Abstract
Extraembryonic ectoderm-derived factors instruct the pluripotent epiblast cells to develop toward a restricted primordial germ cell (PGC) fate during murine gastrulation. Genes encoding Bmp4 of the Dpp class and Bmp8b of the 60A class are expressed in the extraembryonic ectoderm and targeted mutation of either results in severe defects in PGC formation. It has been shown that heterodimers of DPP and 60A classes of bone morphogenetic proteins (BMPs) are more potent than each homodimers in bone and mesoderm induction in vitro, suggesting that BMP4 and BMP8B may form heterodimers to induce PGCs. To investigate how BMP4 and BMP8B interact and signal for PGC induction, we cocultured epiblasts of embryonic day 6.0–6.25 embryos with BMP4 and BMP8B proteins produced by COS cells. Our data show that BMP4 or BMP8B homodimers alone cannot induce PGCs whereas they can in combination, providing evidence that two BMP pathways are simultaneously required for the generation of a given cell type in mammals and also providing a prototype method for PGC induction in vitro. Furthermore, the PGC defects of Bmp8b mutants can be rescued by BMP8B homodimers whereas BMP4 homodimers cannot mitigate the PGC defects of Bmp4 null mutants, suggesting that BMP4 proteins are also required for epiblast cells to gain germ-line competency before the synergistic action of BMP4 and BMP8B.
Keywords: embryogenesis, mouse, stem cells, cell fate determination
Primordial germ cells (PGCs) are progenitors of all gametes. Maternal factors play critical roles in germ cell specification in Drosophila, Caenorhabditis elegans, zebrafish, and frogs (1–5). By contrast, mice appear to use rather different developmental mechanisms. Embryological studies using lineage tracing by Lawson and Hage (6) revealed that proximal epiblast within one or two cell diameters of the extraembryonic ectoderm at embryonic day 6.0–6.5 (E6.0–6.5) eventually generated PGCs. Furthermore, descendents of a labeled cell in the proximal epiblast were found among PGCs and extraembryonic mesoderm cells (in particular the allantois), suggesting that the PGC fate is not fixed before E6.5 and PGCs and allantois share common precursors. Epiblast transplantation experiments showed that epiblast cells at different topological sites before E6.5 gave rise to PGCs only if they were positioned in close proximity to the extraembryonic ectoderm (7). Therefore, it was hypothesized that factors produced by the extraembryonic ectoderm are required for the generation of PGC precursors that eventually migrate toward the primitive streak and then segregate into PGC and allantois lineages. This hypothesis was further supported by epiblast cocultures with extraembryonic ectoderm (8).
Bone morphogenetic proteins (BMPs) are members of the transforming growth factor type β superfamily of growth factors that function as homodimers or heterodimers to signal through heteromeric receptor complexes and downstream SMAD proteins (9–11). Although significant progress has been made in delineating their signal pathways in general, the specificity of different BMPs and their putative receptor complexes remains elusive. Recent studies have shown that members of the DPP class (BMP2 and BMP4) and 60A class (BMP5, BMP6, BMP7, BMP8A, and BMP8B) are often coexpressed in numerous tissues or cell types during embryogenesis (12–14). Furthermore, heterodimeric BMPs between these two groups are about 20 times more potent than homodimers in bone and mesoderm induction (15–18). Therefore, it is possible that heterodimers of DPP and 60A members are functionally more potent in vivo or may even be the only functional form in certain biological systems. This notion is indirectly supported by genetic studies in zebrafish. As shown by Schmid et al. (14), homozygous null mutations in either Bmp7 (snailhouse) or Bmp2b (swirl) in zebrafish yield a phenotype in dorsoventral patterning identical to the double homozygotes. However, the absolute requirement for the production of heterodimers in vivo has not yet been corroborated by biochemical data. Furthermore, these observations in zebrafish can also be fully explained by the requirement of two parallel signaling pathways of BMP7 and BMP2B through separate receptor complexes and SMAD proteins.
Bmp4 and Bmp8b have overlapping expression in the extraembryonic ectoderm and both are required for PGC generation (19, 20). Therefore, the nature of their interaction in this particular system becomes an intriguing biological question. Three models can be proposed to explain their possible relationships. The first model is that BMP4 and BMP8B homodimers function similarly or equally through the same receptor complexes in a dosage-dependent manner (an equal-homodimer model) and a total of three active alleles are required to produce sufficient BMPs for PGC induction. Second, BMP4 and BMP8B may form heterodimers to signal through the receptor complexes (an obligatory heterodimer model). According to this model, in the absence of either Bmp gene, no heterodimers are formed and consequently, no PGCs can be induced from the proximal epiblast. Third, BMP4 and BMP8B homodimers may signal through two separate receptor complexes (a two-pathway model). Because both pathways are required, the absence of either will also nullify the whole PGC formation process. Therefore, the second and third models should have the same consequence when one gene is inactivated. Furthermore, whether these two proteins act directly on epiblast cells or indirectly through surrounding endoderm is also an important issue.
Purified BMP8B homodimers and BMP4/BMP8B heterodimers are not yet available. To distinguish between the above possibilities we have developed a system to express BMP4 and BMP8B homodimers and possible heterodimers in COS-7 cells. Different COS-7 cells were used to coculture the isolated mouse epiblasts or whole embryos. Our data reveal that BMP4 and BMP8B function as homodimers to induce PGCs and that BMP4 proteins are also required for epiblast cells to gain germ-line competency before the synergistic action of BMP4 and BMP8B.
Materials and Methods
Preparation of Expression Vectors.
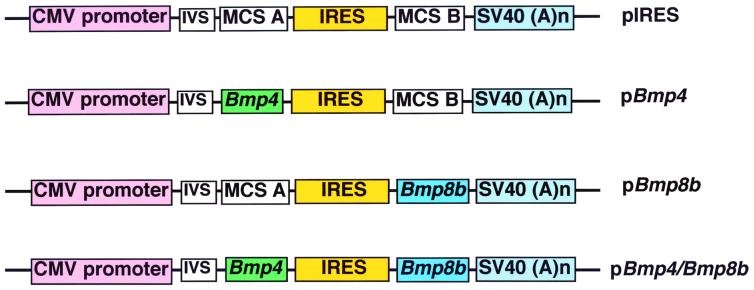
As shown in Fig. 1, pIRES vector was used as the parental vector (CLONTECH #6028–1) that contains a neomycin resistance gene to permit selection in the presence of G418 (not shown). The coding region of human Bmp4 cDNA (21) was inserted into the XhoI site of multiple cloning site (MCS) A to generate pBmp4. Murine Bmp8b cDNA (22) was inserted between XbaI and NotI sites of MCS B to create pBmp8b. Bmp4 cDNA fragment was inserted into the XhoI site of MCS A in pBmp8b to create pBmp4/Bmp8b.
Figure 1.
Schematic representation of expression vectors (only the relevant elements are shown). pIRES, the parental vector obtained from CLONTECH (#6028–1), which contains the immediate early promoter of cytomegalovirus (CMV promoter), an intron (IVS), MCS A, internal ribosome entry site (IRES), and MCS B followed by simian virus (SV40) polyadenylation signals. pBmp4, vector that contains human Bmp4 cDNA in MCS A. pBmp8b, vector that contains murine Bmp8b cDNA in MCS B. pBmp4/Bmp8b, vector that contains human Bmp4 cDNA in MCS A and murine Bmp8b cDNA in MCS B.
Establishment of BMP-Expressing COS-7 Cells and Epiblast Cultures.
COS-7 cells were purchased from ATCC (CRL-1651) and maintained in complete growth medium (DMEM with 4 mM l-glutamine, 4.5 g glucose/liter, 1.5 g sodium bicarbonate/liter, 100 units penicillin/ml, 100 μg streptomycin/ml, and 10% FBS). Exponentially growing COS-7 cells were harvested by trypsin digestion, washed in PBS twice by low-speed centrifugation, and resuspended in cold PBS at 1 × 107 cells/ml. About 5–10 μg DNA (pIRES, pBmp4, pBmp8b, or pBmp4/Bmp8b) was transfected into 1 ml of resuspended COS-7 cells by electroporation with Gene Pulser II (Bio-Rad) at 250 V and 950 μF. Subsequently, transfected cells were maintained in complete medium for 24 h and then in complete medium supplemented with G418 at 360 μg/ml for 14–30 days to obtain a mixture of resistant cells for each DNA construct. COS cells were seeded onto 35-mm culture dishes 2–3 days to reach 80–100% confluency, inactivated with medium containing 10 μg/ml mitomycin C for 3 h, and washed with PBS twice, maintained in complete medium (DMEM with 4 mM l-glutamine, 4.5 g glucose/liter, 1.5 g sodium bicarbonate/liter, 100 units penicillin/ml, 100 μg streptomycin/ml, and 15% FBS), and then used within 16 h for cocultures with epiblast masses or embryos. Epiblast masses with or without endoderm were isolated as described (23). About 5–10 epiblasts or embryos were cocultured in each 35-mm dish. Cocultures were terminated at 24, 48, 72, and 96 h for pilot studies. Culture time of 72 h was chosen for all subsequent cultures as PGCs in the cultured epiblast masses could be readily detected by alkaline phosphatase (ALP) staining (24) and there was a good survival rate of inactivated COS cells.
In Situ Hybridization.
Whole-mount in situ hybridization was performed essentially as described (20, 23). The coding regions of murine Oct4 and Mvh (mouse vasa homolog) were used as template for the preparation of antisense riboprobes (25, 26).
Results
Induction of PGCs by pBmp4/Bmp8b COS-7 Cells.
Embryological studies have established that signals from extraembryonic ectoderm instruct proximal epiblast cells to develop toward germ cell lineage (6, 7). Genetics studies have shown that both Bmp4 and Bmp8b are required for PGC generation during gastrulation by serving as the signals from extraembryonic ectoderm (19, 20). However, it is not known how many signals from extraembryonic ectoderm are required and whether BMP4 and BMP8B are sufficient to induce PGCs. To address these questions, we isolated mouse embryos at E6.0–6.25 and removed the extraembryonic ectoderm and proximal epiblast that contained the PGC precursors. The remaining embryonic portions including the majority of the epiblast cells and visceral endoderm were cultured on top of COS cells transfected with pIRES or pBmp4/Bmp8b (Fig. 2A). As shown in Fig. 2 B–E, after coculture with pIRES COS cells for 72 h, only 12% of the epiblast masses contained discernible PGCs as revealed by ALP staining and cell morphology (24). However, in the presence of pBmp4/Bmp8b COS cells, 65% of the epiblast masses contained PGCs. Furthermore, the average number of PGCs per epiblast was 3.3 ± 2.3 (mean ± SEM) in the former group and 34.2 ± 7.2 in the latter (P < 0.001). Therefore, there was a 54-fold increase in the number of PGCs after culture with pBmp4/Bmp8b COS cells compared with the control cells. To further show that these ALP-positive, PGC-like cells were indeed PGCs, we performed whole-mount in situ hybridization with antisense RNA probes against murine Oct4 and Mvh (25, 26). Clusters of strong Oct4-positive cells (comparable to those observed ALP-positive cells) were detected in the epiblasts cocultured with pBmp4/Bmp8b COS cells, not in those cocultured with pIRES COS cells, indicating that these ALP-positive cells indeed had typical PGC characteristics. The absence of Oct4 expression in non-PGC cells after epiblast culture also was observed by Yoshimizu et al. (8). No obvious Mvh-positive cells were detected in the cultured epiblasts, suggesting that these PGCs were comparable to those of the early embryos (before colonizing gonads) because Mvh expression begins in germ cells after colonization (26).
Figure 2.
Induction of PGCs by BMP4/BMP8B-expressing COS cells. (A) Diagram showing coculture of epiblast masses with COS cells. Mouse embryos (F1 hybrid of 129 SvEv × C57BL/6) at E6.0–6.25 were isolated, in which the extraembryonic ectoderm (red; xe) and epiblast (light blue; ep) are enclosed by endoderm (dark blue; xn for extraembryonic endoderm and en for embryonic endoderm). The extraembryonic ectoderm and proximal epiblast cells were removed with a sharp tungsten needle (red dashed line). The epiblast masses with visceral endoderm were then transferred onto pIRES or pBmp4/Bmp8b COS cells. (B) Percentage of embryos containing PGCs. About 12% of the control epiblast masses (3/25) formed PGCs after coculture with pIRES COS cells, whereas 13 of 20 epiblast masses (65%) contained PGCs after coculture with pBmp4/Bmp8b COS cells. (C) Average number of PGCs per PGC-containing embryo. The average number of PGCs was 3.3 ± 2.3 (mean ± SEM) for epiblast masses cocultured with pIRES COS cells, whereas 34.2 ± 7.2 PGCs were detected in epiblast masses cocultured with pBmp4/Bmp8b COS cells. (D) An example of an epiblast mass cocultured with pIRES COS cells after staining for ALP. No obvious PGC-like cells were observed. (E) An example of an epiblast mass cocultured with pBmp4/Bmp8b COS cells. More than 30 PGCs (red arrows) were found in a cluster and its vicinity. These cells have typical PGC characteristics (in terms of size and shape, having strong ALP staining in the cell membrane, and with a darkly stained spot in the cytoplasm). χ2 was used for statistical analysis. (Bar = 40 μm in D and E.)
Induction of PGCs by BMP4 and BMP8B Homodimers Independent of Endoderm.
Although pBmp4/Bmp8b COS cells can effectively induce PGCs from epiblast cells, it was not known which forms of the proteins (homodimers or heterodimers) were functional. Furthermore it was not clear whether the functional BMPs acted indirectly through the visceral endoderm or directly on epiblast cells to induce PGCs. To address these questions, we cocultured the isolated epiblast cell masses denuded of endoderm with different COS cells (Fig. 3A). As shown in Fig. 3B, culture with pBmp4 or pBmp8b COS cells alone only resulted in background levels of PGCs comparable to those seen with pIRES COS cells. However, pBmp4/Bmp8b COS cells and a mixture of equal number of pBmp4 COS cells and pBmp8b COS cells were similarly potent in PGC induction. PGC numbers in the latter two groups (an average of 41–51 PGCs/embryo) were significantly higher than those of the former three groups (6–15 PGCs/embryo) (P < 0.001). Because the dimerization process of BMPs or transforming growth factor type βs occurs intracellularly during protein synthesis (27, 28), the mixture of BMP4- and BMP8B-producing COS cells will not result in the formation of their heterodimers in medium. Thus, these data clearly indicate that the combination of BMP4 and BMP8B homodimers is not only required but also sufficient to induce PGCs and that these two proteins signal through separate receptor complexes. Furthermore, because the endoderm was removed in these cultures, it is evident that BMP4 and BMP8B homodimers act on epiblast cells directly and not through the visceral endoderm.
Figure 3.
BMP4 and BMP8B homodimers synergistically induce PGCs from the epiblast independent of endoderm. (A) A diagram showing that the extraembryonic ectoderm and proximal epiblast cells of E6.0–6.25 embryos were removed with a tungsten needle (red dashed line). Subsequently, the isolated epiblast masses with endoderm attached were subjected to trypsin and pancreatin digestion (23). The endoderm was removed by gentle pipetting several times and the epiblast masses were transferred on top of COS cells for coculture. (B) Percentage of embryos containing PGCs after coculture with COS cells. About 15.7–18.5% (9/57, 10/60, or 10/54, respectively) of the epiblast masses contained PGCs after coculture with pIRES, pBmp4, or pBmp8b COS cells with an average of 6–15 PGCs/embryo. No significant differences were observed among these three groups. Similar to the results in Fig. 2, about 71% of the epiblast masses cocultured with pBmp4/Bmp8b COS cells contained PGCs (an average of 51 ± 4.5 PGCs/embryo). Interestingly, about 66% of the epiblast masses cocultured with a mixture of equal number of pBmp4 COS cells and pBmp8b COS cells contained PGCs (an average of 41 ± 5.0 PGCs/embryo). No significant difference was detected between the latter two groups. However, the differences of the latter two groups with the former three were very significant (P < 0.001). ANOVA was used as the statistical method.
Mitigation of PGC Defects of Bmp8b Null Mutants by BMP8B Homodimers.
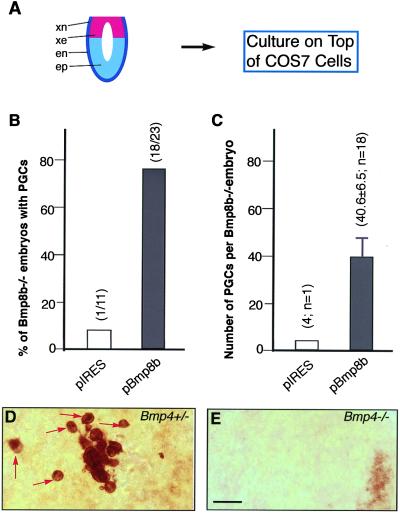
Epiblast cells cannot give rise to PGCs properly in the absence of BMP4 or BMP8B proteins. If indeed these two proteins function as individual homodimers, it is anticipated that BMP4 or BMP8B homodimers produced by COS cells should be able to rescue the PGC defects of the respective null mutants. Therefore, we further cultured Bmp8b mutant embryos at E6.0–6.25 with pBmp8b COS cells (Fig. 4A). As shown in Fig. 4B, Bmp8b homozygous embryos on an 87% C57BL/6 background formed few or no PGCs after coculture with pIRES COS cells, whereas about 78% of them produced an average of 40 PGCs after coculture with pBmp8b COS cells. Thus, this result reveals that BMP8B homodimers can rescue the PGC defects of Bmp8b null mutants and further validates the two-pathway model for BMP signaling in PGC generation.
Figure 4.
Rescue of PGC defects of Bmp8b and Bmp4 mutant embryos. (A) Diagram showing that Bmp8b or Bmp4 homozygous embryos at E6.0–6.25 were cocultured with COS cells. (B and C) PGCs were induced from Bmp8b−/− embryos after coculture with pBmp8b COS cells. Only one of 11 Bmp8b−/− embryos contained four PGCs after coculture with pIRES COS cells. However, 78% of the Bmp8b−/− embryos (18/23) produced an average 40.6 ± 6.5 PGCs/embryo after coculture with pBmp8b COS cells (P < 0.001, χ2 was used). Bmp8b mutants for this study were maintained on an 87.5% C56BL/6 background. (D) A Bmp4+/− embryo after coculture with pBmp4 COS cells contained about 20 PGCs (red arrows) in a small region. (E) A Bmp4−/− embryo contained no PGCs after coculture with pBmp4 COS cells. Bmp4 mutants were maintained on a mixed genetic background of 129 Sv × Black Swiss. (Bar = 40 μm in D and E.)
Inability of BMP4 Homodimers to Rescue Bmp4 Null Mutant Phenotype.
Because BMP4 and BMP8B function as individual homodimers through separate receptor complexes and BMP8B homodimers are capable of rescuing Bmp8b null phenotype in PGC generation, it was anticipated that pBmp4 COS cells were able to mitigate the PGC defects of Bmp4 null mutants. To test this possibility, we cocultured E6.0–6.25 embryos from crosses of Bmp4 heterozygotes on an outbred background (129/Sv × Swiss Black) (29). Surprisingly, none of the Bmp4 homozygous null embryos generated any PGCs after coculture with pBmp4 COS cells (Fig. 4E; n = 6) whereas about 60% of control wild-type (3/5) and Bmp4 heterozygous embryos (8/13) contain discernible PGCs (Fig. 4D). Moreover, Bmp4 homozygous epiblasts (free of endoderm) failed to generate any PGCs even with a mixture of pBmp4 and pBmp8b COS cells (n = 11; data not shown) whereas about 60% of the heterozygous controls contained PGCs (10/17). These data suggest that Bmp4 homozygous epiblast cells are refractory to BMP4 and BMP8B induction.
Discussion
Although high levels of conservation exists in body plan and axis formation across species, germ cell fate specification in mammals (at least in mice) appears to be unique in which maternally derived factors have not been shown to play any roles (1–3). Consistent with data of embryological studies that extraembryonic ectoderm signals the proximal epiblast to develop toward a PGC fate, genetic studies have revealed that BMP4 and BMP8B serve as the extraembryonic ectoderm-derived factors to induce PGCs (6–8, 19, 20). Because previous studies also indicated that heterodimers of DPP and 60A classes of BMPs are more potent in bone and mesoderm induction in vitro (15–18), the possibility has been raised that these heterodimers may function as the major BMPs in vivo. Furthermore, in the field of BMP biology, it is rather unclear as to whether BMPs of different classes share the same receptor complexes or not to exert their biological activities.
The fact that Bmp4 and Bmp8b double heterozygotes have a phenotype identical to that of Bmp4 heterozygotes does not contradict a heterodimer model for the function of these two BMPs (20). Nevertheless, results from our embryo cultures clearly point to a two-pathway model, showing that BMPs of DPP and 60A classes signal synergistically through separate receptor complexes to elicit a single biological process, PGC induction. Genetic studies indicated that Dpp and Screw homodimers act cooperatively to specify the formation of amnioserosa (30), providing evidence for a two-pathway model in Drosophila. The two-pathway model is also consistent with our recent findings that BMP4 and visceral endoderm-derived BMP2 have additive effect, whereas BMP8B and BMP2 or BMP4 do not have additive effect in PGC induction (31). Therefore, the closely related BMP2 and BMP4 (sharing 90% of sequence identity) are likely to function through same or similar receptor complexes whereas the distantly related BMP8B uses separate receptor complexes. It has been shown in cell cultures that BMP2 and BMP4 bind with high affinity to type I receptor ALK3 that is phosphorylated by the recruited type II receptor TALK (9, 10, 32, 33). Among the known type I receptors (ALK2, ALK3, and ALK6) for BMPs, only ALK3 is expressed in the epiblasts (34–38). If BMP4 signals through ALK3 during PGC generation, the receptor complexes responsible for BMP8B signaling in PGC formation (which is currently unknown) may not contain ALK3 because type I receptors are the high-affinity subunits and determine the specificity of ligand binding. Therefore, other BMP type I receptor subunits that should be expressed in the proximal epiblast cells are to be identified.
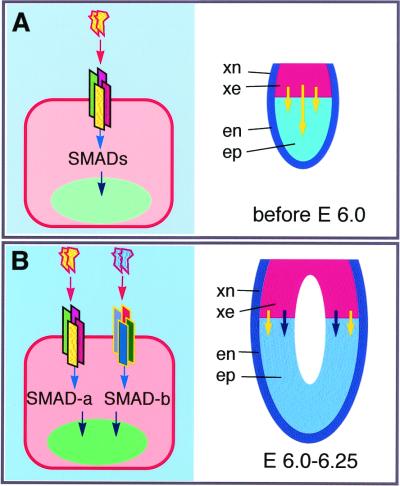
Despite our findings that BMP4 and BMP8B function as individual homodimers and BMP8B homodimers are able to rescue the Bmp8b mutant phenotype, BMP4 homodimers cannot rescue the PGC defects of Bmp4 null mutant embryos of E6.0–6.25. These data strongly suggest that BMP4 is required at multisteps of PGC fate determination whereas BMP8B is mainly required at the later stage. As previously shown, Bmp4 RNA is detected in the inner cell mass at E3.5 and its expression switches to and stays in the extraembryonic ectoderm from E5.5 to 7.5 (19, 20, 39). Therefore, before E6.0–6.25, the inner cell mass and its epiblast derivatives should have already been exposed to BMP4 stimulation. Such stimulation appears to be essential for epiblast cells to gain competency for PGC formation and is not restricted to the proximal epiblast cells because the distal epiblast cells are able to generate PGCs after transplantation, explant culture, or coculture with pBmp4/Bmp8b COS cells. Therefore, our current model is that BMP4 signals through its own receptor complexes and downstream SMAD proteins to instruct the early epiblast cells to gain germ-line competency. Such competency is essential for the subsequent synergistic action of BMP4 and BMP8B (through distinct receptor complexes and SMAD proteins) for PGC induction (Fig. 5). Further studies to identify specific receptor complexes and downstream signaling components will aid in dissecting the molecular mechanisms of PGC fate specification in mammals.
Figure 5.
A sequential model for BMP4 and BMP8B signaling in PGC induction from epiblast cells during mouse embryogenesis. (A) Requirement of BMP4 signaling for the establishment of germ-line competency of the epiblast before E6.0. (Left) BMP4 proteins (indicated by a yellow head-shaped pair) bind to a tetrameric receptor complex on an early epiblast cell and signal through SMAD proteins. (Right) In the normal embryo, BMP4 proteins produced by extraembryonic ectoderm (or inner cell mass) instructs the epiblast cells to become germ-line competent. (B) Induction of PGCs by the synergistic action BMP4 and BMP8B at E6.0–6.25. (Left) A germ-line-competent epiblast cell containing two separate receptor complexes receives BMP4 and BMP8B (a blue head-shaped pair) proteins. These two tetrameric receptor complexes transduce signals through distinct SMAD proteins (SMAD-a and SMAD-b) from the cytoplasm to the nucleus to instruct the cell to become PGCs. (Right) At embryo level, BMP4 and BMP8B produced by the extraembryonic ectoderm synergistically signal the germ-line-competent proximal epiblast cells to become PGCs.
Acknowledgments
We thank Drs. Ray Dunn, Brigid Hogan, Kirstie Lawson, and Lucy Liaw for helpful discussions. This work is supported by grants from the National Institute of Child Health (HD-36218 and HD-39154) and Basil O'Connor Starter Scholar Research Award (to G.-Q.Z.).
Abbreviations
- PGC
primordial germ cell
- BMP
bone morphogenetic protein
- En
embryonic day n
- ALP
alkaline phosphatase
- MCS
multiple cloning site
- IRES
internal ribosome entry site
References
- 1.Eddy E M. Int Rev Cytol. 1975;43:229–280. doi: 10.1016/s0074-7696(08)60070-4. [DOI] [PubMed] [Google Scholar]
- 2.Wylie C. Cell. 1999;96:165–174. doi: 10.1016/s0092-8674(00)80557-7. [DOI] [PubMed] [Google Scholar]
- 3.McLaren A. Genes Dev. 1999;13:373–376. doi: 10.1101/gad.13.4.373. [DOI] [PubMed] [Google Scholar]
- 4.Weidinger G, Wolke U, Koprunner M, Klinger M, Raz E. Development (Cambridge, UK) 1999;126:5295–5307. doi: 10.1242/dev.126.23.5295. [DOI] [PubMed] [Google Scholar]
- 5.Kloc M, Bilinski S, Chan A P, Allen L H, Zearfoss N R, Etkin L D. Int Rev Cytol. 2001;203:63–91. doi: 10.1016/s0074-7696(01)03004-2. [DOI] [PubMed] [Google Scholar]
- 6.Lawson K A, Hage W J. Ciba Found Symp. 1994;182:68–84. doi: 10.1002/9780470514573.ch5. [DOI] [PubMed] [Google Scholar]
- 7.Tam P P, Zhou S X. Dev Biol. 1996;178:124–132. doi: 10.1006/dbio.1996.0203. [DOI] [PubMed] [Google Scholar]
- 8.Yoshimizu T, Obinata M, Matsui Y. Development (Cambridge, UK) 2001;128:481–490. doi: 10.1242/dev.128.4.481. [DOI] [PubMed] [Google Scholar]
- 9.Hogan B L. Genes Dev. 1996;10:1580–1594. doi: 10.1101/gad.10.13.1580. [DOI] [PubMed] [Google Scholar]
- 10.Heldin C H, Miyazono K, ten Dijke P. Nature (London) 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 11.Massague J, Chen Y G. Genes Dev. 2000;14:627–644. [PubMed] [Google Scholar]
- 12.Lyons K M, Hogan B L, Robertson E J. Mech Dev. 1995;50:71–83. doi: 10.1016/0925-4773(94)00326-i. [DOI] [PubMed] [Google Scholar]
- 13.Furuta Y, Piston D W, Hogan B L. Development (Cambridge, UK) 1997;124:2203–2212. doi: 10.1242/dev.124.11.2203. [DOI] [PubMed] [Google Scholar]
- 14.Schmid B, Furthauer M, Connors S A, Trout J, Thisse B, Thisse C, Mullins M C. Development (Cambridge, UK) 2000;127:957–967. doi: 10.1242/dev.127.5.957. [DOI] [PubMed] [Google Scholar]
- 15.Aono A, Hazama M, Notoya K, Taketomi S, Yamasaki H, Tsukuda R, Sasaki S, Fujisawa Y. Biochem Biophys Res Commun. 1995;210:670–677. doi: 10.1006/bbrc.1995.1712. [DOI] [PubMed] [Google Scholar]
- 16.Israel D I, Nove J, Kerns K M, Kaufman R J, Rosen V, Cox K A, Wozney J M. Growth Factors. 1996;13:291–300. doi: 10.3109/08977199609003229. [DOI] [PubMed] [Google Scholar]
- 17.Suzuki A, Kaneko E, Maeda J, Ueno N. Biochem Biophys Res Commun. 1997;232:153–156. doi: 10.1006/bbrc.1997.6219. [DOI] [PubMed] [Google Scholar]
- 18.Nishimatsu S, Thomsen G H. Mech Dev. 1998;74:75–88. doi: 10.1016/s0925-4773(98)00070-7. [DOI] [PubMed] [Google Scholar]
- 19.Lawson K A, Dunn N R, Roelen B A, Zeinstra L M, Davis A M, Wright C V, Korving J P, Hogan B L. Genes Dev. 1999;13:424–436. doi: 10.1101/gad.13.4.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ying Y, Liu X M, Marble A, Lawson K A, Zhao G Q. Mol Endocrinol. 2000;14:1053–1063. doi: 10.1210/mend.14.7.0479. [DOI] [PubMed] [Google Scholar]
- 21.Jones C M, Lyons K M, Lapan P M, Wright C V, Hogan B L. Development (Cambridge, UK) 1992;115:639–647. doi: 10.1242/dev.115.2.639. [DOI] [PubMed] [Google Scholar]
- 22.Zhao G Q, Hogan B L. Mech Dev. 1996;57:159–168. doi: 10.1016/0925-4773(96)00543-6. [DOI] [PubMed] [Google Scholar]
- 23.Hogan B, Beddington R, Constantini F, Lacy E. Manipulating the Mouse Embryo: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. [Google Scholar]
- 24.Ginsburg M, Snow M H, McLaren A. Development (Cambridge, UK) 1990;110:521–528. doi: 10.1242/dev.110.2.521. [DOI] [PubMed] [Google Scholar]
- 25.Scholer H R, Ruppert S, Suzuki N, Chowdhury K, Gruss P. Nature (London) 1990;344:435–439. doi: 10.1038/344435a0. [DOI] [PubMed] [Google Scholar]
- 26.Fujiwara Y, Komiya T, Kawabata H, Sato M, Fujimoto H, Furusawa M, Noce T. Proc Natl Acad Sci USA. 1994;91:12258–12262. doi: 10.1073/pnas.91.25.12258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Massague J. Cell. 1987;49:437–438. doi: 10.1016/0092-8674(87)90443-0. [DOI] [PubMed] [Google Scholar]
- 28.Massague J. Annu Rev Cell Biol. 1990;6:597–641. doi: 10.1146/annurev.cb.06.110190.003121. [DOI] [PubMed] [Google Scholar]
- 29.Winnier G, Blessing M, Labosky P A, Hogan B L. Genes Dev. 1995;9:2105–2116. doi: 10.1101/gad.9.17.2105. [DOI] [PubMed] [Google Scholar]
- 30.Dorfman R, Shilo B Z. Development (Cambridge, UK) 2001;128:965–972. doi: 10.1242/dev.128.6.965. [DOI] [PubMed] [Google Scholar]
- 31.Ying Y, Zhao G Q. Dev Biol. 2001;232:484–492. doi: 10.1006/dbio.2001.0173. [DOI] [PubMed] [Google Scholar]
- 32.ten Dijke P, Yamashita H, Sampath T K, Reddi A H, Estevez M, Riddle D L, Ichijo H, Heldin C H, Miyazono K. J Biol Chem. 1994;269:16985–16988. [PubMed] [Google Scholar]
- 33.ten Dijke P, Miyazono K, Heldin C H. Curr Opin Cell Biol. 1996;8:139–145. doi: 10.1016/s0955-0674(96)80058-5. [DOI] [PubMed] [Google Scholar]
- 34.Dewulf N, Verschueren K, Lonnoy O, Moren A, Grimsby S, Vande Spiegle K, Miyazono K, Huylebroeck D, Ten Dijke P. Endocrinology. 1995;136:2652–2663. doi: 10.1210/endo.136.6.7750489. [DOI] [PubMed] [Google Scholar]
- 35.Mishina Y, Suzuki A, Ueno N, Behringer R R. Genes Dev. 1995;9:3027–3037. doi: 10.1101/gad.9.24.3027. [DOI] [PubMed] [Google Scholar]
- 36.Natsume T, Tomita S, Iemura S, Kinto N, Yamaguchi A, Ueno N. J Biol Chem. 1997;272:11535–11540. doi: 10.1074/jbc.272.17.11535. [DOI] [PubMed] [Google Scholar]
- 37.Gu Z, Reynolds E M, Song J, Lei H, Feijen A, Yu L, He W, MacLaughlin D T, van den Eijnden-van Raaij J, Donahoe P K, Li E. Development (Cambridge, UK) 1999;126:2551–2561. doi: 10.1242/dev.126.11.2551. [DOI] [PubMed] [Google Scholar]
- 38.Mishina Y, Crombie R, Bradley A, Behringer R R. Dev Biol. 1999;213:314–326. doi: 10.1006/dbio.1999.9378. [DOI] [PubMed] [Google Scholar]
- 39.Coucouvanis E, Martin G R. Development (Cambridge, UK) 1999;126:535–546. doi: 10.1242/dev.126.3.535. [DOI] [PubMed] [Google Scholar]