
DNA-encoded miRNA inhibitors, transferred by either nonviral or viral vector technologies, are cost-effective and can be delivered efficiently to specific cells or tissues. Depending on the vector type and the proliferation status of the target cells, DNA-encoded inhibitors may be continuously expressed, facilitating a prolonged miRNA-suppressive response. Expression from inducible and tissue-specific promoters may allow tailored inhibitor expression, and coexpression of genetic reporters may be utilized to track inhibitor expression in cells or live animals (Medina and Slack 2009). A series of designs has been used for vector-encoded miRNA inhibitors. Initial studies described the use of tandemly arranged miRNA target sequences embedded in the 3′ UTR of a reporter gene which was delivered by the use of an adenoviral vector (Care et al. 2007). The efficiency of this type of miRNA inhibition transcript, later termed a miRNA “Sponge” (Ebert et al. 2007), was further improved by introducing a bulge in the central part of the target sites to prevent endonucleolytic target cleavage, prolonging the interaction with the miRNA (Ebert et al. 2007). Additionally, Ebert et al. showed functionality of such “Bulged Sponge” inhibitors expressed as short transcripts from an RNA Polymerase III (Pol III) promoter. A similar approach was used to express short transcripts containing only a single or two miRNA target sites from an RNA Pol III promoter (Scherr et al. 2007; Haraguchi et al. 2009). Scherr et al. demonstrated miRNA suppression by an “antagomiR” inhibitor containing a single perfect miRNA target site (Scherr et al. 2007), whereas Haraguchi et al., by screening of several structural designs, showed effective miRNA inhibition by a “Tough Decoy” inhibitor consisting of a hairpin containing a large, internal bulge exposing two miRNA target sites with imperfect base-pairing with the miRNA (Haraguchi et al. 2009), a design that has later been proven to be effective in synthetic inhibition molecules (Haraguchi et al. 2012). Both the Tough Decoy and the Bulged Sponge inhibitors have been shown to effectively inhibit whole families of miRNAs through base-pairing to the seed sequence, but inhibition efficiency is assumed to be dependent on features also governing natural miRNA target recognition and may, therefore, rely on the overall degree of complementarity (Ebert et al. 2007; Haraguchi et al. 2009). As opposed to such decoy approaches, Wang et al. employed the RNAi machinery to trigger the degradation of miRNAs by producing RNA Pol III-driven small hairpin RNAs (shRNAs) directed toward the sequence of the mature miRNA (Wang et al. 2011).
Lentiviral vectors derived from human immunodeficiency virus type 1 (HIV-1) are widely used molecular tools for gene transfer. The inherent capacity of these vectors to integrate into target cell genomic DNA, in a reaction catalyzed by the viral integrase enzyme, gives rise to persistent transgene expression in both dividing and nondividing cells (Naldini et al. 1996). Moreover, pseudotyping with the surface glycoprotein from vesicular stomatitis virus enables efficient transduction of a wide range of cell types. Together, such properties render lentiviral vectors an attractive platform for delivery of miRNA inhibitor expression cassettes with a variety of potential applications in in vivo and in vitro settings. Until now, lentiviral vectors expressing antagomiRs, Tough Decoys, and Bulged Sponges have been used for efficient miRNA inhibition (Scherr et al. 2007; Gentner et al. 2009; Haraguchi et al. 2009; Mullokandov et al. 2012), but it remains unclear how different miRNA inhibitors compare to each other for functional suppression of miRNAs. Moreover, complicating issues related to recombination and targeting by endogenous miRNAs due to the presence of single or multiple miRNA-complementary sequences in the lentiviral RNA genome have not been previously addressed.
In this study, we perform a side-by-side comparison of seven types of DNA-encoded miRNA inhibitors expressed from transfected plasmid DNA and transduced lentiviral vectors. The highest levels of miRNA-suppressive activity are monitored with Bulged Sponge and Tough Decoy inhibitors. These two types of inhibitors generally show comparable miRNA targeting capacities despite the presence of only two miRNA target sites in the Tough Decoy, as opposed to eight in the Bulged Sponge. Notably, however, we demonstrate that lentiviral vectors encoding Tough Decoy RNAs are significantly less vulnerable to targeting by endogenous miRNAs and, therefore, less prone to reductions in transductional titers than Bulged Sponge-encoding vectors. Importantly, we report that the miRNA inhibition capacities of Tough Decoy inhibitors are maintained and, in some cases, further improved by fusing the Tough Decoy with RNA transcripts produced from a RNA Pol II promoter. Altogether, our findings support the use of RNA Pol II transcripts as carriers of Tough Decoy inhibitors in future applications of DNA-encoded inhibitors for miRNA suppression.
RESULTS
Side-by-side comparison of vector-encoded miRNA inhibitors demonstrates superior activity of Tough Decoy inhibitors
To identify potent DNA-encoded miRNA inhibitors, we set out to directly compare the potency of seven different inhibitor designs that have recently been employed as inhibitors of miRNAs. The panel of inhibitors included (1) a short antagomiR RNA transcript with perfect miRNA complementarity (Scherr et al. 2007), (2) an antagomiR stabilized by two terminal hairpins (Vermeulen et al. 2007), (3) a Tough Decoy inhibitor composed of an RNA hairpin with two bulged miRNA binding sites exposed in the central part of the stem (Haraguchi et al. 2009), (4) a miRNA-directed shRNA (Wang et al. 2011), (5) a Sponge RNA transcript with eight miRNA binding sites, each with perfect complementarity to the miRNAs (Ebert et al. 2007; Gentner et al. 2009), (6) a Bulged Sponge RNA with central bulges in the eight target sites (Ebert et al. 2007; Gentner et al. 2009), and (7) a “mask” RNA that binds to and protects the mRNA from recognition by the miRNA (Fig. 1A; Choi et al. 2007). To directly compare the miRNA inhibition potencies, we constructed two sets of lentiviral vectors (see Fig. 1B for schematic representation of the vector design) expressing inhibitors directed against miR-16 and miR-203, respectively. miR-16 is highly expressed in virus-producing HEK-293T cells, whereas endogenous expression of miR-203 is only minuscule in HEK-293T cells, as determined by quantitative real-time PCR (qPCR) (Fig. 1C), allowing us to study the effects of endogenous miRNA expression on production and transfer of inhibitor-encoding lentiviral vectors. miRNA inhibitors were expressed from the human H1 RNA Polymerase III promoter which facilitates high expression of well-defined short RNA transcripts that are neither capped nor polyadenylated. miRNA inhibition potencies were evaluated using a dual-luciferase assay in which one of the reporter genes encoding Renilla luciferase (RLuc) was fused to a perfect miRNA target site, making it responsive to the activity of the specific miRNA. For plasmid delivery of the inhibitor-encoding expression cassettes, lentiviral vector plasmid DNA expressing each of the miRNA inhibitors was separately cotransfected into HEK-293 cells with plasmid DNA expressing the RLuc reporter. For lentiviral delivery, we transduced HEK-293 cells with lentiviral vectors at a total dose of 25 ng p24 capsid protein corresponding to an estimated multiplicity of infection (MOI) of ∼80. One day after transduction, cells were transfected with the RLuc reporter plasmid. As HEK-293 cells express very low levels of miR-203, a miR203-encoding plasmid, pT2/CMV-pri-miR-203.SV40-neo, was included in the transfection mix in all assays evaluating the capacity of suppressing miR-203. For all assays, luciferase activities were measured 2 d after transfection of the RLuc reporter plasmid, and RLuc activities were normalized to firefly luciferase (FLuc) activities to normalize for potential variations in transfection efficiencies. RLuc:FLuc ratios were normalized to the ratio obtained with a control vector not expressing any type of inhibitor (Vehicle).
FIGURE 1.
Overview of miRNA inhibition strategies. (A) Schematic representations of the seven miRNA inhibitors compared in the current study. (i) AntagomiR with a single perfect miRNA target site (Scherr et al. 2007); (ii) a single perfect miRNA target site stabilized with terminal hairpin structures (Vermeulen et al. 2007); (iii) a Tough Decoy with two bulged miRNA target sites (Haraguchi et al. 2009); (iv) an shRNA targeting the mature miRNA (only the guide strand of the processed shRNA is depicted) (Wang et al. 2011); (v) a miRNA sponge with eight consecutive miRNA perfect target sites (Ebert et al. 2007; Gentner et al. 2009); (vi) a sponge with eight consecutive bulged miRNA target sites (Ebert et al. 2007; Gentner et al. 2009); (vii) a miRNA mask that binds and protects the miRNA target sites on the mRNA (Choi et al. 2007). (B) Schematic representation of the lentiviral plasmid used for expressing miRNA inhibitors from the H1 RNA Pol III promoter (shown in white). (CMV) Cytomegalovirus promoter, (Ψ) viral RNA packaging signal, (RRE) rev response element, (cPPT) central polypurine tract, (PGK) phosphoglycerate kinase promoter, (Puror) puromycin resistance gene (puromycin N-acetyl-transferase [PAC]), (WPRE) woodchuck hepatitis virus posttranscriptional regulatory element. (C) miRNA levels in HEK-293T, HEK-293, and HeLa cells. qPCR threshold cycles (Ct) were determined for miR-16 and miR-203 in HEK-293T, HEK-293, and HeLa cells. Additionally, Ct values for the U48 small nuclear RNA (RNU48) were determined as a reference small RNA. Data are depicted as mean + SEM.
For plasmid transfections, highly variable miRNA suppression capacities were measured for the seven types of inhibitors (Fig. 2A,B). Notably, the mutual hierarchy among the inhibitors was comparable for the two miRNAs. Whereas neither the shRNA nor the mask was functional, the antagomiR and stabilized antagomiR displayed a significant, but only modest, inhibitory effect on the two miRNAs. Higher levels of miRNA inhibition were measured for both sponges, with the Bulged Sponge as the most efficient inhibitor of the two. Consistently for the two targeted miRNAs, the Tough Decoy inhibitor was the most potent inhibitor of miRNA activity, resulting in levels of RLuc expression that were increased 4.3-fold and 3.4-fold relative to the negative control for miR-16 (P = 1.6 × 10−7) and miR-203 (P = 1.1 × 10−7), respectively. To further validate this mutual ranking of the three most potent inhibitors, we performed a titration assay in which increasing dosages of the three inhibitor-encoding plasmids were transfected into HEK-293 cells and observed a clear dose-response correlation for all inhibitors (data not shown). At all concentrations of inhibitor-encoding plasmid, Tough Decoy inhibitors performed better than both Sponges, and Bulged Sponges performed marginally better than Sponges with perfect miRNA complementarity (data not shown). When the inhibitors were expressed from transduced lentiviral vectors, we observed that only vectors encoding Tough Decoy and Bulged Sponge inhibitors were able to suppress the activity of the target miRNAs with statistical significance relative to the negative control (Fig. 2C,D). Notably, we found for both miR-16 and miR-203 that lentiviral vectors encoding Tough Decoy inhibitors resulted in levels of RLuc expression that were significantly higher than those obtained by lentiviral transfer of Bulged Sponge inhibitors (P = 0.018 and P = 0.033 for miR-16 and miR-203, respectively). Together, these experiments demonstrate that, among seven different types of miRNA inhibitors, the most potent interference of miRNA activity was obtained by Tough Decoy inhibitors expressed from both plasmid DNA and lentiviral vectors.
FIGURE 2.
Tough Decoy inhibitors perform best among seven miRNA inhibition strategies when delivered by plasmid transfection or lentiviral transduction. A dual-luciferase assay was used to screen the potency of seven vector-encoded miRNA inhibitors targeting miR-16 (A,C) or miR-203 (B,D). miRNA inhibitor expression cassettes were delivered either by plasmid transfection (A,B) or by lentiviral transduction (C,D). A miR203-encoding plasmid was included in all assays evaluating miR-203 inhibitors. Data are depicted as mean + SEM. (*) P < 0.05, (**) P < 0.01, (***) P < 0.001, (****) P < 0.0001.
Variable transductional titers of inhibitor-encoding lentiviral vectors
The transduction efficiency of lentiviral vectors encoding miRNA inhibitors is potentially affected by the presence of an inhibitor cassette which may disturb the performance of the vector in both producer and recipient cells. Not only could complex secondary structures of the inhibitor affect transcription, reverse transcription, and packaging of the viral genome, but lentiviral vector RNA is also potentially subjected to degradation due to recognition of the inhibitor sequence by the complementary miRNA. Also, it cannot be excluded that miRNA inhibition may affect the virus-producing cells or that the presence of the inhibitor expression cassette in the 3′ LTR may have a negative impact on virus production since inserts within the 3′ LTR reduce viral titers proportionally to the length of the insert (Urbinati et al. 2009).
To address the impact of the different inhibitors on vector transfer, we first determined transductional titers as measured by the number of puromycin-resistant colony-forming units obtained in vector-transduced HeLa cells (Fig. 3A). Marked variations in titers, ranging from 2 × 107 CFU/mL for LV/Mask-16 to 2 × 104 CFU/mL for LV/Sponge-16, were observed among the miR-16 inhibitor-encoding lentiviral vectors, whereas titers for all miR-203-inhibiting vectors were high and did not vary significantly (titers ranging from 1 × 107 to 4 × 107 CFU/mL). Together with the fact that miR-16 was highly expressed and miR-203 was only vaguely expressed in virus producer and recipient cells (HEK-293T and HeLa cells, respectively) (Fig. 1C), these data demonstrated that vector transduction for some of the inhibitors was strongly influenced by endogenous miRNAs. Notably, we found that the titers among the two most potent inhibitor-encoding vectors, LV/Tough Decoy-16 and LV/Bulged Sponge-16, varied 100-fold. Such substantial titer variations between miR16-targeting vectors were potentially caused by the design of the lentiviral vector (Fig. 1B) in which the presence of only a single polyadenylation signal in the 3′ LTR rendered the puromycin N-acetyl-transferase (PAC) mRNA transcript susceptible to miRNA-mediated regulation due to the miRNA target sites in the inhibitor. Hence, these data suggest that vectors encoding a classical Sponge configuration were highly vulnerable to endogenous miRNAs, whereas Tough Decoy-containing transcripts were less sensitive to targeting by their complementary miRNA.
FIGURE 3.
Transfer of Bulged Sponge-encoding lentiviral vectors are markedly affected by endogenous miRNAs targeting the vector. (A) Reduced transductional titer of Sponge-encoding vectors as determined by the number of puromycin-resistant colony-forming units per mL (CFU/mL). (B) Reduced integration capacity of Bulged Sponge-encoding lentiviral vectors as determined by the number of lentiviral integrations quantified by qPCR. Data are depicted as mean + SEM. (*) P < 0.05, (**) P < 0.01, (***) P < 0.001, (****) P < 0.0001.
To examine the transductional titer by a method that did not depend on PAC expression, we next determined the titer for the vectors encoding the Bulged Sponges and Tough Decoys by quantifying by qPCR the number of lentiviral DNA copies integrated in the genome of transduced HeLa cells (Fig. 3B). By this method, which yields an absolute measurement of the production and transfer efficiency of lentiviral vectors, we observed that the titer of LV/Bulged Sponge-16 was reduced sevenfold compared to LV/Tough Decoy-16 (P = 0.0059), indicating that vector RNA was targeted by endogenous miR-16 expressed in the virus-producing cells. In contrast, for vectors encoding miR-203 inhibitors, a small but not statistically significant reduction in titer (threefold; P = 0.13) was evident for the Bulged Sponge-encoding vector compared to the vector expressing the Tough Decoy inhibitor. Furthermore, we observed that the yield of lentiviral vectors expressing miR-16-directed inhibitors varied in a similar fashion, although to a lesser extent, as determined by measuring the viral p24 capsid protein (data not shown). On the contrary, the yield of all miR203-targeting vectors was not affected by the inhibitor design, indicating that the overall production of viral particles, evident for Sponge-encoding vectors in particular, was negatively affected in an endogenous miRNA-dependent fashion. Together, our findings suggest that lentiviral vectors carrying miRNA inhibitor cassettes are vulnerable to regulation by endogenous miRNAs but also demonstrate that Tough Decoy-encoding vectors, relative to Sponge inhibitors, are less prone to such regulation despite their substantial miRNA-suppressive potential.
Integrity of the inhibitor-encoding cassettes during lentiviral transfer
The ability of the reverse transcriptase to switch between the two lentiviral RNA templates during reverse transcription makes direct repeat sequences prone to rearrangement by either duplication or deletion (Gilboa et al. 1979; An and Telesnitsky 2001; ter Brake et al. 2008; McIntyre et al. 2009). To evaluate the integrity of the inhibitor cassette after lentiviral integration, we transduced cells at a very low MOI to obtain clones with single lentiviral integrations and performed PCR-based analyses of the inhibitor cassette on genomic DNA from pooled and single clones. For PCRs performed on pooled clones, only Sponge and Bulged Sponge inhibitors gave rise to multiple bands, suggesting that deletions and duplications had occurred during lentiviral transfer of the Sponges (data not shown). We then sequenced 15 individual clones carrying the Bulged Sponge inhibitor and found a single clone harboring a vector in which one of the eight miRNA target sites was lost, whereas one other clone had gained an additional miRNA target site (data not shown). These findings demonstrated that the integrity of Sponge-encoding cassettes was reduced relative to all other vectors, most probably due to rearrangements during reverse transcription.
miRNA inhibition by RNA Pol II-transcribed Tough Decoy inhibitors
Tough Decoy inhibitors are deliberately designed to be expressed from RNA Pol III promoters as short transcripts that have structural features resembling pre-miRNAs. This will allow active nuclear export through Exportin-5 recognition of the small terminal 3′ overhang and the dsRNA stem (Lund et al. 2004; Haraguchi et al. 2009; Okada et al. 2009). However, RNA Pol III promoters lack temporal and spatial control and do not allow controlled expression of miRNA inhibitors. In contrast, numerous RNA Pol II promoters allow tissue-specific expression, and both inducible and repressible RNA Pol II promoters exist. Bulged Sponge inhibitors expressed as RNA Pol II transcripts in fusion with eGFP mRNA have been found to suppress miRNA activity to a similar extent as short, RNA Pol III-driven Bulged Sponge inhibitors (Ebert et al. 2007). It has not been explored whether the highly structured Tough Decoy inhibitors retain their miRNA-suppressive potential when embedded in a long RNA Pol II transcript. Such an approach would benefit from the higher degree of flexibility conferred by the RNA Pol II promoters and, in addition, mono-cistronic expression of a reporter mRNA and a miRNA inhibitor would allow easy tracking of inhibitor expression while minimizing vector composition.
To investigate if fusion with an RNA Pol II transcript is a feasible approach also for Tough Decoy inhibitors, we fused Tough Decoy and Bulged Sponge inhibitors directed toward five miRNAs, miR-16, miR-203, miR-143, miR-145, and miR-195, to the 3′ UTR of the eGFP gene as shown in Figure 4A. The fusion RNA variants (depicted in Fig. 4B) were expressed from a phosphoglycerate kinase (PGK) promoter in the context of a lentiviral vector plasmid. In dual-luciferase assays based on plasmid transfection, we observed for miR-16, miR-203, miR-145, and miR-195 an increased level of miRNA inhibition by the PGK-driven fusion RNA transcripts containing Tough Decoys and Bulged Sponges relative to the inhibitors expressed from the H1 promoter (Fig. 4C,D,F,G). Notably, the miRNA suppression activity of the miR16-directed Tough Decoy fused to eGFP was almost doubled relative to the H1-driven Tough Decoy transcripts (P = 9.0 × 10−6) and was significantly higher than the activity of the eGFP-fused Bulged Sponge-16 (P = 1.9 × 10−5). The efficiency of eGFP-fused Tough Decoys directed against miR-203, miR-145, and miR-195 was comparable to the efficiency of their Bulged Sponge counterparts (P = 0.19, P = 0.40, and P = 0.91, respectively). Only for miR143-directed inhibitors did we measure decreased efficiencies of the eGFP-fused inhibitors compared to the H1-transcribed inhibitors (Fig. 4E). For this particular miRNA, we also observed higher potency of the Bulged Sponge compared to the Tough Decoy. To further strengthen the concept of expressing Tough Decoy inhibitors as part of longer RNA Pol II transcripts, we generated three additional RNA Pol II and RNA Pol III vector pairs expressing Tough Decoy inhibitors targeting miR-146a, miR-221, and miR-222. For all three miRNAs, equal inhibition potencies were seen for RNA Pol II- and RNA Pol III-driven Tough Decoys (data not shown).
FIGURE 4.
Tough Decoy inhibitors retain their functionality when fused to an RNA Pol II eGFP transcript. (A) Schematic overview of the lentiviral vector plasmid used for expression of miRNA inhibitors in fusion with eGFP. (B) Schematic representations of the expressed fusion transcripts containing the Bulged Sponge and Tough Decoy inhibitors, respectively. (C–G) Dual-luciferase assays comparing Tough Decoy and Bulged Sponge inhibitors transcribed from RNA Pol II and Pol III promoters targeting miR-16 (C), miR-203 (D), miR-143 (E), miR-145 (F), and miR-195 (G). Data are depicted as mean + SEM. (*) P < 0.05, (**) P < 0.01, (***) P < 0.001, (****) P < 0.0001.
In summary, among the eight miRNAs that we have targeted with Tough Decoy inhibitors, four miRNAs (miR-16, miR-145, miR-195, and miR-203) were more potently suppressed by inhibitors embedded in the eGFP transcript, whereas only miR-143 was most efficiently targeted by the H1-driven Tough Decoy transcript. We conclude that the miRNA inhibition potential of Tough Decoy inhibitors is maintained in the context of RNA Pol II transcripts.
miRNA inhibition by lentivirally delivered RNA Pol II-transcribed Tough Decoy inhibitors
To explore the efficacy of lentivirally delivered RNA Pol II-driven inhibitors, we produced lentiviral vectors encoding Tough Decoy and Bulged Sponge inhibitors targeting miR-16 and miR-203 and performed the dual-luciferase assay on HEK-293 cells transduced with equal amounts of vector particles (25 ng p24 capsid protein per well corresponding to an estimated multiplicity of infection [MOI] of ∼80). Once again, Tough Decoy-16 suppressed the miRNA with higher potency than the Bulged Sponge (Fig. 5A), whereas the efficiencies of the two inhibitors were equal in the case of miR-203 (Fig. 5B), resembling the data obtained when the inhibitors were delivered by plasmid transfection. Determination of lentiviral vector titers based on quantification of integrating viral vectors demonstrated that the transductional titer for the vector encoding eGFP-Bulged Sponge-16 was reduced five times relative to the corresponding vector expressing the Tough Decoy-16 fusion transcript (P = 0.0062) (Fig. 5C). On the contrary, vectors encoding miR203-targeting inhibitors did not differ in transductional titers (P = 0.39). Again, we observed that the yield of the eGFP-Bulged Sponge-16-expressing vector, as measured by p24 capsid ELISA, was slightly reduced relative to the vector encoding the eGFP-Tough Decoy-16 transcript (data not shown).
FIGURE 5.
Lentiviral vectors encoding RNA Pol II-transcribed Tough Decoys are functional and not prone to reductions in transductional titers. (A,B) Dual-luciferase assays evaluating the potency of lentivirally delivered RNA Pol II-transcribed Tough Decoys and Bulged Sponge inhibitors targeting miR-16 (A) or miR-203 (B). (C) Transductional titers of lentiviral vectors encoding RNA Pol II-transcribed miR16- and miR203-directed Tough Decoy and Bulged Sponge inhibitors as determined by the number of lentiviral integrations quantified by qPCR. (D,E) Endogenous miRNA levels were quantified by qPCR following lentiviral delivery of RNA Pol II-driven Tough Decoy and Bulged Sponge inhibitors. For miR-16 inhibition, MCF-7 cells were transduced (D), and for miR-203 inhibition, HaCaT cells were transduced (E). (F) TNFα mRNA levels were quantified by qPCR in HaCaT cells transduced with lentiviral vectors encoding RNA Pol II-driven miR-203 inhibitors. Data are depicted as mean + SEM. (*) P < 0.05, (**) P < 0.01, (***) P < 0.001, (****) P < 0.0001.
We next investigated if miRNA inhibition could be monitored by miRNA levels as detected by qPCR. In cells transduced with lentiviral vectors encoding eGFP-Tough Decoy and eGFP-Bulged Sponge inhibitor transcripts targeting miR-16 and miR-203, we measured a marked reduction in the level of miRNA that was detectable by qPCR (Fig. 5D,E). For miR-16, the Tough Decoy gave rise to the lowest levels of detectable miR-16, whereas the opposite was observed for miR-203 for which the Bulged Sponge gave rise to the lowest levels of detectable miR-203.
We have previously established that mRNA encoding the proinflammatory cytokine TNFα is a target of miR-203 in keratinocytes (Primo et al. 2012). To confirm a biological effect of the two RNA Pol II-driven inhibitors, we transduced a human keratinocyte cell line (HaCaT) with lentiviral vectors encoding miR203-directed Tough Decoy and Bulged Sponge inhibitors. By targeting miR-203, which is expressed at low levels in the lentiviral vector producer cells, similar transfer efficiencies of the two vectors could be achieved (Fig. 5C). As shown in Figure 5F, both types of RNA Pol II-expressed inhibitors caused derepression of the TNFα mRNA level, leading to levels of TNFα mRNA that were significantly higher than the levels obtained with a lentiviral control vector that did not express an inhibitor (P = 0.014 and P = 0.0048 for LV/eGFP-Tough Decoy-203 and LV/eGFP-Bulged Sponge-203, respectively). Notably, the increase in the level of TNFα mRNA was similar for the two inhibitors.
In summary, these findings validate the capacity of lentivirus-delivered RNA Pol II-driven Tough Decoy inhibitors in the management of miRNAs. The effect of miRNA inhibition is evident as decreased miRNA levels and increased levels of a target mRNA as detected by qPCR. Yet, with only two miRNA target sites, vectors encoding Tough Decoy inhibitors seem less prone to miRNA targeting than vectors carrying a Bulged Sponge expression cassette.
Analysis of miRNA targeting of the inhibitor-encoding transcript
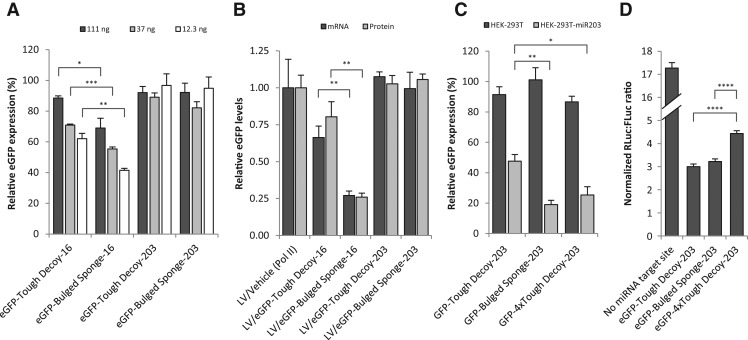
To further analyze targeting of the RNA Pol II-transcribed eGFP-inhibitor fusion RNAs by their complementary miRNAs, we measured eGFP protein levels by flow cytometry in HEK-293 cells transfected with different amounts of plasmids encoding eGFP-fused Tough Decoy and Bulged Sponge inhibitors (Fig. 6A). For decreasing doses of transfected plasmid encoding miR-16 inhibitors, we detected decreasing levels of eGFP compared to the standard plasmid encoding unfused eGFP. This demonstrated increased relative suppression of eGFP expression by miR-16 at low doses of the eGFP-inhibitor fusion transcript. Notably, this effect was more pronounced for eGFP-Bulged Sponge-16 than for eGFP-Tough Decoy-16 (P ≤ 0.04), most likely reflecting that the higher number of target sites in eGFP-Bulged Sponge-16 rendered the transcript more susceptible to miR16-mediated repression. When the eGFP gene was fused to Tough Decoy and Bulged Sponge inhibitors targeting miR-203, the eGFP protein levels were not affected at the three plasmid doses, reflecting the minute amount of miR-203 in HEK-293 cells (Fig. 6A).
FIGURE 6.
Analysis of miRNA targeting of the inhibitor-encoding transcript. (A) Flow cytometric analysis of eGFP expression in HEK-293 cells 2 d after transfection with different amounts of plasmids encoding RNA Pol II-transcribed eGFP fused with Tough Decoy or Bulged Sponge inhibitors targeting either miR-16 or miR-203. eGFP intensity (relative eGFP expression) is presented relative to the eGFP intensity measured in cells transfected with the same amount of the standard unfused eGFP-encoding plasmid. (B) Analysis of eGFP-inhibitor transcript levels and eGFP protein levels following lentiviral delivery of eGFP-inhibitors. Transcript and protein levels were evaluated by qPCR and flow cytometry, respectively. (C) Flow cytometric analysis of eGFP expression in naive HEK-293T cells and HEK-293T cells overexpressing miR203 (HEK-293T-miR203) 2 d after transfection with 12.3 ng of the plasmid encoding RNA Pol II-transcribed eGFP fused with Tough Decoy, Bulged Sponge, or 4 × Tough Decoy (eight miRNA target sites in four consecutive Tough Decoys) inhibitors targeting miR-203. eGFP intensity (relative eGFP expression) is presented relative to the eGFP intensity measured in cells transfected with the same amount of the standard unfused eGFP-encoding plasmid. (D) Dual-luciferase assay comparing Tough Decoy, Bulged Sponge, and 4 × Tough Decoy inhibitors targeting miR-203 and transcribed from RNA Pol II promoters. Data are depicted as mean + SEM. (*) P < 0.05, (**) P < 0.01, (***) P < 0.001, (****) P < 0.0001.
To compare the levels of eGFP-inhibitor fusion RNA transcripts after lentiviral delivery, we transduced HEK-293 cells with equal amounts of lentiviral particles (equal p24 amounts and the same p24:cell ratio as in the inhibition assay depicted in Fig. 5A,B) and measured eGFP mRNA and protein levels by qPCR and flow cytometry, respectively. As shown in Figure 6B, both mRNA and protein levels were reduced for the two miR16-targeting vectors compared to the control, LV/Vehicle, in which eGFP was not fused to an inhibitor. Notably, eGFP mRNA and protein levels were significantly lower for LV/eGFP-Bulged Sponge-16 relative to LV/eGFP-Tough Decoy-16 (P = 0.0089 and P = 0.0068 for mRNA and protein levels, respectively), lending support to the notion that miRNA recognition of the Bulged Sponge induced a higher level of mRNA degradation, resulting in reduced eGFP expression. This view was supported by the fact that eGFP mRNA and protein levels were unaffected in cells transduced with vectors encoding miR-203 inhibitors (Fig. 6B) due to the lack of miR-203 in the transduced cells. This finding also signified that transcription, stability, and translation of the inhibitor-containing transcript was not negatively affected by the presence of the inhibitor per se and that any difference in miRNA inhibition potency resulted from inherent mechanistic differences between the inhibitors and, in the case of lentiviral delivery, from reduced transfer of the vector.
To further analyze the observed difference in the degree of miRNA-targeting between the eGFP-Tough Decoy and eGFP-Bulged Sponge, we constructed an RNA Pol II Tough Decoy vector (eGFP-4 × Tough Decoy-203) carrying four consecutive Tough Decoys, i.e., a total of eight miR-203 target sites. To be able to measure eGFP levels with and without stable expression of miR-203, we established a HEK-293T cell line stably transfected with a miR-203 expression cassette (HEK-293T-miR203) and verified high miR-203 expression by qPCR (miR-203 Ct= 21.8, U48 Ct= 25.7) (cf. Fig. 1C). Transfection of eGFP-Tough Decoy-203 and eGFP-Bulged Sponge-203 in naive HEK-293T gave rise to similar eGFP protein levels, and comparable levels were also observed for eGFP-4 × Tough Decoy-203 (P ≥ 0.20), indicating similar transcription, stability, and translation of the three inhibitor-containing transcripts in cells with very low levels of miR-203 (Fig. 6C). In HEK-293T-miR203 cells, we detected less targeting of eGFP-Tough Decoy-203 compared to eGFP-Bulged Sponge-203 (P = 0.0054), as previously observed with miR16-targeting vectors. Interestingly, we detected comparable eGFP levels from vectors encoding eGFP-Bulged Sponge-203 and eGFP-4 × Tough-203 Decoy (P = 0.36), signifying that target sites embedded in a Tough Decoy configuration were targeted just as strongly by the miRNA as target sites in a Bulged Sponge configuration.
We next addressed the question of whether this increase in targeting of the 4 × Tough Decoy would increase the turnover rate of the inhibitor transcript to an extent that would decrease inhibition potency, or if the increased targeting would lead to increased miRNA inhibition. A luciferase assay showed that, in spite of the equal number of target sites and equal miRNA targeting, the 4 × Tough Decoy-203 performed significantly better than the Bulged Sponge-203 (P = 0.000001) (Fig. 6D), implying that the configuration of the miRNA binding sites in the Tough Decoy enhanced miRNA inhibition relative to the Bulged Sponge configuration.
In summary, our findings demonstrate that Tough Decoy inhibitors embedded in RNA Pol II RNA transcripts are strong inhibitors of miRNA activity. Thus, Tough Decoy inhibitors fused to gene transcripts stand out as an optimal platform for development and use of lentivirally delivered miRNA inhibitors.
DISCUSSION
In the past few years, the molecular miRNA toolbox has expanded considerably with numerous strategies to express and inhibit miRNA function. Several approaches have been pursued to antagonize miRNA activity with RNA molecules intracellularly transcribed from a vector of choice, but direct and compiled comparisons of these approaches have not been undertaken. In the current study, we compared the efficiency of seven different vector-encoded miRNA inhibitor designs expressed from the H1 RNA Pol III promoter. In our experimental setup, neither the shRNA nor the mask design was efficacious. The five remaining inhibitor designs, all based on expressing decoy RNAs, displayed variable inhibition potencies. The stabilized antagomiR with terminal hairpins has previously been shown to increase the inhibitory effect of synthetic antagomiRs (Vermeulen et al. 2007), but such increase in potency was not apparent in our study of vector-encoded inhibitors. As previously observed, the Bulged Sponge performed better than the Sponge (Ebert et al. 2007; Gentner et al. 2009), supporting the notion that miRNA sequestration by the Bulged Sponge was prolonged by prevention of fast endonucleolytic cleavage. The Tough Decoy inhibitor harbors identical bulged target sites, but, in spite of the fewer number of target sites (two vs. eight), the Tough Decoy performed with higher potency than the Bulged Sponge in inhibition studies of both miR-16 and miR-203. These results recapitulate what has earlier been observed in head-to-head comparisons of Tough Decoy and Bulged Sponge inhibitors targeting miR-21 (Haraguchi et al. 2009), miR-122 (Xie et al. 2012), and let-7 (Xie et al. 2012).
Expressed Bulged Sponge inhibitors perform equally well both in the context of short RNA Poll III transcripts and as part of an RNA Pol II transcript of a reporter gene (Ebert et al. 2007). We envisioned that Tough Decoy inhibitors would be functional also in the context of longer RNA transcripts expressed from an RNA Pol II promoter, and we, therefore, fused the inhibitor sequence to the 3′ UTR of eGFP. In plasmid transfection studies, these Tough Decoy RNA Pol II transcripts generally performed with comparable or better potencies than short RNA Pol III Tough Decoy-containing transcripts. Out of eight head-to-head comparisons of Tough Decoy inhibitors expressed as RNA Pol III or Pol II transcripts targeting different miRNAs, only a single Tough Decoy performed with reduced potency as part of an RNA Pol II transcript, whereas equal or improved potencies were observed for the remaining seven Tough Decoys when expressed as part of RNA Pol II transcripts. However, since inhibitor potency is highly dependent on inhibitor concentration, as we observed in the titration experiments (data not shown), a direct comparison between the RNA Pol II and Pol III systems is difficult since it is strongly influenced by the choice of promoter. Gentner et al. have shown for a miR16-targeting Sponge that, when expressed from the strong spleen focus-forming virus (SFFV) promoter, only three lentiviral vector copies were required to achieve the same miRNA inhibition level as 15 vector copies when expressed from the PGK promoter (Gentner et al. 2009). Thus, in our study, miRNA inhibition could likely be improved by using stronger promoters such as the RNA Pol III U6 promoter or, e.g., the RNA Pol II SFFV promoter. This could be combined with using increased amounts of lentiviral vectors, e.g., by concentrating vectors by ultracentrifugation. However, we conclude that embedding the highly structured Tough Decoy inhibitor in a long RNA Pol II-driven RNA transcript is a feasible and highly potent approach to suppress miRNA activity. This is an important finding, demonstrating the feasibility of expressing a transgene and the Tough Decoy inhibitor from a monocistronic transgene cassette. This may simplify vector composition and reduce the overall vector size, which can be crucial for small viral vectors with confined packaging capacity such as recombinant adeno-associated virus (rAAV) vectors. RNA Pol III promoters are limited in numbers and are ubiquitously expressed lacking temporal and spatial control, although inducible RNA Pol III promoters based on expression of Cre and Tet transactivators were previously established (van de Wetering et al. 2003; Tiscornia et al. 2004; Ventura et al. 2004; Wiznerowicz et al. 2006). Obviously, a wealth of RNA Pol II promoters, in contrast, includes numerous promoters with tissue-specific expression profiles as well as inducible promoters. RNA Pol II-transcribed Tough Decoy inhibitors may, therefore, be genetically tailored for expression in certain cell types and tissues or, alternatively, for induced expression allowing confined and regulated miRNA inhibition. Recent studies have exploited the hepatotropism of rAAV9 and adenoviral vectors to express RNA Pol III-transcribed Tough Decoys targeting miR-122 specifically in the liver to reduce cholesterol levels and suppress replication of hepatitis C virus, respectively (Sakurai et al. 2012; Xie et al. 2012). These vectors were administered systemically, and confined liver expression of the Tough Decoys might be improved by the use of liver-specific RNA Pol II promoters, such as the α-1 antitrypsin (hAAT) promoter fused to the ApoE locus control region (Miao et al. 2000).
Our data showed that four out of five eGFP-Tough Decoy fusions targeting different miRNAs performed equal to or better than their Bulged Sponge counterparts. In addition, lentivirally delivered eGFP-Tough Decoy fusion transcripts inhibited the activity of miR-16 and miR-203 to levels that were comparable with suppression obtained with RNA Pol III-transcribed Tough Decoy inhibitors (cf. Figs. 2C,D and 5A,B). As the Tough Decoy hairpin maintains its functionality in the context of an RNA Pol II transcript, these findings suggest that the high potency of Tough Decoy inhibitors is not caused by features unique to the small RNA, such as enhanced nuclear export promoted by Exportin-5. However, the reduced activity observed for RNA Pol III- and Pol II-transcribed Tough Decoy inhibitors targeting miR-143 and RNA Pol III-transcribed Tough Decoy inhibitors targeting miR-195 relative to their Bulged Sponge counterparts may indicate that adverse secondary structures occasionally compromise Tough Decoy functionality. Issues have been raised recently that, for certain targeted miRNAs, the sequence composition of the two opposing target sites in the Tough Decoy hairpin may favor mutual annealing leading to partial structural collapse of the Tough Decoy, resulting in impaired potency (Haraguchi et al. 2012).
Inhibition efficiency following lentiviral delivery of miRNA inhibitors depends on a range of parameters, including vector integrity during vector production, viral encapsidation of vector RNA, efficiency of reverse transcription, transcriptional efficiency of the inhibitor cassette, as well as inhibitor binding and sequestration of the miRNA. Initial analyses of transductional titers by the use of the puromycin resistance gene (PAC) in the vectors indicated that PAC expression for some of the miR16-targeting vectors was strongly regulated via the miRNA inhibitor sequence which was present in the 3′ UTR of PAC. Such targeting by endogenous miRNAs was highly dependent on the inhibitor design carried by the vector. Notably, a large difference was evident between Tough Decoy-16 and the Bulged Sponge-16, with the latter giving rise to a 100-fold reduction in colony-forming units compared to the Tough Decoy-encoding vector. This indicated that the Tough Decoy design was much less vulnerable for targeting by miRNAs despite its robust miRNA-suppressive potential. To investigate the integral effect of miRNA targeting of the viral genome in producer and recipient cells, we quantified the number of integrating vectors for both Pol III- and Pol II-encoded inhibitors. A seven- and fivefold reduction in titers (for RNA Pol III and Pol II, respectively) was evident for vectors encoding Bulged Sponge-16 compared to vectors encoding Tough Decoy-16. These findings indicated that miRNAs in the virus-producing cells and possibly the recipient cells contributed to a decrease in viral transductional titers by targeting vector RNA. In line with this notion, vectors encoding miR203-targeting inhibitors did not display a statistically significant difference in vector production and the number of integrating vectors, most possibly as a reflection of the limited amount of miR-203 in the virus-producing cells. Following lentiviral delivery of the RNA Pol II-driven Tough Decoy and Bulged Sponge inhibitors, we were able to detect decreased levels of the targeted miRNA by qPCR. These results indicate that the inhibitors cause degradation of the targeted miRNAs, which has previously been reported for Tough Decoys (Xie et al. 2012). However, alternatively it has also been shown that transcribed RNA transcripts antisense to the miRNA can sequester the miRNAs, rendering them unavailable for miRNA cDNA synthesis or probing during Northern Blotting (Scherr et al. 2007). Indeed, some reports have not been able to show decreased miRNA levels following miRNA inhibition with Bulged Sponges or Tough Decoys (Ebert et al. 2007; Gentner et al. 2009; Sakurai et al. 2012). One report found decreased miRNA levels upon miRNA inhibition with Tough Decoys as detected by qPCR, but Northern blotting at high temperatures to avoid dsRNA formation failed to validate this decrease (Haraguchi et al. 2009). Thus, we believe that the observed decrease in miR-16 and miR-203 levels might not quantitatively reflect reduced miRNA levels but rather a combination of miRNA degradation and miRNA sequestration.
Analysis of inhibitor levels following plasmid transfection or lentiviral delivery showed similar transcript levels for the Tough Decoy and Bulged Sponge inhibitors targeting miR-203. This indicated that neither of the two inhibitors negatively affected transcription of the eGFP fusion transcript. However, both variants of miR16-targeting inhibitors showed reduced transcript levels compared to wild-type eGFP, which we attribute to targeting of the transcript by endogenous miR-16. Interestingly, when delivered by lentiviral transduction, eGFP-Tough Decoy-16 transcript levels were reduced by 34%, whereas the eGFP-Bulged Sponge-16 levels were reduced by 73% compared to unfused eGFP transcripts, implying that Tough Decoy-containing transcripts were more resistant to miRNA-mediated regulation. Analysis of eGFP protein expression showed that the reduction of protein levels matched the reduced transcript levels. This supports recent claims that mRNA destabilization is the major regulatory mechanism induced by miRNAs (Guo et al. 2010; Huntzinger and Izaurralde 2011). To further characterize the significance of the structural differences between the Tough Decoy and the Bulged Sponge, we constructed a 4 × Tough Decoy vector carrying a total of eight miRNA target sites in four consecutive Tough Decoys. This transcript resembles that of the Bulged Sponge, both carrying the same number of identical target sites, but embedded in different structural contexts. We showed that miRNA-targeting of the 4 × Tough Decoy was similar to the Bulged Sponge, implying that regulation of eGFP levels via the miRNA target sites in the inhibitor sequence is dependent on the number of miRNA target sites rather than on differences in the spatial configuration of the target sites between the Tough Decoy and the Bulged Sponge. We next addressed the question of whether such targeting of the inhibitor transcript would give rise to increased miRNA inhibition or rather would prevent inhibitor levels to reach a certain threshold level required for efficient inhibition. Analysis of the inhibition potency of the 4 × Tough Decoy showed a significant improvement in miRNA inhibition relative to the Tough Decoy and the Bulged Sponge inhibitors, indicating that miRNA inhibition is proportional to the degree of miRNA-targeting of the inhibitor. However, since the 4 × Tough Decoy and the Bulged Sponge are targeted equally, this indicates that the number of target sites is not the only parameter important for inhibition efficiency.
In summary, our data demonstrate that Tough Decoy and Bulged Sponge inhibitors are robust inhibitors of miRNA action. The fact that Tough Decoy inhibitors contain only two miRNA recognition sites is not reflected by reduced activity compared to Bulged Sponge inhibitors containing eight miRNA target sites. Our findings suggest that the Tough Decoy inhibitor contains inherent structural features that support miRNA inhibition. It may be speculated that the structure allows prolonged interaction with the miRNA, thus slowing the enzymatic turnover rate of the miRNA-loaded RISC. We demonstrate high activity of RNA Pol II-transcribed Tough Decoy inhibitors, supporting the use of this platform for future applications of DNA-encoded miRNA inhibitors, including applications that require spatial and/or temporal control of inhibitor expression.
MATERIALS AND METHODS
Plasmids
For expression of miRNA inhibitors, a lentiviral third generation SIN-vector was used. For Pol III-expressed inhibitors, we used the pCCL-PGK-Puro-H1-MCS plasmid, described in Bak et al. (2011), in which the H1 promoter drives the expression of the miRNA inhibitors from an expression cassette located in the U3 region. We also generated a variant of this plasmid encoding eGFP (pCCL-PGK-eGFP-H1-MCS) by exchanging the XhoI/BamHI-flanked puromycin resistance gene with the XhoI/BamHI-flanked eGFP from the lentiviral SIN-vector pCCL-PGK-eGFP (originally designated pCCL.WPS.PGK-eGFP.WHV; a kind gift from Dr. Patrick Aebischer, Swiss Federal Institute of Technology, EPFL, Lausanne, Switzerland). To monitor miRNA activity, a perfect target site for the miRNA was fused to the Renilla luciferase 3′ UTR in the psiCHECK-2 vector (Promega) by cloning annealed oligonucleotides carrying the target sequence into the XhoI and NotI restriction sites of psiCHECK-2. miRNA sequences were derived from miRBase (mirbase.org). For cloning of antagomiRs, stabilized antagomiRs, shRNAs, and masks, phosphorylated and annealed oligonucleotides were cloned into pCCL-PGK-Puro-H1-MCS (digested with AvrII/AscI) (see Supplemental Table T1 for oligonucleotide sequences). For cloning of Tough Decoys, Sponges, and Bulged Sponges, DNA sequences containing the inhibitor sequences flanked by NheI/AscI sites were synthesized and cloned into pUC57 by GenScript. The inhibitor sequences were released from the purchased pUC57 plasmids with NheI/AscI and cloned into AvrII/AscI-digested pCCL-PGK-Puro-H1-MCS or pCCL-PGK-eGFP-H1-MCS. For the construction of lentiviral vectors encoding Pol II inhibitors, sequences including the Tough Decoys and Bulged Sponges were PCR-amplified from the lentiviral vectors encoding the H1-driven inhibitors using primers 5′-AAAAGGTACCGTATGAGACCACCCTAGCCC-3′ (s) and 5′-AAAAGGTACCCAGAGAGACCCAGTACAAGC-3′ (as). The PCR fragments were cloned into pCCL-PGK-eGFP in the sense orientation using the KpnI restriction site located between the WPRE region and U3. pCCL-PGK-eGFP-4 × Tough Decoy-203 was cloned analogously by sequential cloning of the Tough Decoy-203 sequence, PCR-amplified with primers introducing additional restriction sites for sequential insertion of the four Tough Decoy sequences. For miR-203, miR-143, miR-145, miR-146a, miR-221, and miR-222, endogenous miRNA levels were reported to be very low by the smiRNAdb miRNA expression database (http://www.mirz.unibas.ch/cloningprofiles/), and a plasmid expressing the miRNA/miRNA cluster was included in these inhibition assays. The miRNA expression plasmids were constructed by PCR amplification of the genomic region containing the miRNA region/cluster and subsequent insertion of NotI-digested PCR fragment into NotI-digested Sleeping Beauty transposon vector, pT2/CMV-eGFP.SV40-neo (Staunstrup et al. 2012). All constructs were verified by sequencing. The 293T-miR203 cell line stably overexpressing miR-203 was constructed by first exchanging the neomycin resistance gene in the transposon vector encoding miR-203 (pT2/CMV-premiR203.SV40-neo) with a hygromycin resistance gene. Next, 293T cells were transfected with this transposon plasmid and a plasmid encoding a Sleeping Beauty transposase (SB100X) and subsequently subjected to selection with hygromycin. Clones were isolated, and a single clone with marked overexpression of miR-203, as detected by miRNA quantitative Real-Time PCR (qPCR), was selected for further experiments.
Cell culturing
HeLa, HEK-293, HEK-293T, HaCaT, and MCF-7 cells were cultured at standard conditions at 37°C in 5% (v/v) CO2 and maintained in Dulbecco’s modified Eagle’s medium (Cambrex) supplemented with 10% fetal calf serum, penicillin (100 U/mL), streptomycin (0.1 mg/mL), and L-glutamine (265 mg/L).
Production of lentiviral vectors
Lentiviral vectors were produced in HEK-293T cells which were seeded in 10-cm dishes (4 × 106 cells/well). Twenty-four hours after seeding, cells were transfected with a standard calcium phosphate protocol with 3.75 μg pMD.2G, 3 μg pRSV-Rev, 13 μg pMDLg/pRRE, and 13 μg lentiviral transfer vector. Twenty-four hours post-transfection, the medium was changed, and 48 h post-transfection, the viral supernatant was harvested, filtered through 0.45-μm filters to remove cellular debris (Sarstedt), and stored at −70°C. For all transductional experiments, polybrene (Sigma-Aldrich) was added to the virus and supplementary medium at a final concentration of 8 μg/mL. All lentiviral vector preparations were produced in biological triplicates, and the yield of each separate vector preparation was determined by measurements of p24 capsid protein using an HIV-1 p24 Antigen ELISA Kit (ZeptoMetrix) according to the manufacturer’s protocol.
Titer assays
For colony-forming transductional titer determination, cells were seeded in six-well plates (5 × 104 cells/well). On the following day, cells were transduced with viral vectors in serial dilutions. One day post-transduction, puromycin selection was applied (1 µg/mL) for 1 wk before counting the number of colonies. Viral titers (colony-forming units per mL) were calculated by dividing the number of colonies with the dilution factor. For titer assays quantifying the number of lentiviral integrations, cells were seeded in six-well plates (5 × 104 cells/well) and transduced on the following day with different amounts of lentiviral vectors. Additional medium was added to each well to ensure that the total volume in each well was 2 mL. The following day, the medium was changed, and 4 d after transduction, DNA was extracted from each well by a standard NaCl/EtOH precipitation protocol. Quantitative PCR was performed with TaqMan Universal Master Mix II (Applied Biosystems) using primers and probe specific for the WPRE sequence present in the lentiviral vector. Primers and probe specific for albumin were used to normalize for the amount of gDNA. Primers and probes were as follows: WPRE_forward: 5′-GGCACTGACAATTCCGTGGT-3′, WPRE_reverse: 5′-AGGGACGTAGCAGAAGGACG-3′, WPRE_probe: 5′-ACGTCCTTTCCATGGCTGCTCGC-3′ (FAM-BHQ), Alb_forward: 5′-GCTGTCATCTCTTGTGGGCTGT-3′, Alb_reverse: 5′-ACTCATGGGAGCTGCTGGTTC-3′, Alb_probe: 5′-CCTGTCATGCCCACACAAATCTCTCC-3′ (FAM-BHQ). Total proviral copy numbers were determined using the standard curve method. Briefly, a standard curve for each target was made from serial dilutions of plasmids containing the two targets. For WPRE, the pCCL-PGK-Puro-H1-MCS plasmid was used, and for albumin, we used pAlbumin, which was a kind gift from Didier Trono (School of Life Sciences, Ecole Polytechnique Fédérale de Lausanne [EPFL], Lausanne, Switzerland) (plasmid obtained from Addgene, plasmid #22037). The standard curves were used to calculate the copy number of WPRE sequences per cell which, in turn, was used, together with the number of transduced cells and inoculating vector volume, to calculate the number of integrating viral genomes per mL. All titer assays were performed on three separate virus productions of the same vector.
Validation of the integrity of the inhibitor expression cassette
HeLa cells were seeded in six-well plates (5 × 104) and transduced the following day at an MOI << 1 to obtain a population expected to have a single lentiviral integration per cell. The following day, puromycin selection was applied (1 µg/mL), and the cells were allowed to expand, after which DNA was extracted by a standard NaCl/EtOH precipitation protocol. The extracted gDNA was used as the template for 35 cycles of PCR amplification using DreamTaq polymerase (Fermentas) with primers spanning the miRNA inhibitor. Forward primer: 5′-CGATAAGCTTGATATCGAAT-3′. Reverse primer: 5′-CGGCCGCGGCGCGCCTTTCC-3′. As positive controls, the corresponding plasmids with the inhibitor cassettes were used as template.
Dual-luciferase reporter assay
For cotransfection experiments with psiCHECK plasmids and inhibitor-encoding plasmids, cells were seeded in 96-well white plates (3 × 103 cells/well) 1 d before transfection. Cotransfections were performed with a total of 40 ng or 43 ng of DNA, depending on the necessity of including a miRNA expression plasmid (34 ng inhibitor-encoding plasmid, 6 ng psiCHECK plasmid, and 3 ng miRNA-encoding plasmid) using FuGENE-6 (Roche) according to the manufacturer’s protocol. For titration studies of the inhibitor-encoding plasmid, total DNA levels were kept fixed at 40 or 43 ng DNA/well using the pUC19 plasmid as supplementary DNA. Forty-eight hours post-transfection, Renilla- and firefly-luciferase activities were analyzed by the use of the Dual-Glo Luciferase Assay System (Promega) according to the manufacturer’s protocol. Briefly, medium was decanted from the cells, new medium was added (75 µL/well), and the cells were left to equilibrate to room temperature for 20 min. Dual-Glo Reagent was added to the medium (75 µL/well) and after 10 min, firefly luminescence was measured. Dual-Glo Stop & Glo Reagent was then added to the wells, and after another 10 min, Renilla luminescence was measured. All luminescence readings were performed in a multisample plate-reading luminometer (Berthold) with 10-sec integration time. Renilla luciferase activities were normalized to firefly luciferase activities and presented relative to a negative vehicle control set to 1. In transduction studies of inhibitor-encoding lentiviral vectors, cells were seeded in 96-well white plates 1 d before transduction (3 × 103 cells/well) and transduced with the same amount of lentiviral vectors (25 ng CA-p24/well) that were diluted at least twofold in DMEM and supplemented with 8 μg/mL polybrene (Sigma-Aldrich). One day post-transduction, the medium was changed, and cells were transfected with 6 ng psiCHECK plasmid and, in the case of the miR-203 assay, 3 ng miR203-expressing plasmid (pT2/CMV-pri-miR-203.SV40-neo) using FuGENE-6 (Roche) according to the manufacturer’s protocol. Luciferase activities were measured 48 h post-transfection as described above. All dual-luciferase assay experiments were performed at least in biological triplicates and the data summarized as mean + SEM.
miRNA, mRNA, and protein quantification
For eGFP mRNA quantification, total RNA was isolated using the E.Z.N.A. Total RNA Kit I (Omega Bio-tek). For miRNA and TNFα quantification, total RNA, including small RNAs, was purified using the SV Total RNA Isolation System (Promega) or the mirVana miRNA Isolation Kit (Ambion). First-strand cDNA synthesis was performed using the Maxima First Strand cDNA Synthesis Kit for qPCR (Fermentas) according to the manufacturer’s guidelines, and miRNA cDNA synthesis was performed using the TaqMan MicroRNA Reverse Transcription Kit according to the manufacturer’s guidelines (Applied Biosystems). All qPCR reactions were performed using the TaqMan Universal Master Mix II (Applied Biosystems). For eGFP quantification, we used the following primers and probes: eGFP_forward: 5′-CTGCTGCCCGACAACCAC-3′, eGFP_reverse: 5′-TGTGATCGCGCTTCTCGTT-3′, eGFP_probe: 5′-ACCTGAGCACCCAGTCCGCCCT-3′ (FAM-Tamra). For TNFα and the reference gene RPLP0, we purchased primers and probe (gene expression assay ID Hs00174128_m1 and Hs99999902_m1, respectively) from Applied Biosystems. For miRNA qPCR, we used TaqMan MicroRNA Assays for the specific miRNA and U48 as a reference small RNA (Applied Biosystems). Relative mRNA and miRNA levels were determined using the relative standard curve method. Briefly, a standard curve for each gene was made from serial dilutions of the cDNA. The standard curve was then used to calculate relative amounts of target RNA in the samples. Mean RNA values were calculated and the data summarized as mean ± SEM. For flow cytometric determination of eGFP levels, cells were prepared for flow cytometric analysis by formaldehyde fixation and analyzed for eGFP fluorescence on a BD FACSCalibur (Becton, Dickinson).
Statistical analyses
All P-values were calculated by a two-tailed Student’s t-test to test the null hypothesis of no difference between two compared groups. The assumption of equal variances was tested by the F-test. Viral transductional titers were compared by statistical analysis on log-transformed data. In all statistical analyses, P-values <0.05 were considered significant.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
This work was made possible by support from the Lundbeck Foundation, the Novo Nordisk Foundation, The Danish Heart Foundation, Kgl. Hofbuntmager Aage Bangs Fond, Grosserer A. V. Lykfeldts Legat, Agnes og Poul Friis Fond, and Aase og Ejnar Danielsens Fond. R.O.B. is the recipient of a PhD fellowship from the Faculty of Health Sciences at Aarhus University funded by the Lundbeck Foundation and Aarhus University.
REFERENCES
- Ameres SL, Martinez J, Schroeder R 2007. Molecular basis for target RNA recognition and cleavage by human RISC. Cell 130: 101–112 [DOI] [PubMed] [Google Scholar]
- An W, Telesnitsky A 2001. Frequency of direct repeat deletion in a human immunodeficiency virus type 1 vector during reverse transcription in human cells. Virology 286: 475–482 [DOI] [PubMed] [Google Scholar]
- Bak RO, Stenderup K, Rosada C, Petersen LB, Moldt B, Dagnaes-Hansen F, Jakobsen M, Kamp S, Jensen TG, Dam TN, et al. 2011. Targeting of human interleukin-12B by small hairpin RNAs in xenografted psoriatic skin. BMC Dermatol 11: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Care A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, Bang ML, Segnalini P, Gu Y, Dalton ND, et al. 2007. MicroRNA-133 controls cardiac hypertrophy. Nat Med 13: 613–618 [DOI] [PubMed] [Google Scholar]
- Chang K, Elledge SJ, Hannon GJ 2006. Lessons from Nature: MicroRNA-based shRNA libraries. Nat Methods 3: 707–714 [DOI] [PubMed] [Google Scholar]
- Choi WY, Giraldez AJ, Schier AF 2007. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science 318: 271–274 [DOI] [PubMed] [Google Scholar]
- Davis S, Lollo B, Freier S, Esau C 2006. Improved targeting of miRNA with antisense oligonucleotides. Nucleic Acids Res 34: 2294–2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickins RA, Hemann MT, Zilfou JT, Simpson DR, Ibarra I, Hannon GJ, Lowe SW 2005. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat Genet 37: 1289–1295 [DOI] [PubMed] [Google Scholar]
- Ebert MS, Neilson JR, Sharp PA 2007. MicroRNA sponges: Competitive inhibitors of small RNAs in mammalian cells. Nat Methods 4: 721–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Lendeckel W, Tuschl T 2001. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev 15: 188–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentner B, Schira G, Giustacchini A, Amendola M, Brown BD, Ponzoni M, Naldini L 2009. Stable knockdown of microRNA in vivo by lentiviral vectors. Nat Methods 6: 63–66 [DOI] [PubMed] [Google Scholar]
- Gilboa E, Mitra SW, Goff S, Baltimore D 1979. A detailed model of reverse transcription and tests of crucial aspects. Cell 18: 93–100 [DOI] [PubMed] [Google Scholar]
- Guimaraes-Sternberg C, Meerson A, Shaked I, Soreq H 2006. MicroRNA modulation of megakaryoblast fate involves cholinergic signaling. Leuk Res 30: 583–595 [DOI] [PubMed] [Google Scholar]
- Gumireddy K, Young DD, Xiong X, Hogenesch JB, Huang Q, Deiters A 2008. Small-molecule inhibitors of microrna miR-21 function. Angew Chem Int Ed Engl 47: 7482–7484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP 2010. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466: 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi T, Ozaki Y, Iba H 2009. Vectors expressing efficient RNA decoys achieve the long-term suppression of specific microRNA activity in mammalian cells. Nucleic Acids Res 37: e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi T, Nakano H, Tagawa T, Ohki T, Ueno Y, Yoshida T, Iba H 2012. A potent 2′-O-methylated RNA-based microRNA inhibitor with unique secondary structures. Nucleic Acids Res 40: e58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E, Izaurralde E 2011. Gene silencing by microRNAs: Contributions of translational repression and mRNA decay. Nat Rev Genet 12: 99–110 [DOI] [PubMed] [Google Scholar]
- Hutvágner G, Simard MJ, Mello CC, Zamore PD 2004. Sequence-specific inhibition of small RNA function. PLoS Biol 2: E98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing Q, Huang S, Guth S, Zarubin T, Motoyama A, Chen J, Di Padova F, Lin SC, Gram H, Han J 2005. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell 120: 623–634 [DOI] [PubMed] [Google Scholar]
- Lee YS, Kim HK, Chung S, Kim KS, Dutta A 2005. Depletion of human micro-RNA miR-125b reveals that it is critical for the proliferation of differentiated cells but not for the down-regulation of putative targets during differentiation. J Biol Chem 280: 16635–16641 [DOI] [PubMed] [Google Scholar]
- Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U 2004. Nuclear export of microRNA precursors. Science 303: 95–98 [DOI] [PubMed] [Google Scholar]
- McDermott AM, Heneghan HM, Miller N, Kerin MJ 2011. The therapeutic potential of microRNAs: Disease modulators and drug targets. Pharm Res 28: 3016–3029 [DOI] [PubMed] [Google Scholar]
- McIntyre GJ, Yu YH, Tran A, Jaramillo AB, Arndt AJ, Millington ML, Boyd MP, Elliott FA, Shen SW, Murray JM, et al. 2009. Cassette deletion in multiple shRNA lentiviral vectors for HIV-1 and its impact on treatment success. Virol J 6: 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina PP, Slack FJ 2009. Inhibiting microRNA function in vivo. Nat Methods 6: 37–38 [DOI] [PubMed] [Google Scholar]
- Miao CH, Ohashi K, Patijn GA, Meuse L, Ye X, Thompson AR, Kay MA 2000. Inclusion of the hepatic locus control region, an intron, and untranslated region increases and stabilizes hepatic factor IX gene expression in vivo but not in vitro. Mol Ther 1: 522–532 [DOI] [PubMed] [Google Scholar]
- Mullokandov G, Baccarini A, Ruzo A, Jayaprakash AD, Tung N, Israelow B, Evans MJ, Sachidanandam R, Brown BD 2012. High-throughput assessment of microRNA activity and function using microRNA sensor and decoy libraries. Nat Methods 9: 840–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage FH, Verma IM, Trono D 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272: 263–267 [DOI] [PubMed] [Google Scholar]
- Okada C, Yamashita E, Lee SJ, Shibata S, Katahira J, Nakagawa A, Yoneda Y, Tsukihara T 2009. A high-resolution structure of the pre-microRNA nuclear export machinery. Science 326: 1275–1279 [DOI] [PubMed] [Google Scholar]
- Primo MN, Bak RO, Schibler B, Mikkelsen JG 2012. Regulation of pro-inflammatory cytokines TNFα and IL24 by microRNA-203 in primary keratinocytes. Cytokine 60: 741–748 [DOI] [PubMed] [Google Scholar]
- Sakurai F, Furukawa N, Higuchi M, Okamoto S, Ono K, Yoshida T, Kondoh M, Yagi K, Sakamoto N, Katayama K, et al. 2012. Suppression of hepatitis C virus replicon by adenovirus vector-mediated expression of tough decoy RNA against miR-122a. Virus Res 165: 214–218 [DOI] [PubMed] [Google Scholar]
- Scherr M, Venturini L, Battmer K, Schaller-Schoenitz M, Schaefer D, Dallmann I, Ganser A, Eder M 2007. Lentivirus-mediated antagomir expression for specific inhibition of miRNA function. Nucleic Acids Res 35: e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staunstrup NH, Madsen J, Primo MP, Li J, Liu Y, Kragh PM, Li R, Schmidt M, Purup S, Dagnaes-Hansen F, et al. 2012. Development of transgenic cloned pig models of skin inflammation by DNA transposon-directed ectopic expression of human β1 and α2 integrin. PLoS One 7: e36658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Brake O, t Hooft K, Liu YP, Centlivre M, von Eije KJ, Berkhout B 2008. Lentiviral vector design for multiple shRNA expression and durable HIV-1 inhibition. Mol Ther 16: 557–564 [DOI] [PubMed] [Google Scholar]
- Tiscornia G, Tergaonkar V, Galimi F, Verma IM 2004. CRE recombinase-inducible RNA interference mediated by lentiviral vectors. Proc Natl Acad Sci 101: 7347–7351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbinati F, Arumugam P, Higashimoto T, Perumbeti A, Mitts K, Xia P, Malik P 2009. Mechanism of reduction in titers from lentivirus vectors carrying large inserts in the 3′LTR. Mol Ther 17: 1527–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wetering M, Oving I, Muncan V, Pon Fong MT, Brantjes H, van Leenen D, Holstege FC, Brummelkamp TR, Agami R, Clevers H 2003. Specific inhibition of gene expression using a stably integrated, inducible small-interfering-RNA vector. EMBO Rep 4: 609–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA 2007. Switching from repression to activation: MicroRNAs can up-regulate translation. Science 318: 1931–1934 [DOI] [PubMed] [Google Scholar]
- Ventura A, Meissner A, Dillon CP, McManus M, Sharp PA, Van Parijs L, Jaenisch R, Jacks T 2004. Cre-lox-regulated conditional RNA interference from transgenes. Proc Natl Acad Sci 101: 10380–10385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen A, Robertson B, Dalby AB, Marshall WS, Karpilow J, Leake D, Khvorova A, Baskerville S 2007. Double-stranded regions are essential design components of potent inhibitors of RISC function. RNA 13: 723–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Han L, Zhang A, Wang G, Jia Z, Yang Y, Yue X, Pu P, Shen C, Kang C 2011. Adenovirus-mediated shRNAs for co-repression of miR-221 and miR-222 expression and function in glioblastoma cells. Oncol Rep 25: 97–105 [PubMed] [Google Scholar]
- Wiznerowicz M, Szulc J, Trono D 2006. Tuning silence: Conditional systems for RNA interference. Nat Methods 3: 682–688 [DOI] [PubMed] [Google Scholar]
- Xie J, Ameres SL, Friedline R, Hung JH, Zhang Y, Xie Q, Zhong L, Su Q, He R, Li M, et al. 2012. Long-term, efficient inhibition of microRNA function in mice using rAAV vectors. Nat Methods 9: 403–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young DD, Connelly CM, Grohmann C, Deiters A 2010. Small molecule modifiers of microRNA miR-122 function for the treatment of hepatitis C virus infection and hepatocellular carcinoma. J Am Chem Soc 132: 7976–7981 [DOI] [PubMed] [Google Scholar]
- Zeng Y, Wagner EJ, Cullen BR 2002. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol Cell 9: 1327–1333 [DOI] [PubMed] [Google Scholar]
- Zeng Y, Cai X, Cullen BR 2005. Use of RNA polymerase II to transcribe artificial microRNAs. Methods Enzymol 392: 371–380 [DOI] [PubMed] [Google Scholar]
- Zhang S, Chen L, Jung EJ, Calin GA 2010. Targeting microRNAs with small molecules: From dream to reality. Clin Pharmacol Ther 87: 754–758 [DOI] [PMC free article] [PubMed] [Google Scholar]