Summary
Clathrin is a trimeric protein involved in receptor-mediated-endocytosis, but can function as a non-trimer outside of endocytosis. We have discovered that the subcellular distribution of a clathrin cysteine mutant we previously studied is altered and a proportion is also localized to nuclear spaces. MALS shows C1573A hub is a mixture of trimer-like and detrimerized molecules. The X-ray structure of the trimerization domain reveals that that without light chains, a helix harboring cysteine-1573 is reoriented. We propose clathrin has a detrimerization switch, which suggests clathrin topology can be altered naturally for new functions.
Keywords: Clathrin, endocytosis, clathrin monomer, clathrin dimer, detrimerization, p53 transactivation, cancer
Introduction
Three-legged clathrin molecules interlock into a closed polyhedral lattice around transport vesicles during receptor-mediated endocytosis (RME). The ability of clathrin to self-assemble relies entirely on its unique pinwheel shape. This process is spontaneous in vitro, but in cells the formation of a clathrin basket is subject to a host of accessory proteins. Adaptor proteins, non-visual arrestins, and Hsc70 associate with the N-terminal β-propeller domain of clathrin heavy chain [1, 2]. The β-propeller domain is joined to the filamentous heavy chain leg (543–1576) that is subdivided into the distal and proximal domains with a flexible knee joint between them [3, 4]. To complete the clathrin triskelion, the C-terminal tails of three identical heavy chain legs are brought together in the clathrin trimerization domain [5, 6]. The three heavy chain legs each have an attendant clathrin light chain that binds to the proximal region. In vertebrates and invertebrates there are two types of light chain subunits (LCa and LCb [7]), but yeast clathrin has only one kind of light chain [8]. The light chain subunit is proposed to modify the flexibility of the knee joint to regulate lattice formation [4], mediates the binding of Huntingtin-Interacting Proteins (HIPs) [9, 10], and stabilizes the global three-legged clathrin structure by making contact with the trimerization domain [11, 12].
It has become clear that clathrin function extends beyond conventional endocytosis. For example, clathrin lattice formation has been observed to initiate the development of tubulobulbar complexes in rat testis [13]. The stability of kinetochore fibers and the integrity of the centrosome depend on the association of clathrin with TACC3 and ch-TOG proteins [14]. There is also a novel connection between clathrin and human cancer. Rare B cell and T cell lymphomas contain oncogenic elements that are linked to clathrin heavy chain in ways that produce trimeric or dimeric oncogenic fusions [15]. The correlation between clathrin and cancer may be more fundamental because clathrin influences the transactivation of tumor suppressor p53 [16]. To do this, clathrin must be monomeric and inside the nucleus [16, 17]. However, there is no evidence in vivo that trimeric clathrin can be reduced to a monomer unless its trimerization domain is artificially deleted. This is a problem because there is no information to as to whether detrimerized clathrin can be produced naturally to meet specific cellular needs.
To investigate whether the trimeric clathrin can be detrimerized without resorting to deletion, we reexamined our previous observations that clathrin trimer stability was sensitive to a conserved cysteine [12, 18]. In this report, we determined the subcellular distribution of C1573A clathrin hub (aa1074-1675) in a variety of cell types. Confocal microscopy experiments show that WT hub has a diffuse distribution pattern in the cytoplasm, but a proportion of C1573A hub also localizes to nuclear spaces. A longer C1573A clathrin construct (aa865-1675) produces a similar distribution pattern when exogenously expressed in cells. To test the hypothesis that mutating cysteine-1573 alters the trimeric state of clathrin hub, multi-angle light scattering (MALS) experiments were performed on purified WT and C1573A hub samples to identify any molar mass and size differences. MALS experiments were also performed on samples with light chains (LCb) because we previously observed that LCb could reverse the destabilizing effect of the C1573A mutation [18]. The MALS data indicate WT hub is trimeric in solution but there is a mixture of different detrimerized forms of C1573A hub. The detrimerized molecules identified by MALS are driven to be trimer-like when LCb was added.
Finally, to gain atomic-level insights into the contribution of light chain on trimeric clathrin stability, we crystallized a minimum clathrin trimerization domain without any light chain and solved the structure at 3.9 Å. Our X-ray model has the characteristic features of the trimerization domain compared to lower resolution models that have bound light chain [4, 6], but there are obvious topological differences without light chain. Top and side views show leg geometry is distorted. The trimerization domain model shows Helix 7j (1563-1575) is shifted to a new position when superimposed over lower resolution models that have light chains. Together, the experimental and structural evidence presented here suggest clathrin contains a topology switch. This raises the possibility that there is a natural mechanism that can restructure clathrin for functions beyond endocytosis.
Materials and methods
Plasmids
pET15b encoding bovine fragment 1521-1654 (Ctxd with C1528A and T1585L) with an N-terminal 6-histidine tag and pCDNA3.1-bovine hub (1074-1675) WT and 865-1675 WT clathrin with an N-terminal human influenza hemagglutinin (HA) tag were constructed using standard cloning methods. C1573A mutations were created by standard site-directed mutagenesis. Plasmid constructs were verified by DNA sequencing.
Protein expression and purification
All clathrin constructs were expressed in Rosetta 2(DE3) plysS cells and grown at 37 °C in LB and induced at 30 °C with IPTG. Cell pellets were resuspended and sonicated in cold lysis buffer (10 mM Na2HPO4, 10 mM imidazole, 0.5 M NaCl, pH 7.4 with Triton X-100, β-mercaptoethanol, and protease inhibitors). Crude Ctxd lysate was passed through a Ni-saturated chelating sepharose affinity column (GE Healthcare), and then through a POROS 20 HQ anion-exchange column (Perseptive Biosystems) in 10 mM HEPES, 20 mM imidazole, 1% (v/v) glycerol, 10 mM β-mercaptoethanol at pH 8.5. Ctxd was polished using a Superdex 200 column. WT and C1573A hub constructs were polished as previously described [12].
Samples for MALS were dialyzed at 4 °C against 10 mM Tris, pH 7.9 or in buffer also containing 60 mM NaCl and 1 mM EDTA.
Crystallization
Protein at 25 mg/ml was crystallized by the hanging drop method in 2.0 M NaCl, 80mM imidazole, pH 7.0 with PEG 3350. Crystals grew in the trigonal space group R3 (a = 255.78 Å, b = 255.78, c = 312.99, and a = 90°, b = 90°, g = 120°, hexagonal setting), with 24 protomers (8 trimers) in the asymmetric unit. Resolution limit was significantly improved by soaking 2–3 week old crystals in 87.7 mM imidazole, 36% (v/v) glycerol cryogenic buffer containing n-tetradecyl-β-D-maltoside for 20 minutes exposed to open air before plunging in liquid nitrogen.
Data collection
A data set was collected at 100 °K in 0.3° oscillations using a NOIR-1 CCD detector at the MBC 4.2.2 beamline at the Advanced Light Source, Lawrence Berkeley National Laboratory. Data were processed using HKL2000.
Phasing and refinement
Crystal data were phased by molecular replacement using Phaser [19]. A trimer model from the cryo-EM structure (PDB code 1XI4) failed, but a solution was found using search models (histidine tag not included) with manually altered leg orientations. The model was built with O [20] and refined using CNS [21]. Refinement was carried out on partially twinned data (twinning operator= k, h, -l and twinning fraction= 0.484, determined by CNS). Because there were 24 protomers (8 trimers) in the asymmetric unit, non-crystallographic symmetry (NCS) restraints were used throughout the refinement. To minimize distortions, simulated annealing was performed with harmonically restrained backbone atoms (harmonic restraint constant = 10). This was interspersed with model rebuilding with reference to 2Fo - Fc and Fo - Fc maps. Electron density of the histidine tags (not in the search model) of two legs became visible in Fo - Fc maps. The clathrin trimerization domain structure refined against all the data with an Rcryst of 33.9% and an Rfree of 38.7% (see Supplemental Crystallographic Table).
Multi-angle light scattering measurements
WT hub and C1573A hub constructs were purified in 10 mM Tris, pH7.9 buffer or in the same buffer containing 60 mM NaCl and 1 mM EDTA. All experiments were performed using 70 μg of WT or C1573A hub. For experiments with light chain, 70 μg WT or C1573A were incubated with 53.3 μg histidine-tagged Lcb for 20 minutes prior to analysis. Measurements were performed on a Wyatt Technology DAWN HELEOS-II multi-angle light scattering (MALS) instrument coupled to a Wyatt Technology Optilab rEX refractive index (RI) detector. 100 μl of sample (57.7 μg WT or C1573A hub) was loaded onto a Superose 6 10/300GL size exclusion (SEC) column at a flow rate of 0.5 ml/min. SEC-MALS data were analyzed using Wyatt Technology ASTRA software to obtain molar mass (M) and radius of gyration.
Cell culture
All reagents from Gibco (Life Technologies) unless indicated otherwise. Cells were maintained at 37 °C, in a 5% CO2 humidified incubator. HeLa (cervical cancer) and HEK293T (human embryonic kidney) cells were maintained in DMEM with 10% fetal bovine serum (FBS, Sigma-Aldrich) 100 U penicillin/100 mg streptomycin. H1299 (non-small cell lung cancer) cells were maintained in RPMI-1640 media with 10% FBS, 10 mM HEPES, 100 U penicillin/100 mg streptomycin, 2 mM Glutamax, and 1 mM sodium pyruvate. MCF7 (breast cancer) cells were maintained in MEM with 10% FBS, 100 U penicillin/100 mg streptomycin, 2 mM glutamine, 1 mM sodium pyruvate, 0.1% sodium bicarbonate, and 120 ng/ml insulin (Sigma).
Confocal microscopy
H1299, MCF7, HeLa, and 293T cells were plated onto poly-l-lysine coated coverslips and transfected with pcDNA3.1(−)HA-hub (aa1074-1675) WT or C1573A and pCDNA3.1(−)HA-865-1675 WT or C1573A plasmids using FugeneHD (Roche). After 24 h, cells were fixed, permeabilized, and stained with anti-HA (Covance) and anti-mouse AlexaFluor 488 (Molecular Probes, Invitrogen) antibodies. Nuclei were counterstained with Draq5 (Cell Signaling Technologies) and coverslips mounted using ProLong Gold (Invitrogen). Images were obtained using a Leica Sp5 scanning confocal system. At least 100 cells over four independent experiments were analyzed using Leica LAF software. Signal within the nucleus above background was scored nuclear positive. Statistical significance was determined using one-way ANOVA.
Results and discussion
Subcellular localization of C1573A hub
Previously we showed that cysteine-1573 was a key determinant of trimeric clathrin structure [12, 18]. To determine if mutating this conserved cysteine could influence the cellular distribution of mutant clathrin, we analyzed the subcellular distribution of C1573A clathrin hub (aa1074-1675) by confocal microscopy in a variety of cell types. HA-tagged hub WT and C1573A were expressed in HEK293T, HeLa, H1299, and MCF7 cells and cellular distribution was visualized by confocal microscopy. Figure 1A shows HA-tagged WT hub distribution is diffuse and is located in the cytoplasm. The observed staining pattern is consistent with findings that full-length clathrin without the N-terminal domain is diffusely distributed in the cytoplasm [22]. In contrast, C1573A hub was not only cytosolic, but there was also a proportion inside the nucleus. The differential distribution between WT and C1573A hub was observed in every cell type analyzed (Fig. 1A, B). To determine if the relatively small size of hub facilitated nuclear entry, we tried a larger clathrin construct. Figure 1C shows a fraction of HA-865-1675 C1573A in H1299 is detected in nuclear spaces. A limitation of our work is that the C1573A constructs cannot interact with membranes because the N-terminal domain is absent. Therefore, we cannot rule out the possibility that C1573A constructs can enter the nucleus because of their inability to associate with membrane. Further studies will be needed to determine what full-length C1573A clathrin will do. Despite the limitations, the data here demonstrate that cysteine-1573 impacts the subcellular distribution of both hub and clathrin 865-1675. When this conserved cysteine is mutated to alanine, our C1573A constructs are detected in nuclear spaces compared to WT. This nuclear localization prompted us to ask whether the cysteine mutation altered the trimeric structure of clathrin.
Figure 1. Mutation in helix 7j causes nuclear localization of clathrin hub.
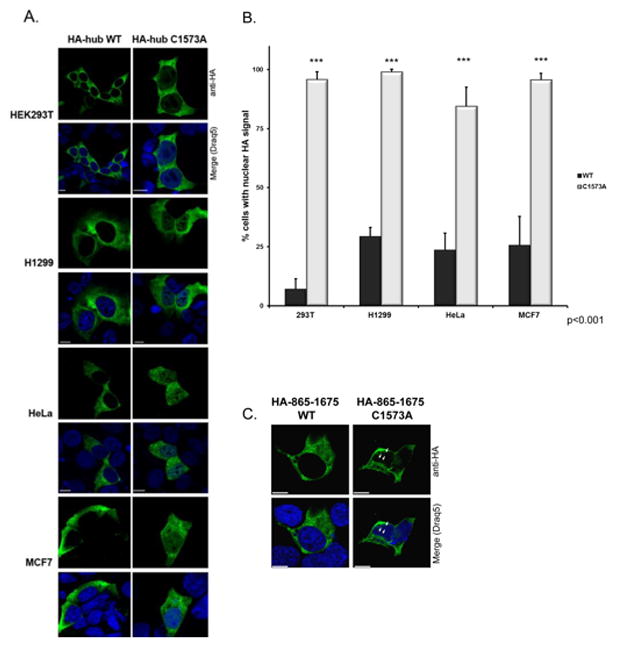
A) Immunofluorescence staining of HA-hub WT and HA-hub C1573A in HEK293T, H1299, HeLa, and MCF7 cells indicates HA signal (green) is localized to membrane/cytoplasm in the HA-hub WT (leftmost panels) and C1573A (right panels). HA signal is also present in the nuclei (counterstained with Draq5 in blue) of nearly all cells expressing HA-hub C1573A (right panels) but not WT cells. B) Number of cells with nuclear HA signal is much higher (over 90% of cells) in cells expressing HA-hub C1573A compared to HA-hub WT cells. Error bars are SEM. ***p<0.001. C) Immunofluorescence in H1299 of HA-clathrin 865-1675 WT (left) and C1573A (right). HA signal is present in the nuclear spaces in HA-clathrin 865-1675 C1573A but not WT cells. Scale bar is 10 μm.
Multi-angle light scattering detects detrimerized C1573A
To assess the multimerization of WT and C1573A hub, MALS experiments were performed on purified protein. A size exclusion column placed before the light scattering instrument resolves different oligomeric states. The blue dashed lines in Fig. 2 reflect the relative concentrations of hub forms, hub/LCb, or unbound LCb. The red curve represents the measured light scattering from one of 18 detectors positioned at different angles. The signals are fit to a linear regression (Debye plots). The slope of the line yields the radius of gyration and the inverse of the y-intercept gives the molar mass values in Table 1. Initially measurements were done on preparations in buffer containing 60mM sodium chloride. Although the WT hub results were consistent between repeats in this buffer, C1573A results varied significantly between samples (see Fig. 2B). Because the activity of clathrin hub can be sensitive to ionic strength [23], MALS experiments were repeated in buffer without any salt (Fig. 2A and Table 1). The WT hub MALS data yield a molar mass of 222.8 ±13.47 kDa that agrees with the calculated molar mass of trimeric hub (213 kDa). The analysis identified trimer-like C1573A hub molecules with a radius of gyration of of 16.8 ±1.6 nm compared to 12.1 ±1.0 nm for WT hub (see Table 1 and Fig. 2). The largest population of C1573A hub is an apparent dimer with a molar mass of 158.83 ±10.17 kDa. The fraction of detrimerized C1573A species can be time sensitive. For example, C1573A in experiment 3 (several days old) was ~44% monomer (molar mass of 74.2 kDa) and only ~7% and ~11% were trimer- or dimer-like. SDS-PAGE analysis confirmed this was not due to degradation (data not shown). We conclude that WT hub exists predominantly as a trimer in solution both with and without light chain, but without light chain C1573A hub is a complex mixture of aggregated, trimer-like, and detrimerized molecules.
Figure 2. Multi-angle light scattering shows detrimerized C1573A hub.
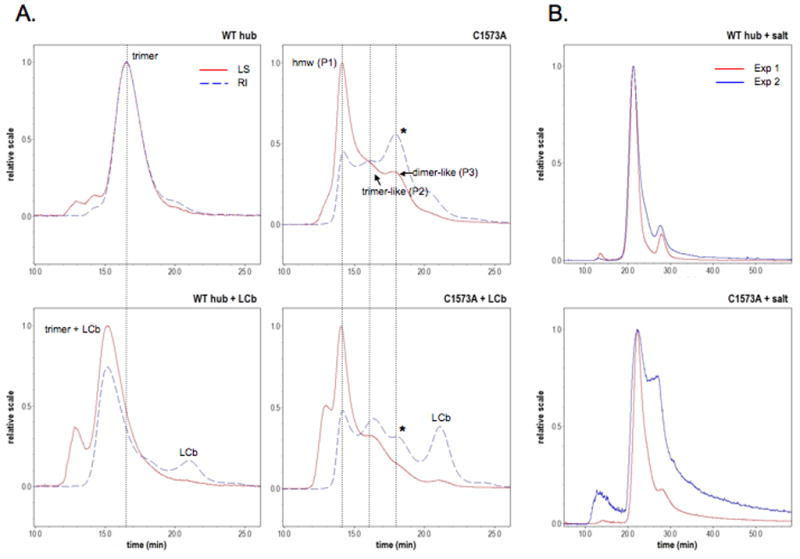
A) Light scattering (LS) and refractive index (RI) profiles of WT hub and C1573A hub with and without LCb. Intensities of scattered light (at 90 degrees in red) are correlated to particle size. Refractive indices (blue dashed lines) reflect the relative amounts of different hub forms resolved by the Superose 6 column. Positions of high molecular weight (hmw), trimer-like, and dimer-like forms in Table 1 are indicated by dotted lines. Addition of LCb alters the amount of different C1573A oligomers (see asterisks right panels and Table 1). For meaningful comparisons, the amount of WT or C1573A hub in the plus LCb experiments was not changed (2-fold molar excess LCb was added to WT or C1573A hub). All experiments were repeated three times with independently purified hub samples. B) C1573A hub light scattering varied significantly in buffer containing 60mM sodium chloride.
Table 1.
Analysis of WT and C1573A hub by multi-angle light scattering
| MM P1a (kDa) | Fb (%) | Rc (nm) | MM P2a | F (%) | R (nm) | MM P3a | F (%) | R (nm) | MM P4a | F (%) | R (nm) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| C1573A | ||||||||||||
| Exp 1 | 551.1 | 25.3 | 23.6 | 276.0 | 21.4 | 16.4 | 167.4 | 42.9 | 13.0 | - | - | - |
| Exp 2 | 529.9 | 28.9 | 22.8 | 270.5 | 20.8 | 15.5 | 161.5 | 39.5 | 11.8 | - | - | - |
| Exp 3d | 492.9 | 3.3 | 27.8 | 232.8 | 6.7 | 18.6 | 147.6 | 10.6 | 10.6 | 74.2 | 44.3 | 8.4 |
| Mean SD | 524.63 ±29.46 | n/d | 24.7 ±2.7 | 259.77 ±23.52 | n/d | 16.8 ±1.6 | 158.83 ±10.17 | n/d | 11.8 ±1.2 | - | - | - |
|
| ||||||||||||
| C1573A + LCb | ||||||||||||
| Exp 1 | 515.3 | 25.8 | 25.7 | 209.5 | 31.6 | 16.4 | 129.6 | 13.7 | 16.9 | - | - | - |
| Exp 2 | 468.3 | 27.9 | 22.9 | 207.0 | 33.6 | 15.3 | 128.0 | 12.6 | 16.1 | - | - | - |
| Exp 3d | 536.9 | 5.5 | 28.5 | 199.9 | 27.9 | 15.0 | 110.6 | 52.9 | 14.2 | - | - | - |
| Mean SD | 506.83 ±35.08 | n/d | 25.7 ±2.8 | 205.47 ±4.98 | n/d | 15.6 ±0.74 | 122.73 ±10.54 | n/d | 15.7 ±1.4 | - | - | - |
| MM P1a (kDa) | Fb (%) | Rc (nm) | MM P2a | F (%) | R (nm) | |
|---|---|---|---|---|---|---|
|
| ||||||
| WT hub | ||||||
| Exp 1 | 210.2 | 100 | 13.2 | - | - | - |
| Exp 2e | 221.2 | 76.9 | 11.4 | 80.5 | 23.1 | 8.2 |
| Exp 3e,f | 237.0 | 65.4 | 11.8 | 111.2 | 20.1 | 11.0 |
| Mean SD | 222.8 ± 13.47 | n/d | 12.1 ± 1.0 | n/d | n/d | n/d |
|
| ||||||
| WT hub + LCb | ||||||
| Exp 1 | 262.0 | 75.9 | 12.1 | 164.9 | 9.9 | 10.3 |
| Exp 2e | 240.5 | 68.9 | 11.3 | 93.7 | 31.1 | 7.7 |
| Exp 3e | 261.2 | 60.1 | 10.7 | 102.4 | 23.9 | 9.2 |
| Mean SD | 254.57 ± 12.19 | n/d | 11.3 ± 0.7 | n/d | n/d | n/d |
Molar mass moments of peaks in Fig. 2.
Means of mass fractions not calculated to show sample dependent variability.
Average rms radius moments (radius of gyration) in nanometers.
Stored several days before measurements.
Sample in 10 mM Tris, 60 mM NaCl, 1 mM EDTA, pH 7.9 buffer.
Minor shoulder not tabulated.
We previously reported that the C-terminal region of LCb supports the stability of the trimerization domain [18]. Here the addition of LCb caused a decrease in the fraction of detrimerized C1573A and an increase in the amount of trimer-like C1573A hub (see Table 1). This is consistent with our proposal that light chains stabilize clathrin by locking down the geometry of legs in the trimerization domain [18]. To better understand how light chain stabilizes clathrin, we solved the crystal structure of the trimerization domain without it.
Crystal structure of the central trimerization domain
We designed a fragment (designated Ctxd is the tripod core and a segment of the proximal domain, aa1521-1654 from Bos taurus) capable of trimerization, but cannot bind clathrin light chain (Fig. 3). The numbering scheme from bovine clathrin is used throughout (accession number NP 776448). Cysteine-1528 was mutated to alanine to avoid heterogeneity because this residue mediates non-specific aggregation [18]. Threonine-1585 is replaced by leucine in a muscle-specific isoform of clathrin (CHC22) that does not bind light chain [24], and we found that changing this threonine to leucine enhanced the stability of clathrin hub. We mutated this threonine in Ctxd to facilitate native trimerization. At first, Ctxd crystals did not diffract beyond ~9 Å. However after evaluating hundreds of conditions, small molecule additives, and detergents, we discovered exposing crystals to open air dramatically increased diffraction to 3.9 Å. This suggests high solvent content (estimated to be ~73% after dehydration) is partly responsible for low crystal resolution. Synchrotron data were collected on the best crystals (Advanced Light Source MBC 4.2.2) and the structure was solved by molecular replacement. The combination of limited crystal quality (best crystals were partially twinned), disordered N- and C-terminal regions, and the sheer size of the asymmetric unit (8 trimers or 24 protomers) made model refinement at 3.9 Å a challenge and this is reflected in the final R-values (Rcryst=33.9% and Rfree=38.7%, see Supplemental Crystallographic Table). The N-terminal portion of Leg B residues and 1625-1654 were not included in the final model because these were too highly disordered. Calculated Fo - Fc maps yielded extra electron density for histidine tags on Legs A and C (see Supplemental Fig. 2). This supports the molecular replacement (MR) solution because histidine tags were not in the initial MR search models. To avoid over-interpreting the medium resolution X-ray model, we only describe gross topological features and any changes associated with them.
Figure 3. Trimerization domain without light chains.
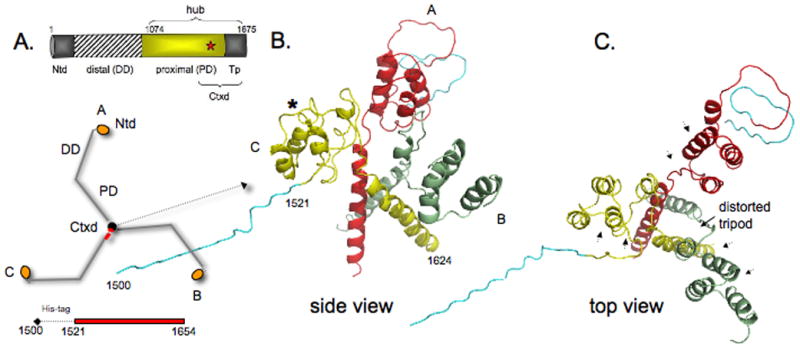
A) Domain map of clathrin heavy chain and schematic of how three identical heavy chain legs A, B, and C of clathrin form the three-legged pinwheel structure: Ntd, N-terminal domain; DD, distal domain; PD, proximal domain (light chain binding region); Tp, tripod core; Ctxd, tripod core with a segment of the proximal domain. Red star denotes cysteine-1573. Histidine-tagged Ctxd construct (1521-1654, red bar) forms a trimer. B and C) side and top views show yellow leg C (numbered 1500-1624, histidine-tag (teal) 1500-1520) is tilted down and away from the red and green legs and the 3-fold symmetry of the helix tripod is distorted. The histidine-tag and clathrin residues 1521-1527 of leg B could not be modeled because of poor density in this region. Asterisk shows Helix 7g is unfolded in the yellow leg and in the other two legs. Arrowheads in C indicate where light chain segments would be.
The structure shown in Fig. 3 has the features of the trimerization domain in lower resolution models [4, 6], but there are distinct differences. The yellow colored leg (labeled C with the histidine-tag) is slipped down and away from the red (A) and green (B) heavy chain legs. The deformed structure is also evident when viewed from the top (Fig. 3C and Supplemental Fig. 1). To see if crystal packing could account for the gross changes in the topology, we examined the environment about a crystallographic 3-fold axis (Supplemental Fig. 1). The packing interactions immediately around Legs A, B, and C are not equivalent, which raises the possibility that some of the observed structural distortions without light chain may be due to uneven crystal packing forces.
The Ctxd structure is informative because it allows us to predict what can happen to trimeric clathrin if light chain is absent, or if the C-terminal end of light chain is displaced from the trimerization domain. However, our crystal structure cannot reconcile any debate over light chain function in cells. Based on structural evidence here we suggest that the clathrin trimerization domain is intrinsically flexible and that light chains are required to maintain the clathrin triskelion. The idea that the trimerization domain can fluctuate is intriguing in light of small-angle neutron scattering experiments that suggest the trimerization region is flexible [25].
We observed that Helix 7g (1521-1529) is disrupted in every leg of the 8 trimers in the asymmetric unit (see asterisk in Fig. 3B). We do not know if this is caused by post-growth crystal dehydration or is the result of crystal packing. The fact that Helix 7g is unraveled could mean that molecular contacts made with neighboring Helix 7h (1534-1543) are insufficient to stabilize Helix 7g. Together, our structure suggests there are regions in the clathrin filamentous leg that are more plastic than others. This idea would be in-line with data indicating clathrin heavy chain is flexible [26].
The three long helices that form a tripod is a prominent feature of the clathrin trimerization domain. The tripod helices, modeled to be straight and rigid in lower resolution structures [4, 6], are somewhat bowed in Ctxd. We do not believe this is due to model bias because Rfree worsens when we try to straighten them. The helical structure breaks up and changes direction after glutamate-1624. Weak electron density prohibited building the last 30 residues (1625-1654). The disorder at the end of each tripod helix was also observed in the low-resolution structure of clathrin hub with light chains [4].
Potential features of a detrimerization switch
To see how a trimerization domain without light chain compares to those with bound light chain, a leg from assembled clathrin or clathrin hub (both with light chains) was superimposed over a leg from Ctxd (PDB code 3QIL). The analyses show that the short Tx2 helix in assembled clathrin (PDB code 1XI4) [6] (see Fig. 4A) and clathrin hub (PDB code 3LVG) [13] is unfolded in every leg in the asymmetric unit. This is potentially significant because Ambivalent Structure Predictor (ASP) [27] predicts 1590NIMDFAMP1597 (Tx2 underlined) is a conformational switch (labeled pCS in Fig. 4A). In addition, Helix 7j is shifted away from 1590NIMDFAMP1597 towards Helix 7h (1532-1544) (see exploded view, Fig. 4B). Helix 7j in our structure is offset about 10 Å relative to the grey helix from 1XI4. Apparently Helix 7j from clathrin hub (colored blue) occupies an intermediate position in the superposition. To confirm the orientation of Helix 7j in our model, the helix was omitted from each of the 24 protomers in the asymmetric unit. Calculated Fo - Fc maps returned electron density where Helix 7j (1563-1575) was originally located. We observed that Helix 7h in our model is displaced compared to its position in 1XI4 (see Fig. 4). This shift could be due to the fact that Helix 7h is adjacent to Helix 7g that has unfolded. Alternatively, Helix 7h could have moved over in response to the repositioning of Helix 7j. We cannot rule out the possibility that it is some combination of the two. We do not believe that the C1528A and T1585L mutations we introduced to improve the sample can cause the movements in structure. Cysteine-1528 is in a disordered segment far from Helix 7j and T1585 is in a helix (Tx1) that is not moved.
Figure 4. Identification of a labile helix outside the trimerization domain.

A) Comparison of leg A of the Ctxd model (3QIL, red) and a leg from 1XI4 [6] (grey) shows Helix 7h (1532-1544) and Helix 7j (1563-1575) do not superimpose. The short Tx2 helix in 1XI4 is unstructured in Ctxd. This region contains a putative conformational switch (pCS). Star symbols indicate location of C1528A (red) and T1585L (green) point mutations. B) Exploded views show the different positions of Helix 7j in our crystal structure without light chain, in clathrin hub with light chain (3LVG), and assembled clathrin with light chain (1X14). Helix 7j in 3QIL (red) is shifted away by ~10 Å (arrow) from the same helix in 1XI4 (grey). Blue helix from PDB 3LVG [4] occupies an intermediate position. Cysteine-1573 (pink) is located near the bottom of Helix 7j. Histidine tags are colored teal.
The stability of Tx2 in 1590NIMDFAMP1597 could be influenced by Helix 7j because Tx2 is unraveled when Helix 7j is moved away (Fig. 4A). The position of glycine-1567 near the middle of Helix 7j may also be significant because glycine is a helix breaker [28]. The assembled clathrin and clathrin hub models both have light chains, but Ctxd has none. The fact that Helix 7j is shifted by a distance that is about the diameter of an α-helix could mean that the position of Helix 7j is sensitive to the orientation of light chain. This is supported by the observation that C-terminal segments of light chain are close to Helix 7j [4]. The fact that structural features presented in Fig. 4 are conserved in lower and higher invertebrates and invertebrates (see Supplemental Fig. 3) suggest they may be relevant for function.
Conclusions
We assessed the subcellular distribution of the C1573A mutant by confocal microscopy in cells and discovered that a proportion of HA-hub C1573A and the longer construct HA 865-1675 C1573A were additionally localized to the nucleus. MALS experiments indicate that a portion of the C1573A hub population is detrimerized and this pool can be made trimeric upon addition of clathrin light chain. To better understand how cysteine-1573 and light chain impacted the oligomerization of clathrin, we solved the crystal structure of the clathrin trimerization domain without any light chain. The model of the isolated trimerization domain reveals structural features that suggest clathrin has a detrimerization switch. Specifically, comparisons of Ctxd to structures with light chain reveal Helix 7j is shifted. This raises the question whether light chain may be able to determine the position of cysteine-1573 by influencing the orientation of Helix 7j. We conclude that the positioning of cysteine-1573 is important for the stability of the trimerization domain. Our findings open the possibility that there exists a natural mechanism for restructuring clathrin for new functions.
Supplementary Material
Highlights.
C1573A clathrin hub is localized to nuclear spaces.
Cysteine-1573 in Helix 7j is essential for normal clathrin topology.
Trimerization domain crystal structure is distorted without clathrin light chain.
X-ray model suggests a detrimerization switch may mediate clathrin structure/function.
Acknowledgments
We thank Frances Brodsky for constructs and Peter Hwang for preliminary diffraction experiments. This work was supported by NIH grant R01 GM064387 to J.A.Y.
Footnotes
Protein Data Bank accession code
The coordinates have been deposited in the RCSB Protein Data Bank with accession code 3QIL.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kirchhausen T. Adaptors for clathrin-mediated traffic. Annu Rev of Cell and Dev Biol. 1999;15:705–32. doi: 10.1146/annurev.cellbio.15.1.705. [DOI] [PubMed] [Google Scholar]
- 2.Brodsky FM, Chen CY, Knuehl C, Towler MC, Wakeham DE. Biological basket weaving: formation and function of clathrin-coated vesicles. Annu Rev Cell Dev Biol. 2001;17:517–568. doi: 10.1146/annurev.cellbio.17.1.517. [DOI] [PubMed] [Google Scholar]
- 3.Ybe JA, Brodsky FM, Hofmann K, Lin K, Liu SH, Chen L, Earnest TN, Fletterick RJ, Hwang PK. Clathrin self-assembly is mediated by a tandemly repeated superhelix. Nature. 1999;399:371–375. doi: 10.1038/20708. [DOI] [PubMed] [Google Scholar]
- 4.Wilbur JD, Hwang PK, Ybe JA, Lane M, Sellers BD, Jacobson MP, Fletterick RJ, Brodsky FM. Conformation switching of clathrin light chain regulates clathrin lattice assembly. Dev Cell. 2010;18:841–848. doi: 10.1016/j.devcel.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu SH, Wong ML, Craik CS, Brodsky FM. Regulation of clathrin assembly and trimerization defined using recombinant triskelion hubs. Cell. 1995;83:257–267. doi: 10.1016/0092-8674(95)90167-1. [DOI] [PubMed] [Google Scholar]
- 6.Fotin A, Cheng Y, Sliz P, Grigorieff N, Harrison SC, Kirchhausen T, Walz T. Molecular model for a complete clathrin lattice from electron cryomicroscopy. Nature. 2004;432:573–579. doi: 10.1038/nature03079. [DOI] [PubMed] [Google Scholar]
- 7.Kirchhausen T, Scarmato P, Harrison SC, Monroe JJ, Chow EP, Mattaliano RJ, Ramachandran KL, Smart JE, Ahn AH, Brosius J. Clathrin light chains LCa and LCb are similar, polymorphic, and share repeated heptad motifs. Science. 1987;236:320–324. doi: 10.1126/science.3563513. [DOI] [PubMed] [Google Scholar]
- 8.Newpher TM, Idrissi FZ, Geli MI, Lemmon SK. Novel function of clathrin light chain in promoting endocytic vesicle formation. Mol Biol Cell. 2006;17:4343–52. doi: 10.1091/mbc.E06-07-0606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Legendre-Guillemin V, Metzler M, Lemaire JF, Philie J, Gan L, Hayden MR, McPherson PS. Huntingtin interacting protein 1 (HIP1) regulates clathrin assembly through direct binding to the regulatory region of the clathrin light chain. J Biol Chem. 2005;280:6101–6108. doi: 10.1074/jbc.M408430200. [DOI] [PubMed] [Google Scholar]
- 10.Niu Q, Ybe JA. Crystal structure at 2.8 A of Huntingtin-interacting protein 1 (HIP1) coiled-coil domain reveals a charged surface suitable for HIP1 protein interactor (HIPPI) J Mol Biol. 2008;375:1197–1205. doi: 10.1016/j.jmb.2007.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J, Wang Y, O’Halloran TJ. Clathrin light chain: importance of the conserved carboxy terminal domain to function in living cells. Traffic. 2006;7:824–832. doi: 10.1111/j.1600-0854.2006.00438.x. [DOI] [PubMed] [Google Scholar]
- 12.Ybe JA, Perez-Miller S, Niu Q, Coates DA, Drazer MW, Clegg ME. Light chain C-terminal region reinforces the stability of clathrin heavy chain trimers. Traffic. 2007;8:1101–1110. doi: 10.1111/j.1600-0854.2007.00597.x. [DOI] [PubMed] [Google Scholar]
- 13.Young JS, Guttman JA, Vaid KS, Shahinian H, Vogl AW. Cortactin (CTTN), N-WASP (WASL), and clathrin (CLTC) are present at podosome-like tubulobulbar complexes in the rat testis. Biol Reprod. 2009;80:153–161. doi: 10.1095/biolreprod.108.070615. [DOI] [PubMed] [Google Scholar]
- 14.Royle SJ. The role of clathrin in mitotic spindle organization. J Cell Sci. 2012;125:19–28. doi: 10.1242/jcs.094607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blixt MK, Royle SJ. Clathrin heavy chain gene fusions expressed in human cancers: analysis of cellular functions. Traffic. 2011;12:754–761. doi: 10.1111/j.1600-0854.2011.01183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enari M, Ohmori K, Kitabayashi I, Taya Y. Requirement of clathrin heavy chain for p53-mediated transcription. Genes Dev. 2006;20:1087–99. doi: 10.1101/gad.1381906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohmori K, Endo Y, Yoshida Y, Ohata H, Taya Y, Enari M. Monomeric but not trimeric clathrin heavy chain regulates p53-mediated transcription. Oncogene. 2008;27:2215–2227. doi: 10.1038/sj.onc.1210854. [DOI] [PubMed] [Google Scholar]
- 18.Ybe JA, Ruppel N, Mishra S, VanHaaften E. Contribution of cysteines to clathrin trimerization domain stability and mapping of light chain binding. Traffic. 2003;4:850–856. doi: 10.1046/j.1600-0854.2003.0139.x. [DOI] [PubMed] [Google Scholar]
- 19.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones TA, Bergdoll M, Kjeldgaard MO. A macromolecule modelling environment. In: Bugg C, Ealick S, editors. Crystallography and Modelling Methods in Molecular Design. New York: Springer-Verlag; 1990. pp. 189–199. [Google Scholar]
- 21.Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 22.Willox AK, Royle SJ. Functional analysis of interaction sites on the N-terminal domain of clathrin heavy chain. Traffic. 2012;13:70–81. doi: 10.1111/j.1600-0854.2011.01289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ybe JA, Greene B, Liu SH, Pley U, Parham P, Brodsky FM. Clathrin self-assembly is regulated by three light-chain residues controlling the formation of critical salt bridges. EMBO J. 1998;17(5):1297–1303. doi: 10.1093/emboj/17.5.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu S-H, Towler MC, Chen E, Chen C-Y, Song, Apodaca G, Brodsky FM. A novel clathrin homolog that co-distributes with cytoskeletal components functions in the trans-Golgi network. Embo J. 2001;20:272–284. doi: 10.1093/emboj/20.1.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferguson ML, Prasad K, Boukari H, Sackett DL, Krueger S, Lafer EM, Nossal R. Clathrin triskelia show evidence of molecular flexibility. Biophys J. 2008;95:1945–1955. doi: 10.1529/biophysj.107.126342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kotova S, Prasad K, Smith PD, Lafer EM, Nossal R, Jin AJ. AFM visualization of clathrin triskelia under fluid and in air. FEBS Lett. 2010;584:44–48. doi: 10.1016/j.febslet.2009.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Young M, Kirshenbaum K, Dill KA, Highsmith S. Predicting conformational switches in proteins. Protein Sci. 1999;8:1752–1764. doi: 10.1110/ps.8.9.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chakrabartty A, Kortemme T, Baldwin RL. Helix propensities of the amino acids measured in alanine-based peptides without helix-stabilizing side-chain interactions. Protein Sci. 1994;3:843–852. doi: 10.1002/pro.5560030514. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


