Abstract
The paper presents some novel approaches to the empirical analysis of diversity and similarity (overlap) in biological or ecological systems. The analysis is motivated by the molecular studies of highly diverse mammalian T-cell receptor (TCR) populations, and is related to the classical statistical problem of analyzing two-way contingency tables with missing cells and low cell counts. The new measures of diversity and overlap are proposed, based on the information-theoretic as well as geometric considerations, with the capacity to naturally up-weight or down-weight the rare and abundant population species. The consistent estimates are derived by applying the Good-Turing sample-coverage correction. In particular, novel consistent estimates of the Shannon entropy function and the Morisita-Horn index are provided. Data from TCR populations in mice are used to illustrate the empirical performance of the proposed methods vis a vis the existing alternatives.
Keywords: Contingency tables, antigen receptors, richness and diversity estimation, Renyi’s entropy, Renyi’s divergence
1 Introduction
The recent successes of the Panvax study (see, e.g. Mohebtash et al 2011), have invigorated the scientific efforts to obtain a vertebrate cancer vaccine and, consequently, reignited the interest in systematic analysis of T-cell populations. In vertebrates, T-cell populations are typically analyzed in terms of their capacities to recognize the so-called antibody generating molecules or antigens. An antigen is a foreign molecule which, when introduced into the body of a vertebrate, triggers the antibody production by the immune system. This immune system response is initiated when T-cells recognize and respond to antigens via their T-cell receptors (TCRs). TCRs are heterodimer proteins with two chains: α and β in αβ T-cells and γ and δ in γδ T-cells. The genes encoding these proteins are generated by the so-called V(D)J DNA recombination during thymic T-cell development. In this process, T-cell precursors randomly recombine different V, D, and J gene segments and assemble the mature gene encoding a TCR chain. By enumeration of all such possible recombinations alone, one concludes that there are 1018 distinct TCR chains in humans (Janeway, 2005) and 1015 in mice (Davis and Bjorkman, 1988). The experimentally observed numbers of different recombinations seem to confirm this order of magnitude (Arstila et al, 1999; Memon et al, 2012). In the presence of such a large number of different antigen receptor chain types, the statistical analysis of the samples obtained from different TCR populations presents a formidable challenge, due to the unavoidable issue of chain types under-sampling, even with the use of modern high-hroughput methods of TCR data collection (Salameire et al, 2009; Van Den Berg et al, 2011).
Generally, the data consisting of several samples from different TCR populations may be broadly characterized as an empirical two-way contingency table with columns representing different population samples and rows representing different chain types (referred to as TCR species below). In such table the data under-sampling is reflected in the low observed cell counts and an unknown total number of non-empty rows (species). The descriptive summary of a single column (TCR population) in the table is typically based on the notion of a diversity, whereas the descriptive comparison of two or more columns relies on the concepts of either a pairwise or multi-way overlap or similarity. The choices of the appropriate measures of diversity and overlap are fundamental for summarizing and analyzing TCR data with proper accounting for the uncertainty caused by the TCR under-sampling. Unfortunately, the under-sampling issue seems to be largely ignored in most TCR studies (Hsieh et al, 2012) with little discussion of the possible effects of the under-sampling bias on the data analysis results. Indeed, in most TCR studies the statistical methodology is borrowed from the field of macro-ecological systems (see, e.g., Baum and McCune 2006), where the under-sampling problem is not as severe. Consequently, the ecological indices applied to TCR data tend to under-report the true size of the repertoires, possibly distorting the true relations between T-cell populations (Gras et al, 2008).
In order to address this problem, the current paper proposes a new mathematical and statistical framework which naturally incorporates the under-sampling uncertainty into analyzing TCR populations, by means of appropriately weighting the empirical species counts. Our framework combines the information theoretic ideas for measuring diversity and overlap with the statistical approaches of adjusting the empirical (plug-in) estimates for the under-counting rare species in populations. Consequently, the estimators proposed here incorporate the empirically observed abundance patterns in order to quantify and compare different TCR populations. For the diversity analysis, our approach specializes in some specific cases to the earlier proposed methods of Chao and Shen (2003) and Vu et al (2007), combining the empirical Shannon entropy with the so-called Horvitz-Thompson and the Good-Turing coverage corrections (cf. Section 2 below). For the overlap analysis, in the contingency table framework described above, our method may be viewed as an extension of the two-way mutual information (Kullback-Leibler) statistic or the Peason chi-square statistic. In addition to the information-based measures, we also consider here some geometric ones, like e.g. the extended Morisita-Horn index. Whereas our results are motivated by specific examples of TCR data, they are readily applicable also to a more classical analysis of two-way tables (see, for example, the standard reference text by Agresti 2002), whenever the issues of low cell counts or under-sampling are of concern.
The paper is organized as follows. The remainder of the current section briefly reviews the basic concepts related to biodiversity and comparison of finite populations, focusing especially on the entropy-based measures applicable to TCR data. We discuss in particular the concepts of diversity and diversity measure as well as an effective number of species and the similarity (overlap) between pairs of populations. In Section 2 we discuss the sample-adjusted methods based on the notion of a sample coverage, as well as state the consistency results (in Theorems 1,2 and 4) for the proposed estimates. In Section 3 we illustrate the ideas developed in Section 2 via analyzing data from a recent mouse TCR study. Section 4 contains conclusions and summary of our main points. The proofs of the consistency results and some data-related figures are provided in the Appendix.
Throughout the paper, when appropriate, we consider the contingency table model of TCR data arranged into a two-way (m n) table [cij], with columns representing n different populations of T-cells and rows representing m antigen receptor types (species). In statistical terms, we consider therefore n independent multinomial distributions p1 = (c11/∑ ci,…cm,/∑ci1),… pn = (c1,n/∑ ci,n,…,cm,n/∑ cin)with the union of their supports being over m < ∞ points. We denote by um the vector of uniform probabilities on the set {1,2,…m} and by Δm–1 the probability simplex in . The summation symbol ∑ used without index indicates here and elsewhere that the summation is with respect to the subscript i.
1.1 Diversity Measures
In the ecological literature, the term ‘diversity’ typically means the ordered abundance of population species. Despite the fact that this meaning is not completely universal (see, e.g. Spellerberg and Fedor 2003) we adapt it for the purpose of current discussion. Formally, consider a set of m < ∞ species (TCRs) and a population c = (c1,…, cm) . Following Valiant (2008), we have the following.
Definition 1
For a given population c = (c1,…,cm) of m species (TCRs), its diversity or fingerprint is the vector Fc = (v1,…, vmaxi ci) where vk = |{i : ci = k}|. Any nonnegative, real function with values D(Fc) is called a measure of diversity or an index of diversity.
Since the dimension of the fingerprint Fc varies, it is convenient to define the function D on the set of all non-negative infinite sequences of natural numbers, with some additional constraints allowing for partial ordering (see next subsection). Such constraints may be formalized via the following definition of index monotonicity which we shall need later on. Let and note that corresponds to a vector with v1 = m and vi = 0 for i > 1. In this notation, we have
Definition 2
The diversity index D is called monotone if is nondecreasing in m.
Many nonparametric measures of diversity considered in ecological literature are rooted in the information theory, see e.g. Tóthmérész (1995); Ricotta (2005); Keylock (2005). Probably the best known example of a monotone diversity index is the (Shannon) entropy function H1. This index has an appealing property that, for given m, the diversity of all normalized populations p ∈ Δm–1 is maximized by the uniform vector um ∈ Δm–1 and that D(um) = D.
Example 1 (Shannon’s entropy)
For any population c with a fingerprint Fc, define its Shannon entropy diversity index by
Alternatively, in terms of the normalized population p ∈ Δm–1, we have
| (1.1) |
Note that H1 is monotone, since H1 = logm, and that H1(p) ≤ log m.
A useful extension of the above example, which is of interest in the following sections, is the so-called Renyi entropy (Rényi, 1961).
Example 2 (Renyi’s entropy)
The Renyi entropy of order α ∈ [0,∞] is given by
| (1.2) |
for p ∈ Δm–1, with the limiting cases of interest H0(p) = logm, H1(p) = –∑pi log pi and H∞(p) = –log(maxi pi).
The case α = 2 in the above example is sometimes known as the Rao quadratic entropy (Nayak, 1986) with the quantity SI(p) = exp(–H2(p)) known as the Simpson index (Keylock, 2005). Although the Simpson index is not monotone in the sense of our Definition 2, it is easy to see that its inverse ISI(p) = exp(H2(p)) has that property. For that reason, it is sometimes more convenient to consider ISI instead of SI.
It is clear that the Renyi entropy of order α < 1 puts more weight on the rare species (rare TCRs) and the Renyi entropy of order α > 1 puts more weight on the abundant ones. As discussed e.g. in Tóthmérész (1995), it is often natural to analyze the overall population diversity as a function of the parameter α (the so-called diversity profile of a population). Since the profile considers an entire class of indices of differently weighted abundances, it provides more extensive information than a single index.
In addition to diversity profiling, one may also consider a Horvitz-Thompson-type correction for under-sampling. The idea was first proposed by Chao and Shen (2003) for the Shannon entropy H1, but it naturally extends to the Renyi entropy index given in the previous example and motivates the following.
Example 3
A more general class of measures of diversity based on (1.2), which incorporates the Horvitz-Thompson-type of adjustment with the sample of size n is
| (1.3) |
where α ∈ [0, 1) ∪ (1, ∞]. Note that (p) in not defined, and that we may take = Hα, where Hα is given by (1.2).
1.1.1 Effective Number of Species
Different monotone diversity indices may be compared with each other by applying the concept of an effective number of species or ENS, introduced by Jost (2006). We state a precise definition below and note that, unlike the actual number of species, the ENS may take non-integer values.
Definition 3
(ENS) Let c be an arbitrary population and D a monotone diversity measure. For any y of the form y = m + α (0 ≤ α ≤ 1) define D := (1 – α)D(Fm) + αD(Fm+1). The effective number of species for the pair (c,D) is the smallest solution y = y0 of the equation
Except for the populations with the uniform profile , the effective number of species is typically less than the number of species m. A simple example follows.
Example 4
For D = Hα the effective number of species (say, k) in a population p ∈ is given by the Hill number kα = for integer k, and by the linear interpolation between the values of kα otherwise. It follows, in particular, that for D = and n sufficiently large, we have kα ≈ exp ( (p)).
The limiting case α = 1 in the above example was suggested in Jost (2006).
The concept of diversity and effective number of species is useful in characterizing a single population, however, in order to compare two or more populations one takes a different approach, based on the idea of an overlap (or similarity) measure, which is discussed next.
1.2 Overlap Measures
The two-way and multi-way TCR repertoire comparisons are of interest whenever the data from multiple TCR repertoires are collected. The standard methods used for such comparisons (Chen et al, 2003; Komatsu et al, 2009; Pacholczyk et al, 2007, 2006) rely on calculating species overlap indices. For the purpose of current discussion, we define the concept of an overlap and an overlap measure as follows.
Definition 4
Consider n populations c1, c2,…cn, each with at most m species, so that ci ∈ for i = 1…,n. Let supp(ci) denote the support of ci. The overlap between vectors c1,…,cn is then Sn = supp(ck). Any function such that shall be called an overlap measure or an overlap index.
There has been a large number of different measures of overlap proposed in the ecological and social networks literature over last 50 years. Perhaps the two oldest and most widely used overlap indices are the Jaccard index and the Sørensen index.
Example 5 (Jaccard and Sørensen indices)
For the pairs of populations (c1, c2) ∈ the Jaccard index (J) of similarity and the closely related Sørensen index (L) are defined as follows
Both J and L indices, as well as their various modifications, seem to be widely used and accepted in both the ecological and immunological literature since their introduction in the late 40′s (see, e.g., Chao et al 2005; Hsieh et al 2006; Chen et al 2003; Komatsu et al 2009; Staveley-O’Carroll et al 1998; Butz and Bevan 1998).
In the modern theory of contingency tables, measuring overlap often relies on the information-based criteria (the standard mutual information statistic being an example). In this paper, we find it particularly useful to consider the following Renyi divergence measure, which is also of interest in the context of independence testing in two-way tables (see, e.g. Agresti 2002).
Example 6 (Renyi divergence)
For a pair of normalized populations (p1, p2) ∈ Δm–1 × Δm–1, their Renyi divergence of order α ∈ [0,∞] is given by
Note that in the limiting cases we have , which is the Kullback-Leibler divergence, and .
An alternative family of overlap indices may be derived geometrically, based on an angle (or any appropriate angular measure) between two population vectors in . The greater the angle, the more dissimilar (less overlapping) two populations tend to be. One of the more popular geometric angular measures is the Morisita-Horn index (Magurran, 2005), which gives the cosine of an angle between a pair of standardized population vectors.
Example 7 (Morisita-Horn index and Bhattacharyya’s coefficient)
Formally, the Morisita-Horn index (MH) between a pair of population vectors is defined as
or, more succinctly, in terms of the inner products of the normalized populations p1, p2,
MH index has the property that it is non-negative and bounded by unity, attaining its minimum/maximum when c1⊥c2 and c1 = c2, respectively. Unfortunately, it also suffers from being overly sensitive to the high abundance components (frequent species) of c1 and c2. For that reason, in populations with prevalent low abundances (rare species), it is often more suitable to use a different index, known as the Bhattacharyya (BC) coefficient, defined as the cosine of an angle between the vectors and , i.e.
Note from Example 6 that we have the relation .
1.2.1 PG Index
It is straightforward to extend the ideas presented in Example 7 to a general geometric index parametrized by two nonnegative parameters, and therefore able to put weight on rare (resp. abundant) receptors in a more flexible way. We refer to it a power-geometric or PG index.
Example 8 (PG index)
For any pair (p1, p2) ∈ Δm–1 × Δm–1 and α, β ∈ (0,∞) its PG index of overlap is defined as
| (1.4) |
The PG index extends both the MH and BC indices above, as it gives the cosine of an angle between the vectors pα := and . When α < 1 and β < 1, the PG index is less affected than the Morisita-Horn index by the overlap of the most abundant species, whereas the opposite is true when α > 1 and β > 1. It follows that, similarly to the Renyi entropy, the PG index puts more weight on rare or abundant species, depending on the values of the parameters α, β. We study the properties of estimates based on the PG index in the next section.
In analogy with a diversity profile, we refer to the function α → PGα,α as an overlap profile or a similarity profile. Following the idea of the adjusted Renyi entropy of Example 3, we also define the Horvitz-Thompson adjusted PG index as
with . Note that PG1,1 is simply the Morisita-Horn index in Example 7. In the following sections, when it is not ambiguous, we sometimes also write PG(α,β) for PGα,β.
2 Sample Adjusted Estimates
As discussed earlier, in the context of TCR populations the issue of under-sampling bias may be particularly severe due to the naturally occurring diversity of TCR repertoires on one hand, and the limitations in data collection (e.g., cost of sequencing at very high depth, see, e.g., Nielsen et al 2011) on the other. Due to these concerns, we propose here the ‘sample-adjusted’ versions of both the diversity and overlap indices, build upon the concepts of Renyi entropy and divergence and combined with the idea of a sample coverage. As illustrated in the next section, it seems that for highly under-sampled data this approach compares favorably with many of the existing ones described in the previous section. We start with the following.
Definition 5 (Sample coverage)
Let X = (X1,…,Xm) denote a multinomial random variable Mult(n,p) and set = 1 if Xi > 0 and = 0 otherwise. The X-based sample coverage is given by
| (2.1) |
The sample coverage may be interpreted as the (posterior) probability of discovering a new multinomial class in the next sample. For that reason, in many fields like, e.g., ecological biodiversity studies, the concept of a sample coverage is mostly used to estimate the probability of discovering a new species in a population of plants or animals. Outside biodiversity modeling, some recent applications of coverage were proposed, for instance, for analyzing genetic data (cf. Mao and Lindsay 2002). Note that the definition above readily extends to the case when m = ∞.
Whereas the sample coverage in itself is not available without knowing the population parameters, the following empirical estimate, known as the Good-Turing coverage estimator, (Good, 1953) offers a viable substitute. This empirical sample coverage is given by
where the symbol f1 denotes the number of components (classes) of X observed exactly once in the sample of size n. The properties of the above estimate were originally studied by Esty (1986, 1983) who in particular showed its asymptotic consistency ( → 0 as n → ∞) and normality. Recently, a necessary and sufficient for the asymptotic normality of was given by Zhang and Zhang (2009).
2.1 Adjusted Diversity Measures
The idea of applying coverage adjustment to estimate diversity via entropy analysis was first introduced in Chao and Shen (2003). In our approach described in this section, we take the original idea a step further via an additional coverage correction applied to the geometric and information-based weighted diversity and overlap measures introduced in Section 1. The estimates constructed in this way put more weight on the less frequent species and are therefore expected to be more robust against the under-sampling bias. As shown below, as long as the sample coverage converges to unity reasonably fast, these adjusted estimates are consistent under mild regularity conditions.
To describe our approach, we start by combining a notion of the sample coverage with that of the Renyi entropy (cf. Example 3). The resulting coverage-adjusted version of the Renyi diversity index (1.3) is
| (2.2) |
Note that the integer value interpolated from the values kC := exp(HC(p)) = for C < 1 may be viewed as the corresponding adjusted effective number of species.
From the above definition it is clear that the diversity index HC is maximal when only singletons are observed (i.e. each species in the sample is observed exactly once, that is f1 = n), in which case the effective number equals to the observed number of species. If the sample coverage equals 1 (i.e. all species are observed) then HC index puts equal weight on frequent and non-frequent species and is simply the Shannon entropy H1.
Since typically neither C nor p are available, an appropriate empirical version, asymptotically equivalent to HC, needs to be considerd, with the obvious candidates being and . Here we concentrate on the latter, with the required consistency result given in the following Theorem 1 where, in order to avoid trivialities, it is assumed that the probability vectors are possibly infinite, i.e., p ∈ Δ∞. The results stated in Theorem 1 are related to those of Antos and Kontoyiannis (2001) and Vu et al (2007) who showed that the adjusted Shannon entropy estimator (see below for notation) is consistent in estimating H1(p). The assertions of the theorem below extend this fact to the class of Renyi entropies Hα(p). The proof is deferred to the Appendix.
Denote by the plug-in maximum likelihood estimator (MLE), based on the sample of size n, of the probability vector and set
Theorem 1
Let α ∈ (0,∞) and assume that Hα(p) < ∞. If α < 1 or if α > 1 and for some r > 0, then
If α = 1 then
and, on the set { infinitely often},
where .
Note that it follows from the above that for α = 1 and = 1 (i.e. no singletons in the sample), we may simply take as the consistent estimator of H1(p). In general, with no additional information about the population p, the choice of α = 1 is typical. However, sometimes other choices of α might be also appropriate, particularly if one wishes to either over-emphasize or de-emphasize the rare (or frequent) species.
Remark 1
The result of Theorem 1 provides an important insight into the empirical diversity profile analysis (see Section 1). The obvious profile estimate based on the sample of size n, which mimics the behavior of the function h(α) = Hα (·) around α = 1, is , where . When n is large, the theorem above states that with probability one, for any α > 0, although not necessarily uniformly in α.
2.2 Adjusted Overlap Measures
We now turn our attention to the analysis of overlap (similarity) between the populations of T-cell receptors. We examine two somewhat different approaches to similarity estimation. The first one, based on the PG-index introduced earlier (see Example 8) is analogous to the adjusted Renyi entropy approach discussed above in the context of diversity measures. The second one is based on the relative mutual information function of a contingency table and in the following sections is referred to as the I-index. We start with the description of the coverage-modified PG-index.
2.2.1 PG Index
Recall the modified PG-index given in (1.4). In analogy with the Renyi entropy adjustment, in the notation of the previous subsection, we may now consider as the sample-coverage and Horvitz-Thompson adjusted geometric measure of overlap. The adjusted PG-index is seen to assign more weight to the observed rare species when computing high dimensional angle between normalized population vectors. Our main result for the new measure of overlap is the following strong consistency theorem. The proof is again deferred to the Appendix.
Theorem 2
Let α,β ∈ (0,∞) and let (i = 1, 2) be given by (2.2), with their respective sample coverage estimates (i = 1, 2). Assume that and , as well as for some r1 > 0, if α > 1 and for some r2 > 0, if β > 1. Then
Note that by taking α = β = 1 in the above result, it follows in particular that the statistic is a consistent estimator of the Morisita-Horn index described in Example 7.
2.2.2 Information Index
The second proposed adjusted overlap index is based on the generalized mutual information statistic in two-way tables, and may be therefore viewed as an information-theoretical extension of the standard Pearson chi-square statistic (see e.g., Agresti 2002). Unlike the PG index, this new information index (or I-index) is also applicable for measuring overlap across multiple populations. In order to describe it, we return now to the two-way contingency table settings. Recall from the Introduction that we consider a two-way (m × n) table as a nonnegative matrix C = [cij] with columns representing n different population c1, c2,…, cn of TCRs and rows representing m receptors. Let be a corresponding normalized matrix with columns p1, p2,…,pn. Denote also pio = ∑jpij, poj = ∑ipij and the corresponding row and column marginals as Po = (po1,…,pon) ∈ Δn–1, Po = (p1o,…,pmo) ∈ Δm–1, as well as Q = Po ⊗ Po := [pio poj]. Note that P,Q ∈ Δmn–1. The idea behind the I-index is to measure the ‘strength’ of the dependence between marginals of the contingency table, instead of e.g., quantifying the pairwise similarity of its columns-specific frequencies. The new index is also scaled to take values in the unit interval. Sometimes (e.g. for clustering purposes) it is more convenient to work with its complement, which measures the lack of overlap or the dissimilarity among n columns. Formally, these two new measures are defined as follows.
Definition 6 (I-index)
For any real m × n matrix C of nonnegative entries, the I-index of order α ∈ (0, 2) is defined as
and the corresponding dissimilarity measure as
.
Remark 2
Let us note that in the case of α = 1 the above definition yields
which is the mutual information index scaled by the Shannon entropy of the column-marginal Po.
It follows from the definition that when α > 1 the I-index puts more weight on the entries of P with positive dependence (i.e. when pij ≥ piopoj) and when α < 1, it puts more weight on the entries with negative dependence (i.e. when pij ≤ piopoj). This feature makes it potentially useful for analyzing the dependence structure of a contingency table (see, for example, Agresti 2002).
The basic properties of the I-index (or, equivalently of Qα) are summarized in the following proposition.
Proposition 1
For α ∈ (0, 2) the following holds
0 ≤ Qα(C) ≤ 1,
Qα(C) = 0 iff p1 = p2 = ⋯ = pn,
if the vectors c1, c2,…,cn form an orthogonal system, then Qα(C) = 1.
Proof
First let us argue (ii). If p1 = p2 = ⋯ = pn then C = [cij] is of column rank one, which is equivalent to P = Q and thus Fα(P,Q) = 0 and Qα(C) = 0. On the other hand, if Iα(P,Q) = 0 then P = Q and so the matrix C is of rank one, and thus p1 = p2 = … = pn. For the proof of (iii) consider the fact that if c1, c2,…cn are orthogonal, then ci,j = ∑k ci,k for i = 1,…,m and j = 1,…,n and so it is easy to see that Fα(P,Q) = H2–α(Po), implying Qα(c1, c2,…,cn) = 1. Finally, the proof of (i) for α = 1 follows from the properties of the mutual information (see Remark 2). For any other α ∈ (0, 1) ∪ (1, 2) note that it suffices to prove it for Iα. Note also that if α < 1 then (pij/pio)α–1 ≥ 1 for i = 1,2,…,m and j = 1,…,n and if α > 1 then (pij/pio)α–1 ≤ 1, which establishes that Iα ≥ 0. To show that Iα ≤ 1, consider 1 < α < 2 and . The result now follows, due to the monotonicity of the log function. The case 0 < α < 1 is handled similarly.
Since the definition of the I-index so far does not involve any under-sampling corrections, the consistency of Iα (hence also Qα) follows, after some elementary algebra, from the results on the consistency of the plug-in Renyi entropy estimators obtained by Antos and Kontoyiannis (2001) (under the term ‘plug-in’ we understand here and elsewhere that the population p is replaced with its sample MLE ). For completeness, we state the result here.
Theorem 3
Let be the empirical MLE of P ∈ Δ∞ based on the random sample of size n. Then
Here P may be interpreted as a two-way normalized table of infinite dimension. For the proof, see Antos and Kontoyiannis (2001).
Recall that the order of the I-index determines the weight put on the positive and negative dependencies. In our setting, the positive dependence between the distributions intuitively means that if a receptor in one population is more abundant, it also tends to be more abundant in the remaining populations, with the opposite being true for the negative dependance. In terms of the overlap profiles, this implies that a large value of I-index with α < 1 (resp. α > 1) indicates negative (resp. positive) dependance between P and Q. This is in contrast to the diversity profiles discussed earlier, where the value of the Renyi entropy index α, in relation to unity, was associated with up- or down- weighting the rare (abundant) species.
As in the case of the PG index, the coverage adjustment of the I-index may be accomplished by taking , where now is the Good-Turing estimator of the sample coverage for the entire table (that is, the estimator of the sample coverage based on C). Due to the fact that , the corrected index emphasizes the negative type of dependence in the presence of under-sampling, implying that we infer the higher overlap between populations based on the observed overlap between the rare species. Thus, since the Iα index is non-increasing in α, tends to overestimate overlap when under-sampling is likely, which is generally desirable in the context of TCR populations. It is of interest to note that the Horvitz-Thompson type correction for the I-index, although possible, does not work as well as for the PG index, due to the different type of normalizations applied in these two cases. In particular, the naive implementation of the Horvitz-Thompson correction along the lines of (1.4) in case of the I-index may have undesirable effects, like e.g., cause the loss of α-monotonicity property and make the Iα values fall outside the unit interval. For these and other reasons, only the Good-Turing type correction of the I-index is considered below. The required consistency result is formally stated in the following theorem, with the proof provided in the Appendix.
Theorem 4
Let be the empirical MLE of P ∈ Δ∞ and let be the Good-Turnig sample coverage estimator, both based on the random sample of size n from P. If ∑i,j pij logr(1/pij) < ∞ for some r > 1, then
| (A.1) |
3 Example: TCR Data Analysis
To illustrate the applicability of our proposed indices vis a vis some standard ones, and to assess their performance, we analyze two TCR datasets obtained from high-throughput sequencing experiments conducted in the molecular immunology lab of Dr Leszek Ignatowicz at Georgia Health Science University. Each dataset consists of the counts of different TCRs in thymic T-cells derived from the transgenic ‘TCRmini’ mice (for a detailed description of the ‘TCRmini’ animal model, see Pacholczyk et al 2007; Rempala et al 2011) and represents a different stage in T-cells evolution. One dataset consists of the so-called ‘regulatory’ T-cells, expressing the FoxP3 protein (via the green fluorescent protein or GFP), whereas another one consists of the so-called ‘naive’ T-cells, which do not express the FoxP3 marker. In what follows, we shall refer to them as GFP+ and GFP− populations, and denote by (see Definition 1 of Section 1) and , respectively. The total number of species (i.e., all sequenced T-cell receptors) in each population is co1 = ∑ci1 = 244, 035 and co2 = ∑ci2 = 232, 210, respectively and the number of distinct species (i.e., T-cell receptor types) is m1 = 3,904 and m2 = 5,048, respectively. The number of the species overlapping between populations equals 1,371, with the total number of overlapping species equal to 45,508.
The values of the several diversity and overlap indices calculated for both datasets are listed in Table 1. As expected from the principles of T-cells evolution, the observed GFP+ population of functionally active regulatory T-cells is seen as having a significantly higher diversity than GFP− population of inactive, naive T-cells. The diversity indices calculated are the Shannon entropy (H1) and the inverse Simpson index (ISI). The effective number of species (ENS) in Table 1 is calculated based on the H1 diversity measure. Note that in the case of ISI the effective number of species is simply the value of the index itself. The overlap indices presented in Table 1 indicate that there seems to be a relatively low similarity between two populations, as measured by the traditional Sørensen (L) and Morisita-Horn (MH) indices (see, Examples 5 and 7). The similarity appears somewhat higher, when measured with the Chao-Jaccard (CJ) index. The Chao-Jaccard index (Chao et al, 2005) is a version of the Jaccard index (J) given in Example 5, incorporating an additional adjustment for the effect of under-sampling. This adjustment apparently slightly biases its performance in the current analysis. The value of the last overlap measure, the I-index of order α = 1 (I1-index) is, as expected, similar to the value of the Sørensen index L. In addition to the values listed in Table 1, for the illustration purposes we have also calculated additional values for the PG indices with several different pairs of parameters. These are PG(0.25, 0.16) = 0.26, PG(0.61, 0.40) = 0.24, PG(0.83, 0.70) = 0.34, PG(0.94, 0.91) = 0.26. Due to the varying coverage, the differences between those values and the value of MH = PG(1, 1) = 0.21 in Table 1 represent the effect of the rare species on the geometric angular measure of overlap between the two populations.
Table 1.
Numerical values of diversity and overlap indices for the two TCR datasets. Note that MH = PG(1, 1).
| Diversity | GFP − | GFP + | Overlap |
|---|---|---|---|
| m | 3,904 | 5,048 | MH = 0.21 |
| H 1 | 4.97 | 6.3 | CJ = 0.72 |
| ENS | 144 | 546 | L = 0.31 |
| ISI | 26 | 95 | I1-ind = 0.38 |
3.1 Experimental Design
In order to compare the performance of various measures of diversity and overlap, we sampled repeatedly with replacement from both T-cell populations in several scenarios, with sample sizes varying from n = 100 () to n = 100,000 (), and compared the resulting estimates to their respective population values presented in Table 1. The performance of all the estimators was then assessed by comparing their respective rates of convergence to the true population values.
3.2 Diversity Analysis
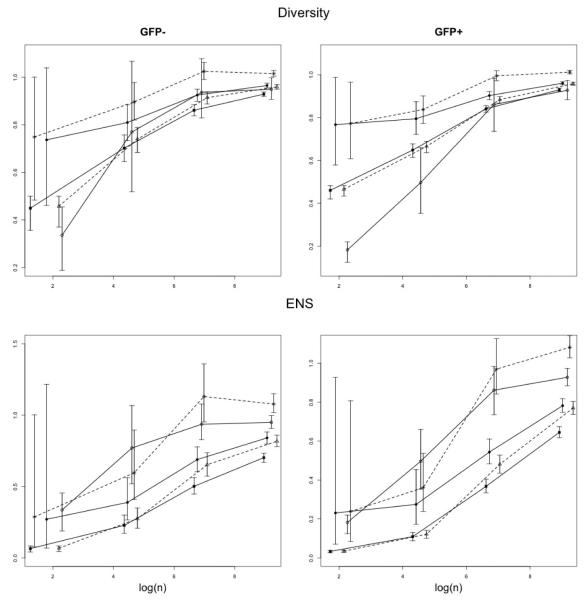
For the five diversity estimators and the corresponding ENS estimators, the results for GFP− and GFP+ are summarized in Table 2 and 3, respectively. For the four entropy-based estimators (i.e., all except ISI), the numerical values of their means and 95% confidence bounds, relative to the true population-based value of H1 or ENS from Table 1, are reported for different sample sizes n, based on B = 500 repetitions. The same characteristics are also reported for ISI, relatively to the population-based ISI value from Table 1 (note that in this case ISI is also ENS). For better visual comparison, the values in Tables 2 and 3 are also plotted against log n in Figure A.1 (see Appendix). As seen both from the top plots in Figure A.1 and from the respective entries of the tables, in all the scenarios considered the diversity estimator based on the proposed coverage-adjusted Renyi entropy () enjoys the relative values closest to unity and the smallest variability (shortest CI). In terms of the ENS estimation, for the GFP− dataset the estimator of ISI index is seen as performing slightly better than the coverage adjusted Renyi entropy ENS, however, in this particular case, the base for the relative ISI values is much smaller (26) than the one for the entropy estimators (144). For the GFP+ dataset, in which the ISI values is much larger (95), both ENS estimators are seen to perform similarly, significantly outperforming the remaining estimators. For the diversity estimation, in both datasets the closest competitor to the proposed adjusted Renyi entropy is seen to be the Chao-Shen estimator (). Both of these estimators are coverage-and-Horvitz-Thompson-adjusted, which seems to give them a distinct edge in the low coverage ( of 30% or less) scenarios. In both datasets it is seen that the sample coverage of about 70-80% is needed for a reasonably accurate estimation of the true population values.
Table 2. Diversity and ENS for GFP−.
The mean and 95% confidence intervals for relative inverse Simpson’s index (ISI) and several entropy-based indices discussed in the paper, reported for different size (n) sub-samples drawn from GFP− dataset. The presented values (based on B = 500 repetitions) are reported relatively to the values in the complete dataset. For each pair of labeled rows, the top row gives the relative values of the indices and the bottom one gives the corresponding relative values of the effective numbers of species (ENS). In each scenario, the relative values closest to one are italicized. “Plug-in H1” refers to the naive, empirical estimate of H1.
| n = 102 | n = 103 | n = 104 | n = 105 | |
|---|---|---|---|---|
| Stat/ENS | ||||
| ISI | 0.34 (0.19,0.45) | 0.77 (0.52,1.07) | 0.94 (0.83,1.08) | 0.95 (0.90,0.99) |
| 0.34 (0.19,0.45) | 0.77 (0.52,1.07) | 0.94 (0.83,1.08) | 0.95 (0.90,0.99) | |
|
| ||||
| 0.46 (0.37,0.50) | 0.74 (0.68,0.78) | 0.92 (0.89,0.94) | 0.96 (0.95,0.97) | |
| 0.07 (0.04,0.08) | 0.27 (0.21,0.35) | 0.65 (0.57,0.74) | 0.83 (0.78,0.86) | |
|
| ||||
| 0.75 (0.49,1.00) | 0.90 (0.82,0.98) | 1.02 (1.00,1.06) | 1.01 (1.00,1.02) | |
| 0.29 (0.077,1.00) | 0.60 (0.41,0.90) | 1.13 (0.95,1.35) | 1.07 (1.01,1.15) | |
|
| ||||
| 0.73 (0.46,1.04) | 0.80 (0.73,0.89) | 0.92 (0.90,0.95) | 0.96 (0.95,0.97) | |
| 0.27 (0.06,1.22) | 0.39 (0.26,0.56) | 0.69 (0.60,0.78) | 0.84 (0.79,0.88) | |
|
| ||||
| Plug-in H1 | 0.45 (0.36,0.50) | 0.70 (0.65,0.76) | 0.86 (0.84,0.88) | 0.93 (0.92,0.94) |
| 0.06 (0.04,0.08) | 0.23 (0.17,0.30) | 0.50 (0.44,0.56) | 0.70 (0.67,0.73) | |
Table 3. Diversity and ENS for GFP+.
The relative diversity and ENS for GFP+ population based on B = 500 repetitions of the sub-sampling, for each n value. The same layout as in Table 2.
| n = 102 | n = 103 | n = 104 | n = 105 | |
|---|---|---|---|---|
| Stat/ENS | ||||
| ISI | 0.18 (0.12,0.22) | 0.50 (0.35,0.66) | 0.86 (0.73,0.98) | 0.93 (0.88,0.97) |
| 0.18 (0.12,0.22) | 0.50 (0.35,0.66) | 0.86 (0.73,0.98) | 0.93 (0.88,0.97) | |
|
| ||||
| 0.47 (0.43,0.48) | 0.67 (0.64,0.69) | 0.88 (0.87,0.89) | 0.96 (0.95,0.97) | |
| 0.04 (0.03,0.04) | 0.12 (0.10,0.14) | 0.48 (0.44,0.53) | 0.77 (0.74,0.80) | |
|
| ||||
| 0.77 (0.60,0.96) | 0.84 (0.77,0.90) | 1.00 (0.97,1.01) | 1.01 (1.00,1.02) | |
| 0.24 (0.09,0.80) | 0.36 (0.24,0.54) | 0.97 (0.84,1.12) | 1.08 (1.03,1.14) | |
|
| ||||
| 0.76 (0.57,0.99) | 0.80 (0.72,0.87) | 0.90 (0.88,0.92) | 0.96 (0.95,0.97) | |
| 0.23 (0.07,0.93) | 0.27 (0.17,0.45) | 0.54 (0.48,0.61) | 0.78 (0.75,0.82) | |
|
| ||||
| Plug-in H1 | 0.46 (0.42,0.48) | 0.65 (0.61,0.68) | 0.84 (0.82,0.86) | 0.93 (0.92,0.94) |
| 0.03 (0.02,0.04) | 0.10 (0.09,0.13) | 0.36 (0.33,0.40) | 0.64 (0.61,0.67) | |
Fig. A.1. Diversity and ENS plots.
Plots of relative values of the diversity (upper panel) and effective number of species or ENS (lower panel) estimators from Tables 2 and 3 against log sample size (log n). Joined by the solid lines are, from the top: (i) ISI, (ii) Plug-in, (iii) . Joined by the dashed lines are, from the top: (i) , and (ii) . For better visibility, the x-coordinates of the plotted symbols were slightly shifted so as to avoid vertical overlap.
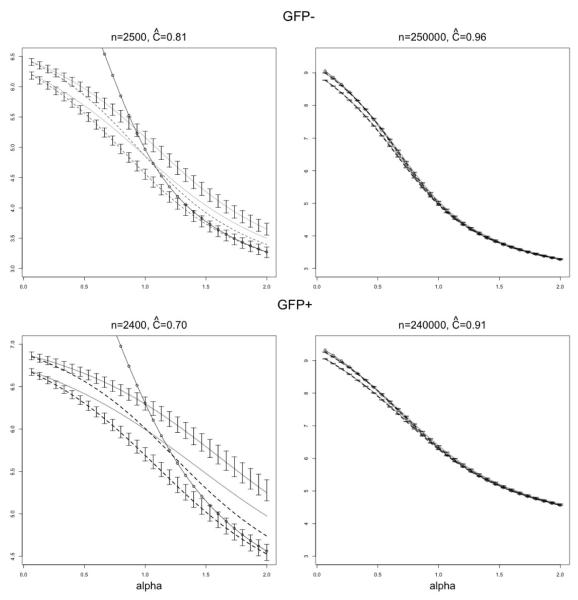
In the final part of the diversity analysis, we have also analyzed the diversity profiles of GFP− and GFP+ datasets, using four different estimators discussed earlier, namely , , and Hα (plug-in). In this analysis only two sampling scenarios were considered, with B = 500 repetitions as before, but with the sample sizes equal to 10% and 100% of the respective total populations in TCR datasets. The resulting plots of the means from B repetitions are presented in Figure A.2 in the Appendix. Despite the fact that some of the confidence bounds around the means are supressed for better visibility, it seems clear from the plots that the reliable (high accuracy, high precision) estimates of the profiles are only available with very large coverage (90% or more). For lower coverage, all profile estimators seem to suffer from the particularly severe downward bias for the index values α < 1.
Fig. A.2. Diversity profile plots.
Due to the low accuracy of the estimators, only high-coverage scenarios are considered. The true population-level diversity profiles for GFP− (upper panels) and GFP+ (lower panels) are compared with the means of the four profile estimators calculated from B = 500 repetitions for different sample sizes (and coverage). The horizontal axis gives the order α of the Renyi entropy. In each panel, the upper grey line is the average of , the lower grey line is the average of , the upper dashed line is the average of , and the lower dashed line is the average of Hα plug-in estimator. The 95% CI bounds are supplied for the profile estimators around the means of , and Hα plug-in estimators. The dash-circle line is the true diversity profile for each of the datasets.
3.3 Overlap Analysis
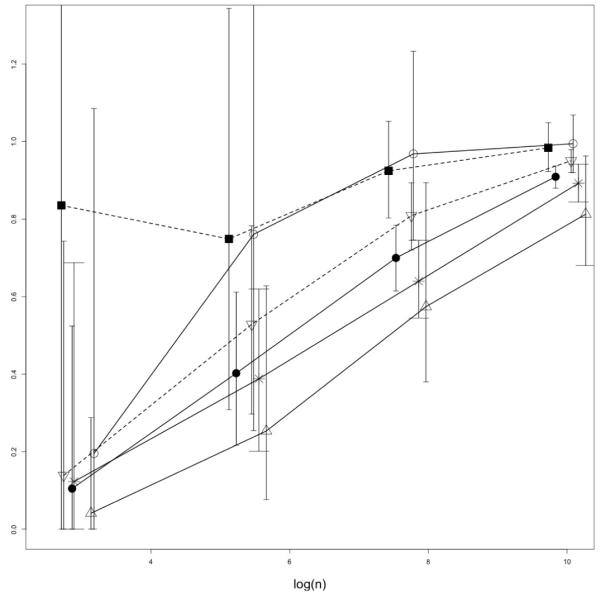
For the purpose of the overlap analysis, we have considered pairs of samples from both TCR populations and compared the values of the sample estimators with the population indices summarized in Table 1. As before, the analysis was performed based on several scenarios with varying sample sizes, each with B = 500 repetitions, and the estimator values were taken relative to the true values of the respective parameters. The results are summarized in Table 4 and additionally plotted in Figure A.3 in the Appendix. As seen from the inspection of the table entries and the plots, the mean values of the sample-adjusted PG index () are the closest ones to the true population level values uniformly across all sample sizes n considered, except for one scenario (n = 100,000). The related MH index performs on average reasonably well most of the time, but it seems that overall the PG index has a distinct advantage on average against the competitors, particularly in the low coverage scenarios. However, both PG and MH indices achieve this at the expense of high variability (long confidence bounds). The I-index appears to perform better on average than the Sørensen L with high enough coverage, and both significantly outperform the CJ index. The I-index and its sample-corrected version are also seen as much less variable than the geometric indices. Overall, when both the accuracy and precision (i.e. bias and variance) of the competing estimators are considered, it seems that the sample-corrected PG- and I-indices perform consistently better that their competitors.
Table 4. Overlap measures.
The relative values of several overlap estimators for two TCR datasets with different subsample sizes (n). For each n the means and 95% CI bounds for each index are reported (based on B = 500 repetitions) relatively to their respective values computed from the complete dataset.
| n = 102 | n = 103 | n = 105 | n = 106 | |
|---|---|---|---|---|
| Stat | ||||
| PG | 0.84 (0.00,4.2) | 0.76 (0.31,1.31) | 0.92 (0.80,1.04) | 0.99 (0.92,1.05) |
| I1-ind | 0.10 (0.00,0.59) | 0.40 (0.18,0.62) | 0.69 (0.62,0.78) | 0.91 (0.88,0.95) |
| -ind | 0.14 (0.00,0.74) | 0.53 (0.30,0.78) | 0.81 (0.72,0.90) | 0.95 (0.92,0.98) |
| L | 0.12 (0.00,0.73) | 0.38 (0.20,0.59) | 0.64 (0.53,0.74) | 0.88 (0.84,0.94) |
| CJ | 0.04 (0.00,0.30) | 0.24 (0.06,0.62) | 0.56 (0.37,0.85) | 0.81 (0.68,1.01) |
| MH | 0.17 (0.00,1.07) | 0.74 (0.23,1.43) | 0.96 (0.73,1.22) | 0.99 (0.92,1.09) |
Fig. A.3. Overlap plots.
The estimated relative values of overlap indices for GFP− and GFP− TCR populations plotted against log sample size (log n). The plotted points are mean observed values, relative to the true population parameters from Table 1 based on B = 500 repetitions. The bars represent 95% confidence intervals as in Table 4. Joined by the solid lines are: (i) MH (open circles), (ii) L (stars) and (iii) CJ (triangles) and I1-plug-in index (filled circles). Joined by the dashed lines are (i) (squares), and -index (upside-down triangles). For better visibility, the x-coordinates of the plotted symbols were slightly shifted so as to avoid vertical overlaps.
4 Summary and Discussion
In this work we have attempted to mathematically formalize, in terms of the multinomial counts and the related contingency table models, some of the important concepts of the biodiversity theory, particularly the notions of a diversity and an overlap index and an effective number of species. We have focused especially on the properties of the entropic diversity and overlap indices, which seem to be commonly used in the literature for the purpose of analyzing the under-sampled population data, like, for instance, TCR immunological data. In this context, we have proposed new measures of diversity and overlap, which are based, respectively, on the Renyi diversity index and on the angular, geometric overlap index of the Morisita-Horn type, which we dubbed “power-geometric” or PG. Both of these measures have the capacity to naturally up-weight or down-weight the rare or the abundant species in a population, as deemed appropriate, which makes them especially appealing for highly diverse data, like TCR. We have also shown here that these proposed measures may be efficiently approximated via sample estimators with the under-sampling bias correction. The correction is accomplished by incorporating the so-called Good-Turing and Horvitz-Thompson adjustments into the empirical plug-in estimators. For some important special cases of the entropic diversity analysis, this approach specializes to the Chao-Shen correction of the Shannon entropy estimator, but, in general, our method is seen to produce even more efficient estimators than the Chao-Shen correction. This is clearly seen in the biological data examples discussed, as well as in other simulation results which we have conducted with synthetic data but did not report here. Similarly, in the case of the overlap analysis, the same method suggests a highly efficient estimator of the popular Morisita-Horn index, which, for small sample sizes and/or low coverage, appears to have a distinct edge over the standard plug-in estimator currently widely used in the literature.
For the overlap analysis of multiple populations, we proposed here a method based on the Renyi divergence function, which enjoys similar properties to the Renyi diversity index in terms of its ability to down- or up-weight the rare or abundant species, as desired. The resulting statistic has many properties similar to the mutual information index. In particular, its zero value also characterizes the marginal independence in a contingency table and hence may be used for testing purposes. In numerical comparisons, the I-index performed well among the information-based estimators, especially when corrected for under-sampling.
The results and examples presented here indicate that when measuring diversity and overlap for highly heterogeneous populations by means of any of the entropic or geometric indices discussed here, the incorporation of an under-sampling correction into the empirical estimator is, overall, beneficial: it typically improves the index performance for samples with small coverage and does not significantly degrade it for samples with large coverage.
Further empirical and theoretical studies of all the estimates proposed are in order, as they might help to develop a systematics approach to optimizing the under-sampling adjustment, in order to achieve good efficiency. One possible direction could be to combine the coverage-based correction proposed here with the Good-Turning probability mass function estimate of the underlying probability distribution (Orlitsky et al, 2003). Based on the recent results obtained in Orlitsky et al (2004) it appears that such an estimate might enjoy some optimality properties (for instance, in the “mini-max” sense) in the suitable class of contingency tables, although it is not immediately clear whether this would translate into optimality properties for the diversity and overlap measures considered here.
One issue not addressed in the current paper is the construction of statistical tests for numerically comparing the proposed estimates of diversity and overlap over multiple populations (or their pairs). In general, the comprehensive solution to this problem requires the analysis of the weak limits of our estimates under broad assumptions. Such weak limit results could be the used to derive both the asymptotic confidence intervals, as well as any consistency results for the suitable resampling plans. Although the detailed analysis of this problem relies on different mathematical tools (notably, the results from the general theory of empirical processes) and is therefore outside our present scope, the general strategy to be pursued seems relatively straightforward. Namely, under the assumptions that guarantee the convergence of the appropriate empirical processes to their standard Gaussian limits, the normal limits are expected for a broad class of functions of empirical counts, including also the estimates discussed here (see e.g. Esteban and Morales 1995, for some examples). The joined asymptotic normality results for pairs of estimates should be sufficient to establish, for instance, the consistency of the resampling-based tests under modest assumptions on the underlying distributions. As we continue to work on the topic, we hope to comprehensively address this and other issues (like estimation optimality) in our future papers.
Acknowledgements
The authors would like to thank Prof. Leszek Ignatowicz for allowing the use of his experimental data on TCR populations and for helpful discussions and comments on the early drafts of the paper. We are also grateful to the reviewers for their valuable suggestions and for pointing out some additional references.
Research partially supported by US NIH grant R01CA-152158 (GAR, MS) and US NSF grant DMS-1106485 (GAR).
A Appendix: Proofs
In this section we prove Theorems 1 and 2. Recall that for the purpose of consistency analysis we consider populations with possibly an infinite number of species (i.e. the number of receptors m ≤ ∞) and we let the sample size n increase to infinity. We write Xn = O(an) (resp. Xn = o(an)) to denote the fact that the random sequence Xn and a deterministic sequence an satisfy with probability one supn Xn/an < ∞ (resp. Xn/an → 0).
Auxiliary Results
Denote and . In order to prove the main results, we need the following
Lemma 1 Let α ∈ (0,∞) and p be a vector of probabilities (possibly of infinite length) for which Sα(p) < ∞.
If α > 1 and ∑pi logr 1/pi < ∞ for some r > 0, then .
If α < 1 then .
If α = 1 and ∑pi log 1/pi < ∞, then .
Additionally, in the above we may replace by . That is, under any of the hypothesis in (i) – (iii), we also have
| (A.2) |
Proof First, we consider the consistency of . By the results of Antos and Kontoyiannis (2001, Section 2), the plug-in estimator of the power sum is strongly consistent for each α ∈ (0,∞), that is,
| (A.3) |
Moreover, the assumption that ∑k pi logr 1/pi < ∞ for some r > 0 is sufficient (following Vu et al 2007) for
| (A.4) |
In view of (A.2) it suffices to show that under (i)–(iii) we have
| (A.5) |
To this end, consider first α > 1 and note that the following holds with probability one
We now establish that both majorizing terms (I) and (II) vanish asymptotically a.s. To this end note that since a.s., then
due to the consistency of the plug-in power sum estimator of order α and the sample coverage estimator. Apropos (II), set and consider
The function is decreasing in x for x ∈ (0, 1) and thus, for n sufficiently large, the first term (IIa) is majorized by
For the second term, once again due to a.s., we have
| (A.6) |
for 0 < β < α – 1. This establishes (II) → 0 a.s. and hence also (A.4) for α > 1.
Consider now the case when 0 < α ≤ 1. Note that, since implies that ∑pi log1–α 1/pi < ∞, the relation (A.3) holds true with r = 1–α for α < 1 and is forced by our assumption with r = 1 when α = 1. Moreover, (A.5) still holds and the majorizing terms (I) and (IIa) may be handled identically as above. For the remaining term (IIb), note that for 0 < α ≤ 1 and
Note also that
Asymptotically, the first term above vanishes a.s. in view of the result of Antos and Kontoyiannis (2001) and the third one vanishes a.s. due to the summability assumption and the fact that . On the other hand, the middle term is bounded a.s. by the asymptotically vanishing terms
in view of the result of Antos and Kontoyiannis (2001). Hence from (A.6) it follows that (IIb) → 0 a.s. and the parts (i) – (iii) of Lemma (1) are established.
Finally, we also establish (A.1). Note that without loss of generality we may assume that infinitely often) = 1.
Assume first that α > 1 and ∑pi logr 1/pi < ∞ for some r < 0, and choose β such that 1 < β < α and α – α – 1 < 0. Due to the almost sure convergence of to 1 we may without loss of generality assume that for each a.s. We have
The maximum is attained at the point
thus
since, under the assumption that for some r < 0 ∑pi logr 1/pi < ∞, we know that a.s., by the first part of the lemma.
For α < 1 and under the assumption that , it follows from the inequality log x ≤ nx1/n valid for x > 0, n ≥ 1, that ∑pi logr (1/pi) < ∞, for each r > 0. For any r > 1 we have therefore
since n(1/logr n) → 1, n → ∞ for any r > 1.
Now, for α = 1 under the assumption that the entropy of p is finite, we have similarly as above that
| (A.7) |
since n1/log n → e, and a.s. n → ∞. Hence, under the assumptions of the lemma, we have (for any α > 0) , a.s. and (A.1) follows.
With the above lemma in hand, we are now ready for the proof of the Theorem 2, which becomes relatively straightforward.
Proof of Theorem 2
Note that it suffices to show that the estimators of the power sums of the type and are strongly consistent. The result in each case follows by Lemma 1.
The next step is to prove Theorem 1.
Proof of Theorem 1
Note that for α ≠ 1 the assertions follow from Lemma 1 by continuity of the bivariate function g(x, y) := (x – 1)−1 log y. For the remaining case α = 1, the first assertion a.s. follows by an argument similar to that used in the proof of the lemma and hence we forgo the details. To argue the second assertion, note that we may assume without loss of generality that P( infinitely often) = 1 and that in view of the result in Antos and Kontoyiannis (2001) which asserts that a.s., it suffices to show
To this end, note that by Cauchy’s mean value theorem and (iii) of Lemma 1
| (A.8) |
for some φn such that . Note that 1 – φn = O(log–r n) due to (A.3) and consequently, from the proof of Lemma 1, it follows that its assertions also holds with φn in place of . In particular, in view of (A.1) with α = 1,
| (A.9) |
Re-write Δn as follows
| (A.10) |
where in the last inequality we applied the bound . It is obvious that a.s. For the term (I), consider the following.
since a.s., in view of (A.8) and a.s., as well as n1–φn = exp [O(log1–r n)] → 1 a.s. The remaining expression (II) needs to be handled similarly to the analogous term considered in the proof of Lemma 1. First note that
and therefore it suffices to consider (II)’ instead. To this end, set πn := log n/n and note that
The first term (IIa) is majorized by
For the second term (IIb), set
| (A.11) |
Note also that
Asymptotically, the first term above vanishes a.s. in view of the result of Antos and Kontoyiannis (2001) and the third one vanishes a.s. due to the finite entropy assumption and the fact that .
On the other hand, the middle term is bounded a.s. by the asymptotically vanishing terms
in view of the result of Antos and Kontoyiannis (2001). Hence from (A.11) it follows that (IIb) → 0 a.s. and therefore Δn ≤ (I) + (II) + (II) → 0 a.s. in (A.9) and the required result (A.7) is established.
Proof of Theorem 4
We only consider the more difficult case of α = 1. The case of any other α ≠ 1 may be handled by the arguments similar to those used in the proof of Lemma 1. Without loss of generality assume that infinitely often) = 1, since otherwise the result follows by the consistency of the ‘plug-in’ estimate of the I-index (Theorem 3). Note that it suffices to prove that
| (A.12) |
and
| (A.13) |
where . For the proof of the above assertions, we again use Cauchy’s mean value theorem. To argue (A.12), let us note that there exists a φn with such that almost surly,
where . By the assumption that Pij pij logr 1/pij < ∞ for some r > 1, we have as before that . a.s. Since 1/n ≤ τij ≤ n, therefore
Similarly, we obtain
where dn := max{1–nφn−1, n1−φn–1}. Since the entropy H1(P) is finite and dn n → ∞ then the assertion (A.12) follows. To argue (A.13) let us note again that there exists a φn (possibly different from the one considered above) with such that
By the elementary algebra
and
which completes the proof.
Contributor Information
Grzegorz A. Rempała, Department of Biostatistics and Cancer Research Center Georgia Health Sciences University, Augusta, GA 30912 grempala@georgiahealth.edu
Michałl Seweryn, Department of Biostatistics, Georgia Health Sciences University, Augusta, GA 30912 Department of Mathematics and Computer Science, University of Łódz, Poland mseweryn@georgiahealth.edu; msewery@math.uni.lodz.pl.
References
- Agresti A. Wiley Series in Probability and Statistics. 2nd edn Wiley; 2002. Categorical Data Analysis. [Google Scholar]
- Antos A, Kontoyiannis I. Convergence properties of functional estimates for discrete distributions. Random Structures & Algorithms. 2001;19(3-4):163–193. [Google Scholar]
- Arstila T, Casrouge A, Baron V, Even J, Kanellopoulos J, Kourilsky P. A direct estimate of the human αβ T-cell receptor diversity. Science. 1999;286(5441):958. doi: 10.1126/science.286.5441.958. [DOI] [PubMed] [Google Scholar]
- Baum P, McCune J. Direct measurement of t-cell receptor repertoire diversity with amplicot. Nature methods. 2006;3(11):895–901. doi: 10.1038/NMETH949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butz EA, Bevan MJ. Massive expansion of antigen-specific cd8+ T-cells during an acute virus infection. Immunity. 1998;8(2):167–75. doi: 10.1016/s1074-7613(00)80469-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao A, Shen T. Nonparametric estimation of Shannon’s index of diversity when there are unseen species in sample. Environmental and Ecological Statistics. 2003;10(4):429–443. [Google Scholar]
- Chao A, Chazdon RL, Colwell RK, Shen TJ. A new statistical approach for assessing similarity of species composition with incidence and abundance data. Ecology Letters. 2005;8:148–159. [Google Scholar]
- Chen W, Jin W, Hardegen N, Lei KJ, Li L, Marinos N, McGrady G, Wahl SM. Conversion of peripheral cd4+cd25- naive T-cells to cd4+cd25+ regulatory t cells by tgf-beta induction of transcription factor foxp3. J Exp Med. 2003;198(12):1875–86. doi: 10.1084/jem.20030152. DOI 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MM, Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. Nature. 1988;334(6181):395–402. doi: 10.1038/334395a0. DOI 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- Esteban MD, Morales D. A summary on entropy statistics. Kybernetika. 1995;31(4):337–346. [Google Scholar]
- Esty W. A normal limit law for a nonparametric estimator of the coverage of a random sample. The Annals of Statistics. 1983:905–912. [Google Scholar]
- Esty W. The efficiency of Good’s nonparametric coverage estimator. The Annals of Statistics. 1986:1257–1260. [Google Scholar]
- Good I. The population frequencies of species and the estimation of population parameters. Biometrika. 1953;40(3-4):237–264. [Google Scholar]
- Gras S, Kjer-Nielsen L, Burrows S, McCluskey J, Rossjohn J. T-cell receptor bias and immunity. Current opinion in immunology. 2008;20(1):119–125. doi: 10.1016/j.coi.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Hsieh CS, Zheng Y, Liang Y, Fontenot JD, Rudensky AY. An intersection between the self-reactive regulatory and nonregulatory T-cell receptor repertoires. Nat Immunol. 2006;7(4):401–10. doi: 10.1038/ni1318. DOI 10.1038/ni1318. [DOI] [PubMed] [Google Scholar]
- Hsieh CS, Lee HM, Lio CWJ. Selection of regulatory T-cells in the thymus. Nat Rev Immunol. 2012;12(3):157–67. doi: 10.1038/nri3155. DOI 10.1038/nri3155. [DOI] [PubMed] [Google Scholar]
- Janeway Cea. Garland Science. 6th edn New York: 2005. Immunobiology: The Immune System in Health And Disease. [Google Scholar]
- Jost L. Entropy and diversity. Oikos. 2006;113(2):363–375. [Google Scholar]
- Keylock C. Simpson diversity and the shannon–wiener index as special cases of a generalized entropy. Oikos. 2005;109(1):203–207. [Google Scholar]
- Komatsu N, Mariotti-Ferrandiz ME, Wang Y, Malissen B, Waldmann H, Hori S. Heterogeneity of natural foxp3+ T-cells: a committed regulatory T-cell lineage and an uncommitted minor population retaining plasticity. Proc Natl Acad Sci U S A. 2009;106(6):1903–8. doi: 10.1073/pnas.0811556106. DOI 10.1073/pnas.0811556106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magurran AE. Biological diversity. Curr Biol. 2005;15(4):R116–8. doi: 10.1016/j.cub.2005.02.006. DOI 10.1016/j.cub.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Mao C, Lindsay B. A poisson model for the coverage problem with a genomic application. Biometrika. 2002;89(3):669–682. [Google Scholar]
- Memon SA, Sportès C, Flomerfelt FA, Gress RE, Hakim FT. Quantitative analysis of T-cell receptor diversity in clinical samples of human peripheral blood. J Immunol Methods. 2012;375(1-2):84–92. doi: 10.1016/j.jim.2011.09.012. DOI 10.1016/j.jim.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohebtash M, Tsang KY, Madan RA, Huen NY, Poole DJ, Jochems C, Jones J, Ferrara T, Heery CR, Arlen PM, Steinberg SM, Pazdur M, Rauckhorst M, Jones EC, Dahut WL, Schlom J, Gulley JL. A pilot study of muc-1/cea/tricom poxviral-based vaccine in patients with metastatic breast and ovarian cancer. Clin Cancer Res. 2011;17(22):7164–73. doi: 10.1158/1078-0432.CCR-11-0649. DOI 10.1158/1078-0432.CCR-11-0649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak T. An analysis of diversity using Rao’s quadratic entropy. Sankhyā: The Indian Journal of Statistics, Series B. 1986;48:315–330. [Google Scholar]
- Nielsen R, Paul J, Albrechtsen A, Song Y. Genotype and SNP calling from next-generation sequencing data. Nature Reviews Genetics. 2011;12(6):443–451. doi: 10.1038/nrg2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlitsky A, Santhanam N, Zhang J. Always Good–Turing: Asymptotically optimal probability estimation. Science. 2003;302(5644):427–431. doi: 10.1126/science.1088284. [DOI] [PubMed] [Google Scholar]
- Orlitsky A, Santhanam N, Zhang J. Universal compression of memoryless sources over unknown alphabets. Information Theory, IEEE Transactions on. 2004;50(7):1469–1481. [Google Scholar]
- Pacholczyk R, Ignatowicz H, Kraj P, Ignatowicz L. Origin and T-cell receptor diversity of foxp3+cd4+cd25+ T-cells. Immunity. 2006;25(2):249–59. doi: 10.1016/j.immuni.2006.05.016. DOI 10.1016/j.immuni.2006.05.016. [DOI] [PubMed] [Google Scholar]
- Pacholczyk R, Kern J, Singh N, Iwashima M, Kraj P, Ignatowicz L. Nonself-antigens are the cognate specificities of foxp3+ regulatory T-cells. Immunity. 2007;27(3):493–504. doi: 10.1016/j.immuni.2007.07.019. DOI 10.1016/j.immuni.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rempala GA, Seweryn M, Ignatowicz L. Model for comparative analysis of antigen receptor repertoires. J Theor Biol. 2011;269(1):1–15. doi: 10.1016/j.jtbi.2010.10.001. DOI 10.1016/j.jtbi.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rényi P. On measures of information and entropy. Proceedings of the 4th Berkeley Symposium on Mathematics, Statistics and Probability; 1960.1961. pp. 547–561. [Google Scholar]
- Ricotta C. Through the jungle of biological diversity. Acta Biotheoretica. 2005;53(1):29–38. doi: 10.1007/s10441-005-7001-6. [DOI] [PubMed] [Google Scholar]
- Salameire D, Le Bris Y, Fabre B, Fauconnier J, Solly F, Pernollet M, Bonnefoix T, Leroux D, Plumas J, Jacob MC. Efficient characterization of the tcr repertoire in lymph nodes by flow cytometry. Cytometry A. 2009;75(9):743–51. doi: 10.1002/cyto.a.20767. DOI 10.1002/cyto.a.20767. [DOI] [PubMed] [Google Scholar]
- Spellerberg I, Fedor P. A tribute to claude shannon (1916–2001) and a plea for more rigorous use of species richness, species diversity and the ?Shannon–Wiener index. Global Ecology and Biogeography. 2003;12(3):177–179. [Google Scholar]
- Staveley-O’Carroll K, Sotomayor E, Montgomery J, Borrello I, Hwang L, Fein S, Pardoll D, Levitsky H. Induction of antigen-specific T-cell anergy: An early event in the course of tumor progression. Proc Natl Acad Sci U S A. 1998;95(3):1178–83. doi: 10.1073/pnas.95.3.1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóthmérész B. Comparison of different methods for diversity ordering. Journal of Vegetation Science. 1995;6(2):283–290. [Google Scholar]
- Valiant P. PhD thesis. MIT; 2008. Testing symmetric properties of distributions. [Google Scholar]
- Van Den Berg HA, Molina-París C, Sewell AK. Specific t-cell activation in an unspecific t-cell repertoire. Sci Prog. 2011;94(Pt 3):245–64. doi: 10.3184/003685011X13139280383942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu VQ, Yu B, Kass RE. Coverage-adjusted entropy estimation. Statistics In Medicine. 2007;26(21):4039–4060. doi: 10.1002/sim.2942. DOI 10.1002/sim.2942. [DOI] [PubMed] [Google Scholar]
- Zhang CH, Zhang Z. Asymptotic normality of a nonparametric estimator of sample coverage. Annals of Statistics. 2009;37:2582–2595. [Google Scholar]