Summary
Background
Many cells are remarkably proficient at tracking very shallow chemical gradients, despite considerable noise from stochastic receptor-ligand interactions. Motile cells appear to undergo a biased random walk: spatial noise in receptor activity may determine the instantaneous direction, but because noise is spatially unbiased it is filtered out by time-averaging, resulting in net movement up-gradient. How non-motile cells might filter out noise is unknown.
Results
Using yeast chemotropic mating as a model, we demonstrate that a polarized patch of polarity regulators “wanders” along the cortex during gradient tracking. Computational and experimental findings suggest that actin-directed membrane traffic contributes to wandering by diluting local polarity factors. The pheromone gradient appears to bias wandering via interactions between receptor-activated Gβγ and polarity regulators. Artificially blocking patch wandering impairs gradient tracking.
Conclusions
We suggest that the polarity patch undergoes an intracellular biased random walk that enables noise filtering by time-averaging, allowing non-motile cells to track shallow gradients.
Introduction
Many cells track extracellular chemical gradients to direct cell migration (chemotaxis) or growth (chemotropism). Whereas swimming bacteria compare chemical concentrations at different times (temporal gradient-sensing)[1], eukaryotes compare chemical concentrations on the up- and down-gradient sides of the cell (spatial gradient-sensing)[2]. These cells can track remarkably shallow gradients [3–6]. Because the instantaneous noise stemming from stochastic receptor-ligand interactions would often exceed the tiny gradient signal [6, 7], cells are thought to use time-averaging to filter the consistent spatial signal from the random noise [8]. Chemotaxing cells can perform time-averaging via a biased random walk [7]: although noise influences instantaneous trajectory, over time random fluctuations in different directions cancel out, leaving net movement up-gradient.
As a biased random walk requires movement, non-motile gradient-tracking cells presumably employ some other form of time-averaging. The mating response of the yeast Saccharomyces cerevisiae provides a genetically tractable system to study chemotropic gradient tracking [9]. Haploid yeast of opposite mating type orient polarized growth up pheromone gradients to find and fuse with mating partners. How do such non-motile cells filter noise to orient growth up-gradient?
The direction of growth in yeast is dictated by the conserved Rho-family GTPase Cdc42, which orients actin cables that direct vesicle delivery and growth towards the polarization site [10]. Cdc42 clusters in a cortical “polarity patch” with other polarity regulators, including the Cdc42 guanine nucleotide exchange factor (GEF), Cdc24, and the scaffold protein, Bem1, which is thought to facilitate a positive-feedback loop that contributes to Cdc42 polarity [11]. In a pheromone gradient, the polarity patch is generally located on the up-gradient side of the cell, resulting in growth in that direction. Pheromone binds to G-protein-coupled receptors, generating free Gβγ that can recruit Cdc24 via the scaffold protein Far1 [12, 13]. Thus, ligand-bound receptors can spatially bias Cdc42 activation.
In shallow gradients, the internal Cdc42 gradient must be much steeper than the external pheromone gradient to result in polarized growth and mating projection formation. In fact, Cdc42 becomes highly polarized even in uniform pheromone, when it is oriented towards a “default site” specified by the bud-site-selection protein Rsr1 [14]. Polarity is established even when all spatial information is removed by exposing rsr1Δ cells to uniform pheromone, implying that mating polarity can be initiated by random stochastic fluctuations that are then amplified by positive feedback. Positive feedback provides a mechanism to generate a steep Cdc42 gradient from a shallow (or even flat) pheromone gradient.
Positive feedback is self-reinforcing, so, once specified, growth orientation should remain stable. However, cells that initiate polarized growth in the “wrong” direction re-orient to better align with the gradient over time [4, 15]. How is positive feedback overcome to allow repositioning or redefining of the cell’s front?
We find that in cells exposed to non-saturating concentrations of pheromone, the polarity patch wanders around the cell cortex. The degree of wandering depends on the pheromone concentration, and cells engineered to block wandering can no longer effectively re-orient to track a pheromone gradient. Our findings suggest that vesicle traffic overcomes positive feedback to promote an intracellular “random walk” by the polarity patch, which is biased by the pheromone gradient to enable non-motile cells to correct errors in growth orientation and effectively track shallow gradients.
Results
A wandering polarity patch in sub-saturating pheromone
Cdc42 and other polarity proteins localize to the tip of the pointy mating projection in saturating pheromone [9]. Because the population of cell-surface receptors is saturated, such cells cannot track gradients. Cells actually tracking a gradient form broad rather than pointy projections [4, 15–17]. Similarly, cells in uniform, non-saturating pheromone formed broad projections (Fig. 1A). (To precisely control pheromone concentration, all of our strains lack the Bar1 protease that degrades α-factor.)
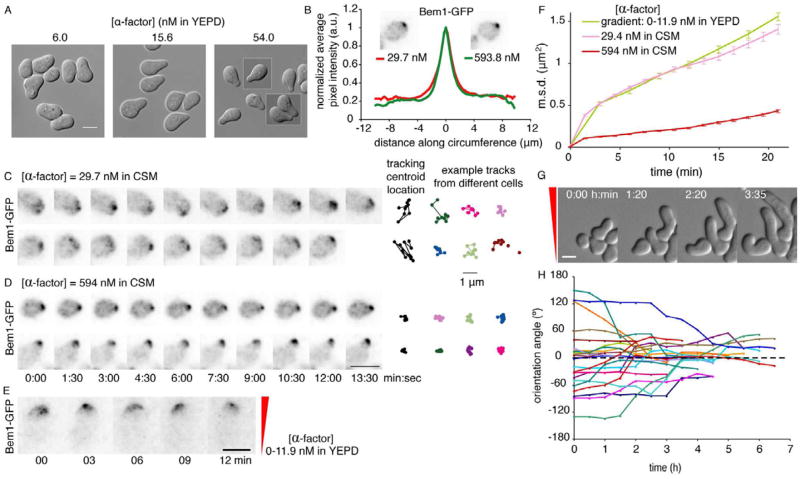
Fig. 1. A wandering polarity patch in cells exposed to sub-saturating pheromone.
A) Mating projection morphology is correlated with pheromone concentration. DIC images of MATa bar1 cells (DLY10065) treated with the indicated concentration of α-factor for 2.5 h. B) Polarity protein distribution is unaffected by pheromone concentration. Fluorescence images and averaged cortical distributions of Bem1-GFP (methods, Fig. S1A–B) at the indicated concentrations. C,D) The polarity patch wanders in sub-saturating pheromone. BEM1-GFP cells (DLY10065) were filmed at the indicated α-factor concentrations; 15 min patch centroid tracks (black, same cells; color, other cells) shown at right (methods, Fig. S1D–F). E) Wandering occurs on the up-gradient side of the cell (gradient: 0–11.9 nM α-factor in YEPD, indicated in red). F) Mean squared displacement (m.s.d.: error bars show s.e.m.) for the centroid of the wandering Bem1-GFP patch in cells (DLY10065) treated with pheromone as indicated. G,H) Cells gradually reorient growth to align with the gradient. DIC images (G) and growth orientation angles (H) at the indicated times (gradient as in E). Perfect alignment is 0°, dashed line. Scale bars, 5 μm. All fluorescence images are deconvolved, inverted maximum projections.
Variations in cell morphology could stem from differences in polarity protein distribution [18]. We monitored polarity using Bem1-GFP, which is fully functional, unlike GFPC-dc42 [19]. The distribution of Bem1-GFP did not differ significantly in cells exposed to low or high concentrations of pheromone (Fig. 1B, Fig. S1). Instead, the Bem1-GFP patch fluctuated in intensity and “wandered” around the cortex in cells exposed to low pheromone (Fig. 1C, movie S1). Other components of the polarity patch, including the GEF Cdc24 and activated Cdc42, behaved similarly (Fig. S1C). In contrast, Bem1-GFP was stably localized to the projection tip in cells exposed to high pheromone (Fig. 1D, movie S1). To quantify patch wandering, we tracked the centroid of the Bem1-GFP patch (Fig. S1, Fig. 1C–D). The average area covered by the patch centroid during wandering (mean squared displacement or m.s.d.) grew more rapidly at low pheromone concentrations (Fig. 1F), confirming that the patch exhibits dose-dependent wandering. Cells exposed to high concentrations of pheromone exhibited little wandering even before forming a projection (Fig. S1G). Cells tracking a gradient of pheromone in a microfluidics device also exhibited a wandering polarity patch, predominantly on the up-gradient side of the cell (Fig. 1E, movie S2). The degree of wandering in gradient-treated cells was comparable to that in cells exposed to low uniform pheromone (Fig. 1F).
Highly polarized Cdc42 would direct secretion to a narrow window, but if Cdc42 shifts to a new location, secretion should follow. Over time, this would result in distributed growth over a broad region, despite the tightly polarized instantaneous distribution of polarity regulators. If the wandering patch spent more time on one side of the projection tip, then growth would “turn” in that direction. Indeed, when cells initially grew a projection mis-aligned with the pheromone gradient, they slowly re-oriented growth up-gradient (Fig. 1G,H and movie S2)[4, 8, 15], suggesting that polarity patch wandering may facilitate error correction during gradient tracking.
These findings indicate that there is a process that overcomes positive feedback to perturb polarity patch integrity and promote wandering. Moreover, high-concentration pheromone reduces wandering, raising the question of how wandering is constrained by pheromone. One obvious candidate is the bud-site-selection protein Rsr1, which uses internal cues to specify the “default” site to which cells polarize in uniform pheromone [14].
Rsr1 and Gβγ-Far1-Cdc24 constrain wandering
Mating projections of RSR1 cells appeared pointier than those of rsr1Δ cells (Fig. 2A)[20]. Maximum curvature (a quantitative measure of pointiness: Fig. S2A–C) increased with pheromone concentration, with RSR1 cells pointier than rsr1Δ cells at all concentrations (Fig. 2B). M.s.d. grew more slowly in RSR1 cells than rsr1Δ cells (Fig. 2C), suggesting that Rsr1 constrains the wandering, yielding pointier mating projections.
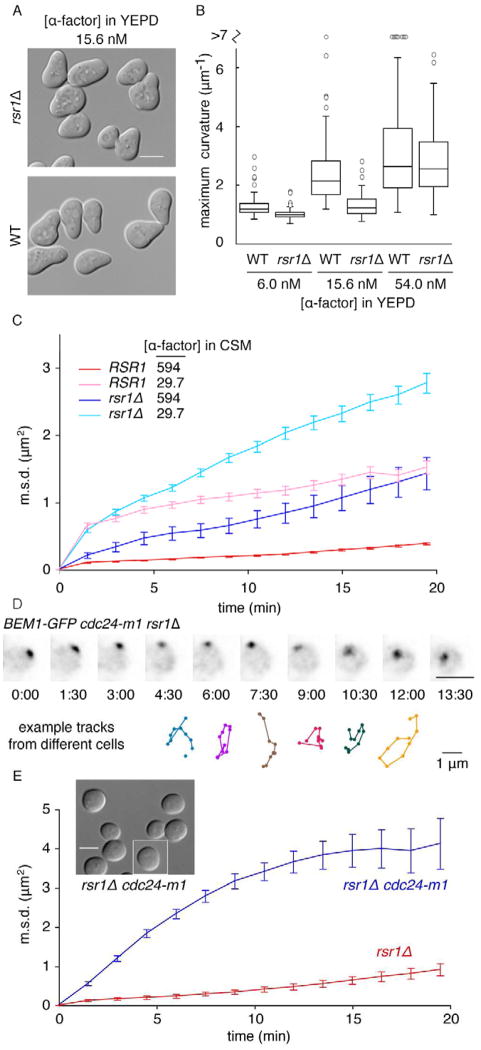
Fig. 2. Wandering is constrained by Rsr1 and by Gβγ-Far1 at high pheromone concentration.
A) RSR1 cells make pointier projections than rsr1Δ cells. DIC images of WT (DLY10065) and rsr1Δ (DLY10066) cells treated with 15.6 nM α-factor in YEPD for 2.5 h. B) Maximum curvature (methods, Fig. S2A–C) for the same cells at different pheromone concentrations. Box: median and quartiles (n>67 cells). C) The patch wanders more in rsr1Δ cells than in RSR1 cells. Wandering m.s.d. for Bem1-GFP in WT (DLY10065) and rsr1Δ (DLY11740) cells at the indicated α-factor concentrations (nM) (n>19 cells). RSR1 traces from Fig. 1F. D) The patch wanders in cdc24-m1 rsr1Δ cells (DLY11638) even at high pheromone concentration (594 nM in CSM). Deconvolved, inverted maximum projections of Bem1-GFP at different times (min:sec). Colored tracks: 15 min centroid trajectories from representative cells. E) Wandering m.s.d. for cdc24-m1 rsr1Δ (DLY11638) and rsr1Δ (DLY10066) cells in 0.3 μM α-factor (n>24 cells). Inset: DIC images of cdc24-m1 rsr1Δ cells. Scale bars, 5 μm.
Even rsr1Δ cells exhibited reduced wandering and pointier projections at higher pheromone concentrations (Fig. 2B–C). What underlies concentration-dependent restraint of patch wandering? Upon ligand binding, pheromone receptors catalyze local release of free Gβγ, which recruits Far1-Cdc24 from the cytoplasm, leading to local Cdc42 activation [12, 13]. A mutation that prevents the Far1-Cdc24 interaction (cdc24-m1) [12] resulted in dramatically wandering patches in rsr1Δ cells, even in saturating concentrations of pheromone (Fig. 2D–E, movie S3), consistent with previous reports [21]. Vesicle-trafficking markers co-localized with wandering polarity markers in cdc24-m1 rsr1Δ cells, confirming that secretion follows a wandering polarity patch (Fig. S2D–F). Dramatic patch wandering resulted in widely distributed growth, precluding projection formation [21](Fig. 2E inset). Thus, Rsr1 and Gβγ-Far1-Cdc24 each provide parallel constraints on wandering at high pheromone concentration. An additional, unknown mechanism also contributes to dose-dependent wandering because even cdc24-m1 rsr1Δ “unconstrained” cells exhibited less wandering at higher concentrations of pheromone (Fig. S2G). All constraints are relaxed in sub-saturating pheromone, suggesting that wandering is a physiologically important phenomenon at pheromone concentrations relevant to gradient tracking.
Vesicle traffic provides a potential mechanism to drive wandering
We compared the “unconstrained” wandering in rsr1Δ cdc24-m1 mutants with simulated diffusion on the surface of a sphere (see Supplement Computational Methods). For diffusion, the m.s.d. initially increases with slope 4Dt (where D is the diffusion coefficient and t is time) but then plateaus at 2R2 as it is constrained by cell geometry (where R is the radius of the sphere). With a diffusion coefficient matched to the initial m.s.d. slope of the wandering patch, m.s.d. initially grew more steeply than predicted for diffusion (Fig. 3A), suggesting that wandering direction is not entirely random (see below).
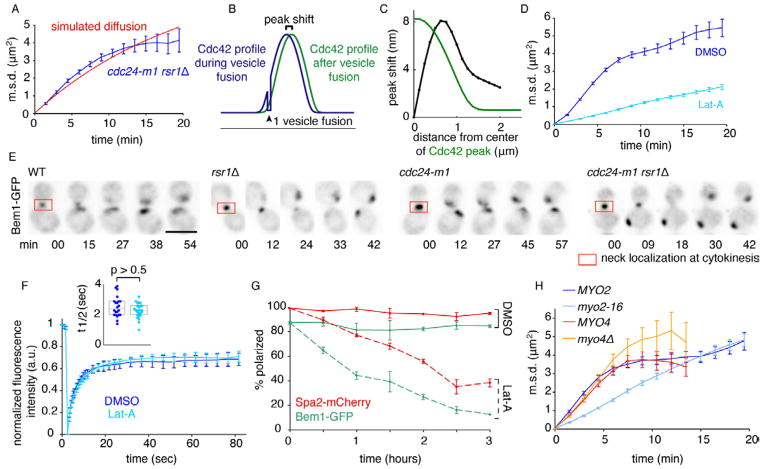
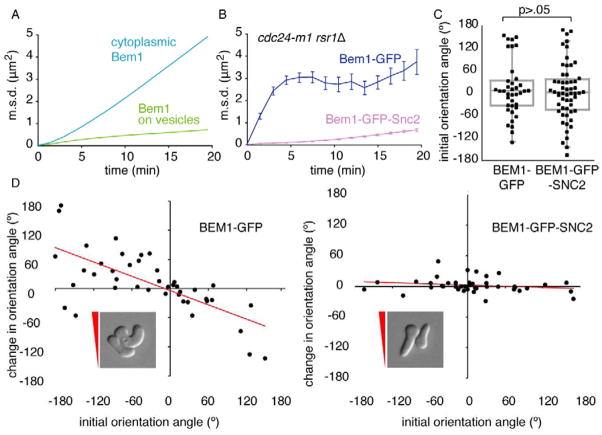
Fig. 3. Wandering, but not polarity establishment, requires F-actin and Myo2.
A) Comparison of wandering (blue: from Fig. 2E) with simulated diffusion (red: D=0.0016 μm2/s) on the surface of a 4 μm diameter sphere. B) Schematic: fusion of a vesicle off-center from the polarity peak would shift the peak away from the fusion site. X-axis: cortical position, Y-axis: [Cdc42]. C) Peak shift (black) resulting from simulated fusion of a single vesicle depends on the distance between the fusion site and the center of the peak (for comparison, the shape of the Cdc42 concentration profile is shown in green). D) Wandering is reduced upon actin depolymerization. cdc24-m1 rsr1Δ (DLY11306) cells in 59.4 nM α-factor were treated with 200 μM Lat-A (sufficient to dismantle actin patches: Fig. S3A) or DMSO (control). E) Polarity establishment is actin-independent. WT (DLY11742), rsr1Δ (DLY11740), cdc24-m1 (DLY11094), and cdc24-m1 rsr1Δ (DLY11079) cells were treated with 0.3 μM α-factor and 200 μM Lat-A. Deconvolved, inverted maximum projections of Bem1-GFP at different times, showing cells completing cytokinesis and then polarizing. Bar, 5μm. F) Cdc42 recycles rapidly. Average kinetics (mean +/− s.e.m.) of GFP-Cdc42 FRAP in DLY13898 cells treated with α-factor (3 μM in CSM) and DMSO or Lat-A as indicated (n>11 cells). Inset: distribution of fitted t1/2. Each dot is one cell. Box: median and quartiles. p-value from Student’s t-test. G) Polarity is gradually lost in Lat-A. DLY11742 cells were pretreated for 90 min with 0.3 μM α-factor, then exposed to DMSO or 200 μM Lat A. The % of cells (mean +/− s.e.m.) with polarized Bem1-GFP or Spa2-mCherry was scored (n>100 cells). H) Wandering is reduced in myo2-16 mutants. cdc24-m1 rsr1Δ MYO2 (DLY11079) and cdc24-m1 rsr1Δ myo2-16 (DLY12404) cells were imaged in α-factor (0.6 μM in CSM) at 34°C (restrictive for myo2-16 mutants: Fig. S3B), and cdc24-m1 rsr1Δ MYO4 (DLY11638) and cdc24-m1 rsr1Δ myo4Δ (DLY14160) cells were imaged in α-factor (0.3 μM in CSM) at 30°C (n>34 cells).
A potential wandering mechanism is suggested by recent work modeling cell polarity in vegetative yeast cells, where actin-independent positive feedback establishes and maintains polarity. Positive feedback involves GTP-Cdc42-mediated recruitment of a cytoplasmic complex containing Bem1, a Cdc42 effector, and the GEF Cdc24. Cdc24 activates neighboring Cdc42, leading to recruitment of more Bem1 complex, etc. [11]. Because vesicles deliver membrane that lacks Bem1 complexes, vesicle fusion transiently dilutes the local polarity regulators, perturbing the polarity patch [22].
Using a recently-developed mathematical model [22], we asked how vesicle fusion might affect polarity patch location. If a vesicle fuses off-center from the polarity peak, diluting polarity regulators there, the polarity peak is predicted to shift away from the vesicle fusion site because positive feedback is stronger on the opposing side. The degree of peak shift depends on where the vesicle fuses relative to the peak center (Fig. 3B–C). Thus, vesicle fusion would lead to a shift in peak position, away from the vesicle.
Wandering is actin-dependent
To test whether actin-directed vesicle traffic contributes to wandering, we treated cdc24-m1 rsr1Δ cells with Latrunculin-A (Lat-A) at concentrations sufficient to dismantle F-actin structures (Fig. S3A)[23]. Wandering was greatly reduced following exposure to Lat-A (Fig. 3D), suggesting that F-actin makes a major contribution to wandering. The residual wandering in Lat-A treated cells suggests that an actin-independent process also contributes.
Although Lat-A inhibited patch wandering, it did not disable polarity per se. This was surprising given previous reports that polarity is actin-dependent in pheromone-treated cells [24]. However, cells effectively established a polarized patch of Bem1-GFP upon combined exposure to pheromone and Lat-A (Fig. 3E). Furthermore, actin-independent “symmetry-breaking” polarization occurred even in strains lacking positional information from Rsr1, or the Gβγ-Far1-Cdc24 complex, or both (Fig. 3E). Fluorescence Recovery After Photobleaching (FRAP) revealed a rapid turnover of GFP-Cdc42 at the polarity patch that was unaffected by Lat-A (Fig. 3F), indicating that Cdc42 polarization is dynamically maintained in an actin-independent manner.
When cells pre-treated with pheromone were exposed to Lat-A, the proportion of cells displaying polarized Bem1-GFP or Spa2-mCherry (another polarity marker [25]) declined steadily over 3 h (Fig. 3G). Thus, Lat-A causes a gradual loss of polarity, for unknown reasons. This observation can explain the discrepancy between our results and those of Ayscough and Drubin (1998), who examined polarity 3 h after Lat-A addition.
The type V myosin Myo2 transports vesicles along actin cables to the plasma membrane [26, 27], and myo2-16 mutants at restrictive temperature (Fig. S3B) exhibited reduced wandering compared to isogenic MYO2 control cells (Fig. 3H). In contrast, myo4Δ mutants lacking a type V myosin that traffics ER and mRNAs [26] did not impair wandering (Fig. 3H). The contributions of both F-actin and Myo2 to effective patch wandering led us to ask whether vesicle-mediated polarity perturbation would yield realistic wandering behavior.
Simulated wandering of a polarity patch
To what degree would dilution of local polarity regulators upon secretory vesicle fusion lead to wandering? We began by asking how much a single vesicle fusion would shift the peak of Cdc42 for peaks of different size (amount of Cdc42 in the peak), shape (full width at half-maximum Cdc42 concentration) or dynamics (simulated FRAP of Cdc42). We varied model parameters to obtain peaks that differed in each category. In all cases, off-center vesicle fusion led to a shift of the polarity peak away from the fusion site: small peaks were shifted more than larger peaks, narrow peaks were shifted more than broader peaks, and highly dynamic peaks were shifted less than peaks with slower turnover (Fig. 4A). For a peak matching the size, shape, and FRAP dynamics observed for GFP-Cdc42 in pheromone-treated cells (Fig. 4B), a single vesicle would shift the peak by up to ~10 nm (Fig. 4C).
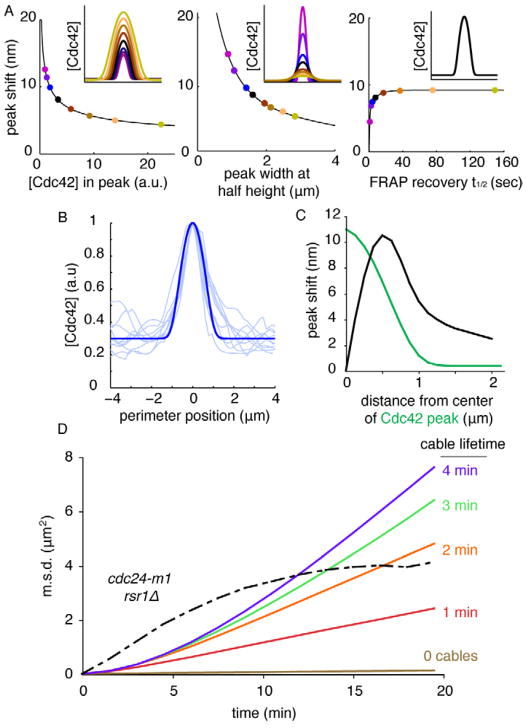
Fig. 4. Simulated wandering due to vesicle traffic.
A) The polarity peak shift resulting from simulated off-center fusion of a single vesicle (methods, Supplement) depends on peak size (left, dots color-matched to inset peaks), shape (middle), and dynamics (right, all peaks have same size/shape: inset). Inset x-axis is 8.9 μm along cell perimeter, y-axis is Cdc42 concentration. B) GFP-Cdc42 intensity profiles from 10 polarized cells (light blue, linescans acquired as in Fig. S1B) were used to adjust model parameters to obtain a matching peak (dark blue). C) Peak shift (as in Fig. 3C) for a polarity peak matching the observed peak size, shape, and dynamics in vivo. D) Simulated wandering due to vesicle traffic from models with 10 actin cables (and cable lifetime as indicated) or random vesicle fusion in the polarity window (0 cables), compared to wandering in cdc24-m1 rsr1Δ mutants (black: from Fig 2E).
Yeast cells secrete about 50 vesicles/min (Supplemental Methods). To model stochastic vesicle traffic (including both exocytosis and endocytosis), we first defined a “window” representing the polarity patch [28]. Vesicle fusion occurred with equal probability anywhere in the window, and endocytosis occurred preferentially in the window [29]. With the window defined as the 2% of membrane harboring the highest [Cdc42], the peak wandered very little (Fig. 4D: 0 cables). However, in vivo, vesicles are delivered to the window by a limited number of actin cables. To simulate actin-directed transport, we assumed that vesicle fusion only occurred at actin cable termini. We considered models with 5–15 cables [30], which were attached at random sites within the window. Vesicles fused with equal probability at any of the currently active cable positions. Cables detached from the cortex with a probability 1/τ per min (τ = average cable lifetime), and were then free to attach to a new random site in the window. This formulation allows actin cables to track wandering polarity sites. Wandering behavior in the actin-containing simulations was much more extensive (Fig. 4D, movie S4). Wandering was relatively insensitive to the number of actin cables (Fig. S4E), but sensitive to cable lifetime (Fig. 4D). Thus, the dilution of polarity factors arising from actin-directed vesicle trafficking would suffice to cause a significant degree of wandering.
Comparison of the simulated wandering with wandering in vivo (Fig. 4D) suggests two conclusions. First, wandering patch m.s.d. plateaued before reaching the level expected from the constraint due to cell geometry. This may reflect the presence of additional constraints on wandering even in rsr1Δ cdc24-m1 mutants. Second, with the assumptions employed above, the simulated wandering did not recapitulate the rapidity with which wandering m.s.d. grew in rsr1Δ cdc24-m1 cells. Moreover, in our simulations the polarity peak remained intact and did not display the large fluctuations in intensity observed in vivo (compare movies S1 and S4). Thus, either vesicle traffic is more perturbing than we considered, or other factors must also contribute to wandering (see Discussion).
Persistence in the direction of polarity patch wandering
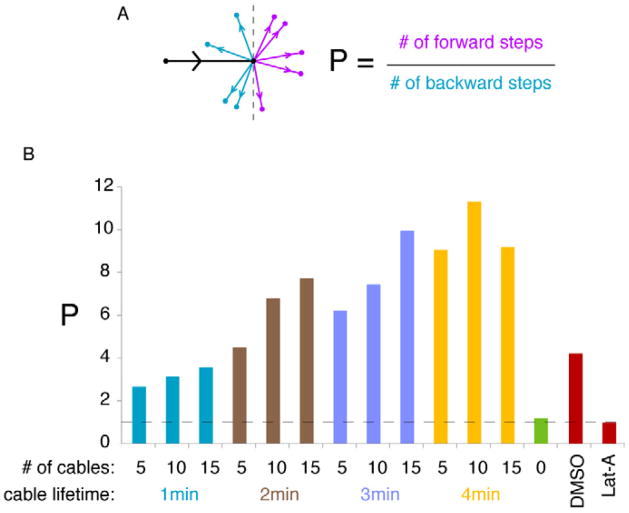
Models with actin cables exhibited more persistent movement of the polarity site than the model in which vesicles fuse randomly in the polarity window (Fig. 5B). Persistence arises because, over the lifetime of an actin cable, vesicles have a propensity to fuse at the same location. Thus, if a cable terminus is off-center from the polarity peak, successive vesicle fusions guided by that cable would shift the peak further away from the cable. To quantify the tendency of wandering trajectories to keep going in the same direction, we tracked polarity patch centroid positions at 1.5 min intervals and classified each step as moving “forward” or “backward” relative to the previous step (Fig. 5A). We defined a persistence measure P as the ratio of forward steps to backward steps. Random movement yields P = 1, matching the model in which vesicle fusion occurred anywhere in the window (Fig. 5B: no cables). However, actin-containing models all had P > 1, with P increasing with cable lifetime (Fig. 5B).
Fig. 5. Wandering displays actin-dependent persistence in models and in cells.
A) Schematic: the black arrow indicates movement of the patch centroid in a 1.5 min imaging or simulation interval, and the other arrows show possible directions of movement in the subsequent interval (purple, forwards; blue, backwards). Persistence, P, is the ratio of forward to backward steps. B) P values for simulated wandering in models with the indicated numbers of actin cables and cable lifetimes (green: model with random vesicle fusion and no cables), as well as experimental P values from movies (red: from Fig. 3D, n>54 cells). Dotted line: P=1 (no persistence). n > 550 steps for each condition.
To assess whether patch wandering exhibited persistence in vivo, we applied the same analysis to wandering patches in rsr1Δ cdc24-m1 cells. Strikingly, wandering trajectories showed significant persistence in the presence of actin, but not in its absence (Fig. 5B).
Patch wandering is critical for error correction during gradient tracking
If dilution of Bem1 and associated polarity factors is responsible for wandering, then concentrating Bem1 on secretory vesicles should reduce or eliminate wandering. Simulations in which vesicles carried a high concentration of Bem1 displayed very little wandering, as expected (Fig. 6A). Bem1 is normally cytoplasmic, but can be tethered to membranes by fusion to the exocytic v-SNARE, Snc2, which is highly concentrated on secretory vesicles [31]. We tracked the polarity patch in Bem1-Snc2 and control cells using Spa2-mCherry, which colocalizes with Bem1 and secretory markers in wandering patches (Fig. S2D–F). Consistent with model predictions, Bem1-Snc2 dramatically suppressed wandering in rsr1Δ cdc24-m1 cells (Fig. 6B, movie S5), as well as in wild-type cells exposed to non-saturating pheromone, resulting in pointy projections (Fig. S5A).
Fig. 6. Concentrating Bem1 on vesicles blocks wandering and impairs gradient tracking.
A) Simulated wandering is blocked by concentrating Bem1 on vesicles. Simulations were conducted using the model with 5 actin cables and 2 min cable lifetime (to match observed persistence: Fig. 5B), with (green) or without (blue) Bem1 concentrated on vesicles. B) Wandering in vivo is blocked by Bem1-Snc2. Wandering of Spa2-mCherry (which co-localizes with Bem1 and secretory vesicles: Fig. S2D–F) was tracked in cdc24-m1 rsr1Δ strains with Bem1-GFP (DLY11638) or Bem1-GFP-Snc2 (DLY11759) (n>17 cells) treated with 59.4 nM α-factor. C) Initial growth orientation in a gradient is not affected by Bem1-Snc2. CDC24 RSR1 strains with Bem1-GFP (DLY10065) or Bem1-GFP-Snc2 (DLY10063) were exposed to a gradient of pheromone (0–11.9 nM α-factor in YEPD) and the first detectable outgrowth in DIC was scored (each dot is one cell, perfect alignment is 0°, p-value from Kolmogorov-Smirnov test, n>37). D) Re-orientation is impaired by Bem1-Snc2. Change in orientation angle after 3 h in the pheromone gradient (Fig. S5B) is plotted against initial orientation angle (n>41). Perfect reorientation would yield a line with slope −1, zero reorientation would yield a line with slope 0. Insets: cells with “incorrect” initial orientation, red triangle indicates direction of gradient.
If a wandering polarity patch is necessary for gradient tracking, then preventing wandering should impair cells’ ability to correct errors in growth orientation. We exposed control and Bem1-Snc2 cells to a gradient of pheromone and measured individual projection orientation angles over time. Initial orientation angles were similarly distributed in Bem1-Snc2 and control cells (Fig. 6C), but the growth orientation of Bem1-Snc2 cells did not improve with time (Fig. S5B, movie S6). Whereas control cells that initially oriented down-gradient re-oriented to track the gradient, Bem1-Snc2 cells did not re-orient (Fig. 6D), forming straight projections even when initial growth was in the “wrong” direction (Fig. 6D inset, movie S6). These results suggest that polarity patch wandering is necessary for gradient tracking.
Discussion
Actin-independent positive feedback promotes cell polarity during mating
Yeast cells exposed to uniform pheromone develop a polarized cell “front” (polarity patch), even if they lack Rsr1 [14]. Thus, pheromone-treated cells can break symmetry in the absence of spatial cues, suggesting that they employ positive feedback to develop a polarity site from starting stochastic fluctuations. We found that symmetry-breaking polarization occurred even in Lat-A-treated cells, implying that positive feedback is actin-independent. A previous report demonstrated that polarization of the pheromone receptor can occur in the absence of F-actin [32]. However, we found that polarity establishment occurred in cdc24-m1 cells, where polarity is spatially uncoupled from Gβγ and the receptor [12], suggesting that actin-independent feedback occurs at the level of polarity regulators, downstream of pheromone receptors. Given that the same polarity regulators polarize in budding and mating cells, our findings suggest that similar molecular mechanisms may underlie polarity establishment in both cases.
The polarity patch wanders in cells treated with sub-saturating pheromone
Positive feedback in the polarity machinery explains how cells can form a “front” even in shallow gradients. However, positive feedback could also be a problem: stochastic noise in receptor occupancy could set off the hair-trigger amplification mechanism to establish a “front” in the wrong direction. Furthermore, because positive feedback is self-reinforcing, an established polarity site should be difficult to reorient. And yet, cells that initially misalign growth with respect to the gradient do reorient over time [4, 15]. Thus, cells must possess the capability to overcome positive feedback in the polarity machinery to reorient cell growth.
We report that when cells are exposed to non-saturating concentrations of uniform pheromone, the polarity patch fluctuates in intensity and “wanders” around the cortex. Imaging at 1.5 min intervals, the patch generally “moves” only a short distance while appearing to remain intact, but sometimes the patch splits into 2–3 distinct spots and can even disappear entirely, reappearing elsewhere. These behaviors suggest that cells possess a mechanism that can counteract positive feedback, allowing patch repositioning.
Although our findings are the first to document wandering behavior at sub-saturating pheromone, wandering was previously described in cdc24-m1 rsr1Δ mutants (but not wild-type, or single-mutant cdc24-m1 or rsr1Δ cells) exposed to saturating pheromone [21]. This suggests that Rsr1 and Gβγ-Far1-Cdc24 provide parallel mechanisms to restrain wandering in high pheromone. Our findings imply that both of these restraints become weaker at lower concentrations of pheromone, and that wandering is a physiologically important phenomenon for gradient tracking.
Previously, another study documented persistent wave-like movement of broad polarity-factor crescents in vegetative rsr1Δ cells [33]. Subsequent work did not recapitulate this observation, but instead suggested that the wave-like motion might be linked to stress from toxic probes and filming conditions [19]. Nevertheless, Ozbudak et al. [33] proposed ideas to explain wave-like motion that may apply to wandering in pheromone-treated cells (see below).
Vesicle trafficking contributes to polarity patch wandering
Once a polarity patch is assembled, actin cables are oriented towards the patch, resulting in delivery and fusion of secretory vesicles at that site. Because most polarity regulators are thought to be absent from secretory vesicles (including Bem1, the GEF Cdc24, and all known Cdc42 effectors), the fusion of a vesicle would transiently dilute the local polarity regulators. Even if Cdc42 itself were concentrated on vesicles, mathematical modeling suggests that vesicle traffic would still perturb the polarity patch [22]. Our simulations indicate that actin-independent positive feedback reinforces the polarity peak on the side opposite a vesicle fusion event, resulting in a net shift of the polarity peak away from the fusion site. This effect promotes wandering behavior in models with stochastic actin-directed vesicle fusion, suggesting that vesicle traffic underlies wandering.
If polarity patch wandering is driven by directed vesicle traffic, then wandering should depend on actin cables, on the vesicle-trafficking myosin Myo2, and on vesicle fusion proteins. We found that eliminating F-actin or reducing Myo2 function severely reduced wandering (we were unable to test secretory fusion mutants because they lost the polarity patch at restrictive temperature). This hypothesis further predicts that wandering would be eliminated by concentrating polarity regulators on secretory vesicles. We found that fusing Bem1 (a scaffold that binds to many other polarity proteins) to the v-SNARE Snc2 eliminated wandering.
Another prediction is that actin-mediated vesicle traffic should move a polarity patch in a persistent manner [33]. If a polarity peak shifted to one side, there would be a temporal lag before actin cables relocated to the new peak position. During the lag, vesicles would continue to traffic preferentially to the “old” peak position, resulting in continual off-center fusion events on the same side of the peak, driving motion in the same direction. Simulating stochastic actin cable attachment to and detachment from the polarity peak, we found that the degree of directional persistence depended on cable lifetime. Statistical analysis of wandering revealed a significant level of persistence in vivo. In aggregate, the modeling and in vivo data strongly support the hypothesis that vesicle trafficking promotes wandering of the polarity patch.
Other sources of wandering
Our simulations of wandering did not quantitatively recapitulate the rapid wandering observed in cdc24-m1 rsr1Δ mutants, nor did they exhibit the polarity patch break-ups or disappearances observed in vivo. Thus, either vesicle traffic is more perturbing than we considered in our models, or some other process also contributes to wandering.
One potential factor that could enhance the perturbing effect of vesicle traffic is the presence of Cdc42-directed GTPase Activating Proteins (GAPs) on vesicles. GAPs contributed to wave-like polarity patch motion in vegetative cells [33], and the GAP Bem3 may be concentrated on secretory vesicles [34]. If vesicles carry GAPs, their fusion would disrupt the polarity patch to a greater degree than the dilution process we modeled. Of course, it is also possible that some non-vesicular Myo2 cargo contributes to wandering.
Although the majority of the wandering (as well as the patch break-up and disappearance events) was actin-dependent, there was nevertheless a residual level of wandering in Lat-A-treated cdc24-m1 rsr1Δ cells. This seems likely to have arisen from stochastic molecular noise in protein interactions, which would not be captured by our deterministic model. Molecular noise may have limited power to promote wandering on its own, but it may synergize with vesicle-driven wandering, and the combination may suffice to account for the wandering observed in vivo.
A wandering polarity patch provides the opportunity for noise filtering
It has long been appreciated that formation of a pointy mating projection requires a much higher concentration of pheromone than that required to induce cell-cycle arrest [35]. We found that projection morphology was correlated with the degree of patch wandering: extensive wandering distributed growth around the cortex, generating fat, rounded projections, whereas restrained wandering maintained a constant direction of growth, generating pointy projections. Thus, cell shape reflects the integral of polarity patch location over time, and net growth reflects a time-average of the polarity patch position. These observations suggest the appealing hypothesis that the wandering polarity patch provides the same time-averaging benefit, in terms of noise filtering, as that due to the biased random walk performed by motile cells tracking shallow gradients.
If patch wandering enables more effective gradient tracking, then preventing wandering should reduce cells’ ability to track gradients. Indeed, when wandering was blocked by concentrating Bem1 on vesicles, gradient tracking was greatly impaired. Although we cannot rule out the possibility that the Bem1-v-SNARE fusion had additional side-effects (Fig. S5C), in sum our findings suggest that polarity patch wandering provides the mechanistic basis for the gradient-induced reorientation and error correction exhibited by yeast cells tracking gradients.
How is wandering biased by pheromone gradients?
If a wandering polarity patch undergoes a “biased random walk” resulting in net growth up-gradient, then wandering must be biased by the extracellular pheromone gradient. As discussed above, both Rsr1 and the Gβγ-Far1-Cdc24 connection can restrain wandering in a pheromone concentration-dependent manner. Intriguingly, genetic manipulations that impaired gradient tracking also prevented rsr1Δ cells from forming pointy mating projections [36]. Since our findings indicate that forming a pointy projection requires effective restraint of patch wandering, this correlation suggests that constraining wandering is important for gradient tracking.
Both active Rsr1 and Gβγ-Far1 bind to, and perhaps activate, the GEF Cdc24 [12, 13, 37–39], but in a pheromone gradient, Gβγ-Far1 is dominant over Rsr1 [40]. Thus, cells in a pheromone gradient may have two pools of active cortical Cdc24: one recruited to the polarity patch by positive feedback, and one recruited to locations with free Gβγ (i.e. ligand-activated pheromone receptors). If the polarity patch is not correctly aligned up-gradient, the pool of Cdc24 localized by Gβγ will be offset from the positive-feedback-localized pool, resulting in two polarity clusters. Mathematical modeling suggested that two polarity clusters far apart from each other in the same cell would compete for cytoplasmic Bem1 complexes and one patch would grow at the expense of the other. However, if the clusters were close to each other, then they would merge into one peak at an intermediate location [11, 31, 41]. Such competition and merging may enable an off-center peak of Gβγ-localized Cdc24 to bias wandering of the positive-feedback-generated peak towards the highest concentration of ligand-bound receptors. The details of pheromone-induced bias of patch wandering remain to be determined and offer a rich ground for future studies.
Conclusion
Cells tracking shallow gradients must amplify the gradient to properly localize polarity factors and they must filter out noise from stochastic receptor-ligand interactions. We show that yeast cells employ actin-independent positive feedback to amplify the pheromone gradient resulting in highly polarized growth. Additionally, cells possess actin-dependent mechanisms that perturb such positive feedback and allow re-positioning of the polarity site. Computational and experimental findings suggest that the same vesicle trafficking that enables polar growth also causes polarity patch wandering. This intrinsically-driven wandering is restrained by increasing pheromone concentration and provides a potential mechanism to improve the alignment between polar growth direction and the pheromone gradient. In principle, an intracellular wandering polarity patch that performs a biased random walk could enable noise filtering and effective tracking of shallow gradients, even for non-motile cells.
Experimental Procedures
Yeast strains and pheromone treatment
Standard molecular genetic procedures were employed to generate yeast strains, listed in Supplementary Information online. α-factor (Genway Biotech, San Diego, CA) treatment in complete synthetic medium (CSM: MP Biomedicals, Solon, OH) plus 2% dextrose appeared less effective than in YEPD, so concentrations are not comparable between media.
Live-Cell Microscopy
Cells were grown to mid-log phase in CSM to OD600 = 0.1, and pre-treated with α-factor for 1–2 h prior to imaging. Cells were mounted on a slab of CSM plus α-factor, which was solidified with 2% agarose (Denville Scientific, Inc., Metuchen, NJ). Slab edges were sealed with petroleum jelly, and cells were filmed at 30°C unless otherwise noted. Images were acquired and deconvolved essentially as described [19], except images from Movie S1 and Fig. S1C, which were acquired using an Andor XD revolution spinning disk confocal microscope with an Andor Ixon3 897 512 EMCCD camera.
In some cases, two strains were mixed on the same slab to compare polarity patch behavior under identical conditions. Genotype was distinguished using Spa2-mCherry, which was present in only one strain and did not affect wandering (not shown).
Image Analysis
Polarity patch tracking was performed on deconvolved images using Volocity software (Improvision, Inc., Waltham, MA). For each timelapse, a single threshold was applied to select pixels that visually overlapped with polarity patches (Fig. S1). For each cell, three-dimensional coordinates of the thresholded region centroid were collected for all timepoints. During filming, stage positions exhibited some drift. Before plating on the slab, cells were mixed with 0.2 μm Tetraspeck fluorescent beads (Invitrogen, Eugene, OR). One bead was selected as the “origin,” and used to standardize all positional coordinates from that stage position.
Centroid tracks were used to calculate m.s.d. For all pairs of timepoints ti, tj and corresponding centroid positions pi, pj, the distance between pi and pj was squared, and the average of these was calculated for each time interval T=tj−ti. Trajectories from cells in which patches broke up or disappeared were separated so that only centroid tracks from continuous trajectories were used to calculate the m.s.d.
Latrunculin-A Treatment
Cells harboring the actin-patch marker Abp1-mCherry were used to confirm the efficacy of Lat-A treatment (Fig. S3). To track wandering, cells were pre-treated for 1 h with 60 nM α-factor, collected by centrifugation, and resuspended in CSM + 200 μM Lat-A (Invitrogen, Carlsbad, CA) before plating on a Lat-A slab, prepared as follows: 2 μl of 20 mM Lat-A was added to a 200 μl slurry of unheated agarose in CSM with α-factor. The slurry was heated in a boiling water bath until the agarose had melted and used to prepare a slab as above. Control cells were treated with DMSO instead of Lat-A.
To film polarity establishment (Fig. 3E), cells were mixed with 0.3 μM α-factor and 200 μM Lat-A and plated on a Lat-A slab (as above) 10 min before filming.
To document long-term loss of polarity, cells were grown in YEPD and pre-treated for 90 min with 0.3 μM α-factor. The culture was split and treated with either 200 μM Lat-A or DMSO and incubated at 30°C. Samples were collected every 30 min and fixed for 5 min with 3.7% formaldehyde (Sigma-Aldrich, St. Louis, MO).
Photobleaching
For FRAP analysis, cells were grown and prepared on a Lat-A slab as above except that the cells (BAR1) were treated with 3 μm α-factor. FRAP analysis was performed at 30°C on an inverted Olympus IX71 microscope with an Evolve back-thinned EM-CCD camera connected to a Deltavision Imaging System (Applied Precision, Issaquah, WA). Images were acquired using a 100X (1.40NA) oil immersion objective and the Deltavision SoftWoRx Resolve 3D capture program. Photobleaching was achieved by a single 0.1 s laser pulse at 488 nm (25% laser power) and 30 subsequent images were acquired at adapative time intervals based on an expected recovery t1/2 of 3 s. Mean fluorescence intensity was monitored in a 1.9 μm diameter circle normalized to pre-bleach peak intensity. t1/2 for each cell was determined from a double-exponential fit to each curve.
Microfluidics
A microfluidics device was operated as previously described [16]. A gradient of 0–11.9 nM α-factor in YEPD was used for all experiments. Media containing α-factor was mixed with 2 μg/mL sulforhodamine 101 (Sigma-Aldrich, St. Louis, MO) to visualize the gradient. DIC images were analyzed using ImageJ software (http://rsbweb.nih.gov/ij/index.html). A line was drawn from the projection tip into the interior of the cell, normal to the tip of the projection. The angle between the line and the gradient was measured every 30 min. Orientation measurements were restricted to cells within the linear part of the gradient and only cells with unobstructed access up the gradient were scored. Scoring was terminated if a cell budded or was “bumped” by another cell, changing orientation.
Computational modeling of positive feedback and vesicle trafficking
A mathematical model combining an actin-independent reaction-diffusion polarity system with vesicle exo- and endocytosis [22] was adapted as described in Supplementary Information online.
Supplementary Material
Highlights.
A wandering polarity site distributes growth in yeast cells responding to pheromone
Wandering, but not polarity establishment, is actin-dependent
A mathematical model incorporating polarity and vesicle traffic exhibits wandering
Blocking wandering impairs gradient tracking
Acknowledgments
We thank N. Buchler and S. Kornbluth for critical readings of the manuscript; S. Johnson for technical expertise with microscopy; A. McClure for assistance with data analysis; J. Kelley for assistance with microfluidics; A. Layton for assistance with computational modeling; A. Johnson for analysis of Bem1-Snc2 arrest recovery kinetics; and D. Stone and Lew lab members for stimulating discussions. This work was supported by NIH grants GM79271 and GM84071 (T.C.E.), and GM62300 (D.J.L.) as well as a supplement for collaborative science (D.J.L. & T.C.E.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wadhams G, Armitage J. Making sense of it all: bacterial chemotaxis. Nat Rev Mol Cell Biol. 2004;5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- 2.Swaney KF, Huang C-H, Devreotes PN. Eukaryotic Chemotaxis: A Network of Signaling Pathways Controls Motility, Directional Sensing, and Polarity. Annual Review of Biophysics. 2010;39 doi: 10.1146/annurev.biophys.093008.131228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zigmond SH. Ability of polymorpho-nuclear leukocytes to orient in gradients of chemotactic factors. J Cell Biol. 1977;75:606–616. doi: 10.1083/jcb.75.2.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Segall JE. Polarization of yeast cells in spatial gradients of alpha mating factor. Proc Natl Acad Sci U S A. 1993;90:8332–8336. doi: 10.1073/pnas.90.18.8332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mato JM, Losada A, Nanjundiah V, Konijn TM. Signal input for a chemotactic response in the cellular slime mold Dictyostelium disoideum. Proc Natl Acad Sci U S A. 1975;75:4991–4993. doi: 10.1073/pnas.72.12.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miyanaga Y, Matsuoka S, Yanagida T, Ueda M. Stochastic signal inputs for chemotatic response in Dictyostelium cells revealed by single molecular imaging techniques. BioSystems. 2007;88:251–260. doi: 10.1016/j.biosystems.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 7.van Haastert PJM, Postma M. Biased random walk by stochastic fluctuations of chemoattractant-receptor interactions at the lower limit of detection. Biophys J. 2007;93:1787–1796. doi: 10.1529/biophysj.107.104356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chou CS, Bardwell L, Nie Q, Yi TM. Noise filtering tradeoffs in spatial gradient sensing and cell polarization response. BMC Systems Biology. 2011;5:196. doi: 10.1186/1752-0509-5-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arkowitz RA. Chemical Gradients and Chemotropism in Yeast. Cold Spring Harb Perspect Biol. 2009;1 doi: 10.1101/cshperspect.a001958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bi E, Park HO. Cell Polarization and Cytokinesis in Budding Yeast. Genetics. 2012;191:347–387. doi: 10.1534/genetics.111.132886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson JM, Jin M, Lew DJ. Symmetry breaking and the establishment of cell polarity in budding yeast. Current Opinion in Genetics & Development. 2011;21:740–746. doi: 10.1016/j.gde.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nern A, Arkowitz RA. A Cdc24p-Far1p-Gbetagamma protein complex required for yeast orientation during mating. J Cell Biol. 1999;144:1187–1202. doi: 10.1083/jcb.144.6.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Butty AC, Pryciak PM, Huang LS, Herskowitz I, Peter M. The role of Far1p in linking the heterotrimeric G protein to polarity establishment proteins during yeast mating. Science. 1998;282:1511–1516. doi: 10.1126/science.282.5393.1511. [DOI] [PubMed] [Google Scholar]
- 14.Madden K, Snyder M. Specification of sites for polarized growth in Saccharomyces cerevisiae and the influence of external factors on site selection. Mol Biol Cell. 1992;3:1025–1035. doi: 10.1091/mbc.3.9.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moore TI, Chou CS, Nie Q, Jeon NL, Yi TM. Robust spatial sensing of mating pheromone gradients by yeast cells. PLoS One. 2008;3:e3865. doi: 10.1371/journal.pone.0003865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hao N, Nayak S, Behar M, Shanks RH, Nagiec MJ, Errede B, Hasty J, Elston TC, Dohlman HG. Regulation of Cell Signaling Dynamics by the Protein Kinase-Scaffold Ste5. Molecular Cell. 2008;30:649–656. doi: 10.1016/j.molcel.2008.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Falconnet D, Niemisto A, Taylor RJ, Ricicova M, Galitski T, Shmulevich I, Hansen CL. High-throughput tracking of single yeast cells in a microfluidic imaging matrix. Lab on a Chip. 2011;11:466–473. doi: 10.1039/c0lc00228c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Slaughter BD, Das A, Schwartz JW, Rubinstein B, Li R. Dual modes of Cdc42 recycling fine-tune polarized morphogenesis. Developmental Cell. 2009;17:823–835. doi: 10.1016/j.devcel.2009.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howell Audrey S, Jin M, Wu CF, Zyla Trevin R, Elston Timothy C, Lew Daniel J. Negative Feedback Enhances Robustness in the Yeast Polarity Establishment Circuit. Cell. 2012;149:322–333. doi: 10.1016/j.cell.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elia L, Marsh L. A Role for a Protease in Morphogenic Responses during Yeast Cell Fusion. The Journal of Cell Biology. 1998;142:1473–1485. doi: 10.1083/jcb.142.6.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nern A, Arkowitz RA. G proteins mediate changes in cell shape by stabilizing the axis of polarity. Mol Cell. 2000;5:853–864. doi: 10.1016/s1097-2765(00)80325-1. [DOI] [PubMed] [Google Scholar]
- 22.Savage NS, Layton AT, Lew DJ. Mechanistic mathematical model of polarity in yeast. Molecular Biology of the Cell. 2012;23:1998–2013. doi: 10.1091/mbc.E11-10-0837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ayscough KR, Stryker J, Pokala N, Sanders M, Crews P, Drubin DG. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ayscough KR, Drubin DG. A role for the yeast actin cytoskeleton in pheromone receptor clustering and signalling. Curr Biol. 1998;8:927–930. doi: 10.1016/s0960-9822(07)00374-0. [DOI] [PubMed] [Google Scholar]
- 25.Sheu YJ, Santos B, Fortin N, Costigan C, Snyder M. Spa2p interacts with cell polarity proteins and signaling components involved in yeast cell morphogenesis. Mol Cell Biol. 1998;18:4053–4069. doi: 10.1128/mcb.18.7.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pruyne D, Legesse-Miller A, Gao L, Dong Y, Bretscher A. Mechanisms of polarized growth and organelle segregation in yeast. Annu Rev Cell Dev Biol. 2004;20:559–591. doi: 10.1146/annurev.cellbio.20.010403.103108. [DOI] [PubMed] [Google Scholar]
- 27.Schott D, Ho J, Pruyne D, Bretscher A. The COOH-Terminal Domain of Myo2p, a Yeast Myosin V, Has a Direct Role in Secretory Vesicle Targeting. J Cell Biol. 1999;147:791–808. doi: 10.1083/jcb.147.4.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marco E, Wedlich-Soldner R, Li R, Altschuler SJ, Wu LF. Endocytosis optimizes the dynamic localization of membrane proteins that regulate cortical polarity. Cell. 2007;129:411–422. doi: 10.1016/j.cell.2007.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Layton AT, Savage NS, Howell AS, Caroll SY, Drubin D, Lew DJ. Modeling vesicle traffic reveals unexpected consequences for Cdc42p-mediated polarity establishment. Curr Biol. 2011;21:184–194. doi: 10.1016/j.cub.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu JH, Crevenna AH, Bettenbuhl M, Freisinger T, Wedlich-Soldner R. Cortical actin dynamics driven by formins and myosin V. Journal of Cell Science. 2011;124:1533–1541. doi: 10.1242/jcs.079038. [DOI] [PubMed] [Google Scholar]
- 31.Howell AS, Savage NS, Johnson SA, Bose I, Wagner AW, Zyla TR, Nijhout HF, Reed MC, Goryachev AB, Lew DJ. Singularity in Polarization: Rewiring Yeast Cells to Make Two Buds. Cell. 2009;139:731–743. doi: 10.1016/j.cell.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suchkov DV, DeFlorio R, Draper E, Ismael A, Sukumar M, Arkowitz R, Stone D. Polarization of the yeast pheromone receptor requires its internalization but not actin-dependent secretion. Mol Biol Cell. 2010;21:1737–1752. doi: 10.1091/mbc.E09-08-0706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ozbudak EM, Becskei A, van Oudenaarden A. A System of Counteracting Feedback Loops Regulates Cdc42p Activity during Spontaneous Cell Polarization. Dev Cell. 2005;9:565–571. doi: 10.1016/j.devcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 34.Knaus M, M-P, PG, van Drogen F, Springer S, Jaquenoud M, Peter M. Phosphorylation of Bem2p and Bem3p may contribute to local activation of Cdc42p at bud emergence. Embo J. 2007;26:4501–4513. doi: 10.1038/sj.emboj.7601873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moore SA. Comparison of Dose-Response Curves for Alpha Factor-induced Cell Division Arrest, Agglutination, and Projection Formation of Yeast Cells. Journal of Biological Chemistry. 1983;258:13849–13856. [PubMed] [Google Scholar]
- 36.Strickfaden SC, Pryciak PM. Distinct Roles for Two G{alpha} G Interfaces in Cell Polarity Control by a Yeast Heterotrimeric G Protein. Mol Biol Cell. 2008;19:181–197. doi: 10.1091/mbc.E07-04-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park HO, Bi E, Pringle JR, Herskowitz I. Two active states of the Ras-related Bud1/Rsr1 protein bind to different effectors to determine yeast cell polarity. Proc Natl Acad Sci U S A. 1997;94:4463–4468. doi: 10.1073/pnas.94.9.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wiget P, Shimada Y, Butty AC, Bi E, Peter M. Site-specific regulation of the GEF Cdc24p by the scaffold protein Far1p during yeast mating. Embo J. 2004;23:1063–1074. doi: 10.1038/sj.emboj.7600123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shimada Y, Wiget P, Gulli MP, Bi E, Peter M. The nucleotide exchange factor Cdc24p may be regulated by auto-inhibition. Embo J. 2004;23:1051–1062. doi: 10.1038/sj.emboj.7600124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valtz N, Peter M, Herskowitz I. FAR1 is required for oriented polarization of yeast cells in response to mating pheromones. J Cell Biol. 1995;131:863–873. doi: 10.1083/jcb.131.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goryachev AB, Pokhilko AV. Dynamics of Cdc42 network embodies a Turing-type mechanism of yeast cell polarity. FEBS Lett. 2008;582:1437–1443. doi: 10.1016/j.febslet.2008.03.029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.