Abstract
Fas/Fas ligand (FasL) system is one of the key apoptotic signaling entities in the extrinsic apoptotic pathway. De-regulation of this pathway, i.e. by mutations may prevent the immune system from the removal of newly-formed tumor cells, and thus lead to tumor formation. The present study investigated the association between −1377 G/A (rs2234767) and −670 A/G (rs1800682) polymorphisms in Fas as well as single nucleotide polymorphisms INV2nt −124 A/G (rs5030772) and −844 C/T (rs763110) in FasL in a sample of Iranian patients with breast cancer. This case-control study was done on 134 breast cancer patients and 152 normal women. Genomic DNA was extracted from whole blood samples. The polymorphisms were determined by using tetra-ARMS-PCR method. There was no significant difference in the genotype distribution of FAS rs2234767 polymorphism between cases and controls. FAS rs1800682, FASL rs5030772, and FASL rs763110 genotypes showed significant associations with an increasing risk of breast cancer (odds ratio OR = 3.18, P = 0.019; OR = 5.08, P = 0.012; OR = 2.40, P = 0.024, respectively). In conclusion, FAS rs2234767 was not associated with breast cancer risk. Though, FAS rs1800682, FASL rs5030772, and FASL rs763110 polymorphisms were associated with the risk of breast cancer in the examined population.
Introduction
Breast cancer is the main cause of cancer-related death among women in western countries, with the sporadic form of the disease constituting more than 90% of all breast cancers [1], [2]. Breast cancer is also one of the most frequent malignancies among Iranian women [3], [4]. In addition to environmental factors and body weight, genetic invariability affects not only the chance of cancer development but also its aggressiveness, therapy response and overall prognosis [5], [6], [7]. Apoptosis is one of physiological forms of cell death that plays an important role in maintaining tissue homeostasis. Abnormal regulation of apoptosis contributes to the pathogenesis of cancer [8], [9]. The acquired capability to oppose apoptotic stimuli is one of the main characteristics of a malignant cell. Alterations of apoptotic pathways are key events in the development of variety of human diseases including cancer [10], [11].
The FAS/FASL signaling system activates a major extrinsic cell death pathway, that is of particular importance for the regulation of acquired immune response, maintaining immuno-privileged sites and fulfils other regulatory functions [12], [13]. Fas (TNFRSF6/CD95/APO-1) is a cell surface receptor, expressed in a variety of tissues [14]. FasL, also known as TNFSF6 or CD95L, is a member of the tumor necrosis factor superfamily and the natural ligand to Fas [15]. The Fas interaction with the FasL may trigger the death signal cascade, and eventually the cell expressing FAS would die. Studies have shown that decreased expression of Fas and/or elevated expression of FasL could be detected in many kinds of human tumors [16], [17].
The FAS/FASL system may have two opposite effects on cancer. The expression of FAS on tumor cells, if downstream signaling pathways are functional, may assist FAS-triggered killing of tumors by immune-effector cells [15], [18]. The expression of FASL on tumor cells may repel specific antitumor immune response, thus turning tumor into an immuno-privileged site [19], [20]. The role anticancer-drug-induced FAS/FASL system in tumor-cell killing has been championed by some [21], it is currently however often disregarded as experimental artifact [22], [23].
Two polymorphisms have been identified in the FAS promoter region: one in the silencer region, G to A substitution at nucleotide position −1377 (rs2234767), and the other in the enhancer region, A to G substitution at nucleotide position −670 (rs1800682). These two polymorphisms are located within the stimulatory protein-1 (Sp1) and the signal transducers and activators of transcription 1 (STAT1) transcription factor binding sites, respectively [24], [25]. Because these sequence variations in the FAS gene promoter region may influence Fas expression and deregulate cell death signaling, they could contribute to carcinogenesis [25]. The human FASL gene is located on chromosome 1q23, consists of four exons spanning ∼8 kb, and encodes 281amino acids [26]. There are two reported polymorphisms: C to T changes at nucleotide position −844 (FASL-844 C/T, rs763110) in the promoter region [27] and A to G change at nucleotide position −124 of intron 2 (FASL INV2nt −124 A/G, rs5030772). FASL −844 C/T is located in a putative binding motif for a transcription factor, CAAT/enhancer-binding protein h, and the −844 C allele may increase basal expression of FASL compared with the −844 T allele [27], suggesting that the FASL −844 C/T polymorphism may influence FASL expression and FasL mediated signaling, and ultimately, the susceptibility to cancer. So far, the functional relevance of the FASL IVS2nt −124 A/G polymorphism has not been reported.
Numerous studies investigated the role of FAS and FASL gene polymorphisms in the etiology of various cancers including cervix, breast, bladder, lung, prostate, head and neck and esophagus [25], [28], [29]. However, the role of FAS and FASL gene polymorphisms in breast cancer has not been conclusively established. Thus, the present study, that assesses the association between −1377 G/A and −670 A/G polymorphisms in FAS as well as single nucleotide polymorphisms −844 C/T and INV2nt −124 A/G in FASL adds an important information about the role of this death pathway in breast cancer.
Materials and Methods
Cancer patients and controls
This case control study was performed in 134 patients with histologically confirmed breast cancer and in 152 population based healthy women who participate in a screening project for metabolic syndrome with no history of cancer. The local Ethical Committee of Zahedan University of Medical Sciences approved the study protocol and written informed consent was obtained from all subjects. Two ml of venous blood drawn from each subject and genomic DNA was extracted from peripheral blood as described previously [30].
T-ARMS-PCR assay
Tetra-amplification refractory mutation system–polymerase chain reaction (T-ARMS-PCR) is a rapid and simple technique for detection of single nucleotide polymorphism [30], [31], [32]. In the present study we designed T-ARMS-PCR assay for the detection FAS and FASL polymorphisms. The primers used are listed in Table 1. Two allele-specific (inner) primers have been designed in opposed directions and, in combination with the common primers, can simultaneously amplify both the wild-type and the mutant alleles in a single-tube PCR (see result section for schematic depiction).
Table 1. Tetra-ARMS-PCR primers and conditions for Fas, −1377 A/G (rs2234767), −670 A/G (rs1800682) and FasL Ivs2nt 124 A/G (rs5030772), FASL −844 C/T (rs763110).
| Primers | Sequence (5′→3′) | Annealing temp. (°C) |
| Fas −1377 A/G (rs2234767) | ||
| FO | CCTTCCCTCACACCCCTTTTCCTTCC | 64 |
| RO | CTTTGGCATCGTCCACCAAGCTCTG | |
| FI | AGTGTGTGCACAAGGCTGGCCCA | |
| RI | TTAGTGCCATGAGGAAGACCCTGTGC | |
| Fas −670 A/G (rs1800682) | ||
| FO | GGGGCTATGCGATTTGGCTTAAGTTGTT | 62 |
| RO | GTAGTTCAACCTGGGAAGTTGGGGAGGT | |
| FI | TTTTTCATATGGTTAACTGTCCATTCCCGG | |
| RI | GCAACATGAGAGGCTCACAGACGGTT | |
| FasL Ivs2nt 124 A/G (rs5030772) | ||
| FO | GGTCTTCTTGGATTAGTCACCCAACTT | 57 |
| RO | CACTTTCCTCAGCTCCTTTTTTTCAG | |
| FI | CTGCAGTTCAGACCTACATGATTAGTCTG | |
| RI | TTAAAACCGTAAATGGCAACAGTCTAAAAT | |
| FASL −844 C/T (rs763110) | ||
| FO | TGCAGTTAACTACGATAGCACCACTGCA | 63 |
| RO | AGGAGGAAAACATCTGTTGCCATCATCT | |
| FI | GGCAAACAATGAAAATGAAAACATGGC | |
| RI | AACCCACAGAGCTGCTTTGTATTGCA |
PCR was performed using commercially available PCR premix (AccuPower PCR PreMix, BIONEER, Daejeon, Korea) according to the manufacturer's instructions. For detection of polymorphisms of FAS −670 A/G (rs1800682) and −1377 A/G (rs2234767), 1 µL template DNA (∼100 ng/µL), 1 µL of each primer (10 pmol/µL), and 15 µL DNase-free water were added into a 20 µL PCR tube containing the AccuPower PCR PreMix. The same PCR condition was used for detection of FASL (rs763110), except that 0.6 µL of each primer and 16.6 µL water were added. For FASL (rs5030772), 1 µL template DNA, 1 µL of outer primer, 0.5 µL of inner primer, 0.5 µL of DMSO and 15.5 µL was added.
PCR cycling was performed at 95°C for 5 min, followed by 30 cycles of a denaturation for 30 s at 95°C, annealing for 30 s at different temperatures (Table 1), extension s at 72°C (30 s for rs1800682 and rs2234767; 40 s for rs5030772 and rs763110) and a final extension for 10 min at 72°C in a thermocycler (Corbett research, Australia). The amplified products were separated by electrophoresis on a 2% agarose gel containing 0.5 µg/ml ethidium bromide.
The genotyping analysis was randomly repeated for 10% of samples for confirmation for quality control and the results were 100% concordant. To confirm the genotyping results, selected PCR-amplified DNA samples (n = 3, respectively, for each genotype) were examined by DNA sequencing and the results determined by T-ARMS-PCR were concordant with those determined by sequencing (see the result section for more details).
Real-time RT–PCR (qRT–PCR) assay
We investigated the expression levels of FAS in 10 tumor samples and 16 adjacent normal tissue breast normal tissues. Briefly, total RNA was extracted from the tissues using TRIzol reagent. Then complementary DNA (cDNA) synthesis from total RNA was catalyzed in a final volume of 25 µL by cDNA synthesis kit (Bioneer, K-2045, Korea) according to the manufacturer's protocol. Expressions for the Fas and the housekeeping GAPDH genes were investigated using real-time quantitative PCR (qRT-PCR) (Applied Biosystems). FAS (F: 5′-TGAAGGACATGGCTTAGAAGTG-3′, R: 5′-GGTGCAAGGGTCACAGTGTT-3′) and GAPDH primers (F: 5′-GAAGGTGAAGGTCGGAGTC-3′, R: 5′- GAAGATGGTGATGGGATTTC-3′) were used [33]. Samples were assayed in a 25 µL reaction mixture including 2 µL of cDNA, 12.5 µL of 2× Master Mix (Fermentas, Cat No. K0221), 1 µL of each specific primer (10 µM), and 8.5 µL of RNase-free water. qRT-PCR reactions were performed with an initial incubation at 95°C for 10 minutes followed by 40 cycles at 95°C for 15 seconds and 60°C for 60 seconds.
Statistical analysis
The statistical analysis of the data was performed using the SPSS 18.0 software. Genotypes and alleles were compared between groups by use of χ2 test. The strength of the association between the polymorphisms and cancer risk was measured by odds ratios (ORs) with 95% confidence intervals (CIs). Binary logistic regression was used for all analysis variables to estimate risk as odds ratio (OR) with 95% confidence intervals (CIs) using age as covariate. According to our findings, sample power was calculated for FAS and FASL polymorphisms by comparison of each genotype with the sum of other related genotypes (Table 2) at each polymorphic region by using STATA 10 software and is shown in table 3.
Table 2. Clinical and pathological characteristics of breast carcinoma patients.
| Characteristic | Number |
| Stage | |
| I | 28 |
| II | 63 |
| III | 26 |
| IV | 15 |
| Unknown | 2 |
| Grade | |
| I | 23 |
| II | 72 |
| III | 16 |
| IV | 18 |
| Unknown | 5 |
| Estrogen Receptor | |
| Positive | 70 |
| Negative | 61 |
| Unknown | 3 |
| Progesterone Receptor | |
| Positive | 69 |
| Negative | 58 |
| Unknown | 7 |
| HER2 | |
| Positive | 51 |
| Negative | 80 |
| Unknown | 3 |
| Histology | |
| Ductal carcinoma | 101 |
| Lobular carcinoma | 7 |
| Other | 22 |
Table 3. Frequency distribution of Fas rs2234767, rs1800682 and FasL rs5030772, rs763110 gene polymorphisms in normal and breast cancer individuals.
| Polymorphisms | Breast cancer n (%) | Normal n (%) | *OR (95% CI) | p-value | Study power % |
| FAS | |||||
| −1377A/G (rs2234767) | |||||
| AA | 8 (6.0) | 11 (7.2) | 1.00 | - | 4 |
| AG | 106 (79.1) | 115 (75.7) | 1.49 (0.48–4.65) | 0.429 | 8 |
| GG | 20 (14.9) | 26 (17.1) | 1.24 (0.34–4.49) | 0.747 | 5 |
| Alleles | |||||
| A | 122 (45.5) | 140 (46.0) | 1.00 | - | 3 |
| G | 146 (54.5) | 164 (54.0) | 1.03 (0.74–1.42) | 0.877 | 3 |
| −670 A/G (rs1800682) | |||||
| AA | 55 (41.0) | 63 (41.4) | 1.00 | - | 3 |
| AG | 55 (41.0) | 78 (51.3) | 1.04 (0.56–1.92) | 0.901 | 37 |
| GG | 24 (18.0) | 11 (7.3) | 3.18 (1.21–8.33) | 0.019 | 73 |
| Alleles | |||||
| A | 165 (61.6) | 204 (67.1) | 1.00 | - | 25 |
| G | 103 (38.4) | 100 (32.9) | 1.27 (0.90–1.79) | 0.189 | 25 |
| FASL | |||||
| Ivs2nt 124 A/G (rs5030772) | |||||
| AA | 77 (57.4) | 92 (60.5) | 1.00 | - | 6 |
| AG | 40 (29.9) | 55 (36.2) | 0.87 (0.47–1.59) | 0.650 | 17 |
| GG | 17 (12.7) | 5 (3.3) | 5.08 (1.04–18.22) | 0.012 | 78 |
| Alleles | |||||
| A | 194 (72.4) | 239 (78.6) | 1.00 | - | 50 |
| G | 74 (27.6) | 65 (21.4) | 1.40 (0.95–2.06) | 0.096 | 50 |
| −844 C/T (rs763110) | |||||
| CC | 42 (31.3) | 62 (40.8) | 1.00 | - | 34 |
| CT | 51 (38.1) | 64 (42.1) | 1.17 (0.58–2.36) | 0.663 | 10 |
| TT | 41 (30.6) | 26 (17.1) | 2.40 (1.12–5.14) | 0.024 | 73 |
| Alleles | |||||
| C | 135 (50.4) | 188 (61.8) | 1.00 | - | 76 |
| T | 133 (49.6) | 116 (38.2) | 1.61 (1.14–2.24) | 0.007 | 76 |
Adjusted for age, age at menarche, menopausal status, body mass index (BMI).
Results
The study group consists of 134 histopathologically confirmed female cases with breast cancer (age; 47.97±13.27 years), and 152 healthy females (age; 43.53±13.52 years). The clinicopathologic characteristics of patients are summarized in Table 2. The present study examined the association between −1377 G/A and −670 A/G polymorphisms in FAS as well as single nucleotide polymorphisms −844 C/T and INV2nt −124 A/G in FASL (Fig. 1). Should any of the studied polymorphisms be significantly associated with ant of the type of breast cancer, it would provide important indirect information about the role of death pathway in breast cancer. The study was performed applying Tetra-amplification refractory mutation system–polymerase chain reaction. The stretches of genes targeted for amplification are depicted in figure 2 and the corresponding primers used for the T-ARMS-PCR are shown in Table 1. The amplified products were separated by electrophoresis on a 2% agarose gel containing 0.5 µg/ml ethidium bromide (please see figure 3 as an example). For quality-control purposes, the genotyping analysis was randomly repeated for 10% of samples for confirmation for quality control and the results were 100% concordant. To confirm the genotyping results, selected PCR-amplified DNA samples (n = 3, respectively, for each genotype) were examined by DNA sequencing and the results determined by T-ARMS-PCR (Fig. 3) were concordant with those determined by sequencing (Fig. 4). Table 3 summarizes the genotype and allele frequencies of the examined polymorphisms in the FAS gene promoter region (−1377 G/A, rs2234767; and −670 A/G, rs1800682), as well as two in the FASL gene (−844 C/T in the promoter region, rs763110; and −124A/G in the second intron, rs5030772). Significances of associations (p-values) of those alleles with breast cancer risk are also shown in Table 3 (second-from the right column).
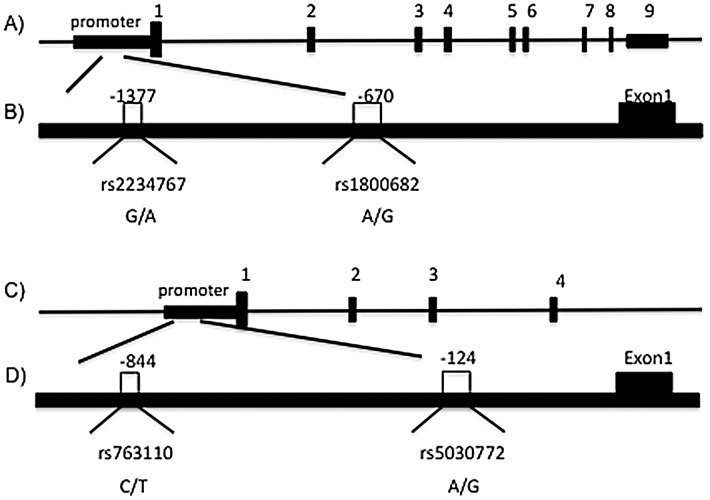
Figure 1. Maps of the human Fas and FasL genes with polymorphism positions indicated.
(A) Map of the human Fas gene. Exons 1–9 are numbered and represented by black boxes. (B) position of the single nucleotid variations within the core promoter of the Fas gene, a G>A polymorphism at position −1377 and a A>G polymorphism at position 670. (C) Map of the human Fas Ligand gene. Exons 1–4 are numbered and represented by black boxes. (D) Position of single nucleotide polymorphisms (SNPs) within the Fas Ligand promoter, a C>T substitution at position −844 and a A<G substitution at position −124.
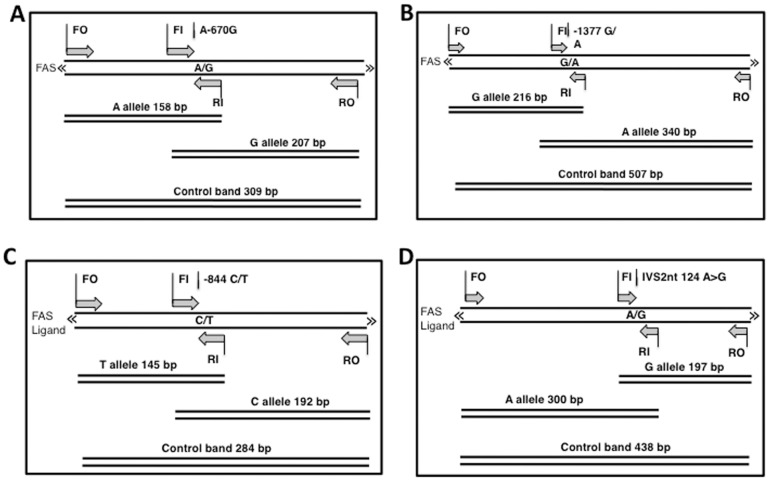
Figure 2. Schematic representation of Tetra-Primer Amplification Refractory Mutation System.
T-ARMS-PCR was used for the detection of SNPs of Fas rs1800682 (A), Fas rs2234727 (B), FasL rs763110 (C) and FasL rs5030772 (D). Two forward and two reverse specific primers are used to produce three potential products. Product sizes were 158 bp for A allele, 207 bp for G allele, and 309 bp for two outer primers (control band) for Fas rs1800682 (A). Product sizes were 216 bp for G allele, 340 bp for A allele, and 507 bp for control band for Fas rs2234727 (B). Product sizes were 145 bp for T allele, 192 bp for C allele, and 284 bp for control band for FasL rs763110 (C). Product sizes were 197 bp for G allele, 300 bp for A allele, and 438 bp for control band for FasL rs5030772 (D).
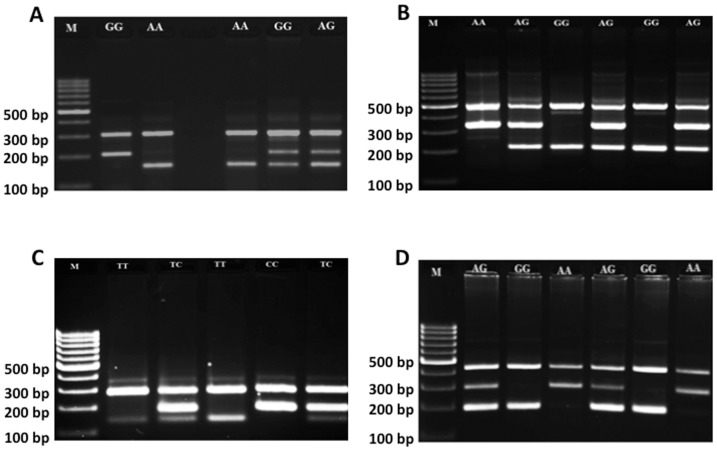
Figure 3. Electrophoresis pattern of tetra-ARMS-PCR for detection of polymorphisms.
Agarose gel electrophoresis was used to detect band-pattern of tetra-ARMS-PCR for Fas rs1800682 (A), Fas rs2234727 (B), FasL rs763110 (C), and FasL rs5030772 (D). M = DNA marker.
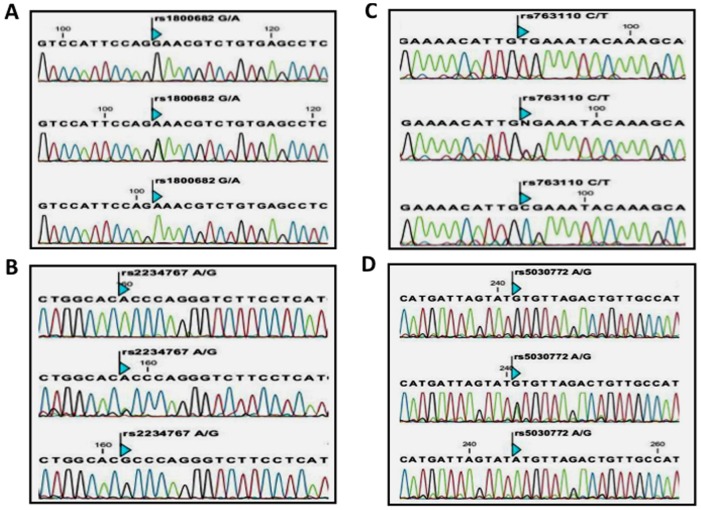
Figure 4. Examples of DNA sequencing results of Fas and FasL prlymorphisms.
DNA sequencing results of Fas rs1800682 (A), Fas rs2234727 (B), FasL rs763110 (C) and FasL rs5030772 (D) for tetra-ARMS-PCR results depicted in figure 3 are shown.
The current study does not demonstrate a significant difference in FAS −1377A/G genotype frequencies between patients and controls (χ2 = 0.49, p = 0.78). Hence no association was observed between −1377A/G polymorphism of FAS and risk of breast cancer (Table 3). On the other hand the genotype distribution of the FAS–670A/G polymorphism was significantly different between patients and control groups (χ2 = 8.25, p = 0.016). There was a significant association between the FAS −670 A/G polymorphism and breast cancer risk and the −670 GG genotype is a risk factor for susceptibility to breast cancer (OR = 3.181; 95% CI = 1.21–8.33; p = 0.019).
There was a significant difference between breast cancer subjects and controls in the distribution of FASL Ivs2nt 124 A/G (rs5030772) genotypes (χ2 = 9.15, p = 0.010) and the GG genotype exhibited significant association with the risk of breast cancer (OR = 5.08; 95% CI = 1.04–18.022; p = 0.012). A significant difference was identified in the distribution of FASL −844 C/T genotype between patients and controls (χ2 = 7.57, p = 0.022) and the TT genotype showed an increased risk for breast cancer (OR = 2.40; 95% CI = 1.12–5.14; p = 0.024).
Genotype distribution of FAS-1377A/G polymorphism in case and control are not consistent with Hardy–Weinberg equilibrium (HWE). The distribution of FAS −670 A/G genotype did not deviate from the HWE in case patients or control subjects. While, genotype distribution of FASL Ivs2nt 124 A/G and −844 C/T deviate from the HWE in case group. Furthermore, haplotypes analysis was performed (Tables 3 and 4). The results showed that rs2234767A/rs1800682A/rs5030772A/rs763110T, rs2234767G/rs1800682G/rs5030772A/rs763110T and rs2234767A/rs1800682A/rs5030772G/rs763110T haplotypes decreased the risk of breast cancer (OR = 0.18, 95%CI = 0.04–0.88, p = 0.04, OR = 0.03, 95%CI = 0.00–0.43, p<0.01, and OR = 0.04, 95%CI = 0.00–0.60, p = 0.02, respectively) in comparison with rs2234767G/rs1800682A/rs5030772A/rs763110C haplotype.
Table 4. Haplotype frequencies of FAS (rs2234767, rs1800682) and FasL (rs5030772, rs763110) genes polymorphisms in normal and breast cancer subjects.
| rs2234767 | rs1800682 | rs5030772 | rs763110 | Case | Control | OR (95%CI) | P |
| G | A | A | C | 0.1032 | 0.1949 | 1.00 | - |
| A | A | A | C | 0.1294 | 0.1699 | 1.42 (0.28–7.09) | 0.67 |
| A | G | A | T | 0.1034 | 0.1054 | 2.57 (0.42–15.60) | 0.30 |
| G | A | A | T | 0.0694 | 0.1353 | 0.69 (0.20–2.36) | 0.55 |
| G | G | A | C | 0.1125 | 0.0755 | 0.56 (0.12–2.60) | 0.46 |
| A | A | A | T | 0.0879 | 0.0346 | 0.18 (0.04–0.88) | 0.04 |
| G | A | G | C | 0.0961 | 0.0115 | 0.88 (0.13–5.96) | 0.90 |
| A | G | A | C | 0.0354 | 0.0783 | 0.28 (0.04–2.12) | 0.22 |
| G | A | G | T | 0.0219 | 0.0591 | 6.28 (0.51–77.32) | 0.15 |
| G | G | A | T | 0.0330 | 0.0195 | 0.03 (0.00–0.43) | <0.01 |
| A | A | G | T | 0.0488 | 0.0604 | 0.04 (0.00–0.60) | 0.02 |
| A | A | G | C | 0.0184 | 0.0149 | 0.11 (0.01–2.07) | 0.14 |
| G | G | G | T | 0.0745 | 0.0081 | 0.03 (0.00–1.02) | 0.05 |
| G | G | G | C | 0.0269 | 0.0084 | 0.14 (0.01–2.41) | 0.18 |
| A | G | G | C | 0 | 0.0242 | 2.51 (0.12–54.32) | 0.56 |
The expression level of Fas was determined in breast cancer and normal tissues by qRT-PCR using the 2−ΔΔCT method. The results showed that the expression levels of FAS mRNA were not significantly different between breast cancer and normal tissues (P = 0.588).
Discussion
Breast tumor cells frequently down-regulate FAS and/or up-regulate FASL expression [34], [35], [36], [37]. Death receptor activation initiates extrinsic apoptotic pathways, which in some cell types may be sufficient to carry apoptosis, whereas in others it also branches towards the intrinsic/mitochondrial pathway, via caspase-8-dependent Bid-cleavage [38], [39], [40]. Numerous polymorphism sites have been identified to influence the development of breast cancer [2], [7]. In the present study we found lack of association between FAS expression and breast cancer. In normal breast epithelium, Fas protein is expressed constitutively, while in primary breast cancer, its expression was found to be less uniform [41]. It has been reported that the lack of Fas in the primary tumor was associated with perilymphatic fat infiltration and metastasis either to the regional lymph nodes or to the bones [42], [43].
Several previous researches have focused on the association between different gene polymorphisms within cell death pathways and the risk of developing malignancies and developing cancers. FAS/FASL polymorphisms commands major attention as it may play different, context-dependent roles in cancer [44], [45]; 1) apoptosis promotion by FasL on T-lymphocytes in Fas-expressing cancer cells, this mechanism could play an important responsibility in cell mediated cytotoxic reactions against malignant cells. 2) malignant cells could escape immune system expression of FasL, thus repelling Fas-expressing immune-effector cells, hence preventing the immune system from recognizing mutated tumor cells (tumor as immuno-privileged site). 3) FasL expressed on T cells, may not only aids them to kill target (cancer) cells, but it was also associated with an enhanced rate of activation-induced cell death in T cells [46]. Various correlations have been drawn from genetic polymorphisms in the death pathway genes FAS and FASL and the risk of cancer development [47], [48], [49], [50], [51], [52].
In the present study, we investigate whether the FAS-1377 G/A, FAS-670 A/G, FASL-844 T/C and FASL Ivs2nt 124 A/G polymorphism in cell death pathway genes were associated with the risk of the development of breast cancer in a sample of Iranian population (south east of Iran). We found that the FAS-1377 G/A did not affect the risk of breast cancer, while FAS −670 G/A, FASL Ivs2nt 124 A/G and FASL −844 C/T gene polymorphisms are risk factors for this disease in our study population. Previous researches have also addressed FAS/FASL polymorphisms in breast cancer but in some of these studies the results only partially corroborated with our findings. Crew et al., have reported no association between breast cancer and FAS −1377 G/A, FAS −670 G/A and FASL −844C/T polymorphisms [53]. On the other hand, Zhang et al have found a significant association between FAS −1377G/A and FASL-844T/C gene polymorphisms and risk of breast cancer, but they reported no association between FAS −670 G/A and breast cancer risk [46]. We observed a significant association between breast cancer risk and FAS −670 G/A genotype, thus our findings show partly-different association pattern that the above mentioned ones. Moreover, they observed a significantly increased breast cancer risk associated with the FAS −1377G/A genotype, but we report lack of such association. They reported decreased risk associated with FASL −844TT (OR, 0.66; 95% CI, 0.43–1.00) genotype, but we found increased risk with mentioned genotype (OR, 2.40; 95% CI, 1.12–5.14). Contrary to our results, Krippl et al., in a study of 500 breast cancer patients and 500 controls in a Caucasian population in Austria, reported a significant association between FAS −1377G/A and increased risk of breast cancer, but they found no associations with FAS −670G/A and FASL −844C/T [54]. The FASL −844 C/T has a considerable impact on the promoter activity of the FASL gene in an in vitro assay system, because the polymorphism affects a binding affinity for the transcription factor C/EBP. A significantly higher basal expression of FasL is associated with the FasL −844 C allele compared with the T allele [27]. Hence, the FASL −844 C allele, which drives a higher expression of FASL in T cells, was associated with an enhanced rate of activation-induced cell death in T cells [46]. It has been reported that FASL −844 T allele has a possible protective effect on cancer risk [55]. Although the exact mechanism for this inverse association in our study was not clear, other polymorphisms of FASL may also play a role. There is no clear explanation for deviation from HWE in our population. The possible reason may be due to genetic drift.
We recognize some limitations of the present study: (i) we have no data on some known risk factors (e.g., family history, previous benign conditions, oral contraceptive or hormone therapy use, etc.), (ii) a relatively small sample size. Nevertheless, we believe that the presented here data provides an important input into the debate regarding the clinical relevance of investigated polymorphisms.
Our work is a valuable addition to the ongoing discussion on the influence of various polymorphisms within the signaling molecules pertaining to cell death pathways, and cancer formation. The so far only partial overlapping of conclusions reached from our data analysis as compared to previously published studies (please see above) may reflect differences in genetic background between the studied populations, and variations in external (i.e. environmental), or other (i.e. obesity) factors that influence studied populations [5], [8], [56]. As cell death pathways are ubiquitous, active in most cells in eucaryotes, our manuscript provides additional voice in the discussion on the role of Fas/FasL system on the clinical outcome of breast- and other cancers. One has to keep in mind that although apoptosis is the dominant, fastest cell death program, autophagy appears to play increasingly important role as an accessory cell death mechanism [57], [58], [59].
Funding Statement
SG was supported by Parker B. Francis Fellowship in Respiratory Disease. ML, WC, and MVJ kindly acknowledge the core/startup support from Linköping University, from Integrative Regenerative Medicine Center (IGEN), and from Cancerfonden (CAN 2011/521). No additional external funding received for this study. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Stecklein SR, Jensen RA, Pal A (2012) Genetic and epigenetic signatures of breast cancer subtypes. Frontiers in bioscience 4: 934–949. [DOI] [PubMed] [Google Scholar]
- 2. Wiechec E (2011) Implications of genomic instability in the diagnosis and treatment of breast cancer. Expert Rev Mol Diagn 11: 445–453. [DOI] [PubMed] [Google Scholar]
- 3. Mousavi SM, Montazeri A, Mohagheghi MA, Jarrahi AM, Harirchi I, et al. (2007) Breast cancer in Iran: an epidemiological review. Breast J 13: 383–391. [DOI] [PubMed] [Google Scholar]
- 4. Babu GR, Samari G, Cohen SP, Mahapatra T, Wahbe RM, et al. (2011) Breast cancer screening among females in iran and recommendations for improved practice: a review. Asian Pacific journal of cancer prevention : APJCP 12: 1647–1655. [PubMed] [Google Scholar]
- 5. Gurevich-Panigrahi T, Panigrahi S, Wiechec E, Los M (2009) Obesity: pathophysiology and clinical management. Curr Med Chem 16: 506–521. [DOI] [PubMed] [Google Scholar]
- 6. Nordgard SH, Alnaes GI, Hihn B, Lingjaerde OC, Liestol K, et al. (2008) Pathway based analysis of SNPs with relevance to 5-FU therapy: relation to intratumoral mRNA expression and survival. Int J Cancer 123: 577–585. [DOI] [PubMed] [Google Scholar]
- 7. Wiechec E, Hansen LL (2009) The effect of genetic variability on drug response in conventional breast cancer treatment. Eur J Pharmacol 625: 122–130. [DOI] [PubMed] [Google Scholar]
- 8. Ghavami S, Hashemi M, Ande SR, Yeganeh B, Xiao W, et al. (2009) Apoptosis and cancer: mutations within caspase genes. J Med Genet 46: 497–510. [DOI] [PubMed] [Google Scholar]
- 9. Wong RS (2011) Apoptosis in cancer: from pathogenesis to treatment. Journal of experimental & clinical cancer research : CR 30: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Evan GI, Vousden KH (2001) Proliferation, cell cycle and apoptosis in cancer. Nature 411: 342–348. [DOI] [PubMed] [Google Scholar]
- 11. Fulda S (2010) Modulation of apoptosis by natural products for cancer therapy. Planta medica 76: 1075–1079. [DOI] [PubMed] [Google Scholar]
- 12. Los M, Wesselborg S, Schulze-Osthoff K (1999) The role of caspases in development, immunity, and apoptotic signal transduction: lessons from knockout mice. Immunity 10: 629–639. [DOI] [PubMed] [Google Scholar]
- 13. Rashedi I, Panigrahi S, Ezzati P, Ghavami S, Los M (2007) Autoimmunity and Apoptosis – Therapeutic Implications. Curr Med Chem 14: 3139–3159. [DOI] [PubMed] [Google Scholar]
- 14. Los M, van de Craen M, Penning CL, Schenk H, Westendorp M, et al. (1995) Requirement of an ICE/Ced-3 protease for Fas/Apo-1-1mediated apoptosis. Nature 371: 81–83. [DOI] [PubMed] [Google Scholar]
- 15. Villa-Morales M, Fernandez-Piqueras J (2012) Targeting the Fas/FasL signaling pathway in cancer therapy. Expert Opin Ther Targets [DOI] [PubMed] [Google Scholar]
- 16. O'Connell J, O'Sullivan GC, Collins JK, Shanahan F (1996) The Fas counterattack: Fas-mediated T cell killing by colon cancer cells expressing Fas ligand. The Journal of experimental medicine 184: 1075–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Peduto Eberl L, Guillou L, Saraga E, Schroter M, French LE, et al. (1999) Fas and Fas ligand expression in tumor cells and in vascular smooth-muscle cells of colonic and renal carcinomas. Int J Cancer 81: 772–778. [DOI] [PubMed] [Google Scholar]
- 18. O'Brien DI, Nally K, Kelly RG, O'Connor TM, Shanahan F, et al. (2005) Targeting the Fas/Fas ligand pathway in cancer. Expert Opin Ther Targets 9: 1031–1044. [DOI] [PubMed] [Google Scholar]
- 19. Brando C, Mukhopadhyay S, Kovacs E, Medina R, Patel P, et al. (2005) Receptors and lytic mediators regulating anti-tumor activity by the leukemic killer T cell line TALL-104. J Leukoc Biol 78: 359–371. [DOI] [PubMed] [Google Scholar]
- 20. Ni X, Hazarika P, Zhang C, Talpur R, Duvic M (2001) Fas ligand expression by neoplastic T lymphocytes mediates elimination of CD8+ cytotoxic T lymphocytes in mycosis fungoides: a potential mechanism of tumor immune escape? Clin Cancer Res 7: 2682–2692. [PubMed] [Google Scholar]
- 21. Los M, Herr I, Friesen C, Fulda S, Schulze-Osthoff K, et al. (1997) Cross-resistance of CD95- and drug-induced apoptosis as a consequence of deficient activation of caspases (ICE/Ced-3 proteases). Blood 90: 3118–3129. [PubMed] [Google Scholar]
- 22. Wesselborg S, Engels IH, Rossmann E, Los M, Schulze-Osthoff K (1999) Anticancer drugs induce caspase-8/FLICE activation and apoptosis in the absence of CD95 receptor/ligand interaction. Blood 93: 3053–3063. [PubMed] [Google Scholar]
- 23. Wieder T, Essmann F, Prokop A, Schmelz K, Schulze-Osthoff K, et al. (2001) Activation of caspase-8 in drug-induced apoptosis of B-lymphoid cells is independent of CD95/Fas receptor-ligand interaction and occurs downstream of caspase-3. Blood 97: 1378–1387. [DOI] [PubMed] [Google Scholar]
- 24. Huang QR, Morris D, Manolios N (1997) Identification and characterization of polymorphisms in the promoter region of the human Apo-1/Fas (CD95) gene. Molecular immunology 34: 577–582. [DOI] [PubMed] [Google Scholar]
- 25. Sibley K, Rollinson S, Allan JM, Smith AG, Law GR, et al. (2003) Functional FAS promoter polymorphisms are associated with increased risk of acute myeloid leukemia. Cancer Res 63: 4327–4330. [PubMed] [Google Scholar]
- 26. Takahashi T, Tanaka M, Inazawa J, Abe T, Suda T, et al. (1994) Human Fas ligand: gene structure, chromosomal location and species specificity. International immunology 6: 1567–1574. [DOI] [PubMed] [Google Scholar]
- 27. Wu J, Metz C, Xu X, Abe R, Gibson AW, et al. (2003) A novel polymorphic CAAT/enhancer-binding protein beta element in the FasL gene promoter alters Fas ligand expression: a candidate background gene in African American systemic lupus erythematosus patients. J Immunol 170: 132–138. [DOI] [PubMed] [Google Scholar]
- 28. Koshkina NV, Kleinerman ES, Li G, Zhao CC, Wei Q, et al. (2007) Exploratory analysis of Fas gene polymorphisms in pediatric osteosarcoma patients. Journal of pediatric hematology/oncology : official journal of the American Society of Pediatric Hematology/Oncology 29: 815–821. [DOI] [PubMed] [Google Scholar]
- 29. Yang M, Sun T, Wang L, Yu D, Zhang X, et al. (2008) Functional variants in cell death pathway genes and risk of pancreatic cancer. Clin Cancer Res 14: 3230–3236. [DOI] [PubMed] [Google Scholar]
- 30. Hashemi M, Moazeni-Roodi AK, Fazaeli A, Sandoughi M, Bardestani GR, et al. (2010) Lack of association between paraoxonase-1 Q192R polymorphism and rheumatoid arthritis in southeast Iran. Genet Mol Res 9: 333–339. [DOI] [PubMed] [Google Scholar]
- 31. Hashemi M, Moazeni-Roodi AK, Fazaeli A, Sandoughi M, Taheri M, et al. (2010) The L55M polymorphism of paraoxonase-1 is a risk factor for rheumatoid arthritis. Genet Mol Res 9: 1735–1741. [DOI] [PubMed] [Google Scholar]
- 32. Hashemi M, Moazeni-Roodi A, Bahari A, Taheri M (2012) A Tetra-Primer Amplification Refractory Mutation System-Polymerase Chain Reaction for the Detection of rs8099917 IL28B Genotype. Nucleosides Nucleotides Nucleic Acids 31: 55–60. [DOI] [PubMed] [Google Scholar]
- 33. Das H, Koizumi T, Sugimoto T, Chakraborty S, Ichimura T, et al. (2000) Quantitation of Fas and Fas ligand gene expression in human ovarian, cervical and endometrial carcinomas using real-time quantitative RT-PCR. Br J Cancer 82: 1682–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gutierrez LS, Eliza M, Niven-Fairchild T, Naftolin F, Mor G (1999) The Fas/Fas-ligand system: a mechanism for immune evasion in human breast carcinomas. Breast cancer research and treatment 54: 245–253. [DOI] [PubMed] [Google Scholar]
- 35. Mottolese M, Buglioni S, Bracalenti C, Cardarelli MA, Ciabocco L, et al. (2000) Prognostic relevance of altered Fas (CD95)-system in human breast cancer. International journal of cancer Journal international du cancer 89: 127–132. [DOI] [PubMed] [Google Scholar]
- 36. Muschen M, Moers C, Warskulat U, Even J, Niederacher D, et al. (2000) CD95 ligand expression as a mechanism of immune escape in breast cancer. Immunology 99: 69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Reimer T, Herrnring C, Koczan D, Richter D, Gerber B, et al. (2000) FasL:Fas ratio–a prognostic factor in breast carcinomas. Cancer research 60: 822–828. [PubMed] [Google Scholar]
- 38. Martin TR, Hagimoto N, Nakamura M, Matute-Bello G (2005) Apoptosis and epithelial injury in the lungs. Proceedings of the American Thoracic Society 2: 214–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dufournet C, Uzan C, Fauvet R, Cortez A, Siffroi JP, et al. (2006) Expression of apoptosis-related proteins in peritoneal, ovarian and colorectal endometriosis. Journal of reproductive immunology 70: 151–162. [DOI] [PubMed] [Google Scholar]
- 40. Ghavami S, Eshraghi M, Kadkhoda K, Mutawe MM, Maddika S, et al. (2009) Role of BNIP3 in TNF-induced cell death–TNF upregulates BNIP3 expression. Biochimica et biophysica acta 1793: 546–560. [DOI] [PubMed] [Google Scholar]
- 41. Leithauser F, Dhein J, Mechtersheimer G, Koretz K, Bruderlein S, et al. (1993) Constitutive and induced expression of APO-1, a new member of the nerve growth factor/tumor necrosis factor receptor superfamily, in normal and neoplastic cells. Laboratory investigation; a journal of technical methods and pathology 69: 415–429. [PubMed] [Google Scholar]
- 42. Bebenek M, Dus D, Kozlak J (2007) Fas/Fas-ligand expressions in primary breast cancer are significant predictors of its skeletal spread. Anticancer Res 27: 215–218. [PubMed] [Google Scholar]
- 43. Bebenek M, Dus D, Kozlak J (2008) Fas expression in primary breast cancer is related to neoplastic infiltration of perilymphatic fat. Adv Med Sci 53: 49–53. [DOI] [PubMed] [Google Scholar]
- 44. Sun T, Miao X, Zhang X, Tan W, Xiong P, et al. (2004) Polymorphisms of death pathway genes FAS and FASL in esophageal squamous-cell carcinoma. Journal of the National Cancer Institute 96: 1030–1036. [DOI] [PubMed] [Google Scholar]
- 45. Liu K, Caldwell SA, Abrams SI (2005) Immune selection and emergence of aggressive tumor variants as negative consequences of Fas-mediated cytotoxicity and altered IFN-gamma-regulated gene expression. Cancer research 65: 4376–4388. [DOI] [PubMed] [Google Scholar]
- 46. Zhang B, Sun T, Xue L, Han X, Lu N, et al. (2007) Functional polymorphisms in FAS and FASL contribute to increased apoptosis of tumor infiltration lymphocytes and risk of breast cancer. Carcinogenesis 28: 1067–1073. [DOI] [PubMed] [Google Scholar]
- 47. Sun T, Miao X, Zhang X, Tan W, Xiong P, et al. (2004) Polymorphisms of death pathway genes FAS and FASL in esophageal squamous-cell carcinoma. J Natl Cancer Inst 96: 1030–1036. [DOI] [PubMed] [Google Scholar]
- 48. Sun T, Zhou Y, Li H, Han X, Shi Y, et al. (2005) FASL −844C polymorphism is associated with increased activation-induced T cell death and risk of cervical cancer. J Exp Med 202: 967–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kurooka M, Nuovo GJ, Caligiuri MA, Nabel GJ (2002) Cellular localization and function of Fas ligand (CD95L) in tumors. Cancer Res 62: 1261–1265. [PubMed] [Google Scholar]
- 50. Lee SH, Shin MS, Lee HS, Bae JH, Lee HK, et al. (2001) Expression of Fas and Fas-related molecules in human hepatocellular carcinoma. Hum Pathol 32: 250–256. [DOI] [PubMed] [Google Scholar]
- 51. Zhang X, Miao X, Sun T, Tan W, Qu S, et al. (2005) Functional polymorphisms in cell death pathway genes FAS and FASL contribute to risk of lung cancer. J Med Genet 42: 479–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhou RM, Wang N, Chen ZF, Duan YN, Sun DL, et al. (2010) Polymorphisms in promoter region of FAS and FASL gene and risk of gastric cardiac adenocarcinoma. J Gastroenterol Hepatol 25: 555–561. [DOI] [PubMed] [Google Scholar]
- 53. Crew KD, Gammon MD, Terry MB, Zhang FF, Agrawal M, et al. (2007) Genetic polymorphisms in the apoptosis-associated genes FAS and FASL and breast cancer risk. Carcinogenesis 28: 2548–2551. [DOI] [PubMed] [Google Scholar]
- 54. Krippl P, Langsenlehner U, Renner W, Koppel H, Samonigg H (2004) Re: Polymorphisms of death pathway genes FAS and FASL in esophageal squamous-cell carcinoma. J Natl Cancer Inst 96: 1478–1479; author reply 1479. [DOI] [PubMed] [Google Scholar]
- 55. Zhang Z, Qiu L, Wang M, Tong N, Li J (2009) The FAS ligand promoter polymorphism, rs763110 (−844C>T), contributes to cancer susceptibility: evidence from 19 case-control studies. Eur J Hum Genet 17: 1294–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wiechec E, Wiuf C, Overgaard J, Hansen LL (2011) High-resolution melting analysis for mutation screening of RGSL1, RGS16, and RGS8 in breast cancer. Cancer Epidemiol Biomarkers Prev 20: 397–407. [DOI] [PubMed] [Google Scholar]
- 57. Alavian SM, Ande SR, Coombs KM, Yeganeh B, Davoodpour P, et al. (2011) Virus-triggered autophagy in viral hepatitis - possible novel strategies for drug development. J Viral Hepat 18: 821–830. [DOI] [PubMed] [Google Scholar]
- 58. Amaravadi RK, Thompson CB (2007) The roles of therapy-induced autophagy and necrosis in cancer treatment. Clin Cancer Res 13: 7271–7279. [DOI] [PubMed] [Google Scholar]
- 59. Ghavami S, Mutawe MM, Sharma P, Yeganeh B, McNeill KD, et al. (2011) Mevalonate Cascade Regulation of Airway Mesenchymal Cell Autophagy and Apoptosis: A Dual Role for p53. PLoS One 6: e16523. [DOI] [PMC free article] [PubMed] [Google Scholar]