Abstract
α-Synuclein has recently been implicated in the pathophysiology of alcohol abuse due to its role in dopaminergic neurotransmission. In these studies, genetic variability in the α-synuclein gene influences its expression which may contribute to susceptibility to chronic alcohol abuse. Real-time PCR was used to quantify α-synuclein mRNA expression in autopsy samples of human dorsolateral prefrontal cortex. Because of the association between length of the α-synuclein-repeat 1 microsatellite marker and expression levels of the gene, this marker was genotyped in a Caucasian sample of 126 controls and 117 alcoholics using capillary gel electrophoresis. The allele and genotype frequencies of α-synuclein-repeat 1 marker differed significantly between alcoholics and controls. Alcoholics had greater frequencies of the shortest allele found (267 bp). The shortest allele of the α-synuclein-repeat 1 marker was associated with decreased expression of α-synuclein in prefrontal cortex. Individuals with at least one copy of the 267 bp allele were more likely to exhibit an alcohol abuse phenotype. These results suggest that individuals with the 267 bp allele may be at increased risk of developing alcoholism and that genetic variation at the α-synuclein-repeat 1 locus may influence α-synuclein expression in the prefrontal cortex.
Keywords: human, mRNA, post-mortem, prefrontal cortex, SNCA-Rep1
INTRODUCTION
Alcoholism is a complex, multi-factorial disorder with substantial health, societal and economic consequences worldwide. The World Health Organization estimates that alcohol abuse contributes 4% to the global burden of disease and results in ~2 million deaths annually (WHO, 2004). The cost to society associated with excessive alcohol consumption, including lost productivity, health-care costs, road accident-related costs and crime-related costs was estimated to be $15 billion in 2004–05 in Australia alone (Collins and Lapseley, 2008). Up to 10% of the population is drinking at hazardous or harmful levels that exacerbate the deleterious effects of alcohol on the brain (AIHW, 2008).
Both genetic and environmental factors influence the complex etiology and pathophysiology of alcohol abuse. Family, adoption, and twin studies provide estimates for an alcoholism heritability of ~50% (Ducci and Goldman, 2008; Mayfield et al., 2008). Large-scale linkage and association studies have identified many genetic variants that influence the risk of alcoholism and related phenotypes (Ducci and Goldman, 2008; Mayfield et al., 2008). It is likely that several genes contribute to each phenotype. Genes that modify tolerance, acute sensitivity to intoxication, dependence, and craving may alter an individual’s susceptibility. Genetic factors may also underlie the neuroadaptive changes that occur in response to chronic alcohol abuse. Alcohol acts on a number of neurotransmission-associated molecular targets, and affects many different cellular processes. Although many candidate genes have been identified, only a few have functional loci that moderate the effects of alcohol. Understanding neurotoxic mechanisms associated with alcohol abuse are key to developing treatment strategies and identifying new therapeutic targets.
Whole-genome linkage analyses have mapped alcohol dependence and related phenotypes to a region on chromosome 4 containing a cluster of genes for alcohol dehydrogenase (ADH1B and ADH1C) and α-synuclein (SNCA) (Ehlers et al., 2004; Reich et al., 1998; Saccone et al., 2000; Williams et al., 1999). The syntenic chromosomal region in rats selectively bred for high alcohol preference has been linked to a quantitative trait locus (QTL) for alcohol consumption (Liang et al., 2003) and congenic rats with this QTL exhibit lowered expression for α-synuclein particularly in the frontal cortex (Liang et al., 2010). The SNCA gene is highly polymorphic, and the association between sequence variation in SNCA and neurodegenerative disorders such as Parkinson’s disease has been well documented. Sequence variation occurs within the coding region as well as in the α-synuclein-repeat 1 (SNCA-Rep1) microsatellite ~10 kb upstream from the translation start site (Touchman et al., 2001). The latter has five known alleles with a varying number of di-nucleotide repeats that range in size from 265 to 273 bp. Recent studies have shown that the length of the SNCA-Rep 1 allele, as well as polymorphisms in the 5′- and 3′-untranslated region (UTR) of the gene influence its expression levels in brain and blood (Fuchs et al., 2008; Linnertz et al., 2009; McCarthy et al., 2011). Furthermore, polymorphisms in the SNCA gene are associated with alcohol-use phenotypes, including alcohol dependence and craving (Bonsch et al., 2005a). Single-nucleotide polymorphisms (SNPs) in the gene are associated with alcohol dependence: a haplotype block in the 3′UTR is more abundant in individuals who crave alcohol (Foroud et al., 2007).
This study investigated the expression level of the most abundant α-synuclein transcript in the frontal cortex of human alcoholics and controls and used a case-control approach to determine the influence of the SNCA-Rep1 microsatellite repeat on the expression level of the gene.
MATERIALS & METHODS
Sample population
Ethical clearance for the project was obtained from the Griffith University Human Ethics Committee (MSC/02/06/HREC). Alcoholics and controls were selected on the basis of alcohol intake. Alcoholics were subdivided based on the presence of complicating diseases. Controls were defined as individuals who consumed less than 20 g of ethanol per day on average. Alcoholics were defined by National Health and Medical Research (NHMRC)/WHO criteria as individuals who consumed more than 80 g of ethanol per day for most of their adult lives. Many of the alcoholics used in this study had consumed over 200 g of ethanol per day and had been drinking for over 20 years. The alcoholic group includes alcoholics with pathologically confirmed cirrhosis of the liver. Cases with a history of poly-drug abuse or other neurological conditions such as Parkinson disease, Wernicke Korsakoff syndrome or Hepatic Encephalopathy were excluded.
Expression studies were performed on cohort of individuals for whom brain tissue was available. Alcoholics and controls were matched as closely as possible for post mortem interval (PMI), gender and age at death. Total RNA was isolated from the dorsolateral prefrontal cortex (Brodmann areas 6 and 8) of 25 controls (mean age 59.0 ± 2.8 y, PMI 31.3 ± 4.8 h) and 35 alcoholics (mean age 52.5 ± 2.6 y, PMI 28.7 ± 3.2 h). These individuals were a subset of those used for genotyping studies.
For genotyping studies, a more extensive population was used to determine which alleles were most frequent in our Australian population. This study population consisted of 126 controls (45 female, 81 male, mean age 63.3 ± 1.6 y) and 117 alcoholics (22 female, 95 male, mean age 56.2 ± 1.4 y). Alcoholics were defined according to the World Health Organization/National Health and Medical Research Council (WHO/NHMRC) criteria by a consumption > 80 g of ethanol per day for most of their adult lives; controls consumed < 20 g ethanol per day. All individuals were Caucasians of European origin.
Expression analysis
Total RNA was extracted from the dorsolateral prefrontal cortex using Trizol™ (Gibco BRL, Invitrogen, Mt Waverley, Vic, Australia) according to the manufacturer’s instructions.
The RNA was then dispensed in 20 μL aliquots and stored at −70°C until required. RNA quantity was measured by absorbance at 260 nm using a Nanodrop (Thermo Scientific, Waltham, MA, USA). The quality of all RNA samples was determined by visual inspection of electropherograms produced by the Agilent 2100 bioanalyzer (Agilent Technologies, Palo Alto, CA). The presence of distinct 18S and 28S ribosomal peaks and the absence of multiple peaks corresponding smaller RNA fragments were indicative of high-quality total RNA samples. Degraded samples characteristically show a decreased 28S rRNA peak area, a rise in the baseline between the 18S and 28S rRNA and an increase in the baseline area below the 18S rRNA that spreads with smaller 28S rRNA fragments. Samples which show degradation on the electropherogram are excluded from further study. Samples were reverse-transcribed as previously described (Ho et al., 2010; MacKay et al., 2011).
Primers were designed using Primer Express® v1.5 Software (Applied Biosystems) and synthesized by Sigma-Aldrich P/L (Castle Hill, NSW, Australia). Each primer set was verified for specificity using the Basic Local Alignment Search Tool (BLAST) from the GenBank non-redundant nucleotide sequence database (Altschul et al., 1997). All assays were designed such that at least one primer spanned an exon boundary to eliminate gDNA amplification. Primer sequences are shown in Table 1.
Table 1.
Primer Sequences
| Target | Method | Primer Sequence |
|---|---|---|
| SNCA-Rep1 | PCR | Forward: 5′-[6FAM]CCTGGCATATTTGATTGCAA-3′ |
| Reverse: 5′-GACTGGCCCAAGATTAACCA-3′ | ||
| α-synuclein | qPCR | Forward: 5′-GTGTGGCAACAGTGGCTGAG-3′ |
| Reverse: 5′-TGGGGCTCCTTCTTCATTCTTG-3′ | ||
| GAPDH | qPCR | Forward: 5′-TGCACCACCAACTGCTTAGC-3′ |
| Reverse: 5′-GGCATGGACTGTGGTCATGAG-3′ |
Quantitative real-time PCR was carried out using a Qiagen Rotor-Gene Q System. Each PCR consisted of 2 μL of a 1/50 dilution of cDNA, 10 μL SYBR® Green PCR Master Mix (Biorad, Australia), and 300 nM of each primer pair in a final volume of 20 μL. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the internal reference. All reactions were performed in duplicate. Primer pairs were optimized according to the method outlined in Livak et al (Livak and Schmittgen, 2001) to ensure that the primer efficiencies were similar. The primer efficiencies for GAPDH and α-synuclein primer pairs were 1.92 and 1.93 respectively.
The amplification plot of fluorescence vs. cycle number was used to set the threshold (T) in the exponential phase of the reaction above the baseline. This was kept constant between runs to allow for analysis between plates. The cycle threshold (CT) was calculated as the cycle number of an amplifying PCR product where it crosses the fixed threshold line. The differences in the mean CT values of the duplicate samples from the GAPDH reference were calculated using Microsoft Excel to give ΔCT values. ΔCT values were used for statistical analysis (Ho et al., 2010; MacKay et al., 2011; Ridge et al., 2008) relative expression values are presented as 2−ΔCT values (Livak and Schmittgen, 2001) plotted in GraphPad v5 (GraphPad Software Inc, La Jolla, CA, USA).
DNA Extraction and Genotyping
Genomic DNA (gDNA) was extracted from human brain tissue by phenol/chloroform extraction followed by ethanol precipitation (Sambrook et al., 1989).
Published primers were used for the microsatellite marker SNCA-Rep1 (Xia et al., 1996). A fluorescent 6-FAM labeled forward primer and an unlabeled reverse primer were obtained from Sigma Genosys (Castle Hill, NSW, Australia). SNCA-Rep1 was genotyped using PCR and fluorescent capillary-gel electrophoresis. PCR was carried out using the Mastercycler® ep gradient S (Eppendorf, North Ryde, NSW, Australia). For each sample, 20 ng of gDNA was amplified with 1x Colourless GoTaq® Flexi buffer, 2.5 mM MgCl2, 0.1 mM dNTPs, and 0.5 U of GoTaq® Flexi Hot Start DNA Polymerase (Promega, Annandale, NSW, Australia), with 0.18 μM each of forward and reverse primers in a final volume of 25 μL. Cycling conditions were: 95°C for 10 min, then 40 cycles of 95°C for 30 s, 58°C for 30 s and 72°C for 30 s followed by a final extension of 72°C for 5 min and 4°C for 5 min. Random samples of PCR product (6 μL) from each plate were electrophoresed on 3% agarose gels for 30 min at 90 V to confirm fragment size.
Following amplification, 0.5 μL of the PCR product was combined with 9.25 μL Hi-Di™ Formamide and 0.25 μL GeneScan™-500 LIZ™ size standard 1/10 dilution (Applied Biosystems, Mulgrave, Vic, Australia) in a final volume of 10 μL and run on an Applied Biosystems Hitachi ABI-3130 Genetic Analyzer. GeneMapper® software v4.0 (Applied Biosystems) was used to determine genotype; all alleles were verified manually.
Statistical Analyses
Only three of the known SNCA-Rep1 alleles were detected: 267, 269 and 271 bp. These were used to determine genotype frequencies. GENEPOP v4.0.10 (1995) online software was used to determine if the SNCA-Rep1 genotype frequencies were in Hardy-Weinberg Equilibrium (HWE) (Raymond and Rousset, 1995). A HWE exact probability test for multiallelic markers using Markov chain sampling was performed on the control samples. SNCA-Rep1 genotype data were analyzed using CLUMP v2.3 (Sham and Curtis, 1995). Subjects with an identified genotype (v.i.) were assigned a value of 1 and termed the risk group; all other genotypes were assigned the value 0 and termed the no-risk group. Logistic regression was performed using SPSS. The model was adjusted for confounding factors such as gender and age.
RESULTS
Analysis of relative expression levels using real-time PCR data is critically dependent on using a housekeeping gene that does not differ in its expression between cases and controls. Previous studies have identified GAPDH to be the most appropriate housekeeping gene for these studies (Ho et al., 2010). Accordingly, the expression of GAPDH was measured using real-time PCR in each sample included in this study. Analysis of the raw CT values showed no significant differences in the expression of GAPDH between controls and alcoholics similar to previous expression studies using the same tissue samples (F1,58 = 0.274, P = 0.63) (Ho et al., 2010; MacKay et al., 2011).
We measured the expression of the most abundant α-synuclein mRNA transcript which encodes the wildtype SNCA140 variant, in the dorsolateral prefrontal cortex of chronic alcoholics as well as age- and sex-matched controls using real-time PCR. The prefrontal cortex is particularly susceptible to alcohol-induced neuronal loss compared with other cortical regions. Overall, alcoholics had lower α-synuclein expression than controls in this brain region (ANOVA, F1,58 = 6.493; P = 0.013; Figure 1). There was no significant difference in expression between males and females overall (F1,58 = 0.71, P = 0.79). However, when SNCA expression was compared in males and females separately, SNCA140 expression was significantly lower in male alcoholics (N=21) than in male controls (N = 12; F1,31 = 5.63, P = 0.024). However, the difference between female controls (N=13) and female alcoholics (N=14) was not significantly different (F1,25 = 1.70, P = 0.20) likely due to reduced case numbers and a high degree of variability in the data.
Fig. 1.
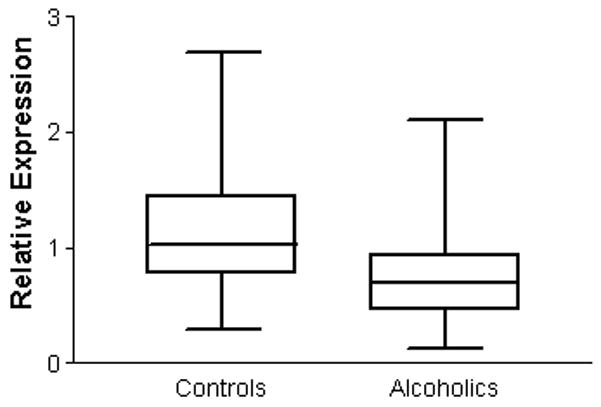
Expression of α-synuclein mRNA in controls and alcoholic cases. ΔCT values were converted to 2−ΔCT values. Data is presented as a box plot showing the range of values from the 25th percentile to the 75th percentile with a line representing the median value. A significant reduction in gene expression was observed in the prefrontal cortex of alcoholics compared with controls (ANOVA, F1,58 = 6.493; P = 0.013).
Since SNCA-Rep1 genotype influences the transcription of the SNCA gene in cell-culture studies (Chiba-Falek and Nussbaum, 2001), we analyzed the allele and genotype frequencies of SNCA-Rep1 in a case-control study. Table 2 shows the genotype and allele frequencies for this marker. No subject in this population had a 265 bp or 273 bp allele. The most common allele in both alcoholics and controls was allele 2 (269 bp). The frequency of the 267 bp allele was higher in alcoholics than in controls, whereas the frequencies of the 269 and 271 bp alleles were lower in alcoholics than in controls. All six of the possible genotypes were observed. Alcoholics had a higher frequency of the 267/267 bp and 267/269 bp genotypes than controls. The population was in HWE (P = 0.149).
Table 2.
Association analysis of genotype and allele frequencies of SNCA-Rep1
| Genotype freq. N (%) | χ2 (P) | Allele freq. N (%) | χ2 (P) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Controls | Alcoholics | Controls | Alcoholics | |||||||||
| SNCA-Rep1 | 267/267 | 6 | (4.8) | 11 | (9.4) | 267 | 57 | (22.6) | 78 | (33.3) | ||
| 9.913 | ||||||||||||
| 267/269 | 42 | (33.3) | 55 | (47) | 269 | 164 | (65.1) | 141 | (60.3) | |||
| (0.0054) | ||||||||||||
| 269/269 | 48 | (38.1) | 37 | (31.6) | 8.393 | 271 | 31 | (12.3) | 15 | (6.4) | ||
| 267/271 | 3 | (2.4) | 1 | (0.85) | (0.042) | |||||||
| 269/271 | 26 | (20.6) | 12 | (10.3) | ||||||||
| 271/271 | 1 | (0.8) | 1 | (0.85) | ||||||||
| Total | 126 | 117 | ||||||||||
The χ2 values and associated P values for SNCA-Rep1 from CLUMP analysis are shown in Table 2. A significant difference in genotype and allele frequency was seen between controls and alcoholics. From the preliminary statistical analysis, the SNCA-Rep1 267 bp allele was identified as a potential risk allele for alcohol abuse. Logistic regression showed that subjects with at least one copy of the SNCA-Rep1 267 bp allele were more likely to have been alcohol abusers (Adjusted odds ratio (ORadj) = 2.21, df = 1, P = 0.012; 95% confidence interval (CI) = 1.19–4.11).
To investigate the influence of SNCA-Rep1 genotype on α-synuclein expression, comparisons of expression levels were made between individuals with and without the 267 bp allele. Individuals with at least one copy of the 267 bp allele showed significantly lower expression of α-synuclein in the prefrontal cortex (Figure 2). Of the 25 control individuals, 18 had the no risk allele (72%) and 7 had the risk allele (28%) and of the 35 alcoholics, 12 had the no risk allele (34.3%) and 23 had the risk allele (65.7%).
Fig. 2.

Effect of SNCA-Rep1 genotype on α-synuclein mRNA expression. ΔCT values were converted to 2−ΔCT values.. Data is presented as a box plot showing the range of values from the 25th percentile to the 75th percentile with a line representing the median value. ‘No risk’ denotes individuals without a copy of the SNCA-Rep1 267bp risk allele and ‘Risk’ denotes individuals with at least one copy of the 267 bp risk allele. A significant reduction in gene expression was observed in the ‘risk’ allele group compared with those in the ‘no risk’ allele group(ANOVA, F1,58 = 13.399; P = 0.001).
DISCUSSION
We found that the expression of α-synuclein is significantly reduced in the dorsolateral prefrontal cortex of alcoholics compared with controls. Previous studies have shown that the expression of this gene can be correlated with the length of the SNCA-Rep1 microsatellite marker, with greater expression in individuals who have the longer alleles. Thus, we investigated the association between the SNCA-Rep1 microsatellite repeat marker and alcohol abuse phenotype, and the effect of SNCA-Rep1 genotype on α-synuclein mRNA expression in this brain region.
We detected only three SNCA-Rep1 alleles in our study subjects, of lengths 267, 269, and 271 bp, suggesting that this cohort, which is of northern European and Anglo-Celtic descent, may have lower allelic variability than some others. In particular, no individual in our study had a 273 bp allele, which has previously been associated with alcohol dependence (Bonsch et al., 2005a; Chiba-Falek and Nussbaum, 2001; Clarimon et al., 2007). In contrast, alcoholics studied here showed a higher frequency of the shortest allele (267 bp), as well as the 267/267 bp and the 267/269 bp genotypes, than controls. Furthermore, individuals with at least one copy of the SNCA-Rep1 267 bp allele were over twice as likely as controls to have abused alcohol. Studies using in vitro models have shown that the expression of α-synuclein is correlated with the length of SNCA-Rep1 allele, with longer alleles associated with increased expression (Chiba-Falek and Nussbaum, 2001). Genetic variation in the 5′- and 3′-UTRs of the SNCA gene has also been correlated with altered expression levels in human brain (Fuchs et al., 2008; Linnertz et al., 2009) although the direction of change (up- or down-regulation) is dependent on the specific polymorphism studied and also on brain region. The results presented here are the first to determine the role of genetic variation in the SNCA gene on the expression levels of α-synuclein in the brain of human alcoholics. In previous studies, Bonsch et al (Bonsch et al., 2005b) showed that alcohol-dependent individuals had longer SNCA-Rep1 alleles than controls and that the length of these alleles were associated with increased expression of α-synuclein mRNA in formed elements in the blood. In our population, alcoholics had lower α-synuclein mRNA expression than controls in the prefrontal cortex, and subjects with at least one copy of the SNCA-Rep1 267 bp risk allele had lower expression, suggesting that alcoholics may express lower levels of α-synuclein constitutively as a result of their SNCA-Rep1 genotype. Our results are similar to recently published findings which show that alcohol-naïve inbred alcohol-preferring rats have lower expression levels of SNCA transcripts in the frontal cortex compared with congenic P.NP rats in which the alcohol preferring QTL on chromosome 4, which contains the SNCA gene, was replaced by the equivalent QTL from the inbred nonpreferring rat line (Liang et al., 2010). Interestingly, the difference in expression was specific for the frontal cortex and emphasizes the need for further studies on the role of α-synuclein in the pathophysiology of chronic alcohol abuse. These data also call into question whether SNCA transcript expression in blood cells accurately reflects expression in the brain. Further studies will need to obtain both blood and cortical samples from the same subjects.
The full range of functions of α-synuclein is unknown, although there is strong evidence that it has roles in dopaminergic transmission (Perez et al., 2002), synaptic dopamine homeostasis (Lotharius et al., 2002), regulation of dopamine storage in vesicles and release into the synapse, and dopamine reuptake into dopaminergic neurons (Sidhu et al., 2004). Polymorphisms that affect the expression of α-synuclein could thus interfere with some or all of these processes. Lee et al (Lee et al., 2001) showed that α-synuclein increases the uptake of dopamine and dopamine-induced apoptosis through its binding and functional coupling to the dopamine transporter. A decrease in α-synuclein might affect the re-uptake of dopamine, possibly thereby altering neuronal signaling. Self and Nestler (Self and Nestler, 1998) suggest that dopaminergic transmission is the main mediator of craving, withdrawal, and the reinforcement pathways in alcohol addiction. Altered expression ofα-synuclein could have effects on neuronal plasticity and dopaminergic reward pathways. These pathways project to the dorsolateral prefrontal cortex, a key area for the executive functions that are disrupted in severe chronic alcohol misuse in human subjects (Koob and Volkow, 2010). There are complex, sometimes reciprocal, interactions between α-synuclein, dopamine, and GABA-mediated transmission in forebrain pathways related to addiction (Hemby, 2004; Rideout et al., 2003; Wu et al., 2010). Our studies have shown that changes in the expression of GABAA subunit transcripts are among the most marked alterations in human alcoholic prefrontal cortex (Buckley and Dodd, 2004; Dodd and Lewohl, 1998; Lewohl et al., 1997a, b; Lewohl et al., 2001) and that these effects may be moderated by genotype, including polymorphisms in the dopamine receptor DRD2 gene (Buckley et al., 2006; Dodd et al., 2004). GABAA receptor and SNCA genes cluster in a region associated with alcohol abuse (Edenberg and Foroud, 2006). Together these interrelationships may contribute to pathogenesis in this critical cortical region (Kril et al., 1997).
In conclusion, we report the expression of α-synuclein is significantly reduced in the dorsolateral prefrontal cortex of alcoholics consistent with alterations in dopaminergic signaling and reuptake. We also report a significant association between the frequency of the 267 bp allele of SNCA-Rep1 and alcohol abuse, and propose that this allele may increase the risk for the development of alcohol abuse. The increased prevalence of the allele was associated with lower expression levels of α-synuclein. Further studies on the expression of β- and γ-synuclein transcripts, as well as the relevant proteins, will shed further light on the involvement of this family in mediating the pathological effects of chronic alcoholism. Future studies should also investigate the effects of other polymorphisms in the SNCA gene, particularly in the 3′ UTR, on the expression of the gene and their association with chronic alcohol abuse.
Acknowledgments
We would like to acknowledge the Australian Brain Bank Network, which supplied the brain tissue for analysis. Allison Eckert and Donna Sheedy provided detailed information on the cases. We thank the next of kin for providing informed consent for the studies. Financial support was provided by the National Institute of Alcoholism and Alcohol Abuse (USA, NIH AA12404; R01 AA012725-04) and the NHMRC.
Footnotes
Author Contribution
PD and JL were responsible for the study concept and design. PJ and RM performed the expression studies and genotyping experiments. PJ, RM, RL and JL assisted with data analysis and interpretation of findings. PJ and JL drafted the manuscript. RM, PD and RL provided critical revision of the manuscript for important intellectual content. All authors critically reviewed content and approved final version for publication.
References
- AIHW. Australia’s health 2008. Canberra: 2008. [Google Scholar]
- Bonsch D, Lederer T, Reulbach U, Hothorn T, Kornhuber J, Bleich S. Joint analysis of the NACP-REP1 marker within the alpha synuclein gene concludes association with alcohol dependence. Hum Mol Genet. 2005a;14:967–971. doi: 10.1093/hmg/ddi090. [DOI] [PubMed] [Google Scholar]
- Bonsch D, Lederer T, Reulbach U, Hothorn T, Kornhuber J, Bleich S. Joint analysis of the NACP-REP1 marker within the α synuclein gene concludes association with alcohol dependence. Human Molecular Genetics. 2005b;14:967–971. doi: 10.1093/hmg/ddi090. [DOI] [PubMed] [Google Scholar]
- Buckley ST, Dodd PR. GABAA receptor ® subunit mRNA expression in the human alcoholic brain. Neurochemistry International. 2004;45:1011–1020. doi: 10.1016/j.neuint.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Buckley ST, Foley PF, Innes DJ, Loh E-W, Shen Y, Williams SM, Harper CG, Tannenberg AEG, Dodd PR. GABAA receptor ® isoform protein expression in human alcoholic brain: interaction with genotype. Neurochemistry International. 2006;49:557–567. doi: 10.1016/j.neuint.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Chiba-Falek O, Nussbaum RL. Effect of allelic variation at the NACP-Rep1 repeat upstream of the alpha-synuclein gene (SNCA) on transcription in a cell culture luciferase reporter system. Hum Mol Genet. 2001;10:3101–3109. doi: 10.1093/hmg/10.26.3101. [DOI] [PubMed] [Google Scholar]
- Clarimon J, Gray RR, Williams LN, Enoch MA, Robin RW, Albaugh B, Singleton A, Goldman D, Mulligan CJ. Linkage disequilibrium and association analysis of α-synuclein and alcohol and drug dependence in two American Indian populations. Alcoholism, Clinical and Experimental Research. 2007;31:546–554. doi: 10.1111/j.1530-0277.2007.00338.x. [DOI] [PubMed] [Google Scholar]
- Collins D, Lapseley H. The costs of tobacco, alcohol and illicit drug use to Australian society in 2004/2005. Canberra: 2008. [Google Scholar]
- Dodd PR, Foley PF, Buckley ST, Eckert AL, Innes DJ. Genes and gene expression in the brain of the alcoholic. Addict Behav. 2004;29:1295–1309. doi: 10.1016/j.addbeh.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Dodd PR, Lewohl JM. Cell death mediated by amino acid transmitter receptors in human alcoholic brain damage: conflicts in the evidence. Ann N Y Acad Sci. 1998;844:50–58. [PubMed] [Google Scholar]
- Ducci F, Goldman D. Genetic approaches to addiction: genes and alcohol. Addiction. 2008;103:1414–1428. doi: 10.1111/j.1360-0443.2008.02203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Foroud T. The genetics of alcoholism: identifying specific genes through family studies. Addiction biology. 2006;11:386–396. doi: 10.1111/j.1369-1600.2006.00035.x. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Gilder DA, Wall TL, Phillips E, Feiler H, Wilhelmsen KC. Genomic screen for loci associated with alcohol dependence in Mission Indians. Am J Med Genet B Neuropsychiatr Genet. 2004;129B:110–115. doi: 10.1002/ajmg.b.30057. [DOI] [PubMed] [Google Scholar]
- Foroud T, Wetherill LF, Liang T, Dick DM, Hesselbrock V, Kramer J, Nurnberger J, Schuckit M, Carr L, Porjesz B, Xuei X, Edenberg HJ. Association of alcohol craving with α-synuclein (SNCA) Alcoholism, Clinical and Experimental Research. 2007;31:537–545. doi: 10.1111/j.1530-0277.2007.00337.x. [DOI] [PubMed] [Google Scholar]
- Fuchs J, Tichopad A, Golub Y, Munz M, Schweitzer KJ, Wolf B, Berg D, Mueller JC, Gasser T. Genetic variability in the SNCA gene influences alpha-synuclein levels in the blood and brain. Faseb J. 2008;22:1327–1334. doi: 10.1096/fj.07-9348com. [DOI] [PubMed] [Google Scholar]
- Hemby SE. Morphine-induced alterations in gene expression of calbindin immunopositive neurons in nucleus accumbens shell and core. Neuroscience. 2004;126:689–703. doi: 10.1016/j.neuroscience.2004.01.056. [DOI] [PubMed] [Google Scholar]
- Ho AM, MacKay RK, Dodd PR, Lewohl JM. Association of polymorphisms in RGS4 and expression of RGS transcripts in the brains of human alcoholics. Brain Res. 2010;1340:1–9. doi: 10.1016/j.brainres.2010.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob GF, Volkow ND. Neurocircuitry of addiction. Neuropsychopharmacology. 2010;35:217–238. doi: 10.1038/npp.2009.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kril JJ, Halliday GM, Svoboda MD, Cartwright H. The cerebral cortex is damaged in chronic alcoholics. Neuroscience. 1997;79:983–998. doi: 10.1016/s0306-4522(97)00083-3. [DOI] [PubMed] [Google Scholar]
- Lee FJ, Liu F, Pristupa ZB, Niznik HB. Direct binding and functional coupling of α-synuclein to the dopamine transporters accelerate dopamine-induced apoptosis. The FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2001;15:916–926. doi: 10.1096/fj.00-0334com. [DOI] [PubMed] [Google Scholar]
- Lewohl JM, Crane DI, Dodd PR. Expression of the alpha 1, alpha 2 and alpha 3 isoforms of the GABAA receptor in human alcoholic brain. Brain Res. 1997a;751:102–112. doi: 10.1016/s0006-8993(96)01396-0. [DOI] [PubMed] [Google Scholar]
- Lewohl JM, Crane DI, Dodd PR. Zolpidem binding sites on the GABA(A) receptor in brain from human cirrhotic and non-cirrhotic alcoholics. Eur J Pharmacol. 1997b;326:265–272. doi: 10.1016/s0014-2999(97)85422-2. [DOI] [PubMed] [Google Scholar]
- Lewohl JM, Huygens F, Crane DI, Dodd PR. GABA(A) receptor alpha-subunit proteins in human chronic alcoholics. J Neurochem. 2001;78:424–434. doi: 10.1046/j.1471-4159.2001.00414.x. [DOI] [PubMed] [Google Scholar]
- Liang T, Kimpel MW, McClintick JN, Skillman AR, McCall K, Edenberg HJ, Carr LG. Candidate genes for alcohol preference identified by expression profiling in alcohol-preferring and -nonpreferring reciprocal congenic rats. Genome Biol. 2010;11:R11. doi: 10.1186/gb-2010-11-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang T, Spence J, Liu L, Strother WN, Chang HW, Ellison JA, Lumeng L, Li TK, Foroud T, Carr LG. α-Synuclein maps to a quantitative trait locus for alcohol preference and is differentially expressed in alcohol-preferring and -nonpreferring rats. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:4690–4695. doi: 10.1073/pnas.0737182100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnertz C, Saucier L, Ge D, Cronin KD, Burke JR, Browndyke JN, Hulette CM, Welsh-Bohmer KA, Chiba-Falek O. Genetic regulation of alpha-synuclein mRNA expression in various human brain tissues. PLoS One. 2009;4:e7480. doi: 10.1371/journal.pone.0007480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lotharius J, Barg S, Wiekop P, Lundberg C, Raymon HK, Brundin P. Effect of mutant α-synuclein on dopamine homeostasis in a new human mesencephalic cell line. Journal of Biological Chemistry. 2002;277:38884–38894. doi: 10.1074/jbc.M205518200. [DOI] [PubMed] [Google Scholar]
- MacKay RK, Colson N, Dodd PR, Lewohl JM. Differential expression of 14-3-3 isoforms in human alcoholic brain. Alcohol Clin Exp Res. 2011 doi: 10.1111/j.1530-0277.2011.01436.x. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield RD, Harris RA, Schuckit MA. Genetic factors influencing alcohol dependence. Br J Pharmacol. 2008;154:275–287. doi: 10.1038/bjp.2008.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy JJ, Linnertz C, Saucier L, Burke JR, Hulette CM, Welsh-Bohmer KA, Chiba-Falek O. The effect of SNCA 3′ region on the levels of SNCA-112 splicing variant. Neurogenetics. 2011;12:59–64. doi: 10.1007/s10048-010-0263-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez RG, Waymire JC, Lin E, Liu JJ, Guo F, Zigmond MJ. A role for α-synuclein in the regulation of dopamine biosynthesis. The Journal of Neuroscience. 2002;22:3090–3099. doi: 10.1523/JNEUROSCI.22-08-03090.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond M, Rousset F. GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. The Journal of Heredity. 1995;86:248–249. [Google Scholar]
- Reich T, Edenberg HJ, Goate A, Williams JT, Rice JP, Van Eerdewegh P, Foroud T, Hesselbrock V, Schuckit MA, Bucholz K, Porjesz B, Li TK, Conneally PM, Nurnberger JI, Jr, Tischfield JA, Crowe RR, Cloninger CR, Wu W, Shears S, Carr K, Crose C, Willig C, Begleiter H. Genome-wide search for genes affecting the risk for alcohol dependence. Am J Med Genet. 1998;81:207–215. [PubMed] [Google Scholar]
- Rideout HJ, Dietrich P, Savalle M, Dauer WT, Stefanis L. Regulation of α-synuclein by bFGF in cultured ventral midbrain dopaminergic neurons. Journal of Neurochemistry. 2003;84:803–813. doi: 10.1046/j.1471-4159.2003.01574.x. [DOI] [PubMed] [Google Scholar]
- Ridge JP, Ho AM-C, Innes DJ, Dodd PR. The expression of NMDA receptor subunit mRNA in human chronic alcoholics. Annals of the New York Academy of Sciences. 2008;1139:10–19. doi: 10.1196/annals.1432.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone NL, Kwon JM, Corbett J, Goate A, Rochberg N, Edenberg HJ, Foroud T, Li TK, Begleiter H, Reich T, Rice JP. A genome screen of maximum number of drinks as an alcoholism phenotype. Am J Med Genet. 2000;96:632–637. doi: 10.1002/1096-8628(20001009)96:5<632::aid-ajmg8>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Gel electrophoresis of DNA. In: Nolan C, editor. Molecular Cloning: A Laboratory Manual. Cold Spring Harbour Press; Cold Spring Harbour: 1989. [Google Scholar]
- Self DW, Nestler EJ. Relapse to drug-seeking: neural and molecular mechanisms. Drug and Alcohol Dependence. 1998;51:49–60. doi: 10.1016/s0376-8716(98)00065-9. [DOI] [PubMed] [Google Scholar]
- Sham PC, Curtis D. Monte Carlo tests for associations between disease and alleles at highly polymorphic loci. Annals of Human Genetics. 1995;59:97–105. doi: 10.1111/j.1469-1809.1995.tb01608.x. [DOI] [PubMed] [Google Scholar]
- Sidhu A, Wersinger C, Vernier P. Does alpha-synuclein modulate dopaminergic synaptic content and tone at the synapse? Faseb J. 2004;18:637–647. doi: 10.1096/fj.03-1112rev. [DOI] [PubMed] [Google Scholar]
- Touchman JW, Dehejia A, Chiba-Falek O, Cabin DE, Schwartz JR, Orrison BM, Polymeropoulos MH, Nussbaum RL. Human and mouse α-synuclein genes: comparative genomic sequence analysis and identification of a novel gene regulatory element. Genome Research. 2001;11:78–86. doi: 10.1101/gr.165801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. Global Status Report on Alcohol. Department of Mental Health and Substance Abuse, WHO; Geneva: 2004. [Google Scholar]
- Williams JT, Begleiter H, Porjesz B, Edenberg HJ, Foroud T, Reich T, Goate A, Van Eerdewegh P, Almasy L, Blangero J. Joint multipoint linkage analysis of multivariate qualitative and quantitative traits. II. Alcoholism and event-related potentials. Am J Hum Genet. 1999;65:1148–1160. doi: 10.1086/302571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N, Joshi PR, Cepeda C, Masliah E, Levine MS. α-Synuclein overexpression in mice alters synaptic communication in the corticostriatal pathway. Journal of Neuroscience Research. 2010;88:1764–1776. doi: 10.1002/jnr.22327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Rohan de Silva HA, Rosi BL, Yamaoka LH, Rimmler JB, Pericak-Vance MA, Roses AD, Chen X, Masliah E, DeTeresa R, Iwai A, Sundsmo M, Thomas RG, Hofstetter CR, Gregory E, Hansen LA, Katzman R, Thal LJ, Saitoh T. Genetic studies in Alzheimer’s disease with an NACP/α-synuclein polymorphism. Annals of Neurology. 1996;40:207–215. doi: 10.1002/ana.410400212. [DOI] [PubMed] [Google Scholar]


