Summary
Differences in the clinical pathology of mammalian prion diseases reflect distinct heritable conformations of aggregated PrP proteins, called prion strains. Here, using the yeast [PSI+] prion, we examine the de novo establishment of prion strains (called variants in yeast). The [PSI+] prion protein, Sup35, is efficiently induced to take on numerous prion variant conformations following transient overexpression of Sup35 in the presence of another prion, e.g. [PIN+]. One hypothesis is that the first [PSI+] prion seed to arise in a cell causes propagation of only that seed’s variant, but that different variants could be initiated in different cells. However, we now show that even within a single cell, Sup35 retains the potential to fold into more than one variant type. When individual cells segregating different [PSI+] variants were followed in pedigrees, establishment of a single variant phenotype generally occurred in daughters, granddaughters or great granddaughters—but in 5% of the pedigrees cells continued to segregate multiple variants indefinitely. The data is consistent with the idea that many newly formed prions go through a maturation phase before they reach a single specific variant conformation. These findings may be relevant to mammalian PrP prion strain establishment and adaptation.
Keywords: Prion, Sup35, Yeast, [PSI+], Variant, Oligomers
Introduction
The mammalian prion is a misfolded infectious form of the PrP protein, which when accumulated in the central nervous system, leads to neurodegenerative disease (Prusiner, 1998). These include Creutzfield-Jacob disease in humans, scrapie in sheep and bovine spongiform encephalopathy in cattle. Pathogenicity is attributed to the conversion of the alpha helical rich cellular prion protein, PrPC, into a beta sheet rich prion form, PrPSc, which is aggregated and protease K resistant. Differences in the conformations of infectious PrPsc (that are each composed of the same PrP polypeptide) are proposed to be responsible for the distinct pathologies of prion strains (Safar et al., 1998).
Several proteins in the yeast Saccharomyces cerevisiae have been shown to be able to form prions and many of these have been shown to confer specific heritable phenotypes (Alberti et al., 2009; Crow and Li, 2011; Derkatch et al., 2001; Du et al., 2008; Patel et al., 2009; Sondheimer and Lindquist, 2000; Suzuki et al., 2012; Wickner, 1994; Wickner et al., 1995). [PSI+], [PIN+] and [URE3], the prion forms of Sup35, Rnq1 and Ure2 respectively, are the best studied prions in yeast (Derkatch et al., 1997; Tuite and Cox, 2003; Wickner, 1994). Similar to their mammalian counterparts, the prion forms of these yeast proteins are amyloid-like (Glover et al., 1997; King et al., 1997; Sondheimer and Lindquist, 2000; Wickner, 1994). In addition, yeast prions also exist as different strains, called variants, that can be distinguished on the basis of distinct phenotypic and biochemical characteristics (Bradley et al., 2002; Bradley and Liebman, 2003; Derkatch et al., 1996; Schlumpberger et al., 2001).
Sup35 is a translational termination factor in its normal soluble form. However, in its prion aggregated [PSI+] state, its ability to terminate translation at stop codons becomes inefficient. Thus the phenotype of [PSI+] is suppression of nonsense mutations and in some variants this can cause toxicity and slow growth (Cox et al., 1988; McGlinchey et al., 2011). Sup35 can be divided into three domains. The C-terminal domain is essential for viability and function whereas the N-terminal domain, also called the prion domain, is necessary to form and join prion aggregates and has a non-prion non-essential function in general mRNA turnover (Derkatch et al., 1996; Hosoda et al., 2003; Ter-Avanesyan et al., 1993). The middle domain (M) is required for the faithful maintenance of certain [PSI+] prion variants (Bradley and Liebman, 2004; Liu et al., 2002).
Overexpression of full length Sup35, or a fragment containing the prion domain of Sup35 (Sup35NM), causes the appearance of [PSI+] in the presence of [PIN+] (Derkatch et al., 1996; Derkatch et al., 1997; Derkatch et al., 2001). This efficient [PSI+] induction phenomenon is explained by a cross-seeding model, which proposes that the preexisting prion, [PIN+], templates the initial conversion of soluble Sup35 protein molecules to the [PSI+] prion form (Choe et al., 2009; Derkatch et al., 2001; Derkatch et al., 2004).
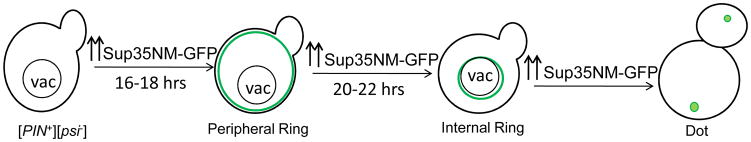
Overexpression of Sup35NM fused to GFP (Sup35NM-GFP) in [psi−] [PIN+] cells causes the appearance of ring or line-like fluorescent aggregates. Cells with such aggregates give rise to [PSI+] progeny (Zhou et al., 2001). These newly appearing ring-like aggregates are initially localized at the cell periphery. After about 20–22 hrs the rings shrink to surround the vacuole and sometimes collapse into a dot found at the perivacuolar region. While these large structures never leave the mother cell, daughters appear to inherit [PSI+] seed that is too small to be seen using a fluorescence microscope. This seed enables the daughter to propagate [PSI+] and directly form a large dot, in the presence of overexpressed Sup35NM-GFP (Fig. 1) (Ganusova et al., 2006; Mathur et al., 2010). Cells that do not form rings were never observed to give rise to [PSI+] daughter cells. Also, overexpression of Sup35NM-GFP in already established [PSI+] cells gives rise to cells with big fluorescent dots, never rings. Thus ring like aggregates are the hallmark of newly appearing [PSI+] following Sup35NM-GFP overexpression in [PIN+] cells. Similarly, when Sup35NM-GFP was constitutively overexpressed in the absence of endogenous SUP35NM, rings appeared that upon continued propagation converted into large dots. These rings overlapped preautophagosomal markers characteristic of the insoluble protein deposit (IPOD). Rings were composed of long fiber bundles and lysates of the ring cells could transmit [PSI+] when the ring fibers were fragmented (Tyedmers et al., 2010).
Figure 1. Stages during de novo induction of [PSI+].
Previous work showed that when Sup35NM-GFP is over expressed in 74D-694 [psi−][PIN+] cells, fluorescent line or ring like structures appear along the cell periphery in about 16–18 hrs. If the induction is prolonged for another 4 hrs, the peripheral ring often becomes an internal ring surrounding the vacuole before collapsing into a dot (Ganusova et al., 2006; Mathur et al., 2010; Zhou et al., 2001).
Interestingly, different [PSI+] variants were obtained following overexpression of Sup35NM even in the same genetic background (Derkatch et al., 1996). Variants of [PSI+] can be distinguished by differences in levels of Sup35 aggregation and hence differences in translation termination efficiency and toxicity, as well as differences in stability, aggregate structure and oligomer size (Derkatch et al., 1996; King and Diaz-Avalos, 2004; Krishnan and Lindquist, 2005; Kryndushkin et al., 2003; McGlinchey et al., 2011; Tanaka et al., 2004; Toyama et al., 2007; Uptain et al., 2001). In addition variants differ in their responses to alterations in chaperone levels and their ability to be transmitted across transmission barriers (Kushnirov et al., 2000a; Kushnirov et al., 2000b). Strong [PSI+] variants have a larger number of aggregates and these aggregates are smaller than the larger, less frequent aggregates found in weak [PSI+] variants. Thus, there are more aggregate ends available in strong [PSI+] variants to recruit soluble Sup35, resulting in the stronger nonsense suppression phenotype (Derkatch et al., 1996). There are also variants of [PSI+] that are intermediate in phenotype between strong and weak [PSI+] and all strong or all weak [PSI+] are not identical (King, 2001; Kochneva-Pervukhova et al., 2001).
Once established, prion variants generally do not interconvert (Derkatch et al., 1996; Kochneva-Pervukhova et al., 2001). However, weak [PSI+] prion variants have been shown to switch to strong [PSI+] in the presence of epigallocatechin-3-gallate (EGCG). Whether this small molecule promotes remodeling of the weak [PSI+] prion or selects for low levels of EGCG-resistant strong [PSI+] prions that were present in some of the weak [PSI+] variant cells is unknown (Roberts et al., 2009). If the latter is true it suggests that prion variants can occasionally spontaneously “mutate” to another variant. When two variants are present in a single cell, and depending upon their relative numbers, the variant that generates seeds more rapidly is believed to cause loss of the other variant, by more efficiently capturing available soluble protein. (Bagriantsev and Liebman, 2004; Bradley et al., 2002; Tanaka et al., 2006).
It remained unknown, if the different [PSI+] variants induced by transiently overexpressing Sup35NM always arose in separate cells, or if more than one variant could arise in a single cell. One hypothesis was that the ring aggregates in each cell that arose during [PSI+] induction were composed of a single [PSI+] variant conformation—resulting from the first seed to appear and grow in that cell, so that different variants could only be initiated in different cells. Here we test this hypothesis and show, to the contrary, that more than one [PSI+] variant can arise from a single ring cell.
Results
[PSI+] variants are not always established at the ring stage
To investigate when different variants of [PSI+] are established during the induction of [PSI+], we transiently overexpressed Sup35NM fused with GFP (Sup35NM-GFP) in 74D-694 [PIN+] [psi−] cells carrying the pCUP1-SUP35NM::GFP (LEU2) plasmid. As shown previously, this transient overexpression gave rise to cells containing bright fluorescent rings indicative of the ability to give rise to [PSI+] (Fig. 2A) (Zhou et al., 2001).
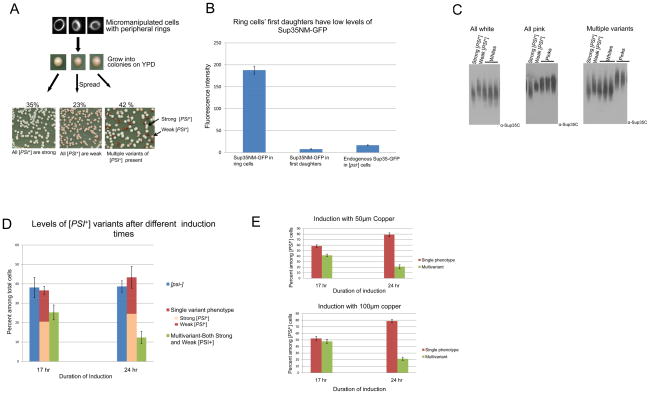
Figure 2. More than one [PSI+] variant can arise from a single cell following de novo induction of [PSI+].
A. One ring cell can give rise to progeny that are all strong [PSI+], all weak [PSI+] or a mixture of strong and weak [PSI+]. [PSI+] was induced de novo by over expressing Sup35NM-GFP in [PIN+] [psi−] cells. Cells with rings were micromanipulated and grown on YPD plates for 3 days where Sup35NM-GFP expression was turned off. The resulting colonies were suspended in water and spread on YPD. The types of [PSI+] variants present in the ring cell progeny were determined from the color of these colonies (>500 ring containing cells were micromanipulated). Only ring cells giving rise to some [PSI+] progeny are depicted. B. Ring cells’ first daughters have low levels of Sup35NM-GFP. Comparison of endogenous Sup35-GFP fluorescence in [psi−] cells with cytoplasmic Sup35NM-GFP fluorescence in ring containing mother cells and their first daughters. Images of GF-658 (MAT α ade1–14 ura3–52 leu2–3,112 trp1–289 his3–200 SUP35-GFP) were taken for the [psi−] cells. Ring cells and daughters of ring cells were imaged from L1749 (MAT a ade1–14 ura3–52 leu2–3,112 trp1–289 his3–200) transformed with pCup-SUP35NM-GFP after induction until ring stage. After induction and the appearance of rings, cells were washed with water and grown in YPD for ~ 3 hrs. Images were then acquired and quantified from three different trials of 10 cells each (see Experimental Procedures). All cells were imaged under identical conditions Bar graphs represent the average fluorescence intensity of Sup35-GFP and Sup35NM-GFP in the respective cells. The level of Sup35-GFP was reduced about 25% relative to the level of untagged Sup35 in Western blots (data not shown). This may reflect differential degradation in the lysate. C. Confirmation that different color colonies have variants with different sized Sup35 oligomers. Cell lysates were prepared from colonies shown at the bottom of 2A. Crude lysates were treated with 2% SDS at room temperature and SDS resistant oligomers were analyzed by SDD-AGE analysis. Lysates from strong [PSI+] (L1762) and weak [PSI+] (L1758) variants were run as controls. Left panel shows results from ring cells whose [PSI+] progeny were all white, center shows results from ring cells whose [PSI+] progeny were all pink and right panel shows results from ring cells whose [PSI+] progeny were both white and pink. D. [PSI+] variant establishment varies with the duration of Sup35NM protein induction. [PIN+] [psi−] cells containing the Sup35NM-GFP plasmid were grown in 50 μM CuSO4 at 30°C for 17 or 24 hrs to induce [PSI+]. Ring aggregate containing cells were then isolated and progeny examined. Ring cells gave rise to [PSI+] variants with a single phenotype more frequently after 24 vs. 17 hrs of Sup35NM-GFP expression. Each bar represents standard error of more than 3 trials of at least 100 viable ring cells. Data includes all ring cells whether or not they gave rise to any [PSI+]. E. Increasing expression of Sup35NM-GFP does not alter the variant establishment. Individual ring cells were micromanipulated after 17 or 24 hrs of induction with 50 μM or 100 μM of CuSO4 and the [PSI+] variant types arising from these cells were determined. No significant difference was observed in the relative frequencies of [PSI+] variants whether 50 or 100 μM CuSO4 was used. Each bar indicates standard error of more than three trials of 100 viable ring cells. Data shown includes only ring cells giving rise to some [PSI+] progeny. (For all experiments starting OD at 0 hrs=0.1, OD at 17 hrs~1.0 and OD at 24 hrs ~2.0 unless otherwise indicated).
Individual ring containing cells were isolated by micromanipulation and grown into colonies on rich, glucose containing medium (YPD) for 3 days. To determine the [PSI+] variant status of the cells in these colonies, they were suspended in water and individual cells were again grown into colonies on YPD. The color of these colonies reflected the [PSI+] variant status of the suspended cells because the ability to read through the premature stop codon in the ade1–14 mutation caused [PSI+] cells to turn from a dark red color to white, in proportion to the level of read through (see Materials and Methods). These viable ring containing cells frequently (~60%) gave rise to some [PSI+] progeny, although ~40% of the ring cells had only [psi−] progeny. Most (95%) of the non-red [PSI+] colonies among the progeny failed to grow when transferred to –Leu, indicating that the plasmid was lost during growth on the non-selective YPD medium. Since it is difficult to distinguish all [PSI+] variant types, we focused on phenotypically distinct strong (white) and weak (pink) [PSI+] variants. We found that of the ring cells isolated after 17 hrs of induction that gave rise to some [PSI+] progeny, ~60 % of the time the [PSI+] progeny were either all strong or all weak [PSI+], but ~40% of the time the [PSI+] progeny clearly included a mixture of variants including both strong and weak [PSI+] (Fig. 2A). The proportion of the two variants in the mixture fell between the range of 50:50 and 90:10.
The de novo induction of [PSI+] is caused by high levels of Sup35NM-GFP and only the ring containing cells, not their daughters have these high levels. Indeed, while the level of Sup35NM-GFP in ring cells is 10X higher than the level of endogenous Sup35, when ring cells are placed on non-inducing media their daughters contain a much lower level of Sup35NM-GFP, less than half of the endogenous Sup35 level (Fig. 2B). This is not surprising because most of the Sup35NM-GFP fluorescence is confined to the ring and is not diffuse within the cytoplasm and therefore not easily transmissible to daughter cells. Sup35NM-GFP levels were estimated by comparing the fluorescence intensities of Sup35NM-GFP in ring containing mothers and their daughters with control [psi−] cells containing endogenous Sup35 tagged with GFP (Satpute-Krishnan and Serio, 2005). Thus, it appears that a heterogeneous mixture of [PSI+] aggregates, capable of giving rise to different variants of [PSI+] can be initially formed in a single cell following Sup35NM-GFP overexpression. Indeed, the mixture of variants does not result from an induced change of one variant type into another because overexpressing of Sup35NM-GFP in the presence of either strong or weak [PSI+] did not cause a phenotypically distinguishable new variant to appear (Figure S1).
[PSI+] aggregates are composed of oligomeric species which are comparatively more stable than normal protein aggregates. These oligomers are SDS resistant at room temperature and their sizes can be estimated on agarose gels by semi-denaturing gel electrophoresis (SDD-AGE) (Bagriantsev and Liebman, 2004; Kryndushkin et al., 2003). We show that [PSI+] cells initially derived from a single ring cell and that all appear alike on plates had a similar oligomer size distribution (Fig. 2C left, center). Also as expected, the pink and white colonies derived from a single ring cell displayed the different oligomer sizes characteristic for weak and strong [PSI+] respectively (Fig. 2C right).
[PSI+] establishment can be altered by increasing the duration of Sup35NM-GFP overexpression
To investigate whether the duration of induction of Sup35 is crucial in determining the resulting [PSI+] variant, we compared the [PSI+] variant status of progeny of individual ring containing cells micromanipulated after 17 vs. 24 hrs of Sup35NM-GFP induction. As shown above, after 17 hrs of induction, ~ 40% of the [PSI+] generating ring cells (corresponding to 25% of total cells) gave rise to both strong and weak [PSI+] progeny. This fraction was reduced to 20% (12.5% of total) when ring cells were micromanipulated after 24 hrs of induction (Fig. 2D). At both times ~ 60 % of the viable ring cells gave rise to some non-red [PSI+] colonies. Likewise at both times, of the cells giving rise to a single [PSI+] phenotype, ~60% were strong [PSI+] and ~40% were weak [PSI+]. Thus, with longer Sup35NM-GFP overexpression, fewer cells retain the ability to give rise to both strong and weak [PSI+] but the level of [psi−] cells is unchanged. In contrast, altering the overexpression levels of Sup35NM by increasing the concentration of copper had no effect in the pattern of [PSI+] variant establishment at either time point (Fig. 2E).
Weak or strong [PSI+] variants usually get established within the first few divisions of ring cells
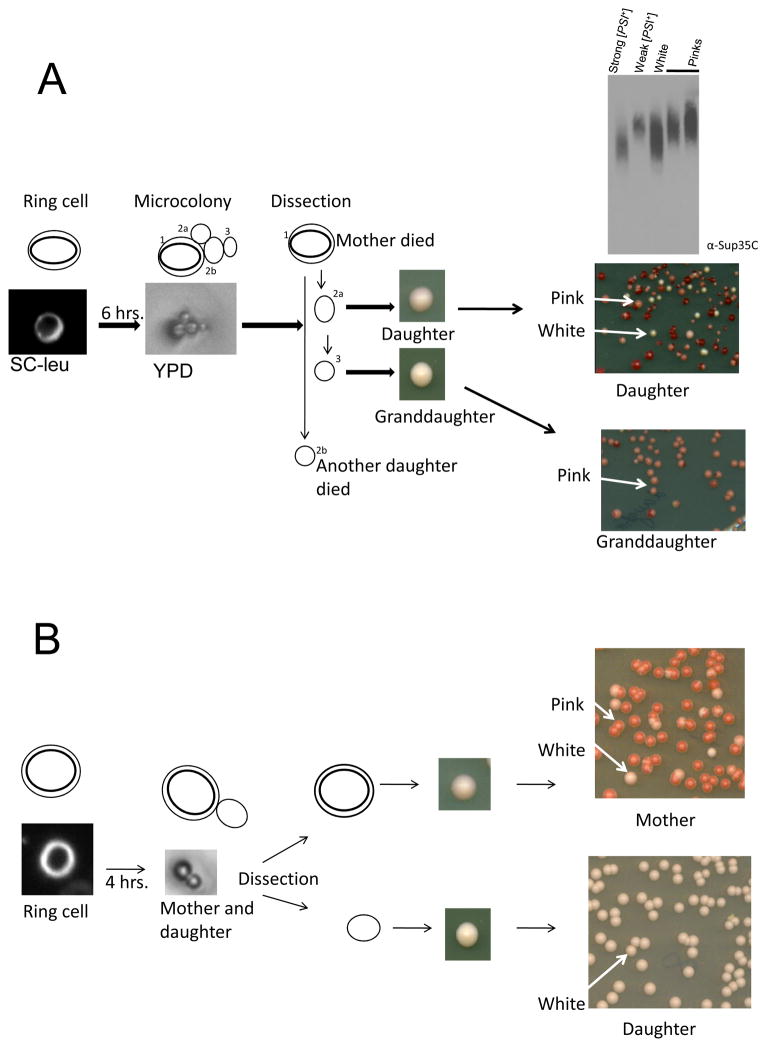
Our findings show that single [PSI+] phenotypes do not always get established with the initial formation of ring aggregates. Thus to determine when a single phenotype gets established, we analyzed pedigrees of ring containing cells. Individual ring cells were micromanipulated after 17 hrs of Sup35NM-GFP overexpression and allowed to divide a few times. These cells were then separated and grown into colonies on YPD where their [PSI+] status was determined by color. We examined 150 unbudded single ring cells: 90 cells did not divide, 19 had all [psi−] daughters, 31 had all strong or weak [PSI+] daughters, and 10 had both weak [PSI+] and strong [PSI+] daughters. In 8 of the pedigrees of the latter 10 ring cells, a daughter, granddaughter or great granddaughter lost the ability to transmit either strong or weak [PSI+] to their daughters. In the example in Figure 3A, a weak [PSI+] variant became established in a granddaughter. In this pedigree the mother and one daughter cell failed to grow into a colony. The progeny of the other daughter included strong and weak [PSI+] but the progeny of a granddaughter were all weak [PSI+]. SDD-AGE analysis of the daughter’s white and pink progeny showed the expected difference in the size of oligomers. Thus, in this case the variant was established in a granddaughter. In another example (Fig. 3B), the ring cell’s progeny include cells with strong and weak [PSI+] variants. In contrast, the daughter had only strong [PSI+] progeny. Thus, the variant was established in the daughter.
Figure 3. Pedigree analysis to determine when [PSI+] variants are established.
Cells with pCup-SUP35NM-GFP induced to form rings in plasmid selection medium plus Cu2+ were micromanipulated on an agar pad, placed on YPD and allowed to divide for a few generations. The resulting cells were dissected (see Materials and Methods) while keeping track of the different generations. The cells were then grown into colonies on YPD which were then spread on YPD to score for the [PSI+] variant in the progeny of each cell in the microcolony. A. Pedigree in which a weak [PSI+] variant was established in a granddaughter. The ring containing mother cell (1) and one daughter cell (2b) did not grow. The other daughter (2a) gave rise to more than one variant. The granddaughter (3) gave rise to only weak [PSI+] progeny. The Sup35 SDS resistant oligomer sizes present in the pink and white colonies from daughter (2a) were compared with standard strong [PSI+] (L1762) and standard weak [PSI+] (L1758). Lysates treated with 2% SDS were subjected to western blotting and probing with Sup35C antibody. B. A pedigree in which strong [PSI+] is established in a daughter. Both weak and strong [PSI+] were detected in the progeny of the initial aggregate containing mother cell. The daughter only gave rise to strong [PSI+] progeny.
Unspecified [PSI+]
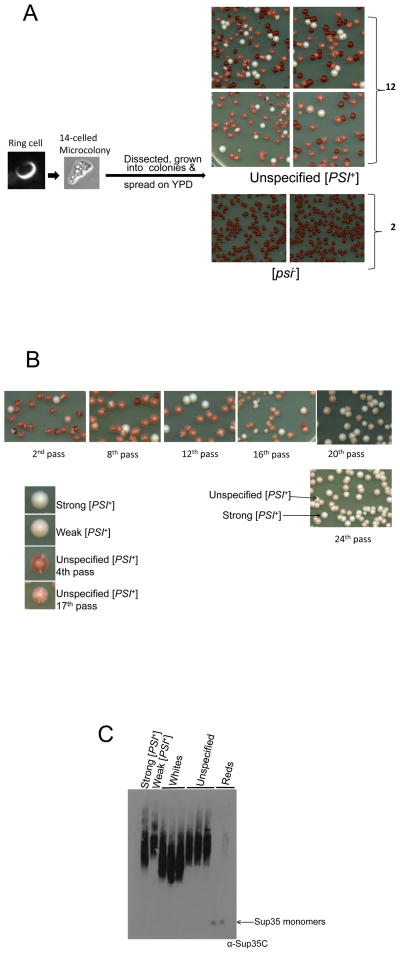
In contrast to the above 8 pedigrees, in 2 pedigrees all [PSI+] daughters, granddaughters and great granddaughters transmitted both strong and weak [PSI+] to some of their progeny. In one such pedigree, a ring cell formed a 14 celled microcolony. When these individual cells were separated and grown into colonies, 2 were [psi−], while most of the colonies from the other 12 appeared to be dark pink with numerous white sectors, although there were also solid red [psi−], some solid white (strong [PSI+]) and occasional solid pink (weak [PSI+]) colonies (Fig. 4A). When the sectored colonies were streaked out, they continued to give rise to mostly pink colonies sectoring white like themselves, as well as some solid red, white and rare pink colonies. Simple observation of the sectoring colonies cannot distinguish if the sectors are pink, white and red or just white and red. However, the fact that subculturing of these colonies did not give large numbers of red colonies suggests that these sectoring colonies are largely pink and white with limited red sectors. We call these sectored colonies ‘unspecified [PSI+]’ and refer to strong or weak [PSI+] as specified [PSI+] variants. Furthermore when cells picked from white sectors were subcultured they gave rise to non-sectoring white colonies, whereas cells picked from pink sectors grew into a mixture of cells with unspecified, some strong, few [psi−] and rarely weak [PSI+]. All of the unspecified [PSI+] colonies had lost the plasmid, so leaky expression of SUP35-GFP from the plasmid cannot be responsible for the unspecified phenotype. Even after 25 sequential subculturings, unspecified [PSI+] continued to give rise to progeny with more than one variant. Interestingly, with each subculturing there were fewer [psi−] colonies. The reduced number of [psi−] colonies correlated with the sectoring colonies becoming lighter and lighter. By the 25th pass the population contained only pink/white sectored (unspecified [PSI+]), white (strong [PSI+]) and pink (weak [PSI+]) colonies but no red ([psi−]) cells (Fig. 4B).
Figure 4. Some [PSI+] variants remain unspecified.
A. Pedigree of an unspecified [PSI+] cell. A ring containing cell was grown into a 14 cell microcolony. Each of the cells in the microcolony were separated, grown into colonies, and spread on YPD. Most of the cells from 12 of the colonies grew into dark pink colonies that had white sectors. There were also some [psi−] red, some solid white (strong [PSI+]) and occasional solid pink (weak [PSI+]) colonies seen. The sectoring colonies are called unspecified [PSI+]. The two other colonies gave rise to only red, [psi−] cells. B. After continued propagation, unspecified [PSI+] cells have a reduced number of [psi−] progeny. On subsequent subculturing, unspecified [PSI+] gave rise to fewer and fewer [psi−] daughters, but never lost the ability to give rise to unspecified strong and weak [PSI+] daughters. When it reached the 24th pass it did not give rise to any [psi−] daughters. Also, the unspecified colony color became lighter with each subculturing but still showed white and pink sectors indefinitely. C. Comparative SDD-AGE analysis of the oligomer size of unspecified [PSI+].
This type of unspecified [PSI+] was ~ 5 % of the total [PSI+] induced de novo. When Sup35NM-GFP was overexpressed overnight in L1749 and plated on YPD, of about 1,250 non-red colonies scored, 60 had the phenotype of unspecified [PSI+]; dark pink with numerous white sectors. They were subcultured on YPD for 2 passes, and 52 of them behaved like the unspecified [PSI+] described above and none contained the plasmid.
Unspecified [PSI+] vs. strong and weak [PSI+] variants
Like other [PSI+] variants, unspecified [PSI+] was cytoducible and was cured by growth in 5 mM GuHCl. Since unspecified [PSI+] sectored colonies contained strong [PSI+] and weak [PSI+] cells, unspecified, strong and weak [PSI+] cytoductants were expected from the cytoduction mixture. Among 50 [PSI+] cytoductants scored from a mating of unspecified [PSI+] donors with the [psi−][PIN+] kar1 recipient (L1998), 5 were unspecified, 10 were weak and 35 were strong [PSI+]. Thus the unspecified [PSI+] can be cytoduced and is also maintained in a different genetic background.
Sup35 oligomers from an unspecified [PSI+] culture correspond to the oligomer size characteristic of weak [PSI+] rather than strong [PSI+] (Fig. 4C). Overexpression of the Hsp104 chaperone cures [PSI+] by an unknown mechanism (Chernoff et al., 1995). We used galactose inducible Hsp104 to increase the level of Hsp104 in unspecified, strong (L1762) and weak (L1758) [PSI+] colonies for 1–24 hrs. Curing was measured by plating the samples on YPD to look for the percentage of red [psi−] colonies (data not shown). As reported previously, curing by excess Hsp104 is more rapid in weak than strong [PSI+] (Derkatch et al., 1996). The time needed for excess Hsp104 to cure unspecified [PSI+] was intermediate.
To compare the microscopic appearance of [PSI+] aggregates in unspecified, strong (L1762), and weak [PSI+] (L1758) strains, we expressed Sup35NM-GFP protein at a low level to stain the aggregates. Either 1–3 big aggregates per cell or numerous tiny aggregates per cell were observed in all three variants.
The unspecified [PSI+] property is independent of [PIN+]
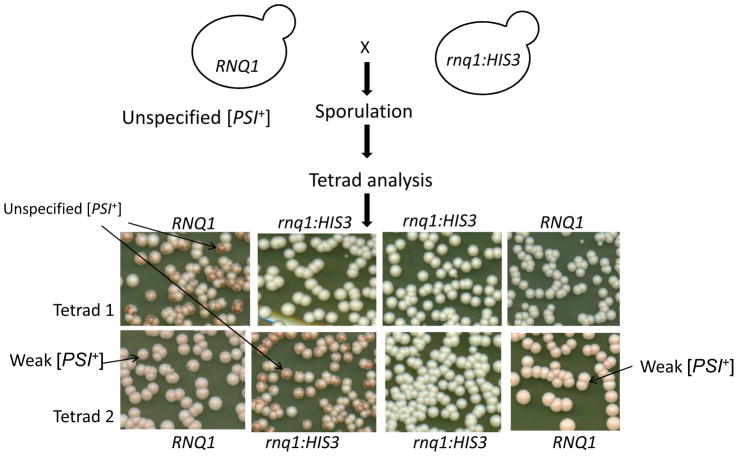
Unspecified [PSI+] cells were shown to retain [PIN+] because Rnq1 was found in the pellet fractions of lysate when subjected to high speed centrifugation (data not shown). To determine if the unspecified [PSI+] character is dependent on the presence of [PIN+], we asked if unspecified [PSI+] could propagate if [PIN+] were lost. We crossed unspecified [PSI+] with a [psi−] rnq1Δ strain and examined the [PSI+] status of meiotic progeny (Fig. 5). The unspecified character segregated in a non-Mendelian fashion and was found in 7 [PIN+] and 8 rnq1Δ (which are necessarily [pin−]) segregates from 14 tetrads with 4 viable spores and 3 tetrads with 3 viable spores, establishing that the unspecified phenotype is independent of the [PIN+] prion.
Figure 5. The unspecified [PSI+] phenotype is not dependent on [PIN+].
Unspecified [PSI+] was crossed with an RNQ1 deletion strain L3102 ([psi−] SUP35-GFP, rnq1::HIS3) and diploids were sporulated. The progeny from two of the 17 tetrads examined are shown where unspecified [PSI+] was found in both a wild type RNQ1 and an rnq1:HIS3 segregant.
Unspecified [PSI+] does not mimic a mixture of weak and strong [PSI+] propagons
Our finding that unspecified [PSI+] cells give rise to both specified strong and weak [PSI+] progeny could be explained if unspecified [PSI+] cells contained a mixture of strong and weak [PSI+] propagons. An alternative explanation is that the [PSI+] propagons in these cells were truly unspecified and could mature into specified strong or weak [PSI+] variants in daughter cells.
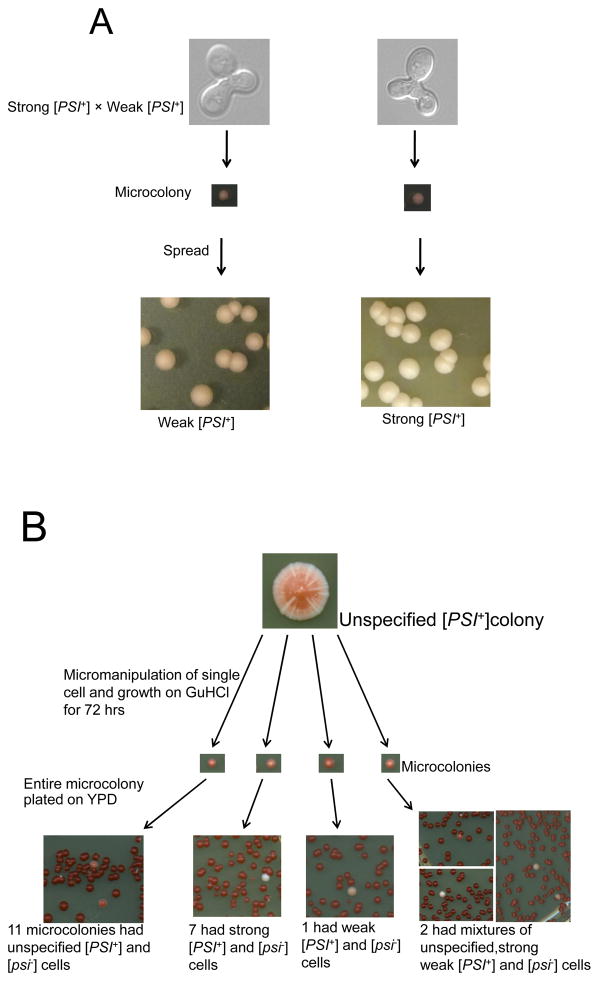
To determine if the unspecified [PSI+] phenotype could be created by mixing strong and weak [PSI+] propagons in a single cell, we micromanipulated zygotes resulting from the mating of strong and weak [PSI+] and examined their progeny. These zygotes were allowed to grow for 17 or 72 hrs and the resulting microcolonies were plated on YPD (Fig. 6A). Among the 76 individual zygotes and their diploid progeny examined, strong [PSI+] prevailed for 53 zygotes while weak [PSI+] prevailed in the remaining 23 zygotes. SDD-AGE analysis of progeny from representative zygotes showed that the pink and white zygote progeny respectively had oligomer sizes similar to weak and strong [PSI+] parents (data not shown). There were <1% pink colonies among white zygote populations and <1% white colonies among pink zygote populations. On subculturing, the colors of pink and white colored colonies remain unchanged. These results suggest that all zygotes arising from crosses of weak and strong [PSI+] cells are not identical. Possibly, depending upon the numbers of propagons in the particular mating cells, weak [PSI+] can sometime take over the population. Previously, we found only strong [PSI+] to prevail in such crosses, but different variants were used and only 8 diploid colonies were examined in the earlier study (Bradley et al., 2002). In any event, none of the progeny from any of the zygotes showed sectored colonies characteristic of unspecified [PSI+].
Figure 6. A mixture of two [PSI+] variants does not mimic unspecified [PSI+].
A. Strong [PSI+] and weak [PSI+] were mated and zygotes were micromanipulated individually. Zygotes were allowed to grow to form microcolonies before being spread on YPD plates to determine the variant present in the progeny cells. Some zygotes gave rise to essentially all strong [PSI+] while other zygotes gave rise to essentially all weak [PSI+] progeny. B. Unspecified [PSI+] prion propagon analysis. Unspecified [PSI+] cells were subjected to propagon analysis by micromanipulating individual cells from unspecified [PSI+] colonies and placing them on medium containing 3mM GuHCl to block their propagons from multiplying. After 72 hrs the whole microcolony was spread on YPD without GuHCl to score for unspecified, weak and strong [PSI+]. The number of non-red colonies reflected the number of propagons in the original cell.
To further compare cells that contain a mixture of weak and strong [PSI+] propagons with unspecified [PSI+] we examined the types of propagons present in these two types of cells. We grew individual cells from an unspecified [PSI+] culture, as well as zygotes formed by mating weak and strong [PSI+] haploids, into colonies on medium containing GuHCl. GuHCl, which does not block cell division, inhibits the shearing of prion aggregates or fibers, thereby stopping propagons from dividing (Eaglestone et al., 2000). Thus only the propagons already present in the initial cell or zygote are distributed to her progeny. Therefore, theoretically as the colony grows, cells equivalent to the number of propagons in the original cell or zygote should each inherit a single propagon while the rest of the cells in the colony will not get any propagons (Cox et al., 2003). Only the cells containing a propagon will become [PSI+] when grown on YPD without GuHCl and the [PSI+] variant present will reflect the variant of the inherited propagon. The type and number of [PSI+] colonies obtained after plating the colonies grown in the presence of GuHCl on YPD, corresponds to the propagon type and number present in the initial cell or zygote when it was first put on GuHCl.
Twenty cells isolated from unspecified [PSI+] colonies were grown into microcolonies on 3 mM GuHCl for 72 hrs and then spread on YPD to score the non-red [PSI+] colonies for their variant type which reflects the type of propagon they inherited from the original cell placed on GuHCl (Eaglestone et al., 2000). This analysis showed that 11 cells contained only unspecified [PSI+] propagons, 7 had only strong [PSI+] propagons, 1 had only weak [PSI+] propagons and 2 contained a mixture of unspecified, strong and weak [PSI+] propagons (Fig. 6B). Although our propagon count was lower than the propagon number previously reported for other strong and weak [PSI+] variants (Cox et al., 2003), in agreement with previous findings, we show strong [PSI+] daughters had a proportionally higher number of propagons than weak [PSI+] daughters. We also found that the propagon number for unspecified [PSI+] was around the same as that for weak [PSI+] (Table 2). In contrast to the above results, analogous experiments on zygotes formed by mating weak and strong [PSI+] haploids showed that they contained only weak and strong [PSI+] propagons and never any unspecified propagons (Table 3).
Table 2.
Propagon study of 21 cells from an unspecified [PSI+] colony
| Cells | No. of propagons that are*
|
Average number of propagons | ||
|---|---|---|---|---|
| Unspecified | Strong | Weak | ||
|
| ||||
| 1 | 10 | 0 | 0 | Unspecified [PSI+]~26 |
| 2 | 14 | 0 | 0 | |
| 3 | 21 | 0 | 0 | |
| 4 | 19 | 0 | 0 | |
| 5 | 33 | 0 | 0 | |
| 6 | 22 | 0 | 0 | |
| 7 | 13 | 0 | 0 | |
| 8 | 11 | 0 | 0 | |
| 9 | 17 | 0 | 0 | |
| 10 | 51 | 0 | 0 | |
| 11 | 68 | 0 | 0 | |
|
| ||||
| 12 | 0 | 156 | 0 | Strong [PSI+] ~77 |
| 13 | 0 | 113 | 0 | |
| 14 | 0 | 89 | 0 | |
| 15 | 0 | 116 | 0 | |
| 16 | 0 | 39 | 0 | |
| 17 | 0 | 4 | 0 | |
| 18 | 0 | 19 | 0 | |
|
| ||||
| 19 | 0 | 0 | 22 | Weak [PSI+]~22 |
|
| ||||
| 20 | 14 | 5 | 2 | Mixture of all 3 ~14 |
| 21 | 5 | 1 | 2 | |
The numbers of unspecified, strong and weak [PSI+] propagons present in each of 21 cells from an unspecified [PSI+] culture were determined as described previously (Cox et al., 2003).
Table 3.
Propagon counts of strong [PSI+] × weak [PSI+] zygotes.
| Strong [PSI+] × Weak [PSI+] zygotes | No. of propagons in zygote that are*
|
Average number of propagons | ||
|---|---|---|---|---|
| Unspecified | Strong | Weak | ||
|
| ||||
| 1. | 0 | 873 | 125 | Strong [PSI+]~554 |
| 2. | 0 | 262 | 99 | Weak [PSI+]~115 |
| 3. | 0 | 638 | 155 | Unspecified [PSI+]=0 |
| 4. | 0 | 445 | 93 | |
| 5. | 0 | 443 | 102 | |
| 6. | 0 | 668 | 140 | |
| 7. | 0 | 498 | 120 | |
| 8. | 0 | 602 | 85 | |
The numbers of unspecified, strong and weak [PSI+] propagons present in each of the eight zygotes were determined as described previously (Cox et al., 2003).
Discussion
[PSI+] induction by overexpression of Sup35NM-GFP is associated with the transient appearance of ring or line-like aggregates (Zhou et al., 2001) and causes the appearance of variants of [PSI+] that differ in such things as levels of nonsense suppression, stability, level of Sup35 aggregation and toxicity (Derkatch et al., 1996; Kryndushkin et al., 2003; McGlinchey et al., 2011; Uptain et al., 2001; Zhou et al., 1999). Since, overexpression of Sup35NM-GFP does not cause ring formation and does not alter the variant phenotype in established [PSI+] cells, it appears that once a prion variant is established it is unaltered by the overexpression of prion protein (Fig. 2F). Here, we used the appearance of the ring or line-like aggregates to investigate the process of prion variant establishment. We show that a single [PSI+] variant is not always specified when ring aggregates are formed (Fig. 2). Rather, we found that the potential to give rise to more than one [PSI+] variant can co-exist in cells with ring aggregates and can be transmitted to daughter cells.
It is important to keep in mind that we only scored differences between phenotypically distinct strong and weak [PSI+] strains. Thus while our work proves that multiple variants can be induced in a single cell, we can only detect a small fraction of such events. This is because progeny that are all strong (or all weak) [PSI+] may still contain distinct variants that differ in more subtle properties.
We considered the possibility that only single [PSI+] variants appeared in mother cells, but were sometimes lost after being transmitted to a daughter, followed by the induction of a new [PSI+] variant in the same mother cell that was then transmitted to a later daughter. However, since ring aggregates have never been observed to disappear and then reappear this seems unlikely. It is also unlikely that [PSI+] could arise de novo in daughters who lost, or never inherited, [PSI+] from their ring cell mothers since daughters do not have high levels of Sup35NM-GFP.
Another novel finding of this work was that 5% of the [PSI+] prions induced were “unspecified” in variant type. While it has been reported previously that newly appearing [PSI+] is often unstable, this instability referred to the loss of the prion—not its conversion into another variant (Derkatch et al., 1996). We did previously describe an ‘undifferentiated [PSI+]’, propagated by the N-domain of Sup35 (in the absence of the M domain, Sup35 1–113, Sup35 1–123), that was capable of forming both strong and weak [PSI+] when cytoduced into wild-type Sup35 recipients (Bradley and Liebman, 2004). In contrast to this earlier work, the unspecified [PSI+] described here is propagated by full length Sup35 including the M domain.
One possible explanation for unspecified [PSI+] is that it is a toxic variant, causing efficient selection for altered non-toxic variants, such as weak or strong [PSI+]. However, toxic or lethal [PSI+] causes excessive nonsense suppression (McGlinchey et al., 2011), while unspecified [PSI+] has a phenotype intermediate between strong and weak [PSI+] and is not associated with any reduction in growth.
Thus, to explain our data we consider two other non-exclusive hypotheses 1) that multiple prion variants are induced in a single cell and 2) that a single newly appearing prion aggregate is capable of changing into different variant conformations. The multiple variant hypothesis proposes that cells with initial ring aggregates contain [PSI+] aggregates with more than one conformation. According to this hypothesis our data could be explained if these different shaped [PSI+] prion conformers sometimes segregated from each other, and sometimes were transferred simultaneously, when transmitted to daughter cells. However, this hypothesis alone does not explain the data since in contrast to the ring containing cells that give rise to considerable proportions of both strong and weak [PSI+] progeny, individual zygotes known to contain a mixture of strong and weak [PSI+] propagons always gave rise to essentially all strong or all weak [PSI+] progeny. In addition, according to this hypothesis, cells with unspecified [PSI+] would have to retain both variants indefinitely.
In contrast, we show that cells with the unspecified [PSI+] generally do not contain a mixture of different prion variant propagons. Indeed, many cells in the unspecified [PSI+] colony contained only unspecified [PSI+] propagons, that by definition gave rise to progeny of more than one variant, (see Table 2). This strongly supports the idea of heritable prion conformations that can frequently undergo different alterations in conformation giving rise to distinct prion variants. We call this phenomenon prion maturation.
The maturation hypothesis proposes that newly formed propagons, carry the dynamic ability to fold into more than one conformation before becoming established as a mature prion with a specific variant shape. In a newly formed [PSI+] cells, because of their immature state, these aggregates could mature into both strong and weak [PSI+] variant conformers. The time required for an immature propagon to mature could vary, sometimes occurring in the ring cell itself, sometimes in its daughters or granddaughters—and sometimes persisting indefinitely. Our finding that increasing the time of overexpression of Sup35NM-GFP in [psi−] [PIN+] cells, ncreases the proportion of ring cells giving rise to only a single variant de novo is consistent with the maturation hypothesis, where the extended period of overexpression could provide the time needed for immature propagons to mature into specified variant conformations.
It seems unlikely that specified- weak and strong propagons present in the mother cell would completely segregate out in daughters and granddaughters since each is likely to inherit many propagons from her mother. Instead, possibly once an immature propagon becomes a specified variant in such daughter or granddaughter cells, the specified propagon could seed the maturation of the remaining unspecified propagons to become specified. However, since individual unspecified [PSI+] cells can contain both specified and unspecified propagons, the presence of specified propagon does not always immediately cause the maturation of other coexisting unspecified [PSI+] variant propagons.
The finding that unspecified propagons exist, as distinct from a mixture of weak and strong [PSI+] propagons, provides strong support for the maturation model - at least for unspecified [PSI+]. This together with the fact that the behavior of unspecified [PSI+] cells is distinct from that of zygotes containing a mixture of strong and weak [PSI+] propagons supports the hypothesis that immature unspecified propagons are present in unspecified [PSI+] cells. While it is possible that the unspecified propagons contain a mixture of strong and weak fibers that continue to propagate together, we prefer the hypothesis that the conformation of the unspecified propagons is distinct from both strong and weak [PSI+]. However, the presence of both weak and strong [PSI+] in some single unspecified [PSI+] cells also supports the multiple variant hypothesis. Thus it appears that a combination of both the maturation and multiple variant hypotheses best explain our results.
Prion “adaptation” has been proposed to explain why primary passage of the mammalian PrP prion across species lines is often associated with prolonged incubation periods, while subsequent intra-species propagation results in shorter incubation periods and increased lethality (Bruce et al., 1994; Collinge et al., 1995; Fraser et al., 1992; Hill and Collinge, 2004; Prusiner et al., 1990; Race et al., 2001). Likewise there is a striking difference in the infectious properties of Chronic Wasting Disease prions after multiple rounds of PMCA (Protein Misfolding Cyclic Amplification) or passages in mice when compared to the original inoculum (Meyerett et al., 2008). Cervid PrPSc can catalyze the conversion of human PrPC to PrPSc but only after several passages with human PrPC either in transgenic mice or in vitro (Barria et al., 2011). To explain these observations, it has been proposed that foreign infecting PrP protein is in a conformation that is incompatible with propagation of a stable prion when transmitted to the host PrP sequence. Therefore, the host PrP protein will propagate unstable prion conformations until a stable conformation is acquired. Thus, the same host protein sequence goes from an unstable conformation to a stable one resulting in the more efficient conversion of PrPc to the toxic PrPSc prion species (Collinge and Clarke, 2007; Collinge, 2012). Alterations in the conformation of mammalian PrP prions (called prion “mutations”) have also been proposed to explain this adaptation phenomenon. It is proposed that such mutations result in a prion population composed of a “cloud” of different conformations. Depending upon the suitability of the environment, the conformation that multiplies fastest will take over the others e.g. during sequential passages in a new species (Weissmann, 2012).
Although the species barrier is caused by incompatibility between two PrP sequences, the adaptation phase is very reminiscent of the prion maturation process hypothesized to explain our results as they both involve evolution of conformations of a single protein (Collinge and Clarke, 2007; Race et al., 2001). It is possible that an “unspecified” conformation first arises in the new host because the new sequence is incompatible with the conformation of the infecting prion, and that adaptation within this species involves the conversion of the “unspecified” conformation into one that is compatible in the new host. This suggests that PrP adaptation may involve an unspecified conformation that can mature into a compatible conformation, as well as a sorting out prions with different conformations.
Experimental Procedures
Yeast strains, plasmids and growth conditions
The [psi−][pin−] and [PIN+] strains used in this study are derivatives of 74-D694 (MATa ade1–14 leu2–3,112 his3-Δ200 trp1–289 ura3–52) and are listed in Table 1. Whenever [PIN+] was used it was always the “high” variant type (Bradley et al., 2002). Saccharomyces cerevisiae strains were grown at 30°C using standard media and cultivation procedures (Sherman, 1986). Complex media contained 2% dextrose (YPD) or 2% glycerol (YPG). Synthetic media (SD) contained 2% dextrose and appropriate amino acids. The lithium acetate method was used for yeast transformation (Gietz and Woods, 2002).
Table 1.
Yeast strains used in this study
| Strains | Description | Source |
|---|---|---|
| 74D-694 | MAT a ade1–14 ura3–52 leu2–3,112 trp1–289 his3–200 | (Derkatch et al., 1996) |
| L1749 | 74D-694 [psi−] High [PIN+] | (Derkatch et al., 1996) |
| L1758 | 74D-694 High [PIN+] Weak [PSI+] | (Bradley et al., 2002) |
| L1762 | 74D-694 High [PIN+] Strong [PSI+] | (Bradley et al., 2002) |
| L1998 | MATα ade2–1 SUQ5 lys1-1 his3–11, 15 leu1 kar1-1 cyhR High [PIN+] | (Bradley et al., 2003) |
| GF658 | MATα ade1–14 trp1–289 his3–200 ura3–52 leu2–3,112 SUP35-GFP [psi−] | (Satpute-Krishnan et al., 2005) |
| L3102 | MATα ade1–14 trp1–289 his3–200 ura3–52 leu2–3,112 [psi−] SUP35-GFP, rnq1::HIS3 | (This study) |
| L2717 | MATα ade1–14 leu2–3112 ura3–52 trp1–289 lys9-A21 Strong [PSI+] | (Vishveshwara et al., 2009) |
Plasmid p1182 (pCUP1-SUP35NM::GFP) carries the selectable marker LEU2 and the Sup35NM-GFP fusion under the CUP1 promoter and is used to induce [PSI+] de novo (Zhou et al., 2001). Strains transformed with pCUP1-SUP35NM::GFP were maintained on synthetic complete medium lacking leucine (–Leu).
Determination of [PSI+] variants arising from single ring containing cells
To induce ring aggregates or [PSI+], Sup35NM-GFP [PIN+] transformants were grown in plasmid selective (–Leu) media supplemented with 50–150 μM CuSO4 overnight.
To isolate unbudded ring cells, since YPD is auto fluorescent, micromanipulation was done on a thin noble agar pad which was then placed on a YPD plate for further growth. After 2–3 days when the colonies grew to 2–3 mm in diameter, a portion of them was spread on YPD at a concentration of ~200 cells per plate.
To distinguish [PSI+] colonies from Mendelian suppressor mutations, individual cells were grown into colonies on 5mM guanidine hydrochloride (GuHCl) and then checked for loss of suppression by spotting on –Ade, YPD and YPG media (Bradley et al., 2003; Tuite et al., 1981). GuHCl eliminates prions by inactivating the chaperone Hsp104, whereas suppressors remain unaffected (Jung and Masison, 2001).
[PSI+] color assay
All yeast strains used have the ade1–14 allele that has a nonsense mutation and is frequently used to score for [PSI+] (Chernoff et al., 1995). Normal [psi−] cells with the functional Sup35 translation termination factor terminate protein translation efficiently at the premature ade1–14 nonsense codon, which causes cells to be Ade− and to accumulate red pigment on rich medium like YPD. In contrast, in [PSI+] cells, aggregation and thus inactivation of Sup35 causes some read through of the ade1–14 premature stop codon, so some full length Ade1 is synthesized giving Ade+ white (strong [PSI+]), pink (weak [PSI+]) or sectored (unspecified [PSI+]) colonies.
We examined the effects of all plasmids used in this study on the color of [PSI+] cells and found no effects. This was important because a Gal-Sup35 plasmid we used in a previous study caused anti-suppression even when the Gal promoter was turned off on glucose. The presence of this plasmid caused strong [PSI+] cells to grow into pink colonies on YPD, that had white sectors whenever the plasmid was lost (Patel and Liebman, 2011).
Fluorescent microscopy and quantification of cytoplasmic Sup35NM-GFP levels
Fluorescent images were acquired with a Zeiss Axioskop 2 microscope and an AxioCam digital camera (Carl Zeiss), and processed with AxioVision software (Carl Zeiss). For quantification, L1749 transformed with pCUP1-SUP35NM::GFP and grown overnight in –Leu medium containing 50 μm CuSO4 was washed and grown in YPD for another 3 hrs. Images were acquired from randomly chosen ring containing cells with a single attached bud and from control [psi−] cells with Sup35 endogenously tagged with GFP (kindly supplied by T. Serio) (Satpute-Krishnan and Serio, 2005). The fluorescence intensity was determined with image J software (Rasband, 1997–2012). An interior region of the cell excluding the vacuole was selected with the “brush” tool. The mean background intensity of an area next to each cell was subtracted from the cell’s mean fluorescence intensity to get the actual value for that cell.
Pedigree analysis
Micromanipulation of individual ring containing cells was done on a thin 2% noble agar pad which was transferred to a YPD plate where it was allowed to divide for 6–10 hrs. The agar pad was then removed from the plate and the cells were examined under the fluorescent microscope and were separated on the agar pad. The pad was then returned to a YPD plate and the separated cells were allowed to form colonies which were then respread on YPD to score for [PSI+] variants on the basis of colony color.
Biochemical analysis of yeast lysates
Cell lysates were prepared from 50 ml of overnight culture, by vortexing cells in 750 μl of lysis buffer (80 mM Tris, 300 mM KCl, 10 mM MgCl2 and 20 % [wt/vol] glycerol, 1:50 diluted protease inhibitor cocktail [Sigma], and 5 mM PMSF) at pH 7.6 with 0.5 mm glass beads (Biospec) at high speed five times for 1 min separated with 1 min cooling in ice. Lysates were precleared of cell debris by centrifuging two times at 600 g for 1 min at 4°C (Kushnirov et al., 2006).
To analyze [PSI+] aggregates by SDD-AGE, ~40 μg of crude lysate was treated with 2 % SDS sample buffer (25 mM Tris, 200 mM glycine, 5 % glycerol, and 0.025 % bromophenol blue) for 7 min at room temperature, electrophoretically resolved in a horizontal 1.5 % agarose gel in a standard tris/glycine/SDS buffer, transferred to a polyvinylidene difluoride membrane and probed with Sup35C antibody as described previously (Bagriantsev et al., 2006).
Cytoduction
Cytoduction was carried out between [RHO+] donors and mitochondrial deficient [rho−] recipients. Either donor or recipient carried a kar1 mutation that inhibits nuclear fusion (Conde and Fink, 1976). Following mating, cytoductants and diploids were selected by growth on synthetic media lacking amino acids specifically required by the donor and using glycerol as the sole carbon source where functional mitochondria are required for growth. Thus the cytoductant would inherit the nucleus from one parent and mitochondria from another. Cytoductants were distinguished from diploids on the basis of their auxotrophic markers.
Propagon analysis
The qualitative and quantitative analysis of propagons per cell was done by using the previously described in vivo propagon dilution method (Cox et al., 2003).
[PSI+] variants study in zygotes
Strong [PSI+] (L2717) (Vishveshwara et al., 2009) and weak [PSI+] (L1758) were mated overnight on YPD plates and individual micromanipulated zygotes were grown overnight on YPD. The resulting microcolonies which were then suspended in water and spread on YPD where pink and white colonies were respectively scored as weak vs. strong [PSI+]. Colonies were confirmed to be diploid by marker analysis.
Supplementary Material
As a control experiment Sup35NM-GFP was over expressed in strong [PSI+] and weak [PSI+] cells. Single cells containing bright fluorescent dots were micromanipulated and allowed to form colonies that were spread on YPD for the color assay (200 strong [PSI+] and 200 weak [PSI+] cells were micromanipulated).
Acknowledgments
We thank Dr. S. L. Lindquist (Massachusetts Institute of Technology) for Rnq1 antibodies, Dr T. R. Serio (Brown University) for strains and Dr. Vidhu Mathur (University of Illinois) for helpful comments on the manuscript. We thank Zi Yang (University of Illinois) for making the L3102 strain. This work was supported by the National Institute of Health (NIH) Grant R01GM056350 to S.W.L. The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of NIH.
Footnotes
Authors declare that they have no conflict of interest.
References
- Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagriantsev S, Liebman SW. Specificity of prion assembly in vivo. [PSI+] and [PIN+] form separate structures in yeast. J Biol Chem. 2004;279:51042–51048. doi: 10.1074/jbc.M410611200. [DOI] [PubMed] [Google Scholar]
- Bagriantsev SN, Kushnirov VV, Liebman SW. Analysis of amyloid aggregates using agarose gel electrophoresis. Methods Enzymol. 2006;412:33–48. doi: 10.1016/S0076-6879(06)12003-0. [DOI] [PubMed] [Google Scholar]
- Barria MA, Telling GC, Gambetti P, Mastrianni JA, Soto C. Generation of a new form of human PrP(Sc) in vitro by interspecies transmission from cervid prions. J Biol Chem. 2011;286:7490–7495. doi: 10.1074/jbc.M110.198465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley ME, Edskes HK, Hong JY, Wickner RB, Liebman SW. Interactions among prions and prion “strains” in yeast. Proc Natl Acad Sci U S A. 2002;99(Suppl 4):16392–16399. doi: 10.1073/pnas.152330699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley ME, Bagriantsev S, Vishveshwara N, Liebman SW. Guanidine reduces stop codon read-through caused by missense mutations in SUP35 or SUP45. Yeast. 2003;20:625–632. doi: 10.1002/yea.985. [DOI] [PubMed] [Google Scholar]
- Bradley ME, Liebman SW. Destabilizing interactions among [PSI(+)] and [PIN(+)] yeast prion variants. Genetics. 2003;165:1675–1685. doi: 10.1093/genetics/165.4.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley ME, Liebman SW. The Sup35 domains required for maintenance of weak, strong or undifferentiated yeast [PSI+] prions. Mol Microbiol. 2004;51:1649–1659. doi: 10.1111/j.1365-2958.2003.03955.x. [DOI] [PubMed] [Google Scholar]
- Bruce M, Chree A, McConnell I, Foster J, Pearson G, Fraser H. Transmission of bovine spongiform encephalopathy and scrapie to mice: strain variation and the species barrier. Philos Trans R Soc Lond B Biol Sci. 1994;343:405–411. doi: 10.1098/rstb.1994.0036. [DOI] [PubMed] [Google Scholar]
- Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- Choe YJ, Ryu Y, Kim HJ, Seok YJ. Increased [PSI+] appearance by fusion of Rnq1 with the prion domain of Sup35 in Saccharomyces cerevisiae. Eukaryot Cell. 2009;8:968–976. doi: 10.1128/EC.00353-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinge J, Palmer MS, Sidle KC, Hill AF, Gowland I, Meads J, Asante E, Bradley R, Doey LJ, Lantos PL. Unaltered susceptibility to BSE in transgenic mice expressing human prion protein. Nature. 1995;378:779–783. doi: 10.1038/378779a0. [DOI] [PubMed] [Google Scholar]
- Collinge J, Clarke AR. A general model of prion strains and their pathogenicity. Science. 2007;318:930–936. doi: 10.1126/science.1138718. [DOI] [PubMed] [Google Scholar]
- Collinge J. Cell biology. The risk of prion zoonoses. Science. 2012;335:411–413. doi: 10.1126/science.1218167. [DOI] [PubMed] [Google Scholar]
- Conde J, Fink GR. A mutant of Saccharomyces cerevisiae defective for nuclear fusion. Proc Natl Acad Sci U S A. 1976;73:3651–3655. doi: 10.1073/pnas.73.10.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox B, Ness F, Tuite M. Analysis of the generation and segregation of propagons: entities that propagate the [PSI+] prion in yeast. Genetics. 2003;165:23–33. doi: 10.1093/genetics/165.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox BS, Tuite MF, McLaughlin CS. The psi factor of yeast: a problem in inheritance. Yeast. 1988;4:159–178. doi: 10.1002/yea.320040302. [DOI] [PubMed] [Google Scholar]
- Crow ET, Li L. Newly identified prions in budding yeast, and their possible functions. Semin Cell Dev Biol. 2011 doi: 10.1016/j.semcdb.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch IL, Chernoff YO, Kushnirov VV, Inge-Vechtomov SG, Liebman SW. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics. 1996;144:1375–1386. doi: 10.1093/genetics/144.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch IL, Bradley ME, Zhou P, Chernoff YO, Liebman SW. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics. 1997;147:507–519. doi: 10.1093/genetics/147.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: the story of [PIN(+)] Cell. 2001;106:171–182. doi: 10.1016/s0092-8674(01)00427-5. [DOI] [PubMed] [Google Scholar]
- Derkatch IL, Uptain SM, Outeiro TF, Krishnan R, Lindquist SL, Liebman SW. Effects of Q/N-rich, polyQ, and non-polyQ amyloids on the de novo formation of the [PSI+] prion in yeast and aggregation of Sup35 in vitro. Proc Natl Acad Sci U S A. 2004;101:12934–12939. doi: 10.1073/pnas.0404968101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Park KW, Yu H, Fan Q, Li L. Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae. Nat Genet. 2008;40:460–465. doi: 10.1038/ng.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaglestone SS, Ruddock LW, Cox BS, Tuite MF. Guanidine hydrochloride blocks a critical step in the propagation of the prion-like determinant [PSI(+)] of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2000;97:240–244. doi: 10.1073/pnas.97.1.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser H, Bruce ME, Chree A, McConnell I, Wells GA. Transmission of bovine spongiform encephalopathy and scrapie to mice. J Gen Virol. 1992;73 (Pt 8):1891–1897. doi: 10.1099/0022-1317-73-8-1891. [DOI] [PubMed] [Google Scholar]
- Ganusova EE, Ozolins LN, Bhagat S, Newnam GP, Wegrzyn RD, Sherman MY, Chernoff YO. Modulation of prion formation, aggregation, and toxicity by the actin cytoskeleton in yeast. Mol Cell Biol. 2006;26:617–629. doi: 10.1128/MCB.26.2.617-629.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- Glover JR, Kowal AS, Schirmer EC, Patino MM, Liu JJ, Lindquist S. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell. 1997;89:811–819. doi: 10.1016/s0092-8674(00)80264-0. [DOI] [PubMed] [Google Scholar]
- Hill AF, Collinge J. Prion strains and species barriers. Contrib Microbiol. 2004;11:33–49. doi: 10.1159/000077061. [DOI] [PubMed] [Google Scholar]
- Hosoda N, Kobayashi T, Uchida N, Funakoshi Y, Kikuchi Y, Hoshino S, Katada T. Translation termination factor eRF3 mediates mRNA decay through the regulation of deadenylation. J Biol Chem. 2003;278:38287–38291. doi: 10.1074/jbc.C300300200. [DOI] [PubMed] [Google Scholar]
- Jung G, Masison DC. Guanidine hydrochloride inhibits Hsp104 activity in vivo: a possible explanation for its effect in curing yeast prions. Curr Microbiol. 2001;43:7–10. doi: 10.1007/s002840010251. [DOI] [PubMed] [Google Scholar]
- King CY, Tittmann P, Gross H, Gebert R, Aebi M, Wuthrich K. Prion-inducing domain 2–114 of yeast Sup35 protein transforms in vitro into amyloid-like filaments. Proc Natl Acad Sci U S A. 1997;94:6618–6622. doi: 10.1073/pnas.94.13.6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King CY. Supporting the structural basis of prion strains: induction and identification of [PSI] variants. J Mol Biol. 2001;307:1247–1260. doi: 10.1006/jmbi.2001.4542. [DOI] [PubMed] [Google Scholar]
- King CY, Diaz-Avalos R. Protein-only transmission of three yeast prion strains. Nature. 2004;428:319–323. doi: 10.1038/nature02391. [DOI] [PubMed] [Google Scholar]
- Kochneva-Pervukhova NV, Chechenova MB, Valouev IA, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD. [Psi(+)] prion generation in yeast: characterization of the ‘strain’ difference. Yeast. 2001;18:489–497. doi: 10.1002/yea.700. [DOI] [PubMed] [Google Scholar]
- Krishnan R, Lindquist SL. Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature. 2005;435:765–772. doi: 10.1038/nature03679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryndushkin DS, Alexandrov IM, Ter-Avanesyan MD, Kushnirov VV. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J Biol Chem. 2003;278:49636–49643. doi: 10.1074/jbc.M307996200. [DOI] [PubMed] [Google Scholar]
- Kushnirov VV, Kochneva-Pervukhova NV, Chechenova MB, Frolova NS, Ter-Avanesyan MD. Prion properties of the Sup35 protein of yeast Pichia methanolica. Embo J. 2000a;19:324–331. doi: 10.1093/emboj/19.3.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnirov VV, Kryndushkin DS, Boguta M, Smirnov VN, Ter-Avanesyan MD. Chaperones that cure yeast artificial [PSI(+)] and their prion-specific effects. Curr Biol. 2000b;10:1443–1446. doi: 10.1016/s0960-9822(00)00802-2. [DOI] [PubMed] [Google Scholar]
- Kushnirov VV, Alexandrov IM, Mitkevich OV, Shkundina IS, Ter-Avanesyan MD. Purification and analysis of prion and amyloid aggregates. Methods. 2006;39:50–55. doi: 10.1016/j.ymeth.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Liu JJ, Sondheimer N, Lindquist SL. Changes in the middle region of Sup35 profoundly alter the nature of epigenetic inheritance for the yeast prion [PSI+] Proc Natl Acad Sci U S A. 2002;99(Suppl 4):16446–16453. doi: 10.1073/pnas.252652099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur V, Taneja V, Sun Y, Liebman SW. Analyzing the birth and propagation of two distinct prions, [PSI+] and [Het-s](y), in yeast. Mol Biol Cell. 2010;21:1449–1461. doi: 10.1091/mbc.E09-11-0927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlinchey RP, Kryndushkin D, Wickner RB. Suicidal [PSI+] is a lethal yeast prion. Proc Natl Acad Sci U S A. 2011;108:5337–5341. doi: 10.1073/pnas.1102762108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerett C, Michel B, Pulford B, Spraker TR, Nichols TA, Johnson T, Kurt T, Hoover EA, Telling GC, Zabel MD. In vitro strain adaptation of CWD prions by serial protein misfolding cyclic amplification. Virology. 2008;382:267–276. doi: 10.1016/j.virol.2008.09.023. [DOI] [PubMed] [Google Scholar]
- Patel BK, Gavin-Smyth J, Liebman SW. The yeast global transcriptional co-repressor protein Cyc8 can propagate as a prion. Nat Cell Biol. 2009;11:344–349. doi: 10.1038/ncb1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel BK, Liebman SW. Corrigendum. Genetics. 2011;188:1023. [Google Scholar]
- Prusiner SB, Scott M, Foster D, Pan KM, Groth D, Mirenda C, Torchia M, Yang SL, Serban D, Carlson GA, et al. Transgenetic studies implicate interactions between homologous PrP isoforms in scrapie prion replication. Cell. 1990;63:673–686. doi: 10.1016/0092-8674(90)90134-z. [DOI] [PubMed] [Google Scholar]
- Prusiner SB. Prions. Proc Natl Acad Sci U S A. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Race R, Raines A, Raymond GJ, Caughey B, Chesebro B. Long-term subclinical carrier state precedes scrapie replication and adaptation in a resistant species: analogies to bovine spongiform encephalopathy and variant Creutzfeldt-Jakob disease in humans. J Virol. 2001;75:10106–10112. doi: 10.1128/JVI.75.21.10106-10112.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasband WS. ImageJ. U. S. National Institutes of Health; Bethesda, Maryland, USA: 1997–2012. http://imagej.nih.gov/ij/ [Google Scholar]
- Roberts BE, Duennwald ML, Wang H, Chung C, Lopreiato NP, Sweeny EA, Knight MN, Shorter J. A synergistic small-molecule combination directly eradicates diverse prion strain structures. Nat Chem Biol. 2009;5:936–946. doi: 10.1038/nchembio.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safar J, Wille H, Itri V, Groth D, Serban H, Torchia M, Cohen FE, Prusiner SB. Eight prion strains have PrP(Sc) molecules with different conformations. Nat Med. 1998;4:1157–1165. doi: 10.1038/2654. [DOI] [PubMed] [Google Scholar]
- Satpute-Krishnan P, Serio TR. Prion protein remodelling confers an immediate phenotypic switch. Nature. 2005;437:262–265. doi: 10.1038/nature03981. [DOI] [PubMed] [Google Scholar]
- Schlumpberger M, Prusiner SB, Herskowitz I. Induction of distinct [URE3] yeast prion strains. Mol Cell Biol. 2001;21:7035–7046. doi: 10.1128/MCB.21.20.7035-7046.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Hicks JB. Methods in Yeast Genetics. Plainview, New York: Cold Spring Harbor Lab; 1986. [Google Scholar]
- Sondheimer N, Lindquist S. Rnq1: an epigenetic modifier of protein function in yeast. Mol Cell. 2000;5:163–172. doi: 10.1016/s1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- Suzuki G, Shimazu N, Tanaka M. A yeast prion, Mod5, promotes acquired drug resistance and cell survival under environmental stress. Science. 2012;336:355–359. doi: 10.1126/science.1219491. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. Conformational variations in an infectious protein determine prion strain differences. Nature. 2004;428:323–328. doi: 10.1038/nature02392. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Collins SR, Toyama BH, Weissman JS. The physical basis of how prion conformations determine strain phenotypes. Nature. 2006;442:585–589. doi: 10.1038/nature04922. [DOI] [PubMed] [Google Scholar]
- Ter-Avanesyan MD, Kushnirov VV, Dagkesamanskaya AR, Didichenko SA, Chernoff YO, Inge-Vechtomov SG, Smirnov VN. Deletion analysis of the SUP35 gene of the yeast Saccharomyces cerevisiae reveals two non-overlapping functional regions in the encoded protein. Mol Microbiol. 1993;7:683–692. doi: 10.1111/j.1365-2958.1993.tb01159.x. [DOI] [PubMed] [Google Scholar]
- Toyama BH, Kelly MJ, Gross JD, Weissman JS. The structural basis of yeast prion strain variants. Nature. 2007;449:233–237. doi: 10.1038/nature06108. [DOI] [PubMed] [Google Scholar]
- Tuite MF, Mundy CR, Cox BS. Agents that cause a high frequency of genetic change from [psi+] to [psi−] in Saccharomyces cerevisiae. Genetics. 1981;98:691–711. doi: 10.1093/genetics/98.4.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuite MF, Cox BS. Propagation of yeast prions. Nat Rev Mol Cell Biol. 2003;4:878–890. doi: 10.1038/nrm1247. [DOI] [PubMed] [Google Scholar]
- Tyedmers J, Treusch S, Dong J, McCaffery JM, Bevis B, Lindquist S. Prion induction involves an ancient system for the sequestration of aggregated proteins and heritable changes in prion fragmentation. Proc Natl Acad Sci U S A. 2010;107:8633–8638. doi: 10.1073/pnas.1003895107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uptain SM, Sawicki GJ, Caughey B, Lindquist S. Strains of [PSI(+)] are distinguished by their efficiencies of prion-mediated conformational conversion. Embo J. 2001;20:6236–6245. doi: 10.1093/emboj/20.22.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishveshwara N, Bradley ME, Liebman SW. Sequestration of essential proteins causes prion associated toxicity in yeast. Mol Microbiol. 2009;73:1101–1114. doi: 10.1111/j.1365-2958.2009.06836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissmann C. Mutation and selection of prions. PLoS Pathog. 2012;8:e1002582. doi: 10.1371/journal.ppat.1002582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner RB. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- Wickner RB, Masison DC, Edskes HK. [PSI] and [URE3] as yeast prions. Yeast. 1995;11:1671–1685. doi: 10.1002/yea.320111609. [DOI] [PubMed] [Google Scholar]
- Zhou P, Derkatch IL, Uptain SM, Patino MM, Lindquist S, Liebman SW. The yeast non-Mendelian factor [ETA+] is a variant of [PSI+], a prion-like form of release factor eRF3. Embo J. 1999;18:1182–1191. doi: 10.1093/emboj/18.5.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Derkatch IL, Liebman SW. The relationship between visible intracellular aggregates that appear after overexpression of Sup35 and the yeast prion-like elements [PSI(+)] and [PIN(+)] Mol Microbiol. 2001;39:37–46. doi: 10.1046/j.1365-2958.2001.02224.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a control experiment Sup35NM-GFP was over expressed in strong [PSI+] and weak [PSI+] cells. Single cells containing bright fluorescent dots were micromanipulated and allowed to form colonies that were spread on YPD for the color assay (200 strong [PSI+] and 200 weak [PSI+] cells were micromanipulated).