The regulation of mature cellular tRNAs has newly emerging significance and scope. Because tRNA structure and function are strongly affected by post-transcriptional modification, changes in the extent of modification of any given tRNA can have rapid and selective impact on translation. This study shows that yeast growth arrest increases cytosine base modification in a manner discriminatory among different tRNA sequences. The findings of this study characterize a post-transcriptional change in the tRNA pool linked to cellular stress.
Keywords: tRNA(His), tRNA modifications, 5-methylcytidine, Trm4, cellular stress
Abstract
tRNAs are highly modified, each with a unique set of modifications. Several reports suggest that tRNAs are hypomodified or, in some cases, hypermodified under different growth conditions and in certain cancers. We previously demonstrated that yeast strains depleted of tRNAHis guanylyltransferase accumulate uncharged tRNAHis lacking the G−1 residue and subsequently accumulate additional 5-methylcytidine (m5C) at residues C48 and C50 of tRNAHis, due to the activity of the m5C-methyltransferase Trm4. We show here that the increase in tRNAHis m5C levels does not require loss of Thg1, loss of G−1 of tRNAHis, or cell death but is associated with growth arrest following different stress conditions. We find substantially increased tRNAHis m5C levels after temperature-sensitive strains are grown at nonpermissive temperature, and after wild-type strains are grown to stationary phase, starved for required amino acids, or treated with rapamycin. We observe more modest accumulations of m5C in tRNAHis after starvation for glucose and after starvation for uracil. In virtually all cases examined, the additional m5C on tRNAHis occurs while cells are fully viable, and the increase is neither due to the GCN4 pathway, nor to increased Trm4 levels. Moreover, the increased m5C appears specific to tRNAHis, as tRNAVal(AAC) and tRNAGly(GCC) have much reduced additional m5C during these growth arrest conditions, although they also have C48 and C50 and are capable of having increased m5C levels. Thus, tRNAHis m5C levels are unusually responsive to yeast growth conditions, although the significance of this additional m5C remains unclear.
INTRODUCTION
During maturation, tRNA molecules are extensively processed and highly modified with a unique set of post-transcriptional modifications. In Saccharomyces cerevisiae, ∼13 nt are modified in each tRNA, and there are 25 unique tRNA modifications that have been found at 36 positions on the tRNA (Phizicky and Hopper 2010). These post-transcriptional modifications are highly conserved in different organisms, underscoring their importance. Many tRNA modifications near the anti-codon have important roles in decoding mRNA (Agris et al. 2007), as evidenced by lethality or slow growth phenotypes (Phizicky and Hopper 2010). For example, loss of 2′-O-methylation at positions 32 and 34 or loss of either m1G or t6A at position 37 results in slow growth (Bjork et al. 2001; Pintard et al. 2002; El Yacoubi et al. 2009; Guy et al. 2012). In addition, a number of modifications in the tRNA body have important roles in stabilizing tRNA (Kadaba et al. 2004; Alexandrov et al. 2006; Kadaba et al. 2006; Chernyakov et al. 2008b; Phizicky and Hopper 2010; Whipple et al. 2011). However, the precise roles of many modifications are not yet known in detail and are still under investigation.
Although it has been generally assumed that modifications made to tRNAs are constitutively added at similar levels, a number of reports have described altered tRNA modification levels under various conditions in different organisms. Bacillus subtilis tRNAs were reported to be hypomethylated during log phase growth (Singhal and Vold 1976), and Bacillus stearothermophilus tRNAs have higher 2′-O-methylation levels at 70°C than at 50°C (Agris et al. 1973). Similarly, several types of cancer cell lines have reduced yW (wybutosine) modification of tRNAPhe (Grunberger et al. 1975; Mushinski and Marini 1979, 1983; Kuchino et al. 1982; Grunberger et al. 1983), and tRNAPhe species from a hepatoma and a breast carcinoma have an unexpected additional 1-methylguanosine (m1G) modification, as well as additional dihydrouridine and 5-methylcytidine (m5C) (Kuchino and Borek 1978). In addition, human and murine hepatoma cells have reduced levels of queosine in their tRNAAsp (Kuchino et al. 1981; Randerath et al. 1984; Pathak et al. 2005), while in Drosophila, queosine levels of several tRNAs increase with age and with the percentage of yeast in the diet (Hosbach and Kubli 1979; Owenby et al. 1979).
Emerging data also suggest growth-dependent changes in tRNA modification levels in the yeast Saccharomyces cerevisiae. Mitochondrial tRNALys(UUU) has reduced 2-thiolation following growth at elevated temperature (Kamenski et al. 2007). In addition, it was recently reported that oxidative stress mediated by hydrogen peroxide results in increased levels of m5C, 2′-O-methylcytidine (Cm), and 2,2-dimethylguanosine (m22G) levels and that a number of other modifications are affected by treatment with several chemicals that induce cellular stress (Chan et al. 2010, 2012).
We previously reported that depletion of the essential tRNAHis guanylyltransferase Thg1 results in the loss of G−1 from tRNAHis, with concomitant accumulation of deacylated tRNAHis and the subsequent accumulation of additional m5C on tRNAHis ∼8 h after the loss of G−1 (Gu et al. 2005). The additional m5C occurs at residues C48 and C50 (∼0.5 mol/mol tRNA at each position) (Gu et al. 2005), adjacent to the known m5C49 modification, and appears to be due to Trm4 methyltransferase, which catalyzes formation of m5C at C34, C40, C48, and C49 in all substrate tRNAs in yeast (Motorin and Grosjean 1999). Interestingly, the additional m5C that accumulates following Thg1 depletion was not found on tRNAGly(GCC), which also has cytidines at these positions, and tRNAHis was observed to accumulate in the nucleus during Thg1 depletion (Gu et al. 2005), presumably by retrograde transport of cytoplasmic tRNA to the nucleus (Shaheen and Hopper 2005; Takano et al. 2005). At the time, we speculated that the additional m5C found in tRNAHis from Thg1-depleted cells might be due to the increased availability of deacylated tRNAHis to Trm4, which is localized to the nucleus (Wu et al. 1998), or possibly due to a regulatory response from the uncharged tRNAHis as the cells arrested growth due to loss of Thg1 (Gu et al. 2005). Alternatively, the lack of the G−1 modification on tRNAHis might trigger the accumulation of m5C, much as Thermus thermophilus cells lacking either m7G46 or ψ55 have altered modifications under certain conditions (Tomikawa et al. 2010; Ishida et al. 2011).
In this work, we explore the conditions in which tRNAHis can be modified with additional m5C. We show that m5C accumulates in tRNAHis under a variety of conditions in which growth is arrested and that this accumulation is not associated with cell death. Although several conditions examined involve nutrient deprivation, the GCN4 pathway is not responsible for the additional m5C. However, the target of rapamycin (TOR) pathway may play a role in the m5C response, since rapamycin treatment results in an increase in tRNAHis m5C levels. Remarkably, we also show that this additional m5C is specific to tRNAHis, relative to two other tRNAs with cytidine residues at the same positions, which are capable of being overmodified with m5C in vivo. We conclude that m5C modification of tRNAHis is unusually sensitive to yeast growth conditions, although the cellular function of this phenomenon remains unclear.
RESULTS
m5C levels are increased in tRNAHis from thg1ts strains but not from a thg1-Δ strain
Our initial goal was to explore the cause of additional m5C on tRNAHis observed following depletion of Thg1 (Gu et al. 2005), which adds G−1 to the 5′ end of tRNAHis (Gu et al. 2003). The secondary structure and modifications of tRNAHis are depicted in Figure 1A. The m5C modification (catalyzed by Trm4) is clearly resolved from other nucleosides by reverse-phase HPLC analysis of tRNAHis purified from wild-type and trm4-Δ strains (Fig. 1B). Values of nucleoside modifications are quantified from HPLC traces and are expressed as moles of modification per mole of tRNA (mol/mol tRNA).
FIGURE 1.
Post-transcriptional modifications of mature tRNAHis. (A) Secondary structure of mature S. cerevisiae tRNAHis. Post-transcriptional modifications are indicated. G−1 is added to the 5′ end of tRNAHis by tRNAHis guanylyltransferase (Thg1). Under normal growth conditions, Trm4 methylates position 49 to form 5-methylcytidine (m5C, circle). When Thg1 is depleted, Trm4 also methylates adjacent cytidines at positions 48 and 50 (squares). (B) Representative HPLC traces of tRNAHis nucleosides. Wild-type (BY4741) and trm4-Δ cells were grown to log phase in SD complete media, and tRNAHis was purified, digested with P1 nuclease, phosphatase-treated, and separated by reverse-phase HPLC. Insets compare the wild type vs. trm4-Δ m5C peak (left), with the Am peak (right) as a control.
Based on our previously published data, it seemed plausible that increased levels of m5C upon Thg1 depletion might be due to the loss of the essential Thg1 protein, the consequent loss of the tRNAHis G−1 residue, and the accumulation of uncharged tRNAHis (Gu et al. 2005). To test this, we examined a thg1-Δ strain that was viable due to overexpression of tRNAHis and the histidyl-tRNA synthetase HTS1 (Preston and Phizicky 2010). We find that this strain has normal levels of m5C on tRNAHis relative to wild type (Table 1, WT). Since this thg1-Δ strain also has approximately 15-fold more tRNAHis than a wild-type strain and the tRNAHis is mostly deacylated (Preston and Phizicky 2010), the availability of uncharged tRNAHis alone is likely not the cause of additional m5C levels. In support of this conclusion, increasing the amount of available tRNAHis by overexpression of tRNAHis in a wild-type strain has no effect on tRNAHis m5C levels (Table 1). Therefore, it is possible that the addition of m5C to tRNAHis is, instead, triggered by growth arrest due to loss of Thg1 function.
TABLE 1.
tRNAHis modification levels in conditions of reduced Thg1 activity

To explore the connection between Thg1 depletion-mediated growth arrest and elevated tRNAHis m5C levels, we measured m5C levels in tRNAHis purified from three different thg1 temperature-sensitive mutants (Y146H, G172D/L233S, and Y8C) before and after shift to 37°C (Table 1). Growth arrest was apparent by 3 h for the thg1-Y146H strain and by 4–5 h for the thg1-G172D/L233S and thg1-Y8C strains (data not shown). Whereas tRNAHis from the wild-type strain and each of the thg1 mutants have near normal amounts of m5C when grown at 24°C (0.92–1.25 mol/mol tRNA), tRNAHis m5C levels increase dramatically to 2.36–2.41 mol/mol tRNA when thg1 mutants are shifted to 37°C for 7 h. tRNAHis from the wild-type strain has normal levels of m5C after the temperature shift, and levels of control modifications (dihydrouridine (D), pseudouridine (ψ), m1G, and Am) remain unchanged in tRNAHis isolated from each strain (Table 1; data not shown).
tRNAHis m5C levels increase when temperature-sensitive strains are grown at nonpermissive temperature
Based on the data above, the increase in m5C levels could be correlated with lack of growth or with cell death, associated with loss of Thg1 function. To determine if an increase in tRNAHis m5C levels is specific to loss of Thg1 function and/or subsequent cell death, we grew a set of temperature-sensitive (ts) strains at permissive or nonpermissive temperatures and analyzed cell viability. We then purified tRNAHis and measured modification levels by HPLC analysis. We grew BY4741 (WT) and the thg1-Y146Hts strain as a control for additional m5C. We also grew three temperature-sensitive strains unrelated to tRNA processing: the fcp1-1ts strain, which is defective for transcription by RNA polymerase II at nonpermissive temperature (Kobor et al. 1999), the abf1-102ts strain, which has a mutation in a DNA binding protein involved in transcriptional regulation, DNA replication, and DNA repair (Buchman et al. 1988; Reed et al. 1999; Miyake et al. 2004), and the cdc48-9ts strain, which has a mutation in an ATPase involved in protein export from the ER to the cytoplasm (Ye et al. 2001). Growth arrest was apparent by 2–3 h for the fcp1-1ts strain, by 3–4 h for the thg1-Y146Hts strain, by 6–7 h for the cdc48-9ts strain, and by over 7 h for the abf1-102ts strain (data not shown).
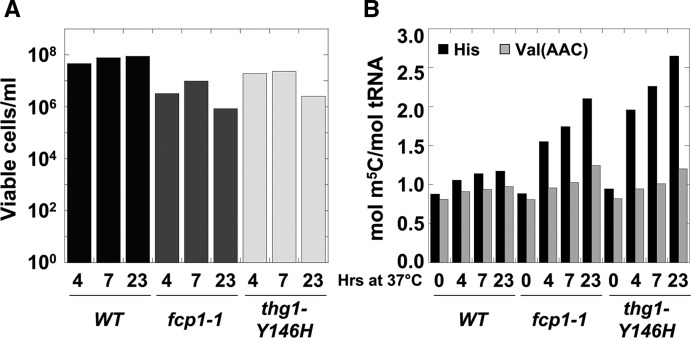
For two of the temperature-sensitive strains grown at 37°C, we observe an increase in m5C levels on tRNAHis following growth arrest and without any significant loss of cell viability (Fig. 2; Table 2). Thus, at the 7 h time point at 37°C, the fcp1-1ts and cdc48-9ts strains are nearly completely viable (Fig. 2A; Table 2; data not shown) and their tRNAHis have 1.75 and 1.51 mol m5C/mol tRNA, respectively, compared to 0.92 and 0.93 mol/mol tRNA at 24°C (Fig. 2B; Table 2; data not shown). At the same time point after shift to 37°C, tRNAHis m5C levels increase only slightly in the wild-type strain (from 0.92 to 1.12 mol/mol tRNA) and in the abf1-102ts mutant (from 0.89 to 1.18 mol/mol tRNA) (data not shown). We ruled out the possibility that the increased tRNAHis m5C levels in the fcp1-1ts strain were somehow due to the absence of the G−1 residue of tRNAHis by direct examination of the 5′ end of tRNAHis. As expected, there was no change in the G−1 status in fcp1-1ts and WT strains and a marked reduction in G−1 of tRNAHis from the thg1-Y146Hts grown at 37°C (data not shown). Thus, the additional m5C on tRNAHis is unrelated to the G−1 addition activity of Thg1, and we conclude that growth arrest, but not cell death, of temperature-sensitive strains results in additional m5C on tRNAHis.
FIGURE 2.
Temperature-sensitive strains have additional m5C on tRNAHis when grown at nonpermissive temperature. (A) Assessment of viable cell titer following growth at 37°C. (B) m5C levels in tRNAHis and tRNAVal(AAC) during growth at 37°C. tRNAHis and tRNAVal(AAC) were purified from BY4741, thg1-Y146H, and fcp1-1 strains, and modification levels were measured by HPLC. m5C levels for tRNAHis (black) and tRNAVal(AAC) (gray) during temperature-sensitive strain growth at 37°C were plotted. m5C values during growth at 24°C were plotted as the 37°C 0-h time point.
TABLE 2.
Temperature-sensitive strains grown at nonpermissive temperature have additional m5C on tRNAHis

We also find evidence that the increased amount of m5C is specific to tRNAHis. tRNAVal(AAC), which also normally has unmodified C48 and C50 residues adjacent to m5C49, has only marginally increased levels of m5C 7 h after temperature shift in the fcp1-1ts mutant (from 0.81 to 1.03 mol/mol tRNA) and in the thg1-Y146Hts mutant (from 0.82 to 1.01 mol/mol tRNA) (Fig. 2B; Table 2).
tRNAHis m5C levels increase in several conditions in which a wild-type strain arrests growth and is still viable
Since the accumulation of m5C in tRNAHis occurs in temperature-sensitive strains following growth arrest, we reasoned that wild-type strains might also have elevated m5C levels in conditions where the cells stop growing. We, therefore, examined modification status of tRNAHis in the BY4741 strain after growth into stationary phase and after nutrient starvation.
As BY4741 is grown in SD complete medium, we find that tRNAHis m5C levels increase from 0.92 mol/mol tRNA in mid-log phase growth to 1.55 mol/mol tRNA in the first day of stationary phase and then gradually increase to 1.81 mol/mol tRNA over the next 5 d of stationary phase (Fig. 3, black bars). In contrast, during this time course, the control modifications (Am, Gm+ m1G, D, and ψ) remain virtually unchanged in the tRNAHis (Fig. 3, light and dark gray bars; data not shown). The increase in m5C seems to correlate with the diauxic shift, which transitions energy production from fermentation to respiration when glucose levels are low (Herman 2002). Accumulation of m5C is not likely due to cell death since cells in stationary phase are known to be viable (Werner-Washburne et al. 1996; Allen et al. 2006).
FIGURE 3.

tRNAHis m5C levels increase when BY4741 is grown to stationary phase. tRNAHis was purified from BY4741 grown in SD complete media in mid-log phase and in stationary phase for the indicated number of days. Nucleoside modifications were analyzed by HPLC, and m5C (black), Am (dark gray), and m1G + Gm (light gray) values are plotted over time. Other control modifications (dihydrouridine and pseudouridine) were unchanged during the time course (data not shown). The increase in m5C levels between log phase growth and Day 1 of stationary phase is highly reproducible (Day 0 log phase, m5C = 0.86 ± 0.03, n = 6; Day 1 stationary phase, m5C = 1.53 ± 0.11, n = 7, mean ± standard deviation) (see Table 3).
We also find that levels of tRNAHis m5C increase following several nutrient starvation treatments of the BY4741 strain, in each case with no significant loss of cell viability (Table 3). We find that m5C levels increase significantly following 6 h of starvation for histidine (from 0.86 to 1.80 mol/mol tRNA) and following 24 h of starvation for histidine (to 2.00 mol/mol tRNA), for leucine (to 1.66 mol/mol tRNA), and for a combination of amino acids and uracil (SD minimal; to 1.68 mol/mol tRNA). m5C levels also significantly increase to 1.53 mol/mol tRNA following 24 h of growth in SD complete medium, because the cells had reached saturation. In contrast, we observe a more modest increase in m5C levels following 24 h of starvation for glucose (to 1.32 mol/mol tRNA), and a minimal increase following 24 h of starvation for uracil (to 1.23 mol/mol tRNA). The BY4741 strain viable cell titer remained constant during all of these treatments (Table 3), although cell growth ceased by ∼3 h in each case (data not shown).
TABLE 3.
tRNAHis m5C levels increase following starvation for amino acids or glucose
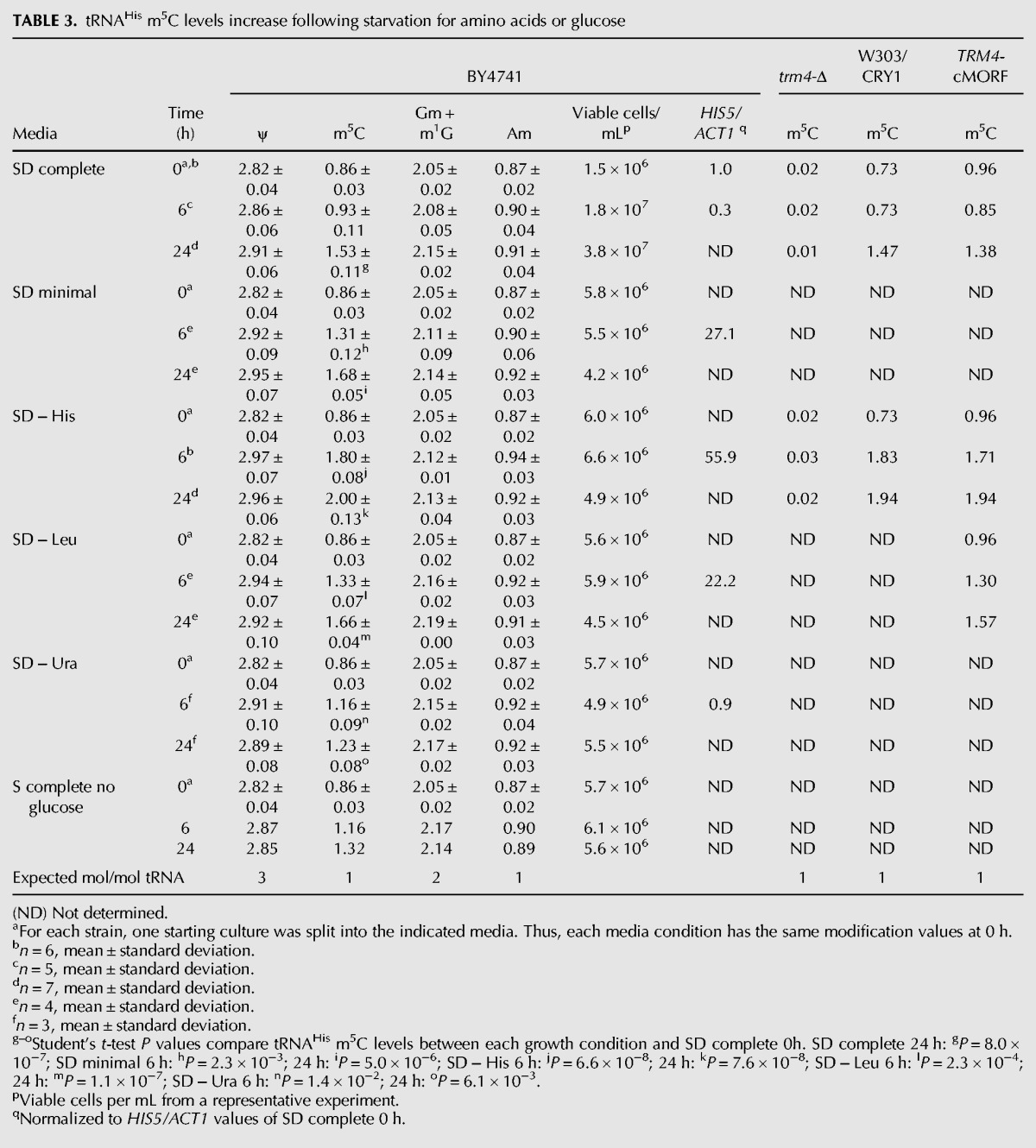
It is likely that Trm4 is responsible for the additional m5C modifications observed in tRNAHis under these conditions, for two reasons. First, Trm4 is the only known S. cerevisiae m5C methyltransferase and can catalyze m5C formation on substrate tRNAs at C34, C40, C48, and C49 (Motorin and Grosjean 1999). Second, trm4-Δ mutants depleted of Thg1 lack all m5C, including the additional m5C found in tRNAHis at C48 and C50 (Gu et al. 2005). Consistent with this, we find that deletion of TRM4 abolishes the accumulation of m5C in tRNAHis that occurs upon histidine starvation of BY4741 (Table 3), suggesting that Trm4 catalyzes formation of the additional m5C on tRNAHis during starvation conditions.
The GCN4 pathway is not responsible for additional m5C levels on tRNAHis
Since we observed significant increases in m5C on tRNAHis after starving BY4741 for amino acids and only a marginal increase following starvation for uracil, we reasoned that additional m5C might result from activation of the general amino acid starvation pathway. Indeed, we had previously shown that depletion of Thg1 leads to activation of the GCN4 pathway, in addition to the accumulation of m5C on tRNAHis (Gu et al. 2005). Thus, as expected, our amino acid starvation conditions lead to increased levels of both HIS5 and LYS1 mRNAs (Table 3; data not shown), which are known to be under GCN4 control (Natarajan et al. 2001).
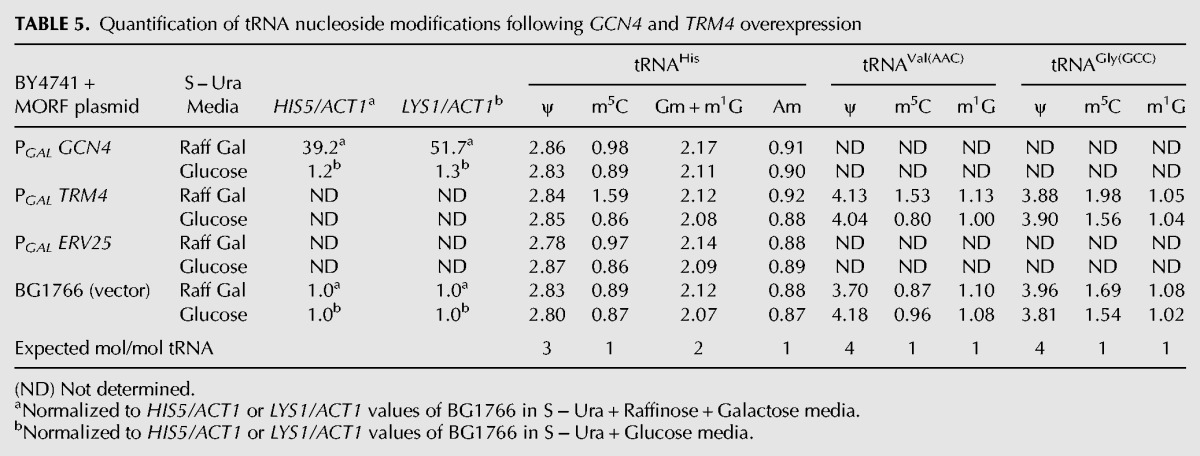
Two lines of evidence suggest that the increased m5C levels observed in tRNAHis during starvation are not due to the GCN4 pathway. First, we find nearly identical levels of elevated m5C in tRNAHis after 5 h of histidine starvation of wild-type (0.86 to 1.71 mol/mol tRNA) and gcn4-Δ strains (0.83 to 1.65 mol/mol tRNA) (Table 4), while control modifications (D, ψ, Gm, m1G, and Am) are virtually unchanged (Table 4; data not shown). Second, induction of the GCN4 pathway by overexpression of GCN4 from the PGAL promoter, using the yeast movable ORF (MORF) collection (Gelperin et al. 2005), does not affect m5C levels of tRNAHis (Table 5). Accordingly, although overexpression of GCN4-MORF in galactose dramatically increases mRNA levels of the known Gcn4 targets HIS5 and LYS1 relative to levels observed in glucose-containing media, there is only a very limited effect on m5C levels of tRNAHis (from 0.89 to 0.98 mol/mol tRNA). Similarly, there is no effect on m5C levels of tRNAHis when the vector control strain is grown in galactose (from 0.87 to 0.89 mol/mol tRNA) or when a control MORF construct (ERV25-MORF) is expressed in galactose (from 0.86 to 0.97 mol/mol tRNA). However, we note that our growth conditions do allow for an increase in tRNAHis m5C, since overexpression of TRM4-MORF increases tRNAHis m5C levels to 1.59 mol/mol tRNA (Table 5). Thus, we conclude that tRNAHis m5C levels are not the result of GCN4 pathway induction.
TABLE 4.
tRNAHis m5C levels during inhibition of starvation pathways

TABLE 5.
Quantification of tRNA nucleoside modifications following GCN4 and TRM4 overexpression
tRNAHis m5C levels increase during prolonged inhibition of the target of rapamycin pathway
We also tested the role of the TOR pathway in the induction of additional m5C. The TOR pathway is another mechanism by which yeast respond to nutrient deprivation. In yeast, Tor1 and Tor2 can form two distinct multiprotein complexes: TORC1 (containing either Tor1 or Tor2), which is inhibited by rapamycin; and TORC2 (containing Tor2), which is insensitive to rapamycin (Loewith et al. 2002). To inhibit TORC1, we sought to treat cells with rapamycin, but BY4741 is relatively insensitive to rapamycin treatment when TOR1 is present (data not shown). Therefore, we used the W303/CRY1 strain, after first showing that BY4741 and W303/CRY1 had virtually identical increases in tRNAHis m5C levels after histidine starvation, while all other modifications (D, ψ, Gm, m1G, and Am) were unaffected by starvation (Table 3; data not shown). Growth arrest triggered by rapamycin treatment (50, 100, and 150 nM) was apparent by 1.5–2 h, and we observed significant decreases in cell viability after treatment with 100 nM and 150 nM rapamycin for longer than 4.5 h (data not shown). However, we did observe increases in m5C levels following several treatments with rapamycin that did not lead to loss of viability. We detected a modest increase in tRNAHis m5C levels when cells were treated with 150 nM rapamycin for 3 h (from 0.89 ± 0.01 to 1.18 ± 0.03 mol/mol tRNA, mean ± standard deviation; n = 2) (data not shown) and greater increases when cells were treated with 100 nM rapamycin for 4.5 h (from 0.84 to 1.39 mol/mol tRNA) (Table 4) or with 50 nM rapamycin for 24 h (from 0.84 to 1.73 mol/mol tRNA) (data not shown), without loss of viability. In each of these cases, we observe no change in all other tRNAHis modifications (Table 4; data not shown). These data suggest that inhibition of the TOR pathway or the resulting G1 cell cycle arrest (Kunz et al. 1993; Barbet et al. 1996; Helliwell et al. 1998) can trigger additional m5C levels on tRNAHis.
Trm4 protein levels do not increase during starvation conditions
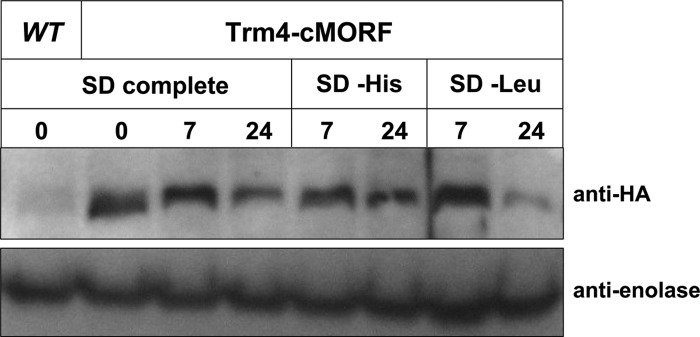
A simple explanation for the increase in tRNAHis m5C levels is that Trm4 is more highly expressed during starvation conditions. To monitor endogenous Trm4 levels during starvation, we first inserted a chromosomal affinity tag derived from the MORF library (Gelperin et al. 2005) immediately 5′ of the TRM4 stop codon to encode a Trm4-cMORF fusion. We then assayed tRNAHis m5C levels before and after amino acid starvation, demonstrating that the tag did not alter levels of m5C in tRNAHis compared to those in a wild-type strain (Table 3), and using cells from the same growth, we extracted cell lysate and measured Trm4 protein levels by Western blotting for the HA epitope. Control modifications (D, ψ, Gm, m1G, and Am) remained constant over the time course of growth (data not shown). However, although tRNAHis m5C levels are elevated at 24 h in both the wild-type and the TRM4-cMORF strain (Table 3), we find that Trm4 protein levels appear to decrease at the 24-h time point (Fig. 4), consistent with experiments that demonstrate reductions in TRM4 mRNA levels (Natarajan et al. 2001) and ribosome occupancy (Ingolia et al. 2009) following starvation.
FIGURE 4.
Trm4-cMORF levels do not increase during starvation. BY4741 and Trm4-cMORF strains were grown in SD complete, SD – His, or SD – Leu media and grown for 24 h at 30°C. Crude extracts were prepared at 0, 7, and 24 h, and 30 μg of protein were loaded in each lane and resolved by SDS-PAGE, followed by Western blotting. Anti-HA antibody was used to detect the cMORF tag on Trm4. BY4741 crude extract was used as a negative control. The same blot was incubated with anti-enolase antibody as a loading control.
Aminoacylation status of tRNAHis is variably affected by different starvation conditions
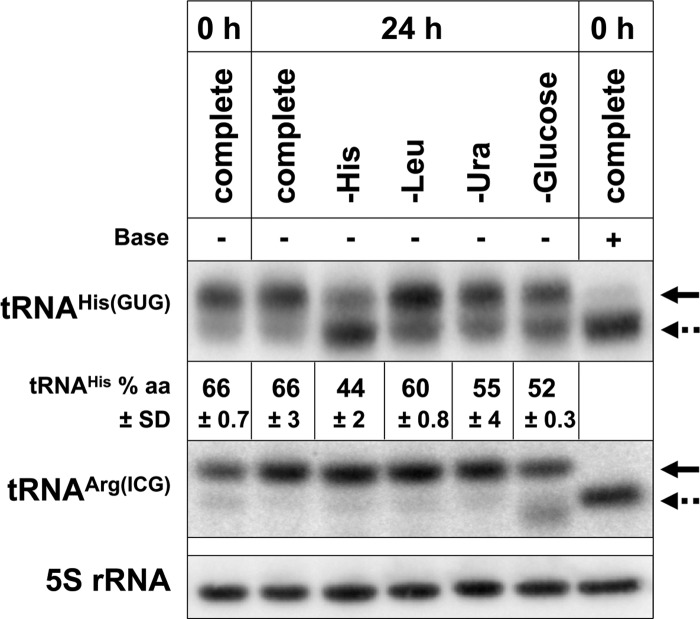
Starvation for a particular amino acid in yeast and in Escherichia coli has been shown to affect the charging status of the corresponding tRNA isoacceptors, with some effects on other tRNAs (Dittmar et al. 2005; Zaborske et al. 2009). Therefore, tRNAHis aminoacylation status may be affected by the starvation conditions that we examined, and changes in tRNAHis aminoacylation levels might correlate with the amount of additional m5C added to tRNAHis. To address this possibility, we starved the BY4741 strain for histidine, leucine, uracil, or glucose, or grew BY4741 to stationary phase and extracted RNA in acidic conditions to preserve tRNA aminoacylation. We performed the experiment with two independent cultures which serve as biological replicates. A Northern blot of samples from one of the replicates is shown in Figure 5. Prior to starvation, when the cultures are growing in log phase (SD complete 0 h), tRNAHis is 66% aminoacylated. Following 24 h of starvation for histidine, leucine, uracil, or glucose, tRNAHis is 44%, 60%, 55%, and 52% aminoacylated, respectively. Furthermore, following 24 h of growth in SD complete medium (stationary phase), tRNAHis is 66% aminoacylated. These data suggest that aminoacylation levels do not directly correlate with additional m5C levels. Although histidine starvation induces the most additional m5C and the tRNAHis is the most deacylated, we observe no change in aminoacylation levels of tRNAHis following entry into stationary phase (Fig. 5), although tRNAHis m5C levels increase by 0.67 mol/mol tRNA (Table 3). In addition, tRNAHis is more aminoacylated following leucine starvation than following uracil or glucose starvation (Fig. 5), but leucine starvation induces more additional m5C than uracil or glucose (Table 3).
FIGURE 5.
Analysis of tRNAHis aminoacylation under different starvation conditions. RNA from the wild-type (BY4741) strain grown in the indicated media was isolated in acidic conditions to preserve aminoacylation, and 2 μg of RNA were resolved on an acidic gel, analyzed by Northern blot for tRNAHis, tRNAArg(ICG), and 5S rRNA, and quantified using ImageQuant software. A control sample was treated with base (+) to detect migration of deacylated tRNA. (Solid arrows) Aminoacyl-tRNA, (dotted arrows) deacyl-tRNA. Each percent aminoacylation value is the mean ± standard deviation (SD) from two independent cultures. A representative Northern blot from one of the two independent cultures is shown.
Increases in m5C levels are unique to tRNAHis
Since additional m5C on tRNAHis occurs at C48 and C50 (adjacent to m5C49) in Thg1-depleted cells (Gu et al. 2005), we examined tRNAGly(GCC) and tRNAVal(AAC), two of the four additional tRNAs known to have m5C49 and unmodified C48 and C50 (Juhling et al. 2009). Consistent with results from Thg1-depleted cells, we find little change in m5C levels of these two tRNA species after starvation of wild-type cells (Table 6). Whereas histidine starvation for 24 h results in a large increase in tRNAHis m5C levels (from 0.86 to 2.00 mol/mol tRNA), tRNAGly(GCC) m5C levels increase modestly (from 1.44 to 1.74 mol/mol tRNA), and tRNAVal(AAC) m5C levels barely increase (from 0.79 to 1.03 mol/mol tRNA) (Fig. 6; Table 6). Similarly, leucine starvation for 24 h results in a substantial increase in tRNAHis m5C levels (from 0.86 to 1.66 mol/mol tRNA) but only modest increases in m5C levels of tRNAGly(GCC) (from 1.44 to 1.70 mol/mol tRNA) and tRNAVal(AAC) (from 0.79 to 0.98 mol/mol tRNA). Control modification levels for each tRNA remain normal during these treatments (Table 6; data not shown). Furthermore, m5C levels only slightly increase in tRNAVal(AAC) when temperature-sensitive strains are shifted to 37°C, relative to the drastic induction of additional m5C in tRNAHis (Fig. 2B; Table 2). Thus, we infer that m5C accumulation occurs preferentially on tRNAHis in conditions where cells stop growing.
TABLE 6.
tRNAHis, tRNAVal(AAC), and tRNAGly(GCC) m5C levels during starvation
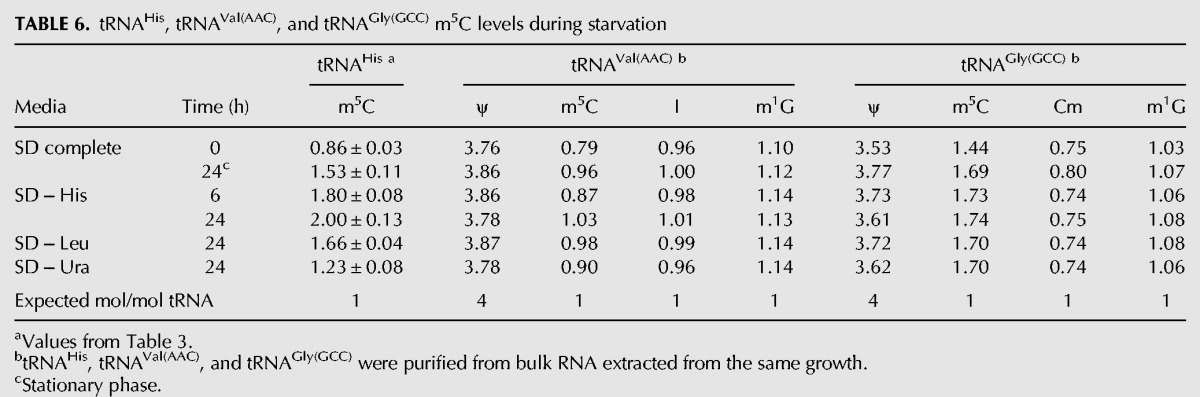
FIGURE 6.

m5C increases specifically on tRNAHis during starvation. tRNAHis (black), tRNAVal(AAC) (dark gray), and tRNAGly(GCC) (light gray) were purified from BY4741 grown in the indicated starvation conditions. Nucleoside modifications were analyzed by HPLC, and m5C values are plotted over time for each tRNA.
In support of this claim, both tRNAVal(AAC) and tRNAGly(GCC) have the capacity to acquire additional m5C after overexpression of TRM4-MORF (Table 5). Strikingly, TRM4 overexpression results in a nearly identical increase in m5C levels of tRNAHis (from 0.86 to 1.59 mol/mol tRNA) and tRNAVal(AAC) (from 0.80 to 1.53 mol/mol tRNA) and a smaller but still significant increase in m5C levels of tRNAGly(GCC) (from 1.56 to 1.98 mol/mol tRNA), whereas all other control modifications (D, ψ, Gm, m1G, I, Cm, and Am) are normal (Table 5; data not shown). Since both tRNAVal(AAC) and tRNAGly(GCC) can be modified with additional m5C in vivo by Trm4 but are not substrates for additional m5C modification (presumably by Trm4) during growth arrest conditions, we conclude that the additional m5C during growth arrest is specific to tRNAHis.
DISCUSSION
We have demonstrated here that m5C levels of tRNAHis are unusually responsive to a number of different conditions in which yeast cells stop growing. We had previously shown that tRNAHis accumulates additional m5C at C48 and C50 in the variable loop, adjacent to m5C49, following depletion of Thg1 and the subsequent growth arrest (Gu et al. 2005). We have extended this analysis to show that tRNAHis has additional m5C when either thg1 temperature-sensitive strains or temperature-sensitive strains unrelated to tRNA function are grown at nonpermissive temperature and when wild-type strains such as BY4741 are grown to saturation or are starved for histidine, amino acids, leucine, or (to a lesser extent) for glucose or uracil. In the majority of these cases, the accumulation of additional m5C on tRNAHis is not caused by loss of cell viability. In addition, we have shown that the accumulation of additional m5C on tRNAHis is not due to induction of the GCN4 pathway and is not due to an increase in Trm4 protein levels during our starvation conditions, although the additional m5C modification is absent in a trm4-Δ mutant. Remarkably, this additional m5C specifically occurs on tRNAHis, and not on two other possible substrates, tRNAVal(AAC) or tRNAGly(GCC), that also have m5C49 flanked by cytidines, although these tRNA species can acquire more m5C when Trm4 is overproduced.
The molecular signals that trigger Trm4 to modify tRNAHis with additional m5C during growth arrest conditions remain unclear. Although tRNAHis localizes to the nucleus following Thg1 depletion (Gu et al. 2005), and Trm4 is a nuclear protein (Wu et al. 1998), our data suggest that modulation of tRNA subcellular dynamics is not the sole contributor to additional m5C levels, since glucose starvation results in rapid accumulation of tRNAs in the nucleus (Shaheen and Hopper 2005; Whitney et al. 2007), but we observe only modest accumulation of m5C relative to that observed with histidine or leucine starvation. Three lines of evidence suggest that tRNA charging status is also not the sole contributor to additional m5C on tRNAHis. First, tRNAHis from a thg1-Δ strain has normal m5C levels, despite the fact that the tRNA is mostly deacylated (Preston and Phizicky 2010). Second, tRNAHis has an additional 0.80 mol m5C/mol tRNA (Table 3) following 24 h of leucine starvation, although tRNAHis aminoacylation status is virtually unaffected (Fig. 5). Third, tRNAHis m5C increases by 0.67 mol/mol tRNA (Table 3) when wild-type cells are grown to stationary phase and tRNAHis aminoacylation levels are unaltered (Fig. 5).
However, it remains possible that both deacylation of tRNAHis and its presence in the nucleus contribute to the additional m5C that we observe. This would be consistent with the observation that more m5C accumulates on tRNAHis during histidine starvation than during other starvation conditions. Furthermore, increased m5C on tRNAHis occurs when tRNAs are known to accumulate in the nucleus: during starvation for amino acids, starvation for glucose, and as cells enter late-log and near-stationary phase (Whitney et al. 2007). Indeed, we previously showed that the additional m5C that accumulates upon Thg1 depletion occurs at a time when tRNAHis is deacylated and is localized to the nucleus (Gu et al. 2005), and it is known that both charged and uncharged tRNA can accumulate in the nucleus during starvation (Whitney et al. 2007). It is not completely clear why different levels of tRNAHis m5C arise during leucine starvation compared to glucose starvation; however, it is clear that different stress conditions have different effects on the cell since they elicit transcriptionally distinct types of growth arrest (Gasch et al. 2000; Boer et al. 2008; Brauer et al. 2008; Klosinska et al. 2011).
The specificity of additional m5C modification for tRNAHis is also puzzling. Although overproduction of Trm4 results in similar levels of additional m5C on tRNAHis, tRNAVal(AAC), and tRNAGly(GGC), the additional m5C is much more specific for tRNAHis after temperature shift of fcp1ts or thg1ts strains, after starvation of a wild-type strain for histidine or leucine, and after growth to stationary phase. Therefore, we presume that, under these conditions, tRNAHis somehow becomes more accessible to Trm4, or Trm4 activity is altered, but it is not yet clear how this is accomplished. This specificity for tRNAHis might possibly be explained by a unique structural property of tRNAHis or by a secondary protein that binds tRNAHis during growth arrest. One possibility is that specificity is related to the unique sensitivity of histidyl-tRNAHis to deacylation in vitro (Chernyakov et al. 2008a; Preston and Phizicky 2010), and the observation that tRNAHis is partially deacylated when isolated (Gu et al. 2005; Preston and Phizicky 2010), which could mean that this tRNA species is a cellular sensor for poor growth conditions.
Although our data demonstrate that the additional m5C levels of tRNAHis during histidine starvation accumulate independently of the GCN4 gene and are not provoked by induction of the GCN4 pathway, it is not clear if the accumulation of m5C of tRNAHis involves the TOR pathway, which also plays a role in stationary phase and quiescence (Herman 2002). We observe a distinct increase in tRNAHis m5C levels when cells are treated with rapamycin, but this increase is modest compared to other treatments. However, because rapamycin treatment results in G1 phase cell cycle arrest (Kunz et al. 1993; Barbet et al. 1996; Helliwell et al. 1998), it is difficult to determine whether the increased m5C is due to inhibition of the TOR pathway or to the resulting growth arrest. It is intriguing to speculate that the TOR pathway is connected to the accumulation of m5C in tRNAHis, because high-throughput studies have reported that deletion of TRM4 results in increased rapamycin sensitivity (Parsons et al. 2004) and that overexpression of TRM4 confers rapamycin resistance (Butcher et al. 2006), although we have not been able to observe such effects (data not shown).
The function of the additional m5C of tRNAHis has not yet been elucidated. Since overexpression of TRM4-MORF appears to increase m5C levels of several tRNA species and causes a slow-growth phenotype (Yoshikawa et al. 2011; data not shown; JM Dewe and EM Phizicky, unpubl.), we infer that excess m5C modification has a deleterious effect on function of one or more tRNAs. Conversely, m5C modification of tRNAs can play a protective role during stress conditions. Amino acid starvation in Tetrahymena and oxidative stress in yeast, humans, and plants results in cleavage of tRNAs in the anti-codon loop (Lee and Collins 2005; Thompson et al. 2008), and the Drosophila m5C methyltransferase Dnmt2 is important for viability during oxidative stress and heat shock conditions (Schaefer et al. 2010), because its modification of C38 in the anti-codon loop protects substrate tRNAs from cleavage by angiogenin in both Drosophila and mice (Goll et al. 2006; Schaefer et al. 2010). Thus, the additional m5C of tRNAHis may have a protective function, similar to the role of m5C49 of tRNAVal(AAC) in reducing the extent of rapid decay of tRNAs lacking m7G46 (Alexandrov et al. 2006; Chernyakov et al. 2008b; Dewe et al. 2012). Intriguingly, while preparing this manuscript, it was reported that hydrogen peroxide treatment leads to alteration of m5C levels in yeast tRNALeu(CAA), with 70% more m5C34 and 20% less m5C48, and these changes were associated with increased expression of a ribosomal protein enriched with TTG codons (Chan et al. 2012). Further experiments will be required to fully understand the biology of m5C, the cause of the additional m5C of tRNAHis, and the role of the additional m5C during growth arrest.
MATERIALS AND METHODS
Strain construction
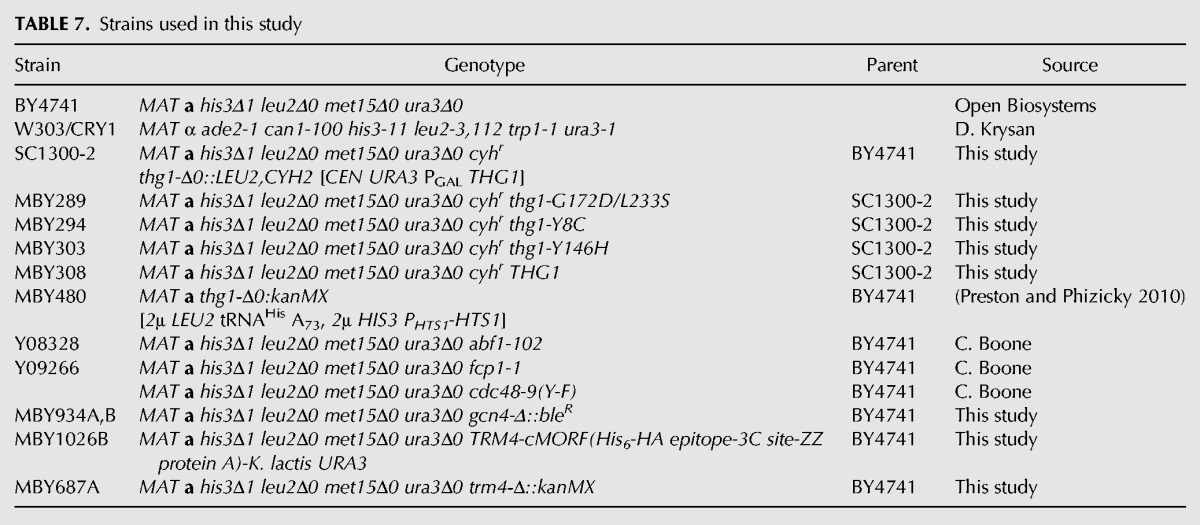
Strains used in this study are listed in Table 7.
TABLE 7.
Strains used in this study
To construct the gcn4-Δ::bleR (MBY934) strain, we amplified the phleomycin resistance cassette from the pUG66 plasmid (Gueldener et al. 2002) with GCN4 Up 179 + Phleo Forward (5′-ATCATGTACCCGTAGAATTTTATTCAAGATGTTTCCGTAACGGCAGCTGAAGCTTCGTAC-3′) and GCN4 Down 312 + Phleo Reverse (5′-GCATTAGCTATAACACGTTAATATGGTGGAGTCAGCTGAGAAGGCATAGGCCACTAGTGG-3′) to add 43-nt sequence upstream of and downstream from GCN4 to the Phleo cassette 5′ and 3′ ends, respectively. We extended the GCN4 homologous regions to 87 nt by amplifying the first PCR product with GCN4 Up 223 (87) Forward (5′-ACTGTCAGTTTTTTGAAGAGTTATTTGTTTTGTTACCAATTGCTATCATGTACCCGTAGA-3′) and GCN4 Down 356 (87) Reverse (5′-CATGAGTACTCCTAAATAGGGCGATATTTTAAAGTTTCATTCCAGCATTAGCTATAACAC-3′). This extended PCR product was transformed into BY4741 as previously described (Sherman 1991) and selected on YPD media containing 8 μg/mL phleomycin (Chernyakov et al. 2008b). The gcn4-Δ::bleR strain was verified by PCR.
The TRM4-cMORF strain, MBY1026B, was constructed in a manner similar to the gcn4-Δ::bleR strain. We used the plasmid, AVA0258, which encodes the cMORF tag (consisting of His6, HA epitope, 3C protease site, ZZ protein A), followed by Kluyveromyces lactis URA3 as a marker for insertion into the chromosome. We PCR-amplified this construct using TRM4 C-ter + TAP 5′ (5′-GAACCTCTACTGAAGCTCCTAGCGCTGCTAATAACCCAGCTTTCTTGTACAAAGTGG-3′; contains 43 nt corresponding to the C terminus of Trm4 without the stop codon and a 5′ portion of the cMORF tag sequence), and TRM4 Downstream (w/stop) + TAP 3′ (5′-CTTTACAGTGGAGGGGATAAGAAACATGATAACTATCATACGACTCACTATAGGG-3′; inserts the Trm4 stop codon after the cMORF tag and URA3 marker and contains 43 nt of the region downstream from TRM4). We then used this PCR product to extend the regions homologous to TRM4 by PCR amplification with TRM4 C-ter extended (TAP) 5′ (5′-GACTGAATCTCCCGCAGAAACTACTACCGGAACCTCTACTGAAGCTC-3′) and TRM4 Downstream extended (TAP) 3′ (5′-AGTATTATATTCTTATTTTTGCCTTTTAATAATATACATTTACTTTACAGTGGAGGGGAT-3′). The resulting strain was verified by PCR.
We constructed thg1 temperature-sensitive (thg1ts) strains, MBY294 (thg1-Y8C), MBY289 (thg1-G172D,L233S), and MBY303 (thg1-Y146H) in two steps. First, we randomly mutagenized a single copy THG1 plasmid, screened for temperature sensitivity in a thg1-Δ strain, and sequenced the resulting alleles. We then inserted these thg1ts alleles and wild-type THG1 into the THG1 locus by transformation of the SC1300-2 strain in which THG1 was deleted with a LEU2,CYH2 cassette in a cyhr background, and selected for cycloheximide resistance, accompanied by leucine auxotrophy. Temperature sensitivity was confirmed and complemented by a wild-type copy of THG1.
Strain starvation
Strains were grown to log phase at 30°C in SD complete media, then spun down and resuspended in the following media conditions: SD complete, SD minimal (contains only yeast nitrogen base and glucose), SD − His, SD − Leu, SD − Ura, or S complete no glucose. These cultures were grown for 24 h at 30°C, and cells were harvested for analysis of tRNA modifications at indicated times.
Temperature-sensitive strain growth
Strains were grown to log phase in YPD at 24°C, diluted to OD600= 0.5 and grown at either 24°C or 37°C for the time indicated.
Rapamycin treatment of W303/CRY1
W303 was pretreated with DMSO by growing in YPD media containing 1% DMSO overnight until the strain reached early log phase. Cells were harvested prior to drug treatment and after growth in the presence of the indicated concentration of rapamycin or 1% DMSO for the indicated amount of time.
Assessment of viable cell titer
For each condition, a given volume of culture was plated onto YPD plates. Cells were grown for 3 d at 30°C or at 24°C for temperature-sensitive strains, and colonies were counted and normalized to the volume plated to calculate viable cells per milliliter of culture.
Bulk RNA isolation and purification of tRNA
Bulk low molecular weight RNA was isolated from 150 to 300 OD of yeast cells that were grown in conditions described above, using a hot phenol extraction method, as described elsewhere (Kotelawala et al. 2008). Total RNA was extracted from stationary phase cells by lysis with glass beads, phenol-chloroform extraction, and ethanol precipitation, as described previously (Letzring et al. 2010). tRNAs were purified using 5′ biotinylated DNA oligomers complementary to the following: nt 48–72 for tRNAHis (5′ Bio tRNAHis: 5′-/Biotin/GCCATCTCCTAGAATCGAACCAGGG-3′) (Preston and Phizicky 2010), nt 52–76 for tRNAGly(GCC) (BioGly: 5′-/Biotin/TGGTGCGCAAGCCCGGAATCGAACC-3′) (Gu et al. 2005), and nt 55–76 for tRNAVal(AAC) (Biotin-tRNAVal1: 5′-/Biotin/TGGTGATTTCGCCCAGGATCGA-3′). For each tRNA purified, 22.5 pmol of oligomer were first bound to streptavidin magnetic particles (Roche). Then, bulk RNA (1–1.25 mg) was added to oligomer-bound beads in the presence of 2.4 M tetraethylammonium chloride (TEACl; Sigma), washed, and the tRNA was melted off the oligomer at 60°C to obtain pure tRNA. The resulting tRNA was desalted and concentrated using Amicon Ultra 4 10,000 MWCO columns (Millipore).
HPLC analysis of tRNA nucleosides
tRNAs (1.25 μg) were digested at 37°C for at least 2 h using 0.5 μg P1 nuclease (MP Biomedicals) in a buffer containing 20 mM NaOAc pH 5.2 and 0.2 mM ZnCl2 and then treated with calf intestinal phosphatase (Roche) for at least 1 h. Nucleosides were resolved by reverse-phase HPLC essentially as described (Gehrke and Kuo 1989), and each nucleoside was identified and quantified as described previously (Jackman et al. 2003; Kotelawala et al. 2008).
Quantitative RT-PCR
RNA was extracted using the hot phenol extraction method (Kotelawala et al. 2008) and treated with RQ1 RNase-free DNase (Promega), followed by a phenol-chloroform extraction, two chloroform extractions, and ethanol precipitation. This RNA was reverse transcribed with Superscript II Reverse Transcriptase (Invitrogen) using Random Primers (Invitrogen). Next, this DNA was PCR-amplified using Fast SYBR Green master mix (Applied Biosystems) and 0.2 μM each of 5′ and 3′ primers specific to ACT1 (ACT1 Set 1 Forward 5′-ACGTTCCAGCCTTCTACGTTTCCA-3′ and ACT1 Set 1 Reverse 5′-ACGTGAGTAACACCATCACCGGAA-3′, HIS5 (HIS5 Set 1 Forward 5′-AATGCCCATGGACCTACTCCAGTT-3′ and HIS5 Set 1 Reverse 5′-ACACCTAGGCACAGATTGTCAGCA-3′), or LYS1 (LYS1 Set 1 Forward 5′-AGCAGACACTACCAACCCTCACAA-3′ AND LYS1 Set 1 Reverse 5′-CTTGGCAGCAAAGAAGGCAAGTGA-3′), with the following amplification scheme: 95°C for 20 sec and then 40 cycles of 95°C for 3 sec and 60°C for 30 sec.
Analysis of aminoacylated RNA
RNA was isolated from 50 OD of cells in acidic conditions (pH 4.5), and 2 μg of RNA were resolved by PAGE under acidic conditions, as previously described (Chernyakov et al. 2008a), and transferred to Hybond N+ membrane (Amersham Biosciences). The membrane was UV cross-linked, hybridized with 5′-labeled oligomers tRNAHis (40–64) (5′-CTAGAATCGAACCAGGGTTTCATC-3′), ArgP1 (5′-TAGCCAGACGCCGTGAC-3′), and 5S RNA (5′-GGT AGATATGGCCGCAACC-3′) to detect tRNAHis, tRNAArg (ICG), and 5S rRNA, and visualized with a Typhoon PhosphorImager (GE Healthcare).
ACKNOWLEDGMENTS
We thank Elizabeth Grayhack for invaluable insight and discussions throughout this work. We also thank Charlie Boone (U. Toronto) for the gift of temperature-sensitive yeast strains and Mark Dumont (U. Rochester) for the gift of enolase antibody. We thank Jason Salter and Jane Jackman for constructing the SC1300-2 strain used to generate the thg1ts strains and Marv Wickens (U. Wisconsin-Madison) for the use of laboratory equipment to complete the experiment in Figure 5. This research is supported by NIH Grant GM52347 to E.M.P. M.A.P. was supported by NIH Training Grant in Cellular, Biochemical and Molecular Sciences 5T32 GM068411.
REFERENCES
- Agris PF, Koh H, Soll D 1973. The effect of growth temperatures on the in vivo ribose methylation of Bacillus stearothermophilus transfer RNA. Arch Biochem Biophys 154: 277–282 [DOI] [PubMed] [Google Scholar]
- Agris PF, Vendeix FA, Graham WD 2007. tRNA’s wobble decoding of the genome: 40 years of modification. J Mol Biol 366: 1–13 [DOI] [PubMed] [Google Scholar]
- Alexandrov A, Chernyakov I, Gu W, Hiley SL, Hughes TR, Grayhack EJ, Phizicky EM 2006. Rapid tRNA decay can result from lack of nonessential modifications. Mol Cell 21: 87–96 [DOI] [PubMed] [Google Scholar]
- Allen C, Buttner S, Aragon AD, Thomas JA, Meirelles O, Jaetao JE, Benn D, Ruby SW, Veenhuis M, Madeo F, et al. 2006. Isolation of quiescent and nonquiescent cells from yeast stationary-phase cultures. J Cell Biol 174: 89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbet NC, Schneider U, Helliwell SB, Stansfield I, Tuite MF, Hall MN 1996. TOR controls translation initiation and early G1 progression in yeast. Mol Biol Cell 7: 25–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjork GR, Jacobsson K, Nilsson K, Johansson MJ, Bystrom AS, Persson OP 2001. A primordial tRNA modification required for the evolution of life? EMBO J 20: 231–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer VM, Amini S, Botstein D 2008. Influence of genotype and nutrition on survival and metabolism of starving yeast. Proc Natl Acad Sci 105: 6930–6935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauer MJ, Huttenhower C, Airoldi EM, Rosenstein R, Matese JC, Gresham D, Boer VM, Troyanskaya OG, Botstein D 2008. Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast. Mol Biol Cell 19: 352–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman AR, Kimmerly WJ, Rine J, Kornberg RD 1988. Two DNA-binding factors recognize specific sequences at silencers, upstream activating sequences, autonomously replicating sequences, and telomeres in Saccharomyces cerevisiae. Mol Cell Biol 8: 210–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher RA, Bhullar BS, Perlstein EO, Marsischky G, LaBaer J, Schreiber SL 2006. Microarray-based method for monitoring yeast overexpression strains reveals small-molecule targets in TOR pathway. Nat Chem Biol 2: 103–109 [DOI] [PubMed] [Google Scholar]
- Chan CT, Dyavaiah M, DeMott MS, Taghizadeh K, Dedon PC, Begley TJ 2010. A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet 6: e1001247 10.1371/journal.pgen.1001247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CT, Pang YL, Deng W, Babu IR, Dyavaiah M, Begley TJ, Dedon PC 2012. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat Commun 3: 937 10.1038/ncomms1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernyakov I, Baker MA, Grayhack EJ, Phizicky EM 2008a. Identification and analysis of tRNAs that are degraded in Saccharomyces cerevisiae due to lack of modifications. Methods Enzymol 449: 221–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernyakov I, Whipple JM, Kotelawala L, Grayhack EJ, Phizicky EM 2008b. Degradation of several hypomodified mature tRNA species in Saccharomyces cerevisiae is mediated by Met22 and the 5′-3′ exonucleases Rat1 and Xrn1. Genes Dev 22: 1369–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewe JM, Whipple JM, Chernyakov I, Jaramillo LN, Phizicky EM 2012. The yeast rapid tRNA decay pathway competes with elongation factor 1A for substrate tRNAs and acts on tRNAs lacking one or more of several modifications. RNA 18: 1886–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmar KA, Sorensen MA, Elf J, Ehrenberg M, Pan T 2005. Selective charging of tRNA isoacceptors induced by amino-acid starvation. EMBO Rep 6: 151–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Yacoubi B, Lyons B, Cruz Y, Reddy R, Nordin B, Agnelli F, Williamson JR, Schimmel P, Swairjo MA, de Crecy-Lagard V 2009. The universal YrdC/Sua5 family is required for the formation of threonylcarbamoyladenosine in tRNA. Nucleic Acids Res 37: 2894–2909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11: 4241–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrke CW, Kuo KC 1989. Ribonucleoside analysis by reversed-phase high-performance liquid chromatography. J Chromatogr 471: 3–36 [DOI] [PubMed] [Google Scholar]
- Gelperin DM, White MA, Wilkinson ML, Kon Y, Kung LA, Wise KJ, Lopez-Hoyo N, Jiang L, Piccirillo S, Yu H, et al. 2005. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev 19: 2816–2826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, Golic KG, Jacobsen SE, Bestor TH 2006. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 311: 395–398 [DOI] [PubMed] [Google Scholar]
- Grunberger D, Weinstein IB, Mushinski JF 1975. Deficiency of the Y base in a hepatoma phenylalanine tRNA. Nature 253: 66–67 [DOI] [PubMed] [Google Scholar]
- Grunberger D, Pergolizzi RG, Kuchino Y, Mushinski JF, Nishimura S 1983. Alterations in post-transcriptional modification of the Y base in phenylalanine tRNA from tumor cells. Recent Results Cancer Res 84: 133–145 [DOI] [PubMed] [Google Scholar]
- Gu W, Jackman JE, Lohan AJ, Gray MW, Phizicky EM 2003. tRNAHis maturation: An essential yeast protein catalyzes addition of a guanine nucleotide to the 5′ end of tRNAHis. Genes Dev 17: 2889–2901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Hurto RL, Hopper AK, Grayhack EJ, Phizicky EM 2005. Depletion of Saccharomyces cerevisiae tRNAHis guanylyltransferase Thg1p leads to uncharged tRNAHis with additional m5C. Mol Cell Biol 25: 8191–8201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueldener U, Heinisch J, Koehler GJ, Voss D, Hegemann JH 2002. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res 30: e23 10.1093/nar/30.6.e23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy MP, Podyma BM, Preston MA, Shaheen HH, Krivos KL, Limbach PA, Hopper AK, Phizicky EM 2012. Yeast Trm7 interacts with distinct proteins for critical modifications of the tRNAPhe anticodon loop. RNA 18: 1921–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell SB, Howald I, Barbet N, Hall MN 1998. TOR2 is part of two related signaling pathways coordinating cell growth in Saccharomyces cerevisiae. Genetics 148: 99–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman PK 2002. Stationary phase in yeast. Curr Opin Microbiol 5: 602–607 [DOI] [PubMed] [Google Scholar]
- Hosbach HA, Kubli E 1979. Transfer RNA in aging Drosophila: II. Isoacceptor patterns. Mech Ageing Dev 10: 141–149 [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida K, Kunibayashi T, Tomikawa C, Ochi A, Kanai T, Hirata A, Iwashita C, Hori H 2011. Pseudouridine at position 55 in tRNA controls the contents of other modified nucleotides for low-temperature adaptation in the extreme-thermophilic eubacterium Thermus thermophilus. Nucleic Acids Res 39: 2304–2318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman JE, Montange RK, Malik HS, Phizicky EM 2003. Identification of the yeast gene encoding the tRNA m1G methyltransferase responsible for modification at position 9. RNA 9: 574–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhling F, Morl M, Hartmann RK, Sprinzl M, Stadler PF, Putz J 2009. tRNAdb 2009: Compilation of tRNA sequences and tRNA genes. Nucleic Acids Res 37: D159–D162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J 2004. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev 18: 1227–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba S, Wang X, Anderson JT 2006. Nuclear RNA surveillance in Saccharomyces cerevisiae: Trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S rRNA. RNA 12: 508–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamenski P, Kolesnikova O, Jubenot V, Entelis N, Krasheninnikov IA, Martin RP, Tarassov I 2007. Evidence for an adaptation mechanism of mitochondrial translation via tRNA import from the cytosol. Mol Cell 26: 625–637 [DOI] [PubMed] [Google Scholar]
- Klosinska MM, Crutchfield CA, Bradley PH, Rabinowitz JD, Broach JR 2011. Yeast cells can access distinct quiescent states. Genes Dev 25: 336–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobor MS, Archambault J, Lester W, Holstege FC, Gileadi O, Jansma DB, Jennings EG, Kouyoumdjian F, Davidson AR, Young RA, et al. 1999. An unusual eukaryotic protein phosphatase required for transcription by RNA polymerase II and CTD dephosphorylation in S. cerevisiae. Mol Cell 4: 55–62 [DOI] [PubMed] [Google Scholar]
- Kotelawala L, Grayhack EJ, Phizicky EM 2008. Identification of yeast tRNA Um44 2′-O-methyltransferase (Trm44) and demonstration of a Trm44 role in sustaining levels of specific tRNASer species. RNA 14: 158–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchino Y, Borek E 1978. Tumour-specific phenylalanine tRNA contains two supernumerary methylated bases. Nature 271: 126–129 [DOI] [PubMed] [Google Scholar]
- Kuchino Y, Shindo-Okada N, Ando N, Watanabe S, Nishimura S 1981. Nucleotide sequences of two aspartic acid tRNAs from rat liver and rat ascites hepatoma. J Biol Chem 256: 9059–9062 [PubMed] [Google Scholar]
- Kuchino Y, Borek E, Grunberger D, Mushinski JF, Nishimura S 1982. Changes of post-transcriptional modification of wye base in tumor-specific tRNAPhe. Nucleic Acids Res 10: 6421–6432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz J, Henriquez R, Schneider U, Deuter-Reinhard M, Movva NR, Hall MN 1993. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell 73: 585–596 [DOI] [PubMed] [Google Scholar]
- Lee SR, Collins K 2005. Starvation-induced cleavage of the tRNA anticodon loop in Tetrahymena thermophila. J Biol Chem 280: 42744–42749 [DOI] [PubMed] [Google Scholar]
- Letzring DP, Dean KM, Grayhack EJ 2010. Control of translation efficiency in yeast by codon-anticodon interactions. RNA 16: 2516–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, Oppliger W, Jenoe P, Hall MN 2002. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell 10: 457–468 [DOI] [PubMed] [Google Scholar]
- Miyake T, Reese J, Loch CM, Auble DT, Li R 2004. Genome-wide analysis of ARS (autonomously replicating sequence) binding factor 1 (Abf1p)-mediated transcriptional regulation in Saccharomyces cerevisiae. J Biol Chem 279: 34865–34872 [DOI] [PubMed] [Google Scholar]
- Motorin Y, Grosjean H 1999. Multisite-specific tRNA:m5C-methyltransferase (Trm4) in yeast Saccharomyces cerevisiae: Identification of the gene and substrate specificity of the enzyme. RNA 5: 1105–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mushinski JF, Marini M 1979. Tumor-associated phenylalanyl transfer RNA found in a wide spectrum of rat and mouse tumors but absent in normal adult, fetal, and regenerating tissues. Cancer Res 39: 1253–1258 [PubMed] [Google Scholar]
- Mushinski JF, Marini M 1983. Tumor-specific tRNA modifications in mouse plasmacytomas and other tumors. Recent Results Cancer Res 84: 121–132 [DOI] [PubMed] [Google Scholar]
- Natarajan K, Meyer MR, Jackson BM, Slade D, Roberts C, Hinnebusch AG, Marton MJ 2001. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol Cell Biol 21: 4347–4368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owenby RK, Stulberg MP, Jacobson KB 1979. Alteration of the Q family of transfer RNAs in adult Drosophila melanogaster as a function of age, nutrition, and genotype. Mech Ageing Dev 11: 91–103 [DOI] [PubMed] [Google Scholar]
- Parsons AB, Brost RL, Ding H, Li Z, Zhang C, Sheikh B, Brown GW, Kane PM, Hughes TR, Boone C 2004. Integration of chemical-genetic and genetic interaction data links bioactive compounds to cellular target pathways. Nat Biotechnol 22: 62–69 [DOI] [PubMed] [Google Scholar]
- Pathak C, Jaiswal YK, Vinayak M 2005. Hypomodification of transfer RNA in cancer with respect to queuosine. RNA Biol 2: 143–148 [DOI] [PubMed] [Google Scholar]
- Phizicky EM, Hopper AK 2010. tRNA biology charges to the front. Genes Dev 24: 1832–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintard L, Lecointe F, Bujnicki JM, Bonnerot C, Grosjean H, Lapeyre B 2002. Trm7p catalyses the formation of two 2′-O-methylriboses in yeast tRNA anticodon loop. EMBO J 21: 1811–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston MA, Phizicky EM 2010. The requirement for the highly conserved G−1 residue of Saccharomyces cerevisiae tRNAHis can be circumvented by overexpression of tRNAHis and its synthetase. RNA 16: 1068–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randerath E, Agrawal HP, Randerath K 1984. Specific lack of the hypermodified nucleoside, queuosine, in hepatoma mitochondrial aspartate transfer RNA and its possible biological significance. Cancer Res 44: 1167–1171 [PubMed] [Google Scholar]
- Reed SH, Akiyama M, Stillman B, Friedberg EC 1999. Yeast autonomously replicating sequence binding factor is involved in nucleotide excision repair. Genes Dev 13: 3052–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, Lyko F 2010. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev 24: 1590–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen HH, Hopper AK 2005. Retrograde movement of tRNAs from the cytoplasm to the nucleus in Saccharomyces cerevisiae. Proc Natl Acad Sci 102: 11290–11295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F 1991. Getting started with yeast. Methods Enzymol 194: 3–21 [DOI] [PubMed] [Google Scholar]
- Singhal RP, Vold B 1976. Changes in transfer ribonucleic acids of Bacillus subtilis during different growth phases. Nucleic Acids Res 3: 1249–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano A, Endo T, Yoshihisa T 2005. tRNA actively shuttles between the nucleus and cytosol in yeast. Science 309: 140–142 [DOI] [PubMed] [Google Scholar]
- Thompson DM, Lu C, Green PJ, Parker R 2008. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA 14: 2095–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomikawa C, Yokogawa T, Kanai T, Hori H 2010. N7-Methylguanine at position 46 (m7G46) in tRNA from Thermus thermophilus is required for cell viability at high temperatures through a tRNA modification network. Nucleic Acids Res 38: 942–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner-Washburne M, Braun EL, Crawford ME, Peck VM 1996. Stationary phase in Saccharomyces cerevisiae. Mol Microbiol 19: 1159–1166 [DOI] [PubMed] [Google Scholar]
- Whipple JM, Lane EA, Chernyakov I, D’Silva S, Phizicky EM 2011. The yeast rapid tRNA decay pathway primarily monitors the structural integrity of the acceptor and T-stems of mature tRNA. Genes Dev 25: 1173–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney ML, Hurto RL, Shaheen HH, Hopper AK 2007. Rapid and reversible nuclear accumulation of cytoplasmic tRNA in response to nutrient availability. Mol Biol Cell 18: 2678–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Brockenbrough JS, Paddy MR, Aris JP 1998. NCL1, a novel gene for a non-essential nuclear protein in Saccharomyces cerevisiae. Gene 220: 109–117 [DOI] [PubMed] [Google Scholar]
- Ye Y, Meyer HH, Rapoport TA 2001. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature 414: 652–656 [DOI] [PubMed] [Google Scholar]
- Yoshikawa K, Tanaka T, Ida Y, Furusawa C, Hirasawa T, Shimizu H 2011. Comprehensive phenotypic analysis of single-gene deletion and overexpression strains of Saccharomyces cerevisiae. Yeast 28: 349–361 [DOI] [PubMed] [Google Scholar]
- Zaborske JM, Narasimhan J, Jiang L, Wek SA, Dittmar KA, Freimoser F, Pan T, Wek RC 2009. Genome-wide analysis of tRNA charging and activation of the eIF2 kinase Gcn2p. J Biol Chem 284: 25254–25267 [DOI] [PMC free article] [PubMed] [Google Scholar]