Abstract
The characterization of biomolecules from ancient samples can shed otherwise unobtainable insights into the past. Despite the fundamental role of transcriptomal change in evolution, the potential of ancient RNA remains unexploited – perhaps due to dogma associated with the fragility of RNA. We hypothesize that seeds offer a plausible refuge for long-term RNA survival, due to the fundamental role of RNA during seed germination. Using RNA-Seq on cDNA synthesized from nucleic acid extracts, we validate this hypothesis through demonstration of partial transcriptomal recovery from two sources of ancient maize kernels. The results suggest that ancient seed transcriptomics may offer a powerful new tool with which to study plant domestication.
Introduction
Small changes at the DNA level frequently result in profound and unpredictable changes at the RNA level. Therefore, transcriptomics - the genome-wide expression profiling of RNA – plays a fundamental role of describing evolutionary change, at the molecular, cellular, and phenotypic levels [1]. Some of the most well-documented effects of transcriptomic change concern crops such as maize (Zea mays spp. mays), that have undergone extensive morphological changes over the past 10,000 years as a consequence of domestication [2]–[5]. The domestication process itself is of key interest in archaeological science, and ancient DNA studies of desiccated maize remains have offered important insights such as characterizing the timing of selection for traits [6], and documenting the spread of cultivation [7].
Kernels are one of the most relevant tissues for study in maize breeding and evolution: they contain the nutritional value, and are well-preserved tissues to study as ancient remains [8]. Hence, the characterization of RNA from ancient maize kernels would greatly complement and extend the scope of domestication studies, while unlike the genome, which is generally fixed for a cell line, the transcriptome can vary as a result of external environmental conditions. However, it is unclear whether transcripts appearing in ancient samples would reflect the true composition of transcripts when the plant was alive. Furthermore, the transcriptome reflects the genes, which are being actively expressed at any given time (excluding mutations), allowing the characterization of the functional evolution associated with domestication.
Additionally, the transcriptome could enable the reconstruction of the tempo, and strength, of up/down regulation of protein expression and its relation with phenotypic features, such as morphology, nutritional value, and even pathogen defense. Current analyses in maize leaf trancriptomics [9], for example, have shown that it is possible to exploit the continuous developmental gradient in one tissue to investigate spatial differentiation. Furthermore, modern transcriptomics has also proven relevant for a series of features in model organisms of major functional and evolutionary interest for domestic crops. This includes desiccation tolerance in seeds [10], storage protein transcripts [11], endosperm development and starch filling [12], and even seed dormancy and germination, as discussed by [13].
Despite significant technical advances in the field of ancient genetics, recently cumulating in the publication of several complete ancient genomes [14]–[17], reports of ancient RNA are limited. This may be due to the dogma that RNA, being notoriously labile in the laboratory, has little chance of post mortem survival, given both the ubiquitous presence of RNases, and that the 2′-hydroxyl group of RNA is particularly susceptible to hydrolytic degradation [18]. Despite this, circumstantial evidence suggests long-term survival of RNA in some tissues, which could potentially be exploited in the genetic context. One such example is desiccated seeds, which require survival of diverse RNA transcripts for successful germination [19], and thus are likely to have mechanisms to limit the rate of RNA degradation (e.g. RNA chaperones [20]). That this RNA may persist over long time periods has been attested by both nucleic acid hybridization and mass spectrometric detection of short pieces of ancient RNA derived from radish, cress and maize seeds dating back as far as 3,300 years [21]–[23] (although these studies could not rule out contamination or DNA degradation as the source of the detected uracil), and the germination of a 2,000 year old date palm seed [24].
We hypothesize that these observations, coupled with the power of RNA-Seq [1], could in principle, make ancient seed transcriptomics feasible. To test our hypothesis we have explored, through second-generation sequencing (SGS) and bioinformatic characterization, RNA recovered from desiccated maize kernels from Turkey House Ruin in Navajo County, Arizona (Figure S1), dated to 723±23 14C YBP (Supplementary Methods S1, Figure S2 and Table S1).
Materials and Methods
The samples
A total of 6 maize kernels were utilized in this study. The kernels (Figure S1) were excavated as a single batch (batch 935) at the Turkey House Ruin, Arizona, in 1915 and have been stored at room temperature at the Arizona State Museum (ASM) since. The maize was examined and identified as white or yellow flint corn and reported to be so uniform that they could have come from 1 ear or from very similar ears [25]. Table 1 contains details of the kernel and sample names.
Table 1. Kernel and sample information and names.
| Kernel | Nucleic Acids | Sample Name | Batch | Location | Age | SGS Platform |
| 9351 | RNA | FLX1 | 935 | Turkey House Ruin, Arizona | 723±23 14C YBP | GS FLX |
| DNA | FLX4 | 935 | Turkey House Ruin, Arizona | 723±23 14C YBP | GS FLX | |
| 9352 | RNA | FLX2 | 935 | Turkey House Ruin, Arizona | 723±23 14C YBP | GS FLX |
| DNA | FLX5 | 935 | Turkey House Ruin, Arizona | 723±23 14C YBP | GS FLX | |
| 9353 | RNA | FLX3 | 935 | Turkey House Ruin, Arizona | 723±23 14C YBP | GS FLX |
| DNA | FLX6 | 935 | Turkey House Ruin, Arizona | 723±23 14C YBP | GS FLX | |
| 9354 | RNA | 935130 | 935 | Turkey House Ruin, Arizona | 723±23 14C YBP | HiSeq2000 |
| 9355 | RNA | 935230 | 935 | Turkey House Ruin, Arizona | 723±23 14C YBP | HiSeq2000 |
| 9356 | DNA | AZ Shotgun [25] | 935 | Turkey House Ruin, Arizona | 723±23 14C YBP | GAIIx |
A total of 6 kernels were utilized for either RNA or DNA extraction only, or co-extracted for DNA and RNA. After nucleic acids were co-extracted, samples were divided into 2 aliquots, and RNA samples were DNase treated and DNA samples were RNase treated (see Methods and Supplementary Methods S1), and labeled with unique sample names.
Nucleic acid extraction
All extractions and library builds were carried out in a dedicated ancient DNA laboratory, physically separated from any modern maize and amplified nucleic acids. Both DNA and RNA were examined in this study, so that a comparison between the DNA and RNA content of the kernels could be made. Nucleic acid extraction from the ancient maize kernels was performed using an optimized protocol, involving initial bleaching of the testa to minimize external contaminant carryover, endosperm digestion in a customized buffer (Supplementary Methods S1), co-extraction of DNA and RNA using organic solvents, DNase treatment, and complimentary DNA (cDNA) synthesis.
Complimentary DNA synthesis
Prior to cDNA synthesis, the RNA extracts were treated using DNase I to eliminate the DNA content, following the manufacturer's protocol (Invitrogen, Carlsbad, CA). First-strand cDNA synthesis was performed using random hexamers and a Superscript III First-Strand cDNA Synthesis Kit (Invitrogen), following the manufacturer's instructions. Second-strand synthesis was performed on the cDNA samples, using the Superscript Double-Stranded cDNA Kit (Invitrogen), following the manufacturer's instructions.
Testing for the presence of DNA and RNA
A series of PCR amplifications were performed to (i) investigate DNA and RNA survival in the kernels, to (ii) verify the efficiency of the extraction method at extracting both DNA and RNA, to (iii) verify the efficacy of the DNase I treatment at removing DNA from the extracts, and (iv) to verify the efficacy of the cDNA synthesis method at reverse transcribing RNA into cDNA. The successful PCR products were not sequenced, thus should only be viewed as indicative of DNA or RNA survival.
Specifically, three primer sets were designed (see Supplementary Methods S1 for primer sequences) to amplify (i) DNA only, (ii) DNA and RNA, and (iii) maize RNA only. cDNA and DNA samples were amplified with all three primer sets. For cDNA samples to be included for sequencing they could only be amplified with the latter two primer sets, thus ensuring that the majority of the cDNA nucleic acid content was RNA. Likewise, for DNA samples to be included, they could only be amplified with the first primer set. Included samples were then converted into libraries for deep sequencing on two different SGS platforms.
Roche GS FLX sequencing
Firstly, 6 extracts (3 cDNA and 3 DNA, from the same 3 kernels – Table 1) were sequenced on the Roche Genome Sequencer (GS) FLX (Roche, Basel, Switzerland). This platform is capable of sequencing reads of up to ∼500 bp (up to ∼1 kb using FLX+ chemistry), allowing us to assess the fragment length distribution of our ancient maize kernel RNA and DNA (see Table 1).
Double stranded cDNA from the three kernels (Table 1), samples FLX1, FLX2 and FLX3, was constructed into libraries and sequenced on one lane of a GS FLX PicoTiterPlate (PTP) each, following the manufacturer's Multiple Identifier (MID) Library Preparation Protocol (excluding fragmentation and short fragment removal) using LR70 chemistry.
GS FLX libraries were also constructed based on the DNA extracted from the same three maize kernels, and samples were labeled FLX4, FLX5 and FLX6 (Table 1). Prior to library construction, the DNA containing elute was treated with RNase A (Invitrogen) following the manufacturer's protocol, and were then sequenced on the GS FLX in the same manner as the cDNA.
Illumina HiSeq 2000 sequencing
Secondly, two cDNA samples were sequenced on two lanes of the Illumina HiSeq2000 platform (Illumina, San Diego, CA, USA). Unlike the GS FLX, the conventional read length limit is ∼100 bp. However, the advantage of this platform is that it is able to generate up to 200 Gb per run, allowing us to fully explore the endogenous nucleic acid content of our ancient maize kernels. Double stranded cDNA, from 2 kernels (935130 and 935230), was converted into Illumina HiSeq2000 libraries using the NEBNext Quick DNA Sample Prep Master Mix Set 2 (NEB, Ipswich, MA, USA), following the manufacturer's protocol (excluding fragmentation and small fragment removal steps), and indexed using Multiplex Adapters (Mutliplexing Sample Preparation Oligonucleotide Kit, Illumina). HiSeq2000 sequencing was performed following the manufacturer's protocol for single-read, 100 bp settings.
Results
GS FLX sequencing
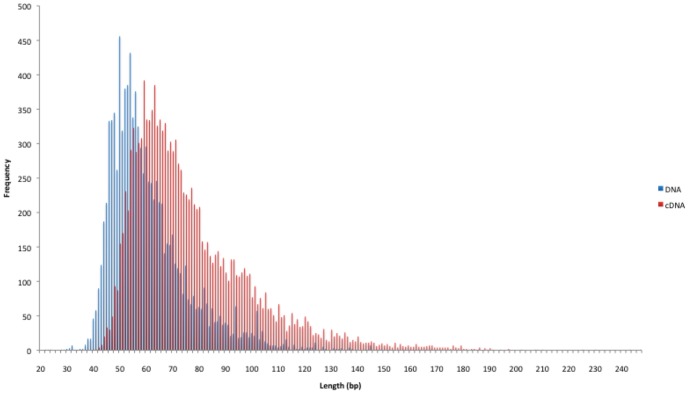
Three cDNA and 3 DNA samples were sequenced on the Roche GS FLX, producing a total of 22,338 sequences from all the cDNA libraries (FLX1-3) and a total of 31,010 sequences from all the DNA libraries (FLX4-6). The sequence lengths obtained ranged from 40–247 bp (Fig. 1) for cDNA, and 22–186 bp for DNA, although these lengths cannot be taken at face value due to (i) automatic filtering out of short reads by the GS FLX sequencer software, and (ii) the longest reads being limited in length by the read capacity of the GS FLX sequencer under LR70 chemistry.
Figure 1. Histogram showing fragment length distribution of maize GS FLX sequence reads from 3 Arizonan DNA (FLX4-6) samples (blue) in comparison to 3 Arizonan cDNA (FLX1-3) samples (red).
HiSeq 2000 sequencing
Two Arizona kernels (935130 and 935230) were constructed into cDNA libraries and deep sequenced on one lane each using the HiSeq2000. A total of ∼260 million reads were generated from the 2 samples. DNA was sequenced in a previously published study [26], using the Illumina Genome Analyzer (GA) IIx.
Data analysis
Endogenous nucleic acid content analysis
In order to explore the endogenous nucleic acids content, GS FLX reads from all cDNA and DNA samples were converted to fastq format and mapped to the B73 maize genome [27], using a combination of BWA (version 0.5.9-r16) [28] and BLAT [29] (Table S2). Potential PCR duplicates were removed using Samtools' rmdup and ambiguous hits were removed by setting a mapping quality filter of 25 and by controlling for XT and XA tags. Even though the endogenous nucleic acid content was high in both cDNA and DNA libraries (48–92% and 42–68%, respectively), the total yield of non-clonal, uniquely mapped reads was low and did not allow a reliable quantification of gene overlap (Table S2). All bam files were manipulated using Samtools as well as Perl and R scripts.
HiSeq 2000 reads were mapped and processed in the same way as GS FLX reads with the exception that, prior to mapping, reads were manipulated with an in-house script (available on request) to remove sequencing adapters and low-quality stretches in the 3′ end of reads. cDNA data was compared with DNA, from kernels from the same batch (Table 1) sequenced in [26].
Endogenous nucleic acid content in HiSeq 2000 libraries (Table S3) was similar to that observed in the GS FLX libraries (Table S2) for both the cDNA and DNA, however the yield of uniquely mapped sequences was ∼200 times higher, allowing a more in-depth analysis of cDNA reads. HiSeq 2000 reads were visualized and contigs, defined as contiguous covered regions in the reference genome, were generated based on the cDNA library with the most reads (935130) using SeqMonk [30]. Additionally, we compared the percentage of reads that mapped to maize, in a non-maize sample (archaeological sunflower seed – results not shown), as to ensure that maize reads detected in our maize samples were not due to contamination occurring in the laboratory. For 23,615,527 indexed reads, we obtained 2711 (0.011%) maize reads, indicating that our high level of maize endogenous reads in both our DNA and RNA samples are not the product of contamination.
To evaluate the level of overlap between cDNA and DNA libraries, contig coordinates were then used to query the same regions in the mapped alignments for the two remaining samples and the read content per contig was recorded.
Using read content quantifications per contig, a Pearson's correlation matrix was calculated to compare cDNA and shotgun experiments (Table 2). A positive correlation was found for cDNA samples (0.8837) whereas negative correlations were found for both cDNA-shotgun comparisons (−0.0027 and −0.0099).
Table 2. Positive correlation matrix of contig read content from cDNA (935130 and 935230) and DNA [25].
| 935130 | 935230 | DNA | |
| 935130 | 1 | 0.08837647 | −0.002722623 |
| 935230 | 0.8837647 | 1 | −0.009936311 |
Finally, mapDamage [30] was used to visualize the fragmentation and misincorporation patterns of both cDNA and DNA libraries. The results of this analysis can be found in Figure S3, S4, and S5.
Identification of functional exon information
In order to extract potential functional information, B73 [27] exon annotations were overlapped with the position of reads. Most of the hits had no associated description; only few annotations with functional description were retrieved with more than three overlapping reads. The top 50 functionally annotated hits for sample 935130 is shown in Table 3 (Table S4) and the full list of exon hits with more than three reads can be found in Tables S5 and S6. The most commonly occurring gene hits were RING zinc finger protein-like, TMV response-related protein, heat shock protein 101 and multidrug resistance associated protein 1. All of these genes are linked with water deficiency and stress response; hence perhaps it is not surprising that we see these genes frequently.
Table 3. Top 50 exon hits with functional annotation from cDNA (935130 and 935230) HiSeq 2000 maize reads (for a detailed version of the table see Supplementary Table S4).
| Sample | Reads | Exon Description |
| 935130 | 3 | cupin, RmlC-type |
| 935130 | 3 | Membrane protein |
| 935130 | 3 | AIR12 |
| 935130 | 3 | Disease resistance gene analog PIC15 Fragment |
| 935130 | 3 | WRKY69 - superfamily of TFs having WRKY and zinc finger domains |
| 935130 | 3 | IQ calmodulin-binding motif family protein |
| 935130 | 3 | cytokinin-O-glucosyltransferase 1 |
| 935130 | 3 | nitrate and chloride transporter |
| 935130 | 3 | invertase cell wall4 (incw4) |
| 935130 | 5 | anthranilate phosphoribosyltransferase-like protein |
| 935130 | 3 | inhibitor of apoptosis-like protein |
| 935130 | 3 | calmodulin-related protein 2, touch-induced |
| 935130 | 3 | meiosis 5 |
| 935230 | 3 | amidophosphoribosyltransferase |
| 935130 | 3 | CDPK protein |
| 935130 | 3 | DNA binding protein |
| 935130 | 3 | WRKY71 - superfamily of TFs having WRKY and zinc finger domains |
| 935130 | 3 | 60S ribosomal protein L19-3 |
| 935130 | 3 | F-box domain containing protein |
| 935130 | 3 | MADS-box transcription factor 26 |
| 935230 | 3 | RING zinc finger protein-like |
| 935130 | 3 | ring canal kelch |
| 935130 | 3 | plant-specific domain TIGR01568 family protein |
| 935130 | 3 | glycerol-3-phosphate acyltransferase 8 |
| 935130 | 3 | ethanolaminephosphotransferase |
| 935230 | 3 | transmembrane BAX inhibitor motif-containing protein 4 |
| 935230 | 3 | elongation factor Tu |
| 935130 | 3 | TMV response-related protein |
| 935130 | 3 | fasciclin-like arabinogalactan protein 8 |
| 935130 | 3 | anther-specific proline-rich protein APG |
| 935230 | 3 | heat-shock protein 101 |
| 935230 | 3 | CCCH transcription factor |
| 935130 | 3 | ubiquitin-protein ligase |
| 935130 | 3 | WRKY DNA-binding protein |
| 935130 | 3 | TMV response-related protein |
| 935130 | 3 | MTD1 |
| 935130 | 3 | Nodulation signaling pathway 2 protein |
| 935130 | 3 | F-box protein |
| 935130 | 3 | cellulose synthase8 |
| 935130 | 4 | metacaspase type II |
| 935130 | 3 | sialyltransferase-like protein |
| 935130 | 3 | 3-methyl-2-oxobutanoate hydroxymethyltransferase |
| 935130 | 3 | indole-3-acetate beta-glucosyltransferase |
| 935130 | 3 | Serine threonine kinase |
| 935130 | 4 | glucan endo-1,3-beta-glucosidase 5 |
| 935130 | 3 | sulfate transporter 3.4 |
| 935230 | 3 | multidrug resistance associated protein 1 |
| 935230 | 3 | hexose carrier protein HEX6 |
| 935130 | 3 | beta-fructofuranosidase, insoluble isoenzyme 2 |
| 935130 | 3 | invertase cell wall3 |
For the detection of exon-exon junctions, the totality of unmapped GS FLX reads were subjected to BLAT searches against B73 maize reference genome [27]. BLAT mapping allows a more accurate estimate of endogenous DNA in our samples since it permits split-read mapping, which is important in regions with genomic rearrangements, and necessary to find exon-exon junctions.
For the DNA (AZ Shotgun) from [26] and both HiSeq 2000 cDNA data sets, sequence duplicates were removed before BLAT search, to avoid redundancy as well as computational time. All reads with two hits in the same chromosome and strand were further analyzed in search of possible splice sites, however, no exon-exon junctions were found.
Repeat content analysis
Repeat content of sequences was explored using RepeatMasker [32] (RM database version 20110419) and repeat content profiles were compared between cDNA and DNA [26] samples. Sequences were retrieved from bam files where clones had been removed and paralogs were still present. Keeping paralogs was important since their removal would cause all the reads within the repeated regions to be excluded from the analysis. For the DNA sample [26] a subsample of 100,000 was randomly selected for this analysis. The dominant elements reported by the RepeatMasker from the two cDNA and one DNA sample can be found in Table 4 (Table S7 for unabridged version).
Table 4. Dominant elements represented in repeat content profiles of cDNA and DNA sequences using RepeatMasker.
| Elements | No. of Elements | Length Occupied (bp) | Percentage of sequences (%) | ||||||
| DNA | 935130 | 935230 | DNA | 935130 | 935230 | DNA | 935130 | 935230 | |
| Retroelements | 45,689 | 57 | 886 | 1,941,117 | 37,987 | 2,637 | 47.88 | 0.05 | 0.01 |
| Total interspersed repeats | 47,989 | 65 | 926 | 2,037,874 | 39,479 | 3,347 | 50.26 | 0.05 | 0.01 |
| Small RNA | 224 | 459,802 | 988,699 | 9,710 | 64,585,935 | 25,848,184 | 0.24 | 89.04 | 72.42 |
As expected, both cDNA libraries showed a depletion of interspersed repeats and an enrichment of smallRNA elements, comprising mainly long and short subunits of rRNA, representing on average ∼80% of the non-clonal mapped reads (Table 4 and Table S7).
Taxonomic characterization of unmapped reads
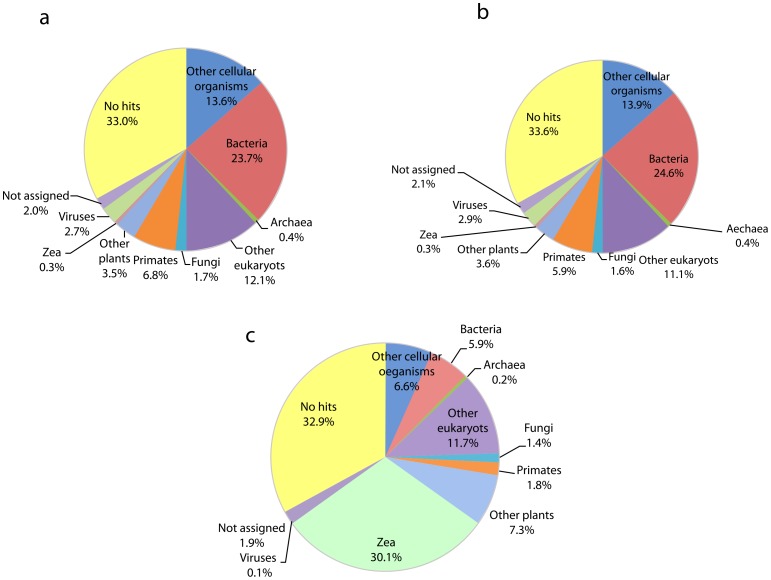
In order to explore the source of the non-maize sequences, 100,000 reads were randomly sampled from the unmapped reads from each sample and a BLAST search (-p blastn) [33] was performed against the nt database. BLAST outputs were then analyzed using MEGAN version .4.62.7 [34] with default parameters for the LCA-assignment algorithm. Approximately 35% of the reads fell in the unassigned or no hits category for all datasets. Among the assigned reads around a third corresponded to prokaryotes (bacteria and archaea) for the cDNA (Fig. 2a and b) and a much smaller fraction for DNA (Fig. 2c). The presence of fungal and primate hits, indicates some environmental and sampling contamination (Fig. 2). Furthermore, Zea mays sequences were also retrieved in the BLAST searches, most likely representing sequences not assembled into the reference B73 genome.
Figure 2. Taxonomic distribution of non-maize HiSeq 2000 reads from cDNA and DNA sets:
(a) represents cDNA sample 935130, (b) represents cDNA sample 935230, and (c) represents a DNA shotgun sample from [25] 100,00 randomly sampled unmapped reads were used to perform BLAST searches and MEGAN was for taxonomic characterization of non-maize reference genome (non-B73) reads.
Discussion
Several observations support the authenticity of the cDNA reads as being derived from ancient maize RNA (as opposed to undigested DNA or modern maize RNA), and that the content contains measurable levels of transcripts other than rRNA, thus are potentially useful for future paleotranscriptomic studies.
Firstly, the majority of the cDNA reads that matched maize were of ribosomal origin (∼80%), as shown by the repetitive elements analysis (based on Illumina data), which is in contrast to the rRNA content of the DNA library, where only 0.24% of the maize-matching reads were rRNA, likely reflecting a portion of rRNA gene content of the maize genome [35]. Moreover, depletion of interspersed repeats in cDNA libraries and enrichment in DNA, also support this observation.
Secondly, the taxonomic distribution of non-maize reads varies considerably when comparing the DNA [26] and the two cDNA samples (Fig. 2), whereas the cDNA samples are highly similar. Such difference is most likely explained by the distinct patterns of DNA and RNA survival of non-endogenous nucleic acid contamination. Furthermore, consistent with previous ancient genomic studies [36], the majority of the non-endogenous reads lacks a significant match to the nt database, a large proportion of which, may represent unsequenced environmental organisms or unassembled regions in the maize genome.
An unexpectedly large fraction of the taxonomic distribution corresponded to Zea (maize that has not been assembled to the B73 reference genome) in the DNA (Fig. 2c) set and but not in the RNA (Fig. 2a and b). In principle, this favors the notion that indeed RNA (from the cDNA samples), rather than DNA, was extracted from the maize kernels since functional and transcribed sequences are most likely to be conserved between B73 genome and other landraces, while random nuclear data is more prone to divergence and consequently, is unable to be mapped to the B73 genome. Alternatively, hits might also correspond to BAC clones not assembled into the maize reference genome but present in the nt database.
Thirdly, a positive correlation (Table 2) was found when comparing contig read content for cDNA samples whereas negative correlations were found for both cDNA-DNA comparisons implying an overlap of enriched regions in both cDNA experiments (Table 2) and suggesting a different nucleic acids source between cDNA and DNA libraries.
Lastly, fragmentation and misincorporation patterns of the cDNA (935130 and 935230) and DNA (AZ Shotgun) libraries, generated using mapDamage [31] (Figures S3, S4, and S5), were compared. For the DNA library, misincorporation as well as fragmentation plots clearly reflected patterns representative of aDNA: excess of purines (A and G) one genomic coordinate before the read, as well as C to T and G to A modifications towards the 5′ and 3′ ends of the reads [37]. In contrast, the cDNA libraries, while strikingly similar, showed none of the aforementioned features of the DNA sample. These results, therefore, imply that non-random mechanisms, different to those in DNA, are responsible for producing such patterns.
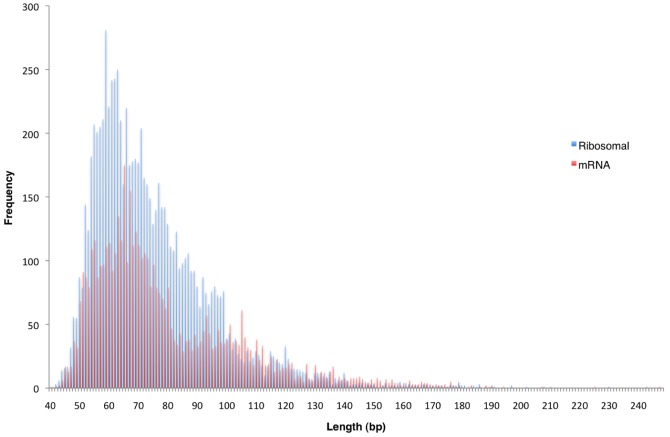
Further tests were performed to characterize the survival of our ancient maize RNA. Firstly, rRNA transcripts in vivo form secondary structures that result in large portions of the transcript existing as a double stranded molecule. A simple comparison of the mean, median and total read length distributions provides a simple means to investigate whether this double strandedness might confer a survival advantage of rRNA over other single stranded RNA transcripts (e.g. mRNAs). For GS FLX reads annotated as maize rRNA in a BLAST search, the mean and median lengths were 75.23 and 70 bp, respectively, while the lengths for mRNAs were 79.56 and 71 bp respectively. Thus, as visually demonstrated in Fig. 3, there appears to be no evidence of preferential survival of rRNA over mRNAs.
Figure 3. Histograms showing fragment length distribution of maize GS FLX sequence reads from cDNA samples (FLX1-3), comparing ribosomal RNA (red) to messenger RNA (blue) fragment lengths.
Additionally, a comparison of the maize DNA versus cDNA GS FLX sequence read lengths was made, showing that the modal read lengths for the cDNA averaged 64 bp compared to 54 bp for the DNA, suggesting that less fragmentation has occurred in RNA than DNA in these maize seeds (Fig. 1). Furthermore, when the modes of rRNA and mRNA were compared from all cDNA samples the modal mRNA length is higher than the modal rRNA value. These results correspond to the requirement for long-term mRNA survival in seeds [19], [38]–[40], and are promising for future studies that might aim to sequence RNA from seeds dating back to the early phases of domestication.
In summary, we demonstrate the long-term survival of, and ability to RNA-Seq, RNA sequences of informative length in archaeological maize kernels. The long-term survival of RNA could be due to the presence of biological mechanisms in seeds for RNA preservation or, as suggested by Venanzi and Rollo [22], may simply reflect the relative level of RNA over DNA. Dormant plant seeds contain mRNA that was transcribed during late embryogenesis and is translated during germination [38]–[40] thus it is perhaps not surprising that mRNA can be detected. However, further research will be required, involving vastly greater sample and data sets, to be able to determine the accuracy of ancient gene expression studies. Furthermore, the authors acknowledge that even if relative abundances of RNA reads reflect transcription when the plant was alive, it would be difficult to compare an ancient seed in one environment to a modern seed in a different environment. Hence, further research would also be required to explore if multiple seeds show similar patterns of transcript abundance that could not otherwise be explained by differences in relative survival compared to modern corn in various environmental settings.
Given (i) the generation here of diverse transcriptomic sequences, (ii) previous documentation of nucleic acid survival in seeds spanning back thousands of years, (iii) the large amounts of well-preserved ancient crop seeds that are held in archaeological collections, often spanning the temporal and geographic range of the species' domestication history, and (iv) the importance of transcriptomal modification during domestication, we suggest that the future study of ancient transcriptomics from dried seeds may offer a powerful new tool with which to complement our understanding of crop domestication.
Supporting Information
Detailed methodology, including nucleic acid extraction method, PCR amplification primer sequences and tests, data analyses and radiocarbon dating.
(DOCX)
Photo of Arizona kernel, batch 935.
(TIF)
Calibration curves for dating of 2 Arizona maize kernels.
(TIF)
Ancient DNA (AZ Shotgun) Fragmentation and Misincorporation Plot.
(TIF)
Ancient RNA (935130) Fragmentation and Misincorporation Plot.
(TIF)
Ancient RNA (935230) Fragmentation and Misincorporation Plot.
(TIF)
Radiocarbon results BP and analytical data, including stable isotope results. All data is acceptable for a material such as this. ‘Used’ is the material analyzed in pretreatment chemistry, whilst ‘yield’ is the amount remaining after the chemical purification procedures applied.
(DOCX)
Fraction of total GS FLX reads mapping to the B73 reference genome for cDNA (4) and DNA (3) libraries. The breakdown of reads mapped with BWA before and after removing sequence duplicates and paralogs is shown. Unmapped reads were then mapped using BLAT to retrieve as many endogenous reads as possible. Estimates of endogenous maize nucleic acid content are highlighted in bold.
(DOCX)
Fraction of total GAIIx (AZ shotgun) and HiSeq (935130 and 935230) reads mapped to the B73 reference genome. BWA (before and after removing sequence duplicates and paralogs) and BLAT mapping values are shown.
(DOCX)
Top 50 exon hits with functional annotation from 935130 cDNA maize read.
(DOCX)
Functionally annotated exon hits for Arizonan kernel 935130.
(DOCX)
Functionally annotated exon hits for Arizonan kernel 935230.
(DOCX)
Repeat content profiles of HiSeq cDNA and GAIIx DNA sequences using RepeatMasker.
(DOCX)
Acknowledgments
The authors thank Kim Magnussen for technical assistance with sequencing.
Funding Statement
The authors thank the Danish Natural Science Research Council and Danish National Research Foundations for financial support. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Wang Z, Gerstein M, Snyder M (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nature Rev 10: 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang RL, Stec A, Hey J, Lukens L, Doebley J (1999) The limits of selection during maize domestication. Nature 398: 236–239. [DOI] [PubMed] [Google Scholar]
- 3. Wright SI, Vroh Bi I, Schroeder SG, Yamasaki M, Doebley JF, et al. (2005) The effects of artificial selection on the maize genome. Science 308: 1310–1314. [DOI] [PubMed] [Google Scholar]
- 4. Yamasaki M, Tenaillon MI, Vroh Bi I, Schroeder SG, Sanchez-Villeda H, et al. (2005) A large-scale screen for artificial selection in maize identifies candidate agronomic loci for domestication and crop improvement. The Plant Cell 17: 2859–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Studer A, Zhao Q, Ross-Ibarra J, Doebley J (2011) Idenitification of functional transposon insertion in the maize domestication gene tb1. Nat Genet 43: 1160–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jaenicke-Despres V, Buckler ES, Smith ES, Gilbert MTP, Cooper A, et al. (2003) Early allelic selection in maize as revealed by ancient DNA. Science 302: 1206–1208. [DOI] [PubMed] [Google Scholar]
- 7. Freitas FO, Bendel G, Allaby RG, Brown TA (2003) DNA from primitive maize landraces and archaeological remains: implications for the domestication of maize and its expansion into South America. J Arch Sci 30: 901–908. [Google Scholar]
- 8. O'Donoghue K, Clapham A, Evershed RP, Brown TA (1996) Remarkable preservation of biomolecules in ancient radish seeds. Proc R Soc Lond B 263: 541–547. [DOI] [PubMed] [Google Scholar]
- 9. Li P, Ponnala L, Gandotra, Wang L, Si Y, et al. (2010) The developmental dynamics of the maize leaf transcriptome. Nat Genet 42: 1060–1067. [DOI] [PubMed] [Google Scholar]
- 10. Buitink J, Leger JJ, Guisle I, Vu BL, Wuilleme S, et al. (2006) Transcriptome profiling uncovers metabolic and regulatory processes occurring during the transition from desiccation-sensitive to desiccation-tolerant stages in Medicago truncatula seeds. Plant J 47: 735–750. [DOI] [PubMed] [Google Scholar]
- 11. Kan Y, Wan Y Beaudoin F, Leader DJ, Edwards K, et al. (2006) Transcriptome analysis reveals differentially expressed storage protein transcripts in seeds of Aegilops and wheat. J Cereal Sci 44: 75–85. [Google Scholar]
- 12. Prioul JL, Méchin V, Lessard P, Thévenot C, Grimmer M, et al. (2008) A joint transcriptomic, proteomic and metabolic analysis of maize endosperm development and starch filling. Plant Biotechnology J 6: 855–869. [DOI] [PubMed] [Google Scholar]
- 13. Holdsworth MJ, Finch-Savage WE, Grappin P, Job D (2007) Post-genomics dissection of seed dormancy and germination. Trends Plant Sci 13: 7–13. [DOI] [PubMed] [Google Scholar]
- 14. Rasmussen M, Li Y, Lindgreen S, Skou Pedersen J, Albrechtsen A, et al. (2010) Ancient human genome sequence of an extinct Palaeo-Eskimo. Nature 463: 757–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Green R, Krause J, Briggs AW, Maricic T, Stenzel U, et al. (2010) A draft sequence of the Neandertal genome. Science 328: 710–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Reich D, Green RE, Kircher M, Krause J, Patterson N, et al. (2010) Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature 468: 1053–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rasmussen M, Guo X, Wang Y, Lohmueller, Rasmussen S, et al. (2011) An aboriginal Australian genome reveals separate human dispersals into Asia. Science 334: 94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lindahl T (1993) Instability and decay of the primary structure of DNA. Nature 362: 709–715. [DOI] [PubMed] [Google Scholar]
- 19. Almoguera C, Jordano J (1992) Developmental and environmental concurrent expression of sunflower dry-seed-stored low-molecular-weight heat-shock protein and Lea mRNAs. Plant Molecular Biol 19: 781–792. [DOI] [PubMed] [Google Scholar]
- 20. Masaki S, Yamada T, Hirasawa T, Todaka D, Kanekatsu M (2008) Proteomic analysis of RNA-binding proteins in dry seeds of rice after fractionation by ssDNA affinity column chromatography. Biotechnol Lett 30: 955–60. [DOI] [PubMed] [Google Scholar]
- 21. Rollo F (1985) Characterization by molecular hybridization of RNA fragments isolated from ancient (1400 B.C.) seeds. Theor Appl Genet 41: 330. [DOI] [PubMed] [Google Scholar]
- 22. Venanzi FM, Rollo F (1990) Mummy RNA lasts longer. Nature 343: 25–26. [DOI] [PubMed] [Google Scholar]
- 23. Rollo F, Venanzi FM, Amici A (1991) Nucleic acids in mummified plant seeds: biochemistry and molecular genetics of pre-Columbian maize. Genet Res 58: 193–201. [DOI] [PubMed] [Google Scholar]
- 24. Sallon S, Solowey E, Korchinsky R, Egli M, Woodhatch I, et al. (2008) Germination, genetics, and growth of an ancient date seed. Science 320: 1464. [DOI] [PubMed] [Google Scholar]
- 25. Bannister B, Dean JS, Robinson WJ (1968) Tree-ring dates from Arizona C-D: Eastern Grand Canyon-Tsegi Canyon-Kayenta Area. Tucson: Lab. Tree-Ring Res [Google Scholar]
- 26. Ávila-Arcos M, Cappellini E, Romero-Navarro JA, Wales N, Moreno-Mayar JV, et al. (2011) Application and comparison of large-scale solution-based DNA capture-enrichment methods on ancient DNA. Scientific Reports 1 doi. 10.1038/srep00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115. [DOI] [PubMed] [Google Scholar]
- 28. Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kent WJ (2002) BLAT - the BLAST-like alignment tool. Genome Res 12: 656–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Babraham Bioinformatics website. Available: http://www.bioinformatics.bbsrc.ac.uk/projects/seqmonk/. Accessed: 2011 Nov.
- 31. Ginolhac A, Rasmussen M, Gilbert MTP, Willerslev E, Orlando L (2011) mapDamage: testing for damage patterns in ancient DNA sequences. Bioinformatics 27: 2153–2155. [DOI] [PubMed] [Google Scholar]
- 32.RepeatMasker website, RepeatModeler Open-1.0. Available: http://www.repeatmasker.org.
- 33. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Bio 215: 403–410. [DOI] [PubMed] [Google Scholar]
- 34. Huson DH, Mitra S, Weber N, Ruscheweyh H, Schuster SC (2011) Integrative analysis of environmental sequences using MEGAN4. Genome Res 21: 1552–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Buescher PJ, Phillips RL, Brambl R (1984) Ribosomal RNA contents of maize genotypes with different ribosomal RNA gene numbers. Biochem Genet 22: 923–930. [DOI] [PubMed] [Google Scholar]
- 36. Miller W, Drautz DI, Ratan A, Pusey B, Qi J, et al. (2008) Sequencing the nuclear genome of the extinct woolly mammoth. Nature 456: 387–390. [DOI] [PubMed] [Google Scholar]
- 37. Briggs AW, Stenzel U, Johnson PLF, Green RE, Kelso J, et al. (2007) Patterns of damage in genomic DNA sequences from a Neandertal. Proc Natl Acad Sci U S A 104: 14616–14621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dure L, Waters L (1965) Long-lived messenger RNA: Evidence from cotton seed germination. Science 147: 410–412. [DOI] [PubMed] [Google Scholar]
- 39. Hammett JR, Katterman FR (1975) Storage and metabolism of poly(adenylic acid)-mRNA in germinating cotton seeds. Biochem 14: 4375–4379. [DOI] [PubMed] [Google Scholar]
- 40. Nakabayashi K, Okamoto M, Koshiba T, Kamiya Y, Nambara E (2005) Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed. Plant J 41: 697–709. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Detailed methodology, including nucleic acid extraction method, PCR amplification primer sequences and tests, data analyses and radiocarbon dating.
(DOCX)
Photo of Arizona kernel, batch 935.
(TIF)
Calibration curves for dating of 2 Arizona maize kernels.
(TIF)
Ancient DNA (AZ Shotgun) Fragmentation and Misincorporation Plot.
(TIF)
Ancient RNA (935130) Fragmentation and Misincorporation Plot.
(TIF)
Ancient RNA (935230) Fragmentation and Misincorporation Plot.
(TIF)
Radiocarbon results BP and analytical data, including stable isotope results. All data is acceptable for a material such as this. ‘Used’ is the material analyzed in pretreatment chemistry, whilst ‘yield’ is the amount remaining after the chemical purification procedures applied.
(DOCX)
Fraction of total GS FLX reads mapping to the B73 reference genome for cDNA (4) and DNA (3) libraries. The breakdown of reads mapped with BWA before and after removing sequence duplicates and paralogs is shown. Unmapped reads were then mapped using BLAT to retrieve as many endogenous reads as possible. Estimates of endogenous maize nucleic acid content are highlighted in bold.
(DOCX)
Fraction of total GAIIx (AZ shotgun) and HiSeq (935130 and 935230) reads mapped to the B73 reference genome. BWA (before and after removing sequence duplicates and paralogs) and BLAT mapping values are shown.
(DOCX)
Top 50 exon hits with functional annotation from 935130 cDNA maize read.
(DOCX)
Functionally annotated exon hits for Arizonan kernel 935130.
(DOCX)
Functionally annotated exon hits for Arizonan kernel 935230.
(DOCX)
Repeat content profiles of HiSeq cDNA and GAIIx DNA sequences using RepeatMasker.
(DOCX)