Eukaryotic translation initiation factor (eIF)4B stimulates recruitment of mRNA to the 43S ribosomal pre-initiation complex (PIC). Yeast eIF4B (yeIF4B), shown previously to bind single-stranded (ss) RNA, consists of an N-terminal domain (NTD), predicted to be unstructured in solution; an RNA-recognition motif (RRM); an unusual domain comprised of seven imperfect repeats of 26 amino acids; and a C-terminal domain. Although the mechanism of yeIF4B action has remained obscure, most models have suggested central roles for its RRM and ssRNA-binding activity. They have dissected the functions of yeIF4B’s domains and show that the RRM and its ssRNA-binding activity are dispensable in vitro and in vivo. The data indicate that the 7-repeats and NTD are the most critical domains, which mediate binding of yeIF4B to the head of the 40S ribosomal subunit via interaction with Rps20.
Keywords: eIF4B, eIF4A, mRNA, translation initiation, ribosome, yeast translation
Abstract
Eukaryotic translation initiation factor (eIF)4B stimulates recruitment of mRNA to the 43S ribosomal pre-initiation complex (PIC). Yeast eIF4B (yeIF4B), shown previously to bind single-stranded (ss) RNA, consists of an N-terminal domain (NTD), predicted to be unstructured in solution; an RNA-recognition motif (RRM); an unusual domain comprised of seven imperfect repeats of 26 amino acids; and a C-terminal domain. Although the mechanism of yeIF4B action has remained obscure, most models have suggested central roles for its RRM and ssRNA-binding activity. We have dissected the functions of yeIF4B’s domains and show that the RRM and its ssRNA-binding activity are dispensable in vitro and in vivo. Instead, our data indicate that the 7-repeats and NTD are the most critical domains, which mediate binding of yeIF4B to the head of the 40S ribosomal subunit via interaction with Rps20. This interaction induces structural changes in the ribosome’s mRNA entry channel that could facilitate mRNA loading. We also show that yeIF4B strongly promotes productive interaction of eIF4A with the 43S•mRNA PIC in a manner required for efficient mRNA recruitment.
INTRODUCTION
Recruitment of mRNA to the eukaryotic ribosome begins with the formation of a ternary complex (TC) comprised of eukaryotic initiation factor (eIF) 2, GTP, and the methionyl initiator tRNA. The TC binds to the 40S ribosomal subunit aided by eIFs 1, 1A, and 3 to form the 43S pre-initiation complex (PIC). The PIC is loaded onto the 5′-end of an mRNA, near the 7-methylguanosine cap, in a manner coordinated by the eIF4F cap-binding complex, eIF4B and eIF3. The PIC then scans along the mRNA until the start codon enters the ribosomal P site and base pairs with the initiator tRNA anticodon. Recognition of the start codon triggers downstream events, including release of eIF1, conversion of eIF2 to its GDP-bound state, and a series of structural rearrangements leading to formation of a “closed” state of the PIC arrested on the mRNA. Release of eIF2•GDP and its associated GTPase activating protein eIF5 then permits the eIF5B-facilitated joining of the 60S ribosomal subunit to form the 80S initiation complex (for review, see Hinnebusch 2011; Aitken and Lorsch 2012).
We previously showed that yeast eIF4F, eIF3, and eIF4B are necessary for rapid and accurate recruitment of natural mRNAs to PICs in a reconstituted Saccharomyces cerevisiae translation initiation system (Mitchell et al. 2010). eIF4F is a heterotrimeric complex comprised of cap-binding protein eIF4E, scaffolding protein eIF4G, and DEAD-box RNA helicase eIF4A (for review, see Parsyan et al. 2011). eIF4F is thought to promote mRNA recruitment and scanning by unwinding secondary structure in the 5′ untranslated regions (UTRs) of mRNAs. eIF3 is a heteromultimeric factor that binds to the ribosome and a number of other factors, and stimulates recruitment of both ternary complex and mRNA to the PIC (Trachsel et al. 1977; Pestova and Kolupaeva 2002; Dmitriev et al. 2003; Mitchell et al. 2010). The third factor required for rapid mRNA recruitment in the yeast system, eIF4B, is an RNA-binding protein (Schreier et al. 1977; Trachsel et al. 1977; Seal et al. 1989; Pestova and Kolupaeva 2002; Dmitriev et al. 2003; Mitchell et al. 2010) often considered an auxiliary factor because both plant and mammalian eIF4B stimulate eIF4A’s RNA helicase activity in vitro (Bi et al. 2000; Rogers et al. 2001; Rozovsky et al. 2008; Nielsen et al. 2011; Ozes et al. 2011; Parsyan et al. 2011). However, in vitro studies suggest that eIF4B is a critical component in the recruitment of mRNAs to PICs (Dmitriev et al. 2003; Mitchell et al. 2010).
The RRM and C-terminal arginine-rich motif (ARM) (Fig. 1A) of mammalian eIF4B bind ssRNA and are required for its ability to stimulate eIF4A helicase activity (Methot et al. 1994; Naranda et al. 1994; Rozovsky et al. 2008), but the mechanism of this regulatory function is unknown. It was also proposed that these RNA-binding domains in eIF4B can promote PIC attachment to mRNA more directly by binding simultaneously to mRNA and 18S rRNA in the 40S subunit (Methot et al. 1996a), and that interaction of the internal “DRYG” repeats with eIF3 can stabilize mRNA–PIC interaction (Methot et al. 1996b). It is unclear, however, which of its domains or potential activities is crucial for the ability of mammalian eIF4B to stimulate mRNA recruitment by the PIC.
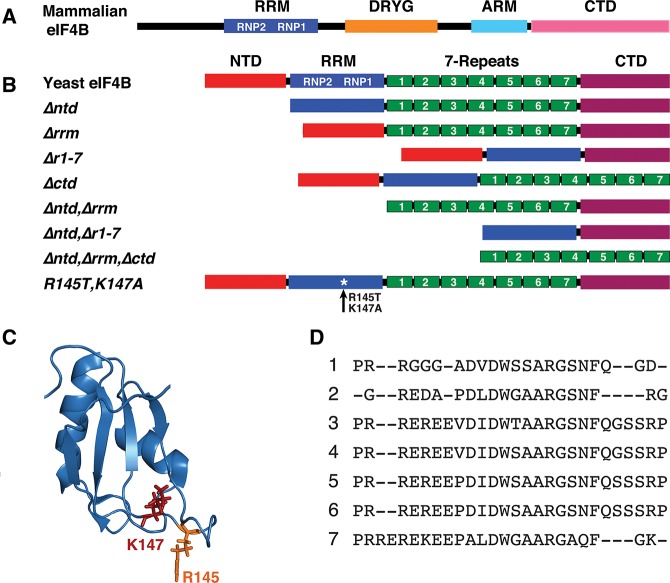
FIGURE 1.
yeIF4B functional domains. Schematic of (A) mammalian eIF4B and (B) yeIF4B domains and mutant constructs. (C) Structure of the human eIF4B RRM, showing positions of conserved residues required for ssRNA binding, generated in PyMOL using pdb 1WI8. (D) Sequence alignment of the 7-repeats of yeIF4B.
Yeast eIF4B (hereafter yeIF4B) was identified both as a suppressor of an eIF4A mutation and by screening an expression library using polyclonal antibodies against yeast eIF4E. yeIF4B has ∼20% sequence identity with mammalian eIF4B, and was shown to stimulate translation in vitro and in vivo (Altmann et al. 1993; Coppolecchia et al. 1993). The gene encoding yeIF4B, TIF3 (formerly called STM1), is not essential, but its deletion results in severe slow growth and cold-sensitive phenotypes, and displays synthetic lethality with initiation factor mutations (Altmann et al. 1993; Coppolecchia et al. 1993; de la Cruz et al. 1997). The absence of yeIF4B in a tif3Δ mutant extract confers cold-sensitive translation in a manner rescued by mammalian eIF4B, indicating that the yeast and mammalian proteins share a conserved critical function. Surprisingly, however, yeIF4B does not stimulate the helicase activity of eIF4A in vitro (Altmann et al. 1995; Rajagopal et al. 2012), and consistent with this, the C-terminal domain (CTD) (Fig. 1A,B) of yeIF4B shows no obvious similarity to that of mammalian eIF4B (Altmann et al. 1993; Coppolecchia et al. 1993). Both yeast and mammalian eIF4B possess eIF4A-independent RNA annealing activities that might promote mRNA–rRNA base-pairing during scanning (Altmann et al. 1995) or PIC binding to mRNA.
The 436-residue yeIF4B can be divided into four putative functional domains (Fig. 1B): (1) an N-terminal domain (NTD) that is predicted to be unstructured in solution and has no obvious similarity to any domains of the mammalian factor; (2) a predicted RRM (Fig. 1C); (3) a segment of seven imperfect repeats of ∼26 amino acids (7-repeats domain) (Fig. 1D) that appear to be unrelated in sequence to the “DRYG” internal repeats of mammalian eIF4B (Altmann et al. 1993; Coppolecchia et al. 1993) and are predicted to be unstructured in solution; and a CTD. Analysis of N- or C-terminally truncated yeIF4B variants led to the conclusion that both the RRM and at least a subset of the seven internal repeats contribute to ssRNA-binding activity, and are crucial for robust RNA annealing activity and stimulation of translation in vitro and in vivo. This led to the proposal that RNA annealing activity is key for the function of yeIF4B in vivo (Niederberger et al. 1998).
Here we dissect the functions of yeIF4B in translation initiation by analyzing the effects of deletions and mutations of its domains on key steps in the process in vitro and in vivo. We report that the 7-repeats domain is critical for productive interaction with the PIC and other components of the initiation machinery in order to promote mRNA recruitment. The NTD also plays a role in accelerating mRNA binding to the PIC. Surprisingly, the factor’s RRM and the ssRNA-binding activity it confers are completely dispensable for function in vitro and in vivo as long as the 7-repeats and NTD are intact. Further, we show that yeIF4B binds to ribosomal protein Rps20 in the head of the 40S subunit, and induces structural changes in the ribosome’s mRNA entry channel. Together, our data suggest that yeIF4B acts directly to modulate the conformation of the 40S head to promote a receptive state of the mRNA entry channel. Our data indicate that yeIF4B also mediates productive interaction of helicase eIF4A with the rest of the initiation machinery to stimulate mRNA recruitment.
RESULTS
To study the roles of domains of yeIF4B in translation in vitro and in vivo, we made constructs in which each domain was deleted, individually or in combination (Fig. 1B). We also altered two residues in the RRM (R145T, K147A) (Fig. 1C) implicated in ssRNA binding in human eIF4B (Naranda et al. 1994).
yeIF4B binds directly to the 40S subunit and ssRNA
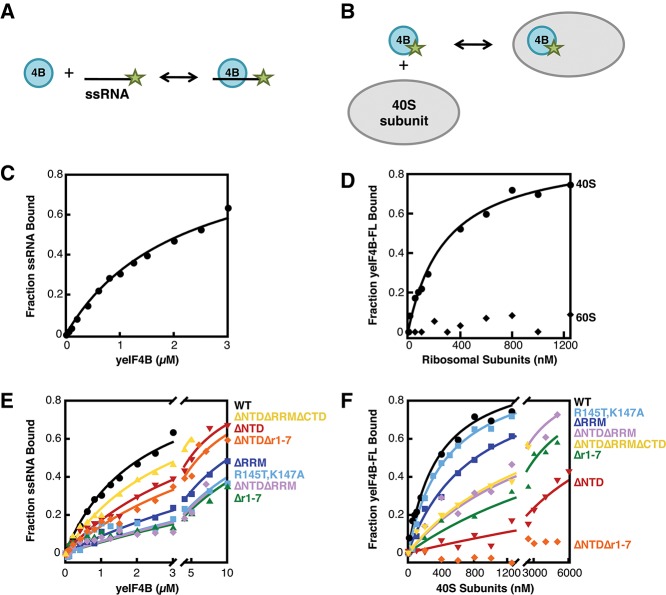
Previous studies suggested that mammalian eIF4B binds to ssRNA and 18S rRNA (Milburn et al. 1990; Hughes et al. 1993; Naranda et al. 1994; Methot et al. 1996a,b), and that yeIF4B also binds ssRNA (Altmann et al. 1995; Niederberger et al. 1998). We developed assays to quantify binding of purified yeIF4B to ssRNA and purified ribosomal subunits in vitro. We measured yeIF4B–ssRNA affinity by monitoring the fluorescence anisotropy of a 3′-fluorescein-labeled 43-nucleotide ssRNA on addition of unlabeled yeIF4B (Fig. 2A). Protein binding to the labeled RNA should decrease the latter’s rate of rotational diffusion and thereby increase fluorescence anisotropy. Consistent with previous evidence that yeIF4B binds ssRNA (Altmann et al. 1995), increasing the concentration of yeIF4B led to an increase in anisotropy. Converting the anisotropy values to the fraction of RNA bound, plotting these values vs. concentration of yeIF4B and fitting with a hyperbolic binding equation yielded a dissociation constant (Kd) of 2.2 µM (Fig. 2C), similar to that previously reported (Altmann et al. 1995). We measured binding of ribosomal subunits to fluorescein-labeled yeIF4B in a similar manner (Fig. 2B). Titrating the concentration of 40S subunits yielded an increase in anisotropy, while no change in anisotropy was observed up to a concentration of 6 µM 60S subunits (Fig. 2D; data not shown), indicating that yeIF4B binds directly and specifically to the 40S subunit with a Kd of 360 nM.
FIGURE 2.
yeIF4B binds to both ssRNA and 40S ribosomal subunits. Binding of yeIF4B to ssRNA was evaluated by monitoring fluorescence anisotropy of ssRNA labeled on its 3′-end with fluorescein as a function of yeIF4B concentration (A,C). Binding of yeIF4B to ribosomal subunits was assessed by measuring the fluorescence anisotropy of yeIF4B labeled on its C terminus with fluorescein at increasing concentrations of 40S or 60S ribosomal subunits (B,D). Data were fit with a hyperbolic binding equation. (E,F) Effects of yeIF4B domain mutations on ssRNA binding (E) and 40S subunit binding (F): WT (black circles); ΔNTD (red inverted triangles); ΔRRM (dark blue squares); R145T, K147A (light blue squares); Δr1-7 (green triangles); ΔNTDΔRRM (purple diamonds); ΔNTDΔr1-7 (orange diamonds); or ΔNTDΔRRMΔCTD (yellow triangles).
The RRM and 7-repeats domains are important for yeIF4B’s ssRNA-binding activity
To determine which domains of yeIF4B are responsible for the factor’s ssRNA-binding activity, we measured the Kd of each protein variant and the fluorescently labeled ssRNA. Deletion of the RRM or mutation of the two key residues K147 and R145 evoked a defect in RNA binding (Fig. 2E) (ΔRRM and R145T, K147A; Kd > 10 μM), consistent with previous reports (Milburn et al. 1990; Methot et al. 1994; Naranda et al. 1994; Altmann et al. 1995; Niederberger et al. 1998). Addition of BSA to 10 µM gave a similar change in anisotropy (data not shown), suggesting that the residual binding observed here is likely nonspecific. Deletion of the repeats also increased the Kd for ssRNA binding by more than fivefold (Kd > 10 μM), while deletion of the NTD had only a twofold effect (Kd = 4.8 μM). The yeIF4B mutant lacking both NTD and RRM behaved similarly to the version lacking just the RRM (Fig. 2E) (ΔNTDΔRRM; Kd > 10 µM). Surprisingly, however, deletion of the NTD from the derivative lacking the 7-repeats actually enhanced binding (Fig. 2E) (ΔNTDΔr1-7; Kd = 6.2 µM) relative to the version lacking only the repeats (Kd > 10 µM). This result suggests that deletion of the 7-repeats impairs RNA binding in part by facilitating inhibition of the RRM’s ssRNA-binding function by the NTD. Together, these data indicate that the RRM and the 7-repeats both play key roles in binding ssRNA, consistent with a previous report (Niederberger et al. 1998).
Ribosome binding by yeIF4B is mediated by the NTD and 7-repeats domain
Next, each version of yeIF4B was tested for its ability to bind the 40S subunit (Fig. 2F). Deletion of the RRM (ΔRRM) or mutation of its two key residues (R145T, K147A) had little effect on binding of yeIF4B to 40S subunits (Kd = 820 and 470 nM, respectively, vs. 360 nM for WT), indicating that the RRM and its ssRNA-binding activity are not critical for interaction with the yeast ribosome. In contrast, deletion of the NTD (ΔNTD; Kd > 10 µM) or 7-repeats (Δr1-7; Kd = 3.2 µM) reduced the affinity of the factor for 40S subunits by ≥9-fold, and a variant lacking both the NTD and 7-repeats (ΔNTDΔr1-7) did not detectably bind 40S subunits. Interestingly, a derivative lacking both the NTD and RRM (ΔNTDΔRRM; Kd = 1.9 µM) bound the 40S subunit ≥5-fold tighter than the version lacking only the NTD, suggesting that the ribosome binding activity of the repeats is partially masked by the RRM in the ΔNTD derivative.
The yeIF4B 7-repeats domain is critical for productive interaction of yeIF4B with the initiation machinery, but the RRM is dispensable for function in vitro
Mammalian eIF4B was identified as a factor that stimulated mRNA binding to the 40S subunit (Trachsel et al. 1977; Benne and Hershey 1978). Using a reconstituted yeast translation initiation system and native gel-shift and toeprinting assays, we recently showed that yeIF4B also stimulates mRNA recruitment to the PIC (Mitchell et al. 2010). In the gel-shift assay, migration of 32P-labeled mRNA during native gel electrophoresis is slowed when mRNA is stably recruited to the PIC. The assay allows both the kinetics (at low concentrations of PICs) and thermodynamics (endpoints) of mRNA recruitment to be followed.
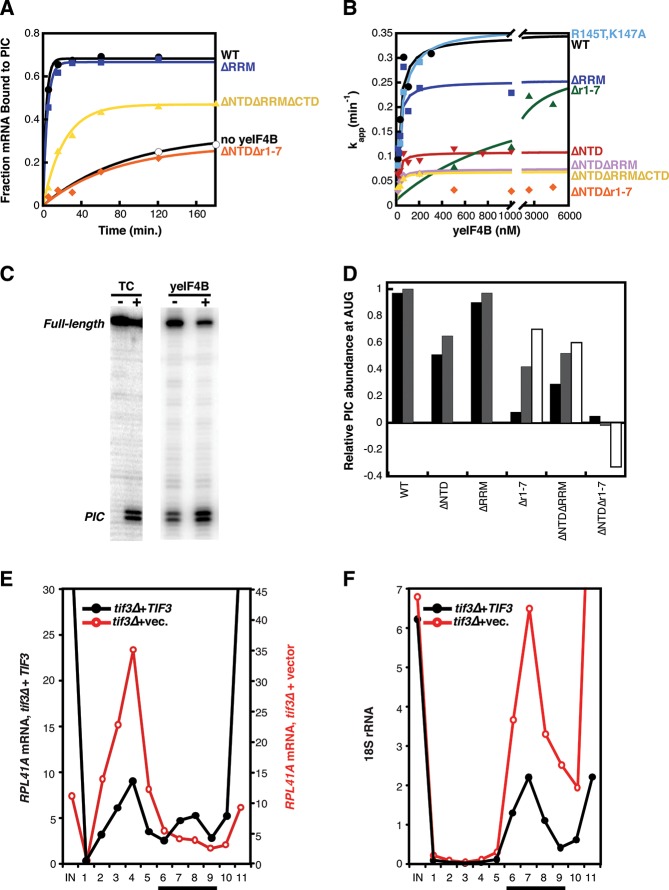
To determine the domains necessary to stimulate mRNA recruitment, we preincubated each of the yeIF4B variants with eIF4F, eIF3, and PICs, then added [32P]-m7Gppp-capped mRNA with the sequence of the native yeast RPL41A transcript (Mitchell et al. 2010). Aliquots were loaded onto a running native gel over time, which served to both stop the reactions and separate the free and bound forms of mRNA. The fraction of mRNA bound to the PIC at each timepoint was quantified using a PhosphorImager and plotted vs. time. The data were fit with a single-exponential rate equation to determine the apparent rate of mRNA recruitment (kapp) (Fig. 3A). The kapp for recruitment was determined at increasing concentrations of WT or mutant yeIF4B and each plot of kapp vs. yeIF4B concentration was fit with a hyperbolic equation to determine the observed maximal rate of recruitment at 30 nM PICs (kmax) and the concentration of yeIF4B yielding the half-maximal rate (K1/2) (Fig. 3B).
FIGURE 3.
The yeIF4B NTD and 7-repeats stimulate mRNA recruitment to the PIC. (A,B) The native gel-shift assay was used to monitor recruitment of 32P-capped-RPL41A mRNA to PICs (30 nM) in the presence of eIF3 (200 nM), eIF4E•eIF4G (30 nM), and eIF4A (1 µM), as a function of concentration of yeIF4B variants (symbols and coloring as in Fig. 2). (A) Examples of time-courses of mRNA recruitment in the presence of different yeIF4B variants. Data are fit with a single-exponential rate equation. (B) Apparent rate constants for mRNA recruitment at 30 nM PICs measured as a function of concentrations of the yeIF4B variants. Data are fit with a hyperbolic equation. (C) Toeprinting analysis of the efficiency of PIC binding to the RPL41A mRNA start codon (as in A) in the presence or absence of 400 nM ternary complex and in the presence or absence of 200 nM yeIF4B. The bands at +16 and +17 nucleotides from the A of the AUG codon correspond to the leading edge of the PIC and are not observed in the absence of ribosomes or ternary complex. The full-length cDNA is also shown. (D) The indicated yeIF4B variants were added to mRNA recruitment reactions at a final concentration of 200 nM (black bars) or 1 µM (gray bars) and analyzed by toeprinting as in C. The Δr1-7, ΔNTDΔRRM, and ΔNTDΔr1-7 mutants had strong defects in mRNA recruitment, so these variants were each added at a third, higher concentration to compensate for the K1/2 defects observed with each (white bars), which was 20 µM, 5.5 µM, or 10 µM, respectively. The fraction of toeprint intensity at the PIC/(PIC + full-length cDNA) was determined for each mutant. This value was normalized such that WT yeIF4B at 1 µM was set to 1, and the fraction with no yeIF4B was set to 0 (Relative PIC abundance at AUG; i.e., 100% stimulation or no stimulation by yeIF4B). (E,F) SDGC of cross-linked WCEs of transformants of tif3Δ strain FJZ052 harboring empty vector or TIF3+ plasmid pFJZ058. Total RNA was extracted from each fraction and RPL41A mRNA (E) and 18S rRNA (F) measured by real-time qPCR. The bar indicates 43S•mRNA PICs.
WT yeIF4B stimulated recruitment at very low concentrations (Fig. 3B) (K1/2 = 34 nM) (Table 1), suggesting that it interacts strongly with the PIC to stimulate mRNA recruitment. The kmax value with saturating WT yeIF4B, 0.31 min−1 (Fig. 3A; Table 1), is 30-fold higher than the rate in the absence of the factor (0.01 min−1), consistent with our previous measurements (Mitchell et al. 2010). Remarkably, the variants containing the R145T, K147A substitutions in the RRM or lacking the RRM entirely stimulated mRNA recruitment as efficiently as the WT factor, with K1/2 values of 33 nM for R145T, K147A, and 15 nM for the ΔRRM variant (Fig. 3B; Table 1). The kmax values for these mutants were also very similar to that for the WT factor: 0.23 and 0.34 min−1 for the ΔRRM and R145T, K147A variants, respectively, vs. 0.31 min−1 for WT. Thus, ssRNA binding and other functions provided by the RRM are dispensable for mRNA recruitment to the PIC in vitro.
TABLE 1.
Summary of in vitro parameters for yeIF4B variants in ssRNA binding, ribosome binding, and mRNA recruitment assays
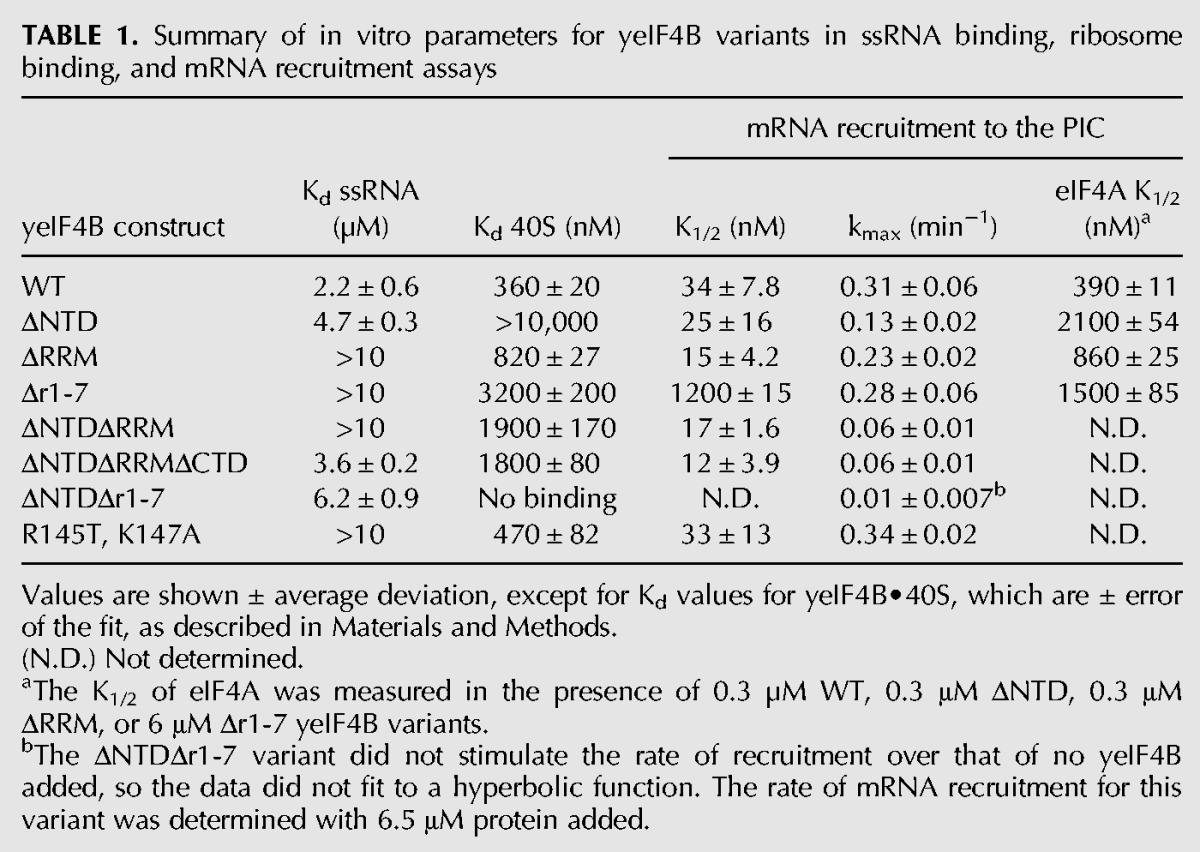
Deletion of the NTD did not affect the K1/2 value for mRNA recruitment relative to WT yeIF4B (Fig. 3B) (ΔNTD; K1/2 = 25 nM) (Table 1), suggesting that this variant interacts strongly with the PIC despite being defective in binding to the 40S subunit alone (Fig. 2F; Table 1). However, kmax for this variant was threefold lower than for the WT factor (0.11 min−1 vs. 0.31 min−1) indicating that the NTD plays a role in mRNA recruitment to the PIC over and above a function in simply binding to components of the initiation machinery.
In contrast, the variant lacking the 7-repeats domain had a K1/2 value 35-fold higher than the WT value (Fig. 3B) (Δr1-7; K1/2 = 1200 nM) (Table 1). Moreover, the variants comprised of the 7-repeats domain alone (ΔNTDΔRRMΔCTD) or 7-repeats plus CTD (ΔNTDΔRRM) stimulated recruitment at very low concentrations (Fig. 3B) (K1/2 = 12 nM) (Table 1), albeit with fivefold reduced kmax values relative to the WT factor. Together, these data suggest that the 7-repeats domain modulates productive interaction between yeIF4B and the PIC.
Consistent with important roles for the NTD and 7-repeats domain in yeIF4B function, the variant lacking both of these regions did not stimulate mRNA recruitment to the PIC at any concentration tested up to 5 µM (Fig. 3B) (ΔNTDΔr1-7). This variant also lost detectable affinity for the 40S subunit (Fig. 2F), suggesting that 40S binding is connected to mRNA recruitment activity.
The NTD and 7-repeats domains are important for assembly of the PIC on the start codon of the mRNA
Primer extension (“toeprinting”) assays allow the position of the PIC on an mRNA to be determined. PICs are allowed to bind mRNA in the presence of recruitment factors (the eIF4 factors and eIF3) and scan to the start codon. Reverse-transcriptase extension of a 32P-labeled DNA primer annealed to the 3′-end of the mRNA will stall 16–17 nucleotides from the P site codon of a PIC stably associated with mRNA, allowing the PIC’s position to be mapped. Previous toeprinting studies showed that the presence of eIF4B enhances positioning of PICs at the start codon of mRNAs (Pestova and Kolupaeva 2002; Dmitriev et al. 2003; Mitchell et al. 2010), consistent with our results (Fig. 3C). Increased abundance of PICs at the AUG could reflect the efficiency of several steps—mRNA recruitment, scanning, and start codon recognition—as well as the overall stability of the complex once it is positioned at the start codon.
As a further test of the roles of yeIF4B’s domains in mRNA recruitment and later steps, we used toeprinting to determine the efficiency of PIC binding to the RPL41A mRNA start codon for each yeIF4B mutant, comparing it to the efficiency observed in the presence (1.0 relative abundance) or absence (0 relative abundance) of WT yeIF4B. These experiments were performed at two to three concentrations of yeIF4B, with at least one being saturating (where possible). The results in Figure 3D mirror those observed in the mRNA recruitment assays (Fig. 3A,B), indicating that the latter reflect formation of bona fide PIC•mRNA complexes. Deletion of the RRM had little effect on the abundance of PICs observed at the start codon, whereas deletion of the 7-repeats resulted in significantly decreased abundance at low concentrations. This effect could be partially suppressed by increasing the concentration of the Δr1-7 variant, consistent with what we observed in native gel assays (Fig. 3A,B). Deletion of the NTD evoked an approximately twofold decrease in abundance of PICs at the AUG, and this effect was exacerbated by also deleting the RRM (Fig. 3D) (ΔNTDΔRRM), in agreement with the native gel results (Fig. 3B). Most striking was the ΔNTDΔr1-7 variant, which, at 0.2 µM, did not promote complex formation on the AUG relative to the amount observed in the absence of yeIF4B; and at higher concentrations actually decreased PIC recruitment to the start codon below the level observed without yeIF4B (negative values of relative PIC abundance). These data suggest that the ΔNTDΔr1-7 protein traps complexes or mRNA in a nonfunctional state, and further indicate that the 7-repeats and NTD are necessary for yeIF4B function.
yeIF4B stimulates mRNA recruitment to the PIC in vivo
Having established that yeIF4B is required for efficient mRNA recruitment to PICs in vitro, we wished to establish that it promotes this step of initiation in vivo. To this end, we used formaldehyde cross-linking of living cells to stabilize native 43S/43S•mRNA PICs. Whole-cell extracts (WCEs) were fractionated by sucrose density gradient sedimentation (SDGS), and RPL41A mRNA and 18S rRNA were assayed in each fraction by quantitative PCR. RPL41A mRNA was analyzed because its short length enables efficient separation of 40S-bound and free mRNP species (Jivotovskaya et al. 2006) and for consistency with the in vitro studies above. In extracts of a tif3Δ mutant complemented by WT TIF3 on a high-copy (hc) plasmid, we observed comparable amounts of RPL41A mRNA in the 40S- and mRNP-containing fractions. In the tif3Δ extract, in contrast, the amount of RPL41A mRNA in free mRNP greatly exceeded the 40S associated fraction, and a defined peak of mRNA in the 40S fractions was not evident (Fig. 3E). The amount of 18S rRNA in the 40S region was considerably higher in the mutant extract (Fig. 3F), presumably reflecting an accumulation of free 40S subunits resulting from the lower rate of translation initiation and attendant reduction in polysomes in tif3Δ cells (Fig. 6C, below; Coppolecchia et al. 1993). Despite the increase in both free 40S subunits (18S rRNA) and free RPL41A mRNP in the mutant extract (Fig. 3E,F), we calculated that the ratio of RPL41A mRNA to 18S rRNA in the 40S fractions was ≥3-fold lower in the tif3Δ vs. WT extract. Western analysis of the same gradient fractions revealed no significant reductions in the levels of eIF1A, eIF2, or eIF3 subunits, relative to 40S subunit protein Rps22, with the exception of a small reduction in the a/Tif32 subunit of eIF3 (data not shown). Thus, it appears that yeIF4B is not required for efficient assembly of 43S PICs, but is important for subsequent attachment of these complexes to mRNA to form 43S•mRNA PICs in vivo.
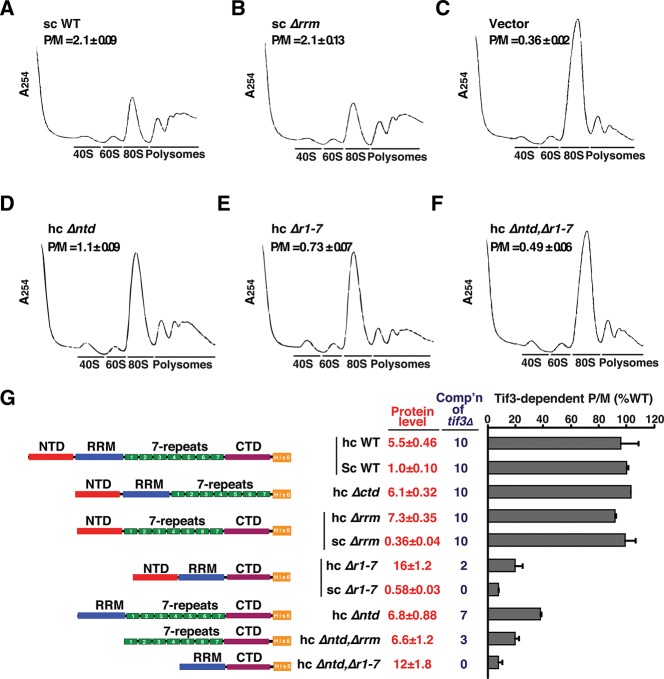
FIGURE 6.
The 7-repeats and NTD are required, but the RRM is dispensable, for WT translation initiation in vivo. (A–F) Polysome profiles of tif3Δ mutants. Strains from Figure 5 were grown in SC-Leu at 30°C to an A600 of ∼1, with cycloheximide added prior to harvesting. WCEs were separated by SDGC and scanned at 254 nm. Mean polysome/monosome ratios (P/M) and SEM from multiple replicates are given. (G) Polysome profiles were determined for the indicated strains as in A–F. The mean P/M ratio for the vector transformants was subtracted from the mean P/M ratios for mutant or sc WT transformants, and the resulting values were normalized to that obtained for sc WT to yield Tif3-dependent P/M ratios. Protein levels and complementation ability (comp’n) from Figure 5 are included for comparison.
yeIF4B promotes productive interaction of eIF4A with the initiation machinery
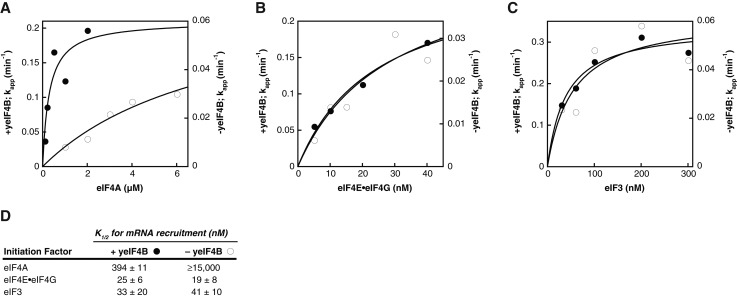
Our in vitro measurements showed that yeIF4B binds 40S subunits with a Kd of 360 nM. The K1/2 of WT yeIF4B in mRNA recruitment is >10-fold lower (Table 1), indicating that yeIF4B interacts productively with the initiation machinery at much lower concentrations than it binds the 40S subunit alone. A potential explanation for these data is that yeIF4B interacts with an additional factor that stabilizes its binding to the PIC. To test this hypothesis, we formed PICs and measured the kinetics of mRNA recruitment at varying concentrations of eIF3, eIF4A, and eIF4E•eIF4G complex in the presence and absence of yeIF4B (Fig. 4). We then plotted the rates of recruitment vs. the concentration of the factor that was varied and fit the data with a hyperbolic equation to derive the K1/2 of each factor. In the presence of yeIF4B, eIF4A stimulated PIC binding to mRNA with a K1/2 of 394 nM, but in the absence of yeIF4B the K1/2 of eIF4A was increased by >38-fold (K1/2 ≥15,000 nM) (Fig. 4A,D), indicating that productive interaction of eIF4A with the PIC is strongly enhanced by yeIF4B.
FIGURE 4.
yeIF4B promotes functional interaction of eIF4A with the PIC. (A–C) Apparent rate constants for recruitment of RPL41A mRNA, measured as in Figure 3A, in the presence (200 nM; filled circles) or absence (open circles) of yeIF4B as a function of concentration of eIF4A (A), eIF4E•eIF4G (B), or eIF3 (C). When not varied, the concentration of mRNA recruitment factors was as in Figure 3A and Materials and Methods. Data are fit with a hyperbolic equation. (D) Mean K1/2 values from at least two experiments. Errors given are average deviations. The highest concentration of eIF4A experimentally achievable was 6 µM, which was not saturating; therefore, the value of K1/2 derived from the fit in the absence of yeIF4B is a lower limit.
We also determined the K1/2 for the eIF4E•eIF4G complex (Fig. 4B) and eIF3 (Fig. 4C) in the presence and absence of yeIF4B. Although the kinetics of mRNA recruitment were decreased significantly by omission of yeIF4B in each case, as expected (Figs. 3A and 4, open circles; note difference in y-axes), the K1/2 values for mRNA recruitment were essentially the same for both factors regardless of whether yeIF4B was added (Fig. 4B,C). Thus, the large effect of yeIF4B on the K1/2 for eIF4A is not observed for eIF4E•eIF4G or eIF3. However, because the K1/2 values for the latter factors are very low, suggesting that binding is tight and that the K1/2 curves may represent stoichiometric titrations, yeIF4B might affect their interactions with the PIC as well.
To determine which domains of yeIF4B were necessary for the decrease in the K1/2 for eIF4A in mRNA recruitment, we also determined K1/2 values for eIF4A in the presence of saturating amounts of the ΔNTD, ΔRRM, or Δr1-7 variants (Table 1) (eIF4A K1/2). Relative to WT yeIF4B, each deletion increased the K1/2 by at least twofold, with deletion of the NTD and 7-repeats domains having the strongest effects. While these data do not unequivocally assign the eIF4A recruitment function to a single domain, they again indicate the partial redundancy of the NTD and 7-repeats.
The 7-repeats domain is paramount, the NTD is important, and the RRM is dispensable for yeIF4B function in translation initiation in vivo
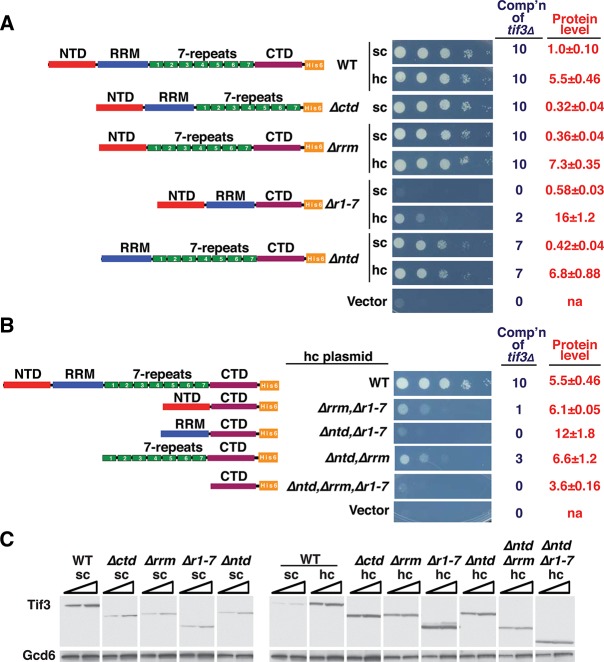
To address the functional requirements for the yeIF4B domains in vivo, we generated a set of tif3 mutant alleles encoding the same mutant proteins analyzed above and tested them for complementation of the slow-growth phenotype (Slg−) of a tif3Δ mutant. The tif3 alleles, under the native promoter, were introduced on single-copy (sc) or hc plasmids, and serial dilutions of the transformants were spotted on solid medium. Growth was scored semiquantitatively (from 0 to 10) by determining the highest dilutions at which colony formation occurred, plus the size of individual colonies at that dilution. Because we came to essentially identical conclusions about the relative levels of complementation at 18°C, 30°C, and 37°C (data not shown), only results of the 18°C tests are presented below, as tif3Δ cells grow most poorly at 18°C (Altmann et al. 1993; Coppolecchia et al. 1993). A C-terminal His6 epitope was included in each construct to enable quantification of protein expression by Western analysis, and we verified that the sc WT TIF3-His6 allele restored growth of the tif3Δ strain to a level indistinguishable from that of the parental TIF3+strain (data not shown).
Consistent with our in vitro measurements of mRNA recruitment, deletion of the 7-repeats domain dramatically impaired complementation of the tif3Δ mutation. In fact, the sc Δr1-7 allele was indistinguishable from empty vector, even though protein expression was reduced by only ∼40% from the WT level. On a hc plasmid the Δr1-7 product was overexpressed by ∼28-fold (∼16-fold relative to WT yeIF4B), and this restored low-level complementation of the tif3Δ mutation, indicating that the NTD can promote growth in the absence of the 7-repeats when the protein concentration is sufficiently high (Fig. 5A,C). Thus, the 7-repeats domain is crucial, but not absolutely essential, for yeIF4B function in vivo. Remarkably, deletion of the entire RRM had no observable effect on complementation, even though the sc Δrrm construct was expressed at only 36% of the WT yeIF4B level (Fig. 5A,C). These results are consistent with findings above that the RRM domain is essentially dispensable for yeIF4B function in mRNA recruitment in vitro. Eliminating the NTD evoked an obvious reduction in complementation of tif3Δ for both the sc and hc alleles, even though expression of this variant from the hcΔntd construct exceeded that of WT yeIF4B (Fig. 5A,C). Thus, in agreement with our in vitro data, the NTD is more important than the RRM, but less important than the 7-repeats for yeIF4B function in vivo. Removing the CTD reduces expression of yeIF4B but, surprisingly, does not affect complementation of tif3Δ (Fig. 5A,C), suggesting that yeIF4B is normally produced in excess of the amount needed for WT growth.
FIGURE 5.
The 7-repeats and NTD are required, but the RRM is dispensable, for full complementation of the tif3Δ growth defect. (A,B) 10-fold serial dilutions of transformants of tif3Δ strain FJZ052 harboring sc or hc plasmids with the indicated TIF3 alleles or empty vector were spotted on SC-Leu medium and incubated for 4 d at 18°C. Growth was scored from 0 (empty vector) to 10 (TIF3+) based on colony size and highest dilution where colonies appeared, and reported as complementation (comp’n) of tif3Δ. (C) WCEs prepared under denaturing conditions from strains in A and B were subjected to Western analysis with antibodies against His6 epitope (to visualize Tif3-His6 proteins) or an eIF2B subunit (Gcd6) as loading control. Intensities of Tif3-His6 signals relative to Gcd6 were normalized to those for sc TIF3+ transformants. Mean and standard errors calculated from replicate determinations are listed under “Protein level” in A and B. (na) Not applicable.
We also examined the effects of removing multiple domains on yeIF4B function in vivo. Deleting the NTD from the hc construct lacking the 7-repeats abolished the residual function observed for the parental hcΔr1-7 allele with only a small reduction in protein expression (hcΔr1-7 in Fig. 5A vs. hcΔntdΔr1-7 in Fig. 5B). This result agrees with our findings above that the ability of the Δr1-7 variant to promote PIC assembly at high concentrations is abolished by eliminating its NTD (Fig. 3B,D). We also found that eliminating the RRM reduces complementation in the absence of the NTD (hcΔntd in Fig. 5A vs. hcΔntdΔrrm in Fig. 5B), which agrees with our findings of reduced activity for the ΔNTDΔRRM double mutant vs. ΔNTD single mutant in mRNA recruitment (Fig. 3B,D). Finally, it is interesting that the hcΔntdΔrrm construct exhibits appreciable complementation (Fig. 5B), indicating that the 7-repeats provide significant yeIF4B function in vivo in the absence of both NTD and RRM. By comparison, the hc construct containing only the NTD and CTD (hcΔrrmΔr1-7) is barely functional, and that containing only the RRM and CTD (hcΔntdΔr1-7) is nonfunctional (Fig. 5B). Thus, the 7-repeats domain is more critical than the NTD, which is more important than the RRM, for yeIF4B function in vivo.
The NTD and 7-repeats domains of yeIF4B stimulate translation in vivo
To establish that the complementation activity of different yeIF4B variants indicates their ability to promote general translation initiation in vivo, we examined the polysome profiles of a subset of transformants analyzed above for their Slg− phenotypes. In agreement with previous results (Coppolecchia et al. 1993), tif3Δ cells contain a reduced amount of total polysomes and increased 80S monosomes, with a polysome to monosome ratio (P/M) approximately sixfold lower than for WT cells (Fig. 6A,C). In agreement with the complementation data (cf. Fig. 6G comp’n values with bar graphs of P/M ratio), deletion of the RRM or CTD had no effect on the P/M ratio, indicating that these domains are dispensable for WT translation initiation in vivo (Fig. 6A,B,G). Furthermore, deleting the 7-repeats produced a P/M ratio only slightly greater than the low value seen in tif3Δ cells, whereas overexpressing this variant (hcΔr1-7) restored ∼20% of the function of WT yeIF4B in polysome formation (Fig. 6E,G). The residual function of this construct was abolished by additionally removing the NTD (Fig. 6G, hcΔr1-7 vs. hcΔntdΔr1-7), supporting the findings above that the Δr1-7 variant can stimulate PIC assembly at high concentrations, whereas the double mutant containing only the RRM and CTD is nonfunctional (Fig. 3A,B,D).
Eliminating the NTD alone substantially reduces the ability of yeIF4B to support polysome assembly in vivo, but not to the same extent observed on eliminating the 7-repeats, even when the latter is overexpressed (cf. hcΔntd and hcΔr1-7 in Fig. 6D,E,G). Thus, the NTD is more important than the RRM, but less critical than the 7-repeats for robust translation initiation in vivo. Finally, removing the RRM from the variant lacking the NTD reduced the P/M ratios (hcΔntd vs. hcΔntdΔrrm in Fig. 6G), in accordance with the diminished function observed in vivo (Fig. 5B) and in vitro (Fig. 3A,B,D) on removing the RRM from the ΔNTD variant. The ability of the resulting construct containing only the 7-repeats and CTD (hcΔntdΔrrm) to support appreciable polysome formation agrees with its ability to stimulate mRNA recruitment in vitro (Fig. 3B,D).
yeIF4B binds to Rps20 in the head of the 40S subunit
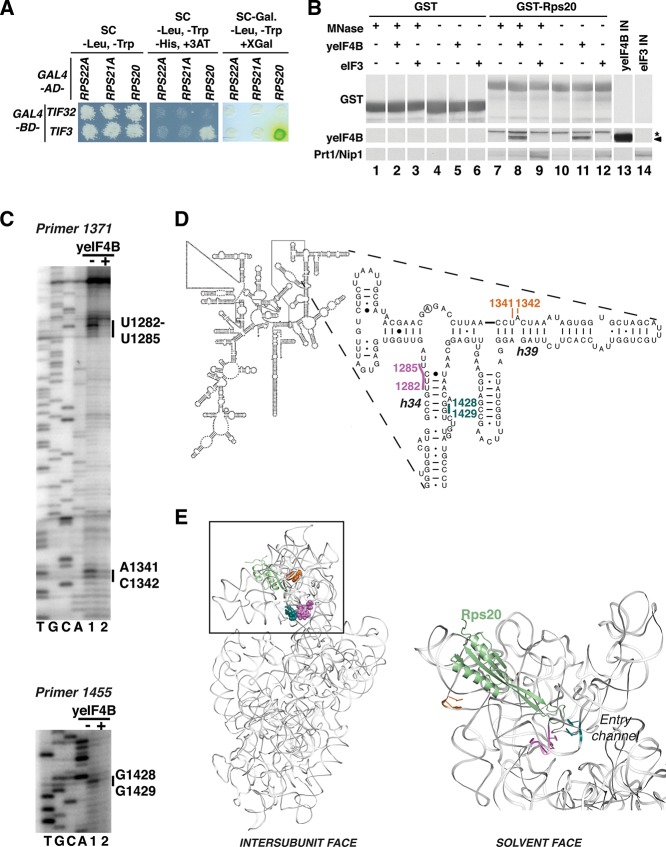
To gain more information about the molecular mechanisms underlying yeIF4B’s stimulation of mRNA recruitment, we probed its interaction with the 40S subunit. We first looked for interactions of yeIF4B with ribosomal proteins by yeast two-hybrid analysis (Fig. 7A). A library of 32 small subunit ribosomal protein (Rps) genes fused to the coding sequence for the Gal4 activation domain (RPSX-GAD) was tested for interaction with Gal4 DNA-binding domain (GBD) fusions to TIF3. Only one ribosomal protein, Rps20, gave results indicating two-hybrid interaction with yeIF4B (Fig. 7A; data not shown).
FIGURE 7.
yeIF4B binds Rps20 in the head of the 40S subunit and induces structural changes in the mRNA entry channel. (A) Interaction of yeIF4B with Rps20 in yeast two-hybrid assays. Transformants of yeast strain Y187 harboring GAL4–BD fusions to TIF3 or TIF32 (encoding eIF3a) were mated with transformants of strain Y190 carrying GAL4–AD fusions to the indicated RPS genes. Diploids selected on SC-Leu,-Trp were replica-plated to this medium (left) and to indicator media for scoring expression of the resident UASGAL-HIS3 and UASGAL-lacZ reporters: SC-Leu,-Trp,-His with 30 mM 3AT (middle) and SC-Leu,-Trp,+XGal (right), respectively. (B) yeIF4B interacts with Rps20 in vitro. A GST fusion to Rps20 (lanes 7–12) or GST alone (lanes 1–6) was expressed in Escherichia coli and 100 µg was immobilized on glutathione Sepharose beads and tested for binding to 10 µg of purified yeIF4B or eIF3. Immobilized proteins were treated with micrococcal nuclease (MNase) prior to binding assays as indicated. A total of 30% of pull-downs and 5% of input samples (IN) were resolved by SDS-(4%–20%) PAGE and subjected to Western analysis with antibodies against GST, yeIF4B, and eIF3 subunits Prt1 and Nip1. (*) A nonspecific band, and the arrowhead indicates yeIF4B. A similar experiment (data not shown) demonstrated that GST–Rps20 does not detectably pull down eIF1. (C–E) Hydroxyl radical footprinting of yeIF4B binding to 40S ribosomal subunits. 40S subunits were incubated with yeIF4B storage buffer or yeIF4B, and cleavage was initiated by adding Fe2+-EDTA, H2O2, and ascorbic acid to generate hydroxyl radicals. The 18S rRNA was extracted from each reaction and analyzed by primer extension and denaturing gel electrophoresis. Sites of protection from hydroxyl radical cleavage are indicated on the gel (C), in the secondary structure of the 18S rRNA (D), and on the 3D structure of the 40S subunit (E). For clarity Rps20 is the only ribosomal protein shown (in green). Panels D and E were generated using the yeast 18S rRNA secondary structure model from the Comparative RNA website (http://www.rna.ccbb.utexas.edu) and the 3.0 Å structure of the yeast 80S ribosome (PDB 3U5B and 3U5C) (Ben-Shem et al. 2011) in PyMOL, respectively.
To provide evidence for direct Rps20–yeIF4B interaction, a GST–Rps20 fusion or GST alone was immobilized on glutathione beads and incubated with purified yeIF4B or eIF3, and the bound fractions were subjected to Western analysis with antibodies against yeIF4B or eIF3 subunits Prt1/b and Nip1/c. As eIF3 was shown previously to interact with Rps20 through its Tif35/g subunit (Cuchalova et al. 2010), it served as a useful positive control. Both yeIF4B and the eIF3 subunits were retained on the beads in the presence or absence of micrococcal nuclease treatment, indicating RNA-independent interaction with GST-Rps20, whereas neither yeIF4B nor eIF3 interacted with GST alone (Fig. 7B). As an additional negative control, we tested the ability of GST–Rps20 to pull down eIF1. As expected, eIF1 did not detectably bind GST–Rps20 (data not shown), indicating that the interaction with yeIF4B is a specific one.
Finally, we used hydroxyl radical footprinting to map the binding site of yeIF4B on the 40S subunit. We treated 40S subunits in the presence or absence of equimolar (1 µM) yeIF4B with Fe2+-EDTA, ascorbic acid, and hydrogen peroxide to generate hydroxyl radicals (Powers and Noller 1995). These radicals cleave exposed regions of the rRNA, whereas binding of proteins to the ribosome can protect the rRNA from cleavage and provide information about interaction sites or induced conformational changes that alter rRNA accessibility. After the cleavage reaction, the 18S rRNA was extracted and hydroxyl radical cleavages were mapped by primer extension (Fig. 7C). Binding of yeIF4B protected two areas of 18S rRNA in the 40S subunit from hydroxyl radical cleavages observed in the absence of yeIF4B (but not observed in the absence of H2O2) (Fig. 8A,B, cf, lanes 0, 1, +/−•OH): in helix (h)34 and h39 (Fig. 7C,D). yeIF4B binding strongly protected nucleotides 1282–1285 and 1428–1429 in h34, which is near the neck of the 40S subunit on the intersubunit face and approaches the mRNA-binding channel. Located on the solvent-exposed face of the 40S head, nucleotides 1341 and 1342 of h39 also exhibited strong protections on yeIF4B binding. Interestingly, the nucleotides protected in both h34 and h39 flank Rps20 (Fig. 7E). No other protections were observed in any other region of the 18S rRNA (data not shown). Taken together, these data indicate that yeIF4B binds to Rps20 in the 40S subunit head.
FIGURE 8.
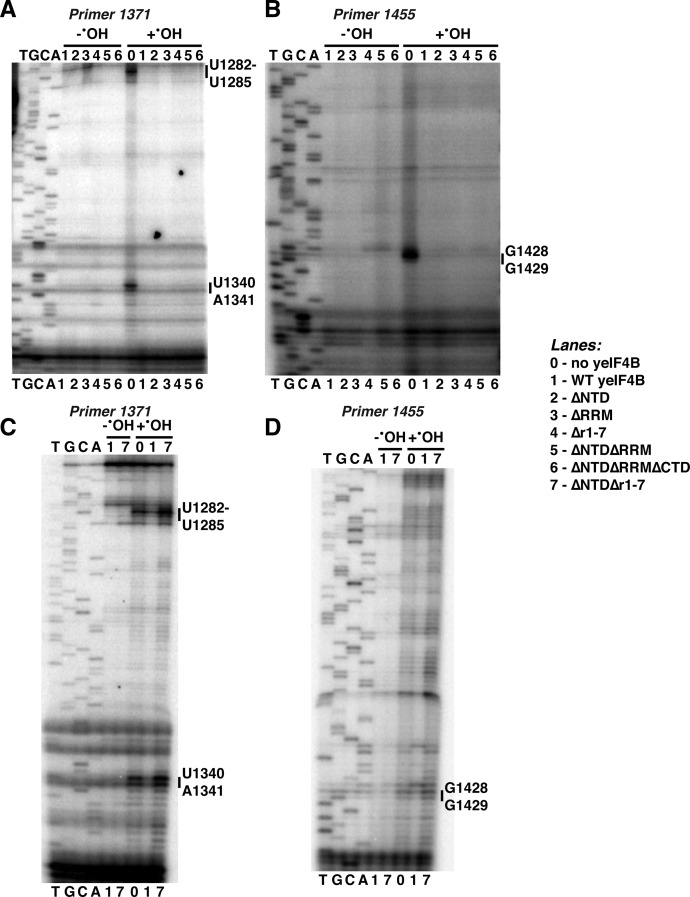
The NTD and 7-repeats domains perform overlapping functions in binding to the 40S ribosomal subunit and promoting structural changes in the mRNA entry channel. Purified 40S subunits were incubated with storage buffer, yeIF4B, or yeIF4B variants as indicated in the figure for lanes 0–7 (concentrations listed in Materials and Methods), and the reactions were then incubated −/+Fe2+-EDTA, hydrogen peroxide, and ascorbic acid to generate hydroxyl radicals, as detailed in Materials and Methods. The 18S rRNA was extracted from ribosomes incubated with (A,B) WT, ΔNTD, ΔRRM, Δr1-7, ΔNTDΔRRM, and ΔNTDΔRRMΔCTD yeIF4B variants or (C,D) WT and ΔNTDΔr1-7 yeIF4B under each condition and analyzed by primer extension with primer 1371 (A,C) and 1455 (B,D) followed by denaturing gel electrophoresis. Sites of protection from hydroxyl radical cleavage are indicated on the gels.
We also investigated 40S interaction with yeIF4B variants lacking functional domains (Fig. 8) by incubating 40S subunits with each variant at saturating concentrations (Fig. 2F) and subjecting the complexes to hydroxyl radicals, as described above. As a negative control, we incubated 40S subunits with high concentrations of the ΔNTDΔr1-7 variant, which does not detectably bind the ribosome (Figs. 2F, 8C,D). We found that each of the variants that bind the ribosome gave the same pattern of protections observed for WT yeIF4B, whereas the ΔNTDΔr1-7 variant did not protect these regions, even at a concentration of 10 µM. Because the same protections were observed by different variants, we postulate that the protections observed upon yeIF4B binding are mostly the result of movement of Rps20 on direct binding to one or more domains of yeIF4B. This movement may result in larger rearrangements in the 40S head near the mRNA entry channel and affect accessibility of h34 and h39 to hydroxyl radical cleavage.
DISCUSSION
We have analyzed the functions of the individual domains in yeIF4B in translation initiation. We report that yeIF4B binds specifically and tightly to the small ribosomal subunit and promotes mRNA binding to the PIC in vitro and in vivo using two distinct domains, the NTD and 7-repeats. We also mapped yeIF4B binding to Rps20 in the head of the 40S subunit and provide data suggesting that yeIF4B acts to modulate the structure of the mRNA entry channel and to promote productive binding of eIF4A to the PIC in order to stimulate mRNA recruitment.
The RRM of yeIF4B is dispensable for function
Niederberger et al. (1998) previously reported that an N-terminal truncation of yeIF4B removing both the NTD and RRM domains greatly compromised the factor’s ability to complement the slow-growth phenotype of the tif3Δ strain, whereas robust complementation was observed with a C-terminally truncated construct containing the NTD, RRM, and the first three repeats of the 7-repeat domain (Niederberger et al. 1998). Because the N-terminal truncation that removed the RRM domain also abolished the ability of yeIF4B to rescue translation in a tif3Δ extract and greatly impaired the RNA strand exchange activity of yeIF4B, it was proposed that RRM-dependent strand-exchange activity is important for yeIF4B’s stimulation of translation initiation in vivo (Niederberger et al. 1998). Our results do not support this proposal, however, as we found that an internal deletion of the RRM has no impact on complementation of the Slg− phenotype or defect in general translation initiation of tif3Δ cells (Figs. 5, 6). In addition, we showed that yeIF4B lacking a functional RRM retains nearly WT activity in ribosome binding and mRNA recruitment in vitro (Fig. 3). Thus, the RRM and its RNA-binding activities are dispensable for yeIF4B function both in vitro and in vivo. Our results also indicate that a model in which the RRM is crucial for tethering yeIF4B to the 18S rRNA and enabling it to bridge the activated eIF4F–mRNP complex and 40S subunit (Methot et al. 1996b) is improbable for yeast.
The conservation of the RRM in eIF4Bs from fungi to mammals (Altmann et al. 1993) suggests that it must confer some evolutionary advantage. One possibility is that the RRM modulates or buttresses the critical interactions mediated by the repeats and the NTD. Consistent with this idea, deleting the RRM exacerbates the effect of deleting the NTD on the mRNA recruitment activity of the factor (Figs. 3, 5, 6, cf. ΔNTD and ΔNTDΔRRM), suggesting that the RRM can enhance the function of the 7-repeats if the NTD is absent.
The effects of double deletions of the domains of yeIF4B on the RNA and 40S binding functions of the factor also suggest a functional interplay among the NTD, RRM, and 7-repeats. Deleting the 7-repeats decreases the affinity for ssRNA by approximately fivefold; however, when the NTD is also deleted the affinity for RNA actually increases approximately twofold (Fig. 2E), suggesting that the NTD inhibits the ssRNA-binding function of the RRM in the absence of the 7-repeats. Similarly, deletion of the NTD reduces the 40S binding affinity of the factor by ≥28-fold, but also deleting the RRM actually increases the binding affinity more than fivefold relative to deleting the NTD alone (Fig. 2F). This suggests that the RRM inhibits the 40S binding function of the 7-repeats when the NTD is removed in the ΔNTD mutant. One model to explain these data is that the NTD and 7-repeats interact with one another in the full-length factor, which prevents each from binding to the RRM; deleting the NTD or 7-repeats allows the other domain to interact nonproductively with the RRM. In addition, the 7-repeats on their own bind ssRNA with nearly the same affinity as WT yeIF4B, whereas the ΔNTDΔRRM mutant has compromised ssRNA-binding activity (Fig. 2E), suggesting that the CTD can also inhibit the ssRNA-binding function of the 7-repeats. An intriguing possibility is that the RRM and CTD play regulatory roles in yeIF4B function.
The NTD and the 7-repeats play key, partially redundant roles in yeIF4B function
Unlike deletion of the RRM, removal of the NTD or 7-repeats domains of yeIF4B resulted in the compromised function of the factor both in vitro and in vivo. The 7-repeats domain clearly plays the most important role; deletion of this domain increased the mutant factor’s K1/2 in the mRNA recruitment assay in vitro by >30-fold and abolished the ability of the factor to complement the general translation initiation defects of tif3Δ cells when expressed at WT levels. Consistent with an important role for the 7-repeats in productive binding of yeIF4B to the initiation machinery, overexpression of the Δr1-7 variant partially restored translation initiation in the tif3Δ strain. Moreover, the repeats by themselves (ΔNTDΔRRMΔCTD) stimulated mRNA recruitment sixfold above the rate observed in the absence of yeIF4B and had a K1/2 similar to WT (Fig. 3B; Table 1). Likewise, overexpressing the variant with only the 7-repeats plus the CTD (ΔNTDΔRRM) partially rescued the translation initiation defect of Δtif3 cells.
The NTD also plays a role in promoting mRNA recruitment to the PIC. Deleting the NTD moderately reduced function in all assays, both in vitro and in vivo. The effect of this deletion does not appear to involve a simple binding defect, as increasing the ΔNTD variant concentration did not restore its function in vitro or in vivo. The fact that increasing the concentration of the Δr1-7 mutant in vitro and in vivo partially suppressed the effects of the deletion, whereas the ΔNTDΔr1-7 mutant was nonfunctional at all concentrations, indicates that the NTD and 7-repeats have partially overlapping functions and that the NTD can stimulate mRNA recruitment even when the 7-repeats are missing. as long as the concentration of the factor is high enough to permit its binding to the PIC.
It is also possible that some of these effects are the result of misfolding of the deletion variants. However, we find this unlikely for several reasons. First, structure prediction programs indicate that the NTD and 7-repeats domain of yeIF4B are intrinsically unstructured (data not shown); therefore, we do not expect these domains to become misfolded in the deletion constructs. In contrast, the RRM has a defined structure, but deletion of this domain results in no change in activity, indicating that it is dispensable for function and that the remaining domains retain their full ability to stimulate translation initiation in the absence of the folded RRM (Figs. 3, 5, 6; Table 1). Likewise, the CTD is also dispensable in vitro and in vivo (Figs. 3, 5, 6; Table 1). Secondly, the deletion variants lacking the NTD or the 7-repeats domain both retain some function. The Δr1-7 variant produces nearly WT rates of mRNA recruitment when high enough concentrations are added (Fig. 3B; Table 1), indicating a binding defect, which suggests that the NTD is still able to accelerate mRNA recruitment in this construct and that it is not misfolded. The ΔNTD variant promotes mRNA recruitment, albeit at reduced rates, and retains the ability to interact with the initiation machinery at low concentrations, indicating that the 7-repeats domain is functional in this variant. This, again, suggests that it is not misfolded. Finally, the 7-repeats on their own stimulate mRNA recruitment in vitro (ΔNTDΔRRMΔCTD) (Fig. 3B; Table 1) and the 7-repeats plus the CTD partially complement the growth defect of the tif3 null strain (Fig. 5B; ΔntdΔrrm) indicating that the repeats retain function even when taken out of the context of the rest of the protein. Taken together, these data strongly suggest that misfolding of the deletion variants is not responsible for the negative effects observed upon deletion of the NTD and 7-repeats domains, although, as discussed above, it seems likely that some higher-order interactions do occur within several of the deletion mutants.
Interactions of yeIF4B with the PIC and eIF4A stimulate mRNA recruitment
The fact that the K1/2 for yeIF4B in mRNA recruitment assays in vitro is 10-fold lower than the Kd for 40S subunits (Table 1) suggests that the factor makes contacts with components of the PIC in addition to the 40S subunit itself. Although both the NTD and 7-repeats mediate interaction of yeIF4B with the 40S subunit, only deletion of the 7-repeats domain confers an increase in the K1/2 value (>30-fold) for yeIF4B, indicating that the 7-repeats mediate these additional interactions.
The fact that the presence of yeIF4B in our mRNA recruitment assays lowers the K1/2 for eIF4A by ≥40-fold (Fig. 4) indicates that these two proteins interact physically or functionally in yeast. This is consistent with previously reported genetic interactions between eIF4A and eIF4B in yeast (Coppolecchia et al. 1993; de la Cruz et al. 1997) and RNA-dependent physical interactions between mammalian eIF4A and eIF4B (Grifo et al. 1983; Hughes et al. 1993; Rozovsky et al. 2008; Nielsen et al. 2011), as well as interactions between the mammalian eIF4B homolog eIF4H and eIF4A (Marintchev et al. 2009). Interaction between eIF4B and eIF4A could be responsible for the low K1/2 of eIF4B in mRNA recruitment assays relative to its Kd for 40S subunits. Other candidates for an interaction partner are eIF3 (Vornlocher et al. 1999), which might help to correctly position regions of one or both factors at the mRNA entry channel (Chiu et al. 2010), and eIF4G (de la Cruz et al. 1997), which would allow yeIF4B to bridge interaction between eIF4F and the PIC.
yeIF4B is a 40S ribosomal subunit-binding protein
Previous work had suggested that mammalian eIF4B interacts with ribosomes in vivo (Hughes et al. 1993; Naranda et al. 1994) and that its RRM binds specifically to 18S rRNA (Methot et al. 1996a). Our data demonstrate that yeIF4B binds specifically and tightly to the 40S ribosomal subunit, although this activity does not require the RRM domain and is instead mediated by the NTD and 7-repeats. The importance of the 40S subunit-binding activity for yeIF4B’s function in mRNA recruitment is suggested by the fact that all of the mutations analyzed that decrease 40S affinity also diminish the factor’s ability to promote mRNA recruitment in vitro and in vivo (Fig. 4; Table 1, cf. 40S subunit Kd values and K1/2 and kmax values). In contrast, there is no correlation between ssRNA-binding affinity and function in mRNA recruitment, arguing against a model in which yeIF4B functions solely as an mRNA-binding protein. The mutant with the most severely impaired 40S subunit binding, ΔNTDΔr1-7, also lacks detectable function in mRNA recruitment in vitro (Fig. 3) and does not stimulate translation in vivo (Figs. 5, 6), whereas it retains significant ssRNA-binding activity. These data strongly suggest that 40S subunit binding is required for yeIF4B to promote mRNA recruitment. The weak RNA-binding activity imparted by the 7-repeats may also be involved in mRNA recruitment; for example, when bound to the PIC one or more of the repeats might interact with the mRNA in or near the entry channel.
yeIF4B binding to Rps20 induces structural changes in the mRNA entry channel
We provide three lines of evidence that yeIF4B binds the head of the 40S subunit. First, yeIF4B interacts with Rps20 in yeast two-hybrid and GST-pulldown analyses. Hydroxyl radical footprinting also indicates that yeIF4B binds near Rps20 in the 40S head. Rps20 has a globular domain projecting from the solvent-exposed face of the 40S head and a long ribosome-binding loop that extends through the head toward the intersubunit face of the neck (Ben-Shem et al. 2011). yeIF4B binding protected two regions of rRNA from hydroxyl radical cleavages (Fig. 7C–E), near each end of Rps20, with the most footprints observed near the A site in h34 on the intersubunit face, and two additional footprints in h39 between Rps20 and protein Asc1 (RACK1 in mammals) near the top of the solvent-exposed face of the 40S subunit (Sengupta et al. 2004; Ben-Shem et al. 2011). The strong protections indicate that these regions either directly contact yeIF4B or become less accessible with rearrangements that occur on yeIF4B binding to Rps20.
Helix 34 extends across the top of the A site and also makes up part of the latch connection between the 40S head and body that is thought to open to facilitate binding of TC and mRNA and allow scanning (Schluenzen et al. 2000; Ogle et al. 2001; Passmore et al. 2007; Ben-Shem et al. 2011). This helix has also been implicated in decoding in both the A and P sites during elongation and initiation, respectively, and in the translocation of mRNA and tRNAs through the ribosome (O’Connor et al. 1997; Matassova et al. 2001; Konstantinidis et al. 2006). Given the importance of the flexibility of this region in allowing conformational changes during each of these processes, it seems possible that yeIF4B binding could induce a rearrangement in h34 that promotes mRNA recruitment.
Helix 34 also makes up one side of the mRNA entry channel (Yusupova et al. 2001). The proximity of the Rps20 ribosome-binding loop to this channel could provide a means for yeIF4B to facilitate mRNA recruitment. yeIF4B could physically extend the mRNA-binding channel itself, directly binding to both the ribosome and mRNA, or it could indirectly alter the accessibility of the entry channel by inducing a movement of the Rps20 ribosome binding loop that pushes it toward h34. Either of these possibilities would explain the observed protections by yeIF4B of bases 1282–1285 and 1428 and 1429 near the mRNA entry channel. However, we favor the latter because yeIF4B variants lacking the 7-repeats or NTD confer the same patterns of protection from hydroxyl radical cleavage. These data seem most consistent with a mechanism in which the 7-repeats and NTD can each bind to Rps20, perhaps on the solvent side of the head where the ribosomal protein’s globular domain lies, and induce a conformational change of the loop that promotes reorganization of the entry channel, facilitating mRNA recruitment. It seems less likely that the 7-repeats and NTD can each bind independently to the entry channel and also directly interact with the mRNA to promote recruitment, especially as the NTD has little influence on the factor’s ssRNA-binding activity. The 7-repeats likely also mediate interactions with other components of the PIC that serve to anchor the factor to the complex and possibly also promote proper organization of other factors or the mRNA. The multiple copies of the repeats may allow this critical domain of the factor to play all of these distinct roles in mRNA recruitment simultaneously.
MATERIALS AND METHODS
Plasmid constructions
All plasmids used are listed in Supplemental Table S1, and all primers for plasmid construction are listed in Supplemental Table S2. To construct TIF3 alleles for analysis in yeast, a DNA fragment containing WT TIF3 with its native upstream (747 bp) and downstream (340 bp) flanking sequences was generated by PCR amplification from genomic DNA using primers PZ024 and PZ025 (listed in Supplemental Table S2, with relevant restriction sites in lowercase) and cloned between the HindIII and BamHI sites of single-copy LEU2, CEN4, ARS1 vector YCplac111 to produce pFJZ049. Coding sequences (CDS) for the His6 tag were inserted immediately before the stop codon by PCR fusion using primers PZ026 and PZ027 (Supplemental Table S2, with His6 CDS in bold) together with primers PZ024 and PZ025 described above, to produce plasmid pFJZ050. TIF3 mutant alleles were generated by PCR fusion using primers listed in Supplemental Table S2 and primers PZ024 and PZ025 described above. TIF3 alleles were subcloned as HindIII–BamHI fragments from sc plasmids described above into high-copy LEU2, 2 µ vector YEplac181. DNA sequences of the entire PCR-amplified regions were verified in these and all other newly constructed plasmids. Supplemental Table S3 contains a summary of the yeIF4B amino acids removed in each variant allele. Plasmids for expression of yeIF4B proteins in Escherichia coli from the T7 promoter were generated by PCR amplifying DNA fragments harboring NdeI and XmaI restriction sites at their termini from yeast plasmids described above containing the CDS of the relevant yeIF4B variants, using the primers listed in Supplemental Table S2 and inserting them between the NdeI and XmaI sites of pTYB2 (NEB) (Supplemental Table S1).
The TIF3 plasmid for yeast two-hybrid analysis pFJZ012 was generated by amplifying a fragment containing full-length TIF3 harboring EcoRI or BamHI sites at the termini using primers PZ007 and PZ012 and inserting it between the EcoRI and BamHI sites of TRP1 plasmid pGBKT7 to generate plasmid pGBKT7-TIF3 with yeIF4B CDS fused to CDS for the Gal4 DNA-binding domain. The complete CDSs of 40S subunit proteins are fused to those for the Gal4 activation domain in derivatives of LEU2 plasmid pGADT7 that were constructed previously (Valasek et al. 2003). Plasmid pED30, containing the complete CDS for Rps20 fused to GST CDS for expression in E. coli was generated by inserting the BamHI–SalI fragment amplified with primer RPS20-f and RPS20-r (Supplemental Table S2) between the BamHI and SalI sites of pGEX-5x-3 (Smith and Johnson 1988).
To construct plasmid pFJZ043 containing the tif3Δ::hisG-URA3-hisG cassette, the 408-bp upstream of and 340-bp downstream from TIF3 were amplified from pFJZ050 with primers PZ022/PZ023 and PZ020/PZ021, respectively. The resulting PCR products were cloned into pHQ221 (Cherkasova et al. 2010) by replacing the resident EcoRI–KpnI and SalI–SphI fragments with the appropriate TIF3 fragments.
Yeast strain constructions
One TIF3 allele in diploid strain BY4743 (MATa/MATa his3-Δ1/his3-Δ1 leu2-Δ0/leu2-Δ0 met15-Δ0/MET15 LYS2/lys2-Δ0 ura3-Δ0/ura3-Δ0) (Winzeler et al. 1999) was disrupted by using the tif3Δ::hisG-URA3-hisG cassette obtained from plasmid pFJZ043, selecting for Ura+ transformants, to produce strain FJZ001. Haploid strains FJZ052 (tif3Δ) (MATα his3-Δ1 leu2-Δ0 met15-Δ0 lys2-Δ0 ura3-Δ0 tif3Δ::hisG) and FJZ046 (TIF3) (MATα his3-Δ1 leu2-Δ0 met15-Δ0 lys2-Δ0 ura3-Δ0 TIF3) were obtained by isolating Ura+ and Ura− ascospore clones, respectively, by tetrad dissection after sporulation of FJZ001, and then eliminating the URA3 marker from the Ura+ isolate by counter-selection on medium containing 5-fluoroorotic acid (5-FOA) (Boeke et al. 1987) to generate the tif3Δ::hisG allele in FJZ052. The presence of tif3Δ in FJZ052 was verified by PCR analysis of chromosomal DNA using primers PZ001, PZ002, and PZ004 (Supplemental Table S2). Additionally, we determined that the Slg− phenotype of FJZ052 could be fully complemented by sc TIF3 plasmid pFJZ049, pFJZ050, or hc plasmid pFJZ058 to yield rates of colony formation at 18°C, 23°C, and 36°C on synthetic complete medium lacking Leu (SC-Leu) plates (Sherman et al. 1974) indistinguishable from that of vector transformants of FJZ046 (data not shown).
Reagents
Ribosomes, initiation factors, capped mRNA, and methionylated initiator tRNA were prepared as described, with the modification that E. coli cells expressing the eIF4E·eIF4G complex and yeast cells were lysed using a Nitrogen mill (Acker et al. 2007; Mitchell et al. 2010). RNA with the sequence 5′-GGAAU(CU)7AUG(CU)10C-3′-6-Fluorescein was purchased from IDT for ssRNA-binding assays. yeIF4B and variants were purified (Mitchell et al. 2010) and labeled at their C-termini with fluorescein using expressed protein ligation as described for eIF1 (Acker et al. 2007). All in vitro reactions were performed in 1× Recon Buffer (30 nM HEPES·KOH at pH 7.4, 100 nM KOAc at pH 7.6, 3 mM Mg(OAc)2, and 2 mM DTT).
Fluorescence anisotropy assays
The fluorescence anisotropies of ssRNA-FL and eIF4B-FL were followed in either a 120-µL or 300-µL reaction, respectively, using an excitation wavelength of 495 nM and emission at 520 nM. The anisotropy of ssRNA-FL increased upon addition of unlabeled eIF4B, with no significant change in overall intensity. Buffer controls were performed by monitoring the anisotropy upon addition of equal volumes of eIF4B storage buffer (20 mM Hepes·KOH at pH 7.4, 100 mM KOAc at pH 7.6, 10% glycerol, 2 mM DTT) instead of eIF4B to ensure no substantial changes in anisotropy were observed. Ribosome binding was followed by monitoring the fluorescence anisotropy of eIF4B-FL upon addition of 40S or 60S subunits. Equal volumes of ribosome storage buffer (20 mM Hepes·KOH at pH 7.4, 100 mM KOAc at pH 7.6, 2.5 mM Mg(OAc)2, 250 mM sucrose, 2 mM DTT) were used as a negative control. Experiments were repeated five times or more for WT eIF4B and twice or more for each variant. In the second of each of the ribosome-binding experiments with deletion variants, 40S subunits were only titrated up to 1 µM to ensure that the data from the first experiments were reproducible and the Kd and error of the fit obtained from the first experiment are reported. No significant change in overall fluorescence intensity was observed at any point in the titrations.
Gel shift assay for mRNA recruitment
Twelve microliter reactions were set up as described (Mitchell et al. 2010) containing 1X Recon Buffer, 1 mM GDPNP·Mg2+, 200 nM eIF2, 200 nM Met-tRNAiMet, 2 mM ATP·Mg2+, 30 nM 40S subunits, 1 µM eIF1, 1 µM eIF1A, 200 nM eIF3, 30 nM eIF4E·eIF4G copurified complex, 1 µM eIF4A, and eIF4B as indicated. These complexes were incubated for 10 min at 26°C before adding 15 nM 32P-m7G-RPL41A. Recruitment of 32P-m7G-RPL41A was monitored following incubations at 26°C for the indicated times by loading samples onto a running 1× THEM (34 mM Tris Base, 57 mM HEPES, 0.1 mM EDTA, 2.5 mM MgCl2) buffered, 4% polyacrylamide (37.5:1, acrylamide:bisacrylamide) gel, and running the gel at maximum 200 volts for 1 h with a circulating water bath set at 22°C. Assays were repeated two or more times for each protein. Addition of 300 nM eIF5 did not affect the kinetics or K1/2 for mRNA recruitment of eIF4A or yeIF4BΔr1-7 (data not shown).
Toeprinting
Toeprinting reactions were performed as previously described (Mitchell et al. 2010) with the modification that AMV reverse transcriptase was purchased from New England Biolabs. Yeast eIF4B concentrations are indicated in the legend to Figure 3. Full-length cDNA and the terminated cDNA product 16–17 nucleotides downstream from the start codon were resolved on sequencing gels and quantified using a PhosphorImager. The toeprint corresponding to the PIC was not present when the ternary complex (Fig. 3C) or 40S subunits (data not shown) were omitted. The fraction of the intensity of the bands for PIC/(PIC + full-length) was calculated in the presence/absence of each variant, and this value was normalized to the fraction observed in the presence of 1 µM eIF4B (1.0) and no eIF4B (0).
Hydroxyl radical footprinting
A 40S·yeIF4B complex was formed by incubating 1 µM 40S subunits with 1 µM yeIF4B in a 50-µL reaction in 1× Recon Buffer at 26°C for 10 min. Proteins were dialyzed into 1× Recon Buffer to remove glycerol, and variants were added to the following concentrations in Figure 8: ΔNTD, 7 µM; ΔRRM, 2 µM; Δr1-7, 2 µM; ΔNTDΔRRM, 2 µM; ΔNTDΔRRMΔCTD, 3 µM; and ΔNTDΔr1-7, 10 µM. After a 10-min equilibration on ice, 25 µL was removed to a tube containing 1 µL of each of the following freshly made reagents: 50 mM Fe(NH4)2(SO4)2, 100 mM EDTA (pH 8.0), 250 mM ascorbate, and 2.5% H2O2. The remaining 25 µL was incubated in the absence of hydroxyl radicals as a control for nuclease contamination (Fig. 8). Reactions were mixed and returned to ice for a 10-min incubation before quenching with an equal volume of 100 mM thiourea. 18S rRNA was extracted using an RNeasy mini kit (Qiagen) and analyzed by primer extension with primers spanning the 18S rRNA (Supplemental Table S2; Shin et al. 2011). The footprints reported in Figure 7 were extremely reproducible (n = 6).
Complementation analysis
Transformants of tif3Δ strain FJZ052 harboring WT or mutant tif3 alleles on the relevant sc or hc LEU2 plasmids were generated (Ito et al. 1983) and examined for rates of colony formation at 18°C, 30°C, and 36°C by spotting serial 10-fold dilutions of saturated cultures on SC-Leu plates and scoring the number and size of colonies at each dilution daily.
Western analysis of yeIF4B expression
WCEs were prepared by extraction with trichloroacetic acid (TCA) (Reid and Schatz 1982) and subjected to Western blot analysis as described previously (Valasek et al. 2007) using antibodies against the His6 epitope (Invitrogen) or Gcd6 (eIF2Bε) (Cigan et al. 1993). The His6 signals were quantified using NIH ImageJ (Rasband, 1997–2012) and normalized with that of Gcd6 signals from the same loadings. All samples were compared with that of the transformant harboring WT TIF3 on a sc plasmid analyzed on the same gel to calculate mutant protein levels as percentages of WT yeIF4B, and standard errors were calculated from three replicates prepared from independent transformants.
Analysis of polysome profiles and native 43S/48S PICs
Transformants of tif3Δ strain FJZ052 harboring the appropriate plasmid-borne WT or mutant tif3 alleles were cultured overnight in SC-Leu medium at 30°C to A600 ∼1, and cycloheximide was added to 50 μg/mL 10 min prior to harvesting. WCEs were prepared in breaking buffer (20 mM Tris-HCl at pH 7.5, 50 mM NaCl, 10 mM MgCl2, 1 mM DTT, 200 μg/mL heparin, 50 μg/mL cycloheximide, and 1 Complete EDTA-free Protease Inhibitor Tablet [Roche]/50 mL buffer). Next, 12.5 A260 units of WCEs were separated by velocity sedimentation on a 4.5%–45% sucrose gradient by centrifugation at 39,000 rpm for 3.2 h at 4°C in an SW41Ti rotor (Beckman). Gradient fractions were scanned at 254 nm to visualize ribosomal species. The cross-linking assay for observing 43S PICs, and the following quantitative real time PCR (qRT-PCR) using reverse-transcribed cDNA for analyzing 43S·mRNA PIC formation were performed as previously described (Chiu et al. 2010).
Yeast-two-hybrid assays
Strains Y190 (MATa leu2-3,-112 ura3-52 trp1-901 his3-Δ200 ade2-101 gal4Δ gal80Δ URA3::GAL-lacZ LYS::GAL(UAS)-HIS3) and Y187 (MATα gal4Δ gal80Δ his3-Δ200 trp1-901 ade2-101 ura3-52 leu2-3,-112 met2-URA3::GAL1(UAS)-GAL1TATA-lacZ) (Harper et al. 1993) were transformed with the appropriate GAL4-AD and GAL4-BD fusion constructs, respectively, or with the corresponding empty vectors. Confluent lawns of Y187 transformants and patches of Y190 transformants were grown on the appropriate selective medium (SC-Trp and SC-Leu, respectively) and replica plated to YPD plates for mating. After incubating at 30°C for 8 h, the YPD plates were replica plated to SC-Leu-Trp. Single colonies were subsequently colony-purified by streaking cells on SC-Leu-Trp, and the resulting diploid clones were patched on SC-Leu-Trp plates and replica plated to SC-Leu-Trp-His + 30 mM 3AT and SC-Leu-Trp. Cell growth was recorded after 2 d at 30°C. β-galactosidase expression was analyzed by culturing patches of yeast cells on a nitrocellulose filter placed on the surface of an SC-Leu-Trp plate, floating the filter on liquid nitrogen, and then covering the filter with an overlay of 1.0% agarose in 0.1 M NaH2PO4 (pH 7.0) containing 40 µg/mL of 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal) and incubating until color developed.
GST-pull down assays
Glutathione S-transferase (GST) pull-down assays were performed as described previously (Chiu et al. 2010), except that purified yeIF4B or eIF3 proteins were used, and incubation with micrococcal nuclease was added to control for indirect interactions bridged by RNA. Briefly, 1–5 mg of immobilized GST-Rps20 and GST were incubated in 100 μL wash buffer I (1× PBS at pH 7.4, 10% glycerol, 1.5 mM DTT, and 0.25% Triton X-100) with 2 mM CaCl2, with or without 1200 units of micrococcal nuclease (Fermentas EN0181). The reactions were stopped by washing with wash buffer I plus 6 mM EDTA, then washing three times with regular wash buffer I.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank the members of our laboratories for useful comments and suggestions. We also thank Michael Altmann (University of Bern) for generously sharing antibodies against yeIF4B and Patrick Linder (University of Geneva) for gifts of yeast strains. This work was supported by the Intramural Research Program of the NIH (A.G.H., F.Z.), NIH grant GM62128 (J.R.L.), and an AHA postdoctoral fellowship (S.E.W.). L.V. was supported by the Czech Science Foundation P305/12/G034.
REFERENCES
- Acker MG, Kolitz SE, Mitchell SF, Nanda JS, Lorsch JR 2007. Reconstitution of yeast translation initiation. Methods Enzymol 430: 111–145 [DOI] [PubMed] [Google Scholar]
- Aitken CE, Lorsch JR 2012. A mechanistic overview of translation initiation in eukaryotes. Nat Struct Mol Biol 19: 568–576 [DOI] [PubMed] [Google Scholar]
- Altmann M, Muller PP, Wittmer B, Ruchti F, Lanker S, Trachsel H 1993. A Saccharomyces cerevisiae homologue of mammalian translation initiation factor 4B contributes to RNA helicase activity. EMBO J 12: 3997–4003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmann M, Wittmer B, Methot N, Sonenberg N, Trachsel H 1995. The Saccharomyces cerevisiae translation initiation factor Tif3 and its mammalian homologue, eIF-4B, have RNA annealing activity. EMBO J 14: 3820–3827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benne R, Hershey JW 1978. The mechanism of action of protein synthesis initiation factors from rabbit reticulocytes. J Biol Chem 253: 3078–3087 [PubMed] [Google Scholar]
- Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M 2011. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science 334: 1524–1529 [DOI] [PubMed] [Google Scholar]
- Bi X, Ren J, Goss DJ 2000. Wheat germ translation initiation factor eIF4B affects eIF4A and eIFiso4F helicase activity by increasing the ATP binding affinity of eIF4A. Biochemistry 39: 5758–5765 [DOI] [PubMed] [Google Scholar]
- Boeke JD, Trueheart J, Natsoulis G, Fink GR 1987. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol 154: 164–175 [DOI] [PubMed] [Google Scholar]
- Cherkasova V, Qiu H, Hinnebusch AG 2010. Snf1 promotes phosphorylation of the α subunit of eukaryotic translation initiation factor 2 by activating Gcn2 and inhibiting phosphatases Glc7 and Sit4. Mol Cell Biol 30: 2862–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu WL, Wagner S, Herrmannova A, Burela L, Zhang F, Saini AK, Valasek L, Hinnebusch AG 2010. The C-terminal region of eukaryotic translation initiation factor 3a (eIF3a) promotes mRNA recruitment, scanning, and, together with eIF3j and the eIF3b RNA recognition motif, selection of AUG start codons. Mol Cell Biol 30: 4415–4434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cigan AM, Bushman JL, Boal TR, Hinnebusch AG 1993. A protein complex of translational regulators of GCN4 mRNA is the guanine nucleotide-exchange factor for translation initiation factor 2 in yeast. Proc Natl Acad Sci 90: 5350–5354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppolecchia R, Buser P, Stotz A, Linder P 1993. A new yeast translation initiation factor suppresses a mutation in the eIF-4A RNA helicase. EMBO J 12: 4005–4011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuchalova L, Kouba T, Herrmannova A, Danyi I, Chiu WL, Valasek L 2010. The RNA recognition motif of eukaryotic translation initiation factor 3 g (eIF3 g) is required for resumption of scanning of posttermination ribosomes for reinitiation on GCN4 and together with eIF3i stimulates linear scanning. Mol Cell Biol 30: 4671–4686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz J, Iost I, Kressler D, Linder P 1997. The p20 and Ded1 proteins have antagonistic roles in eIF4E-dependent translation in Saccharomyces cerevisiae. Proc Natl Acad Sci 94: 5201–5206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmitriev SE, Terenin IM, Dunaevsky YE, Merrick WC, Shatsky IN 2003. Assembly of 48S translation initiation complexes from purified components with mRNAs that have some base pairing within their 5′ untranslated regions. Mol Cell Biol 23: 8925–8933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifo JA, Tahara SM, Morgan MA, Shatkin AJ, Merrick WC 1983. New initiation factor activity required for globin mRNA translation. J Biol Chem 258: 5804–5810 [PubMed] [Google Scholar]
- Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ 1993. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75: 805–816 [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG 2011. Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol Mol Biol Rev 75: 434–467, first page of table of contents [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes DL, Dever TE, Merrick WC 1993. Further biochemical characterization of rabbit reticulocyte eIF-4B. Arch Biochem Biophys 301: 311–319 [DOI] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A 1983. Transformation of intact yeast cells treated with alkali cations. J Bacteriol 153: 163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jivotovskaya AV, Valasek L, Hinnebusch AG, Nielsen KH 2006. Eukaryotic translation initiation factor 3 (eIF3) and eIF2 can promote mRNA binding to 40S subunits independently of eIF4G in yeast. Mol Cell Biol 26: 1355–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis TC, Patsoukis N, Georgiou CD, Synetos D 2006. Translational fidelity mutations in 18S rRNA affect the catalytic activity of ribosomes and the oxidative balance of yeast cells. Biochemistry 45: 3525–3533 [DOI] [PubMed] [Google Scholar]
- Marintchev A, Edmonds KA, Marintcheva B, Hendrickson E, Oberer M, Suzuki C, Herdy B, Sonenberg N, Wagner G 2009. Topology and regulation of the human eIF4A/4G/4H helicase complex in translation initiation. Cell 136: 447–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matassova AB, Rodnina MV, Wintermeyer W 2001. Elongation factor G-induced structural change in helix 34 of 16S rRNA related to translocation on the ribosome. RNA 7: 1879–1885 [PMC free article] [PubMed] [Google Scholar]
- Methot N, Pause A, Hershey JW, Sonenberg N 1994. The translation initiation factor eIF-4B contains an RNA-binding region that is distinct and independent from its ribonucleoprotein consensus sequence. Mol Cell Biol 14: 2307–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Methot N, Pickett G, Keene JD, Sonenberg N 1996a. In vitro RNA selection identifies RNA ligands that specifically bind to eukaryotic translation initiation factor 4B: The role of the RNA remotif. RNA 2: 38–50 [PMC free article] [PubMed] [Google Scholar]
- Methot N, Song MS, Sonenberg N 1996b. A region rich in aspartic acid, arginine, tyrosine, and glycine (DRYG) mediates eukaryotic initiation factor 4B (eIF4B) self-association and interaction with eIF3. Mol Cell Biol 16: 5328–5334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milburn SC, Hershey JW, Davies MV, Kelleher K, Kaufman RJ 1990. Cloning and expression of eukaryotic initiation factor 4B cDNA: Sequence determination identifies a common RNA recognition motif. EMBO J 9: 2783–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell SF, Walker SE, Algire MA, Park EH, Hinnebusch AG, Lorsch JR 2010. The 5′-7-methylguanosine cap on eukaryotic mRNAs serves both to stimulate canonical translation initiation and to block an alternative pathway. Mol Cell 39: 950–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naranda T, Strong WB, Menaya J, Fabbri BJ, Hershey JW 1994. Two structural domains of initiation factor eIF-4B are involved in binding to RNA. J Biol Chem 269: 14465–14472 [PubMed] [Google Scholar]
- Niederberger N, Trachsel H, Altmann M 1998. The RNA recognition motif of yeast translation initiation factor Tif3/eIF4B is required but not sufficient for RNA strand-exchange and translational activity. RNA 4: 1259–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen KH, Behrens MA, He Y, Oliveira CL, Jensen LS, Hoffmann SV, Pedersen JS, Andersen GR 2011. Synergistic activation of eIF4A by eIF4B and eIF4G. Nucleic Acids Res 39: 2678–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor M, Thomas CL, Zimmermann RA, Dahlberg AE 1997. Decoding fidelity at the ribosomal A and P sites: Influence of mutations in three different regions of the decoding domain in 16S rRNA. Nucleic Acids Res 25: 1185–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle JM, Brodersen DE, Clemons WM Jr, Tarry MJ, Carter AP, Ramakrishnan V 2001. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292: 897–902 [DOI] [PubMed] [Google Scholar]
- Ozes AR, Feoktistova K, Avanzino BC, Fraser CS 2011. Duplex unwinding and ATPase activities of the DEAD-box helicase eIF4A are coupled by eIF4G and eIF4B. J Mol Biol 412: 674–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsyan A, Svitkin Y, Shahbazian D, Gkogkas C, Lasko P, Merrick WC, Sonenberg N 2011. mRNA helicases: The tacticians of translational control. Nat Rev Mol Cell Biol 12: 235–245 [DOI] [PubMed] [Google Scholar]
- Passmore LA, Schmeing TM, Maag D, Applefield DJ, Acker MG, Algire MA, Lorsch JR, Ramakrishnan V 2007. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol Cell 26: 41–50 [DOI] [PubMed] [Google Scholar]
- Pestova TV, Kolupaeva VG 2002. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev 16: 2906–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers T, Noller HF 1995. Hydroxyl radical footprinting of ribosomal proteins on 16S rRNA. RNA 1: 194–209 [PMC free article] [PubMed] [Google Scholar]
- Rajagopal V, Park EH, Hinnebusch AG, Lorsch JR 2012. Specific domains in yeast translation initiation factor eIF4G strongly bias RNA unwinding activity of the eIF4F complex toward duplexes with 5′-overhangs. J Biol Chem 287: 20301–20312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid GA, Schatz G 1982. Import of proteins into mitochondria. Yeast cells grown in the presence of carbonyl cyanide m-chlorophenylhydrazone accumulate massive amounts of some mitochondrial precursor polypeptides. J Biol Chem 257: 13056–13061 [PubMed] [Google Scholar]
- Rogers GW Jr, Richter NJ, Lima WF, Merrick WC 2001. Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. J Biol Chem 276: 30914–30922 [DOI] [PubMed] [Google Scholar]
- Rozovsky N, Butterworth AC, Moore MJ 2008. Interactions between eIF4AI and its accessory factors eIF4B and eIF4H. RNA 14: 2136–2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluenzen F, Tocilj A, Zarivach R, Harms J, Gluehmann M, Janell D, Bashan A, Bartels H, Agmon I, Franceschi F, et al. 2000. Structure of functionally activated small ribosomal subunit at 3.3 Å resolution. Cell 102: 615–623 [DOI] [PubMed] [Google Scholar]
- Schreier MH, Erni B, Staehelin T 1977. Initiation of mammalian protein synthesis. I. Purification and characterization of seven initiation factors. J Mol Biol 116: 727–753 [DOI] [PubMed] [Google Scholar]
- Seal SN, Schmidt A, Marcus A 1989. Ribosome binding to inosine-substituted mRNAs in the absence of ATP and mRNA factors. J Biol Chem 264: 7363–7368 [PubMed] [Google Scholar]
- Sengupta J, Nilsson J, Gursky R, Spahn CM, Nissen P, Frank J 2004. Identification of the versatile scaffold protein RACK1 on the eukaryotic ribosome by cryo-EM. Nat Struct Mol Biol 11: 957–962 [DOI] [PubMed] [Google Scholar]
- Sherman F, Stewart JW, Jackson M, Gilmore RA, Parker JH 1974. Mutants of yeast defective in iso-1-cytochrome c. Genetics 77: 255–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin BS, Kim JR, Walker SE, Dong J, Lorsch JR, Dever TE 2011. Initiation factor eIF2γ promotes eIF2-GTP-Met-tRNAi(Met) ternary complex binding to the 40S ribosome. Nat Struct Mol Biol 18: 1227–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DB, Johnson KS 1988. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 67: 31–40 [DOI] [PubMed] [Google Scholar]
- Trachsel H, Erni B, Schreier MH, Staehelin T 1977. Initiation of mammalian protein synthesis. II. The assembly of the initiation complex with purified initiation factors. J Mol Biol 116: 755–767 [DOI] [PubMed] [Google Scholar]
- Valasek L, Mathew AA, Shin BS, Nielsen KH, Szamecz B, Hinnebusch AG 2003. The yeast eIF3 subunits TIF32/a, NIP1/c, and eIF5 make critical connections with the 40S ribosome in vivo. Genes Dev 17: 786–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valasek L, Szamecz B, Hinnebusch AG, Nielsen KH 2007. In vivo stabilization of preinitiation complexes by formaldehyde cross-linking. Methods Enzymol 429: 163–183 [DOI] [PubMed] [Google Scholar]
- Vornlocher HP, Hanachi P, Ribeiro S, Hershey JW 1999. A 110-kilodalton subunit of translation initiation factor eIF3 and an associated 135-kilodalton protein are encoded by the Saccharomyces cerevisiae TIF32 and TIF31 genes. J Biol Chem 274: 16802–16812 [DOI] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906 [DOI] [PubMed] [Google Scholar]
- Yusupova GZ, Yusupov MM, Cate JH, Noller HF 2001. The path of messenger RNA through the ribosome. Cell 106: 233–241 [DOI] [PubMed] [Google Scholar]