Summary
Introduction
The cleavage stage mouse embryo is composed of superficially equivalent blastomeres that will generate both the embryonic inner cell mass (ICM) and the supportive trophectoderm (TE). However, it remains unsettled whether the contribution of each blastomere to these two lineages can be accounted for by chance. Addressing the question of blastomere cell fate may be of practical importance, as preimplantation genetic diagnosis (PGD) requires removal of blastomeres from the early human embryo. To determine if blastomere allocation to the two earliest lineages is random, we developed and utilized a recombination-mediated, non-invasive combinatorial fluorescent labeling method for embryonic lineage tracing.
Results
When we induced recombination at cleavage stages, we observed a statistically significant bias in the contribution of the resulting labeled clones to the trophectoderm or the inner cell mass in a subset of embryos. Surprisingly, we did not find a correlation between localization of clones in the embryonic and abembryonic hemispheres of the late blastocyst and their allocation to the TE and ICM, suggesting that TE-ICM bias arises separately from embryonic-abembryonic bias. Rainbow lineage tracing also allowed us to demonstrate that the bias observed in the blastocyst persists into post-implantation stages, and therefore has relevance for subsequent development.
Discussion
The Rainbow transgenic mice that we describe here have allowed us to detect lineage-dependent bias in early development. They should also enable assessment of the developmental equivalence of mammalian progenitor cells in a variety of tissues.
Introduction
During the cleavage stages of preimplantation development, the embryo undergoes serial cell divisions to produce 2, 4, and 8 seemingly identical cells dubbed blastomeres. After three additional cell divisions, the embryo will have formed a structure known as the blastocyst. The blastocyst consists of two distinct cell populations: the trophectoderm (TE), and the inner cell mass (ICM). The TE comprises the majority of the blastocyst and will become the placenta, while the ICM will give rise to the embryo proper and supportive tissues of the primitive endoderm. Although the embryonic blastomeres appear similar, it is a question of considerable interest whether each has an equal probability of giving rise to either the TE or ICM, or instead possesses an intrinsic lineage bias (reviewed in [1-2]).
Groundbreaking early studies using radioactive tracers and dyes showed that individual blastomeres have the potential to contribute to both the TE and ICM [3-4]. More recently, microinjection of a plasmid encoding Cre recombinase into single blastomeres of embryos containing a Cre-dependent lacZ reporter gene, found no apparent bias in contribution to different regions of the blastocyst [5]. However, distinct patterns of clone contribution to different tissues were observed in postimplantation embryos [5]. The methods employed in these early experiments could only be used to label one blastomere per embryo, and therefore, the interactions between multiple blastomere daughters could not be examined [3-4]. It has also been suggested that perturbations resulting from invasive labeling procedures could affect subsequent behavior of blastomere-derived daughter cells [6-7], preventing findings from being applicable to undisturbed embryos. For instance, an early observation that the earliest dividing 4-cell blastomere contributes disproportionately to the ICM [4] could not be confirmed in later experiments using live imaging [8].
Less-invasive markers, such as membrane-labeling dyes, and intrinsic features of the embryo [9-11], sometimes combined with time-lapse imaging [10, 12], have also been used to assess blastomere fate. These studies primarily focused on the contribution of 2-cell stage blastomere daughters to the embryonic region (Em) of the blastocyst, containing the ICM and the overlying polar TE or the abembryonic region (Ab), containing the mural TE surrounding the blastocoel cavity. In one study, the second polar body was observed to localize consistently to the Em-Ab boundary of the blastocyst, suggesting that different regions of the zygote might have distinct fates [9]. In another, the location of the sperm entry point was proposed to influence Em-Ab orientation of the blastocyst [6]. Consistent with these observations, transplantation of cytoplasm from the animal pole of the zygote to an ectopic location was observed to alter the orientation of the first cleavage division [11]. However, embryos lacking either animal or a vegetal cytoplasm are able to develop to term, suggesting that neither is necessary for development [13]. Furthermore, experiments utilizing time-lapse imaging indicated that bias might only occur in embryos with an intact zona pellucida (ZP), and disappears when the ZP is removed. Thus, bias could also result from extrinsic constraints rather than from intrinsic differences between the blastomeres [12, 14]. All these experiments necessitated pooling results from multiple embryos for statistical analysis, making it possible that a subpopulation of embryos displaying significant bias was not detected [6-7, 10-12, 14-16].
More recent studies have used time-lapse imaging of fluorescently labeled nuclei to track blastomere daughters to the early blastocyst stage and assess their Em-Ab contribution [8, 17-18]. Some observers suggested that certain orientations of cleavage division from the 2-cell stage to the 4-cell stage may produce blastomere daughters with bias to contribute to the Em region [8]. An independent experiment analyzing this relationship found that the observed bias could be eliminated by removal of the ZP, and may be a property of embryonic geometry as opposed to intrinsic differences between cells [17].
Subsequent studies focused on detecting intrinsic differences between blastomeres. Higher levels of histone 3 arginine 26 methylation (H3R26m) in 4-cell blastomeres have been proposed to be associated with an increased contribution of daughter cells to the ICM and polar TE [19-20]. Fluorescence decay after photoactivation also reveled differences between 4-cell blastomeres with regard to accessibility of nuclear DNA to binding by the ICM transcription factor Oct4 [21]. Blastomeres where Oct4 bound to DNA more strongly displayed a tendency to divide asymmetrically, resulting in their daughter cells contributing to the ICM [21-22].
Despite a large number of studies examining the issue of blastomere bias, several important questions remain to be addressed. It is still unclear whether the pattern of contribution of blastomere daughters to distinct regions of the blastocyst (ICM/TE or Em/Ab) can be accounted for by random allocation. Furthermore, the exact nature of the relationship between Em-Ab and TE-ICM contribution, if any, still remains to be determined. Finally, it is important to ascertain what relevance the bias observed at preimplantation stages has for the postimplantation embryo. To provide new insight into the fate of mouse blastomeres, we have developed and employed a non-invasive, heritable, multi-color lineage tracing strategy. Our finings provide new answers to each of these questions and are consistent with the proposal that blastomeres arising from certain cell division patterns may inherit epigenetic factors predisposing them either to an ICM or a TE cell fate.
Results
Generation and characterization of Rainbow mice
Multi-color lineage tracing was achieved through modification of a combinatorial fluorescent labeling method originally developed for tracing neuronal projections, called the Brainbow [23]. In Brainbow mice, stochastic action of Cre recombinase on sets of target (Lox) sites results in expression of varying combinations of fluorescent proteins, creating unique color labels for individual cells (Figure S1A). Greater numbers of Brainbow loci within the genome result in increased numbers of possible color combinations (Figure S1B).
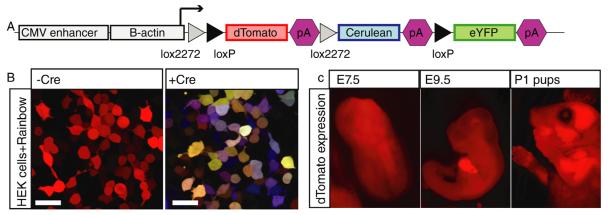
To allow multi-color lineage tracing in many organs at various stages of development, we constructed a transgene in which the broadly-expressed CAGGS promoter [24-26] was placed upstream of the brainbow1.0 construct [23](Figure 1A). After validating the multicolor labeling efficacy of our construct in a heterologous expression system (Figure 1B), we used pronuclear injection to generate transgenic mice. We obtained 3 founder animals with strong, ubiquitous red fluorescent protein (RFP) expression (Rainbow1-3; Figure 1C and Figure S2).
Figure 1. A mouse for Rainbow lineage tracing.
(A) The Rainbow construct. (B) Rainbow recombination after co-transfection with Cre into HEK cells. (C) dTomato expression at E6.5, E9.5, and in P1 Rainbow2 pups. See also Figure S1.
We observed robust expression of RFP in almost all tissues of adult rainbow2 animals (Figure S2A). Flow cytometry demonstrated that more than 80% of splenocytes and 90% of thymocytes, including B- and T-cells, also expressed RFP (Figure S2B). RFP was also observed in transgenic embryos at E3, E4.5, E6.5, E9.5, E14.5 and in neonates (Figure 1C, 2A, 3B).
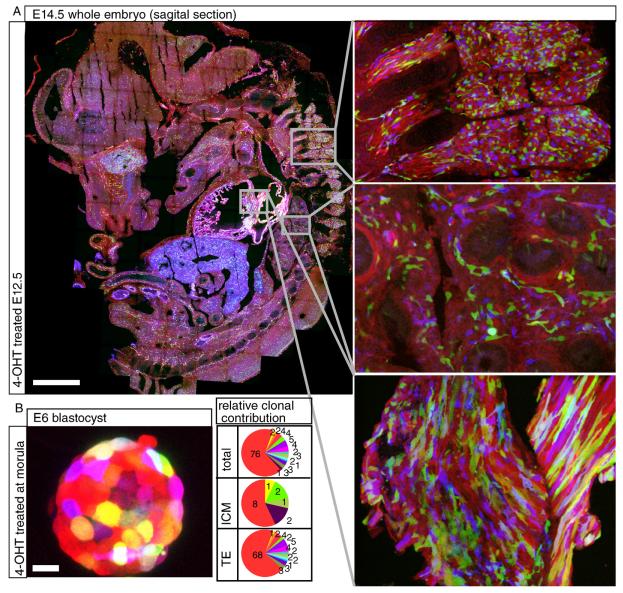
Figure 2. Expression and recombination of the Rainbow transgene.
(A) Sagital section through an E14.5 Rainbow3; CreER embryo that had been treated with 4-OHT at E12.5. Scale bar = 1 mm. Top inset: dorsal root ganglia. Middle inset: lung Bottom insert: heart muscle. (B) Wholemount confocal image of a Rainbow2; CreER blastocyst that had been treated with 4-OHT at the morula stage. Scale bar 20 μm. See also Figure S2.
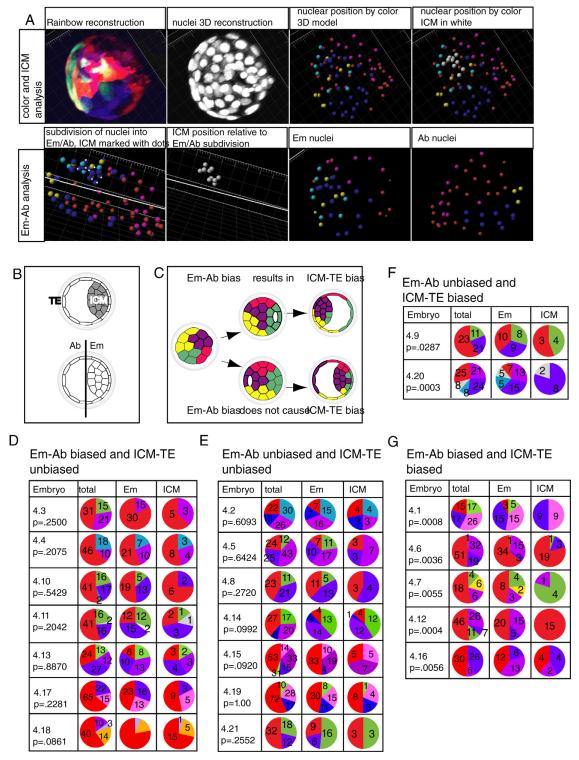
Figure 3. Skewed contribution of cleavage stage blastomeres to TE and ICM lineages.
(A) Experimental outline: Embryos were treated with 4-OHT at different stages of development, and contribution of each recombined clone to the ICM and TE was quantified. Images are confocal projections of whole-mount embryos (B) An untreated Rainbow2; CreER blastocyst, where only dTomato expression can be detected. The structure in the lower right corner is cells hatching from the ZP. (C-D) Representative Rainbow2; CreER embryos treated with 4-OHT at the 4-cell (C) and 8-cell (D) stages. Embryos are labeled as biased or unbiased. Scale bars 20 μm. P = Fisher probability value for each embryo. Pie charts represent proportional contribution of cells in each clone to the overall embryo, ICM, or TE. The number of cells in each population is listed on the corresponding slice of the pie chart. See also Figure S3 and Tables S1-S4.
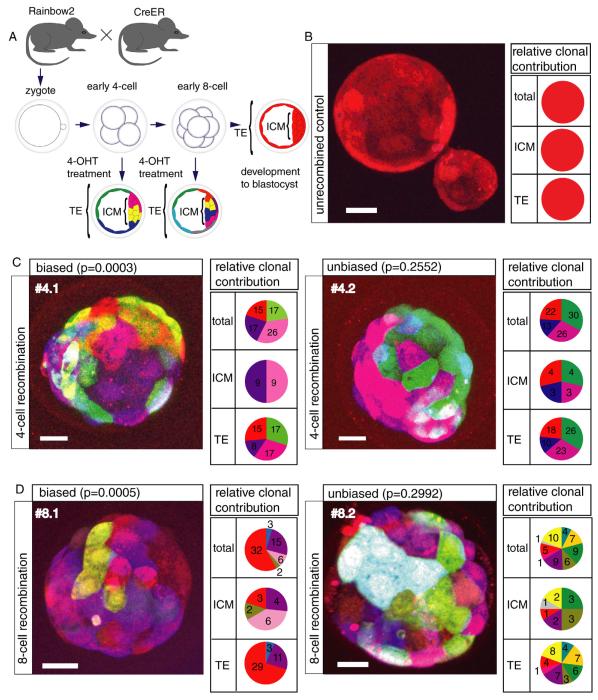
To test whether recombination could be induced in vivo, we crossed Rainbow animals to CAGGS:CreERTM2 (CreER) mice that allow for temporal control of Cre activity through exposure to 4-hydroxytamoxifen (4-OHT), which is the active metabolite of the drug tamoxifen [27]. When pregnant females were treated with tamoxifen at E13.5, and the embryos isolated and sectioned 2 days later, we observed many distinct colors in various tissues (Figure 2A).
To test recombination efficiency in preimplantation embryos, we isolated zygotes from rainbow2 females crossed to CreER males, cultured these embryos until the morula stage, exposed them to 4-OHT, and then quantified the number of colors in the resulting blastocysts (Figure 2B). Recombination was undetectable in the absence of CreER and extremely rare without 4-OHT exposure (4/1157 recombined cells in 12 blastocysts) (Figure 3B). In striking contrast, embryos treated with 4-OHT were observed to contain cells with many distinct color combinations (Figure 2B, 3C-D, Figure S3).
Redundant recombination events causing multiple cells to be labeled with the same color are a concern in these experiments as the progeny of such cells would be indistinguishable from each other. Increasing the number of possible color combinations would decrease the probability of such events. When we quantified the ratios of CFP, YFP, and RFP in rainbow2 embryos we observed at least 21 distinct color combinations within a single imaging session (as many as 27 total colors for all imaging sessions, Figure S1). For the rainbow3 and rainbow1 lines, 10 and 6 distinct color combinations could be observed, respectively. Although the utility of each Rainbow mouse line is likely to differ depending on experimental context, we selected rainbow2 for our experiments due to the large number of colors produced.
Results of recombination induction at the cleavage stages
We next tested whether the Rainbow system could be used to label blastomere daughters. We isolated Rainbow2; CreER zygotes and induced recombination with 4-OHT treatment at the 4-cell stage, 48 hours after administration of human chorionic gonadotrophin, (HCG). Following in vitro culture to the expanded blastocyst stage, confocal image analysis of the resulting blastocysts revealed groups of cells of distinct colors, which appeared to be clones (Figure 3C, Figure S3). When relative ratios of red, blue, and yellow fluorescence in each of these cells were plotted on a 3-dimensional scatterplot, they segregated into distinct groups, confirming the accuracy of color assignments (Figure S1 D).
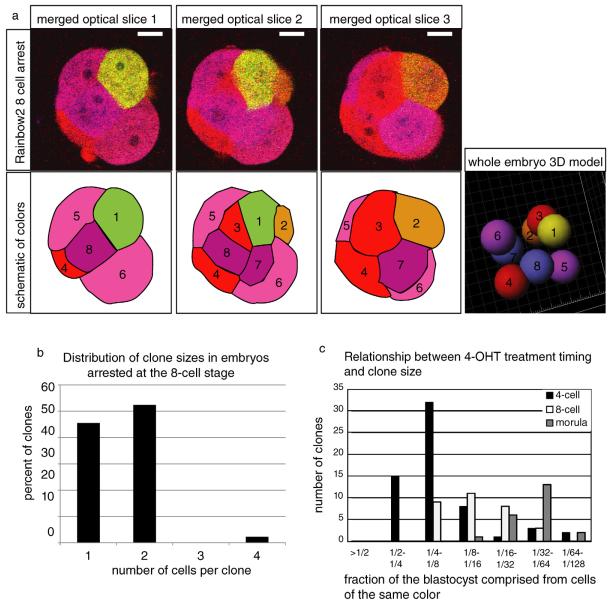
Most clones observed in these embryos comprised 1/4 or 1/8 of the blastocyst (24.8±6.3% of embryo for 24/33 clones; 11.0±2.0% for 7/33 clones; Figure 3C-D, 4C, Figure S3). Cell divisions in the preimplantation mouse embryo up to the 6th cleavage (when our analysis was performed) have been reported to be relatively synchronized [28-29]. Therefore, these clones likely arose as a result of recombination in 4- and 8-cell blastomeres. We tested this assumption by inducing recombination at the 4-cell stage and arresting the embryos at the 8-cell stage with the DNA synthesis inhibitor aphidicolin, to allow accumulation of fluorescent proteins (Figure 4A, B). When analyzed, 91% of the resulting 8-cell embryos (10/11) were observed to contain at least one pair of cells of the same color, indicating that many temporally stable 4-cell stage recombination events had occurred (Figure 4A, B).
Figure 4. Correlation between the timing of recombination and administration of 4-OHT.
(a) Representative confocal optical slices through a pharmacologically arrested 8-cell Rainbow2 embryo that had been treated with 4-OHT at the 4-cell stage. Slice thickness 3 μm, scale bars 20 μm (b) Distribution of clone sizes in embryos experimentally arrested at the 8-cell stage. N=44 clones. (c) Relationship between clone size at the blastocyst stage and timing of 4-OHT treatment. See also Figures S4 and S6 and Tables S3 and S4.
As we had already observed that clone sizes in embryos treated at the morula stage are considerably smaller than in those treated at the 4-cell stage (Figure 2, Figure S6), it seemed likely that the size of the clones observed in the blastocyst was dependent on the timing of 4-OHT treatment. To test this further, we next quantified clone sizes in blastocysts that had been treated with 4-OHT at the late 4-cell/early 8-cell stage (59 hours post HCG), (Figure 3A, D, Figure 4C, Figure S6B). This later exposure would be expected to label 8-cell and 16-cell stage blastomeres and therefore generate clone sizes intermediate in size between those obtained from morula and 4-cell treatments. Indeed, 12/31 clones comprised 12±2.5% of the total cells in the embryo, while 11/31 clones comprised 5.8±1.1%. Only 3/31 clones comprised 1/4 (on average 21.1±2.9%) of these blastocyst; Figure 3D and 4C; Figure S6). On average, these later clones comprised 9±6% of the blastocyst, as opposed to 21±11% for the 4-cell stage treatment. Thus, the contribution of these uniquely labeled clones to the blastocyst was consistent with most recombination events occurring during a cleavage division synchronous with the 4-OHT pulse (Figure 4C).
The above observations are all consistent with Rainbow colors functioning as independent markers indicative of blastomere lineage. Based on this, we formulated a statistical model designed to test for random allocation of labeled cells to the ICM and TE. If cells have no bias to contribute to either lineage, then each clone would be expected to comprise a portion of the ICM and TE that is proportional to that clone’s prevalence within the blastocyst. Under this model, some clones would be expected to contribute disproportionately to the TE simply by chance, as the number of cells in the ICM is much smaller than the number of cells in the TE. In contrast, significant and reproducible deviation from proportional contribution to the TE and ICM would indicate that daughters of different blastomeres are non-randomly allocated to these two lineages.
The null hypothesis in our analysis was that the probability that any given cell will contribute to the ICM is independent of color label. To test this null hypothesis, we used Fisher’s test, which is suitable for analysis of small samples. Non-recombined (red) cells were excluded from our analysis, as we could not readily determine their provenance. Importantly, we were able to assign cells to the TE and ICM with 99% accuracy (456/461 cells of 6 embryos, e.g. Figure S6).
We first used this statistical model to test the hypothesis that blastomere daughters are randomly allocated to the TE and ICM in embryos treated with 4-OHT at the 8-cell stage. We observed a statistically significant bias in 1 of 5 embryos examined (n=31 clones from 5 embryos; p<0.05 by Fisher’s test using independence of color to ICM contribution as the null hypothesis; Figure 3D, Figure S6 and Table S1).
As our analysis included several embryos, each of which represented a separate statistical test, we would expect to observe some bias simply by chance, as a result of testing multiple samples. Therefore, we performed a Monte-Carlo simulation to determine the likelihood that our results were obtained entirely by chance and to compute an aggregate p-value for the data set. The aggregate p-value as computed by Monte-Carlo simulation for embryos treated with 4-OHT at the 8-cell stage was 1.22×10−3. Thus, it is unlikely that bias we observed in this data set could be explained by chance, allowing us to reject the null hypothesis of independence between color labels and ICM contribution.
It is important to note that embryos containing dead blastomeres were excluded from our analysis (Figure S4C), and that the relative proportion of ICM to TE cells remained unchanged, even after prolonged 4-OHT treatment (Figure S4A-B). Thus, our observation of bias could not be explained by either selective death of blastomeres or biological effects of the 4-OHT pulse.
We next performed a similar analysis on embryos that had been treated with 4-OHT at the 4-cell stage. In this data set, we observed a significant deviation from random allocation in 30% of the embryos assayed (n=62 clones from 21 embryos; Figure 3C, Figure S3 and Table S2;). The aggregate p-value for this data set as determined by Monte-Carlo simulation was 3 × 10−9, indicating that our results were highly unlikely to be accounted for by chance. The increased proportion of biased embryos observed after 4-cell 4-OHT treatment, as opposed to the 8-cell 4-OHT treatment, is most likely a result of our enhanced ability to detect bias in the 4-cell treated data set. The earlier 4-OHT treatment produced larger clones, resulting in greater statistical power.
Importantly, embryos with detectable bias were comparable to unbiased embryos with regard to total number of cells (biased: 73±15, unbiased: 72±28), and the percentage of total cells that contributed to the ICM (biased:12%±4%, unbiased: 16%±3%). As we observed a large number of colors that occurred with approximately equal frequency (Tables S3 and 4, Figure S1) it is likely that most recombination events that we analyzed were unique. Thus our results suggest that many 4-cell embryos contain developmentally non-equivalent blastomeres, but previous studies indicate that they are unlikely to be lineage restricted[30], unlike progenitors in other eukaryotes, such as Drosophila imaginal disks[31].
Relationship between spatial localization of clones and their TE-ICM contribution
It has recently been proposed that differences between 4-cell blastomeres can result in distinct propensities to contribute to either Em, or Ab hemispheres of the blastocyst [7, 12, 15, 17, 28, 32-36]. While the ICM is by definition always in the Em hemisphere, Em-Ab localization is a description of spatial allocation, as opposed to lineage contribution (Figure 5B). Thus, analyzing the relationship between ICM-TE and Em-Ab localization of blastomere descendants could indicate whether the orientation of the blastocoel cavity is dependent on the localization of clones with bias to contribute to the ICM (Figure 5C, Figure S5C-F). If this were the case, we would expect to see a strong correlation between clone contribution to the ICM and the Em region of the blastocyst. In contrast, if ICM-TE bias is not a result of Em-Ab bias, we would expect that Em contribution and ICM contribution of labeled cells to be weakly, or uncorrelated (Figure 5C). When we analyzed the relationship between Em-Ab and TE-ICM contribution in blastocyst treated with 4-OHT at the 4-cell stage, the resulting correlations were either small or undetectable (Figure 5A and D-G, Figure S5C-F). Our results are consistent with a model where the orientation of the clone(s) biased to contribute to the ICM does not impact the orientation of the blastocoel cavity, at least at the expanded blastocyst stage (Figure 5C). It is therefore possible that Em contribution is independent from ICM contribution starting from the time that the embryo initiates cavitation. Alternately, morphological changes occurring during blastocyst development could weaken the relationship between Em and ICM contribution at later stages.
Figure 5. Comparison between embryonic-abembryonic contribution and TE-ICM bias.
(a) Analysis of contribution of clones to embryonic and abembryonic regions of a representative Rainbow embryo, showing colors, nuclei, designation of differently colored spots, designation of the ICM and the embryonic-abembryonic axis, and contribution of different clones to the embryonic and abembryonic halves of the embryo. (b) an illustration of the difference between TE/ICM and Em-AB designations (c) Model: Biased contribution of blastomeres to the ICM could be uncoupled from hemisphere bias if the blastocoel cavity forms independently of the location of a clone that preferentially contributes to the ICM. The number of cells in each population is listed on the corresponding slice of the pie chart. (d-g) A comparison of ICM versus embryonic contribution in embryos that had been treated with 4-OHT at the 4-cell stage. “Emb” indicates proportion of contribution to the embryonic hemisphere of the blastocyst. See also Figure S5.
In the three-dimensional reconstruction of Rainbow embryos treated with 4-OHT at the 4-cell stage, substantial mixing among cells of different colors was observed in the ICM. To determine whether adhesive or migratory properties of the daughters of different blastomeres account for variation in their ICM contribution, we attempted to detect a correlation between the amount of cell mixing and ICM contribution. The amount of cell mixing was determined based on the number of discrete discontinuous regions present in the each clone. No correlation between that measure and percent of clone contributing to the ICM was observed (R= −0.07), suggesting that differential clone adhesion and migration are not the major mechanism by which non-random lineage allocation occurs (Figure S5B).
Maintenance of blastomere bias post-implantation
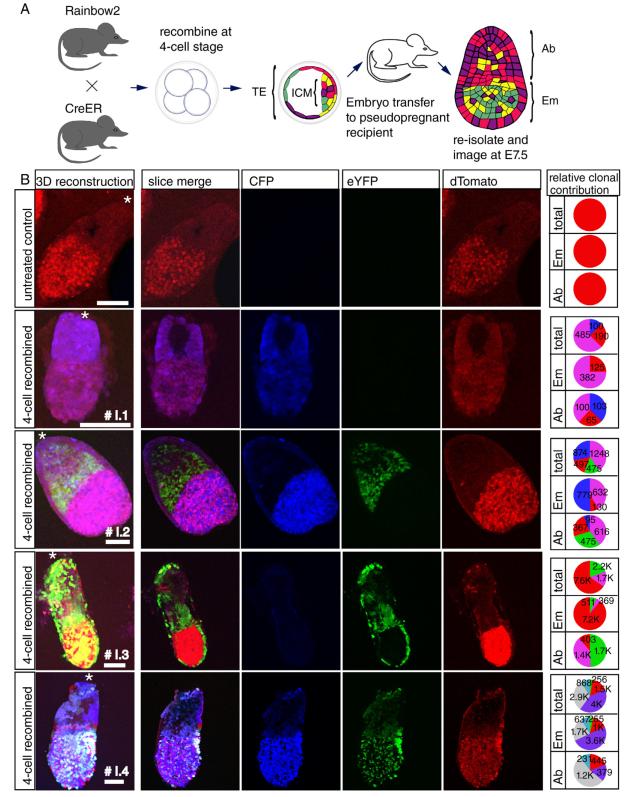
To analyze multiple clones post-implantation, we transferred blastocysts that had been pulsed with 4-OHT at the 4-cell stage into pseudopregnant recipient females. We re-isolated and imaged these embryos at E7.5. Three of four embryos contained 4 or fewer large clones of distinct colors, indicating that unique labeling of 4-cell blastomeres had likely occurred. A considerably smaller fifth clone in one embryo appeared to have arisen from a later recombination event (Figure 6B).
Figure 6. Preferential contribution of clones resulting from 4-OHT treatment at the 4-cell stage to different regions of postimplantation embryos.
(a) Experimental outline: Embryos were induced to undergo recombination at the 4-cell stage, transferred into pseudopregnant recipients, and re-isolated at E7.5 for analysis. (b) Slices and 3D reconstructions of resulting embryos show skewed contribution of clones to either embryonic or abembryonic lineages. Asterisks indicate location of ectoplacental cone. Scale bars=100 μm. The number of cells in each population is listed on the corresponding slice of the pie chart. See also Figure S6.
To test for bias in blastomere contribution to postimplantation embryos, we quantified the number of cells of each color in the postimplantation embryonic domain (primarily derived from the ICM) and the postembryonic abembryonic domain (primarily derived from the TE) of each embryo (Figure 6A). We observed clones that were highly skewed in their contribution to either the embryonic or abembryonic domains in N = 3 of 4 embryos (embryo I.1 blue clone, embryo I.2 green and blue clones, and embryo I.3 purple and green clones, Figure 6B). Other clones in these embryos did not appear biased (embryo I.1 purple clone, embryo I.2 purple clone, Figure 6B).
When 4-OHT treatment was performed at the late 8-cell stage, and these embryos were isolated and analyzed at E7.5, we also observed increased skewing in lineage contribution, including a clone that was specific to the epiblast (N=3 embryos, Figure S6C). Importantly, no recombination was detectable in control embryos that had not been exposed to 4-OHT (Figure 6B). Thus, biased contribution of clones to different lineages is detectable postimplantation.
Discussion
We have developed a multicolor lineage tracing method that can be used to assess the contribution of mammalian progenitor cells to many different tissues (Figure 2, S2). The Rainbow method, like all retrospective lineage tracing, suffers from limitations resulting from lack of prospective information, including a limited ability to detect ectopic and redundant recombination events. However, unlike single-color lineage tracing, our system enables analysis of many clones within the same lineage, as multiple recombination events to the same color would be expected to be rare. This property of the Rainbow mouse considerably increases the information content of the data set. Systems using 4-color labeling have proven to be useful in tissues with limited cell mixing [37-38]. However, stochastic labeling using 4 colors in our experiments would have resulted in a considerable number of cases where two or more blastomeres were labeled with the same color. For instance, in a system with completely stochastic recombination and 4 possible colors, the probability of obtaining two cells of the same color after 3 independent recombination events would be 87%. With 10, 20, or 30 possible colors this probability would decrease to 28%,15%, and 10% respectively. Thus, the nature of our experimental system necessitated the use of combinatorial multi-color labeling. The ability to control the rate of recombination by varying the concentration of 4-OHT, and the relatively synchronized nature of cell divisions during preimplantation development [28-29] allowed us to identify and eliminate embryos with redundant recombination events based on clone size.
Using the Rainbow, we examined the contribution of uniquely labeled clones derived from cleavage stage blastomeres to TE and ICM in individual embryos. We used statistical analysis to show that a subset of blastomere daughters display significant bias in contribution to either the TE or ICM. In a previous study [34], it was observed that 42% of mouse embryos within a population exhibited a division pattern that was associated with subsequent lineage bias of 4-cell blastomeres. Further analysis by the same group showed that certain blastomeres derived from these cell divisions exhibit distinct levels of H3R26me, which in turn was associated with a predisposition for contribution to either TE and or ICM [20]. While the correlation between cell division and TE-ICM contribution observed in these studies was indirect, it is interesting that in our experiments, a comparable 30% of embryos treated with 4-OHT at the 4-cell stage displayed significant bias. It is important to note that cleavage pattern frequencies may be variable depending on mouse strain used and other environmental factors. Therefore, it is not unreasonable to expect that the proportion of biased embryos observed would not be precisely identical between studies [8, 17-18, 10-12, 14-16]. The proportion of biased embryos observed in our study is therefore consistent with the proposal that certain orientations of cleavage divisions may confer lineage bias onto 4-cell blastomeres [19-20, 33-34]. It has yet to be determined how these variations in cleavage patterns result in epigenetic differences, such as varying levels of H3R26me [20, 35] and differing Oct4 DNA-binding dynamics at the 4-cell stage [21].
One striking finding from our study is the lack of detectable correlation between Em and ICM contribution in clones derived from 4-OHT treatment. The only significant relationship to emerge from our analysis was a weak negative correlation between the contribution of a given clone to the Ab region and its contribution to the ICM. This would be expected, as the Em region by definition contains the ICM (Figure 5B). However, if bias to contribute to the ICM were the result of preferential contribution to the Em region, we would also expect a strong positive correlation between ICM contribution and Em contribution. Such a correlation was not observed, suggesting that independent mechanisms may drive blastomere contribution to the Em and ICM at the expanded blastocyst stage. From a mathematical perspective, this lack of correlation is possible because the ICM comprises only a small portion of the Emb region of the embryo.
Our observation that only a small subset of clones contributes exclusively either to the Em or Ab regions of the blastocyst is similar to the results obtained from an earlier live imaging study [17], where it was observed that Em-Ab distribution of the daughters of 2-cell blastomeres was strongly influenced by the orientation of the ZP [17]. This interpretation has been contested[8]. The lack of correlation between Em and ICM contribution in our study suggests that the orientation of the ZP, even if it is a determinant the Em-Ab axis, cannot fully account for TE-ICM bias observed in the expanded blastocyst [17].
It may be possible that analyses of blastomere contribution to Em-Ab regions based on parameters different from nuclear position, such as the volume of cytoplasm, may lead to distinct findings. Alternately, if the orientation of the ICM within the blastocyst were to change as the blastocoel cavity expanded, an earlier correlation between ICM and Em contributions might become undetectable at later blastocyst stages, when our analysis was performed. The mechanisms that generate various patterns of TE-ICM and Em-Ab contribution at different stages of development would need to be examined more carefully, in order to fully understand the mixing that blastocyst lineages undergo during blastocoel expansion.
The genetic labeling employed by the Rainbow system allowed us to confirm a prior observation that 4-cell blastomeres display skewed contribution to embryonic and abembryonic lineages post-implantation [5]. In addition, we were able to demonstrate that individual embryos can contain blastomeres exhibiting biased and unbiased contribution patterns to the TE, ICM and their derivatives. It would be important to elucidate the consequences that this bias, and the epigenetic differences underlying it, could have for later development. For instance, the epigenetic differences between daughters of different blastomeres could also influence their survival in the postimplantation embryo, and skew contribution to various fetal lineages. As studies of blastomere non-equivalence advance into post-implantation development, there is an opportunity to better understand effects of blastomere bias on the resulting fetus. A better understanding of the impact of blastomere bias could in turn help improve outcomes for patients undergoing fertility treatments or the efficiency of embryonic development following PGD.
Experimental Procedures
The Rainbow construct was generated by ligating the brainbow1.0 construct into the pCAGEN plasmid (Adgene). Mice were generated by pronuclear injection with linearized Rainbow plasmid into BDF2 embryos at the Harvard Genome Modification facility. To isolate preimplantation embryos, mice were superovulated with HCG and PMS, and mated to males. Embryos were dissected from oviducts in HCZB media, and cultured in KSOM media under mineral oil at 5% CO2, 37°C. To induce recombination at preimplantation stages, embryos were pulsed for 6 hrs with 0.2 μM 4-OHT (Sigma). For aphidicolin arrest, embryos were cultured in KSOM supplemented with 0.05 μg/mL aphidicolin. For imaging, embryos were mounted in HCZB supplemented with 20 μM DRAQ5 far-red nuclear dye. Embryos were imaged live wholemount on Olympus FV1000 and Zeiss LSM710 microscopes with 440, 515, 568, and 633 lasers. For cell membrane staining, zona-denuded blastocysts were incubated overnight in Vybrant DiO cell labeling solution (Invitrogen), diluted 1:100 in KSOM at 37°C, 5% CO2. For postimplantation analysis, E3.5 blastocysts were transferred into uteri of pseudopregnant females 2.5 dpc. Embryos were dissected from the uterus at E7.5, fixed in 4% paraformaldehyde, and imaged. For recombination at E13.5, 2.5 mg Tamoxifen was administered orally to Rainbow3 females previously mated to CAGG-CreER males. Embryos were recovered 2 days later, cryosectioned and imaged. Most image analysis was performed using the Imaris 6.0 image processing software (Bitplane). Nuclei were detected by far-red staining, and independently assigned a color and a position within the blastocyst. Image J was used for quantification of relative fluorescence intensities. For statistical analysis, each embryo was studied as a contingency table with two outcomes but potentially varied counts of differentiable colors. Since not all clones were large enough to support a Pearson’s Chi-squared tests, a Fisher’s p-value was calculated using R-function fisher.test() to test against the hypothesis of independence between color and outcome. Pearson’s r values for embryonic-ICM correlation were calculated using Microsoft Excel.
Supplementary Material
Highlights.
Mouse lines for Rainbow lineage tracing in many tissues and developmental stages.
A fraction of blastocysts exhibited biased clone contribution to the TE/ICM
TE/ICM bias was uncorrelated with embryonic/abembryonic distribution of clones
Postimplantation embryos had skewed contribution of clones to distinct lineages
Acknowledgements
We thank Harvard GMF and A. McMahon for providing transgenic animals and D. Smith, S. Turney, G. Birkhoff, S. Rompani, and S. Fouquet for technical assistance. This work was supported by NICHD grant #HD045732-03, and NIGMS P01 grant GM099117 to K.E., and HHMI.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Takaoka K, Hamada H. Cell fate decisions and axis determination in the early mouse embryo. Development. 2012;139:3–14. doi: 10.1242/dev.060095. [DOI] [PubMed] [Google Scholar]
- 2.Yamanaka Y, Ralston A, Stephenson RO, Rossant J. Cell and molecular regulation of the mouse blastocyst. Dev Dyn. 2006;235:2301–2314. doi: 10.1002/dvdy.20844. [DOI] [PubMed] [Google Scholar]
- 3.Balakier H, Pedersen RA. Allocation of cells to inner cell mass and trophectoderm lineages in preimplantation mouse embryos. Dev Biol. 1982;90:352–362. doi: 10.1016/0012-1606(82)90384-0. [DOI] [PubMed] [Google Scholar]
- 4.Kelly SJ, Mulnard JG, Graham CF. Cell division and cell allocation in early mouse development. J Embryol Exp Morphol. 1978;48:37–51. [PubMed] [Google Scholar]
- 5.Fujimori T, Kurotaki Y, Miyazaki J, Nabeshima Y. Analysis of cell lineage in two- and four-cell mouse embryos. Development. 2003;130:5113–5122. doi: 10.1242/dev.00725. [DOI] [PubMed] [Google Scholar]
- 6.Piotrowska K, Wianny F, Pedersen RA, Zernicka-Goetz M. Blastomeres arising from the first cleavage division have distinguishable fates in normal mouse development. Development. 2001;128:3739–3748. doi: 10.1242/dev.128.19.3739. [DOI] [PubMed] [Google Scholar]
- 7.Alarcon VB, Marikawa Y. Deviation of the blastocyst axis from the first cleavage plane does not affect the quality of mouse postimplantation development. Biol Reprod. 2003;69:1208–1212. doi: 10.1095/biolreprod.103.018283. [DOI] [PubMed] [Google Scholar]
- 8.Bischoff M, Parfitt DE, Zernicka-Goetz M. Formation of the embryonic-abembryonic axis of the mouse blastocyst: relationships between orientation of early cleavage divisions and pattern of symmetric/asymmetric divisions. Development. 2008;135:953–962. doi: 10.1242/dev.014316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gardner RL. The early blastocyst is bilaterally symmetrical and its axis of symmetry is aligned with the animal-vegetal axis of the zygote in the mouse. Development. 1997;124:289–301. doi: 10.1242/dev.124.2.289. [DOI] [PubMed] [Google Scholar]
- 10.Hiiragi T, Solter D. First cleavage plane of the mouse egg is not predetermined but defined by the topology of the two apposing pronuclei. Nature. 2004;430:360–364. doi: 10.1038/nature02595. [DOI] [PubMed] [Google Scholar]
- 11.Plusa B, Grabarek JB, Piotrowska K, Glover DM, Zernicka-Goetz M. Site of the previous meiotic division defines cleavage orientation in the mouse embryo. Nat Cell Biol. 2002;4:811–815. doi: 10.1038/ncb860. [DOI] [PubMed] [Google Scholar]
- 12.Motosugi N, Bauer T, Polanski Z, Solter D, Hiiragi T. Polarity of the mouse embryo is established at blastocyst and is not prepatterned. Genes Dev. 2005;19:1081–1092. doi: 10.1101/gad.1304805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zernicka-Goetz M. Fertile offspring derived from mammalian eggs lacking either animal or vegetal poles. Development. 1998;125:4803–4808. doi: 10.1242/dev.125.23.4803. [DOI] [PubMed] [Google Scholar]
- 14.Motosugi N, Dietrich JE, Polanski Z, Solter D, Hiiragi T. Space asymmetry directs preferential sperm entry in the absence of polarity in the mouse oocyte. PLoS Biol. 2006;4:e135. doi: 10.1371/journal.pbio.0040135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alarcon VB, Marikawa Y. Unbiased contribution of the first two blastomeres to mouse blastocyst development. Mol Reprod Dev. 2005;72:354–361. doi: 10.1002/mrd.20353. [DOI] [PubMed] [Google Scholar]
- 16.Hiiragi T, Solter D. Fatal flaws in the case for prepatterning in the mouse egg. Reprod Biomed Online. 2006;12:150–152. doi: 10.1016/s1472-6483(10)60854-1. [DOI] [PubMed] [Google Scholar]
- 17.Kurotaki Y, Hatta K, Nakao K, Nabeshima Y, Fujimori T. Blastocyst axis is specified independently of early cell lineage but aligns with the ZP shape. Science. 2007;316:719–723. doi: 10.1126/science.1138591. [DOI] [PubMed] [Google Scholar]
- 18.Morris SA, Teo RT, Li H, Robson P, Glover DM, Zernicka-Goetz M. Origin and formation of the first two distinct cell types of the inner cell mass in the mouse embryo. Proc Natl Acad Sci U S A. 2010;107:6364–6369. doi: 10.1073/pnas.0915063107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parfitt DE, Zernicka-Goetz M. Epigenetic modification affecting expression of cell polarity and cell fate genes to regulate lineage specification in the early mouse embryo. Mol Biol Cell. 2010;21:2649–2660. doi: 10.1091/mbc.E10-01-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Torres-Padilla ME, Parfitt DE, Kouzarides T, Zernicka-Goetz M. Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature. 2007;445:214–218. doi: 10.1038/nature05458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Plachta N, Bollenbach T, Pease S, Fraser SE, Pantazis P. Oct4 kinetics predict cell lineage patterning in the early mammalian embryo. Nat Cell Biol. 2011;13:117–123. doi: 10.1038/ncb2154. [DOI] [PubMed] [Google Scholar]
- 22.Fleming TP. A quantitative analysis of cell allocation to trophectoderm and inner cell mass in the mouse blastocyst. Dev Biol. 1987;119:520–531. doi: 10.1016/0012-1606(87)90055-8. [DOI] [PubMed] [Google Scholar]
- 23.Livet J, Weissman TA, Kang H, Draft RW, Lu J, Bennis RA, Sanes JR, Lichtman JW. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450:56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- 24.Lobe CG, Koop KE, Kreppner W, Lomeli H, Gertsenstein M, Nagy A. Z/AP, a double reporter for cre-mediated recombination. Dev Biol. 1999;208:281–292. doi: 10.1006/dbio.1999.9209. [DOI] [PubMed] [Google Scholar]
- 25.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 26.Pratt T, Sharp L, Nichols J, Price DJ, Mason JO. Embryonic stem cells and transgenic mice ubiquitously expressing a tau-tagged green fluorescent protein. Dev Biol. 2000;228:19–28. doi: 10.1006/dbio.2000.9935. [DOI] [PubMed] [Google Scholar]
- 27.Guo C, Yang W, Lobe CG. A Cre recombinase transgene with mosaic, widespread tamoxifen-inducible action. Genesis. 2002;32:8–18. doi: 10.1002/gene.10021. [DOI] [PubMed] [Google Scholar]
- 28.Jedrusik A, Parfitt DE, Guo G, Skamagki M, Grabarek JB, Johnson MH, Robson P, Zernicka-Goetz M. Role of Cdx2 and cell polarity in cell allocation and specification of trophectoderm and inner cell mass in the mouse embryo. Genes Dev. 2008;22:2692–2706. doi: 10.1101/gad.486108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson MH, McConnell JM. Lineage allocation and cell polarity during mouse embryogenesis. Semin Cell Dev Biol. 2004;15:583–597. doi: 10.1016/j.semcdb.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 30.Tarkowski AK, Wroblewska J. Development of blastomeres of mouse eggs isolated at the 4- and 8-cell stage. J Embryol Exp Morphol. 1967;18:155–180. [PubMed] [Google Scholar]
- 31.Vincent JP, Girdham CH, O’Farrell PH. A cell-autonomous, ubiquitous marker for the analysis of Drosophila genetic mosaics. Dev Biol. 1994;164:328–331. doi: 10.1006/dbio.1994.1203. [DOI] [PubMed] [Google Scholar]
- 32.Louvet-Vallee S, Vinot S, Maro B. Mitotic spindles and cleavage planes are oriented randomly in the two-cell mouse embryo. Curr Biol. 2005;15:464–469. doi: 10.1016/j.cub.2004.12.078. [DOI] [PubMed] [Google Scholar]
- 33.Piotrowska-Nitsche K, Perea-Gomez A, Haraguchi S, Zernicka-Goetz M. Four-cell stage mouse blastomeres have different developmental properties. Development. 2005;132:479–490. doi: 10.1242/dev.01602. [DOI] [PubMed] [Google Scholar]
- 34.Piotrowska-Nitsche K, Zernicka-Goetz M. Spatial arrangement of individual 4-cell stage blastomeres and the order in which they are generated correlate with blastocyst pattern in the mouse embryo. Mech Dev. 2005;122:487–500. doi: 10.1016/j.mod.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 35.Plusa B, Hadjantonakis AK, Gray D, Piotrowska-Nitsche K, Jedrusik A, Papaioannou VE, Glover DM, Zernicka-Goetz M. The first cleavage of the mouse zygote predicts the blastocyst axis. Nature. 2005;434:391–395. doi: 10.1038/nature03388. [DOI] [PubMed] [Google Scholar]
- 36.Rossant J, Tam PP. Blastocyst lineage formation, early embryonic asymmetries and axis patterning in the mouse. Development. 2009;136:701–713. doi: 10.1242/dev.017178. [DOI] [PubMed] [Google Scholar]
- 37.Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, Barker N, Klein AM, van Rheenen J, Simons BD, et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 38.Rinkevich Y, Lindau P, Ueno H, Longaker MT, Weissman IL. Germ-layer and lineage-restricted stem/progenitors regenerate the mouse digit tip. Nature. 2011;476:409–413. doi: 10.1038/nature10346. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.